Abstract
Oat- (Avena sativa) maize (Zea mays) chromosome additions are produced by crossing maize and oat. During early embryo development maize chromosomes are preferentially eliminated, and oat plants are often recovered that retain a single maize chromosome. Each of the 10 maize chromosomes recently has been isolated as a separate oat-maize addition. We describe here the mapping of 400 maize sequences to chromosomes using polymerase chain reaction and DNA from the oat-maize addition material. Fifty of the sequences were from cloned markers that had been previously mapped by linkage analysis, and our results were consistent with those obtained using Southern-blot analysis. Previously unmapped expressed sequence tags and sequence tagged sites (350) were mapped to chromosomes. Maize gene sequences and expression data are rapidly being accumulated. Coupling this information with positional information from high throughput mapping programs provides plant biologists powerful tools for identifying candidate genes of interest.
Two types of information traditionally have been used to describe genes: positional and phenotypic. Positional information, provided by mapping genes, has held an important place in maize (Zea mays) research since the late 1920s (Emerson et al., 1935). The location of a gene along with its phenotype remains an important part of the description of a gene (Freeling and Fowler, 1994). Over the years many tools have been developed to increase the efficiency of mapping. Standard marked chromosome lines, B-A translocation lines, and marked reciprocal translocation lines have been widely used (Coe et al., 1988). The first molecular map in maize, published in 1986, contained 116 markers (Helentjaris et al., 1986). Introduction of recombinant inbred lines made a dramatic improvement in the efficiency of mapping (Burr et al., 1988). A recent map based on immortalized F2 lines has more than 1,700 morphological mutants, isozymes, and cloned sequences (Davis et al., 1999).
The advent of molecular biology has increased the importance of mapping. Positional information now has new uses. Mapping is necessary for chromosome walking, which is being used to clone genes in higher plants (Sakai et al., 1995; Frary et al., 2000). Comparative mapping studies have revealed syntenic relationships between chromosomes across species, genera, and families (Ahn and Tanksley, 1993; Gale and Devos, 1998). Coincident with the development of new uses for positional information has been an explosion in the number of genes and sequences that are available to be mapped. There are more than 70,000 maize expressed sequence tag (EST) sequences in public databases.
EST studies provide much of the information used to describe genes. The sequence of an EST often gives a strong indication of function. Transcript abundance and tissue specificity may also be predicted from EST data (Okubo et al., 1992; Ewing et al., 1999). Positional information, traditionally part of a gene's description, cannot be gleaned from ESTs. Mapping can help determine if two ESTs with similar sequences represent different alleles of one locus or two separate loci. Sequences with a predicted function may be associated with known mutations or quantitative trait loci by their physical location. However, coping with the number of sequences available is increasingly difficult with current mapping technologies.
Standard approaches for mapping sequences in maize require markers that are polymorphic between the parents used to create the mapping population. Sequences may be mapped with this material using RFLPs or simple sequence repeat polymorphisms, if they are available. This is an effective means of mapping, but it becomes unwieldy for locating large numbers of sequences. Southern-blot analysis is time consuming, and not all sequences contain useful simple sequence repeats. Single nucleotide polymorphisms have been suggested for mapping human sequences (Kruglyak, 1997; Wang et al., 1998), and there are several technologies available for high throughput analysis (Chee et al., 1996; Hall et al., 2000). It is unfortunate that a public sequence database adequate to identify large numbers of maize single nucleotide polymorphisms is not yet available.
An alternative approach for mapping sequences relies on the recently developed oat-maize chromosome addition lines. Oat- (Avena sativa) maize additions (OMAs) carry one or more maize chromosomes in an oat genomic background. During early embryo development from crosses of hexaploid oat with maize, most or all of the maize chromosomes are eliminated, and developing embryos can be rescued by in vitro culture. Oat plants carrying one or more maize chromosomes are then identified and characterized (Riera-Lizarazu et al., 1996). A complete panel of additions has now been developed, each addition carrying one of the 10 maize chromosomes. Fertile plants from eight of the 10 chromosome additions have been used to establish lines; plants carrying the remaining two chromosomes are maintained clonally (Kynast et al., 2001). Maize sequences can be detected in OMA material with PCR assays. Our objectives for this study were to evaluate the use of this material for mapping maize sequences to chromosomes and to compare the results from high throughput PCR assays with traditional Southern-blot assays.
