Abstract
Direct infusion high-resolution mass spectrometry (DIHRMS) is a novel, high-throughput approach to rapidly and accurately profile hundreds of lipids in human serum without prior chromatography, facilitating in-depth lipid phenotyping for large epidemiological studies to reveal the detailed associations of individual lipids with coronary heart disease (CHD) risk factors. Intact lipid profiling by DIHRMS was performed on 5662 serum samples from healthy participants in the Pakistan Risk of Myocardial Infarction Study (PROMIS). We developed a novel semi-targeted peak-picking algorithm to detect mass-to-charge ratios in positive and negative ionization modes. We analyzed lipid partial correlations, assessed the association of lipid principal components with established CHD risk factors and genetic variants, and examined differences between lipids for a common genetic polymorphism. The DIHRMS method provided information on 360 lipids (including fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, and sterol lipids), with a median coefficient of variation of 11.6% (range: 5.4–51.9). The lipids were highly correlated and exhibited a range of associations with clinical chemistry biomarkers and lifestyle factors. This platform can provide many novel insights into the effects of physiology and lifestyle on lipid metabolism, genetic determinants of lipids, and the relationship between individual lipids and CHD risk factors.
Keywords: lipidomics, mass spectrometry, protocol, genetics, coronary heart disease
Introduction
Lipids have many different functions, from membrane components to cell signaling, and are associated with a number of chronic diseases, including coronary heart disease (CHD).1−4 Atherosclerosis, the main cause of CHD, is associated with lipid accumulation and aggregation, particularly of cholesterol and its derivatives. Furthermore, excess energy intake can lead to hyperlipidemia, a major risk factor for CHD. Many genetic polymorphisms at genes involved in lipid metabolism and lipid circulation are associated with CHD risk.5,6 Improving our understanding of the mechanisms that underlie these associations will facilitate the development of pharmacological interventions to reduce the risk and burden of CHD and other chronic diseases.
The development of new analytical technologies for the analysis of lipids, particularly in mass spectrometry and data processing,7 has led to the rise of lipidomics,8 with the aim of capturing information on a wide range of lipids in a given biological sample, across a large number of individuals.1 In particular, lipidomic analysis of human blood has the potential to identify the role of specific lipids in diseases, including CHD.3 There are important prerequisites for any method that uses profiles of serum lipids to study lipid metabolism. Since by definition an open-profiling method does not target particular lipids, it must have the ability to measure and discriminate a wide range of lipids with minimal bias across different lipid species and classes.9
This study provides a detailed description of the method used for a large-scale lipidomics analysis in 5662 healthy participants in the Pakistan Risk of Myocardial Infarction Study (PROMIS),10 which is currently among the largest MS-based metabolomics studies. While the vast majority of metabolomics studies have focused on Western populations, our study is one of the first to profile lipid levels in a Pakistani population. Chronic diseases in Pakistan are responsible for 50% of the total disease burden,11 and ischemic heart disease and stroke are among the top ten causes of years of life lost.12
Our newly developed method uses direct nanospray-infusion coupled to high-resolution mass spectrometry (DIHRMS), which yielded the intensities of several thousand features. Although the platform itself is nontargeted and measures all signals within a specified mass-to-charge ratio (m/z) window, we developed a novel peak-picking approach based on a local database of lipid species previously detected in pooled samples, which is scalable to large epidemiological studies, to obtain signals for a targeted set of lipids that can reasonably be expected from human serum.
To demonstrate the utility of the DIHRMS platform to inform lipid research and the genetic determinants of human metabolism, we examined levels of individual lipids associated with rs662799, a common genetic polymorphism in the APOA5–APOC3 gene region that is known to associate with major lipid markers and coronary artery disease (CAD).13,14 We chose to focus on this genetic region as it is well-known and has been thoroughly studied, allowing us to validate the platform and demonstrate how it can provide additional information about the association of lipids with genetic variants. Variants in the APOA5–APOC3 region are associated with elevated triacylglycerol levels,14 but most studies to-date have only looked at APOA5–APOC3 variants in relation to total triacylglycerols. Our lipidomics platform provides details in much higher resolution about the nature of the association of genetic variants in this region with over 50 specific triacylglycerol species, as well as with a wide variety of other lipids from various lipid classes.
Methods
Study Description
PROMIS is a case-control study of first-ever acute myocardial infarction (MI) in patients from nine centers in urban Pakistan, the details of which have been published previously.10 Controls were individuals without a self-reported history of cardiovascular disease (who had no ECG changes consistent with a previous MI) drawn from individuals concurrently identified in the same hospitals as index cases. Participants were excluded if they (1) had previous history of cardiovascular disease; (2) had an infection that occurred in the previous 2 weeks; (3) had documented chronic conditions, such as malignancy, any chronic infection, leprosy, malaria or other bacterial/parasitic infections, chronic inflammatory disorders, hepatitis or renal failure on past medical history; (4) were pregnant; or (5) refused to give consent. Anthropometry measurements, including height, weight, systolic blood pressure (SBP), and diastolic blood pressure (DBP), were measured using standardized procedures and equipment. Nonfasting blood samples were drawn from each participant, centrifuged within 45 min of venipuncture, frozen, and transported on dry ice to Cambridge, UK, from which major lipids and other standard biomarkers were measured (e.g., total cholesterol, low-density lipoprotein cholesterol [LDL-C], high-density lipoprotein cholesterol [HDL-C], total triacylglycerols, hemoglobin A1c [HbA1c], apolipoprotein B, C-reactive protein). Samples were stored at −80 °C until use. DNA was extracted from leukocytes in Pakistan and genotyped at the Wellcome Trust Sanger Institute in Cambridge, UK, on either the Illumina 660-Quad genome-wide array or the Illumina HumanOmniExpress array. Details of the genotyping, imputation, and quality control (QC) have been described previously.10,15
Lipidomics Sample Selection and Batch Design
We selected 5674 PROMIS participants who had genetic data and complete information on age, sex, ethnicity, recruitment center, and date of survey completion. Only non-MI participants were selected to avoid possible confounding from the metabolic factors of patients who had recently experienced an MI. The samples were analyzed on 72 plates, each with a maximum of 80 samples per plate, according to a randomized block design that was developed using the “blockTools” package16 in R v3.1.2. Sex, age, ethnicity, center, and time in years since date of survey were used as factors in the design, and the distance between blocks was minimized for all factors. A QC sample was created by pooling 100 μL of serum from 200 randomly selected samples, which was mixed and aliquoted for use on each plate. A subset of the QC sample was diluted with phosphate-buffered saline solution to two different concentrations, giving three different QC samples per plate (QC1 was undiluted, QC2 was 1:1 diluted, and QC3 was 1:3 diluted). For all samples, including QC samples, 15 μL was aliquoted into 1.2 mL Cryovial tubes.
Lipidomics Sample Extraction
We adapted a method for open-profiling of lipids by DIHRMS.17,18 An automated method for the extraction of lipids was developed using an Anachem Flexus automated liquid handler (Anachem, Milton Keynes, UK). Eighty PROMIS samples, four blanks, and 12 QC samples in 1.2 mL Cryovials were placed on the Flexus, 100 μL of Milli-Q H2O was added to each of the wells and mixed, and then 100 μL of the mixture was transferred to a glass coated 2.4 mL deep well plate (Plate+, Esslab, Hadleigh, UK). Next, 250 μL of MeOH was added containing six internal standards (0.6 μM 1,2-di-O-octadecyl-sn-glycero-3-phosphocholine, 1.2 μM 1,2-di-O-phytanyl-sn-glycero-3-phosphoethanolamine, 0.6 μM C8-ceramide, 0.6 μM N-heptadecanoyl-d-erythro-sphingosylphosporylcholine, 6.2 μM undecanoic acid, 0.6 μM trilaurin), followed by 500 μL of methyl tert-butyl ether (MTBE). The plates were then sealed (using Corning aluminum microplate sealing tape [Sigma-Aldrich Company, UK]) and shaken for 10 min at 600 rpm (4g), after which the plate was transferred to a centrifuge and spun for 10 min at 6000 rpm (4000g). Each well in the resulting plate had two layers, with an aqueous layer at the bottom and an organic layer on top. A 96-head microdispenser (Hydra Matrix, Thermo Fisher Ltd., Hemel Hampstead, UK) was used to transfer 25 μL of the organic layer to a glass coated 240 μL low well plate (Plate+, Esslab, Hadleigh, UK), and 90 μL of MS-mix (7.5 mM NH4Ac IPA:MeOH [2:1]) was added using a Hydra Matrix, after which the plate was sealed and stored at −20 °C until analysis.
