Abstract
Following fertilization, the two specified gametes must unite to create an entirely new organism. The genome is initially transcriptionally quiescent, allowing the zygote to be reprogrammed to a totipotent state. Gradually, the genome is activated through a process known as the maternal-to-zygotic transition (MZT), which enables zygotic gene products to replace the maternal supply that initiated development. This essential transition has been broadly characterized through decades of research in several model organisms. Yet, we still lack a full mechanistic understanding of how genome activation is executed and how this activation relates to the reprogramming of the zygotic chromatin architecture. Recent work highlights the central role of transcriptional activators and suggests that these factors may coordinate transcriptional activation with other developmental changes.
Introduction
During the first hours of animal development, the differentiated germ cells, the egg and the sperm, must be reprogrammed to a totipotent state. This process ensures that the newly formed zygotic genome can subsequently drive the differentiation of all the diverse cell types of the adult animal. This efficient reprogramming relies on maternally supplied RNAs and proteins that have been stockpiled in the oocyte. The ability of these maternal products to drive reprogramming was demonstrated more than half a century ago by John Gurdon. In a ground-breaking experiment, Gurdon generated the first cloned frog by transplanting the nucleus of a somatic cell into an enucleated egg1. Although similar somatic cell nuclear transfer (SCNT) experiments have enabled the successful cloning of mammals2, the process is inefficient, which suggests that additional epigenetic factors prime the paternal and maternal genomes for the transition to totipotency following fertilization.
The zygotic genome remains transcriptionally silent while reprogramming takes place3,4. However, for the embryo to continue developing beyond this initial reprogramming phase, the zygotic genome must be expressed. Transcriptional control is passed to the zygote through a process known as the maternal-to-zygotic transition (MZT), during which the degradation of maternal products is coordinated with zygotic genome activation (ZGA)4 (Box 1). There are many parallels between this rapid and efficient developmental transition and experimental reprogramming in culture5. Thus, understanding the mechanisms that underlie genome activation will inform our efforts to direct cellular reprogramming in culture, offering tremendous potential for both the modelling and treatment of disease.
Box1: Decay of maternal transcripts is coordinated with ZGA.
Transcriptional activation of the zygotic genome is coordinated with the degradation of the maternally deposited transcripts that control the initial stages of development. Like zygotic genome activation (ZGA), maternal mRNA clearance is a gradual process. Some transcripts are eliminated soon after fertilization, whereas others are degraded only after the major onset of transcription4. Depending on the species, 30–40% of maternally deposited mRNAs are eliminated by degradation, and overall the levels of up to 60% of maternal mRNAs are considerably reduced5.
Maternal mRNA silencing is controlled by a variety of different RNA-binding protein complexes, which recognize sequences in the maternal RNAs to promote their degradation via cleavage, deadenylation, and decapping5. Recently, novel regulatory mechanisms, such as suboptimal codon usage161,162 and RNA modifications (including N6-methyladenosine163,164), have been implicated as important determinants of RNA stability during the MZT.
Maternal mRNA clearance is required to remove repressive factors and enable zygotic transcription32,52,165. The massive RNA turnover that takes place during the MZT also permits the establishment of embryonic patterning, as many uniformly distributed maternal transcripts are replaced by spatially restricted zygotic transcripts50. Although maternal clearance is permissive for ZGA, zygotic transcription is required, in turn, for degradation of many maternal transcripts34,50,166,167. One mechanism by which this is accomplished is the early zygotic expression of microRNAs that are required for the clearance of hundreds of maternal transcripts168-170. The coordinated execution of ZGA and maternal RNA degradation creates the monumental transcriptome remodeling that is required to reset cellular identity in the embryo.
The MZT is conserved across the kingdom Animalia. In many animals, this massive transcriptional shift coincides with changes to the cell cycle that are referred to as the mid-blastula transition (MBT)4. Prior to ZGA, embryos undergo rapid cellular divisions, switching between DNA replication (S phase) and division (mitosis) without pausing in gap phase6. As the MZT nears completion, the division cycle slows and a gap phase is introduced, providing cells time to grow prior to the next division. Collectively, these changes prepare the embryo for gastrulation, during which cells begin to migrate and differentiate into the major germ layers of the animal4.
In recent years, technological innovations have dramatically improved our ability to interrogate the processes that govern ZGA (Figure 1, Box 2). New live-imaging methods allow the expression of individual genes to be tracked with unprecedented spatial and temporal precision7,8. Likewise, the increased availability of high-throughput sequencing has fostered numerous assays that allow the transcriptome9, transcription-factor binding10, and chromatin structure11-13 to be analysed genome wide. With the advent of single-cell and low-input sequencing methods14-18, both ZGA and the chromatin remodelling that accompanies it have been studied with increasing resolution.
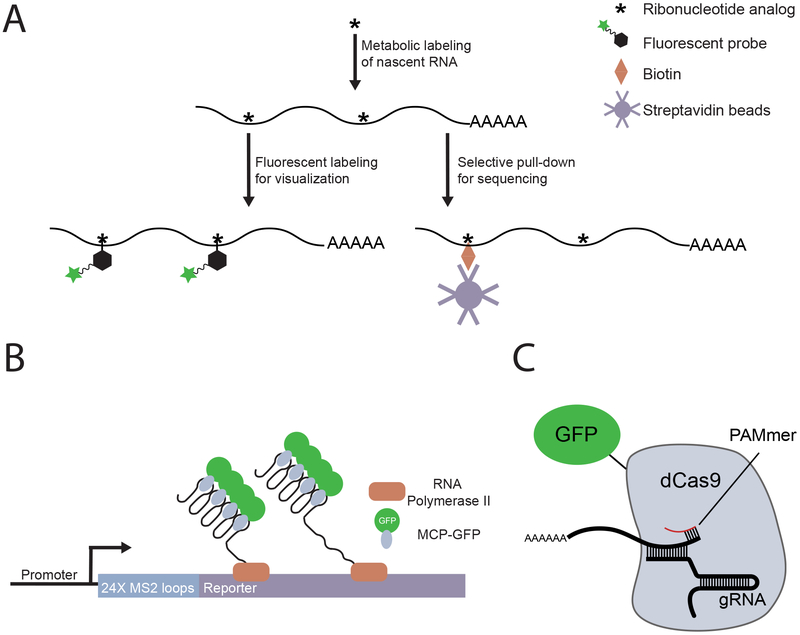
Figure 1: New technologies enable precise detection of zygotic genome activation.
(A)Detection of nascent transcripts by metabolic labeling: Cells are supplied with ribonucleoside or ribonucleotide analogs (for example, 5-ethynyl uridine or 4-thio-UTP) to label actively transcribed RNA. The labeled RNA is functionalized through coupling to a biophysical probe, such as a fluorescent azide for visualization or a biotin group for selective pull-down and sequencing.
(B) Detection of nascent transcripts using MS2-based reporters: A series of MS2 sequences is introduced adjacent to a gene of interest. As the MS2 motifs are transcribed they form RNA stem loops that are bound by a maternally provided MS2 coat protein fused to fluorescent protein (MCP-GFP) to collectively produce a fluorescent spot.
(C) Detection of transcripts by RNA-targeted deadCas9 (dCas9) fused to a fluorescent protein (for example, GFP): The dCas9-GFP fusion protein is targeted to RNA through interaction with a guide RNA (gRNA), which contains sequence that is complementary to the RNA of interest, and a DNA oligo that contains the protospacer adjacent motif (PAM), known as a PAMmer.
Box2: New technologies enable mechanistic insights into zygotic genome activation.
The advent of single-cell sequencing has provided increased resolution with which to detect gene expression in the developing embryo171. Recently, these methods have been combined with advanced computational pipelines to generate transcriptome atlases of frog and zebrafish embryogenesis172-174, allowing the fate of individual cells to be traced from pluripotency to specification. Examining the branch points in these differentiation pathways will lead to the identification of novel lineage-defining transcription factors and expand our understanding of how such factors function. Further, new low-input sequencing techniques have allowed us to profile the transcriptome123,171, methylome64, and chromatin-accessibility landscape14,99 of embryos despite the limited material available for these studies. The first maps of early human embryos have identified fundamental differences in the regulation of human and mouse zygotic genome activation (ZGA)14.
In the early embryo, the ability to detect ZGA by standard methods is limited by an abundance of maternally supplied RNAs. Assays that specifically select for nascent and, therefore, zygotic transcripts offer one way to circumvent this issue. In recent years, new methods utilizing metabolic labeling have improved the ease with which we are able to detect nascent transcription (Figure 1A). One such method, using 4-thio-UTP labeling followed by biotinylation and streptavidin pull-down, was recently used to profile the early zygotic transcriptome in zebrafish49. Other methods incorporate azide-modified uridine analogs into the nascent transcripts, which enables the labelled RNA to be conjugated to a variety of probes through click reactions and Staudinger reactions175. Alternatively, emerging data suggests that attachment of a fluorescent group allows early zygotic transcription to be detected visually35.
Increasingly sensitive, fast, and precise imaging technologies, such as super-resolution microscopy and light-sheet microscopy, have enabled researchers to image living embryos as they develop, and this capability has led to the identification of new structures and processes139,176,177. Advanced imaging methods have also improved the resolution with which ZGA is visualized. Single-molecule fluorescence in situ hybridization (FISH) allowed quantitative measurement of endogenous transcription, whereas live-cell imaging using fluorescently tagged RNA and proteins enabled simultaneous detection of temporal and spatial expression patterns7,8,175,178. These methods have been used to track the activation of individual genes during ZGA179, and have led to the discovery of subtle transcriptional phenomena, such as mitotic memory180 and transcriptional bursting181,182. One effective way to track RNA, is by introducing repeats of the bacteriophage MS2 sequence to the transcribed region of a gene, which allows the expression of this gene to be monitored spatially and temporally183 (Figure 1B). CRISPR/Cas9 technology offers additional ways to image specific transcripts in living embryos8 and can be used in combination with DNA-FISH184 or to introduce RNA reporters at endogenous loci to help avoid potential artifacts of these methods. For example, a catalytically dead version of Cas9 (dCas9) fused to a fluorescent protein can be used to track either specific transcripts or genomic loci in live cells8,185,186 (Figure 1C). Preliminary studies demonstrated that this approach can be used in live embryos to confirm a candidate gene as one of the initially transcribed loci in zebrafish35.
