Abstract
All 10 chromosomes of maize (Zea mays, 2n = 2x = 20) were recovered as single additions to the haploid complement of oat (Avena sativa, 2n = 6x = 42) among F1 plants generated from crosses involving three different lines of maize to eight different lines of oat. In vitro rescue culture of more than 4,300 immature F1 embryos resulted in a germination frequency of 11% with recovery of 379 F1 plantlets (8.7%) of moderately vigorous growth. Some F1 plants were sectored with distinct chromosome constitutions among tillers of the same plant and also between root and shoot cells. Meiotic restitution facilitated development of un-reduced gametes in the F1. Self-pollination of these partially fertile F1 plants resulted in disomic additions (2n = 6x + 2 = 44) for maize chromosomes 1, 2, 3, 4, 6, 7, and 9. Maize chromosome 8 was recovered as a monosomic addition (2n = 6x + 1 = 43). Monosomic additions for maize chromosomes 5 and 10 to a haploid complement of oat (n = 3x + 1 = 22) were recovered several times among the F1 plants. Although partially fertile, these chromosome 5 and 10 addition plants have not yet transmitted the added maize chromosome to F2 offspring. We discuss the development and general utility of this set of oat-maize addition lines as a novel tool for maize genomics and genetics.
Sexual inter-species hybridization is a powerful tool for understanding genome structure and interaction in higher plants. Breaking through inter-species incompatibility also enables chromosome engineering with horizontal (species-to-species) transfer of characters to which plant researchers and breeders have no access when utilizing only intraspecies gene pools. The more remotely related the parental genomes, the more the gene pool may be enriched.
Most of the favored host plants for chromosome engineering are allopolyploids or amphidiploids. Although in rare cases pure diploid species do tolerate alien chromosome additions, e.g. rye (Secale cereale; Kynast, 1986), the redundancy provided by homoeologs in polyploids is expected to better compensate for the loss of genetic information from alien substitution or translocation. Once stabilized as a disomic addition, the alien chromosome pair is transmitted consistently in the majority of the plants (Riley, 1960).
Oat (Avena sativa) and maize (Zea mays) are the most remotely related plant species of which we are aware that can be sexually hybridized and produce stable fertile partial hybrids. When oat × maize fertilizations are accomplished, a primary inter-species hybrid is generated that usually completely eliminates the maize chromosomes in early phases of embryogenesis. Thus, the resulting F1 hybrid becomes an allohaploid oat plant (Rines and Dahleen, 1990; Davis, 1992; Machan et al., 1995; Rines et al., 1996, 1997). Similar observations of uniparental chromosome elimination, i.e. elimination of all maize chromosomes in wide crosses involving maize as the pollen donor, include crosses of Coix lachryma-jobi × maize, Euchlaena perennis (=Zea perennis) × maize, Hordeum vulgare × maize, Saccharum officinarum × maize, Tripsacum andersonii × maize, and wheat (Triticum aestivum) × maize (Longley 1924, 1934; Mangelsdorf and Reeves, 1931; Arnason, 1936; Janaki-Ammal, 1941; Goodspeed and Bradley, 1942; Harada et al., 1954; Stebbins, 1956; Farquharson, 1957; Kandaswami, 1965; De Wet et al., 1983; Zenkteler and Nitzsche, 1984; Laurie and Bennett, 1986, 1988, 1989; O'Donoughue and Bennett, 1988, 1994a, 1994b; Suenaga and Nakajima, 1989; Inagaki and Tahir, 1990; Laurie et al., 1990; Riera-Lizarazu and Mujeeb-Kazi, 1990; Chen et al., 1991; Furusho et al., 1991; Inagaki et al., 1991; Laurie and Reymondie, 1991; Riera-Lizarazu et al., 1992; Inagaki and Mujeeb-Kazi, 1995). In general, when Panicoideae are crossed with Pooideae, chromosomes of the Panicoideae species are rapidly eliminated soon after fertilization in early stages of embryogenesis and chromosomes of the Pooideae species are always retained (Clayton and Renvoize, 1986). In wheat × maize crosses, one or more maize chromosome(s) were eliminated at the first mitosis in about 70% of the F1 embryos (Laurie and Bennett, 1989). By the time the embryos were in the eight-cell stage, all maize chromosomes were lost. The loss of maize chromosomes was attributed to restricted mobility of the maize chromosomes during metaphase and anaphase because of failing spindle fiber attachment to the maize centromeres (Laurie and Bennett, 1988).
A novel situation can occur in oat × maize crosses in that F1 hybrids may retain one or more maize chromosome(s) added to the haploid oat complement (Rines et al., 1995; Riera-Lizarazu et al., 1996). This is in contrast to maize crosses with other species (Rines et al., 1996). There is only one report of a retained maize chromosome in a wheat × maize hybrid, but the extra chromosome was not found to be transmitted to progeny (Comeau et al., 1992). Among maize wide crosses, only oat × maize crosses produced F1 embryos that germinated and grew into vigorous plants with retained maize chromosome(s). In such plants meiotic restitution enhanced the production frequency of completely or partially unreduced F1 gametes resulting in partial fertility. Self-pollination of these plants can generate different types of euploid or aneuploid F2 plants. Disomic alien additions (2n = 6x + 2 = 44) for different maize chromosomes were recovered among these offspring (Rines et al., 1995; Riera-Lizarazu et al., 1996; Kynast et al., 2000). These lines possess one pair of homologous chromosomes of maize added to the entire chromosome complement of hexaploid oats in a doubled haploid, i.e. 100% homozygous, condition. This manner of chromosome isolation into an alien genomic background reduces the large and complex maize genome to one homozygous chromosome pair with a unique combination of homomorphic allelic pairs. Thus, defined individual maize chromosomes, separated from the other maize chromosomes, can become an ideal target for high throughput marker allocation to maize chromosomes and for selective cloning of large maize DNA fragments known to have originated from a particular maize chromosome (Ananiev et al., 1997). By γ-irradiation of oat-maize additions to produce deletions and inter-genomic translocations, the isolated maize chromatin in an oat genomic background can be further reduced to a chromosome segment, thus providing even more refined maize marker mapping or sequence isolation (Riera-Lizarazu et al., 2000).
The addition of a maize chromosome to oat also enables analyses of the expression of homozygous maize allelic pairs in their native chromosomal environment in interaction with the oat host genome.
