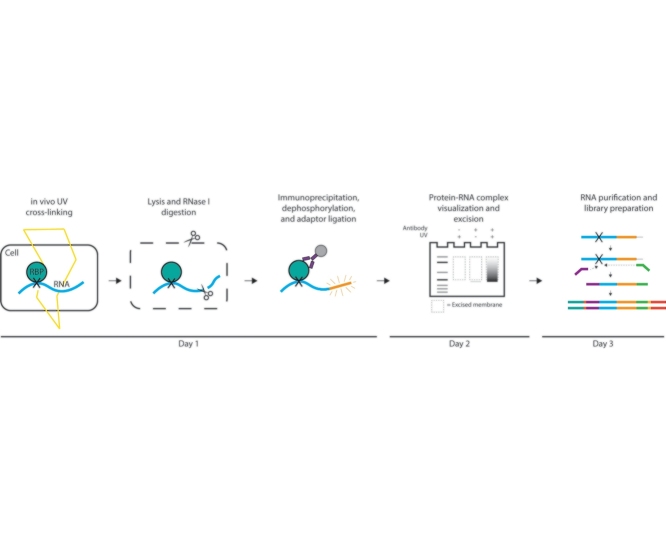
Graphical abstract
Protocol name: Quick-irCLIP – rapid infrared adaptor based individual nucleotide resolution UV cross-linking and immunoprecipitation
Keywords: quick-irCLIP, iCLIP, irCLIP, CLIP, RNA-binding protein, RNA, Protein-RNA interaction
Abstract
RNA-binding proteins (RBPs) are instrumental in the biochemical processing and physiological functioning of non-coding RNAs. Therefore, as interest in non-coding RNAs continues to expand, refining the techniques capable of probing protein-RNA interactions will prove ever more valuable in the characterization of these molecules. To identify the RNAs bound by a given RBP, cross-linking and immunoprecipitation (CLIP) and its iterations have been widely utilized, but these approaches can be complex, labor-intensive, and time consuming. Here, we describe a rapid and technically simple method based upon individual nucleotide resolution CLIP (iCLIP) and infrared CLIP (irCLIP). Termed quick-irCLIP, our protocol circumvents confounding steps, can be completed in less than three days, and is capable of interrogating protein-RNA interactions at single nucleotide resolution.
Specifications Table
| Subject Area: | Biochemistry, Genetics and Molecular Biology |
| More specific subject area: | Protein-RNA Interactions |
| Protocol name: | Quick-irCLIP – Rapid infrared adaptor based individual nucleotide resolution UV cross-linking and immunoprecipitation |
| Reagents/tools: |
|
| Experimental design: | Proteins and RNAs in the cells of interest are cross-linked through UV irradiation. The protein of interest is immunoprecipitated along with its cross-linked RNAs. An infrared RNA adaptor allows for visualization and isolation of protein-RNA complexes following Western blotting. Then the protein is digested, and the RNA is purified and used to create a sequencing library. During library preparation, frequent failure of the reverse transcriptase to read through the cross-link site, allows for the resolution of RNA regions involved in protein-RNA interactions at the nucleotide level. |
| Trial registration: | n/a |
| Ethics: | n/a |
Value of the Protocol
|
Description of protocol
Cross-linking and immunoprecipitation (CLIP) is a popular method used to identify direct protein-RNA interactions. Since its initial inception, the CLIP protocol has accumulated an array of iterations, reflecting various modifications and tweaks. Yet despite the differences between the many versions of CLIP, the general premise remains the same: 1) endogenous protein-RNA interactions are preserved via cross-linking, 2) RNAs are fragmented to dissociate RNA-dependent ribonucleoprotein complexes, 3) protein-RNA complexes are purified and subjected to multiple, stringent washes, 4) proteins are digested and the RNAs are extracted, and 5) RNAs are used for cDNA synthesis which is then used for the construction of the sequencing library. These steps reduce indirect and non-specific interactions, allowing for the isolation of only those RNAs that are in direct contact with the protein of interest. Furthermore, due to the cross-linking of RNA and protein, CLIP can identify the position within the RNA to which the protein binds. For further details regarding the CLIP procedure and its various adaptations see the excellent review by Lee and Ule [1].