In this Review, we discuss the mechanisms that regulate genome activation, focussing on aspects that have been studied in multiple model species (Figure 2) and highlighting the transcription factors at the centre of this process. We will also discuss the dynamics of ZGA, models of ZGA timing, and the interplay between ZGA and chromatin remodelling. For a more comprehensive discussion of maternal RNA degradation5, cell-cycle remodelling6, or chromatin dynamics during the MZT 19,20, we direct you to current reviews.
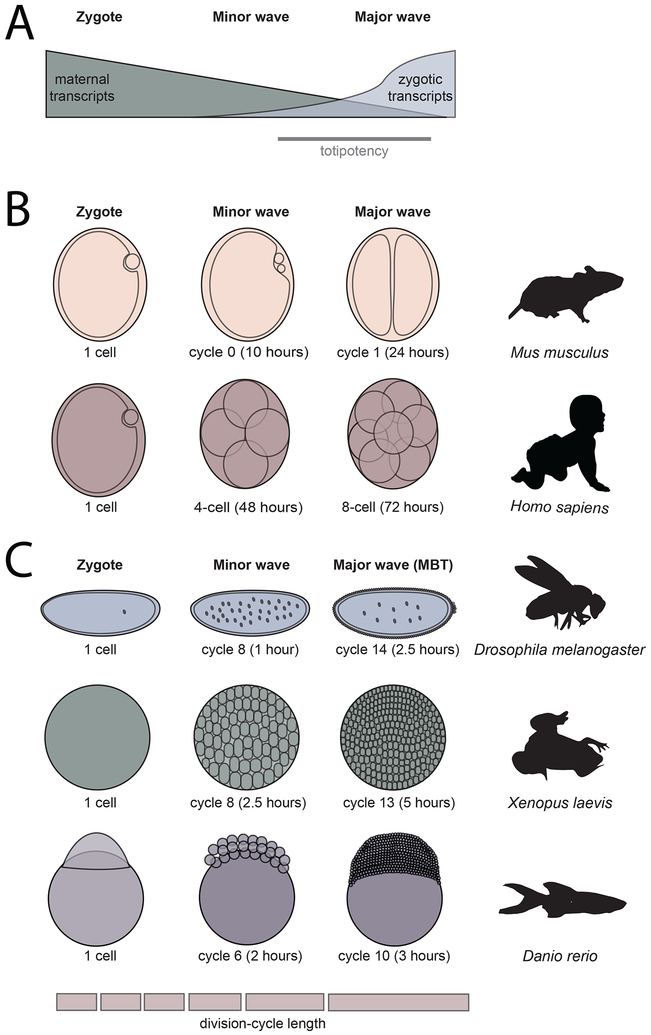
Figure 2. Zygotic genome activation is conserved across animals.
(A) In the first hours of life, animals undergo a process called the maternal-to-zygotic transition (MZT) in which the clearance of maternal products is coordinated with the activation of zygotic transcription. A totipotent state (gray bar) is established during this transition.
(B,C) Key stages of zygotic genome activation are outlined for five model species, indicated on the right. The absolute time (in hours post fertilization) is indicated below. All species begin life as a single-cell zygote. Zygotic transcription initiates in an early minor wave, which is later followed by a major wave of genome activation.
(B) In mice and humans, early cell divisions do not occur as rapidly as in externally fertilized organisms such as frogs, zebrafish and flies (C). Nonetheless, as in other species, genome activation is a gradual process with a minor wave and major wave of transcription.
(C) In frogs, zebrafish, and flies, the rapid division cycles that characterize early development gradually slow over the course of the MZT. In these species, the major wave of genome activation coincides with the mid-blastula transition (MBT). The MBT involves the end of synchronous division cycles, the introduction of a gap phase (G2) to the cell cycle, and additional, species-specific developmental changes.
Models and mechanisms of ZGA timing
ZGA is not a single event, but rather a period over which transcription is gradually activated21-23. It is characterized by two transcriptional waves: a minor wave that occurs during the early cleavage divisions and a major wave that coincides with the first division-cycle pause in many species4. The timing of these waves and the number of division cycles varies widely across animals, but within species the process is tightly controlled and the timing is highly reproducible. Rapidly developing species like worms (Caenorhabditis elegans), frogs (Xenopus laevis), fish (Danio rerio), and flies (Drosophila melanogaster) complete the MZT and enter gastrulation only a few hours after fertilization. By contrast, in slower developing mammals such as mice (Mus musculus) and humans, the MZT takes one or more days4 (Figure 2). This difference between rapidly and slowly developing animals is thought to stem from the nature of the egg6. An egg that is abandoned in a predator-laden environment has different needs than one that is protected in a uterus and externally provided with nutrients. Of course, important differences exist within these simplistic categories. In the fly embryo, for example, cytokinesis is deferred in favour of speed; nuclei divide in a common cytoplasm until the end of the MZT, when cellularization takes place4. Although the particular needs of an egg dictate different modes of embryogenesis, many fundamental processes are conserved, and in all animals the accurate timing of the onset of ZGA depends on several intricately coordinated mechanisms.
Models that explain ZGA timing.
Delayed transcriptional activation of the zygotic genome is thought to be instrumental in allowing the genomes of the sperm and egg to combine and be reprogrammed to totipotency. Nonetheless, the reason for this initial transcriptional quiescence is not fully understood. In the simplest form, two mechanistic models exist to explain the lack of transcription at fertilization: (1) proteins required for transcriptional activation are not present or are inactive; or (2) all the factors required to drive activation are present, but inhibitors prevent expression of the genome
The first major model to explain the timing of ZGA was based on the idea that the early division cycles could regulate activation of the zygotic genome through changes in the ratio of nuclear to cytoplasmic components. In many species, the volume of the embryo does not change during the MZT, that is the volume of cytoplasm within the embryo remains constant. By contrast, during each division cycle both the nuclear volume and the nuclear content, in the form of DNA, increase. Collectively, this leads to a progressive increase in the nucleocytoplasmic ratio (N:C ratio). A landmark study showed that ZGA takes place two cell cycles earlier in polyspermic frog embryos compared with embryos fertilized by a single sperm24. The authors proposed that the embryo’s increasing supply of nuclear material could titrate a maternally supplied repressor to gradually relieve transcriptional repression (Figure 3A). This model was supported by a series of additional experiments in frogs. In these studies, experimentally increasing the DNA content of the embryo by injecting plasmid DNA resulted in premature ZGA25, whereas increasing the cytoplasmic volume using cleavage inhibitors or physical constriction caused a delay in ZGA24. Similarly, transcription was activated early in mutant zebrafish in which the N:C ratio was artificially increased by creating dense patches of DNA through defects in chromosome segregation26. Despite this experimental evidence, the universality of this model was challenged by the discovery that haploid fly embryos execute ZGA with proper timing27. A detailed dissection using compound chromosomes revealed that only a subset of fly genes responds to the N:C ratio28. Similarly, although this ratio influences morphological changes in mice, it does not affect transcription on a global scale29.
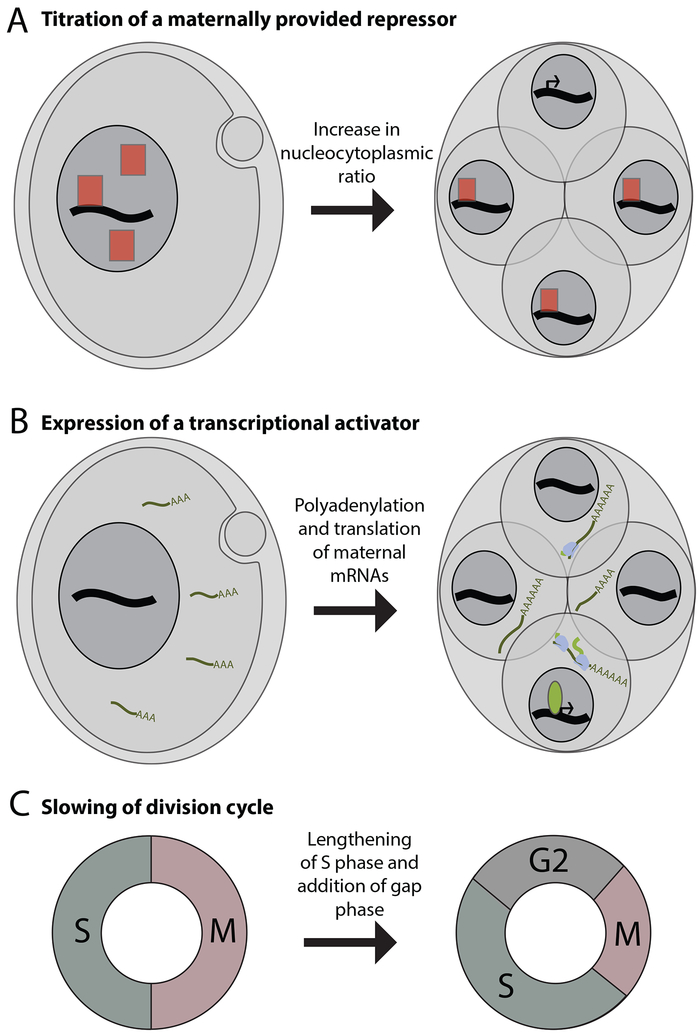
Figure 3. Several mechanisms contribute to the timing of zygotic genome activation.