Direct Infusion High-Resolution Mass Spectrometry
All samples were infused into an Exactive Orbitrap (Thermo, Hemel Hampstead, UK), using a Triversa Nanomate (Advion, Ithaca, US). The Nanomate infusion mandrel was used to pierce the seal of each well before analysis, after which, with a fresh tip, 5 μL of sample was aspirated, followed by an air gap (1.5 μL). The tip was pressed against a fresh nozzle and the sample was dispensed using 0.2 psi nitrogen pressure. Ionization was achieved with a 1.2 kV voltage. The Exactive started acquiring data 20 s after sample aspiration began. The Exactive acquired data with a scan rate of 1 Hz (resulting in a mass resolution of 65 000 full width at half-maximum [fwhm] at 400 m/z). After 72 s of acquisition in positive mode the Nanomate and the Exactive switched over to negative mode, decreasing the voltage to −1.5 kV. The spray was maintained for another 66 s, after which the analysis was stopped, and the tip was discarded before the analysis of the next sample began. The sample plate was kept at 15 °C throughout the analysis. Samples were run in row order and repeated multiple times, if necessary, to ensure accuracy. A technical description with further details of the DIHRMS approach can be found in the Supplementary Methods (Supporting Information).
Lipidomics Data Processing and Peak Picking
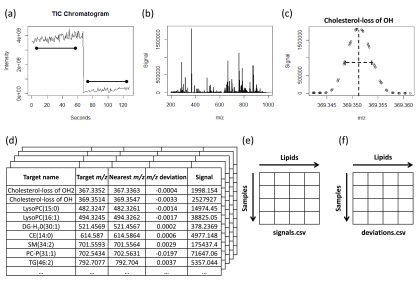
Samples were considered poor quality if there was no signal or the intensity of the total ion current was below 105. We repeated 259 samples that initially met these criteria until suitable signals were obtained for all samples. Once a clean list of 96 raw files, including blanks and QC samples, was obtained for each of the 72 plates, the files were decompressed and converted to mzXML format using the “msconvert” tool in ProteoWizard.19 For each infusion, an average spectrum was calculated from the user-defined time window (Figure 1a). The “xcms” R package7 was used to average 50 spectra per mode using an m/z window of 185–1000 and a retention time window of 20–70 s for positive ionization mode and 95–145 s for negative ionization mode (Figure 1b).
Figure 1.
Schematic of the peak-picking process. (a) XCMS was used to average 50 spectra in positive and negative ionization modes, yielding (b) the average mass spectrum for that particular polarity, for which signals were obtained using a peak-picking algorithm that determined the (c) peak signal at the midpoint of a line drawn at half-height for peaks near signals that corresponded to known lipids. Signals and deviations from known lipids were then (d) combined in a database, and separated into individual files for (e) signals and (f) deviations for each lipid.
To identify lipids prior to data processing, the pooled sample was analyzed on a Velos Elite Orbitrap interfaced with an Advion Nanomate using the previous ionization parameters. Fragmentation was performed using a combination of collision-induced disassociation (CID) and high-energy collision disassociation (HCD) fragmentation. Fragmentation patterns were matched using the LIPID Metabolites and Pathways Strategy (LIPID MAPS) database.20 Peaks were identified relative to a set of m/z values for known lipids downloaded from the LIPID MAPS database, and were retained if they were within a ± 5 ppm window of the target m/z. Lipid subclasses searched in positive ionization mode were cholesterols (Chol), cholesteryl esters (CE), diacylglycerols (DG), triacylglycerols (TG), phosphatic acids (PA), lysophosphosphocholines (LysoPC), phosphatidylcholines (PC), phosphatidylethanolamines (PE), and sphingomyelins (SM). In negative ionization mode, lipid subclasses searched were ceramides (Cer), free fatty acids (FreeFA), PA, PC, PE, phosphatidylglycerols (PG), phosphatidylinositols (PI), phosphatidylserines (PS), and SM. Adducts considered were [M + H]+, [M + NH4]+, and [M + H – 2H2O]+ in positive ionization mode, and [M – H]− and [M + acetate]− in negative ionization mode. Isotope labeling was assessed visually and using the CAMERA package21 within Bioconductor and R. Following annotation in the pooled sample a list of m/z identity pairs, based on expected and possible lipids identified in human serum from the pool, was used to extract small windows of data around the target m/z in the average spectrum (Figure 1c). Note that while different adducts of a lipid are identified as separate entities, this approach focuses on the M ion rather than M + 1 and M + 2 ions, hence “deisotoping” the data by excluding these peaks from the look-up table of identified lipid species. The peak maximum was measured and the two closest points to the half-height on either side were found, resulting in four points. The points with which a horizontal line at half-height intersected a line connecting the two points on either side of the peak (one above the half-height and one below) was used for a peak-width calculation (distance of the line) and a more accurate m/z value for the peak maximum (midpoint of the line). For all m/z identity pairs, the maximum intensity was recorded as well as the deviation of the peaks’ accurate m/z (Figure 1d). A major advantage of this approach is that it could be performed for each sample independently and run in parallel. The final step was the combination of all the signals (Figure 1e) and deviations (Figure 1f) into their respective files. The technical setup yielded average deviations of less than 4 ppm for the detected lipid species.
Lipidomics Data Cleaning and Quality Control
The peak-picking algorithm initially selected all lipids from a list containing 1305 lipids in positive ionization mode and 3772 lipids in negative ionization mode, corresponding to the expected major ions of all known lipids within the m/z range used and including all the previously identified lipid species from the pooled sample in what we refer to as a semi-targeted approach.22 QC was performed to remove lipid signals that were not reliably detectable or did not show a linear response. Lipid signals were removed that were present in fewer than 80% of all QC samples or that had a poor correlation with concentration within the dilution range of QC samples (Pearson correlation r < 0.95). The coefficient of variation (CV) for each lipid signal was then determined and all lipids with CVs of more than 25% were omitted.
For each sample, the sum of the signals of all lipids within each ionization mode that passed the QC steps was calculated. Participants were excluded from analysis in a particular ionization mode if the total signal for the lipids in that mode was less than 5 000 000 (relative units), signifying poor infusion of the sample. Each lipid species was normalized by expressing the signal as a proportion of the total signal for each participant. Since the distributions of most of the lipid signals showed approximate log-normality, natural log-transformation was then applied to each normalized lipid signal. Lipid signals for individual participants were considered outliers and excluded if the normalized, log-transformed signal was more than 10 standard deviations (SD) from the mean for that lipid across all participants. Since the majority of PROMIS participants were not fasting at time of blood draw and their lipid levels could have spiked if they had recently consumed a high-fat meal, 10-SD was used as the cutoff to avoid being overly conservative. It is unlikely that lipids would have true values more than 10-SD from the mean, so any excluded measurements were either below the lower limit of detection or the measurement was false, perhaps due to a contaminant.
Principal Component Analysis
We used principal component analysis (PCA) to determine the main differences in lipid profiles across the participants, excluding 17 lipids (3.8%) with missing signal data in more than 10% of participants. The matrix loadings were orthogonally rotated and the first four principal components were retained based on examination of the scree plot of the eigenvalues. We produced scatter plots comparing the first versus second and third versus fourth principal components, and investigated the association (odds ratios and 95% confidence intervals [CIs]) of these principal components with several established CHD risk factors—overweight, obesity, hypertension, and diabetes—which were defined using well-established cutoff points from published guidelines.23−26
We also conducted genome-wide association analyses using SNPTEST v2.4.127 for the association of these principal components with over 6.7 million genetic variants. Each principal component was adjusted for age group, sex, date of survey, plate (batch), fasting status, and six principal components from the multidimensional scaling matrix to account for population stratification. Beta estimates and standard errors from the association results from the two genetic platforms were combined in a fixed-effect inverse-variance-weighted meta-analysis using METAL (2011–03–25).28
Principal components were also investigated in a subset of triacylglycerols and a subset of lipids associated with rs662799 in the APOA5–APOC3 region.
Assessment of Correlations between Lipids
The partial correlation of each phosphatidylcholine and triacylglycerol with four major lipid markers—total cholesterol, HDL-C, LDL-C, and total triacylglycerols—was assessed with adjustment for age and sex. To examine lipid cross-correlations and distinguish between direct and indirect metabolic interactions,29 we estimated a Gaussian Graphical Model (GGM) on the normalized relative intensities of the lipids using the “genenet” R package,30 which was performed between each pair of lipids while keeping all other lipids constant. The GGM resulted in a set of edges in which each edge connected two detected lipids if their cross-correlation conditioned on all other lipids was significantly different from zero. A similar approach for metabolomics data has been suggested previously.29 We excluded 334 (6%) participants with missing signal data in more than 10% of the lipids and 8 (2%) lipids with missing signal data in more than 20% of the participants from the GGM analysis. To focus on strong effects, we retained only edges in the model that met an FDR cutoff of 0.05 and had a partial correlation coefficient greater than 0.2. This resulted in a network of the partial correlations between all lipids. The results were summarized and combined within each lipid subclass to produce a heat map of the partial correlations between each of the lipid subclasses.