RESULTS
Primary Inter-Species F1 Hybrids
All 10 maize chromosomes were recovered as single additions to the entire haploid complement of oats. These maize chromosome additions were among 379 F1 plants generated from crosses of three different lines of maize to about 60,000 emasculated florets of eight different lines of oat followed by in vitro embryo rescue culture. The time course for in vitro germination of the F1 embryos extended up to 9 weeks with an average of about 2 weeks. After the 9 weeks, embryos were considered dead and no longer maintained if there was no development of shoot and root. By this criterion, the total of 473 germinated F1 embryos account for a germination frequency of about 11% of more than 4,300 immature F1 embryos placed on culture medium.
When the F1 embryos germinated and developed shoots of about 1 cm, plantlets were transferred from constant dark to short days (11 h, constant 20°C). The initial period in light was a very critical stage. Among plantlets that attained the size of 1-cm shoot length, about 20% (94 F1 embryos) suffered from absence of root development. At about 5-cm shoot growth, vigorous plants were transplanted into soil mix and grown as described in “Materials and Methods.” In general, all F1 plantlets showed weak morphology and slow growth after germination. Morphological differences appeared among the F1 plants depending on the added maize chromosome and the oat genetic background. All F1 plants had an enhanced sensitivity to changing growth conditions, such as temperature and photoperiod, compared with normal oats.
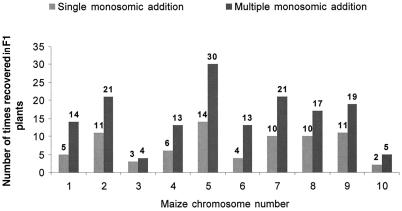
When a plant survived transplantation and started to grow after 1 or 2 weeks, 4- to 5-cm pieces of the leaf blade were clipped and genomic DNA extracted. A total of 379 F1 plants were tested for presence versus absence of maize chromosome(s). A total of 135 F1 plants possessed one or more maize chromosome(s) as indicated by a PCR product generated using primers specific for the maize sequence long terminal repeat (Grande 1-LTR; Fig. 1). Grande 1-LTR is a highly repetitive dispersed repeat present on every maize chromosome. To verify the number of maize chromosomes in maize-positive F1 plants, chromosomes in root tip cells were counted and genomic in situ hybridization (GISH) applied as described in “Materials and Methods.” Two examples are shown in Figure 2. Maize chromosome identities were then determined by PCR of leaf tissue DNA using maize chromosome-specific simple sequence repeat (SSR) markers (Fig. 3). For each maize chromosome several primer sets were selected from the Maize Genome Database (http://www.agron.missouri.edu/) and tested for maize specificity against genomic DNA of the oat lines. Specificity was defined by presence of a PCR product in maize cv Seneca 60 with no detected product from DNA of oat or by PCR products in maize and in oat DNA, but with length polymorphisms in maize compared with oat as detected upon electrophoresis in an agarose gel. An additional criterion for suitability of the marker of interest was that the marker had to be present on both parental chromosomes of the maize Seneca 60 with the above described specificity against oat. Because Seneca 60 is an F1 hybrid, markers were tested also on a representative Seneca 60 F2 progeny set (data not shown). By these criteria only 10 markers (14% of the markers tested) were chosen as most suitable and reliably reproducible (Table I) under our standard SSR assay conditions. Another 25 markers produced maize chromosome-specific bands but were less reproducible or segregating in the Seneca 60 F2 test population. Based on the identification by SSRs, each of the 10 maize chromosomes showed retention in F1 plants both as single additions and in combination with other maize chromosomes (Fig. 4). Maize chromosome 5 was most frequently retained (44 times). Maize chromosome 3 and 10 were most rarely retained (seven times each). Some F1 plants were sectored with distinct chromosome constitutions between tillers of the same plant. The presence and number of maize chromosomes also differed between shoot tissues (PCR analysis) and root tips (GISH analysis) in some F1 plants.
Figure 1.
Identification of maize-positive oat × maize F1 plants. PCR products of genomic DNA of oat, maize, and a selection of 27 oat × maize F1 plants are shown after electrophoresis in 1.5% (w/v) agarose gel. Bands represent 500-bp-long DNA fragments that were amplified with primers for the highly repeated maize-specific sequence tagged site marker Grande 1-LTR, which is dispersed on all maize chromosomes. Maize specificity is shown by product presence in maize DNA (second lane) and absence in oat DNA (first lane). Retained maize chromosomes are indicated in 13 out of the 27 F1 plant DNAs shown. All selected maize-positive F1 plants are then subjected to further analyses for chromosome identification.
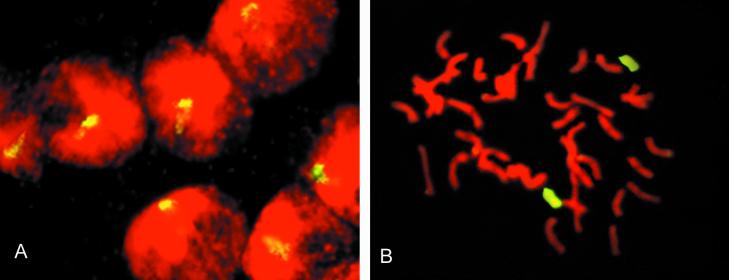
Figure 2.
Visualization of added maize chromatin and chromosomes in oat background. Two examples are shown for GISH by hybridizing fluorescein-labeled maize DNA without unlabeled competitor DNA to root tip meristem cells in interphase and in metaphase. Oat chromosomes are counterstained with propidium iodide (red) and maize chromosomes are yellow-green because of hybridized fluorescein-labeled probe. A, OMAm5.9, an F1 plant with monosomic addition of maize chromosome 5, illustrating that maize chromatin can be scored in F1 interphases. B, OMAd1.7, disomic addition of maize chromosome 1 to hexaploid Starter oat.
Figure 3.
Identification of added maize chromosomes by SSRs. PCR products of genomic DNA of a selection of 10 maize-positive oat × maize F1 plants (A–J), oat, and maize are shown after electrophoresis in 4.5% (w/v) agarose gel. Bands represent DNA fragments that were amplified with SSR markers specific for one of the 10 maize chromosomes. Maize specificity is shown by presence in maize DNA and absence in oat DNA or by length polymorphisms between maize and oat.
Table I.