(A) A maternally supplied repressor (red square) prevents transcription in the early embryo. As the ratios of genetic material (black line) or nuclear volume (grey circle) to cytoplasm increase with each cell division, the repressor is titrated and transcription initiates in cells in which repressor concentration has fallen below a threshold level
(B) The early embryo lacks a key transcriptional activator (green oval). Polyadenylation and translation of maternally supplied mRNA leads to its accumulation. Once a threshold level has been reached, the factor enables expression of its target genes.
(C) The rapid early cell cycles consist of only a DNA replication (S) phase and mitosis (M). At the major wave of zygotic genome activation, the cell cycle slows and a gap phase (G2) is introduced, reducing the time restraint initially placed on transcription.
A counterpart to the N:C ratio driven model, known as the maternal-clock model, posits that fertilization or egg activation initiates a biochemical cascade that serves as a molecular timer. The embryo receives many factors as maternally deposited mRNAs that are often held in a dormant state by inhibitory RNA-binding proteins. Even after this repression is released, it takes time for these transcripts to be polyadenylated, translated, and transported to the nucleus. Thus, in theory, accumulation of any essential factor required to either activate transcription or alleviate repression could contribute to ZGA timing (Figure 3B). A few such factors have been identified to date, including components of the basal transcriptional machinery30,31, the fly maternal clearance factor Smaug32,33, and transcriptional activators in zebrafish and flies34-36.
Of course, these models are not mutually exclusive, and it is becoming increasingly clear that multiple processes coordinately regulate ZGA timing. Indeed, characterisation of the molecules involved in ZGA suggests that titration of maternal repressors, accumulation of transcriptional activators, and division-cycle lengthening collectively create a permissive environment for ZGA to occur.
Titration of maternal repressors.
The N:C ratio model posits the existence of one or more titratable, maternally supplied repressors (Figure 3A). Although gene-specific repressors have long been known37-39, this model predicts the existence of a highly expressed repressor that strongly binds DNA with little sequence specificity. The core histone proteins, H2A, H2B, H3, and H4, form an octamer around which DNA is wrapped to form nucleosomes, and together they fulfil these criteria, as they bind ubiquitously across the genome and inhibit transcription by limiting the ability of regulators to access DNA40. In frogs, levels of the core histone H3 regulate transcription both in vitro and in the embryo, supporting a role for histones in transcriptional repression immediately after fertilization41. Levels of core histones have similarly been shown to regulate ZGA in zebrafish, but in contrast to the original N:C ratio model, it is the levels of soluble, and not DNA-bound histones, that determine the timing of activation42. The density of histones bound to DNA remains steady through the early cleavage cycles; however, a drop in the concentration of unbound nuclear histones coincides with ZGA42. Free histones may initially buffer against premature transcription, but as activators accumulate and histone concentrations are reduced, the balance shifts in favour of activation. Thus, the increase in both nuclear content (DNA) and nuclear volume result in the titration of maternal repressor and transcriptional activation41-44.
Accumulation of transcriptional activators.
In addition to the titration of a maternally deposited repressor, the early embryo may lack one or more essential factors that must accumulate to enable transcription (Figure 3B). A classic example of a factor that is rate-limiting for ZGA is TATA-binding protein (TBP), a general transcription factor that, as part of the TFIID complex, promotes formation of the RNA polymerase II preinitiation complex45. In frogs, translation of TBP is upregulated immediately before the onset of the major wave of transcription and precocious TBP activity results in early ZGA31. In worms, another component of TFIID, TAF-4, is sequestered in the cytoplasm by repressor binding until a phosphorylation cascade triggered by fertilization results in its timely release and the onset of ZGA30. However, a molecular timer based on basal transcription factors does not explain how the appropriate subset of genes is selected for activation. Both general transcription factors that directly license the genome for activation and sequence-specific transcription factors that select the appropriate subset of genes for expression from the zygotic genome must be present for ZGA to occur normally.
Division-cycle lengthening.
In many species, early embryonic development is characterized by a series of rapid division cycles, and the slowing of this cycle is coordinated with the major wave of ZGA (Figure 2). Since transcription is largely shut off when nuclei enter mitosis46, the rapid cleavage cycles of the early embryo leave only brief windows of time for transcription47,48. Fittingly, genes expressed very early in development tend to be short and lack introns47,49-51. The transcriptional output of the genome gradually increases as the division cycle slows, culminating in the major wave of ZGA as cells enter the first G2 pause. These observations suggest that division-cycle slowing could set the pace of genome activation (Figure 3C). In frogs, elongating the early cell cycle results in premature transcription, supporting this model52,53. By contrast, in zebrafish and flies, blocking division-cycle progression does not affect ZGA timing54-56. The relationship between these processes is likely complex. For example, in flies zygotically expressed inhibitors are required to slow the cycle57. Thus, although a rapid division cycle places limits on transcription, division-cycle slowing is interconnected with other mechanisms of ZGA regulation in some species.
Chromatin remodeling in early embryos
In eukaryotes, the genome requires considerable compaction to fit inside the nucleus. DNA is spooled around octamers of histones to form nucleosomes, which are coiled into fibers and looped into higher-order structures58. In addition, post-translational modifications to these histones can influence chromatin structure. This organization modulates the ability of the transcriptional machinery to access the DNA, and thus, has a central role in gene regulation59.
After fertilization, chromatin from the sperm and the egg is unified to create an entirely new genome. Early transcriptional quiescence of the zygotic genome allows the chromatin, which comes from two distinct cell types, to be remodeled to a naive, globally accessible state. It remains unclear whether activation of the zygotic genome is instructive to the changes in the underlying chromatin or whether the changes in chromatin are required for genome activation. However, it is clear that these processes are intimately linked. In this section, we discuss the multiple levels of chromatin reorganization that occur in the early embryo, focusing on the general features that have been studied in multiple species (Figure 4). We highlight events that are shared between the maternal and paternal genomes. Nonetheless, it is important to recognize that these two distinct genomes undergo different processes as they are brought together in the zygote, and that in many species the paternal genome is repackaged through the exchange of histones for protamines19,20,60,61.
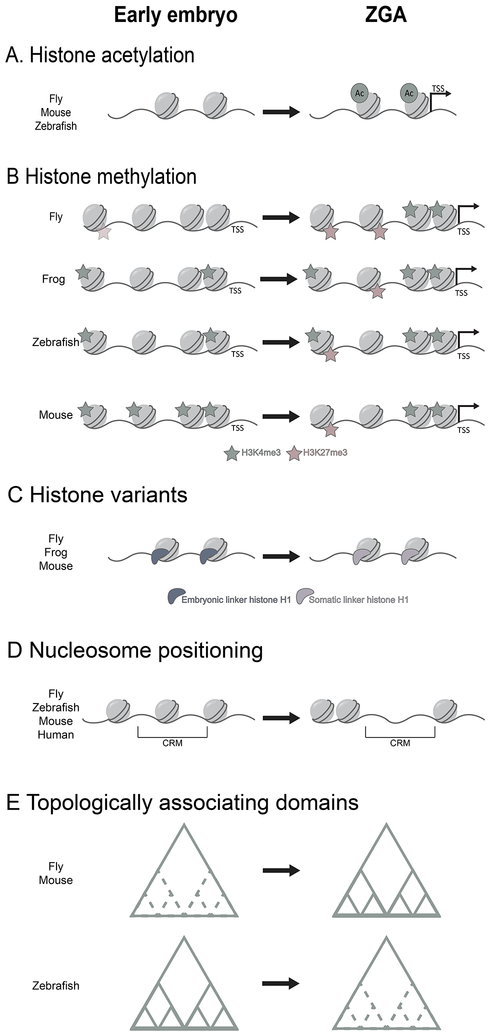
Figure 4. Chromatin is reprogrammed during zygotic genome activation.
(A) In flies, mice, and zebrafish, histone acetylation increases over the course of ZGA, marking genes for activation during this transition. TSS, transcription start site.
(B) In flies, the early embryo may contain low levels of H3K27me3, and both H3K27me3 and H3K4me3 increase sharply during the major wave of genome activation. In frogs, H3K4me3 is present in the early embryo at low levels and increases over the course of ZGA. H3K27me3 is established later, during the major wave of transcription. In early zebrafish, H3K4me3 appears to poise genes for activation. During the major wave of ZGA, H3K4me3 levels increase and H3K27me3 is established. H3K27me3 co-marks histones with H3K4me3, forming bivalent domains. In mice, unusual broad domains of H3K4me3 are restricted to TSS-associated peaks during ZGA with H3K27me3 established later.
(C) In flies, frogs, and mice, embryonic variants of linker histone H1 are replaced with their somatic counterparts at the major onset of ZGA. In flies, it has been shown that incorporation of the somatic H1 variant is instrumental for genome activation to occur.
(D) Defined cis-regulatory elements (CRM), characterized by open chromatin, are established during ZGA in flies, zebrafish, mice, and humans.
(E) In flies and mice, the boundaries of topologically associating domains (TADs) are established concurrently with ZGA. In zebrafish, TADs are present in the early embryo but are lost prior to the major wave of genome activation.
DNA Methylation.
Methylation of cytosine to 5-methylcytosine promotes transcriptional silencing during processes such as genomic imprinting62 and X-chromosome inactivation63. Evidence that human and mouse genomes undergo global demethylation prior to ZGA suggests that this mark could also contribute to the transcriptional silence of the early embryo64-66. By contrast, both frog and zebrafish genomes remain heavily methylated throughout early development67-70. In zebrafish, DNA methylation of the maternal genome is widely reprogrammed as the oocyte-specific methylation pattern is erased and replaced with one similar to that of the inherited paternal genome69,70. Although recent work suggests that DNA methylation may be involved in recruitment of the zebrafish genome-activating transcription factors Nanog and Pou5f3 to distal regulatory elements71,72, methylation at promoters is anti-correlated with accessible chromatin and early zygotic expression of the associated genes68-70. By contrast, methylated promoters are robustly transcribed during ZGA in frogs68. Furthermore, the genomes of worms and flies possess limited amounts of DNA methylation, suggesting that the functions of DNA methylation during ZGA may not be conserved73,74. Thus, DNA methylation is dynamic through the MZT, but the role of this mark in regulating activation of the zygotic genome remains unclear and is species dependent.