RESULTS AND DISCUSSION
Comparing OMA Mapping with RFLP Mapping
RFLP mapping relies on Southern-blot hybridization to detect sequences in a segregating population. Many RFLP markers detect more than one band on a Southern blot and bands may differ in hybridization intensity. These bands reflect the duplicated nature of the maize genome and/or the presence of small gene families. When multiple loci are detected by a single probe, the loci are designated with lowercase letters (a, b, c, etc.) with “a” generally indicating the polymorphic locus corresponding to the band with the strongest hybridization intensity, “b” corresponding to the next strongest, and so on. Even weakly hybridizing bands can be mapped if they are polymorphic in the mapping population. Conversely strongly hybridizing bands cannot be mapped if they are not polymorphic in the mapping population. In sequencing a RFLP probe that detects multiple loci, it may not be possible to know exactly which locus the sequenced gene represents.
OMA mapping uses PCR, or other methods when appropriate, to detect a sequence in a panel of 10 OMA lines where each addition line carries one of the 10 maize chromosomes. Mapping relies on a plus/minus PCR assay; it is not dependent on the segregation of polymorphic markers. A group of 50 previously mapped RFLP probes were remapped to evaluate the effectiveness of using OMA lines for mapping (Fig. 1). PCR primers were designed for these sequences, and tested using maize Seneca 60 DNA, oat Starter-1 DNA, and no template DNA under standard conditions. Primer pairs were discarded that did not efficiently amplify Seneca 60 DNA. Primer pairs that amplified oat sequences were also discarded unless the amplified fragment was a different size than the maize product or amplification of oat sequences was weak and inconsistent. Approximately one-third of the primer pairs met these criteria under our standard conditions. When a pair of primers did not work, either new primers were designed for that sequence or primers were designed for a different sequence.
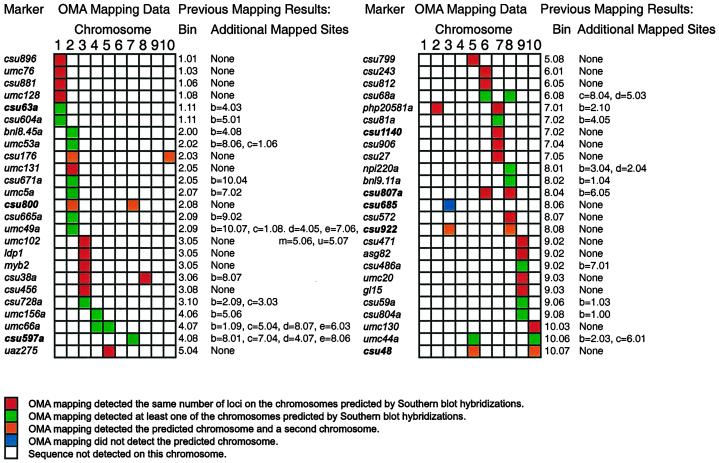
Figure 1.
Remapping markers using OMA materials. Markers were remapped using PCR assays with OMA material. Bin map locations for each marker are given along with the map locations of duplicated sequences. Map locations are from Davis et al. (1999). Markers mapping to more than one location are given letters to distinguish loci and to indicate the strength of hybridization with the cloned probe. csu63a is located on chromosome 1 in bin 11. csu63b is located on chromosome 4 in bin 3; the band on a Southern blot corresponding to csu63b has a weaker signal than the band corresponding to csu63a. Markers in boldface print are discussed in the text.