Fatty Acid Chain Enrichment Analysis
We manually annotated detected lipids with their constituent fatty acid chains using the annotation data from the pooled sample. For each combination of fatty acid chains, we counted the number of GGM edges connecting lipids with that specific combination, which we used to directly estimate P-values of enrichment and depletion. To test whether edges from the GGM were enriched for any combination of fatty acid chains, we permuted the annotation 1000 times using the “BiRewire” R package,31 keeping the number of annotations per lipid and fatty acid chain constant. We then produced a heat map of the partial correlations between lipids based on their constituent fatty acid chains.
Lipidome Scan of APOA5–APOC3 Region
In order to demonstrate the efficacy and potential utility of the platform, a lipidome scan was conducted analyzing the association of lipids with a common polymorphism (rs662799, chr11:116663707) in the APOA5–APOC3 region known to be associated with major lipid markers and CAD.13,14 Within each overall lipid category, the lipid most strongly associated with rs662799 was assessed for correlation with a wide range of major lipids and other circulating biomarkers.
Analyses were performed using Stata v15.1 (StataCorp, 2017) and R v3.3.2 (R Core Team, 2016) except where noted otherwise. Two-sided P-values and 95% CIs are presented.
Results
Description of the Study Population
Lipid profiles obtained using DIHRMS were available for 5662 PROMIS participants following data processing, cleaning, and QC. Demographic and clinical characteristics of these participants are shown in Table 1. The median age was 54 years (range: 27–87 years), the majority of participants (79%) were male, and the average body mass index (BMI) was 26 kg/m2 (range: 14–75 kg/m2). Although all participants in this analysis were healthy controls at baseline, they represent a population at increased risk of MI since 56% of the participants were overweight, 17% were obese, 18% had hypertension, and 38% had diabetes. The subset of PROMIS controls selected for the lipidomics assay were comparable to all PROMIS controls (n = 18 564). However, based on nationwide survey data obtained from the Demographic and Health Survey for Pakistan,32 PROMIS participants were older on average, and a higher proportion consumed tobacco and were overweight, compared with the head of household in the general Pakistani population (Table 1).
Table 1. Demographic and Clinical Characteristics and Coronary Heart Disease Risk Factors of Individuals Assayed by DIHRMS in PROMISa.
| PROMIS
controls assayed by DIHRMS (n = 5662) |
All
PROMIS controls (n = 18 564) |
DHS
Pakistan (n = 13 558) |
||||
|---|---|---|---|---|---|---|
| variable | no. of subjects | mean (SD) or % | no. of subjects | mean (SD) or % | no. of subjects | mean (SD) or % |
| Anthropometric markers | ||||||
| Age at survey (yrs) | 5662 | 54 (9) | 18 564 | 56 (9) | 13 558 | 33 (9) |
| Body-mass index (kg/m2) | 5562 | 26 (5) | 18 290 | 26 (5) | 4698 | 25 (6) |
| Waist-to-hip ratio | 5590 | 0.96 (0.13) | 18 344 | 0.95 (0.06) | – | – |
| Systolic blood pressure (mm Hg) | 5587 | 128 (17) | 18 255 | 128 (17) | – | – |
| Diastolic blood pressure (mm Hg) | 5584 | 81 (9) | 18 247 | 81 (10) | – | – |
| Circulating lipid biomarkers | ||||||
| Total cholesterol (mmol/L) | 5542 | 4.63 (1.33) | 17 935 | 4.68 (1.30) | – | – |
| HDL cholesterol (mmol/L) | 5530 | 0.89 (0.27) | 17 881 | 0.93 (0.28) | – | – |
| LDL cholesterol (mmol/L) | 5439 | 2.77 (1.03) | 17 491 | 2.81 (1.01) | – | – |
| Non-HDL cholesterol (mmol/L) | 5530 | 3.75 (1.31) | 17 884 | 3.75 (1.27) | – | – |
| Loge triacylglycerides (mmol/L) | 5537 | 0.74 (0.53) | 17 920 | 0.69 (0.53) | – | – |
| Categorical variables | ||||||
| Sex | 5662 | 18 564 | 13 558 | |||
| Male | 4466 | 79% | 14 049 | 76% | 12 409 | 92% |
| Female | 1196 | 21% | 4515 | 24% | 1149 | 8% |
| Tobacco consumption status | 5651 | 18 512 | 13 542 | |||
| Current | 1727 | 31% | 5294 | 29% | 1016 | 8% |
| History of diabetes | 5651 | 18 516 | – | |||
| Yes | 780 | 14% | 2435 | 13% | – | – |
| Diabetic drug use status | 5654 | 18 540 | – | |||
| Yes | 561 | 10% | 1847 | 10% | – | – |
| Antihypertensive drug use status | 5655 | 18 539 | – | |||
| Yes | 909 | 16% | 3308 | 18% | – | – |
| CHD risk factors | ||||||
| Overweight | 5562 | 18 290 | 4698 | |||
| Yes | 3116 | 56% | 10 460 | 57% | 1891 | 40% |
| Obese | 5562 | 18 290 | 4698 | |||
| Yes | 926 | 17% | 2951 | 16% | 667 | 14% |
| Hypertension | 5587 | 18 257 | – | |||
| Yes | 987 | 18% | 3240 | 18% | – | – |
| Diabetes | 4212 | 8503 | – | |||
| Yes | 1612 | 38% | 3003 | 35% | – | – |
Definitions: Diabetes = HbA1c ≥ 6.5%; Hypertension = SBP ≥ 140 mm Hg or DBP ≥ 90 mm Hg; Obese = BMI ≥ 30 kg/m2; Overweight = BMI ≥ 25 kg/m2. Abbreviations: BMI, body mass index; CHD, coronary heart disease; DBP, diastolic blood pressure; DHS, Demographic and Health Surveys; SBP, systolic blood pressure; SD, standard deviation. Note: Percentages may not add up to 100% due to rounding. Data for the overall Pakistani population was obtained from the DHS. A dash (−) indicates that data were not available.
DIHRMS Is a Novel, High-Throughput Approach To Rapidly and Accurately Profile Lipid Species
Our DIHRMS method for lipidomics covers a wide range of lipids, including fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, and sterol lipids (Table 2), and does not require prior selection of specific lipids or lipid classes if using the local lipid database alone, in contrast to fragmentation-based approaches using tandem mass spectrometry. The high-throughput nature of the method means that with an analysis time of just over 2 min per sample, it is possible to run a full plate of 96 samples (including blanks and QC samples) within 4 h.
Table 2. Categorization of Lipids in Positive and Negative Ionization Mode Measured by Direct Infusion High-Resolution Mass Spectrometry in PROMISa.
| overall lipid category | lipid main class | lipid subclass | no. (%) of lipids |
|---|---|---|---|
| Fatty acyls (FA) | Fatty acids and conjugates | Free fatty acids (FreeFA) (−) | 22 (5.0%) |
| Glycerolipids (GL) | Diradylglycerols | Diacylglycerols (DG) (+) | 19 (4.3%) |
| Triradylglycerols | Triacylglycerols (TG) (+) | 56 (12.6%) | |
| Glycerophospholipids (GP) | Glycerophosphates | Phosphatic acids (PA) (−) | 20 (4.5%) |
| Phosphatic acids (PA) (+) | 13 (2.9%) | ||
| Glycerophosphocholines | Lysophosphocholines (LysoPC) (+) | 8 (1.8%) | |
| Phosphatidylcholines (PC) (−) | 52 (11.7%) | ||
| Phosphatidylcholines (PC) (+) | 54 (12.2%) | ||
| Glycerophosphoethanolamines | Phosphatidylethanolamines (PE) (−) | 24 (5.4%) | |
| Phosphatidylethanolamines (PE) (+) | 16 (3.6%) | ||
| Glycerophosphoglycerols | Phosphatidylglycerols (PG) (−) | 5 (1.1%) | |
| Glycerophosphoinositols | Phosphatidylinositols (PI) (−) | 25 (5.6%) | |
| Glycerophosphoserines | Phosphatidylserines (PS) (−) | 22 (5.0%) | |
| Sphingolipids (SP) | Ceramides | Ceramides (Cer) (−) | 16 (3.6%) |
| Phosphosphingolipids | Sphingomyelins (SM) (−) | 51 (11.5%) | |
| Sphingomyelins (SM) (+) | 27 (6.1%) | ||
| Sterol lipids (ST) | Sterols | Cholesterols and derivatives (Chol) (+) | 2 (0.5%) |
| Cholesteryl esters (CE) (+) | 12 (2.7%) | ||
| Total lipids | 360 |
(+) denotes lipids measured in positive ionization mode; (−) denotes lipids measured in negative ionization mode. Note the final total takes into consideration the detection of some lipids in both positive and negative mode, multiple adducts, and the possibility of multiple annotations.