Histone modifications.
Post-translational modifications chemically alter histone tails in ways that impact nucleosome stability and the recruitment of transcriptional regulators59. Chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) time course data have revealed widespread changes in the abundance of these marks over the MZT. Here we focus on a subset of modifications that have been broadly studied in multiple organisms: histone acetylation, which is associated with active gene expression, histone H3 lysine 4 trimethylation (H3K4me3), which is associated with active promoters, and H3K27me3, which is associated with repression59.
Histone acetylation is generally associated with chromatin accessibility and active transcription. As might be predicted, in many species histone acetylation increases during the MZT and is associated with genes that are activated during ZGA (Figure 4A). In flies, histone H4 lysine 8 acetylation (H4K8ac), H3K18ac, and H3K27ac are enriched at the transcription start sites (TSS) of genes expressed during the minor and major wave of genome activation75. Similarly, in mice and zebrafish, published16,76 and preliminary35 data indicate that H3K27ac increases on chromatin from the oocyte until the stage at which the genome is activated. Thus, an increase in histone acetylation marking actively transcribed genes appears to be a shared featured of ZGA and may be important in creating accessible regions of chromatin.
H3K4me3 is a canonical mark of activation that is often found at the TSS of genes77. In zebrafish, H3K4me3 is detected at many promoters prior to ZGA and appears to poise genes for activation78,79. Similarly, in frogs H3K4me3 emerges prior to genome activation but increases as the embryo progresses through gastrulation80,81. By contrast, in flies few promoters are marked with H3K4me3 prior to the major wave of ZGA, indicating that early transcription during the minor wave can occur in the absence of this chromatin signature75,82. Despite some differences, a dramatic increase in H3K4me3 accompanies the major onset of transcription in all three of these species (Figure 4B)75,78-80. This trend appears to be reversed in mice, where early embryos possess unusual, broad (5- to 10-kb) domains of H3K4me3. As the genome becomes transcriptionally active these domains are largely restricted to conventional TSS-associated peaks (Figure 4B), and this reprogramming requires transcription16-18. Notably, knockdown of the demethylases responsible for this pruning causes developmental arrest and downregulation of many ZGA genes16,18, suggesting that these broad domains could play a role in preventing premature transcription prior to ZGA.
In contrast to the association of H3K4me3 with active transcription, H3K27me3 is correlated with transcriptional repression. Similarly to H3K4me3, a substantial increase in H3K27me3 is observed in many species during the MZT18,75,78-80,83 (Figure 4B). Thus, the lack of transcriptional activity in early development is not globally imposed by this repressive histone modification. In both flies and worms, H3K27me3 is transmitted from the oocyte to the embryo and may function to regulate gene expression during ZGA83,84. Nonetheless, like in other species there is a dramatic increase in H3K27me3 in flies as the genome is activated75. In most species, the increase in methylation at this residue occurs later in development than the increase in H3K4me318,78-80. In fact in zebrafish, a number of regions that contain H3K4me3 are subsequently modified with H3K27me378,79. The co-occurrence of the generally activating mark H3K4me3 and the repressive H3K27me3 has been termed bivalency and was first identified in embryonic stem cells85. This bivalent mark poises promoters of developmentally regulated genes in pluripotent cells86 and may similarly poise promoters in the zebrafish embryo. Such bivalent domains have not been identified in mice, flies or frogs during the MZT17,18,75,80,87, but other combinations of posttranslational modifications may function to poise regulatory regions in these organisms. For the subset of histone modifications that have been studied in multiple organisms, it is clear that although there are interspecies differences, all examined genomes exhibit global shifts in their histone-modification profile as the embryo proceeds through the MZT. Future studies will be needed to resolve the causal relationship between these modifications and transcriptional activation.
Histone variants.
The constitution of a nucleosome can also be altered by replacing canonical histone proteins with histone variants88-92. The best-studied example is the germline-specific linker histone H1, which has been identified in multiple species. In flies, frogs, and mice, embryonic variants of linker histone H1 are replaced by their somatic counterparts at the major onset of ZGA (Figure 4C)93-95. These embryonic H1 variants are predicted to form less stable nucleosomes than their somatic counterparts and may, therefore, contribute to the naive chromatin environment of the early embryo. The importance of this variant has been demonstrated in flies, where a lack of embryonic H1 results in premature transcription and developmental arrest93.
More recently, a role for the histone variant H2A.Z (H2AFV in zebrafish) has been demonstrated to be instrumental in regulating DNA-methylation dynamics and transcription in zebrafish embryos96. Nucleosomes containing this histone variant and marked by H3K4me1 are anti-correlated with DNA methylation in sperm and function to protect regions of the genome from aberrant methylation. These ‘placeholder’ nucleosomes similarly protect the early embryonic genome immediately after fertilization and play a role in directing the reprogramming of the DNA methylation state of the maternal genome. Misregulation of H2AFV localization results in aberrant gene expression, demonstrating a connection between histone variants, DNA methylation, and genome activation. Although this particular relationship between genome activation and chromatin reprogramming may be specific to zebrafish, placeholder nucleosomes may more generally serve as a platform for maintaining epigenetic information through early cell divisions96.
Nucleosome positioning and chromatin accessibility.
In zebrafish, reorganization of nucleosome positioning coincides with widespread genome activation. Well-positioned nucleosome arrays appear at the promoters of genes, marking many for future activation97. Recently, chromatin accessibility profiling has revealed that defined regulatory regions are established concomitantly with transcriptional activation in flies98, zebrafish71, mice99,100, and humans (Figure 4D)14,101,102. Early mouse embryos possess broad regions of open chromatin that are narrowed down to mark promoters by the major wave of ZGA100. Intriguingly, these broad regions often encompass transposable elements that are transiently expressed during ZGA in mice100,103. This burst of transposable element expression was initially assumed to be a side effect of global chromatin accessibility, however recent work suggests that it may contribute to both chromatin opening104 and early gene expression105,106.
Chromatin domains.
Condensed chromatin fibers form loops that are clustered together into topologically associating domains (TADs)107,108. By keeping specific promoters in the proximity of enhancers or silencers, TADs play an important role in transcriptional regulation109. Recently, chromosome conformation capture (3C) techniques, such as HiC, have been used to assess changes in these 3D chromatin contacts over the course of the MZT110. In fly embryos, TAD boundaries are gradually established in concert with gene activation, but the formation of these boundaries does not depend on transcription (Figure 4E)111,112. Likewise, in mice, the genome lacks tightly defined TADs until after ZGA, and TADs are formed even when transcription is inhibited113,114. Although these studies support the notion that metazoan genomes are largely unstructured during the initial phase of embryogenesis, experiments in zebrafish demonstrate that prior to ZGA the genome is highly structured and that this organization is largely erased as the embryo transitions through the MZT115 (Figure 4E). Together these data suggest that the dynamics of TAD formation during the MZT is not conserved amongst metazoans, and that higher order chromatin structure is formed independently of transcription.
Activators direct gene expression
By binding to specific DNA sequences, transcription factors direct the transcriptional machinery to particular genes. This specificity is of critical importance during ZGA, when it is estimated that 12–15% of the genome is transcriptionally activated4.
The first identified master regulator of ZGA was discovered in flies116. This transcription factor, Zelda (ZLD), is maternally deposited as mRNA and is translationally upregulated in the hour leading up to ZGA36,116-118. ZLD is required for the expression of hundreds of genes during both the major and minor waves of ZGA, and without this essential factor, fly embryos die before completing the MZT116,118. Since ZLD orthologs are limited to the insect clade119, it was initially unclear whether there were factors that function analogously in vertebrates. However, two independent studies identified Nanog, SoxB1, and Pou5f3 (Oct4) as activators of zebrafish ZGA34,120. Interest in these factors was stimulated by their identification as the most highly translated transcription factors in zebrafish embryos immediately after fertilization34. These factors are homologues of the mammalian ‘pluripotency factors’, which are known for their ability to reprogram differentiated cells to a stem cell-like state121. Importantly, although they are not phylogenetically related to ZLD, these transcription factors share several functional characteristics. Like ZLD, the zebrafish activators are translated early in development allowing them to activate the earliest expressed genes and poise hundreds of additional genes for activation during the major onset of ZGA116-118,120.
Advances in low-input sequencing methods have recently led to the first discoveries of mammalian genome activators. One study in human embryos found that OCT4-binding sites are enriched in accessible regulatory regions during ZGA, and knockdown of this factor results in downregulation of hundreds of ZGA genes14. By contrast, OCT4 motifs are not enriched in accessible regions identified by other chromatin profiling methods, leaving the role of OCT4 in human ZGA unclear101,102. Importantly, Oct4 is not involved in ZGA in mice, where it is only required later in embryogenesis14,122. Instead, the binding motif for a lesser known pluripotency factor, Nfy, is enriched in open chromatin during ZGA in mice and Nfy is required for expression of many ZGA genes99. Humans and mice do share at least one family of genome activators known as the DUX transcription factors. Dux (mouse) and DUX4 (human) activate hundreds of ZGA genes in these species, including endogenous retroviral elements (ERV), such as MERVL in mice and HERVL in humans123-125. DUX genes are zygotically expressed as part of the initial wave of ZGA and the mechanisms by which they are activated remain unknown123. Thus, some genome activators are maternally deposited as mRNAs and translationally upregulated, whereas others are regulated at the level of transcription. In all cases, genome activator proteins are not present at fertilization, suggesting their activity must be carefully controlled to prevent premature transcriptional activation.
Activators have been identified in many species (Table 1), but it is clear that additional transcription factors remain to be identified. Nanog, Pou5f3, and SoxB1 are required to drive the minor wave of genome activation in zebrafish, but it is less obvious what activates the major wave of genome activation. By contrast, in mammals the DUX transcription factors are expressed from the zygotic genome, but what drives their initial transcription is unknown. In addition, to date no similar transcriptional activators have been identified in frogs. Together these gaps in knowledge highlight the continued need to identify additional factors that regulate genome activation.