Selected SSR-markers for identification of maize chromosomes
| Maize Chromosome | SSR Marker
|
||
|---|---|---|---|
| No oat band | Polymorphic to oat | Less reproducible | |
| 1 | p-bnlg421 | – | p-bnlg149, p-bnlg109, p-bnlg131 |
| 2 | p-bnlg125 | – | p-bnlg381, p-bnlg198 |
| 3 | – | p-phi047 | p-phi073, p-dupssr23 |
| 4 | – | p-phi079 | p-phi093, p-bnlg490, p-phi072 |
| 5 | p-nc130 | – | p-bnlg105, p-bnlg118 |
| 6 | p-phi070 | – | p-bnlg249, p-bnlg345 |
| 7 | p-phi112 | – | p-phi116, p-phi057 |
| 8 | p-phi080 | – | p-phi014, p-dupssr3, p-dupssr14 |
| 9 | p-phi032 | – | p-bnlg619, p-dupssr19, p-bnlg244, p-phi016 |
| 10 | p-phi059 | – | p-bnlg210, p-phi06, |
Figure 4.
Retention of maize chromosomes. Numbers are shown for the 10 maize chromosomes retained in F1 plants as individual chromosomes and in combination with other maize chromosomes.
The numbers of retained versus eliminated maize chromosomes among various oat × maize F1 genotypes are summarized in Table II, including previous (Riera-Lizarazu et al., 1996) and recent data. The data, however, could not be statistically evaluated because of a high number of F1 embryos that germinated and developed into small plantlets but died before leaf tissue was sufficient for DNA extraction. We observed that the greater the number of maize chromosomes retained the more severe the impact to the plant, with an apparently maximum tolerance of eight maize chromosomes by a haploid oat. Table II shows only those addition plants (379) that developed enough shoot material for DNA extraction and their chromosomes could be counted in root tips. The highest portion (89%) of loss of potential F1 plants was, however, due to the low germination rate of the F1 embryos.
Table II.
Nos. of F1 plants without maize chromosome(s) versus with retained maize chromosome(s)
| Oat Female Parent | Maize Male Parent | Analyzed F1 Plantlets | No. of Maize Chromosome(s) Added to the Oat Complement
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| F1 line | S60 | 18 | 13 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| GAF park | S60 | 69 | 46 | 16 | 5 | 1 | 1 | 0 | 0 | 0 | 0 |
| Kanota | S60 | 11 | 7 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| Kanota | A188 | 9 | 8 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| MN97201 | S60 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Preakness | S60 | 3 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Starter | S60 | 208 | 128 | 45 | 22 | 8 | 3 | 0 | 1 | 0 | 1 |
| Starter | A188 | 2 | – | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Starter | bz1 mum-9 | 7 | – | 4 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| Stout | S60 | 18 | 16 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Sun II | S60 | 33 | 24 | 5 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 379 | 244a | 77 | 36 | 13 | 5 | 0 | 3 | 0 | 1 | |
Nos. of maize-negative F1 plants were not recorded for Starter × A188 and Starter × bz1 mum-9 crosses.
Our data involving multiple chromosome retention revealed no apparent preferential combination of specific maize chromosomes in the F1 plants. However, our data show also that chromosome 3 appears to be the most excluded chromosome from multiple combinations among surviving plants in the oat × maize backgrounds we analyzed. Chromosome 3 was observed only four times in combination with other maize chromosome(s).
Maize Chromosome 1 Additions
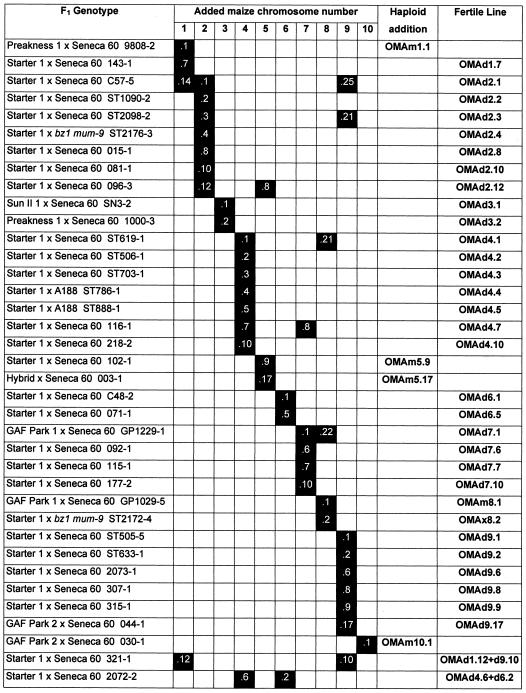
Maize chromosome 1 was observed 19 times among the F1 plants. The chromosome appeared five times as a single addition and 14 times in combination with other maize chromosomes (Fig. 4). Three lines are maintained: OMAm1.1 as a haploid monosomic chromosome 1 addition in the form of a vegetative tiller clone, OMAd1.7 as a fertile disomic chromosome 1 addition line, and OMAd1.12/d9.10 as a double disomic chromosome 1 and 9 addition line (Fig. 5). To identify each addition plant, their designation follows the nomenclature outlined in “Materials and Methods.”
Figure 5.
Currently maintained oat-maize addition stocks.
OMAm1.1 is tillered and semifertile. No positive maize chromosome 1 addition plant was found among more than 200 F2 offspring, indicating a lack of maize chromosome 1 transmission in the oat cv Preakness background. The F1 plant has been kept under short-day condition for more than 2 years by frequently breaking the plant into parts at its shoot base and repotting to generate fresh tillers (tiller clones). The clones are still growing with development of new tillers useful for DNA extraction. The cloned plants flower under either long day or short day and appear to have lost the long-day response common in oats. A severe response to long-day exposure is shorter leaf blades and pollen sterility.
OMAm1.7 looks much like Starter oat but is a little shorter in height and has a tendency toward more erect leaves as in OMAm1.1 (Fig. 6A). Also, this chromosome 1 addition plant shows the same photoperiod response as the OMAm1.1 plant with highest vigor, flowering, and seed set under short-day conditions. A second remarkable character is its chimerism in maize chromosome transmission. The plant had 108 F2 seeds distributed over 11 panicles, with only three F2 plants testing positive for maize chromosome 1. These three plants had 44 chromosomes including a pair of maize chromosomes 1 (OMAd1.7). The three seeds were set on one panicle. All other panicles had offspring negative for maize chromosome 1 (105 seeds) or were sterile. There was no variation among leaves in the tillers with respect to presence versus absence of maize chromosome 1. The F1 plant is still being maintained as a tiller clone with the aim to produce more offspring for analysis of chimerism in plants. The maize-positive F2 plants were fertile and showed sectoring again when the F3 offspring were cytologically analyzed.
Figure 6.
