Abstract
While the microbial community of the small intestine mucus (SIM) may also play a role in human health maintenance and disease genesis, it has not been extensively profiled and whether it changes with diet is still unclear. To investigate the flora composition of SIM and the effects of diet on it, we fed SD rats for 12 weeks with standard diet (STD), high-fat diet (HFD), high-sugar diet (HSD) and high-protein diet (HPD), respectively. After 12 weeks, the rats were sacrificed, SIM and stool samples were collected, and high-throughput 16S rRNA gene sequencing was used to analyze the microbiota. We found that fecal microbiota (FM) was dominated by Firmicutes and Bacteroidetes, while in SIM, Firmicutes and Proteobacteria were the two most abundant phyla and the level of Bacteroidetes dramatically decreased. The microbiota diversity of SIM was less than that of feces. The community composition of SIM varied greatly with different diets, while the composition of FM altered little with different diets. The relative abundance of Bacteroidetes and Allobaculum in SIM were negatively correlated with weight gain. There was no significant correlation between FM and weight gain. In conclusion, the community profile of SIM is different from that of feces and susceptible to diet.
Subject terms: Dysbiosis, Obesity
Introduction
The gastrointestinal tract is chronically exposed to various antigens, mostly of bacterial origin. In the intestine, physical separation of bacteria and the epithelium is largely dependent on mucus which is primarily composed of the highly O-glycosylated mucin 2 (Muc2) secreted by goblet cells in the epithelium. The small intestine harbors a single unattached mucus layer in which different commensal bacteria reside1,2. Resident microbiota in the mucus layer contributes to prevent the invasion and adhesion of luminal bacteria by competing for niches and nutrition1. These bacteria play an active role in shaping and regulating the gut barrier3.
Numerous studies have shown that diet plays an important role in mediating alterations in intestinal flora composition. Studies revealing that high-fat diet (HFD) was responsible for the dysbiosis with decrease in Gram-positive bacteria in the gut lumen4,5. Ingestion of high-fiber diet could raise the levels of Lactobacillus, Bifidobacterium, E. coli6, and bacteria producing short-chain fatty acids7. Rats fed with proteins from beef, pork and fish had a higher level of Firmicutes, while rats fed with casein and soy protein had an increase in the abundance of Bacteroidetes8. During infant period, the microbiota of breast-fed infants is more dominated by Bifidobacterium and Bacteroides compared with formula-fed infants9. The intestinal bacterial structure responds differently to different dietary composition. So far, the majority of microflora analysis results are based on samples taken from the fecal contents or mucosal tissues of the large intestine, especially fecal samples. These samples are often used because they are easily collected. However, the small intestine mucus (SIM) samples are rarely used to investigate the microflora.
The different regions of the intestine harbor distinct bacterial communities10. In humans, Firmicutes and Actinobacteria account for the predominant phyla in duodenal samples, while Bacteroidetes are not detected11. Firmicutes and Bacteroidetes have been identified as the major phyla in the small intestine contents of mice12. This distribution of phyla is distinctly different from the phyla found in both human and mouse feces, which are dominated by Firmicutes and Bacteroidetes13. Also, it has been found that bacterial community in the mucus differs from that in stool and intestinal lumen14,15, and mucus-resident microbiota varies most based on location15. To date, the bacterial community in SIM has not been extensively profiled and whether the microbiota shifts with diets is largely unknown.
Available data indicate that intestinal microbes may affect goblet cell and the mucus layer directly. Changes in goblet cell and in the chemical composition of intestinal mucus are detected in response to diets and alterations of the normal microbiota. The results of a study made by Sharma R et al. (1995), in which germ-free and conventionally maintained rats were fed two different diets and a group of rats born germ-free was inoculated with human flora, showed both rat and human floras reduced the number of cells containing mucins in the small intestine of rats fed on a purified diet, and feeding a commercial diet reduced the volume density of cells containing mucins in the jejunum of conventional rats and the staining density of mucins in the germ-free rats16,17. A study on parenterally and orally fed piglets found an indicator of localized inflammation and goblet cell numbers in the ileum of piglets with total parenteral nutrition18. Another study provided evidence that the dietary composition, microbial flora, as well as the interactions between the dietary constituents and microbial flora changed the mucosal architecture and the mucus composition19. These findings demonstrate that the dietary changes and microbial populations are influential in modifying the amount and proportion of mucins in the small intestine. How the small intestinal mucosa of rats adapts to unbalanced diets, including HFD, high-sugar diet (HSD) and high-protein diet (HPD) remains to be understood.
In this study, we investigated (i) the microbial community of in SIM and feces of rats fed various diets by using 16S rRNA sequencing, (ii) the relationship between changes in microbial composition and weight gain in rats, and (iii) the effects of various diets on the small intestinal mucosa.
Results
Differences of weight gain in rats fed on different diets
The initial body weight of the rats fed standard diet (STD), HFD, HSD or HPD was 129.1 ± 8.14, 127.43 ± 5.40,129.6 ± 6.69 and 130.1 ± 5.24 g, respectively. After 12-week feeding, the body weight of rats in each group increased to 350.6 ± 13.03, 422.1 ± 20.58, 410.9 ± 14.09, and 257.2 ± 13.01 g, respectively. The rats fed HFD or HSD gained significantly more weight than those fed STD (P < 0.01), but there was no significant difference in weight gain between the rats fed HFD and those fed HSD (P = 0.198). The weight gain of the HPD-fed rats was significantly less than the STD-fed rats. (P = 0.000) (Fig. 1).
Figure 1.