Although powerful in their ability to uncover protein-RNA interactions, CLIP methodologies can be technically challenging, labor-intensive, and time consuming. The following protocol synthesizes and deconvolutes previously described iCLIP [2,3] and irCLIP [4] approaches with a simplified library preparation in order to interrogate protein-RNA interactions rapidly and with relative ease. Until the library preparation step, our technique is similar to the other CLIP methods. We use UV light to covalently cross-link protein and RNA. After immunoprecipitation of the protein-RNA complexes, an infrared adaptor is ligated to the RNA. Upon SDS-PAGE and membrane transfer, this infrared adaptor allows for visualization of the complexes with reduced background, increased sensitivity, and without the need for radioisotopes [4]. The RNA is isolated from the membrane and purified. Then, rather than carrying the RNA through an extensive series of steps littered with pitfalls, we prepare a sequencing library from the RNA using a commercially available kit. Due to the polyadenylation step, this kit allows reverse transcription and amplification of all isolated RNAs, including those to which the infrared adaptor has failed to ligate; by not relying on ligation efficiency, this approach may increase yield similar to the sCLIP protocol [5]. Furthermore, while other single nucleotide resolution CLIP methodologies may take 6 or more days to complete, the Quick-irCLIP protocol removes the need for cDNA size selection, circularization, re-linearization, extensive library PCR optimization, and numerous washing and purification steps, allowing its completion in as little as 2.5 days. Thus, the main advantages of the Quick-irCLIP procedure over similar methods are its simplicity and speed, which saves much time, money, and effort.
Before starting
I – Antibody Validation
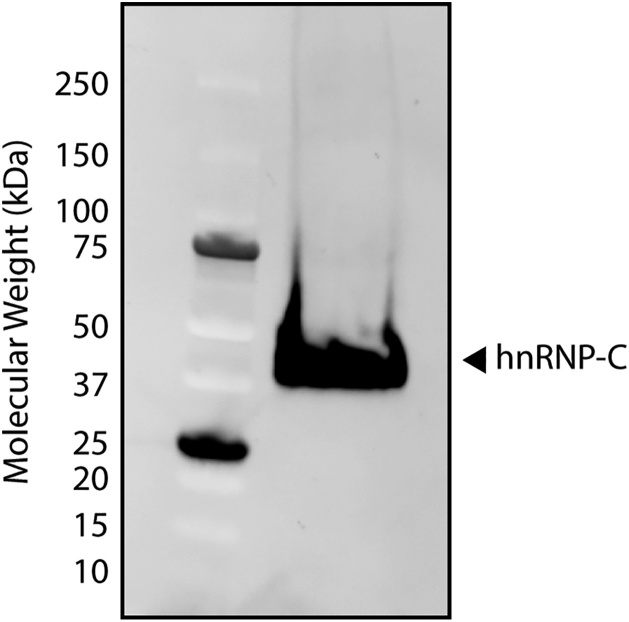
1. When starting the protocol with a new antibody, validate the antibody’s specificity and the antigen’s expression in the cells of interest by western blot. (See Fig. 1 for our validation of the positive control, anti-hnRNP-C antibody.)
Fig. 1.
Validation of hnRNP-C antibody. 30 μg of total protein was run on a 4–15% tris-glycine gel and transferred to a nitrocellulose membrane. After incubations with an anti-hnRNP-C primary antibody and a fluorescently-labeled secondary antibody, the membrane was visualized. Left lane corresponds to the ladder. Right lane corresponds to total protein from ARPE-19 cell line. Arrowhead indicates the expected molecular weight of hnRNP-C.
II – Buffer Preparation
1. Lysis buffer
a. Amount needed per experiment = (3 mL * n) + 7 mL (where n is the number of samples)
b. Components:
• 50 mM Tris-HCl, pH 7.4
• 100 mM NaCl
• 1% Igepal CA-630
• 0.1% SDS
• 0.5% sodium deoxycholate
c. Directions: Filter sterilize and store at 4 °C.
2. Lysis + inhibitor buffer
a. Amount needed per experiment = (2.5 mL * n) + 2.5 mL (where n is the number of samples)
b. Components:
• Lysis buffer
• 1x proteinase inhibitor cocktail
c. Directions: Dilute 100x proteinase inhibitor cocktail in ice-cold lysis buffer. Prepare fresh for each experiment and keep on ice.
3. High-salt wash buffer
a. Amount needed per experiment = (5 mL * n) + 2 mL (where n is the number of samples)
b. Components:
• 50 mM Tris-HCl, pH 7.4
• 1 M NaCl
• 1 mM EDTA
• 1% Igepal CA-630
• 0.1% SDS
• 0.5% sodium deoxycholate
c. Directions: Filter sterilize and store at 4 °C.
4. PNK buffer
a. Amount needed per experiment = (9 mL * n) (where n is the number of samples)
b. Components:
• 20 mM Tris-HCl, pH 7.4
• 10 mM MgCl2
• 0.2% Tween-20
c. Directions: Filter sterilize and store at 4 °C.
5. 5x PNK pH 6.5 buffer
a. Amount needed per experiment = (5 μL * n) (where n is the number of samples)
b. Components:
• 350 mM Tris-HCl, pH 6.5
• 50 mM MgCl2
• 5 mM dithiothreitol
c. Directions: Use nuclease-free water. Dispense to 50 μL, single use aliquots and store at −20 °C. Only use aliquots once.
6. 4x ligation buffer
a. Amount needed per experiment = (6 μL * n) (where n is the number of samples)
b. Components:
• 200 mM Tris-HCl, pH 7.8
• 40 mM MgCl2
• 4 mM dithiothreitol
c. Directions: Use nuclease-free water. Dispense to 50 μL, single use aliquots and store at −20 °C. Only use aliquots once.