Examples of morphological characters in oat-maize additions. A, OMAd1.7, disomic addition of maize chromosome 1 causes an erect leaf phenotype; B, leaf blades of OMA2.1 and OMA4.1, the bluish color of the chromosome 2 addition (bottom blade) contrasts the lighter green color of the chromosome 4 addition (top blade); C, OMAd3.1 shows a crooked panicle (left) compared to the normal panicle of Sun II oat (right); D, OMAm5.17, branched stem with 5 branches (arrows); E, OMAd6.1, leaf blades with necrotic and chlorotic spots (disease lesion mimics); F, OMAd7.1, the stature is reduced, but very similar to GAF Park, the oat parent; G, OMAd9.1, chromosome 9 addition with EPS-syndrome; H, OMAm10.1, grassy type with heavy tillering in young plant age; I, OMAm10.1 after flowering, some tillers form stems with panicles, but the majority of tillers senesce at immature age.
The third hybrid (OMAd1.12/d9.10) was a double monosomic addition for maize chromosomes 1 and 9. The plant looked like regular oat, but was shorter and exhibited accelerated senescence. The plant was fertile with 76 seeds: 10 seeds with both maize chromosomes transmitted, four seeds with no chromosome 1 and only one or two chromosome 9, and 51 seeds without a maize chromosome. Six seeds did not germinate.
Maize Chromosome 2 Additions
Maize chromosome 2 was observed 32 times among the F1 plants. The chromosome appeared 11 times as single additions and 21 times in combination with six of the other maize chromosomes (Fig. 4). Seven different fertile lines are established (OMAd2.1, OMAd2.2, OMAd2.3, OMAd2.4, OMAd2.8, OMAd2.10, and OMAd2.12). Although maize chromosome 2 single additions were found also in GAF Park and Sun II oats, all fertile addition lines are in Starter oat. All F1 plants were vigorous and showed good vegetative growth with high tillering capacity. Compared to haploid Starter, the positive maize chromosome 2 plants were later maturing and had an intense bluish leaf color (Fig. 6B). Stems were more heavily covered by wax than Starter oat. The disomic addition offspring maintained these features with remarkable consistency. Chromosome 2 addition lines have the highest fertility among all oat-maize addition lines produced to date.
Maize Chromosome 3 Additions
Maize chromosome 3 was observed seven times among the F1 plants. Chromosomes 3 and 10 are the most rarely retained maize chromosomes. Chromosome 3 appeared three times as single additions and four times in combination with other maize chromosomes (Fig. 4). The multiple additions and a single addition in Starter oat died at the four-leaf stage. The remaining two lines were fertile—OMAm3.1 derived from Sun II × Seneca 60 and OMAm3.2 derived from Preakness × Seneca 60. Both F1 plants and their positive offspring showed distinct morphology. Their phenotypes include liguleless and a crooked panicle (Fig. 6C). The leaves are larger than those of oat and wrap around the stalk as in maize. Overall, chromosome 3 addition lines were the most morphologically distinct. However, the basic appearance is that of an oat plant. Both F1 plants had moderate fertility with 100% transmission of the maize chromosome.
Maize Chromosome 4 Additions
Maize chromosome 4 was observed 19 times among the F1 plants. The chromosome appeared six times as single additions and 13 times in combination with six of the other maize chromosomes (Fig. 4). One chromosome 4 was found in combination in OMAm4.9, which derived from the cross F1 (MN97201-1 × MN841801-1) × Seneca 60. A second chromosome 4 (OMAm4.15) was found in crosses with Kanota oat. All the other retentions occurred in crosses with Starter. Seven F1 plants were fertile and had transmission of the maize chromosome 4. The lines OMAm4.4 and OMAm4.5 descended from Starter × A188 crosses. All offspring with disomic chromosome 4 additions have light green leaves (Fig. 6B), some lighter than Starter oat. All addition plants were earlier maturing and had small seed. Fertility is moderate and independent of whether Seneca 60 or A188 was the chromosome 4 donor.
Maize Chromosome 5 Additions
Maize chromosome 5 was observed 44 times among the F1 plants. The chromosome appeared 14 times as single additions and 30 times in different combinations involving all other maize chromosomes (Fig. 4). Thus, chromosome 5 is the most frequently retained maize chromosome in oat. All chromosome 5-positive F1 plants tend to develop fewer but thicker tillers. Plants accelerate development after photoperiod induction and senesce earlier than the corresponding haploid oat genotypes. Two single maize chromosome 5 addition F1 plants are being maintained as tiller clones under short-day conditions. OMAm5.9 is an addition to haploid Starter oat. OMAm5.17 is an addition to a haploid F1 (MN97201-1 × MN841801-1) oat line. The most common striking character of these two lines is the development of branched shoots, though more pronounced in OMAm5.17 than in OMAm5.9 (Fig. 6D). The stalk branches were wrapped together in one leaf sheath, concordant with the heavy thick stems. When shifted into long day, OMAm5.9 and OMAm5.17 flowered and set seed. Their phenotypes were similar and of oat type. Moderate fertility provided for more than 160 F2 offspring. Absence of chromosome 5 in all offspring demonstrated a lack of female and male transmission for the maize chromosome 5 in different oat backgrounds tested to date.
Maize Chromosome 6 Additions
Maize chromosome 6 was observed 17 times among the F1 plants. The chromosome appeared only four times as single additions and 13 times in different combinations with the other maize chromosomes (Fig. 4). Two single addition F1 plants were fertile and produced disomic addition offspring (OMAd6.1 and OMAd6.5), which are maintained as established lines. Both lines descended from Starter × Seneca 60 crosses. Both F1 plants tillered in short-day photoperiod. Younger plants were indistinguishable from haploid Starter oat, but older plants developed necrotic and chlorotic spots on the leaf blades similar to disease lesion mimic mutants (Neuffer, 1994). With aging of the plants the symptoms progressively extended over the blades (Fig. 6E) and the leaf sheaths and eventually caused premature drying of entire leaves. Nevertheless, the plants had moderate fertility and transmitted the added maize chromosome to the F2 offspring stably through the following generations of propagating the lines.
Maize Chromosome 7 Additions
Maize chromosome 7 was observed 31 times among the F1 plants. The chromosome appeared 10 times as single additions and 21 times in different combinations involving all other maize chromosomes (Fig. 4). Four of the single addition F1 plants were fertile; OMAm7.1 was derived from a GAF Park × Seneca 60 cross amd OMAm7.6, OMAm7.7, and OMAm7.10 were derived from Starter × Seneca 60 crosses. A common feature of these additions was a reduced stature. Leaf color and panicle shape were very similar to those of their oat parents (Fig. 6F). The cytogenetic behavior was distinct in comparison to other addition plants. Chromosome 7 transmission was distorted. Disomic addition plants produced 16% monosomic and about 12% nonaddition offspring. Through further generations the transmission frequencies seemed to increase, but plants still show a low level of chromosome instability.