Importantly, the DIHRMS method included measurement of neutral lipids such as triacylglycerols and cholesterol esters, which are not covered by the commercial platforms that are currently most widely used in large-scale metabolite phenotyping for genome-wide association studies (GWAS). As a result, the measurements included approximately 125 metabolic features that have not yet been assessed in any of the major GWAS of human metabolism33 and lipids that contain odd-chain fatty acids, which have generally been ignored in preceding metabolic profiling efforts, despite their importance to human health.34
DIHRMS Reveals Detailed Lipid Classes and Their Correlation Structure
Following QC filtering, 207 lipids in positive ionization mode and 238 lipids in negative ionization mode remained for analysis, all with unique mass-to-charge ratios and identifiers. When allowing for species that were detectable as different adducts in positive and negative mode, this equated to the detection of at least 360 different lipid species (note that due to incomplete assignment some peaks were assigned to more than one lipid class). We also excluded 123 participants (2%) as part of the QC process. The extent of missing data according to the number of lipids and participants is shown in Figure S1 of the Supporting Information. We found that 5328 out of 5662 participants (94%) had complete information for at least 90% of the lipids (Figure S1a), and 427 out of 444 lipids (96%) had less than 10% (between 0 and 9%) missing data (i.e., were detected in at least 90% of the samples) (Figure S1b). The median coefficient of variation (CV) was 11.60% (range: 5.4–51.9). The CVs for all 444 retained lipid signals are shown in Figure S2a. The precision was higher in positive mode (median CV 11.48%) than in negative mode (median CV 22.34%). However, the CVs demonstrated that simple normalization yielded reproducible data on a par with other high-throughput metabolic profiling methods.35,36
A scatter plot showing the normalized relative intensities of all the lipids according to their m/z, grouped by participant, is shown in Figure S2b. The wide distribution of the signals across certain lipids, such as cholesterol with loss of −OH, CE(18:2), PC(34:2), and TG(52:2), suggests that the levels of these lipids vary significantly across individuals, whereas the levels of other lipids are more consistent.
While most lipids were correlated with levels of major lipid markers, confirming the validity of the platform, the direction of the correlation varied according to the structure of the individual lipids. The partial correlation coefficients and 95% CIs for each of the 106 phosphatidylcholines and 56 triacylglycerols with four major lipid markers are shown in Figure S3. The majority of phosphatidylcholines were positively correlated with HDL-C and inversely correlated with total triacylglycerols, while there were wide variations in the correlations of phosphatidylcholines with total cholesterol and LDL-C. In contrast, the majority of individual triacylglycerols were positively correlated with total cholesterol and total triacylglycerols and negatively correlated with HDL-C, but did not exhibit a significant correlation with LDL-C.
For many of the individual triacylglycerols, as the number of double bonds increased (i.e., the level of saturation decreased) [e.g., from TG(48:1) to TG(48:2) to TG(48:3), or from TG(50:1) to TG(50:2) to TG(50:3)], the magnitude of the correlation with each of the major lipid markers increased. A likely explanation from previous research in obese individuals is that triacylglycerols in adipose tissue become enriched with monounsaturated fatty acids.37
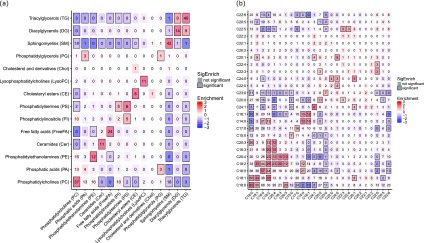
Gaussian Graphical Modeling Highlights 222 Correlated Lipid Species
We applied a Gaussian graphical modeling (GGM) approach that used partial correlations to determine if specific lipids were still strongly correlated after adjusting for all other lipids (Figure 2a). This modeling had two aims: first, for QC, to identify lipids with very high correlations, which were likely to be artifacts of the method; and second, to find relevant correlations that provided information on the biochemical relationship between lipids. The first step in this process was to examine which partial correlates may have arisen from isotopologues with very high correlation coefficients and would be misassignments using our database approach of M ions. From the 314 significant partial correlates, there were ten correlations that were most likely purely M + 1 isotopes (the same lipid that contains one 13C isotope) of other lipid signals and four correlations that were purely M + 2 isotopes (the same lipid that contains two 13C isotopes), based on very high correlations (r > 0.997) and correct isotope ratios. There were 26 correlations where the M + 1 isotope contributed considerably to the signal and four correlations where the M + 2 isotope contributed predominantly to the signal. However, those signals also showed contributions of different lipid signals, for which the correlations were not as high (r < 0.997) or the isotope ratio was incorrect. We identified 36 correlations where the signals came from the same lipid in both positive and negative ionization modes, and two sets of lipids for which the signals overlapped, and the peak-picking algorithm was unable to distinguish the signals. The remaining 222 significant correlations were taken forward for further analysis (Figure 2b).
Figure 2.
Heat map showing relationships between lipid subclasses and constituent fatty acid chains of lipids based on partial correlations derived using Gaussian Graphical Modeling. These heat maps show the relationships between (a) lipid subclasses and (b) constituent fatty acid chains based on the inferred Gaussian Graphical Model (GGM). The number in each cell shows the observed number of GGM edges connecting two lipids in subclasses or constituent fatty acid chains specified on the x- and y-axes. The cells are colored red or blue according to whether the observed number of GGM edges is more or less than expected due to chance alone, and a box is drawn around the cell if there is a significant difference between the numbers of observed versus expected GGM edges.
Principal Component Analysis Classifies Lipids
Next, we assessed the overall technical and biological differences in the lipid signals using PCA. We retained the first four principal components, which explained 55.1% of the variance in the relative intensities of the lipids, based on examination of the scree plot of the eigenvalues (Figure S5). Scatter plots of the matrix loadings of the first versus second and third versus fourth principal components are shown in Figure S6a. The individual lipids are distinguished by color according to the overall lipid category to which they belong.
The first principal component (which explained 31.8% of the variance in the lipid levels) correlated with differences between the positive and negative ionization modes. The dynamic range of the negative mode data was more limited than the positive mode data, and due to the lower ionization efficiency, the data were more prone to ion suppression. These differences between the ionization modes were amplified when the data were expressed relative to total signal intensity. We therefore excluded the first principal component from further data analysis.
The second component (which explained 11.7% of the variance) was dominated by triacylglycerols containing shorter and more saturated fatty acids, which had the highest positive loadings, versus fatty acids (e.g., free linoleic acid) and cholesterol esterified with polyunsaturated fatty acids [e.g., CE(18:2)], which had the strongest negative loadings. The third component (which explained 6.9% of the variance) differentiated saturated phosphatidylcholines [e.g., PC aa (32:0), PC aa (34:0), PC aa (32:0)] from triacylglycerols containing longer, unsaturated fatty acids [e.g., TG(54:5), TG(54:7), TG(56:7)]. The fourth component (which explained 4.8% of the variance) differentiated the odd-chain fatty acid containing sphingomyelins [e.g., SM(39:1), SM(41:1), SM(37:1)] with the highest positive loadings versus saturated free fatty acids and triacylglycerols [e.g., TG(52:2), TG(54:2)] with the strongest negative loadings.
Principal Component Analysis Stratifies Triacylglycerols by Fatty Acid Chains
To provide a more detailed exploration of differences between specific types of lipids, we also examined principal components in a subset of lipids consisting of only triacylglycerols. The corresponding scatter plots of these principal components are shown in Figure S6b. The first principal component of the triacylglycerols distinguished triacylglycerols with an odd number of carbon atoms [e.g., TG(49:3), TG(51:4), TG(55:9)], shown in the blue oval, from those with an even number of carbon atoms [e.g., TG(50:0), TG(52:1), TG(54:3)], shown in the green oval. Previous work has shown that odd-chain fatty acids derive primarily from dairy consumption, while even-chain fatty acids are either synthesized in the liver through de novo lipogenesis or ingested through consumption of saturated fatty foods.34,38,39
The second principal component differentiated triacylglycerols with saturated and monounsaturated fatty acid chains [e.g., TG(44:1), TG(47:1), TG(49:2)], shown in the green and blue ovals at the top of the figure, from triacylglycerols containing ω-3 and ω-6 polyunsaturated fatty acids [e.g., TG(54:4), TG(58:9), TG(57:10)], shown in the orange oval at the bottom of the figure. An important source of these polyunsaturated fatty acids is fish consumption.40 The two pink ovals represent one or more categories that are distinct from dairy consumption, fish consumption, and saturated fat, but further research is needed to determine their biological significance.
We found that the third principal component primarily distinguished triacylglycerols containing polyunsaturated fatty acids from those containing saturated fatty acids. The interpretation of the fourth principal component of the triacylglycerols was not readily apparent.
Association Pattern of APOA5–APOC3 Genetic Variants, Lipids, and Lifestyle Factors
To provide a case study demonstrating the viability of this lipidomics platform with a well-known genetic region, principal components were also examined in a subset of the lipids that were significantly associated with the rs662799 (chr11:116663707) variant in the APOA5–APOC3 gene at P < 8.9 × 10–10. The scatter plots of these principal components are shown in Figure S6c. The first principal component was driven by relative increases in sphingomyelins and decreases in cholesterol esters and triacylglycerols containing unsaturated fatty acids. The second, third, and fourth principal components were very effective at differentiating glycerolipids (i.e., diacylglycerols and triacylglycerols) from the other lipid classes.