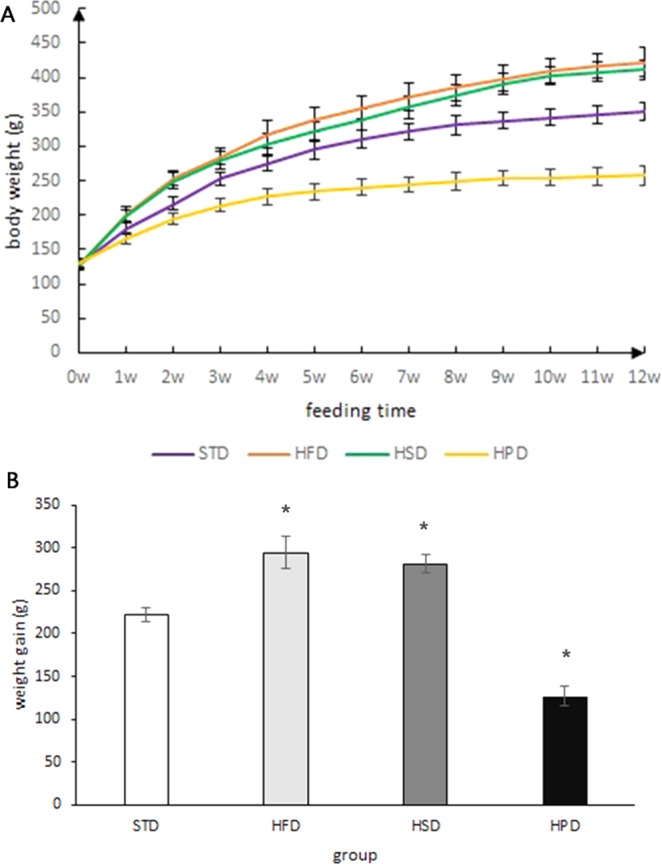
(A) Weight changes in rats fed on different diets at different time points. (B) Weight gain in rats fed on different diets for 12 weeks. (*vs STD group, P < 0.05). There was a significant increase in the rats fed HFD or HSD compared with those fed STD, but there was no significant difference in weight gain between the rats fed HFD and those fed HSD. The rats fed HPD gained the least weight.
The composition of microbiota in SIM and feces of rats fed on STD
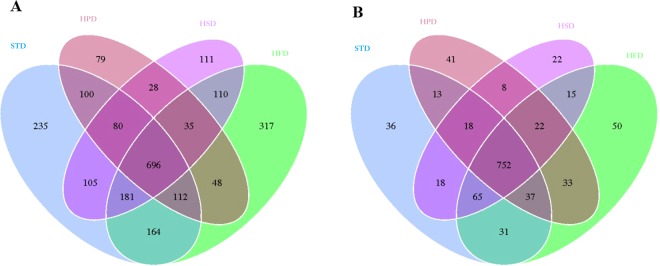
320564 usable raw reads were obtained from 28 SIM samples and paired fecal samples. We used the Venn diagrams to show the interrelationship of OTUs in the fecal samples and SIM samples among different groups (Fig. 2A,B). There were 696 OTUs shared in the SIM samples of each group and the number of OTUs unique to the STD group, HPD group, HSD group and HFD group was 235, 79, 111 and 317, respectively. In comparison, 752 OTUs were shared in the fecal samples of each group and there were 36, 41, 22 and 50 unique OTUs in the fecal samples of STD group, HPD group, HSD group and HFD group, respectively.
Figure 2.
Venn Diagrams based on the shared and unique OTUs. (A) The shared OTUs among the SIM samples of different groups and the unique OTUs in the SIM samples of each group. (B) The shared OTUs among the fecal samples of different groups and the unique OTUs in the fecal samples of each group.
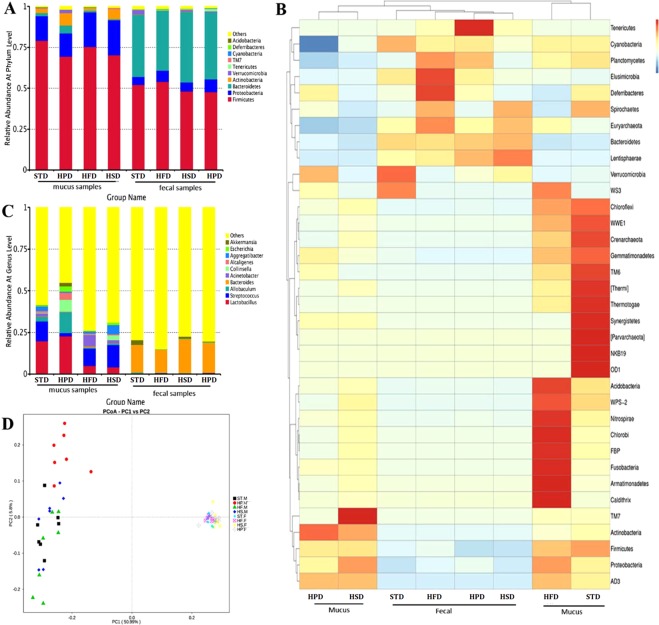
To address the top ten relative abundance of microbial group, the taxonomic classification is performed at phylum, class, order, family, genus and species level, respectively. The microbiota analysis of rats fed STD revealed that there was a dramatic decrease in the relative abundance of Bacteroidetes in SIM samples compared to fecal samples (2.04 ± 1.57% vs 38.11 ± 5.14%, P < 0.01). The relative abundance of Firmicutes, Proteobacteria and Actinobacteria in SIM samples was significantly higher than in fecal samples (79.20 ± 4.25% vs 52.17 ± 3.12%, P < 0.01; 14.94 ± 3.73% vs 4.79 ± 1.00%, P < 0.01; 2.67 ± 0.96% vs 0.14 ± 0.08%, P < 0.01). Verrumcomicrobia were detected only in fecal samples. These results suggested that most of the sequences in SIM samples belonged to Firmicutes and Proteobacteria, accounting for more than 90% of abundance at phylum level, while the rest mainly distributed in Actinobacteria and Bacteroidetes. Fecal microbiota (FM) was dominated by Firmicutes and Bacteroidetes (Fig. 3A). Also, there were great differences in the microflora composition between SIM samples and fecal samples at the genus level. Lactobacillus, Streptococus and Allobaculum were dominant genera in SIM not in fecal samples (19.79 ± 20.85% vs 0.14 ± 0.03%, P < 0.01; 11.84 ± 7.06% vs 0.22 ± 0.14%, P < 0.01; and 2.79 ± 1.63% vs 0.70 ± 1.33%, P < 0.01). Bacteroides was found mainly in fecal samples (Fig. 3B,C).
Figure 3.
(A) Relative abundance of microflora at phylum level in SIM and fecal samples of each group. Firmicutes and Proteobacteria were the most abundant phyla in SIM of each group, while Bacteroidetes and Firmicutes were the major phyla in fecal samples of each group. Actinobacteria was the third phylum in SIM, but not found in fecal samples. (B) Heatmap of the predominant genera identified in SIM and fecal samples of each group. (C) Relative abundance of microflora at genus level in SIM and fecal samples of each group. Lactobacillus, Streptococus, and Allobaculum were the dominant genera in SIM; Bacteroides was the major genus in fecal samples. (D) PCA displayed the distribution of corresponding points of SIM samples and fecal samples of each group.