Maize Chromosome 8 Additions
Maize chromosome 8 was observed 27 times among the F1 plants (Fig. 4). The chromosome appeared 10 times as single additions and 17 times in different combinations involving all other maize chromosomes except maize chromosome 3. All oat lines except MN97201 retained maize chromosome 8. Three positive F1 plants were fertile; however, only two plants transmitted the chromosome to the next generation. OMAm8.1 (GAF Park × Seneca 60) produced one positive F2 offspring with 42 chromosomes. Among its F3 offspring, one positive monosomic addition (2n = 6x + 1 = 43) appeared. The monosomic addition line originates from this genotype. The second fertile F1 plant (OMAm8.2) was derived from a Starter × bz1 mum-9 cross. It produced one maize chromosome 8-positive F2 offspring among 19 offspring plants analyzed. Three F2 seeds did not germinate. Although the maize chromosome identity has been shown by SSR marker application, the chromosomes have not yet been counted. This plant is only 5 weeks old but lacks the vigor of Starter oat.
Maize Chromosome 9 Additions
Maize chromosome 9 was observed 31 times among the F1 plants. The chromosome appeared 12 times as single additions and 19 times in different combinations with the other maize chromosomes (Fig. 4). Five different fertile F1 plants were derived from Starter × Seneca 60 (OMAm9.1, OMAm9.2, OMAm9.6, OMAm9.8, and OMAm9. 9). One fertile F1 plant was derived from Kanota × Seneca 60 (OMAm9.5). One fertile F1 plant was derived from GAF Park × Seneca 60 (OMAm9.17). Six F1 plants produced diploid offspring with disomic additions of chromosome 9. One F1 plant produced a diploid F2 plant with disomic addition of chromosome 9 plus disomic addition of chromosome 1 (OMAd1.12/d9.10). All disomic addition F2 plants inherited a common phenotypic feature, an erratic premature senescence (EPS) syndrome. Single tillers at any stage of development may start to senesce in response to environmental stress while other tillers stay green and healthy (Fig. 6G). Drought and mechanical injury of leaves are among factors that can trigger the syndrome. The expression intensity is dependent on the oat background, but the appearance itself is not. Because of the syndrome severity, the OMAd9.5 line could not be maintained. All F3 offspring died before seed set. The other lines are maintained as disomic addition lines, although there is inconsistent seed set in the plants. In most cases one or two tillers remained green and panicles developed normally. Other tillers died before heading. However, if the EPS syndrome is not expressed, the addition plant looks like normal oat with high fertility.
Maize Chromosome 10 Additions
Maize chromosome 10 was observed seven times among the F1 plants. It appeared only two times as single additions and five times in different combinations (Fig. 4). Six F1 plants died in early stages of development, but one F1 plant (OMAm10.1) could be kept alive. The plant is maintained under short-day conditions in several tiller clones. The plant has a grassy phenotype (Fig. 6H) and shows premature senescence, but not of the EPS syndrome type. When clone parts are shifted to long-day conditions, flowering is induced (Fig. 6I), but there has been no seed set to date.
Maintaining the Oat-Maize Chromosome Addition Stocks
All oat-maize chromosome addition lines maintained at the University of Minnesota are summarized in Figure 5. All stocks are grown in growth chambers under conditions described for regular oat in “Materials and Methods” with the exception of the lines OMAm1.1 (F1), OMAm1.7 (F1), and OMAd1.7 (currently F3). These chromosome 1 additions are being grown in short-day conditions only. All plants are isolated as single plants to guarantee self-breeding and to avoid cross breeding. F1 and F2 plants are isolated as single plants and their respective F2 and F3 offspring are harvested from single panicles. Beginning with the F3 generation, the plants are isolated as single plants and F 4 seeds are harvested and pooled from all panicles for every single plant. For OMAd1.7, line maintenance by harvesting seed from individual panicles will continue because of the sectoring activity observed. The chromosome 7 addition is propagated by individual panicles because of remaining instability for the added maize chromosome. The vegetative maintenance of tiller clones for OMAm1.1, OMAm5.9, OMAm5.17, and OMAm10.1 will continue until disomic addition offspring are generated for maize chromosomes 5 and 10. Experiments attempting to produce doubled haploid plants by application of colchicine are in progress. Genomic DNA of at least one line from each of the 10 different groups of oat-maize chromosome addition stocks has already been distributed to the scientific community and will continue to be available. To date, seeds are available for addition lines involving maize chromosomes 1, 2, 3, 4, 6, 7, 8, and 9.
Besides the total of 35 single additions with all 10 maize chromosomes represented among them, two fertile double disomic addition lines are being maintained. One line is disomic for chromosomes 1 and 9 (OMAd1.12/d9.10) in Starter background. The second line is disomic for chromosomes 4 and 6 (OMAd4.6/d6.2), also in Starter background.
DISCUSSION AND CONCLUSIONS
Oat-maize addition lines are powerful tools for maize genomics (Ananiev et al., 1997). The complete series described herein provides material that enables any maize-specific sequence (relative to oat) to be allocated to maize chromosome. Chromosome duplications can be detected and multigene families mapped. Okagaki et al. (2001) physically mapped more than 400 sequences to maize chromosome using this addition series. The sequences included maize expressed sequence tags, and sequence tagged site and SSR markers. Because of the ability to identify sequences by presence versus absence of an amplification product in the addition lines, the PCR-based assay is straightforward and can be automated for high sample throughput. Oat-maize addition lines have been used as source for developing oat-maize radiation hybrid lines (Riera-Lizarazu et al., 2000). Maize chromosome deletions and intergenomic translocations with oat chromosomes can be induced by γ-irradiation of oat-maize additions. As a result, the reduction of the alien maize chromatin in the oat background to a chromosome segment allows more refined physical mapping or isolation of maize-specific markers and sequences.
A further utility of our addition set is the possibility to isolate single maize chromosomes. By the application of flow cytometry, maize chromosomes can be sorted and chromosome-specific DNA libraries become feasible for all maize chromosomes. As the first example, maize chromosome 9 has been isolated (Li et al., 2001) and further experiments are in progress to isolate the other maize chromosomes to provide chromosome-specific DNA for the construction of chromosome-specific large DNA fragment libraries.
New insights of gene activation and silencing can be achieved by analyses of maize gene activity in an oat host. The very special situation that maize genes are transferred into oat enables special inter-genomic effects to be observed and epistatic alleles to be detected and defined. Muehlbauer et al. (2000) found ectopic expression of the maize lg3 gene in oat-maize chromosome 3 addition plants and were able to relate cell fate features to the hemizygous versus homozygous allele activity.