As shown in Figure S4, out of the lipids that were significantly associated with the APOA5–APOC3 variant, a number of lipids had significantly positive associations with smoking and physical activity, while other lipids had significant inverse associations (the top 20 lipids most significantly associated with each outcome are shown in the figure). A previous study41 showed that two lipid subclasses (phosphocholines and phosphoethanolamines) and 72 individual lipid species are associated with smoking status—our study confirmed many of these associations. Smoking was associated with increased levels of TG(52:2) and decreased levels of TG(52:4), which might suggest that smoking oxidizes unsaturated fatty acids since the latter is more unsaturated. Similar patterns in lipids with differing directions of effect were observed for the association with physical activity, although the majority of the associations were not statistically significant.
Correlation of Lipids with Circulating Biomarkers Relevant to CHD
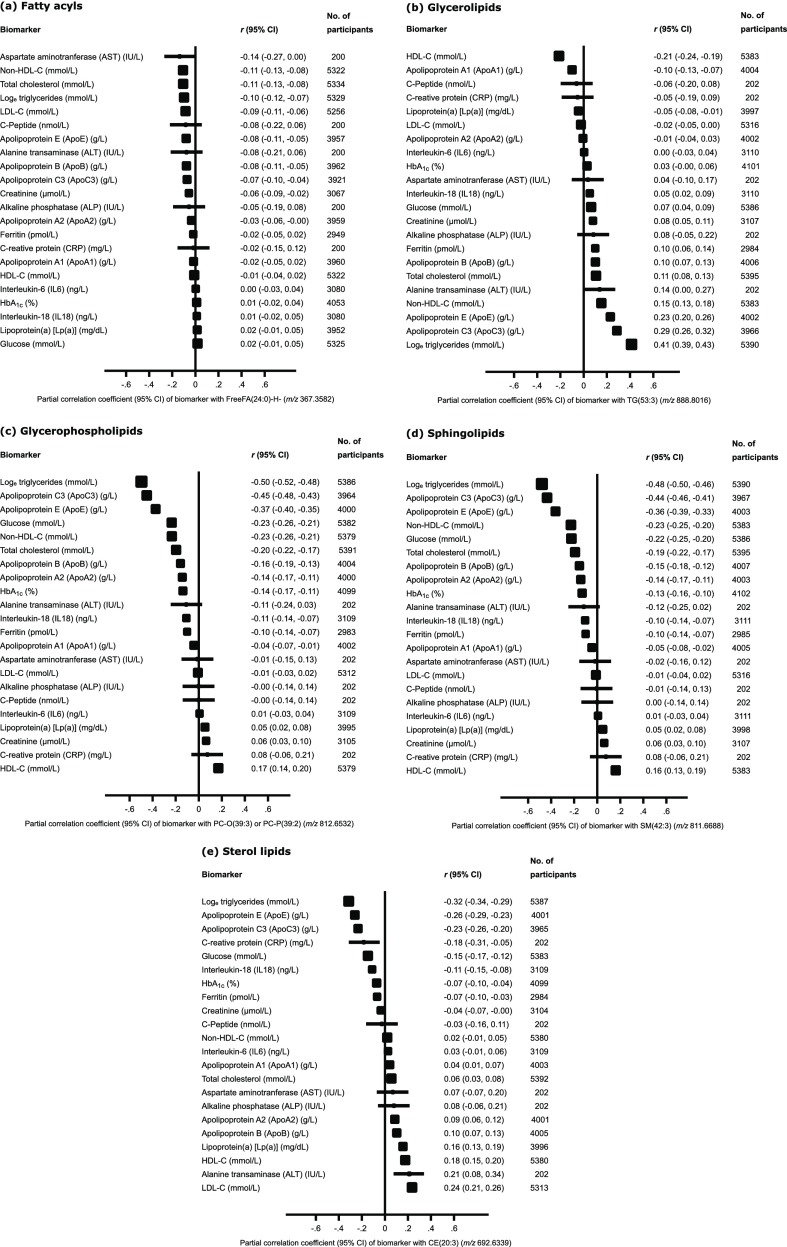
For the lipids within each overall lipid category that were most strongly associated with the APOA5–APOC3 variant [i.e., FreeFA(24:0)-H– (m/z 367.3582) for fatty acids, TG(53:3) (m/z 888.8016) for glycerolipids, PC-O(39:3) or PC-P(39:2) (m/z 812.6532) for glycerophospholipids, SM(42:3) (m/z 811.6688) for sphingolipids, and CE(20:3) (m/z 692.6339) for sterol lipids], the cross-correlation of each lipid metabolite with a wide range of clinical measurements, representing major lipid markers and other circulating biomarkers, was determined (Figure 3a–e).
Figure 3.
Cross-correlations of circulating biomarkers with the lipids within each overall lipid category most strongly associated with rs662799 in the APOA5–APOC3 locus. For the lipids within each overall lipid category that were most strongly associated with rs662799 (chr11:116663707) in the APOA5–APOC3 region, the correlations of these lipids with a range of circulating biomarkers are shown. Analyses were adjusted for age and sex.
FreeFA(24:0)-H– was inversely associated with total cholesterol, non-HDL cholesterol, LDL-C, triacylglycerols, ApoB, ApoC3, ApoE, and several other biomarkers (Figure 3a). Triacylglycerol TG(53:3) (Figure 3b) showed significant positive correlations with levels of total triacylglycerols, as would be expected, but also with ApoB, ApoC3, and ApoE, total cholesterol, non-HDL cholesterol, and several other biomarkers. This triacylglycerol species also exhibited a significant negative correlation with HDL-C and ApoA1. For the sphingolipid [SM(42:3)] (Figure 3d) and sterol lipid [CE(20:3)] (Figure 3e), the strongest inverse associations were found with total triacylglycerols, ApoC3, and ApoE. SM(42:3) had the strongest correlation with HDL-C, while CE(20:3) had the strongest correlation with LDL-C.
Association of Lipids with Various Risk Factors for CHD
The associations of lipid principal components with CHD risk factors are shown in Figure S7. Individuals who were overweight or diabetic—i.e., with high levels of CHD risk factors—were more likely to have lipid profiles similar to those corresponding to positive loadings in the second principal component. In contrast, individuals who were not overweight or diabetic and did not have hypertension—i.e., at reduced risk of CHD—were more likely to have lipid profiles matching positive scores in the third or fourth principal components. For example, a 1-SD increase in the loading scores of the lipids that made up the third principal component resulted in a 20% reduction in the risk of being overweight (OR = 0.80, 95% CI 0.76–0.84) and a 23% reduced risk of having diabetes according to levels of HbA1c (OR = 0.77, 95% CI 0.72–0.81), which is a reflection of long-term blood glucose levels.
Again using the subset of lipids that were significantly associated with the APOA5–APOC3 variant, we also examined the association of the top 20 lipids with the strongest associations with obesity, hypertension, and diabetes (Figure 4). We found that individuals with elevated levels of almost all of these diacylglycerols and triacylglycerols were very likely to have obesity, hypertension, and diabetes, while the majority of sphingomyelins, phosphocholines, and cholesterol esters were predominantly inversely associated with these outcomes. One exception is SM(38:1) (m/z 759.6372), which was positively associated with obesity but inversely associated with hypertension. However, it is important to note that there was no follow-up of the participants in this study and we cannot therefore directly determine associations between lipids and CHD risk; we can only determine associations between lipids and known CHD risk factors.
Figure 4.
Association of lipids with obesity, hypertension, and diabetes. All analyses were adjusted for age and sex. Out of the lipids that were associated with rs662799 in the APOA5–APOC3 locus, results are shown for (a) the top 20 lipids that were most significantly associated with obesity, (b) the top 20 lipids that were most significantly associated with hypertension, and (c) the top 20 lipids that were most significantly associated with diabetes. Definitions: Diabetes = HbA1c ≥ 6.5%; Hypertension = SBP ≥ 140 mm Hg or DBP ≥ 90 mm Hg; Obese = BMI ≥ 30 kg/m2. Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; SBP, systolic blood pressure.
GWAS of Principal Components Derived from Lipid Measurements
Finally, we conducted a GWAS of the second, third, and fourth principal components derived from the lipids measured using DIHRMS (Figure S8 and Table S1 in Supporting Information). We identified 74 unique variants from two loci (APOA5–APOC3 and FADS1–2–3) that reached genome-wide significance (P < 5 × 10–8). The second principal component was only associated with a single variant from the APOA5–APOC3 region (rs662799), the third principal component was associated with 22 variants from the APOA5–APOC3 region, and the fourth principal component was associated with 74 variants from both loci.