Table 1:
Genome-activating transcription factors
| Genome Activator |
Organism | Conservation | Developmental requirement | Chromatin regulation |
|---|---|---|---|---|
| Zelda (ZLD) | Drosophila melanogaster | Insects | Maternal mutants fail to complete cellularization and die before the end of MZT. Zygotic mutants die in late embryogenesis116,144. | In embryos that lack maternal ZLD, nucleosome occupancy is increased127 and chromatin accessibility is lost126 at ZLD-binding sites during ZGA. |
| Pou5f3 | Danio rerio | Jawed vertebrates, paralog of Oct4 | Maternal mutants show a mild delay in epiboly194. Maternal and zygotic mutants exhibit a developmental delay and arrest at gastrulation147, failed endoderm formation195, and patterning defects196. | Knockdown in the embryo results in decreased accessibility at binding sites during ZGA71. |
| Sox19b | Danio rerio | Metazoans | Knockdown of the functionally redundant SOXB1 family (SOX1/2/3/19) results in delayed epiboly, a shortened anterior-posterior axis, and impaired CNS development197. | - |
| Nanog | Danio rerio | Vertebrates | Knockdown impairs endoderm formation, and embryos die at the end of gastrulation198. Mutants lacking maternal and zygotic gene products display impaired epiboly, failure to form axial structures, and massive cell death at the end of gastrulation128,149,150. | Knockdown in the embryo results in decreased accessibility at binding sites during ZGA71. |
| DUX | Mus musculus | Placental mammals | Knockout embryos arrest before the morula or blastocyst stage124. | Induction in mESCs remodels chromatin to resemble that of the early embryo123. |
| NFY | Mus musculus | Eukaryotes | Knockdown in the embryo results in arrest at the morula stage99. | Knockdown in the embryo results in decreased accessibility at binding sites during ZGA99. Displaces nucleosomes from DNA in vitro131. |
| DUX4 | Homo sapiens | Placental mammals | - | Histone H3 is depleted at binding sites following induction in myoblasts132. |
| OCT4 | Homo sapiens | Jawed vertebrates | Null embryos collapse during blastocyst formation199. | Required for open chromatin at binding sites in mESCs136. |
CNS, central nervous system; mESCs, mouse embryonic stem cells; MZT, maternal-to-zygotic transition; ZGA, zygotic genome activation
Potential mechanisms of genome activators.
As discussed above, multiple factors have been identified that are essential for activating transcription from the zygotic genome, but for many it is unclear whether they function predominantly to select genes for expression or whether they also function to directly activate transcription. Indeed, ZLD is bound to thousands of regulatory regions at least one hour before the associated genes are activated117, suggesting that although ZLD binding selects genes to be expressed during ZGA, additional proteins are required to initiate transcription. Indeed, the widespread effect of identified activators of ZGA may stem from their ability to regulate chromatin structure and define regulatory regions (Table 1).
ZLD-binding sites are strongly correlated with regions of chromatin accessibility during the major wave of ZGA in flies117, and embryos that lack ZLD lose accessibility at many of these sites126,127. ZLD binding reduces the local nucleosome occupancy and may promote histone acetylation at the regulatory regions of early expressed genes75,126,127. Likewise, published66 and preliminary data128 suggest that Nanog and Pou5f3 are required for chromatin accessibility at enhancers of developmental genes during ZGA in zebrafish. Notably, recent preliminary work suggests that the binding of all three zebrafish activators (Nanog, Pou5f3, and SoxB1) is required to maintain nucleosome-free sites post-ZGA128. While knockdown experiments demonstrate that Nfy is needed for open chromatin during mouse ZGA99,129, Dux has only been shown to maintain sites of open chromatin in mouse embryonic stem cells (ESCs)123. Fittingly, the Nfy complex contains domains that interact with DNA in a histone-like manner130, allowing the complex to displace nucleosomes from DNA in vitro131. DUX4 can similarly displace histones in myoblasts and recruits the histone acetyltransferases p300/CBP to establish activating histone modifications132. Finally, although there is evidence that many of these factors function as ‘pioneer factors’ (Box 3), the CASE is arguably strongest for mammalian Oct4, which opens chromatin to promote induced pluripotent stem cell (iPSC) reprogramming133,134. Oct4 binds nucleosomal DNA both in vitro and in fibroblasts135, and recent work suggests that both Oct4 and Pou5f3 may function, in part, by recruiting the chromatin remodeller BRG1 (Smarca4a in zebrafish) to stabilize nucleosome positioning71,136. Thus, defining regions of accessible chromatin is a shared function of genome activators.
Box3: Genome activators share characteristics with pioneer factors.
Pioneer factors are specialized transcription factors that are capable of binding to regions of silent chromatin that are inaccessible to most other DNA-binding factors135. This term was first used to describe the mammalian transcription factor FoxA1, a master regulator of liver cell fate187,188. Based on this archetype, pioneer factors open the local chromatin to poise associated genes for rapid activation upon the arrival of additional factors189. Pioneer factors use this ability to facilitate dramatic transcriptional shifts during cell-fate reprogramming. Genome activators are frequently referred to as ‘pioneers’ based on evidence that they can bind to largely inaccessible chromatin and mediate increased chromatin accessibility135. This pioneering activity is a feature of many of the factors that reprogram the genome during ZGA to establish the embryonic transcriptional program126,127,133,135.
Pioneer factors impact chromatin through a variety of mechanisms. Some pioneers function by recruiting chromatin regulators, such as chromatin remodeling complexes and histone-modifying enzymes135,136,190. For FoxA1, simply binding to a nucleosome is enough to open the chromatin191. FoxA1 disrupts interactions between neighboring nucleosomes and displaces histone H1 using a protein domain that structurally resembles this linker histone192,193. This function is reminiscent of the genome activator Nfy, which uses two histone-like domains to displace nucleosomes from the DNA130,131. FoxA1 binds nucleosomal DNA along a single face of the DNA helix, leaving the opposite face in contact with histones192. In fibroblasts, Oct4 and Sox2 similarly bind intact nucleosomes by targeting partial versions of their canonical DNA motifs134, suggesting that this form of interaction could be a common mechanism in reprogramming.
Pioneer factors function as master regulators of cell fate based on their unique ability to convert silent chromatin into active cis-regulatory elements. This function is of critical importance in the early embryo when these elements are established de novo by genome activators in preparation for ZGA. Although genome activators appear to share the ability to regulate chromatin, the mechanisms by which they do so are likely distinct. Thus, to obtain a more nuanced understanding of their function, the mechanism of each factor should be characterized individually.
Genome activators may also influence the establishment of higher-order chromatin structure. Although TADs remain uncharted in human embryos, Oct4 and Sox2 have been shown to bind chromatin reorganization hotspots during iPSC generation and influence insulation strength at TAD borders137. In zebrafish embryos, these activators are enriched at sites bound by the architectural protein cohesin138, a key player in both ZGA and TAD formation107. Recent work suggests that ZLD also contributes to TAD boundary insulation and the formation of long-range contacts between active genes during ZGA111,112. Whereas loops between active genes are identified prior to the major wave of ZGA and are associated with ZLD binding, repressive loops are formed later112. These observations raise the possibility that genome activators help shape 3D genome reorganization by directing the binding of architectural proteins during ZGA.
Recently, single-molecule imaging has revealed that ZLD promotes formation of transient ‘hubs’ of another transcription factor, called Bicoid. Confinement of Bicoid to these hubs creates sites of high local concentration that potentiate its binding to DNA139. Similar methods have revealed that Sox2 forms clusters of enhancers in ESCs140. Intriguingly, the transcriptional activation domains of both ZLD and Sox2 are predicted to be largely unstructured, which may contribute to this clustering function140,141. Based on these and emerging142,143 studies, it is tempting to speculate that these transcription factors could broadly influence gene expression by forming hubs at chromatin boundaries. Nonetheless, the relationship between transcriptional hubs and TADs remains to be defined.
Misexpression of genome activators in development and disease.
Given the potent effects of activators on gene expression, it might come as no surprise that misexpression of these factors has serious consequences for the embryo. In some cases, these factors are essential for development. For instance, both lack of ZLD activity and excessive ZLD activity are lethal to the fly embryo116,144,145, demonstrating the need for precise regulation of this protein. In zebrafish, mutants that lack both maternal and zygotic pou5f3 gene products display numerous developmental defects including delayed gastrulation and an inability to form endoderm146,147. However, the maternal and zygotic functions of this protein overlap such that mutants that lack only maternally provided pou5f3 develop normally146,148. Similarly, zebrafish lacking maternal and zygotic nanog gene products fail to activate a number of zygotically expressed genes and die early in development, due in large part to defects in expression of genes required for the formation of the yolk syncytial layer149,150. However, zebrafish ZGA is only severely disrupted when at least two of the known genome activators are depleted in combination34. Thus, some genome activators function redundantly with other transcription factors or with their zygotic counterparts.
Long before the DUX proteins were implicated in ZGA, they were known for their role in facioscapulohumeral dystrophy (FSHD), an untreatable form of muscular dystrophy that progresses from the face to the lower limbs151. This disease is caused by misexpression of DUX4 in skeletal muscle cells, where it activates aberrant expression of germline- and stem cell-associated genes152-154. Activation of the pluripotency gene network is also a hallmark of cancer, where it may facilitate the proliferative potential of these cells155. Hence, the ability of genome activators to reprogram cells towards pluripotency also makes them potent drivers of tumor development. Accordingly, aberrant expression of Oct4, Nanog, and Sox2 have been associated with numerous forms of cancer156.