7. PK buffer
a. Amount needed per experiment = (250 μL * n) (where n is the number of samples)
b. Components:
• 100 mM Tris-HCl, pH 7.4
• 50 mM NaCl
• 10 mM EDTA
c. Directions: Filter sterilize and store at 4 °C.
8. PK-urea buffer
a. Amount needed per experiment = (250 μL * n) (where n is the number of samples)
b. Components:
• 100 mM Tris-HCl, pH 7.4
• 50 mM NaCl
• 10 mM EDTA
• 7 M urea
c. Directions: Filter sterilize and store at 4 °C
III – Adaptor-Fluorophore Conjugation
1. Resuspend the lyophilized oligonucleotide in 500 μl nuclease-free water.
• Note: The oligonucleotide given in the reagents/tools section reflects the sequence that we used in developing this procedure. For the Quick-irCLIP protocol, this exact sequence is not critical, and the sequence could be modified so long as the new oligonucleotide retained the 5’ phosphate, the internal azide, and the poly(A) stretch.
2. Using the 5’ DNA Adenylation Kit, prepare the adenylation reaction by adding 37.5 μL of 1 mM ATP, 37.5 μL 10x buffer, and 50 μL Methanobacterium thermoautotrophicum (Mth) RNA ligase to 250 μL resuspended oligonucleotide.
• Note: The remaining 250 μL resuspended oligonucleotide can be stored at −20 °C until needed for another adaptor preparation.
3. Incubate the reaction for 2 h at 65 °C.
4. Inactivate the reaction by incubating for 10 min at 85 °C.
5. Transfer the reaction (375 μL) to a new 1.5 mL microcentrifuge tube.
6. Add 40 μL 3 M sodium acetate (pH 5.5) and 1 mL 100% ethanol, mix well by vortexing, and precipitate overnight at −20 °C.
7. Pellet the precipitated, adenylated oligonucleotide by centrifugation at >16,000 RCF for 30 min.
8. Discard supernatant, being careful not to disturb the pellet, and wash with 500 μL ice-cold 80% ethanol.
9. Carefully remove ethanol and allow the pellet to air dry. Do not let the pellet over-dry as this may make resuspension difficult.
10. Resuspend each pellet in 180 μL 1x PBS.
11. Conjugate the infrared dye via “click” chemistry by adding 20 μL of 10 mM IRdye-800CW-DBCO to the 180 μL oligonucleotide solution and incubating for 2 h at 37 °C.
12. Column purify the final adaptor using the QIAquick Nucleotide Removal Kit.
a. To do this, mix the 200 μL “click” reaction with 4.8 mL PNI buffer and dispense 250 μL aliquots across 20 QIAquick nucleotide removal columns.
b. Follow the centrifugation and wash steps as described in the manufacturer’s protocol.
c. Elute using 50 μL nuclease-free water.
d. Pool fractions and quantitate using a Nanodrop (or similar). Concentration should be approximately 10 μM.
13. Aliquot infrared adaptor oligonucleotide and store at −20 °C in the dark
Day 1
I – Sample Collection
1. For each sample, grow cells of interest in a 10 cm dish until 90–100% confluent.
• Note: In general, two negative controls should be included for each experimental sample: a sample that will not be irradiated (“no UV”) and a sample that will not be exposed to the antibody targeting the protein of interest (“no antibody”). Ideally, include a negative control where the protein of interest is knocked out or knocked down. Additionally, when performing the protocol with a new cell type or antibody, it is suggested to include extra experimental samples that can be used to optimize the RNase I concentration and PCR cycle number during the Lysate Preparation and Library Preparation steps, respectively.
• Note: If the RBP of interest is highly expressed and/or interacts with a large number of RNAs, it may be possible to use smaller cell culture plates (e.g. 60 mm plates) for each sample.
2. Remove media from each dish, wash cells once with 10 mL ice-cold 1x PBS, and replace with 1 mL ice-cold 1x PBS.
• Note: At this point, samples should be kept at on ice unless otherwise noted.
3. Place cell culture dishes on an ice-filled tray with dimensions that allow it to fit within the UV cross-linker apparatus.
4. Remove the lids from the cell culture dishes.
5. Irradiate cells at 150 mJ/cm2 at 254 nm.
• Reminder: One or more plates should not be irradiated for use as “no UV” controls.
6. Harvest cells gently using cell scrapers, and transfer cells into 1.5 mL microcentrifuge tubes.
7. Pellet cells via centrifugation at 376 RCF for 1.5 min at 4 °C.
8. Remove supernatant from cell pellets.
• Stopping point: Cell pellets can be snap-frozen and stored at −80 °C until ready for use.