Varied Genetic Associations of Lipids with APOA5–APOC3 Gene Region
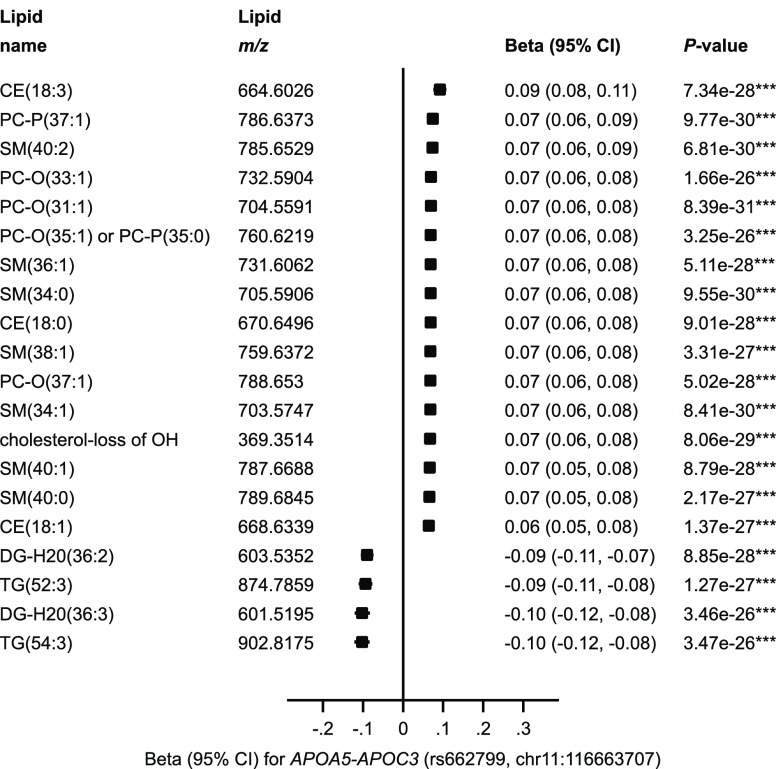
The association of each individual lipid with a common polymorphism (rs662799) in the APOA5–APOC3 cluster revealed differences in the magnitude and direction of the association according to overall lipid category (Figure 5). The four glycerolipids (diacylglycerols and triacylglycerols) had inverse associations with the effect allele for his variant, while the cholesterol esters, phosphatidylcholines, sphingomyelins, and cholesterol had positive associations.
Figure 5.
Association of the top 20 most significantly associated lipids with rs662799 in the APOA5–APOC3 locus. Note: *P < 0.001; **P < 5 × 10–8; ***P < 8.9 × 10–10.
Discussion
In this study, we employed DIHRMS in combination with a novel peak-picking algorithm to measure and characterize approximately 360 lipids in 5662 participants from PROMIS. Our robust approach has three major practical advantages: (1) The automated sample preparation of one plate is possible in 1.5 h, making this approach applicable to large-scale lipid profiling; (2) the method is extremely fast, so that with an analysis time of just over 2 min per sample it is possible to run several hundred samples per day; (3) as a consequence of the high throughput, the cost per sample is greatly reduced; and (4) by virtue of the simplicity of the method it is also robust, with low CVs achievable for many of the lipid species detected. Moreover, the two-dimensional nature of the DIHRMS data limits the need for data alignment algorithms, which are necessary for three-dimensional data such as those generated using LC-based approaches.7 A slight disadvantage is that the use of high-resolution mass spectrometry alone complicates species identification compared to fragmentation-based approaches.17 However, the automated approach and rapid analysis speed opens up the possibility of applying this method to much larger epidemiological studies than was previously possible.17,18
Our determination of the correlation of each phosphatidylcholine and triacylglycerol with major lipid markers demonstrated distinct differences between individual lipids and their relation to lipid markers measured using clinical chemistry (e.g., total cholesterol, total triacylglycerols, LDL-C, HDL-C). Likewise, the GGM indicated that there were significantly more correlations between lipids of the same subclass and lipids with the same constituent fatty acids than would be expected due to chance alone.
From the results of the overall PCA, the second principal component revealed a contrast between free fatty acid levels versus small, saturated triacylglycerols. Several factors could have contributed to this differentiation. Volunteers were recruited at different hospitals and blood samples were taken directly after consent. Thus, there was significant variation in the time since participants had eaten their last meal, which would have strongly affected both the free fatty acid and triacylglycerol pool content in blood plasma through the activity of lipases acting on both blood and adipose tissue triacylglycerols. These enzymes are in part regulated by insulin, thus influencing the association between the second principal component and increased risk of being overweight or diabetic. Because physiological status also influences lipase activities, this could have obscured the genetic associations, with only one SNP demonstrating a genome-wide significant association with the second principal component. Genetic variants in the APOA5–APOC3 region have been associated with type 2 diabetes,42 fatty liver disease,43 hypertriacylglycerolmia,44,45 and dyslipidaemia,46 and associated with metabolites such as 1-linoleoylglycerol in previous metabolomics studies.47 However, as only one variant was significantly associated with the second principal component compared to 74 variants that were associated with the third and fourth principal components, this suggests that the second principal component was largely driven by dietary and physiological patterns rather than genetic differences.
Although the third principal component was most closely characterized by unsaturated triacylglycerols, it did not show any significant associations with genetic variants in the FADS1–2–3 locus, and was only associated with variants in the APOA5–APOC3 region. In contrast, variants in both the FADS1–2–3 and APOA5–APOC3 regions were significantly associated with the fourth principal component. The loadings of the fourth principal component showed that the triacylglycerols containing linoleic acid (18:2), as well as linoleic acid as a free fatty acid, had negative loading scores, while sphingomyelins containing odd-chain fatty acids and desaturated phospholipids had positive loading scores. The association between SNPs within the FADS1–2–3 region with sphingomyelins has been described previously,48 although not explained, and has not previously been described for odd-chain fatty acid-containing sphingomyelins. The effect on the triacylglycerols also explained the association with SNPs in the APOA5–APOC3 locus for the fourth principal component. Both the third and fourth principal components showed inverse associations with the relative risk for being overweight and having diabetes, while only the fourth principal component also showed an inverse association with the relative risk for hypertension. This last observation is striking as sphingomyelins have thus far been implicated in hypertension as precursors to ceramide production, but odd-chain fatty acid-containing sphingomyelins have mostly been unexplored, and in the present study sphingolipid metabolism was associated with perceived positive outcomes in terms of common clinical chemistry measures, including inverse associations with total triacylglycerols, ApoC3, ApoE, total cholesterol, and HbA1c, and a positive association with HDL-C.
Our results from this study complement previous studies that have examined the role of lipids in predicting levels of CVD risk factors and determining risk of CVD. A tandem mass spectrometry-based approach for targeted lipidomics has demonstrated the predictive nature of the approach for general anthropometric characteristics41 and both cardiovascular events and death.49 An analysis of 310 lipid species detected in blood plasma from the Action in Diabetes and Vascular Disease: Preterax and Diamicron MR-Controlled Evaluation (ADVANCE) trial found that the addition of seven lipid species to a base model consisting of fourteen traditional risk factors and medications to predict cardiovascular events increased the C-index from 0.680 (95% CI 0.678–0.682) to 0.700 (95% CI 0.698–0.702; P < 0.0001).49 The prediction of cardiovascular death was similarly improved by the inclusion of four lipid species into the base model.49
Evidence strongly supports that proteins in the APOA5–APOC3 region modulate lipoprotein lipase function and influence liver uptake of remnants, leading to elevated triacylglycerol levels and increased risk of CHD.14,50 However, the details of which specific triacylglycerols, or indeed other lipid species, are influenced by APOA5–APOC3 activity has not been thoroughly understood. The lipidome scan of a common polymorphism in the APOA5–APOC3 region revealed that differences in apolipoprotein metabolism lead to several specific changes in the lipid profile. In particular, increased APOA5–APOC3 activity leads to decreases in levels of specific triacylglycerols, mainly those containing monounsaturated fatty acids. These findings highlight the opportunities of this approach in larger studies, which can include sufficient coverage of rare genetic variants, to help understand how polymorphisms in APOA5–APOC3 can lead to changes in triacylglycerol levels. Increased activity of the APOA5–APOC3 variant also resulted in increased concentrations of cholesterol esters, sphingomyelins, and phosphatidylcholines, which suggests that variants in the APOA5–APOC3 locus have a nonspecific effect on all lipid classes.
A full genome-wide association analysis for each lipid, investigation of novel loci, and identification of lipids that may have a causal effect on risk of CHD was beyond the scope of the present study. However, future work resulting from this lipidomics platform will focus on a GWAS of each of the individual lipids and the subsequent identification of novel associations of lipids with CHD-related loci. While the present study was cross-sectional in nature, we will also explore opportunities to replicate these analyses in prospective cohort studies, which have the opportunity to follow up participants and assess the role of lipid metabolism in disease onset.