The composition of microbiota in SIM and feces of rats fed on unbalanced diets
In addition to the rats fed STD, the composition of SIM microflora was also different from that of FM in the rats fed unbalanced diets. Firmicutes, Proteobacteria and Actinobacteria were the top three abundant phyla in SIM samples of all the unbalanced diet groups, while in fecal samples of these groups, Bacteroidetes and Firmicutes were the major phyla. In the rats fed on HFD, the abundance of Firmicutes and Actinobacteria in SIM was higher than that in feces (75.39 ± 18.21% vs 47.69 ± 5.60%, 1.55 ± 0.57% vs 0.08 ± 0.04%, respectively, P < 0.05), and at the genus level, the abundance of Lactobacillus and Streptococcus was also higher in SIM than that in feces (4.98 ± 3.37% vs 0.41 ± 0.16%, 10.55 ± 7.61% vs 0.18 ± 0.06%, respectively, P < 0.05). In the rats fed on HSD, higher abundance of Proteobacteria, Actinobacteria, Lactobacillus, Streptococcus, Allobaculum, and lower level of Bacteroidetes were present in SIM than that in feces (P < 0.05).
The community composition in SIM samples had different alterations with different diets. In comparison with the rats fed STD, the rats fed HFD or HSD had a decreased abundance of Bacteroidetes in SIM (P < 0.05) and the rats fed HPD had an increased abundance (4.70% ± 6.29% vs 2.03% ± 1.57%, P < 0.05). The relative abundance of Actinobacteria decreased in SIM samples of HFD group (1.55% ± 0.57%vs 2.68 ± 0.96%, P < 0.05), and rose in HSD group and HPD group (P < 0.05). The analysis results of FM showed that the rats fed HPD had fewer abundance of Firmicutes than those fed STD (48.21% ± 7.92% vs 54.05% ± 5.44%, P < 0.05); Higher level of Proteobacteria was detected in the rats fed HPD or HFD compared with those fed STD (P < 0.05); there was no significant difference in the proportion of Proteobacteria between HSD-fed rats and STD-fed rats (P > 0.05); the rats fed HSD had a higher abundance of Bacteroidetes than those fed STD (43.14% ± 6.71% vs 38.12% ± 5.14%, P < 0.05).
At the genus level, the composition of flora in SIM samples also shifted with unbalanced diets. The microbiota analysis on SIM samples revealed that there was a dramatic decrease in the relative abundance of Streptococcus in the rats fed HPD compared to those fed STD(1.93% ± 2.18% vs 11.84% ± 7.06%, P < 0.05); there was no significant difference in the level of Streptococcus between the rats fed HFD or HSD and those fed STD (P > 0.05); the abundance of Lactobacillus was higher in HPD group than STD group (22.71% ± 15.47% vs 19.79% ± 20.85%, P < 0.05) and decreased in HFD group and HSD group compared with STD group (P < 0.05); the level of Allobaculum also increased in HPD group and decreased in HFD group and HSD group. The relative abundance of Acinetobacter rose in HFD group and decreased in HPD group. In contrast, the community profile in fecal samples did not change notably with the diets, either at the phylum level or at the genus level, except that the abundance of Verrumcomicrobia and Akkemansia decreased in various unbalanced diet groups. (Fig. 3A–C)
Alpha diversity and beta diversity of microbiota in SIM and feces
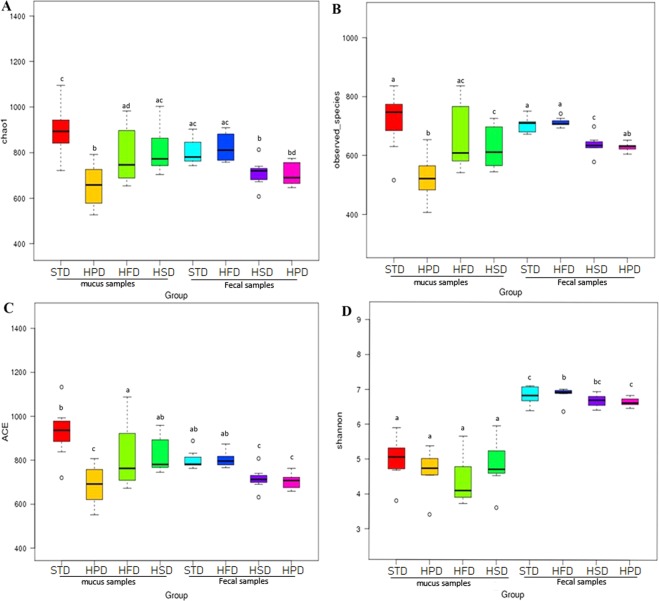
The microbial alpha diversity was estimated based on the originally observed count values prior to any pre-processing, Chao1 index and ACE index were used as measures of community richness. The Shannon index was used as a measure of taxa richness and evenness. Chao1 indexes of microflora in SIM and fecal samples of STD, HFD, HSD and HPD group were as follows: SIM 897.05 ± 117.76, 793.51 ± 132.66, 812.99 ± 104.84, and 655.25 ± 102.63; FM 806.31 ± 60.16, 824.72 ± 65.19, 709.12 ± 63.22, and 707.73 ± 53.30. ACE indexes of SIM microflora were different from that of FM in each group (P < 0.05, Mann-Whitney test). In each group, Shannon indexes of SIM microflora were significantly less than that of FM (STD 4.98 ± 0.66 vs 6.84 ± 0.28, HFD 4.41 ± 0.70 vs 6.87 ± 0.22, HFD 4.86 ± 0.77 vs 6.69 ± 0.19 and HPD 4.67 ± 0.64 vs 6.66 ± 0.13, P < 0.05, respectively) (Fig. 4), indicating microbiota diversity of SIM was lower than FM.
Figure 4.
Box plot of alpha diversity showing differences between each other group. (A) Chao1 index (B) The observed species (C) ACE index (D) Shannon index. The letters a, b, c and d are used to clarify whether the difference between any pair of groups calculated by Wilcoxon analysis was statistically significant (p < 0.05), and there was a significant difference in flora diversity between the two groups sharing no common letter markers.