Conclusions and perspective
In the transcriptional silence that follows fertilization, the genome is reprogrammed to prepare the embryo to give rise to a new animal. The transition from silence to widespread gene expression requires precise regulation. This is accomplished through several coordinated mechanisms, in which transcription factors play a central role. After fertilization, translational upregulation promotes accumulation of genome-activating transcription factors. In one model of ZGA timing, these factors begin to successfully compete with histones for DNA binding after reaching a critical threshold. Competition of this nature would integrate a readout of the N:C-ratio (histone concentration) with a readout of the embryo’s molecular clock (activator levels). However, this tidy model is likely oversimplified. Given that histones are newly synthesized in the early embryo157, their levels should also be regulated by molecular clock-based mechanisms. Regardless, competition for access to regulatory regions of individual genes would help explain differences in the timing of gene activation.
The same mechanisms that drive this transcriptional shift in the embryo function in other cellular reprogramming contexts, including the creation of iPSCs. Transcriptional profiling has revealed a remarkable overlap in the gene networks activated during iPSC generation and ZGA105,158. This overlap can be explained, in part, by the discovery that known master regulators of pluripotency also serve as genome activators in the embryo. These transcription factors direct chromatin remodeling in both of these reprogramming contexts, helping to erase the previous cell identity while creating a new one. These parallels demonstrate the ability of work in stem cell models to inform our understanding of embryogenesis, and vice versa. Despite our progress, the causal relationships between the major processes that accompany ZGA remain unclear, and the mechanisms by which many genome activators function have yet to be defined. For instance, although TADs form independently of transcription111,113,114 and the artificial creation of chromatin loops drives gene expression in at least some contexts159, it has yet to be determined whether TAD formation is required for ZGA. Likewise, it is uncertain whether other chromatin changes are required for transcription or whether they are simply the byproduct of transcriptional activity160. Defining how transcription factors function during the initial stages of development will uncover the connections between chromatin remodeling, the mechanisms that govern ZGA, and other fundamental features required for developmental reprogramming.
Acknowledgements
We would like to thank members of the Harrison lab and the reviewers for helpful feedback on the manuscript. KNS was supported in part by the National Institutes of Health (NIH) National Research Service award T32 GM007215. MMH was supported by grant R01GM11694 from the National Institute of General Medical Sciences and a Vallee Scholar Award.
Glossary
- Totipotent:
The property of a cell with the capacity to form all the cells of an organism, including extraembryonic tissues.
- Chromatin:
The complex of DNA, RNA and protein that comprise the chromosomes of eukaryotes.
- Zygotic:
Relating to the diploid, fertilized egg cell (zygote) that results from the fusion of an egg and a sperm.
- Germ layers:
The three layers of cells (ectoderm, mesoderm, and endoderm) that are formed during gastrulation in the early embryo and differentiate to give rise to all of the organs and tissues of the body.
- Cleavage divisions:
The rapid, modified cell cycle of the early embryo, which consists of only M (mitosis) and S (replication) phases and omits G1 and G2 gap phases. These cycles occur in the absence of cell growth and therefore result in no change in the size of the embryo.
- Nucleocytoplasmic ratio (N:C):
The ratio of nuclear content to the cytoplasmic content in a cell or embryo.
- Polyspermic:
Refers to an egg that has been fertilized by more than one sperm, and, thus, contains three or more copies of each chromosome.
- Haploid:
Having a single set of chromosomes. Most animals have diploid somatic cells (with two paired sets of chromosomes) but produce haploid gametes.
- Compound chromosomes:
Chromosomes formed by the attachment of two homologs through a single centromere that are therefore inherited together through mitosis and meiosis. These can be used to generate embryos deficient for an entire chromosome.
- Protamines:
Small, basic proteins that are used in the place of histones to help package DNA in the sperm of some species.
- Demethylation:
The process by which a demethylase enzyme removes a methyl group from a molecule.
- Transposable elements:
DNA sequences that can move from one position within the genome to another.
- Topologically associating domains (TAD):
Three-dimensional chromosome structures within which regions of DNA physically interact with each other with higher frequency than with regions outside.
- MERVL:
A family of endogenous retroviruses (ERV) expressed in mouse embryos during zygotic genome activation. The human versions are known as HERVL.
Footnotes
Competing interests
The authors have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Genetics thanks Ksenia Kuznetsova, Nadine Vastenhouw, and the other anonymous reviewer(s), for their contribution to the peer review of this work.
References
- 1.Gurdon JB The Developmental Capacity of Nuclei taken from Intestinal Epithelium Cells of Feeding Tadpoles. Development 10, (1962). [PubMed] [Google Scholar]
- 2.Campbell KHS, McWhir J, Ritchie WA & Wilmut I Sheep cloned by nuclear transfer from a cultured cell line. Nature 380, 64–66 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Newport J & Kirschner M A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30, 687–696 (1982). [DOI] [PubMed] [Google Scholar]
- 4.Tadros W & Lipshitz HD The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Yartseva V & Giraldez AJ The Maternal-to-Zygotic Transition During Vertebrate Development: A Model for Reprogramming. Curr. Top. Dev. Biol. 113, 191–232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan K, Seller CA, Shermoen AW & O’Farrell PH Timing the Drosophila Mid-Blastula Transition: A Cell Cycle-Centered View. Trends Genet. 32, 496–507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand E et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–45 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Nelles DA et al. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell 165, 488–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Gerstein M & Snyder M RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furey TS ChIP–seq and beyond: new and improved methodologies to detect and characterize protein–DNA interactions. Nat. Rev. Genet. 13, 840–852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giresi PG, Kim J, McDaniell RM, Iyer VR & Lieb JD FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17, 877–885 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide in Current Protocols in Molecular Biology (John Wiley & Sons, Inc., 2001). doi: 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belton J-M et al. Hi–C: A comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao L et al. Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell 173, 248–259.e15 (2018).This paper profiled chromatin accessibility across early human embryogenesis by sequencing DNase I hypersensitive sites and implicates Oct4 as an activator of ZGA.
- 15.Mezger A et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat. Commun. 9, 3647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl JA et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Liu X et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Eckersley-Maslin MA, Alda-Catalinas C & Reik W Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Xu Q & Xie W Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 28, 237–253 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Collart C et al. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development 141, 1927–1939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey SA et al. Identification of the zebrafish maternal and paternal transcriptomes. Development 140, 2703–2710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lott SE et al. Noncanonical Compensation of the Zygotic X Transcription in Early Drosophila melanogaster Development Revealed through Single-Embryo RNA-Seq. PLoS Biol. 9, e1000590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newport J & Kirschner M A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686 (1982). [DOI] [PubMed] [Google Scholar]
- 25.Prioleau MN, Huet J, Sentenac A & Mechali M Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell 77, 439–449 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Dekens MPS, Pelegri FJ, Maischein H-M & Nusslein-Volhard C The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development 130, 3907–3916 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Edgar BA, Kiehle CP & Schubiger G Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44, 365–72 (1986). [DOI] [PubMed] [Google Scholar]
- 28.Lu X, Li JM, Elemento O, Tavazoie S & Wieschaus EF Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development 136, 2101–2110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee DR, Lee JE, Yoon HS, Roh S Il& Kim MK Compaction in preimplantation mouse embryos is regulated by a cytoplasmic regulatory factor that alters between 1- and 2-cell stages in a concentration-dependent manner. J. Exp. Zool. 290, 61–71 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Guven-Ozkan T, Nishi Y, Robertson SM & Lin R Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell 135, 149–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veenstra GJ, Destree OH & Wolffe AP Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol. Cell. Biol. 19, 7972–7982 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benoit B et al. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development 136, 923–932 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tadros W et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell 12, 143–55 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Lee MT et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 (2013).Using ribosome profiling, the authors identified Pou5f3 (previously Pou5f1), Nanog, and Sox2 as the most highly translated transcription factors in the early zebrafish embryo and then showed that depletion of these factors together resutls in a failure to initiate zygotic genome activation.
- 35.Chan SH et al. Brd4 and P300 regulate zygotic genome activation through histone acetylation. bioRxiv 369231 (2018). doi: 10.1101/369231 [DOI] [Google Scholar]
- 36.Harrison MM, Botchan MR & Cline TW Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev. Biol. 345, 248–55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stancheva I & Meehan RR Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 14, 313–27 (2000). [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Ruzov A et al. Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development 131, (2004). [DOI] [PubMed] [Google Scholar]
- 39.Pritchard DK & Schubiger G Activation of transcription in Drosophila embryos is a gradual process mediated chardby the nucleocytoplasmic ratio. Genes Dev. 10, 1131–1142 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Luo RX & Dean DC Chromatin Remodeling and Transcriptional Regulation. JNCI J. Natl. Cancer Inst. 91, 1288–1294 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Amodeo AA, Jukam D, Straight AF & Skotheim JM Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc. Natl. Acad. Sci. 112, E1086–E1095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joseph SR et al. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife 6, (2017).This paper showed a decrease in the concentration of unbound histones in the nuclei of zebrafish embryos that corresponds with ZGA and presented evidence that this decrease permits transcription factors to compete for DNA binding and activate transcription.