II – Determining Protein Concentration
1. Prepare lysis + inhibitor buffer.
2. Resuspend cell pellets in 1 mL lysis + inhibitor buffer and rotate at 4 °C for 30 min.
3. Assess protein concentration of the lysates by performing a Bradford Assay.
4. Bring all samples to the same protein concentration.
• Note: Dilute the more concentrated samples with lysis + inhibitor buffer, and remove excess to bring all samples to a total volume of 1 mL. To maximize RNA yield it is advisable to use as much protein as possible while holding the amount of protein constant between samples. Since protein yields will vary between cell types and samples, it is not possible to recommend a single concentration. The determination of protein concentration is only used to equalize the amount of protein between samples.
5. Keep lysates on ice, and proceed to Section III – Bead Preparation and Antibody Conjugation.
III – Bead Preparation and Antibody Conjugation
1. For each sample, add 100 μL protein G (or protein A) magnetic beads to a 1.5 mL microcentrifuge tube.
2. Place the microcentrifuge tube in a magnetic rack, remove the supernatant, and wash the beads twice with lysis buffer.
• Note: For these and subsequent bead washes, resuspend beads in 900 μL buffer, then discard the supernatant.
3. Resuspend the beads in 100 μL lysis buffer per sample.
4. Add 2–10 μg antibody per sample to the beads.
• Note: Before adding antibody to the beads, if a “no antibody” negative control is to be included, transfer 100 μL beads to a new 1.5 mL microcentrifuge tube (to which no antibody will be added).
5. Rotate tube(s) at 4 °C for 30–60 min, and proceed to Section IV – Lysate Preparation.
• Note: Conjugated beads will be used in step 1 of Section V – Immunoprecipitation.
IV – Lysate Preparation
1. Prepare a 1:1000 dilution of RNase I (10 units/μl stock) in ice-cold lysis buffer.
2. Add 10 μL RNase I dilution and 2 μL Turbo DNase to each lysate sample.
• Note: Concentration of RNase I can be increased or decreased if the RNA fragment length is determined to be too long or too short upon visualization of the protein-RNA complexes. When using a new cell type or antibody, it is advisable to include samples treated with 1:10, 1:500, 1:1000, and 1:2000 RNase I dilutions. Ideal digestion produces RNA fragments between 50 bp and 300 bp.
3. Incubate samples for 3 min at 37 °C while shaking at 1100 rpm, then immediately transfer cells to ice for 3 min.
4. Centrifuge lysates at >18,000 RCF for 10 min at 4 °C.
5. Transfer lysates to new 1.5 mL microcentrifuge tubes without disturbing the pelleted cell debris.
6. For each sample, load 500 μL lysate onto a Proteus Clarification spin column. Then, centrifuge columns at >18,000 RCF for 1 min. Transfer the flow-through to new 2 mL microcentrifuge tubes.
7. Repeat the previous step with the remainder of the lysates and combine the flow-through of the respective samples.
8. Add 1 mL lysis + inhibitor buffer to each sample, and keep on ice.
• Note: At this point, 20 μl of lysate can be set aside for use as a pre-immunoprecipitation sample for assessing immunoprecipitation efficiency.
V – Immunoprecipitation
1. Once the bead incubation (from step 5 of Section III – Bead Preparation and Antibody Conjugation) has completed, discard the supernatant and wash beads once with high-salt wash buffer.
2. Wash the beads twice with lysis buffer.
3. Resuspend beads in 100 μL lysis buffer per sample.
4. Add 100 μL resuspended beads to cell lysates.
• Note: The volume of the samples should be 2.1 mL.
5. Rotate bead/lysate mixture for 1 h at 4 °C.
6. Place the bead/lysate mixtures in a magnetic rack and discard supernatant.
• Note: At this point, 20 μl of supernatant can be set aside for use as a post-immunoprecipitation sample. Western analysis of this sample and the pre-immunoprecipitation sample (collected above) will allow for the determination of immunoprecipitation efficiency.
7. Wash beads twice with high-salt wash buffer. Rotate the second wash for 5 min at 4 °C, then discard the supernatant.
8. Wash beads twice with PNK wash buffer.
9. Resuspend beads in PNK wash buffer.
VI – Dephosphorylation
1. Prepare dephosphorylation reaction mix. Below are the volumes needed per sample.
• 15 μL nuclease-free water
• 4 μL 5x PNK buffer pH 6.5
• 0.5 μL T4 polynucleotide kinase (10 units/μLstock)
• 0.5 μL RNase inhibitor
2. Remove PNK wash buffer from beads, then remove microcentrifuge tubes from the magnetic rack for 30 s. Return the tubes to the magnetic rack and remove any buffer still remaining.