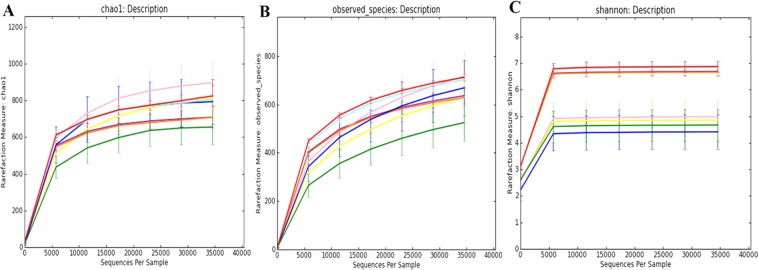
Moreover, Chao1 index, the observed species and Shannon index reached saturation, and the rarefaction curve of each sample also entered the plateau phase (Fig. 5), indicating the sequencing method was appropriate to evaluate the microbial diversity in the present study. Wilcoxon analysis revealed that in the rats fed STD, the community richness in SIM did not significantly differ from fecal community richness, while the community diversity in SIM was significantly less than fecal community diversity. The observed species, Chao1 index, ACE index and Shannon index were shown in Table 1. The results unveiled the diversity of microbiota in SIM samples was greater at all levels than that of FM.
Figure 5.
Rarefaction curves which was used to judge whether further sampling would likely yield additional taxa by whether the curve reached a plateau value. (A) Rarefaction curves of Chao 1 index. (B) Rarefaction curves of observed species. (C) Rarefaction curves of Shannon index.
Table 1.
Alpha diversity.
| Group | Observed Species(x ± SD) | Chao1 (x ± SD) | Shannon (x ± SD) | ACE (x ± SD) |
|---|---|---|---|---|
| STDG-M | 716.714 ± 108.552 | 897.047 ± 117.761 | 4.978 ± 0.664 | 930.605 ± 128.437 |
| HFDG-M | 668.714 ± 123.302 | 793.514 ± 132.661 | 4.411 ± 0.699 | 826.018 ± 155.618 |
| HSDG-M | 629.429 ± 77.743 | 812.994 ± 104.839 | 4.855 ± 0.770 | 829.541 ± 85.448 |
| HPDG-M | 524.857 ± 81.321 | 655.250 ± 102.634 | 4.668 ± 0.643 | 686.901 ± 98.776 |
| STDG-F | 702.857 ± 28.145 | 806.307 ± 60.156 | 6.842 ± 0.277 | 802.140 ± 43.780 |
| HFDG-F | 712.429 ± 16.339 | 824.718 ± 65.188 | 6.870 ± 0.223 | 804.078 ± 38.419 |
| HSDG-F | 636.429 ± 35.804 | 709.124 ± 63.220 | 6.688 ± 0.189 | 715.710 ± 53.080 |
| HPDG-F | 628.429 ± 14.831 | 707.733 ± 53.300 | 6.659 ± 0.127 | 703.627 ± 36.135 |
STDG: standard chow group, HFDG: High-fat diet group, HSDG: High-sugar diet group, HPDG: High-protein diet group, M: the small intestine mucus samples, F: the fecal samples.
Beta diversity was analyzed using Principle component analysis (PCA), as was shown in Fig. 3D. The distribution of points corresponding to SIM samples was discrete among different dietary groups, indicating the community composition of SIM varied greatly with different diets. Also, the distribution of corresponding points of fecal samples in each group tended to cluster, which meant the composition of FM altered little with different diets. Furthermore, there were apparent distances between corresponding points of SIM samples and those of fecal samples in each group, which suggested there were notable differences in microbial composition between SIM samples and fecal samples. These results demonstrated that microbiota of SIM, being different from FM, was susceptible to diet.
To compare the composition of SIM microbiota with that of FM, and to investigate the community structures in response to different diets, Anosim and MPRR analysis were performed. Significant differences were observed between every two groups (P < 0.05), and there were significant differences in the microbial structure between SIM and fecal samples of each group (P < 0.05) (Tables 2 and 3).
Table 2.
Anosim analysis.
| Group | R-value | P-value |
|---|---|---|
| STD.F-STD.M | 1 | 0.001 |
| HFD.F-HFD.M | 1 | 0.002 |
| HSD.M-HSD.F | 1 | 0.001 |
| HPD.M-HPD.F | 1 | 0.001 |
| SDT.M-HPD.M | 0.3168 | 0.004 |
| STD.M-HFD.M | 0.2012 | 0.022 |
| STD.M-HSD.M | 0.2536 | 0.017 |
| HFD.M-HPD.M | 0.6628 | 0.001 |
| HFD.M-HSD.M | 0.3528 | 0.008 |
| HSD.M-HPD.M | 0.6433 | 0.001 |
| STD.F-HPD.F | 0.5967 | 0.001 |
| STD.F-HFD.F | 0.1987 | 0.009 |
| STD.F-HSD.F | 0.1395 | 0.054 |
| HFD.F-HSD.F | 0.691 | 0.001 |
| HFD.F-HPD.F | 0.6142 | 0.001 |
| HSD.F-HPD.F | 0.553 | 0.004 |
R value was ranged from −1 to 1, R. > 0, significant differences between groups, R < 0, significant differences between samples within the group. P < 0.05 was considered statistically significant.
Table 3.
MRPP analysis.
| Group | A | Observed-delta | Expected-delta | Significance |
|---|---|---|---|---|
| STD.F-STD.M | 0.3604 | 0.4437 | 0.6937 | 0.001 |
| HFD.F-HFD.M | 0.4555 | 0.3661 | 0.6723 | 0.003 |
| HPD.F-HPD.M | 0.3384 | 0.4692 | 0.7092 | 0.001 |
| HSD.F-HSD.M | 0.4112 | 0.4067 | 0.6908 | 0.002 |
| STD.F-HPD.F | 0.08692 | 0.3304 | 0.3618 | 0.002 |
| STD.F-HFD.F | 0.03949 | 0.3354 | 0.3492 | 0.002 |
| STD.F-HSD.F | 0.01504 | 0.3276 | 0.3326 | 0.106 |
| HFD.F-HSD.F | 0.07977 | 0.3165 | 0.3440 | 0.002 |
| HFD.F-HPD.F | 0.05896 | 0.3193 | 0.3393 | 0.001 |
| HPD.F-HSD.F | 0.06054 | 0.3115 | 0.3316 | 0.002 |
| STD.M-HPD.M | 0.06111 | 0.5825 | 0.6204 | 0.006 |
| STD.M-HFD.M | 0.0594 | 0.4744 | 0.5043 | 0.024 |
| STD.M-HSD.M | 0.04877 | 0.5228 | 0.5496 | 0.046 |
| HPD.M-HSD.M | 0.1274 | 0.5643 | 0.6467 | 0.002 |
| HPD.M-HFD.M | 0.1786 | 0.516 | 0.6283 | 0.001 |
| HFD.M-HSD.M | 0.06402 | 0.4562 | 0.4874 | 0.019 |
Observe Delta reflects the variety within the group. Expect delta represents variety between groups. A > 0, significant differences between groups. A < 0, significant differences within the group. P < 0.05 was considered statistically significant.