- 43.Jevtić P & Levy DL Nuclear Size Scaling during Xenopus Early Development Contributes to Midblastula Transition Timing. Curr. Biol. 25, 45–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jevtić P & Levy DL Both Nuclear Size and DNA Amount Contribute to Midblastula Transition Timing in Xenopus laevis. Sci. Rep. 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahn S Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11, 394–403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gottesfeld JM & Forbes DJ Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22, 197–202 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Rothe M, Pehl M, Taubert H & Jackle H Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature 359, 156–159 (1992). [DOI] [PubMed] [Google Scholar]
- 48.Shermoen AW & O’Farrell PH Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 67, 303–310 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heyn P et al. The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Rep. 6, 285–292 (2014). [DOI] [PubMed] [Google Scholar]
- 50.De Renzis S, Elemento O, Tavazoie S & Wieschaus EF Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol 5, e117 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swinburne IA & Silver PA Intron delays and transcriptional timing during development. Dev. Cell 14, 324–30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collart C, Allen GE, Bradshaw CR, Smith JC & Zegerman P Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 341, 893–896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimelman D, Kirschner M & Scherson T The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell 48, 399–407 (1987). [DOI] [PubMed] [Google Scholar]
- 54.Farrell JA & O’Farrell PH Mechanism and regulation of Cdc25/Twine protein destruction in embryonic cell-cycle remodeling. Curr. Biol. 23, 118–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang M, Kothari P, Mullins M & Lampson MA Regulation of zygotic genome activation and DNA damage checkpoint acquisition at the mid-blastula transition. Cell Cycle 13, 3828–3838 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCleland ML & O’Farrell PH RNAi of Mitotic Cyclins in Drosophila Uncouples the Nuclear and Centrosome Cycle. Curr. Biol. 18, 245–254 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sung H, Spangenberg S, Vogt N & Großhans J Number of Nuclear Divisions in the Drosophila Blastoderm Controlled by Onset of Zygotic Transcription. Curr. Biol. 23, 133–138 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Jiang C & Pugh BF Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10, 161–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bannister AJ & Kouzarides T Regulation of chromatin by histone modifications. Cell Res 21, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steger K & Balhorn R Sperm nuclear protamines: A checkpoint to control sperm chromatin quality. Anat. Histol. Embryol. 47, 273–279 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Zhou L & Dean J Reprogramming the genome to totipotency in mouse embryos. Trends Cell Biol. 25, 82–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li E, Beard C & Jaenisch R Role for DNA methylation in genomic imprinting. Nature 366, 362–365 (1993). [DOI] [PubMed] [Google Scholar]
- 63.Panning B X-chromosome inactivation: the molecular basis of silencing. J. Biol. 7, 30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo H et al. The DNA methylation landscape of human early embryos. Nature 511, 606–610 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Shen L et al. Tet3 and DNA Replication Mediate Demethylation of Both the Maternal and Paternal Genomes in Mouse Zygotes. Cell Stem Cell 15, 459–471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Messerschmidt DM, Knowles BB & Solter D DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 28, 812–828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Veenstra GJC & Wolffe AP Constitutive genomic methylation during embryonic development of Xenopus. Biochim. Biophys. Acta - Gene Struct. Expr. 1521, 39–44 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Bogdanovic O et al. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Res. 21, 1313–27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potok ME, Nix DA, Parnell TJ & Cairns BR Reprogramming the Maternal Zebrafish Genome after Fertilization to Match the Paternal Methylation Pattern. Cell 153, 759–772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang L et al. Sperm, but Not Oocyte, DNA Methylome Is Inherited by Zebrafish Early Embryos. Cell 153, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu G, Wang W, Hu S, Wang X & Zhang Y Inherited DNA methylation primes the establishment of accessible chromatin during genome activation. Genome Res. gr.228833. 117 (2018). doi: 10.1101/gr.228833.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin Y et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science (80-. ). 356, eaaj2239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takayama S et al. Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 24, 821–830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kelly WG Transgenerational epigenetics in the germline cycle of Caenorhabditis elegans. Epigenetics Chromatin 7, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X-Y, Harrison MM, Villalta JE, Kaplan T & Eisen MB Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bogdanovic O et al. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 22, 2043–2053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eissenberg JC & Shilatifard A Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339, 240–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vastenhouw NL et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464, 922–926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lindeman LC et al. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev. Cell 21, 993–1004 (2011). [DOI] [PubMed] [Google Scholar]
- 80.Akkers RC et al. A Hierarchy of H3K4me3 and H3K27me3 Acquisition in Spatial Gene Regulation in Xenopus Embryos. Dev. Cell 17, 425–434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hontelez S et al. Embryonic transcription is controlled by maternally defined chromatin state. Nat. Commun. 6, 10148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen K et al. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. Elife 2, e00861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zenk F et al. Germ line–inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science (80-. ). 357, (2017). [DOI] [PubMed] [Google Scholar]
- 84.Gaydos LJ, Wang W & Strome S H3K27me and PRC2 transmit a memory of repression across generations and during development. Science (80-. ). 345, 1515–1518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bernstein BE et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 125, 315–326 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Mikkelsen TS et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng H et al. Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals. Mol. Cell 63, 1066–1079 (2016). [DOI] [PubMed] [Google Scholar]
- 88.Weber CM & Henikoff S Histone variants: dynamic punctuation in transcription. Genes Dev. 28, 672–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Whittle CM et al. The Genomic Distribution and Function of Histone Variant HTZ-1 during C. elegans Embryogenesis. PLoS Genet. 4, e1000187 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin C-J, Conti M & Ramalho-Santos M Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 140, 3624–3634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang P, Wu W & Macfarlan TS Maternal histone variants and their chaperones promote paternal genome activation and boost somatic cell reprogramming. BioEssays 37, 52–59 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gaume X & Torres-Padilla M-E Regulation of Reprogramming and Cellular Plasticity through Histone Exchange and Histone Variant Incorporation. Cold Spring Harb. Symp. Quant. Biol. 80, 165–175 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Perez-Montero S, Carbonell A, Moran T, Vaquero A & Azorin F The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev. Cell 26, 578–590 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Smith RC, Dworkin-Rastl E & Dworkin MB Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 2, 1284–95 (1988). [DOI] [PubMed] [Google Scholar]
- 95.Fu G et al. Mouse Oocytes and Early Embryos Express Multiple Histone H1 Subtypes1. Biol. Reprod. 68, 1569–1576 (2003). [DOI] [PubMed] [Google Scholar]
- 96.Murphy PJ, Wu SF, James CR, Wike CL & Cairns BR Placeholder Nucleosomes Underlie Germline-to-Embryo DNA Methylation Reprogramming. Cell 172, 993–1006.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y et al. Canonical nucleosome organization at promoters forms during genome activation. Genome Res 24, 260–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blythe SA & Wieschaus EF Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. Elife 5, e20148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu F et al. Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 165, 1375–1388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu J et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Li L et al. Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol. 20, 847–858 (2018). [DOI] [PubMed] [Google Scholar]
- 102.Wu J et al. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 1 (2018). doi: 10.1038/s41586-018-0080-8 [DOI] [PubMed] [Google Scholar]
- 103.Svoboda P et al. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 269, 276–285 (2004). [DOI] [PubMed] [Google Scholar]
- 104.Jachowicz JW et al. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 49, 1502–1510 (2017). [DOI] [PubMed] [Google Scholar]
- 105.Macfarlan TS et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ishiuchi T et al. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 22, 662–671 (2015). [DOI] [PubMed] [Google Scholar]
- 107.Dixon JR, Gorkin DU & Ren B Chromatin Domains: The Unit of Chromosome Organization. Mol. Cell 62, 668–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bonev B & Cavalli G Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Dekker J & Mirny L The 3D Genome as Moderator of Chromosomal Communication. Cell 164, 1110–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hug CB & Vaquerizas JM The Birth of the 3D Genome during Early Embryonic Development. Trends Genet. 0, (2018). [DOI] [PubMed] [Google Scholar]
- 111.Hug CB, Grimaldi AG, Kruse K & Vaquerizas JM Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 169, 216–228.e19 (2017).This paper detailed the first profile of chromatin conformation changes in the early embryo during the MZT and demonstrated that TAD boundaries are established in concert with zygotic genome activation in Drosophila.
- 112.Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang J-M & Cavalli G Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Mol. Cell 71, 73–88.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 113.Ke Y et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 170, 367–381.e20 (2017). [DOI] [PubMed] [Google Scholar]
- 114.Du Z et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nat. 2017 5477662 547, 232 (2017). [DOI] [PubMed] [Google Scholar]
- 115.Kaaij LJT, van der Weide RH, Ketting RF & de Wit E Systemic Loss and Gain of Chromatin Architecture throughout Zebrafish Development. Cell Rep. 24, 1–10.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liang H-L et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403 (2008).This paper showed that Zelda is an essential factor for zygotic genome activation in Drosophila and in so doing identified the first major activator of the zygotic genome in any species.
- 117.Harrison MM, Li X-Y, Kaplan T, Botchan MR & Eisen MB Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7, e1002266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nien CY et al. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet 7, e1002339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ribeiro L et al. Evolution and multiple roles of the Pancrustacea specific transcription factor zelda in insects. PLOS Genet. 13, e1006868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leichsenring M, Maes J, Mossner R, Driever W & Onichtchouk D Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341, 1005–1009 (2013).The authors demonstrated that Pou5f3 (previously Pou5f1) is bound to chromatin before the onset of zygotic transcription and is instrumental in activating early gene expression.
- 121.Takahashi K & Yamanaka S A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 17, 183–193 (2016). [DOI] [PubMed] [Google Scholar]
- 122.Le Bin GC et al. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development 141, 1001–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hendrickson PG et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49, 925–934 (2017).This paper, along with the co-published paper (De Iaco et al 2017), identified the Dux transcription factors as regulators of mammalian zygotic genome activation and support this conclusion by manipulating DUX4/Dux levels in mouse embryos and cell lines.
- 124.De Iaco A et al. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49, 941–945 (2017).This paper, along with the co-published paper (Hendrickson et al 2017), identified the Dux transcription factors as regulators of mammalian zygotic genome activation and support this conclusion by manipulating DUX4/Dux levels in mouse embryos and cell lines.
- 125.Whiddon JL, Langford AT, Wong C-J, Zhong JW & Tapscott SJ Conservation and innovation in the DUX4-family gene network. Nat Genet 49, 935–940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schulz KN et al. Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res 25, 1715–1726 (2015).This paper, along with co-published paper (Sun et al 2015), demonstrated that Zelda possesses an essential characteristic of pioneer transcription factors, the ability to establish or maintain regions of accessible chromatin.
- 127.Sun Y et al. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25, 1703–1714 (2015).This paper, along with co-published paper (Schulz et al 2015), demonstrated that Zelda possesses an essential characteristic of pioneer transcription factors, the ability to establish or maintain regions of accessible chromatin.