Addition lines allow the analysis of meiotic behavior such as pairing, chromosome orientation, nondisjunction, and mobility as well as chromosome structure in mitosis and interphase. GISH hybridization using labeled maize DNA enables distinct visualization of an added maize chromosome pair in an alien background. By using the oat-maize chromosome 9 addition line, Bass et al. (2000) showed that homologous chromosomes pair and synapse during the telomere bouquet stage after a telomere-mediated reorganization of the early prophase I nucleus as the first step in the homolog search process.
Maize genes and mobile elements may possibly be transferred into oat, which would facilitate gene tagging in oat, and eventually the combination of genetic information from plants with fundamental physiological differences, such as the long-day C3-type oat and the short-day C4-type maize, will raise new questions in the fields of plant physiology and biochemistry. This first complete set of oat-maize chromosome addition lines can significantly contribute to answering such questions.
MATERIALS AND METHODS
Plant Material
Single-plant selections of eight different oat (Avena sativa) cultivars and experimental lines, including Starter, GAF Park, Sun II, Kanota, Preakness, Stout, MN97201, and an F1 (MN97201-1 × MN841801-1), were used in crosses with maize (Zea mays) sweet corn hybrid Seneca 60, dent corn inbred A188, or the experimental line bz1 mum-9, which carries mobile transposable elements. Parental oat and maize plants were grown with multiple planting dates in separate growth chambers to obtain synchronized flowering time for crossing the two species. Oat plants grow better at cool temperatures, i.e. 18°C to 20°C day, 14°C to 15°C night. Photoperiod is critical for the oat lines used; all are of spring-habit type. Plants were grown under short days (11-h light, 13-h dark) to favor vegetative growth and tillering and after 6 to 8 weeks they were shifted to long days (16-h light, 8-h dark) to induce flowering. Light of 300 to 400 μE m−2 s−1 at canopy height was provided from a mixture of fluorescent and incandescent lamps. Introducing transition phases of 30 min of 50% light intensities on each end of the light cycles to simulate sunup and sundown seemed to generate more vigorous plants with higher seed sets. Maize was grown at slightly warmer conditions, i.e. 20°C to 25°C day, 16°C to 18°C night, under long-day conditions with similar light intensities as described for oats.
In Vitro Embryo Rescue Culture
Primary florets of the upper two-thirds part of oat panicles were hand-emasculated either by complete anther removal or by clipping florets. After 48 h these florets were pollinated with freshly shed maize pollen as described in Rines and Dahleen (1990). Forty-eight hours after pollination the upper part of the oat plant was sprayed with 100 mg L−1 2,4-dichlorophenoxyacetic acid. After a further 12 to 14 d, immature embryos were rescued and cultivated on modified Murashige and Skoog medium as described in detail by Rines et al. (1997). Plantlets with about 6-cm shoot height, green shoots, and good roots were transplanted into a mixture of two parts soil and one part potting mix in a growth chamber with the temperature and photoperiod regimes as described for the oat parents.
PCR
To discriminate haploid oat plants without maize chromosomes from those haploid oat plants with one or more retained maize chromosomes, 4- to 5-cm pieces from the first leaf blade of the F1 plants were cut and the genomic DNA extracted according to Murray and Thompson (1980). The DNA samples were screened for the presence of maize chromatin by PCR in the presence of the forward primer 5′-AAA GAC CTC ACG AAA GGC CCA AGG-3′ and the reverse primer 5′-AAA TGG TTC ATG CCG ATT GCA CG-3′ with the following program: 1× (94°C for 5 min), 25× (94°C for 30 s → 58°C for 30 s → 72°C for 30 s), 1× (72°C for 5 min → 4°C). When maize chromosome(s) are present in the corresponding F1 plant, the PCR amplifies a genomic DNA fragment of 500 bp that is part of the long terminal repeats of the maize-specific retrotransposon Grande 1 (Monfort et al., 1995; SanMiguel et al., 1996; Vincent and Martinez-Izquierdo, (GenBank Accession No. X97604); Ananiev et al., 1998). Copies of the retrotransposon Grande 1 are highly dispersed and located on each maize chromosome. PCR under the described conditions does not produce a product from genomic DNA of oats.
To identify the added maize chromosome(s) in a maize-positive F1 plant, the appropriate genomic DNA sample was PCR-amplified in the presence of primers for maize chromosome-specific SSR markers with the following program: 1× (94°C for 5 min), 35× (94°C for 40 s → 62°C for 40 s → 72°C for 45 s), and 1× (72°C for 10 min → 4°C).
SSR markers with known map coordinates were selected from the Maize Genome Database.
Cytology
To observe somatic chromosomes and interphase nuclei from meristem cells, root tips (1.5–2 cm) were obtained by carefully removing the plant and soil from a pot containing a 3- to 4-leaf stage maize-positive F1 plant and snipping off exposed root tips. Root tips were pretreated in a solution containing 0.05% (w/v) colchicine, 0.0025% (w/v) 8-hydroxyquinoline, and 1.5% (v/v) dimethylsufoxide at room temperature for 3.5 to 4 h followed by 16-h ice water treatment to arrest cells at metaphase with good maize and oat chromosome morphology. Root tips were fixed in a 3:1 (v/v) mixture of ethanol and glacial acetic acid at room temperature for 2 d and stored at −20°C.
For chromosome counting, root tips were hydrolyzed in 1 n HCl at 60°C for 12 min and stained in Schiff's reagent at room temperature for 20 min (Feulgen and Rössenbeck, 1924). The meristem cells were squashed in 2% (w/v) aceto-orcein.
For GISH, root tips were washed by gentle shaking in distilled water for 20 min and macerated in 45% (v/v) acetic acid at room temperature for 5 min. The meristem cells were squashed in 45% (v/v) acetic acid. GISH followed the general procedures of Pickering et al. (1997) except that total genomic DNA of maize cv Seneca 60 was labeled with ChromaTide Oregon Green 488 (Molecular Probes, Eugene, OR) and probed onto slides without using an unlabeled competitor DNA. The hybridization was at 80% stringency for 6 h. Post-hybridization washes were at 85% maximum stringency. Chromosomes were counterstained with propidium iodide. Signals were visualized using a microscope (Eclipse 800, Nikon, Tokyo) equipped for epifluorescence. Images were captured with a CCD camera and processed with PhotoShop 5.5 software (Adobe Systems Inc., Mountain View, CA).
A biometrical evaluation of differences in retention among the 10 maize chromosomes is not possible because of the limited number of maize chromosomes recovered in the F1 plants.