Using OMA mapping, 25 of the 50 markers were fully consistent with results from previous RFLP mapping (Davis et al., 1999). OMA mapping detected the same number of loci on the same chromosomes as predicted by Southern-blot analysis. Twenty-two of these markers mapped to a single chromosome, and three were present on two chromosomes. Examples are shown in Figure 2, A and B. Marker p-csu1140 was mapped onto chromosome 7 by Southern-blot analysis, and PCR detected this sequence only on the chromosome 7 addition line (Fig. 2A). Similarly, Southern-blot analysis and OMA mapping placed p-csu807 on chromosomes 6 and 8 (Fig. 2B). It should be noted that OMA mapping places sequences onto chromosomes. Therefore, it is not necessarily true that the locus of the sequence mapped to a chromosome by OMA mapping using PCR is the locus mapped by Southern-blot analysis. In addition, OMA mapping did not determine how many copies of a sequence are present on a chromosome. Results from the remaining 25 markers fell into three classes.
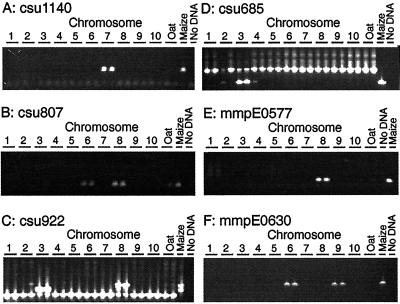
Figure 2.
Mapping sequences to chromosome. A, csu1140 mapped to chromosome 7. B, csu807 Mapped to chromosomes 6 and 8. A weak product in one of the oat controls was not detected in OMA materials. Weak and inconsistent amplification of oat sequences is a problem with OMA mapping. C, csu922 Mapped to chromosomes 3 and 8. Previous work had mapped this sequence to chromosome 8. PCR assays detected a previously unmapped duplication. A sequence in oats that gave a smaller PCR product was also seen. A false positive is visible in one of the two reactions from the chromosome 2 addition line. D, csu685 Mapped to chromosome 3. A larger oat sequence was also amplified; this product was seen in oat control lanes and OMA lanes. The smaller maize product was seen with the chromosome 3 OMA line. Faint bands associated with chromosomes 2 and 4 were not reproducible. E, mmpE0577 Mapped to chromosome 8. F, MmpE0630 mapped to chromosomes 6 and 9.
The first class consisted of 20 markers where Southern-blot analysis and OMA mapping were partially consistent. These markers mapped to two or more loci by Southern-blot analysis. OMA mapping detected the marker on at least one of the predicted chromosomes. In 19 of the 20 cases, the chromosomes detected by OMA mapping included the major locus, indicated by “a” on the RFLP map. For example, p-csu63 maps by Southern-blot analysis to chromosome 1 (csu63a) and chromosome 4 (csu63b); the locus mapped using OMA lines was on chromosome 1. The remaining marker, p-csu597, was mapped to five loci by Southern-blot analysis. The major locus, csu597a, is located on chromosome 4, but OMA mapping placed this marker on chromosome 7, coincident with csu597c. The fragment amplified by PCR from the chromosome 7 addition line was cloned and sequenced. Figure 3 summarizes the sequence comparison between the cloned PCR fragment from chromosome 7 and the published sequence; in the sequenced region 196 out of 199 bp were identical.
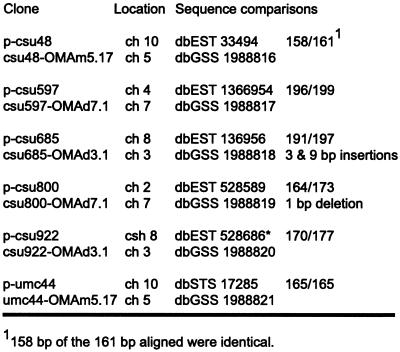
Figure 3.
Sequence comparisons of markers mapping to unexpected chromosomes. Comparisons between sequences amplified from OMA material and sequences reported in GenBank indicate that the amplified sequences originated from a duplicate locus. Sequences from csu48 and csu800 were each interrupted by an intron, and csu922 was interrupted by two introns. Primer sequences were not included in the comparisons.