Correlation analysis between weight gain and gut microbiota
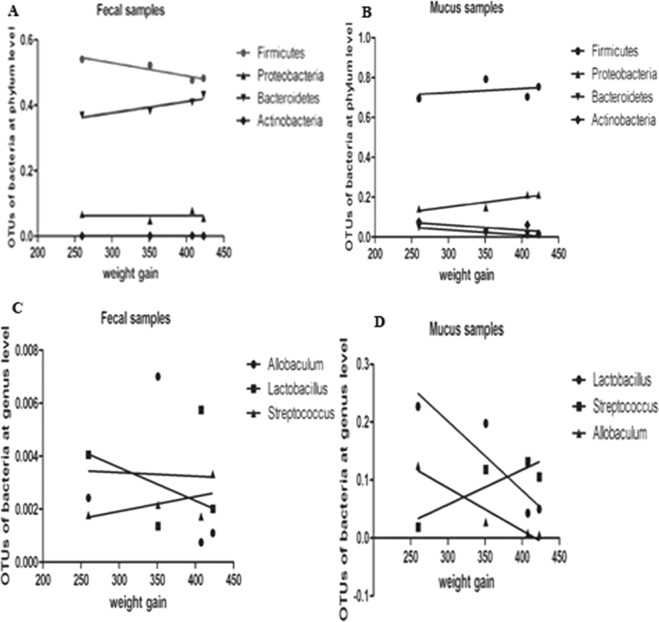
Correlation analysis was used to explore the relationship between weight gain and microbiota in the SIM and feces. The bacterial flora analysis among various groups revealed the relative abundance of Bacteroidetes in SIM samples decreased in the HFD-fed rats and HSD-fed rats fed, and increased in the rats fed HPD. Correlation analysis further unveiled a significant negative correlation between the relative abundance of Bacteroidetes in SIM samples and weight gain (P = 0.04, r = −0.46). The relative abundance of Firmicutes, Proteobacteria and Actinobacteria in SIM samples was not significantly correlated with weight gain (P = 0.21, P = 0.06, P = 0.26, respectively) (Fig. 6A). No significant correlation was found between the relative abundance of Bacteroidetes, Firmicutes, Proteobacteria and Actinobacteria in fecal samples and weight gain(P > 0.05) (Fig. 6B). At the genus level, the relative abundance of Allobaculum in SIM decreased with the increase in body weight of the rats. Further correlation analysis found that Allobaculum were negatively correlated with weight gain (P = 0.033, r = −0.65). Streptococcus and Lactobacillus were not significantly correlated with weight gain (Fig. 6).
Figure 6.
Correlation analysis between gut microbiota and weight gain. (A) The abundance of Bacteroidetes, Proteobacteria, Firmicutes and Actinobacteria in fecal samples had no significant correlation with weight gain. (B) Bacteroidetes in SIM samples was negatively correlated with weight gain (P = 0.04, r = −0.46). (C) The abundance of Allobaculum, Streptococcus and Lactobacillus in fecal samples was not correlated with weight gain. (D) In SIM samples, the abundance of Allobaculum was negatively correlated with weight gain (P = 0.033, r = −0.65), while Streptococcus and Lactobacillus were not significantly correlated with weight gain.
Effects of various diets on the goblet cell number in the small intestinal epithelium of rats
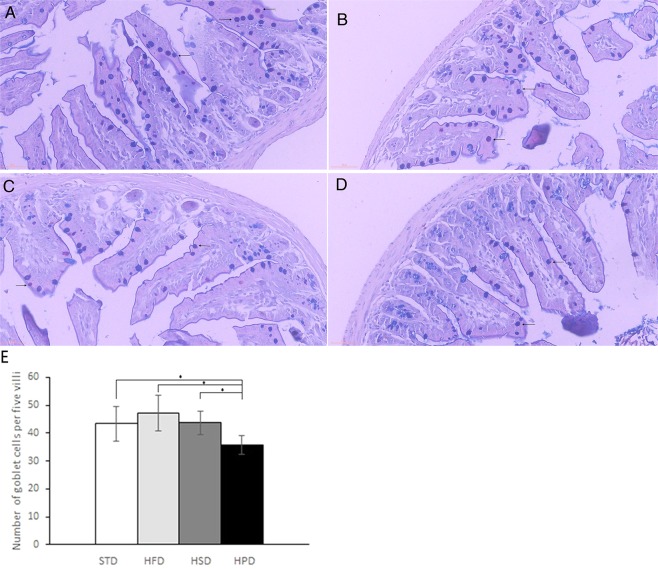
To understand the effects of various diets on the SIM structure, AB/PAS-positive cells in the epithelium were counted (Fig. 7A–D). No significant difference in the number of mucus-positive cells was observed between HFD group and STD group (47.2 ± 6.3 vs 43.3 ± 6.2 cells per 5 villi, P = 0.357) as well as between HFD and HSD group (47.2 ± 6.3 vs 43.7 ± 4.3 cells per 5 villi, P = 0.332). There was also no significant difference between HSD and STD group (P = 0.924). The mucus-positive cell number in HPD group (35.7 ± 3.2 cells per 5 villi) was significantly lower than that in STD group, HFD group and HSD group (P < 0.05) (Fig. 7E). Goblet cell number in villous epithelium of small intestine revealed that both acidic and neutral mucins in small intestinal epithelium were less abundant in response to HPD feeding than to STD feeding, and had no significant change in respond to HFD and HSD compared to STD.
Figure 7.
Representative images of AB-PAS staining of ileum tissues of STD group (A), HFD group (B), HSD group (C) and HPD group (D). (E) Number of goblet cells per 5 villi in the small intestinal epithelium. The mucus-positive cell number was significantly lower in HPD group than STD, HFD and HSD group, *P < 0.05.