Nomenclature and Conventions
Because of the nature of the process of generating oat-maize chromosome addition plants, and the fact that each recovered line represents its own distinct retention event for every maize chromosome, we developed a nomenclature for identification of the addition lines as follows: Each line is named as OMAxy.z. OMA stands for oat, maize, and addition, respectively. The name includes three more alpha numericals: x, y, and z, with the z separated from the y by a period. The x would be an m in the case where the addition is monosomic. The x would be a d in the case where the addition is disomic. The x would be mt or dt in the cases where the additions are monotelosomic or ditelosomic, respectively. The y is the number of the maize chromosome that is added, thus 1 to 10 for the set. For a future possibility, the y may be a B, if a B chromosome of maize origin is added to the oat genome. The z is the identification number of the particular version of the maize chromosome present that traces back to the original single recovery event.
ACKNOWLEDGMENTS
We thank Drs. Burle Gengenbach and John Gronwald (University of Minnesota) for reviewing the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. 9872650) and is a joint contribution of the Minnesota Agricultural Experiment Station and USDA-ARS. Mention of trademark or proprietary product does not constitute a guarantee or warranty by the University of Minnesota or USDA-ARS and does not imply approval over other products that also may be suitable.
LITERATURE CITED
- Ananiev EV, Phillips RL, Rines HW. Complex structure of knob DNA on maize chromosome 9: retrotransposon invasion into heterochromatin. Genetics. 1998;149:2025–2037. doi: 10.1093/genetics/149.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev EV, Riera-Lizarazu O, Rines HW, Phillips RL. Oat-maize chromosome addition lines: a new system for mapping the maize genome. Proc Natl Acad Sci USA. 1997;94:3524–3529. doi: 10.1073/pnas.94.8.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason TJ. Cytogenetics of hybrids between Zea mays and Euchlaena mexicana. Genetics. 1936;21:40–60. doi: 10.1093/genetics/21.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HW, Riera-Lizarazu O, Ananiev EV, Bordoli SJ, Rines HW, Phillips RL, Sedat JW, Agard DA, Cande WZ. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J Cell Sci. 2000;113:1033–1042. doi: 10.1242/jcs.113.6.1033. [DOI] [PubMed] [Google Scholar]
- Chen FQ, Hayes PM, Rivin CJ. Wide hybridization in Hordeum vulgare × Zea mays. Genome. 1991;34:603–605. [Google Scholar]
- Clayton WD, Renvoize SA. Genera graminum: Grasses of the World. Kew Bulletin Additional Series XIII. London: Her Majesty's Stationery Office; 1986. [Google Scholar]
- Comeau A, Nadeau P, Plourde A, Simard R, Maes O, Kelly S, Harper L, Lettre J, Landry B, St-Pierre C-A. Media for the in ovulo culture of proembryos of wheat and wheat-derived inter-specific hybrids and haploids. Plant Sci. 1992;81:117–125. [Google Scholar]
- Davis DW. Characterization of oat haploids and their progeny. MS thesis. St. Paul: University of Minnesota; 1992. [Google Scholar]
- De Wet JMJ, Fletcher GB, Hilu KW, Harlan JR. Origin of Tripsacum andersonii (Gramineae) Am J Bot. 1983;70:706–711. [Google Scholar]
- Farquharson LI. Hybridization of Tripsacum and Zea. J Hered. 1957;48:295–299. [Google Scholar]
- Feulgen R, Rössenbeck M. Mikroskopisch-chemischer Nachweis einer Nucleinsäure vom Typus der Thymonucleinsäure und die darauf beruhende selektive Färbung von Zellkernen in mikroskopischen Präparaten. Z Physiol Chem. 1924;135:203–248. [Google Scholar]
- Furusho M, Suenaga K, Nakajima K. Production of haploid barley plants from barley × maize and barley × Italian ryegrass crosses. Jpn J Breed. 1991;41:175–179. [Google Scholar]
- Goodspeed TH, Bradley MV. Amphidiploidy. Bot Rev. 1942;8:271–316. [Google Scholar]
- Harada K, Murakami M, Fukushima A, Nakazima M. Studies on the intergeneric hybridization between the genus Zea and Coix (Maydeae): breeding study on the forage crops: I. Studies on the intergeneric hybridization between the genus Zep and Coix (Maydeae) Sci Rep Saikyo Univ Agric. 1954;6:139–145. [Google Scholar]
- Inagaki M, Al-Ek W, Tahir MN. A comparison of haploid production frequencies in barley crossed with maize and Hordeum bulbosum L. Cereal Res Commun. 1991;19:385–390. [Google Scholar]
- Inagaki M, Mujeeb-Kazi A. Comparison of polyhaploid production frequencies in crosses of hexaploid wheat with maize, pearl millet and sorghum. Breed Sci. 1995;45:157–161. [Google Scholar]
- Inagaki M, Tahir MN. Comparison of haploid production frequencies of wheat varieties crossed with Hordeum bulbosum L. and maize. Jpn J Breed. 1990;40:209–216. [Google Scholar]
- Janaki-Ammal EK. Intergeneric hybrids of Saccharum. J Genet. 1941;41:217–253. [Google Scholar]
- Kandaswami PA. Proceedings Internationall Society of Sugar Cane Technology Congress. Puerto Rico. 1965. Note on Saccharum Zea hybrids; p. 12. [Google Scholar]
- Kynast RG. Untersuchungen zur Übertragung genetischer Information des hexaploiden Weizens (Triticum aestivum L. em. Thell.) in den diploiden Kulturroggen (Secale cereale L.). PhD thesis. Halle, Germany: Martin Luther University Halle-Wittenberg; 1986. [Google Scholar]
- Kynast RG, Okagaki RJ, Odland WE, Russell CD, Livingston SM, Rines HW, Phillips RL. Towards a radiation hybrid map for maize chromosomes. Maize Genet Coop Newsl. 2000;74:60–61. [Google Scholar]
- Laurie DA, Bennett MD. Wheat × maize hybridization. Can J Genet Cytol. 1986;28:313–316. [Google Scholar]
- Laurie DA, Bennett MD. Chromosome behavior in wheat × maize, wheat × sorghum and barley × maize crosses. In: Brandham PE, editor. Kew Chromosome Conference 3. London: Her Majesty's Stationary Office; 1988. pp. 167–177. [Google Scholar]
- Laurie DA, Bennett MD. The timing of chromosome elimination in hexaploid wheat × maize crosses. Genome. 1989;32:953–961. [Google Scholar]