- 128.Veil M, Yampolsky L, Gruening B & Onichtchouk D Pou5f3, SoxB1 and Nanog remodel chromatin on High Nucleosome Affinity Regions at Zygotic Genome Activation. bioRxiv 344168 (2018). doi: 10.1101/344168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oldfield AJ et al. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 55, 708–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nardini M et al. Sequence-Specific Transcription Factor NF-Y Displays Histone-like DNA Binding and H2B-like Ubiquitination. Cell 152, 132–143 (2013). [DOI] [PubMed] [Google Scholar]
- 131.Coustry F, Hu Q, de Crombrugghe B & Maity SN CBF/NF-Y Functions Both in Nucleosomal Disruption and Transcription Activation of the Chromatin-assembled Topoisomerase IIα Promoter. J. Biol. Chem. 276, 40621–40630 (2001). [DOI] [PubMed] [Google Scholar]
- 132.Choi SH et al. DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 44, 5161–5173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Soufi A, Donahue G & Zaret KS Facilitators and Impediments of the Pluripotency Reprogramming Factors’ Initial Engagement with the Genome. Cell 151, 994–1004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Soufi A et al. Pioneer Transcription Factors Target Partial DNA Motifs on Nucleosomes to Initiate Reprogramming. Cell 161, 555–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zaret KS & Carroll JS Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25, 2227–2241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.King HW & Klose RJ The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stadhouders R et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 50, 238–249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Meier M et al. Cohesin facilitates zygotic genome activation in zebrafish. Development 145, dev156521 (2018). [DOI] [PubMed] [Google Scholar]
- 139.Mir M et al. Dense Bicoid hubs accentuate binding along the morphogen gradient. Genes Dev. 31, 1784–1794 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu Z et al. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. Elife 3, e04236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hamm DC, Bondra ER & Harrison MM Transcriptional activation is a conserved feature of the early embryonic factor Zelda that requires a cluster of four zinc fingers for DNA binding and a low-complexity activation domain. J. Biol. Chem. 290, 3508–18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mir M et al. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. bioRxiv 377812 (2018). doi: 10.1101/377812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dufourt J et al. Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. bioRxiv 282426 (2018). doi: 10.1101/282426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Staudt N, Fellert S, Chung H-R, Jäckle H & Vorbrüggen G Mutations of the Drosophila zinc finger-encoding gene vielfältig impair mitotic cell divisions and cause improper chromosome segregation. Mol. Biol. Cell 17, 2356–65 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hamm DC et al. A conserved maternal-specific repressive domain in Zelda revealed by Cas9-mediated mutagenesis in Drosophila melanogaster. PLOS Genet. 13, e1007120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Onichtchouk D & Driever W Zygotic Genome Activators, Developmental Timing, and Pluripotency. in Current topics in developmental biology 116, 273–297 (2016). [DOI] [PubMed] [Google Scholar]
- 147.Lunde K, Belting H-G & Driever W Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Curr. Biol. 14, 48–55 (2004). [DOI] [PubMed] [Google Scholar]
- 148.Onichtchouk D et al. Oct4/Pou5f1 controls tissue-specific repressors in early zebrafish embryo. J. Stem Cells Regen. Med. 6, 82 (2010). [PubMed] [Google Scholar]
- 149.Veil M et al. Maternal Nanog is required for zebrafish embryo architecture and for cell viability during gastrulation. Development 145, dev155366 (2018). [DOI] [PubMed] [Google Scholar]
- 150.Gagnon JA, Obbad K & Schier AF The primary role of zebrafish nanog is in extra-embryonic tissue. Development 145, dev147793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vanderplanck C et al. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One 6, e26820 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lemmers RJLF et al. A Unifying Genetic Model for Facioscapulohumeral Muscular Dystrophy. Science (80-. ). 329, 1650–1653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Geng LN et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Young JM et al. DUX4 Binding to Retroelements Creates Promoters That Are Active in FSHD Muscle and Testis. PLoS Genet. 9, e1003947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Iglesias JM, Gumuzio J & Martin AG Linking Pluripotency Reprogramming and Cancer. Stem Cells Transl. Med. 6, 335–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu A, Yu X & Liu S Pluripotency transcription factors and cancer stem cells: small genes make a big difference. Chin. J. Cancer 32, 483–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Adamson ED & Woodland HR Histone synthesis in early amphibian development: histone and DNA syntheses are not co-ordinated. J. Mol. Biol. 88, 263–85 (1974). [DOI] [PubMed] [Google Scholar]
- 158.Wang Y et al. Unique molecular events during reprogramming of human somatic cells to induced pluripotent stem cells (iPSCs) at naïve state. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Deng W et al. Reactivation of Developmentally Silenced Globin Genes by Forced Chromatin Looping. Cell 158, 849–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Henikoff S & Shilatifard A Histone modification: cause or cog? Trends Genet. 27, 389–396 (2011). [DOI] [PubMed] [Google Scholar]
- 161.Bazzini AA et al. Codon identity regulates mRNA stability and translation efficiency during the maternal‐to‐zygotic transition. EMBO J. 35, 2087–2103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mishima Y & Tomari Y Codon Usage and 3′ UTR Length Determine Maternal mRNA Stability in Zebrafish. Mol. Cell 61, 874–885 (2016). [DOI] [PubMed] [Google Scholar]
- 163.Ivanova I et al. The RNA m6A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Mol. Cell 67, 1059–1067.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhao BS et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Edgar BA & Datar SA Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila’s early cell cycle program. Genes Dev. 10, 1966–77 (1996). [DOI] [PubMed] [Google Scholar]
- 166.Bashirullah A et al. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 18, 2610–2620 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hamatani T, Carter MG, Sharov AA & Ko MSH Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6, 117–31 (2004). [DOI] [PubMed] [Google Scholar]
- 168.Giraldez AJ et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–9 (2006). [DOI] [PubMed] [Google Scholar]
- 169.Lund E, Liu M, Hartley RS, Sheets MD & Dahlberg JE Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15, 2351–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bushati N, Stark A, Brennecke J & Cohen SM Temporal Reciprocity of miRNAs and Their Targets during the Maternal-to-Zygotic Transition in Drosophila. Curr. Biol. 18, 501–506 (2008). [DOI] [PubMed] [Google Scholar]
- 171.Yan L et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 172.Briggs JA et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science eaar5780 (2018). doi: 10.1126/science.aar5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Farrell JA et al. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wagner DE et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sawant AA et al. A versatile toolbox for posttranscriptional chemical labeling and imaging of RNA. Nucleic Acids Res. 44, e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Strom AR et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mir M et al. Single Molecule Imaging in Live Embryos Using Lattice Light-Sheet Microscopy. in Methods in molecular biology (Clifton, N.J.) 1814, 541–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bothma JP, Norstad MR, Alamos S & Garcia HG LlamaTags: A Versatile Tool to Image Transcription Factor Dynamics in Live Embryos. Cell 173, 1810–1822.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Campbell PD, Chao JA, Singer RH & Marlow FL Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development 142, 1368–1374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ferraro T et al. Transcriptional Memory in the Drosophila Embryo. Curr. Biol. 26, 212–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bothma JP et al. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. U. S. A. 111, 10598–603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Senecal A et al. Transcription Factors Modulate c-Fos Transcriptional Bursts. Cell Rep. 8, 75–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Garcia HG, Tikhonov M, Lin A & Gregor T Quantitative Imaging of Transcription in Living Drosophila Embryos Links Polymerase Activity to Patterning. Curr. Biol. 23, 2140–2145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Deng W, Shi X, Tjian R, Lionnet T & Singer RH CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc. Natl. Acad. Sci. U. S. A. 112, 11870–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Qin P et al. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat. Commun. 8, 14725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.O’Connell MR et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gualdi R et al. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 10, 1670–82 (1996). [DOI] [PubMed] [Google Scholar]
- 188.Lee CS, Friedman JR, Fulmer JT & Kaestner KH The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944–947 (2005). [DOI] [PubMed] [Google Scholar]
- 189.Zaret KS & Mango SE Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev 37, 76–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Blythe SA, Cha S-W, Tadjuidje E, Heasman J & Klein PS β-Catenin Primes Organizer Gene Expression by Recruiting a Histone H3 Arginine 8 Methyltransferase, Prmt2. Dev Cell 19, 220–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cirillo LA et al. Opening of Compacted Chromatin by Early Developmental Transcription Factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289 (2002). [DOI] [PubMed] [Google Scholar]
- 192.Clark KL, Halay ED, Lai E & Burley SK Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420 (1993). [DOI] [PubMed] [Google Scholar]
- 193.Taube JH, Allton K, Duncan SA, Shen L & Barton MC Foxa1 Functions as a Pioneer Transcription Factor at Transposable Elements to Activate Afp during Differentiation of Embryonic Stem Cells. J. Biol. Chem. 285, 16135–16144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lachnit M, Kur E & Driever W Alterations of the cytoskeleton in all three embryonic lineages contribute to the epiboly defect of Pou5f1/Oct4 deficient MZspg zebrafish embryos. Dev. Biol. 315, 1–17 (2008). [DOI] [PubMed] [Google Scholar]
- 195.Reim G, Mizoguchi T, Stainier DY, Kikuchi Y & Brand M The POU domain protein spg (pou2/Oct4) is essential for endoderm formation in cooperation with the HMG domain protein casanova. Dev. Cell 6, 91–101 (2004). [DOI] [PubMed] [Google Scholar]
- 196.Reim G & Brand M Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development 133, 2757–70 (2006). [DOI] [PubMed] [Google Scholar]
- 197.Okuda Y, Ogura E, Kondoh H & Kamachi Y B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo. PLoS Genet. 6, e1000936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Xu C et al. Nanog-like regulates endoderm formation through the Mxtx2-Nodal pathway. Dev. Cell 22, 625–38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fogarty NME et al. Genome editing reveals a role for OCT4 in human embryogenesis. Nature 550, 67–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]