Discussion
Gut microbiota plays profound roles in host health and disease. Considerable research has focused on understanding the communities in the contents and mucosal tissue of the large intestine, especially FM5,20–24. Some information is also available about the microflora composition in the contents of small intestine20. However, there are few studies exploring the community profile in SIM. In this study, we investigated and compared the microbial structure of SIM and feces. The results showed that the microflora composition of SIM was markedly different from that of feces; Firmicutes and Proteobacteria were the two most abundant phyla in SIM, accounting for more than 90% relative abundance of the community, and the third abundant phylum was Actinobacteria; FM was dominated by Firmicutes, Bacteroidetes and Proteobacteria. It has been shown that Bacteroidetes, Firmicutes and Proteobacteria were the major phyla in the small intestine contents as well as in the cecal and colonic contents; anaerobic bacteria, including Alkaliphilus, Butyricimonas, Clostridium and Parabacteroides spp, accounted for the most abundant genera in the small intestine contents20. One research investigating gut microbiota in rhesus macaques has also found that the community in fecal samples could not stand for that in the small intestine contents, and in mucosal samples, facultatively anaerobic clades, such as Helicobacter in the large intestine and Pasteurella in the small intestine were more abundant15. In this study, we also found that the predominant genera in SIM, including Lactobacillus, Streptococcus and Allobaculum, belonged to facultative anaerobes, while the relative abundance of these bacteria in feces was very low. This may be attributed to the fact that the mucous layer lies on top of the epithelial surface, which may contribute to the diffusion of oxygen from the blood to the mucus resulting in the higher oxygen content in the mucous layer compared to the gut lumen. Moreover, we found the community diversity of SIM was less than that of feces, which was also confirmed by the previous study25. ß diversity analysis also suggested there were notable differences in microbial composition between SIM samples and fecal samples. All the findings indicated that there is a characteristic microflora community in SIM, not only different from that in colonic contents but also from that in small intestine contents.
The microbial community profile of SIM is different from that of small intestine contents and feces, which may be due to the environment where the microflora survives. Intestinal contents mainly come from food and provide nutrition for the bacteria in the contents. Microflora composition in intestinal contents is influenced by diet. For example, the composition of fecal flora in Western-style diet population differs from that in high-fiber diet population22. In comparison, the microflora resident in the mucus layer is mainly nourished by mucin. Intestinal contents are able to contact with the mucus with the flow along the gastrointestinal tract, resulting in bacteria in the mucus can be inoculated into the luminal contents. In this way, the microbial community in the intestinal contents comprises bacteria coming from diet and SIM, and fecal microbiota may include bacteria in the mucus along the whole gastrointestinal tract.
So far, it is not clear yet whether diet has impacts on the community profile in SIM. We found the abundance of Bacteroidetes and Actinobacteria in SIM decreased in the rats fed HFD; HSD-fed rats had increased level of Actinobacteria and decreased level of Bacteroidetes; increased abundance of Actinobacteria and Bacteroidetes was present in the rats fed HPD; the community richness of SIM in the rats fed on HFD or HPD increased. These results suggested that different dietary structure lead to different alterations in the composition of microflora in SIM. Furthermore, higher level of Proteobacteria and lower abundance of Firmicutes were found in SIM of rats fed HPD, and a higher abundance of Bacteroidetes and Proteobacteria were present in SIM of HSD-fed rats and HFD-fed rats, respectively. Thus, it can be seen that the response of SIM microflora to a same diet is distinct from that of colonic contents microflora. The result of ß diversity displayed the composition of SIM microbes varied greatly with different diets while the alterations of FM composition was not obvious with diets, demonstrating that microbiota of SIM was susceptible to diet compared to FM. Therefore, contrary to the more stable FM26,27, the SIM microbiota most likely reflect the subject dietary variation. However, there is limited information about the effects of different diets on SIM flora, and the exact mechanism of the effects of various unbalanced diets on the bacterial flora resident in SIM is still unclear. Studies have shown that undigested protein and peptide can be fermented by colonic bacteria into ammonia, indole, phenol and hydrogen sulfide28. These substances not only change the pH value of colonic contents but also are harmful to some bacteria, which contributes to the alterations in the composition of microbiota in colonic contents29,30. Since the small intestine is the main part responsible for digestion and absorption due to its function and anatomical structure, further research is needed to study whether the different effects of various diets on the microbiota structure in SIM are also performed by the metabolic products derived from the dietary ingredients.
We observed more weight was gained in the rats fed on HFD or HSD while less was gained in the rats fed on HPD. Correlation between fecal microbiota and weight gain has been found in other studies. In this study, we found a negative correlation between the abundance of Bacteroidetes in SIM and the rats weight gain, and the level of Firmicutes and Proteobacteria in SIM had no correlation with weight gain. This finding differed from the results on association between fecal flora and weight gain. Previous studies have found the abundance of Bacteroidetes decreased in obese individuals31,32, which is consistent with our results that the HFD -fed or HSD-fed rats with more weight gain had lower abundance of Bacteroides in SIM and the HPD -fed rats with less weight gain had higher level of Bacteroides in SIM. In addition, the bacteria associated with weight gain were different among the rats fed different diets. The level of Actinobacteria in SIM was positively correlated with weight gain in the rats fed HPD and negatively correlated in the rats fed HFD. There was no correlation between fecal Actinobacteria abundance and weight gain of the rats in each group. These results further indicate the association between the bacterial flora in SIM and weight changes is distinct from that between FM and weight change. Furthermore, we found the number of goblet cells significantly decreased in the rats fed HPD. Intestinal mucus is primarily composed of Muc 2, which is secreted by goblet cells in the epithelium1. Decreased number of goblet cells results in a decrease in mucin secretion, which in turn reduces the thickness of the mucus layer. Further study may be needed to make clear whether HPD plays a role in the structural changes of microflora in SIM by affecting goblet cells.
In conclusion, microbial composition in SIM is distinct from that in feces and susceptible to different dietary composition. A decrease in the number of goblet cells may be a contributor to alterations in microflora composition in SIM associated with HPD. Bacteroidetes and Allobaculum in SIM was negatively correlated with weight gain. Further study may be needed to make clear whether HPD plays a role in the structural changes of microflora in SIM by affecting goblet cells.
Methods
Animals
4-week-old female Sprague-Dawley (SD) rats were purchased from housed in a specific-pathogen-free (SPF) environment in the Laboratory Animal Center of People’s Hospital of Hunan province (Changsha, China) in a 12-hour light/dark cycle. After a 1-week adaptation period, the rats were randomly assigned to four groups and shifted to one of the following sterile diets: STD, HFD, HPD or HSD for 12 weeks. The study design was shown in Fig. S3. The diets were purchased from Huafukang Biotechnology Co. Ltd (Beijing, China) and the compositions of diet were mentioned in Table 4.