Conclusions
We show that fast, reproducible, and detailed lipid profiling is possible on very large data sets, revealing that lipid profiles are influenced by physiological, lifestyle, and genetic factors. This lipidomics platform, then, is a useful tool that may prove valuable to identify lipids that could become new biomarkers used for clinical application in areas such as CHD screening,51 risk prediction,52 and drug development.53
Acknowledgments
We acknowledge the contributions of the following individuals: Michael Eiden for his assistance with the batch design, Philip Haycock and Nasir Sheikh for aliquoting and ordering the samples on the plates, and Lee Matthews for performing the lipid profiling. PROMIS has received approval by the relevant ethics committee of each of the institutions involved in participant recruitment and the Center for Non-Communicable Diseases in Karachi, Pakistan. Informed consent was obtained from each participant recruited into the study, including for use of samples in genetic, biochemical, and other analyses. Fieldwork, genotyping, and standard clinical chemistry assays in PROMIS were principally supported by grants awarded to the University of Cambridge from the British Heart Foundation, UK Medical Research Council, Wellcome Trust, EU Framework 6–Funded Bloodomics Integrated Project, Pfizer, Novartis, and Merck. The MRC/BHF Cardiovascular Epidemiology Unit is underpinned by program grants from the UK Medical Research Council (G0800270), British Heart Foundation (SP/09/002), UK National Institute for Health Research Cambridge Biomedical Research Centre, European Research Council (268834), and European Commission Framework Programme 7 (HEALTH-F2–2012–279233). AK and JLG are funded by the UK Medical Research Council under the Lipid Dynamics and Regulation supplementary grant (MC_PC_13030) and Lipid Programming and Signaling program grant (MC_UP_A090_1006) and Cambridge Lipidomics Biomarker Research Initiative (G0800783). DSP and DSt are funded by the Wellcome Trust (105602/Z/14/Z).
Glossary
Abbreviations
- BMI
body mass index
- CHD
coronary heart disease
- CI
confidence interval
- CV
coefficient of variation
- DBP
diastolic blood pressure
- DHS
Demographic and Health Surveys
- DIHRMS
direct infusion high-resolution mass spectrometry
- DNA
deoxyribonucleic acid
- EA
effect allele
- GGM
Gaussian graphical model
- GRCh37
Genome Reference Consortium human genome build 37
- GWAS
genome-wide association study
- HbA1c
hemoglobin A1c
- HDL-C
high-density lipoprotein cholesterol
- LDL-C
low-density lipoprotein cholesterol
- LIPID MAPS
LIPID Metabolites and Pathways Strategy
- m/z
mass-to-charge ratio
- MI
myocardial infarction
- mzXML
mass-to-charge ratio eXtensible Markup Language
- NEA
noneffect allele
- PCA
principal component analysis
- PROMIS
Pakistan Risk of Myocardial Infarction Study
- QC
quality control
- SBP
systolic blood pressure
- SD
standard deviation
- SE
standard error
- SNP
single nucleotide polymorphism.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00786.
Supporting methods, table, and figures (PDF)
Author Contributions
¶ ELH and AK contributed equally to this work. AMW and JLG jointly directed this work. AK generated the lipidomics data. ELH had full access to the data and performed statistical analyses. ELH, AK, and JLG drafted the manuscript and interpreted the results. All authors contributed important intellectual content to the study, and have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): EBF and DZ are employees and shareholders of Pfizer, Inc. JD has received research funding from the British Heart Foundation, the National Institute for Health Research Cambridge Comprehensive Biomedical Research Centre, the Bupa Foundation, diaDexus, the European Research Council, the European Union, the Evelyn Trust, the Fogarty International Centre, GlaxoSmithKline, Merck, the National Heart, Lung, and Blood Institute, the National Institute for Health Research, the National Institute of Neurological Disorders and Stroke, NHS Blood and Transplant, Novartis, Pfizer, the UK Medical Research Council, and the Wellcome Trust. DSa has received funding from Pfizer, Regeneron Pharmaceuticals, Genentech, and Eli Lilly. JLG has received funding from Agilent, WatersGlaxoSmithKline, Medimmune, Unilever, AstraZeneca, the Medical Research Council, the Biotechnology and Biological Sciences Research Council, the National Institute of Health, the British Heart Foundation, and the Wellcome Trust. All other authors declare no competing interests.
Supplementary Material
References
- Griffin J. L.; Atherton H.; Shockcor J.; Atzori L. Metabolomics as a tool for cardiac research. Nat. Rev. Cardiol. 2011, 8 (11), 630–643. 10.1038/nrcardio.2011.138. [DOI] [PubMed] [Google Scholar]
- Shah S. H.; Kraus W. E.; Newgard C. B. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation 2012, 126 (9), 1110–1120. 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann C.; Pechlaner R.; Willeit P.; Langley S. R.; Mangino M.; Mayr U.; Menni C.; Moayyeri A.; Santer P.; Rungger G.; et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation 2014, 129 (18), 1821–1831. 10.1161/CIRCULATIONAHA.113.002500. [DOI] [PubMed] [Google Scholar]
- Pechlaner R.; Kiechl S.; Mayr M. Potential and caveats of lipidomics for cardiovascular disease. Circulation 2016, 134 (21), 1651–1654. 10.1161/CIRCULATIONAHA.116.025092. [DOI] [PubMed] [Google Scholar]
- Willer C. J.; Schmidt E. M.; Sengupta S.; Peloso G. M.; Gustafsson S.; Kanoni S.; Ganna A.; Chen J.; Buchkovich M. L.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45 (11), 1274–1283. 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. J.; Peloso G. M.; Yu H.; Butterworth A. S.; Wang X.; Mahajan A.; Saleheen D.; Emdin C.; Alam D.; Alves A. C.; et al. Exome-wide association study of plasma lipids in > 300,000 individuals. Nat. Genet. 2017, 49 (12), 1758–1766. 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A.; Want E. J.; O’Maille G.; Abagyan R.; Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78 (3), 779–787. 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Shevchenko A.; Simons K. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010, 11 (8), 593–598. 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- Dejaegher B.; Heyden Y. V. Ruggedness and robustness testing. J. Chromatogr A 2007, 1158 (1–2), 138–157. 10.1016/j.chroma.2007.02.086. [DOI] [PubMed] [Google Scholar]
- Saleheen D.; Zaidi M.; Rasheed A.; Ahmad U.; Hakeem A.; Murtaza M.; Kayani W.; Faruqui A.; Kundi A.; Zaman K. S.; et al. The Pakistan Risk of Myocardial Infarction Study: a resource for the study of genetic, lifestyle and other determinants of myocardial infarction in South Asia. Eur. J. Epidemiol. 2009, 24 (6), 329–338. 10.1007/s10654-009-9334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abegunde D. O.; Mathers C. D.; Adam T.; Ortegon M.; Strong K. The burden and costs of chronic diseases in low-income and middle-income countries. Lancet 2007, 370 (9603), 1929–1938. 10.1016/S0140-6736(07)61696-1. [DOI] [PubMed] [Google Scholar]
- Wang H.; Naghavi M.; Allen C.; Barber R. M.; Bhutta Z. A.; Carter A.; Casey D. C.; Charlson F. J.; Chen A. Z.; Coates M. M.; Coggeshall M.; Dandona L.; Dicker D. J.; Erskine H. E.; Ferrari A. J.; Fitzmaurice C.; Foreman K.; Forouzanfar M. H.; Fraser M. S.; Fullman N.; Gething P. W.; Goldberg E. M.; Gra M. C. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388 (10053), 1459–1544. 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013, 45 (1), 25–33. 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordestgaard B. G.; Varbo A. Triglycerides and cardiovascular disease. Lancet 2014, 384 (9943), 626–635. 10.1016/S0140-6736(14)61177-6. [DOI] [PubMed] [Google Scholar]
- Saleheen D.; Soranzo N.; Rasheed A.; Scharnagl H.; Gwilliam R.; Alexander M.; Inouye M.; Zaidi M.; Potter S.; Haycock P.; et al. Genetic determinants of major blood lipids in Pakistanis compared with Europeans. Circ.: Cardiovasc. Genet. 2010, 3 (4), 348–357. 10.1161/CIRCGENETICS.109.906180. [DOI] [PubMed] [Google Scholar]
- Moore R. T.blockTools: Blocking, Assignment, and Diagnosing Interference in Randomized Experiments, Version 0.6–1; 2014.
- Han X.; Gross R. W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003, 44 (6), 1071–1079. 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- Graessler J.; Schwudke D.; Schwarz P. E.; Herzog R.; Shevchenko A.; Bornstein S. R. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One 2009, 4 (7), e6261. 10.1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30 (10), 918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M.; Fahy E.; Cotter D.; Brown A.; Dennis E. A.; Glass C. K.; Merrill A. H. Jr.; Murphy R. C.; Raetz C. R.; Russell D. W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–32. 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl C.; Tautenhahn R.; Böttcher C.; Larson T. R.; Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84 (1), 283–289. 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger O.; Armando A. M.; Brown A. H.; Milne S. B.; Myers D. S.; Merrill A. H.; Bandyopadhyay S.; Jones K. N.; Kelly S.; Shaner R. L.; et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 2010, 51 (11), 3299–3305. 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Definition and Diagnosis of Diabetes Mellitus and Intermediate Hyperglycemia: Report of a WHO/IDF Consultation; World Health Organization: Geneva, Switzerland, 2006.