A second class was composed of four markers, all of which had been mapped to a single locus by Southern-blot analysis. OMA mapping detected the predicted chromosome, but also detected a second chromosome. By Southern-blot analysis, p-csu922 maps to chromosome 8. OMA mapping detected the sequence on chromosome 8 and also chromosome 3 (Fig. 2C). The PCR products from three of the four markers in this group were cloned and sequenced. Sequence identities were 95% or greater between the published sequences for p-csu48, p-csu800, and p-csu922 and sequences from the cloned PCR products (Fig. 3). The inability of Southern-blot analysis to map these loci seems more likely due to a lack of polymorphisms rather than the inability to detect these loci, as sequence comparisons of the cases examined determined that the PCR products had derived from a related sequence. However, the region amplified by PCR corresponded to only a small part of the cloned probe. Whether or not other portions of the probe were also conserved has not been determined.
The third class consisted of one marker, p-csu685, which maps on chromosome 8 by Southern-blot analysis. OMA mapping did not detect this sequence on chromosome 8 but instead detected a sequence on chromosome 3 (Fig. 2D). Cloning and sequencing the chromosome 3 PCR product revealed that the PCR fragment has two small insertions of 3 and 9 bp and six single-base pair mis-matches (Fig. 3) relative to the published p-csu685 sequence.
Reliance on PCR assays for detection of sequences instead of Southern-blot hybridizations offers significant increases in productivity. Reliance on PCR assays also has disadvantages, some of which are apparent in Figure 2. False positives can be seen in one of the two PCR reactions in the chromosome 3 OMA with p-csu922 (Fig. 2C) and in the chromosome 2 and chromosome 4 OMAs with p-csu685 (Fig. 2D). False negatives have also been observed (data not shown). Oat sequences may be amplified in the PCR reactions. In Figure 2B there is a faint band in one of the two oat DNA controls. Performing duplicate PCR reactions allows one to repeat questionable PCR reactions. However, random chance dictates that some markers will be misplaced on the map. Nevertheless, 48 of the 50 markers tested mapped to the major chromosomal location predicted by RFLP mapping arguing that OMA mapping is an accurate means of rapidly mapping sequences to chromosome.
Mapping EST and Sequence Tagged Sites Sequences to Chromosome
Simplicity is a major advantage of mapping sequences using OMA lines. In the future, many of the steps now done by hand will be automated, and with these technologies it is feasible to map the large numbers of sequences available.
Three hundred EST sequences and 50 sequence tagged sites (STS) sequences were mapped to chromosomes. Figure 2E illustrates the mapping of the EST mmpE0577 (GenBank accession no. AI783424) mapped to chromosome 8. Approximately 75% of the sequences mapped to a single chromosome. Twenty percent of the sequences mapped to two chromosomes; EST mmpE0630 (GenBank accession no. AI941963) mapped to chromosomes 6 and 9 (Fig. 2F). The remaining sequences mapped to between 3 to 9 chromosomes. The percentage of duplicated markers varied among chromosomes. Approximately two-thirds of the markers present on chromosomes 4, 5, and 6 were classified as duplicated. One-third of the markers on chromosomes 1, 2, 3, 7, or 9 were duplicated elsewhere in the genome. Chromosomes 8 and 10 were intermediate with 50% of their markers present on another chromosome (Fig. 4). Detailed summaries of the data, along with primer sequences, are available in the online manuscript at the Plant Physiology website (http://www.plantphysiol.org/) and at the University of Minnesota plant genome research website (http://www.agro.agri.umn.edu/rp/genome/).
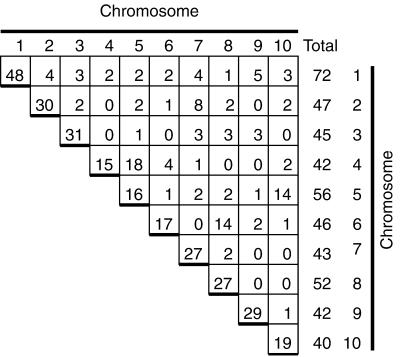
Figure 4.