Table 4.
Dietary Composition.
| Feed | Standard | High protein | High sugar | High fat |
|---|---|---|---|---|
| Casein | 200 | 619.9 | 200 | 271.9 |
| Corn starch | 547 | 122.6 | 75.5 | 0 |
| Dextrin | 0 | 0 | 0 | 133.2 |
| Sucrose | 100 | 99.3 | 571 | 135.9 |
| Soya oil | 70 | 69.5 | 70 | 70 |
| Lard | 0 | 0 | 0 | 275.2 |
| Cellulose | 50 | 49.6 | 50 | 68 |
| Pectin | 0 | 0 | 0 | 0 |
| Minerals | 35 | 35 | 35 | 35 |
| Vitamins | 10 | 10 | 10 | 10 |
| L- cystine | 3 | 9.3 | 3 | 4.1 |
| Choline | 2.5 | 2.5 | 2.5 | 4.2 |
| TBHQ | 0.014 | 0.014 | 0.014 | 0.07 |
| Total | 1017.514 | 1017.714 | 1017.014 | 1007.57 |
Sampling
Groups of rats (n = 7) were housed in individual ventilated cages, and sacrificed at 17-week-old of age. Small intestinal mucus samples were collected by aspirating and scraping from the duodenum to the terminal ileum into sterile EP tubes on the benchtop. Feces were collected with sterile forceps from the terminal portion of the colon into sterile tubes. Immediately after collection, the tubes were flash-frozen in liquid nitrogen and were stored at −80 °C until processed20. The small intestines were removed and the ileum tissues were fixed in paraformaldehyde and embedded in paraffin as previously described33.
DNA extraction
Bacterial genomic DNA was extracted from the fecal and SIM samples using cetyltrimethylammoniumbromide (CTAB) method. The steps of DNA extraction were as follows: The sample was added with 2% w/v CTAB (HiMedia, India) containing freshly prepared lysozyme followed by incubation at 65 °C for 30 min, then phenol (pH 8.0)/chloroform/isoamyl alcohol (25:24:1) was added. Whole content was vortexed and centrifuged at 12,000 rpm/10 min, 4 °C, as described previously34. An equal amount of chloroform: isoamyl alcohol (24:1) was added to the liquid fraction, mixed thoroughly and incubated at room temperature for 10 min. Whole content was centrifuged at 12,000 rpm/10 min, 4 °C. Isopropanol (Sigma-Aldrich) were added into the supernatant and incubated for 6 h at −20 °C. Pellet was seen visually after the incubation. Entire content was centrifuged at 12,000 rpm/10 min, 4 °C. Pellet obtained was washed thrice with 70% ethanol (Fisher Scientific) by centrifugation at 12,000 rpm/10 min, 4 °C. Ethanol was removed completely after washings, and DNA pellet was dried without heating, Double distilled water was added to DNA pellet at an elevated temperature of 55 °C for 10 min. DNA concentration and purity were monitored on 1% agarose gels. DNA was stored at −20 °C.
Bacterial metagenomes and 16S rRNA sequencing
Libraries and sequencing were carried out by the novogene Biotechnology Center at Beijing (China). Amplified products of the V4 region of the 16S rRNA gene originating from the BAC fraction were sequenced with IlluminaHiSeq2500 PE250 (Noher, Beijing, China) using barcoded 515 F (GTGCCAGCMGCCGCGGTAA) and 806 R (GGACTACHVGGGTWTCTAAT) primers, and the average read length was 250 bp. All PCR reactions were carried out with Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, USA). SYB green contained 1ψloading buffer (Solarbio, Beijing, China), and PCR products were resolved on 2% agarose gel electrophoresis. Band of 400–450 bp was chosen for further experiment. The composition of a PCR reaction was as follows: 0.3 μM primers, 0.3 mM dNTPs, 0.5 U polymerase enzyme, and 50 ng DNA template. The PCR program consisted of the following steps: initial denaturation at 98 °C for 1 min, 30 cycles at 98 °C for 10 s, 50 °C for 30 s, and extension at 72 °C for 60 s.
Amplicons were sequenced on a HiSeqIllumina platform (Illumina, San Diego, CA), generating 250 bp paired end reads. The paired reads were merged using FLASH (V1.2.7,http://ccb.jhu.edu/software/FLASH/). Quality filtering on the raw tags were performed under specific filtering conditions to obtain the high-quality clean tags according to the QIIME (V1.7.0, http://qiime.org/index.html) quality-controlled process. Chimeric sequences were removed using UCHIME algorithm (UCHIME Algorithm, http://www.drive5.com/usearch/manual/uchime_algo.html) against the reference database (Gold database, http://drive5.com/uchime/ uchime_download.html)0.19 Reads were clustered into operational taxonomic units (OTUs) using Uparse software (Uparse v7.0.1001, http://drive5.com/uparse/). Sequences with ≥97% similarity were assigned to the same OTUs. Representative sequence for each OTU was screened for further annotation. For each representative sequence, the Green Gene Database (http://greengenes.lbl. gov/cgi-bin/nph-index.cgi) was used based on RDP classifier (Version 2.2, http://sourceforge.net/projects/rdp-classifier/) algorithmto annotate taxonomic information. Alpha diversity is applied in analyzing complexity of species diversity for a sample through 6 indices, including Observed-species, Chao1, Shannon, Simpson, ACE, Good-coverage. All these indices in our samples were calculated with QIIME (Version 1.7.0) and displayed with R software (Version 2.15.3). Beta diversity analysis was used to evaluate differences of samples in species complexity, Beta diversity on both weighted and unweighted unifrac were calculated by QIIME software (Version 1.7.0). Cluster analysis was preceded by PCA, which was applied to reduce the dimension of the original variables using the FactoMineR package and ggplot2 package in R software (Version 2.15.3).
Histology
Paraffin sections (5 μm) of ileum were attached to poly-L-lysine-coated glass slides. After overnight incubation at 37 °C, slides were de-waxed and hydrated step-wise using 100% xylene followed by several solutions of distilled water containing decreasing amounts of ethanol. Sections were stained with Alcian Blue Periodic acid Schiff (AB-PAS) Stain Kit (Solarbio, China)35,36. Mucous cells in small intestinal epithelium were counted (counting the number of cells in 5 adjacent villi per quadrant/4 quadrants/per section/2 sections per animal/7 animals per group) using Motic EasyScanner and DSAssistant software (Changsha Central Hospital, China).