3. Remove beads from the magnetic rack and resuspend in 20 μL dephosphorylation reaction mix.
4. Incubate samples for 20 min at 37 °C while shaking at 1100 rpm.
5. Wash beads once with PNK wash buffer.
6. Wash beads once with high-salt wash buffer. Rotate this wash for 5 min at 4 °C then discard the supernatant.
7. Wash beads once with PNK wash buffer.
8. Resuspend beads in PNK wash buffer.
VII – Adaptor Ligation
1. Prepare ligation reaction mix. Add components in the order indicated. Below are the volumes needed per sample.
• 8.5 μL nuclease-free water
• 5 μL 4x ligation buffer
• 0.5 μL T4 RNA ligase I (10 units/μL stock)
• 0.5 μL RNase inhibitor
• 1.5 μL infrared adaptor oligonucleotide
• 4 μL polyethylene glycol 400
2. Remove PNK wash buffer from beads, then remove microcentrifuge tubes from the magnetic rack for 30 s. Return the tubes to the magnetic rack and remove any buffer still remaining.
3. Remove beads from the magnetic rack and resuspend in 20 μL ligation reaction mix.
4. Incubate samples overnight at 16 °C while shaking at 1100 rpm.
• Stopping point: Do not incubate samples longer than 18 h.
Day 2
I – SDS-PAGE of Protein-RNA Complexes
1. Wash beads once with PNK wash buffer
2. Wash beads twice with high-salt wash buffer. Rotate the first wash for 5 min at 4 °C, then discard the supernatant.
3. Wash beads once with PNK wash buffer. Transfer beads to new 1.5 mL microcentrifuge tubes, then discard the supernatant.
4. Resuspend beads in PNK wash buffer.
5. Prepare 1x MOPS-SDS buffer
• 475 mL water
• 25 mL 20x NuPAGE MOPS-SDS buffer
6. Assemble the protein gel apparatus with a 4–12% NuPAGE Bis-Tris gel and 1x MOPS-SDS running buffer.
7. Add 500 μL antioxidant to the upper chamber of the gel apparatus.
8. Prepare 1x sample loading buffer. Below are the amounts needed per sample.
• 13 μL nuclease-free water
• 5 μL 4x NuPAGE protein loading buffer
• 2 μL sample reducing agent
9. Remove PNK wash buffer from beads, and remove microcentrifuge tubes from the magnetic rack for 30 s. Then, return the tubes to the magnetic rack and remove any buffer still remaining.
10. Resuspend beads in 20 μL 1x sample loading buffer.
11. Incubate samples at 80 °C for 5 min to dissociate protein-RNA complexes from beads. The protein ladder does not need to be heated.
12. Briefly centrifuge the samples to collect any precipitation, and place the tubes on a magnetic rack to separate the beads.
13. Load 20 μL of sample supernatant and 5 μL Dual Color Precision Plus Protein Standard to 4–12% Bis-Tris gel. If possible, leave blank lanes between samples to facilitate extraction of protein-RNA complexes from the membrane.
14. Run the gel for 50 min at 180 V.
II – Transfer and Visualization of Protein-RNA Complexes
1. Prepare fresh transfer buffer.
• 425 mL deionized water
• 50 mL methanol
• 25 mL 20x transfer buffer
2. Following SDS-PAGE, carefully remove gel from cassette, and assemble the blotting ‘sandwich’ and apparatus as per manufacturer’s instructions.
3. Transfer protein-RNA complexes for 1.5 h at 30 V.
4. Remove nitrocellulose transfer membrane and transfer to a light-protected box containing 1x PBS.
5. Visualize the protein-RNA complexes using near infrared imager and return to light-protected box until ready to isolate complexes.
• Note: Every 20 nucleotides of RNA bound to the protein will add approximately 7 kDa to the molecular weight of the protein-RNA complex. As such, the infrared adaptor adds approximately 15 kDa to the molecular weight of the complex.
6. Print a grayscale blot image at 100% scale on acetate film.
III – Protein-RNA Complex Excision
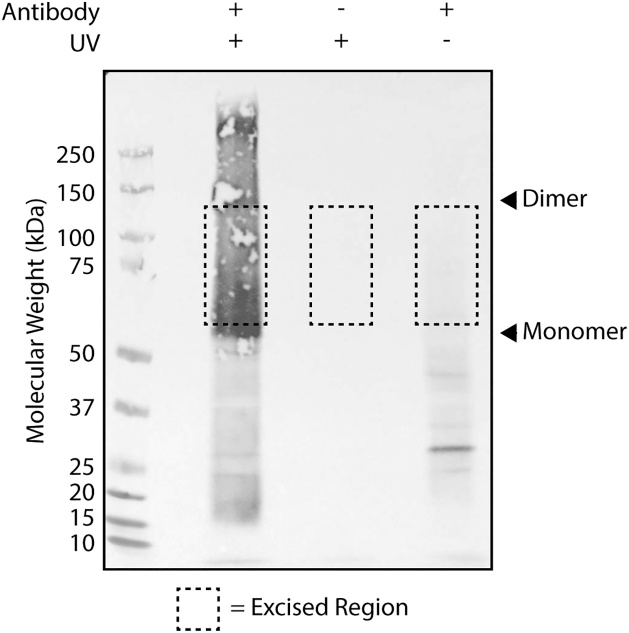
1. Create a cutting mask from the printed image by excising a box from around the protein-RNA signal, starting just above the molecular weight of the protein of interest. Create similarly-sized boxes for the no UV and no antibody controls (See Fig. 2).