- World Health Organization . Use of Glycated Haemoglobin (HbA1c) in the Diagnosis of Diabetes Mellitus: Abbreviated Report of a WHO Consultation; World Health Organization: Geneva, Switzerland, 2011. [PubMed]
- Mancia G.; Fagard R.; Narkiewicz K.; Redon J.; Zanchetti A.; Bohm M.; Christiaens T.; Cifkova R.; De Backer G.; Dominiczak A.; et al. 2013 ESH/ESC guidelines for the management of arterial hypertension: the Task Force for the Management of Arterial Hypertension of the European Society of Hypertension (ESH) and of the European Society of Cardiology (ESC). Eur. Heart J. 2013, 34 (28), 2159–2219. [DOI] [PubMed] [Google Scholar]
- World Health Organization . Obesity and overweight [Fact sheet] https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight (accessed Jan 9, 2019).
- Marchini J.; Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010, 11 (7), 499–511. 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Willer C. J.; Li Y.; Abecasis G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26 (17), 2190–2191. 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J.; Suhre K.; Illig T.; Adamski J.; Theis F. J. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst. Biol. 2011, 5, 21. 10.1186/1752-0509-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opgen-Rhein R.; Strimmer K. From correlation to causation networks: a simple approximate learning algorithm and its application to high-dimensional plant gene expression data. BMC Syst. Biol. 2007, 1, 37. 10.1186/1752-0509-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbi A.; Iorio F.; Dawson K. J.; Wedge D. C.; Tamborero D.; Alexandrov L. B.; Lopez-Bigas N.; Garnett M. J.; Jurman G.; Saez-Rodriguez J. Fast randomization of large genomic datasets while preserving alteration counts. Bioinformatics 2014, 30 (17), i617–23. 10.1093/bioinformatics/btu474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Population Studies (NIPS) Pakistan ICF International. Pakistan Demographic and Health Survey 2012–13; NIPS/Pakistan and ICF International: Islamabad, Pakistan, 2013.
- Kastenmüller G.; Raffler J.; Gieger C.; Suhre K. Genetics of human metabolism: an update. Hum. Mol. Genet. 2015, 24 (R1), R93–R101. 10.1093/hmg/ddv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B. J.; Seyssel K.; Chiu S.; Pan P. H.; Lin S. Y.; Stanley E.; Ament Z.; West J. A.; Summerhill K.; Griffin J. L.; et al. Odd chain fatty acids; new insights of the relationship between the gut microbiota, dietary intake, biosynthesis and glucose intolerance. Sci. Rep. 2017, 7, 44845. 10.1038/srep44845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K.; Shin S.-Y.; Petersen A.-K.; Mohney R. P.; Meredith D.; Wägele B.; Altmaier E.; Deloukas P.; Erdmann J.; et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011, 477 (7362), 54–60. 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illig T.; Gieger C.; Zhai G.; Römisch-Margl W.; Wang-Sattler R.; Prehn C.; Altmaier E.; Kastenmüller G.; Kato B. S.; Mewes H. W.; et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 2010, 42 (2), 137–141. 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew Tan C.; Virtue S.; Murfitt S.; Roberts L. D.; Phua Y. H.; Dale M.; Griffin J. L.; Tinahones F.; Scherer P. E.; Vidal-Puig A. Adipose tissue fatty acid chain length and mono-unsaturation increases with obesity and insulin resistance. Sci. Rep. 2016, 5, 18366. 10.1038/srep18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhi N. G.; Koulman A.; Sharp S. J.; Imamura F.; Kroger J.; Schulze M. B.; Crowe F. L.; Huerta J. M.; Guevara M.; Beulens J. W.; et al. Differences in the prospective association between individual plasma phospholipid saturated fatty acids and incident type 2 diabetes: the EPIC-InterAct case-cohort study. Lancet Diabetes Endocrinol. 2014, 2 (10), 810–818. 10.1016/S2213-8587(14)70146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiden M.; Koulman A.; Hatunic M.; West J. A.; Murfitt S.; Osei M.; Adams C.; Wang X.; Chu Y.; Marney L.; et al. Mechanistic insights revealed by lipid profiling in monogenic insulin resistance syndromes. Genome Med. 2015, 7, 63. 10.1186/s13073-015-0179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhi N. G.; Imamura F.; Sharp S. J.; Koulman A.; Schulze M. B.; Zheng J.; Ye Z.; Sluijs I.; Guevara M.; Huerta J. M.; et al. Association of plasma phospholipid n-3 and n-6 polyunsaturated fatty acids with type 2 diabetes: the EPIC-InterAct case-cohort study. PLoS Med. 2016, 13 (7), e1002094. 10.1371/journal.pmed.1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J. M.; Wong G.; Barlow C. K.; Greeve M. A.; Kowalczyk A.; Almasy L.; Comuzzie A. G.; Mahaney M. C.; Jowett J. B.; Shaw J.; et al. Plasma lipid profiling in a large population-based cohort. J. Lipid Res. 2013, 54 (10), 2898–2908. 10.1194/jlr.P035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C. J.; Sanna S.; Jackson A. U.; Scuteri A.; Bonnycastle L. L.; Clarke R.; Heath S. C.; Timpson N. J.; Najjar S. S.; Stringham H. M.; et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008, 40 (2), 161–169. 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q.; Baker S. S.; Liu W.; Arbizu R. A.; Aljomah G.; Khatib M.; Nugent C. A.; Baker R. D.; Forte T. M.; Hu Y.; et al. Increased apolipoprotein A5 expression in human and rat non-alcoholic fatty livers. Pathology 2015, 47 (4), 341–348. 10.1097/PAT.0000000000000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L. A.; Olivier M.; Hubacek J. A.; Cohen J. C.; Cox D. R.; Fruchart J. C.; Krauss R. M.; Rubin E. M. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 2001, 294 (5540), 169–173. 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- Talmud P. J.; Hawe E.; Martin S.; Olivier M.; Miller G. J.; Rubin E. M.; Pennacchio L. A.; Humphries S. E. Relative contribution of variation within the APOC3/A4/A5 gene cluster in determining plasma triglycerides. Hum. Mol. Genet. 2002, 11 (24), 3039–3046. 10.1093/hmg/11.24.3039. [DOI] [PubMed] [Google Scholar]
- Johansen C. T.; Wang J.; Lanktree M. B.; Cao H.; McIntyre A. D.; Ban M. R.; Martins R. A.; Kennedy B. A.; Hassell R. G.; Visser M. E.; et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 2010, 42 (8), 684–687. 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.-Y.; Fauman E. B.; Petersen A.-K.; Krumsiek J.; Santos R.; Huang J.; Arnold M.; Erte I.; Forgetta V.; Yang T.-P.; et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014, 46 (6), 543–550. 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draisma H. H.; Pool R.; Kobl M.; Jansen R.; Petersen A. K.; Vaarhorst A. A.; Yet I.; Haller T.; Demirkan A.; Esko T.; et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat. Commun. 2015, 6, 7208. 10.1038/ncomms8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshehry Z. H.; Mundra P. A.; Barlow C. K.; Mellett N. A.; Wong G.; McConville M. J.; Simes J.; Tonkin A. M.; Sullivan D. R.; Barnes E. H.; et al. Plasma lipidomic profiles improve upon traditional risk factors for the prediction of cardiovascular events in type 2 diabetes. Circulation 2016, 134 (21), 1637–1650. 10.1161/CIRCULATIONAHA.116.023233. [DOI] [PubMed] [Google Scholar]
- Nordestgaard B. G. Triglyceride-rich lipoproteins and atherosclerotic cardiovascular disease: new insights from epidemiology, genetics, and biology. Circ. Res. 2016, 118 (4), 547–563. 10.1161/CIRCRESAHA.115.306249. [DOI] [PubMed] [Google Scholar]
- Roberts L. D.; Koulman A.; Griffin J. L. Towards metabolic biomarkers of insulin resistance and type 2 diabetes: progress from the metabolome. Lancet Diabetes Endocrinol. 2014, 2 (1), 65–75. 10.1016/S2213-8587(13)70143-8. [DOI] [PubMed] [Google Scholar]
- Meikle P. J.; Wong G.; Barlow C. K.; Kingwell B. A. Lipidomics: potential role in risk prediction and therapeutic monitoring for diabetes and cardiovascular disease. Pharmacol. Ther. 2014, 143 (1), 12–23. 10.1016/j.pharmthera.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Wishart D. S. Applications of metabolomics in drug discovery and development. Drugs R&D 2008, 9 (5), 307–322. 10.2165/00126839-200809050-00002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.