Statistical analysis
Data on weight gained and the percentage of classified sequence reads were expressed as mean ± standard deviation. Student’s t-test, Mann-Whitney test and one-way ANOVA were used for statistical analysis, and Wilcox was used to elevate α and β diversity. Spearman’s test was applied to analyze the relationship between weight gain and gut microbiota. P value < 0.05 was considered to be significantly different.
Ethical approval and informed consent
All animal protocols and experiments were approved by the institutional animal care committee of 2ndxiangya hospital at Central South University, and all the methods were carried out in accordance with the relevant guidelines and regulations.
Supplementary information
Acknowledgements
The authors thank all the participants and professionals for their contributions to this study.
Author Contributions
Conceptualization, Yu Meng and Fanggen Lu.; methodology, Yu Meng and Fanggen Lu.; software, Xiaojun Li; validation, Yu Meng, Jie Zhang and Chunlian Wang; formal analysis, Xiaojun Li; investigation, Yu Meng; resources, Chunlian Wang; data curation, Jie Zhang; writing—original draft preparation, Yu Meng; writing—review and editing, Xiaojun Li; visualization, Chunlian Wang; supervision, Fanggen Lu; project administration, Fanggen Lu; funding acquisition, Fanggen Lu.
Data Availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44994-7.
References
- 1.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. The American journal of clinical nutrition. 2001;73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- 2.Atuma C, et al. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. American journal of physiology. Gastrointestinal and liver physiology. 2001;280:G922–929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 3.Natividad JM, Verdu EF. Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacological research. 2013;69:42–51. doi: 10.1016/j.phrs.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Hildebrandt MA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(1716–1724):e1711–1712. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cani PD, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, et al. Dietary Fiber Intake Regulates Intestinal Microflora and Inhibits Ovalbumin-Induced Allergic Airway Inflammation in a Mouse Model. PloS one. 2016;11:e0147778. doi: 10.1371/journal.pone.0147778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwiertz A, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Y, et al. Intake of Meat Proteins Substantially Increased the Relative Abundance of Genus Lactobacillus in Rat Feces. PloS one. 2016;11:e0152678. doi: 10.1371/journal.pone.0152678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azad MB, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ: Canadian Medical Association journal = journal de l’Association medicale canadienne. 2013;185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Aidy S, et al. The small intestine microbiota, nutritional modulation and relevance for health. Current opinion in biotechnology. 2015;32:14–20. doi: 10.1016/j.copbio.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Angelakis E, et al. A Metagenomic Investigation of the Duodenal Microbiota Reveals Links with Obesity. PloS one. 2015;10:e0137784. doi: 10.1371/journal.pone.0137784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YS, et al. High-throughput 16S rRNA gene sequencing reveals alterations of mouse intestinal microbiota after radiotherapy. Anaerobe. 2015;33:1–7. doi: 10.1016/j.anaerobe.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science translational medicine. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan SH, et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Applied and environmental microbiology. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasuda K, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell host & microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma R, Schumacher U. Morphometric analysis of intestinal mucins under different dietary conditions and gut flora in rats. Digestive diseases and sciences. 1995;40:2532–2539. doi: 10.1007/BF02220438. [DOI] [PubMed] [Google Scholar]
- 17.Sharma R, Schumacher U. The influence of diets and gut microflora on lectin binding patterns of intestinal mucins in rats. Laboratory investigation; a journal of technical methods and pathology. 1995;73:558–564. [PubMed] [Google Scholar]
- 18.Ganessunker D, et al. Total parenteral nutrition alters molecular and cellular indices of intestinal inflammation in neonatal piglets. JPEN. Journal of parenteral and enteral nutrition. 1999;23:337–344. doi: 10.1177/0148607199023006337. [DOI] [PubMed] [Google Scholar]
- 19.Sharma R, et al. Rat intestinal mucosal responses to a microbial flora and different diets. Gut. 1995;36:209–214. doi: 10.1136/gut.36.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onishi JC, et al. Bacterial communities in the small intestine respond differently to those in the caecum and colon in mice fed low- and high-fat diets. Microbiology. 2017;163:1189–1197. doi: 10.1099/mic.0.000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ott SJ, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajilic-Stojanovic M, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environmental microbiology. 2009;11:1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salonen A, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. The ISME journal. 2014;8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verbeke KA, et al. Towards microbial fermentation metabolites as markers for health benefits of prebiotics. Nutrition research reviews. 2015;28:42–66. doi: 10.1017/S0954422415000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sitkin SI, et al. Metabolic Dysbiosis Of the Gut Microbiota And Its. Biomarkers. Eksperimental’naia i klinicheskaia gastroenterologiia = Experimental & clinical gastroenterology. 2016;12:6–29. [PubMed] [Google Scholar]
- 30.Ma N, et al. Contributions of the Interaction Between Dietary Protein and Gut Microbiota to Intestinal Health. Current protein & peptide science. 2017;18:795–808. doi: 10.2174/1389203718666170216153505. [DOI] [PubMed] [Google Scholar]
- 31.Nirmalkar Khemlal, Murugesan Selvasankar, Pizano-Zárate María, Villalobos-Flores Loan, García-González Cristina, Morales-Hernández Rosa, Nuñez-Hernández Jorge, Hernández-Quiroz Fernando, Romero-Figueroa María, Hernández-Guerrero César, Hoyo-Vadillo Carlos, García-Mena Jaime. Gut Microbiota and Endothelial Dysfunction Markers in Obese Mexican Children and Adolescents. Nutrients. 2018;10(12):2009. doi: 10.3390/nu10122009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nature medicine. 2017;23:859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 33.Lan A, et al. High-protein diet differently modifies intestinal goblet cell characteristics and mucosal cytokine expression in ileum and colon. The Journal of nutritional biochemistry. 2015;26:91–98. doi: 10.1016/j.jnutbio.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Zoetendal EG, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. The ISME journal. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamabayashi S. Periodic acid-Schiff-alcian blue: a method for the differential staining of glycoproteins. The Histochemical journal. 1987;19:565–571. doi: 10.1007/BF01687364. [DOI] [PubMed] [Google Scholar]
- 36.Sovran B, et al. Age-associated Impairment of the Mucus Barrier Function is Associated with Profound Changes in Microbiota and Immunity. Scientific reports. 2019;9:1437. doi: 10.1038/s41598-018-35228-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).