Fig. 2.
Visualization of positive control (hnRNP-C) protein-RNA complexes. As indicated, samples were UV irradiated and/or immunoprecipitated using anti-hnRNP-C antibody. Following infrared adaptor labeling, SDS-PAGE, and membrane transfer, protein-RNA complexes were visualized in the far red and colorimetric channels. Channels were overlaid to create a cutting mask. Dashed boxes indicate the regions excised. Arrowheads indicate hnRNP-C monomer and dimer, which are supershifted due to bound RNA.
2. Remove the nitrocellulose membrane from PBS, and wrap it in plastic wrap. Affix the wrapped membrane to cutting surface.
• Note: We use a tempered glass plate as our cutting surface. The plate is wrapped in aluminum foil and baked overnight at 250 °C to degrade RNases.
3. Align the cutting mask with the membrane using the signal from the protein standards as guides. Affix the cutting mask to the cutting surface, and use the windows in the mask to assist in locating and excising the membrane sections possessing protein-RNA complexes.
4. Remove and discard co-excised plastic wrap from the membrane sections.
5. Cut membrane sections into small pieces, and use a 30-gauge needle to assist in transferring the pieces to new 1.5 mL microcentrifuge tubes.
• Note: The exact size of the membrane pieces is not important so long as the pieces are small enough to fit in the bottom of a 1.5 mL microcentrifuge tube.
IV – Protein Digestion and RNA Purification
1. Prepare proteinase K digest mix. Below are the amounts needed per sample.
• 200 μL PK buffer
• 10 μL proteinase K (20 μg/μL stock)
2. Add 200 μL proteinase K digest mix to each tube of nitrocellulose membrane pieces.
3. Incubate samples for 20 min at 37 °C while shaking at 1100 rpm.
4. Add 200 μL PK-urea buffer to each sample.
5. Incubate samples for an additional 20 min at 37 °C while shaking at 1100 rpm.
6. Transfer supernatant to phase lock heavy columns along with 400 μL neutral phenol-chloroform. Discard the membrane sections.
7. Incubate samples for 5 min at 30 °C while shaking at 1100 rpm.
8. Centrifuge samples for 5 min at >18,000 RCF at room temperature.
9. Without touching the gel matrix, transfer the aqueous upper phases to new low-binding 1.5 mL microcentrifuge tubes. Discard the phase lock heavy columns.
10. Centrifuge samples for 1 min at >18,000 RCF at room temperature.
11. Transfer samples to new low-binding 1.5 mL microcentrifuge tubes.
12. Add 0.75 μL glycogen, 40 μL 3 M sodium acetate (pH 5.5), and 1 mL 100% ethanol to each sample.
13. Vortex samples briefly, and precipitate overnight at −20 °C.
• Stopping point: Samples can be kept at −20 °C for several days.
Day 3
I – Library Preparation
1. Centrifuge samples for 20 min at >18,000 RCF at 4 °C.
2. Remove supernatant, leaving approximately 50 μL around the RNA/glycogen pellet.
3. Add 1 mL ice-cold 80% ethanol to each sample, taking care to not overly agitate the pellet.
4. Carefully remove the supernatant, and air dry pellet at room temperature.
• Note: Do not let the pellet over-dry as this may make resuspension difficult.
5. Resuspend pellet in 7 μL nuclease-free water and transfer to 200 μL PCR tubes.
• Note: Pellet can be resuspended in 8 μL nuclease-free water to allow 1 μL to be run on a Bioanalyzer in order to assess RNA concentration and quality.
• Stopping point: Resuspended RNA can be stored at -80 °C for several days.
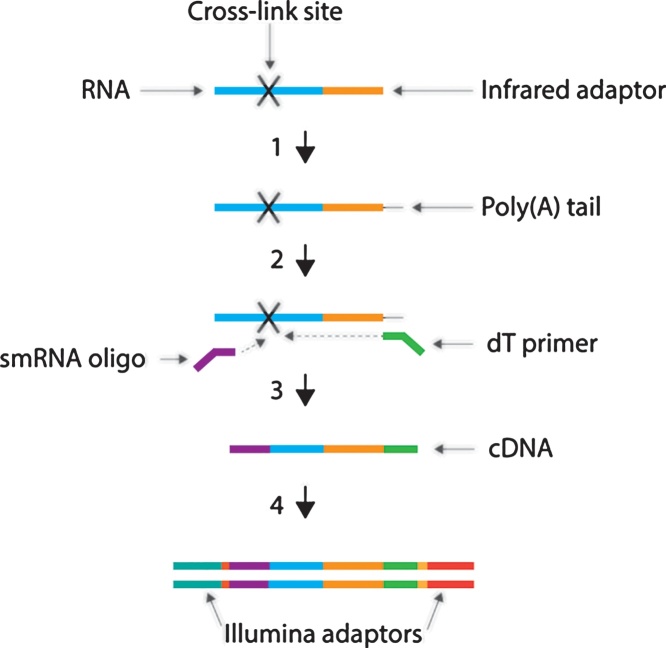
6. Create sequencing libraries following the protocol described in the SMARTer® smRNA-Seq Kit for Illumina User Manual. It is very important to read through the entire manual before beginning library preparation. Fig. 3 illustrates the principle steps of the procedure. Briefly, the protocol entails:
Fig. 3.