- Laurie DA, O'Donoughue LS, Bennett MD. Wheat × maize and other wide sexual hybrids: their potential for genetic manipulation and crop improvement. In: Gustafson JP, editor. Gene Manipulation in Plant Improvement II. New York: Plenum Press; 1990. pp. 95–126. [Google Scholar]
- Laurie DA, Reymondie S. High frequencies of fertilization and haploid seedling production in crosses between commercial hexaploid wheat varieties and maize. Plant Breed. 1991;106:182–189. [Google Scholar]
- Li LJ, Arumuganathan K, Rines HW, Phillips RL, Riera-Lizarazu O, Sandhu D, Zhou Y, Gill KS (2001) Flow cytometric sorting of maize chromosome 9 from an oat-maize chromosome addition line. Theor Appl Genet (in press)
- Longley AE. Chromosomes in maize and maize relatives. J Agr Res. 1924;28:673–682. [Google Scholar]
- Longley AE. Chromosomes in hybrids between Euchlaena perennis and Zea. J Agric Res. 1934;58:789–806. [Google Scholar]
- Machan F, Nesvadba Z, Ohnoutkova L. Production of haploid plants of new wheat and oat donors through wheat × maize and oat × maize crosses. Genet a Slecht. 1995;31:1–10. [Google Scholar]
- Mangelsdorf PC, Reeves RG. Hybridization of maize, Tripsacum and Euchlaena. J Hered. 1931;22:329–343. [Google Scholar]
- Monfort A, Vicient CM, Raz R, Puigdomenech P, Martinez-Izquierdo JA. Molecular analysis of a putative transposable retroelement from the Zea genus with internal clusters of tandem repeats. J DNA Res. 1995;2:255–261. doi: 10.1093/dnares/2.6.255. [DOI] [PubMed] [Google Scholar]
- Muehlbauer GJ, Riera-Lizarazu O, Kynast RG, Martin D, Phillips RL, Rines HW. A maize-chromosome 3 addition line of oat exhibits expression of the maize homeobox gene liguleless 3 and alterations of cell fate. Genome. 2000;43:1055–1064. [PubMed] [Google Scholar]
- Murray MG, Thompson W. Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer MG. Disease lesion mutants. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 291–296. [Google Scholar]
- O'Donoughue LS, Bennett MD. Wide hybridization between relatives of bread wheat and maize. In: Miller TE, Koebner RMD, editors. Proceedings of the Seventh International Wheat Genetics Symposium. England: Cambridge; 1988. pp. 397–402. [Google Scholar]
- O'Donoughue LS, Bennett MD. Comparative responses of tetraploid wheats pollinated with Zea mays L. and Hordeum bulbosum L. Theor Appl Genet. 1994a;87:673–680. doi: 10.1007/BF00222892. [DOI] [PubMed] [Google Scholar]
- O'Donoughue LS, Bennett MD. Durum wheat haploid production using maize wide-crossing. Theor Appl Genet. 1994b;89:559–566. doi: 10.1007/BF00222448. [DOI] [PubMed] [Google Scholar]
- Okagaki RJ, Kynast RG, Livingston SM, Russell CD, Rines HW, Phillips RL. Mapping maize sequences to chromosome using oat-maize chromosome addition materials. Plant Physiol. 2001;125:1228–1235. doi: 10.1104/pp.125.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering RA, Hill AM, Kynast RG. Characterization by RFLP analysis and genomic in situ hybridization of a recombinant and a monosomic substitution plant derived from Hordeum vulgare L. × H. bulbosum L. crosses. Genome. 1997;40:195–200. doi: 10.1139/g97-028. [DOI] [PubMed] [Google Scholar]
- Riera-Lizarazu O, Mujeeb-Kazi A. Maize (Zea mays L.)-mediated wheat (Triticum aestivum L.) polyhaploid production using various crossing methods. Cereal Res Commun. 1990;18:339–345. [Google Scholar]
- Riera-Lizarazu O, Mujeeb-Kazi A, William MDHM. Maize (Zea mays L.)-mediated wheat (Triticum aestivum L.) polyhaploid production in some Triticeae using a detached tiller method. J Genet Breed. 1992;46:335–346. [Google Scholar]
- Riera-Lizarazu O, Rines HW, Phillips RL. Cytological and molecular characterization of oat × maize partial hybrids. Theor Appl Genet. 1996;93:123–135. doi: 10.1007/BF00225737. [DOI] [PubMed] [Google Scholar]
- Riera-Lizarazu O, Vales MI, Ananiev EV, Rines HW, Phillips RL. Production and characterization of maize-chromosome 9 radiation hybrids derived from an oat-maize addition line. Genetics. 2000;156:327–339. doi: 10.1093/genetics/156.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R. The meiotic behavior, fertility and stability of wheat-rye chromosome addition lines. Heredity. 1960;14:89–100. [Google Scholar]
- Rines HW, Dahleen LS. Haploid oat plants produced by application of maize pollen to emasculated oat florets. Crop Sci. 1990;30:1073–1078. [Google Scholar]
- Rines HW, Riera-Lizarazu O, Maquieira SB, Phillips RL. Wide crosses for haploids. In: Scoles G, Rossnagel B, editors. Proceedings of the Fifth International Oat Conference and Eighth International Barley Genetic Symposium, Saskatoon, Saskatchewan, Canada. 1996. pp. 207–212. [Google Scholar]
- Rines HW, Riera-Lizarazu O, Nunez VM, Davis DM, Phillips RL. Oat haploids from anther culture and from wide hybridizations. In: Jain SM, Sopory SK, Veilleux RE, editors. In Vitro Production of Haploids in Higher Plants 4. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 205–221. [Google Scholar]
- Rines HW, Riera-Lizarazu O, Phillips RL. Disomic maize chromosome-addition oat plants derived from oat × maize crosses. In: Oono K, Takaiwa F, editors. Modification of Gene Expression and Non-Mendelian Inheritance. Tsukuba, Japan: National Institute of Agrobiological Resources; 1995. pp. 235–251. [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Artificial polyploids as a tool in plant breeding: genetics in plant breeding. Brookhaven Symp Biol. 1956;9:37–52. [Google Scholar]
- Suenaga K, Nakajima K. Efficient production of haploid wheat T. aestivum through crosses between Japanese wheat and maize Zea mays. Plant Cell Rep. 1989;8:263–266. doi: 10.1007/BF00274125. [DOI] [PubMed] [Google Scholar]
- Zenkteler M, Nitzsche W. Wide hybridization experiments in cereals. Theor Appl Genet. 1984;68:311–315. doi: 10.1007/BF00267883. [DOI] [PubMed] [Google Scholar]