Diagram of library preparation procedure. Membrane-isolated RNA is first polyadenylated (1). The RNA is then reverse transcribed, however, the enzyme frequently fails to read through the cross-link site, leading to termination followed by the addition of non-templated nucleotides bound by the smRNA oligonucleotide (2). Template switching and extension by reverse transcriptase completes the addition of an adaptor to the 3’ end of the first-strand cDNA (3). In the final step, PCR amplification is used to add full-length Illumina adaptors to the cDNA ends (4).
a. Polyadenylation of the input RNA
i. Prepare polyadenylation reaction mix. Below are the volumes needed per sample.
• 0.25 μL poly(A) polymerase (2 units/μL stock)
• 0.25 μL RNase inhibitor (40 units/μL stock)
• 2.5 μL smRNA mix 1
ii. Add 3 μL polyadenylation reaction mix to 7 μL resuspended RNA samples.
iii. Incubate the samples at 16 °C for 5 min, then immediately transfer the samples onto ice.
b. cDNA synthesis
i. Add 1 μL of 3’ smRNA dT primer to each sample.
ii. Incubate the samples at 72 °C for 3 min, then immediately transfer the samples onto ice.
iii. Prepare reverse transcription reaction mix. Below are the volumes needed per sample.
• 6.5 μL smRNA Mix 2
• 0.5 μL RNase inhibitor (40 units/μL stock)
• 2 μL PrimeScript RT (200 units/μL stock)
iv. Add 9 μL reverse transcription reaction mix to each sample.
v. Incubate the samples with the following program:
• 42 °C–60 min
• 70 °C–10 min
• 4 °C–hold
vi. Transfer the samples onto ice.
c. PCR and clean-up
i. Prepare PCR reaction mix. Below are the volumes needed per sample.
• 24 μL nuclease-free water
• 50 μL2x SeqAmp PCR buffer
• 2 μL SeqAmp DNA polymerase
ii. Add 76 μL PCR reaction mix to each sample, then add 2 μL forward primer and 2 μL reverse primer.
iii. Incubate the samples with the following program:
1. 98 °C–1 min
2. 7–17 cycles:
a. 98 °C–10 s
b. 60 °C–5 s
c. 69 °C–10 s
3. 4 °C–hold
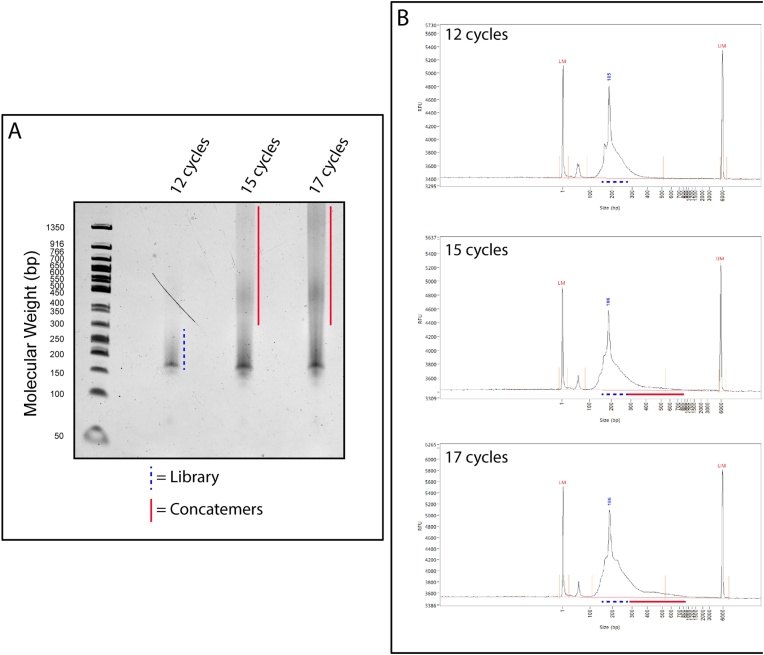
• Note: Regarding this PCR, it is important to use an optimal number of cycles. Too few cycles will result in insufficient library amplification, while too many cycles will result in the production of concatemers, which may not be apparent when using the Bioanalyzer (See Fig. 4). If RNA concentrations have been determined, they can be used to approximate the number of cycles needed for the final PCR based on Table 1 in the SMARTer® smRNA-Seq Kit for Illumina User Manual. Otherwise, 12 cycles can be used initially. In either case, the optimal number of cycles may need to be adjusted based on the cell type and protein of interest.
Fig. 4.
Visualization library preparations performed with varying PCR cycles. A. 20 μL of sequencing libraries were run on a 6% TBE gel. As indicated, libraries were amplified using 12, 15, or 17 cycles in the final PCR. Each library was prepared from samples that were UV irradiated and immunoprecipitated using anti-hnRNP-C antibody. B. Bioanalyzer traces of the libraries shown in A. Although not as apparent as the gel, concatemers are present in the 15 and 17 cycle traces. Dashed blue line indicates presence of the library. Solid red line indicates presence of concatemers.
d. Purify the PCR reactions using the provided NucleoSpin Gel and PCR Clean-up kit
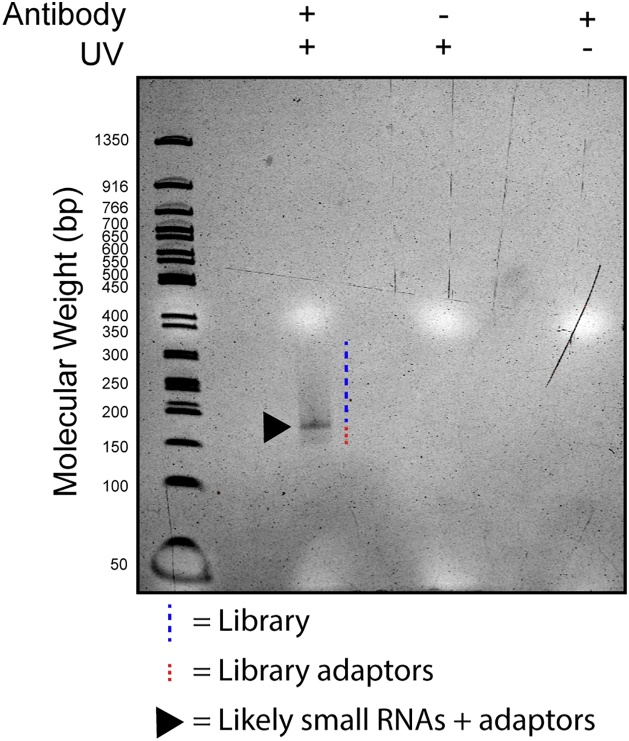
7. Upon completion of the libraries, validate quality by running 1 μL of each preparation on a bioanalyzer or by running 3 μL of each preparation on a 6% TBE gel (See Fig. 5).
Fig. 5.
Visualization of positive control (hnRNP-C) library preparation. 3 μL of sequencing libraries were run on a 6% TBE gel. As indicated, libraries were prepared from samples that were UV irradiated and/or immunoprecipitated using anti-hnRNP-C antibody. Each preparation was performed using 12 cycles for the final PCR. Dashed blue line indicates presence of the library. Dashed red line corresponds to library adaptors. Arrowhead indicates band that likely corresponds to small RNAs ligated with infrared adaptor and taken through the library preparation steps.
8. Once validated, the libraries can be sequenced, aligned, and analyzed according to user preferences.
• Note: Bioinformatic trimming should be conducted prior to alignment. The first three nucleotides on the 5’ end of the first sequencing read (Read 1) should be trimmed, as these nucleotides are added from the template switching oligonucleotide of the library preparation kit. Additionally, the 3’ ends of the reads may need to be trimmed of poly(A) and/or adaptor sequences.
Funding
Funded by: National Institutes of Health/National Eye Institute, United States of America, Grant number: EY028553.
References
- 1.Lee F.C.Y., Ule J. ’Advances in CLIP technologies for studies of protein-RNA interactions. Mol. Cell. 2018;69:354–369. doi: 10.1016/j.molcel.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Sibley C.R. Individual nucleotide resolution UV cross-linking and immunoprecipitation (iCLIP) to determine protein-RNA interactions. Methods Mol. Biol. 2018;1649:427–454. doi: 10.1007/978-1-4939-7213-5_29. [DOI] [PubMed] [Google Scholar]
- 3.Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17 doi: 10.1038/nsmb.1838. 909-U166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarnegar B.J., Flynn R.A., Shen Y., Do B.T., Chang H.Y., Khavari P.A. irCLIP platform for efficient characterization of protein-RNA interactions. Nat. Methods. 2016;13:489–492. doi: 10.1038/nmeth.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kargapolova Y., Levin M., Lackner K., Danckwardt S. sCLIP—an integrated platform to study RNA-protein interactomes in biomedical research: identification of CSTF2tau in alternative processing of small nuclear RNAs. Nucleic Acids Res. 2017;45:6074–6086. doi: 10.1093/nar/gkx152. [DOI] [PMC free article] [PubMed] [Google Scholar]