Summary of mapping EST and STS sequences. Results from mapping 300 EST and 50 STS markers are tabulated. The column of numbers at the far right of the figure give the total number of markers mapped to each chromosome, whereas the numbers on the diagonal gives the number of unique markers mapped to each chromosome. For example, a total of 72 markers were mapped to chromosome 1; of these markers 48 of them were only present on chromosome 1. The number of markers that mapped to each pair of chromosomes is also shown. Chromosome pair 4 and 5 had 18 markers in common. The row and columns do not sum up to the chromosome totals because duplicated sequences are counted more than once. Data from five sequences are not included; each was found on five or more chromosomes.
Detection of Duplicated Sequences
Remapping the 50 markers previously mapped by Southern-blot analysis provides a direct measure of the ability of PCR to detect duplicated sequences. Twenty-three of the 50 markers were identified as duplicated using Southern-blot analysis (Davis et al., 1999); 10 markers were classified as duplicate using PCR assays with OMA material. Southern-blot hybridization mapped 89 loci with the 50 markers. PCR assays mapped 60 loci with the 50 markers; this assumes that each chromosomal assignment made by PCR represents one locus. The 60 chromosomal assignments may reflect more than 60 loci as duplicated sequences may be on the same chromosome. However, this is un-likely as only two markers, p-csu728 and p-csu597, had been mapped to two loci on one chromosome. Some duplicated sequences identified by PCR had not been classified as duplicated by Southern-blot analysis, but overall fewer sequences were classified as duplicated by PCR assays (Fig. 1). Both PCR assays and Southern-blot assays can underestimate the amount of duplication in the maize genome.
PCR conditions may account for the inability of PCR assays to detect more duplicated sequences. The touchdown PCR program used was designed to allow primer sets with a wide range of annealing temperatures to work (Chin et al., 1996). Reducing the stringency of the amplification cycle may permit the detection of more duplicated sequences. However, reaction conditions must be balanced between having too few primers work satisfactorily and the problem of amplifying oat sequences. Reduced stringency may allow oat sequences to be amplified. Increasing stringency can reduce the number of primer sets that amplify oat sequences, but fewer maize sequences might amplify. An added complication arises from the sequence polymorphism present in maize. The maize inbred line B73 was the source of many of the sequences used, and some primers amplified products from B73 but not from Seneca 60 (data not shown), the maize donor for the OMA lines. Furthermore, Seneca 60 is a hybrid; several primers that amplified Seneca 60 genomic DNA did not amplify the allele present in the OMA mapping panel (data not shown). Different PCR cycling conditions are being tested in an attempt to optimize these tradeoffs.
Of 300 EST and 50 STS sequences tested, 91 were classified as duplicated. There are 45 possible pairwise combinations of 10 chromosomes; duplications were detected involving 33 of the combinations (Fig. 4). Duplications were particularly evident between specific chromosome pairs. Eighteen markers were in common between chromosomes 4 and 5; 14 markers were in common between chromosomes 6 and 8. Fourteen markers were also in common between chromosomes 5 and 10. Evidence for duplications in the maize genome was found here even though PCR did not detect as many duplicated sequences as Southern-blot analysis.
Earlier studies based largely on Southern-blot analysis found evidence for extensive duplication between segments of maize chromosomes (Helentjaris et al., 1988; Ahn and Tanksley, 1993; Gale and Devos, 1998; Wilson et al., 1999). These studies have provided evidence for duplicated segments on chromosomes 4 and 5 and on chromosomes 6 and 8 (Gale and Devos, 1998; Wilson et al., 1999). Chromosomes 5 and 10, however, were not reported to share duplicated segments (Gale and Devos, 1998; Wilson et al., 1999). It is possible that the 14 markers found to be shared between chromosomes 5 and 10 here are, in fact, scattered about the chromosome as the order of markers along a chromosome cannot be determined by OMA mapping. OMA mapping provides a means to increase the number of markers mapped to chromosome, and the OMA lines provides a means of recovering duplicated sequences from specific chromosomes for further analysis.
Future Directions
OMA materials are appropriate for mapping sequences to chromosomes, but this approach cannot order sequences along a chromosome. For certain purposes this is sufficient. Some duplicated genes will be identified. Individual members of multigene families may be mapped and cloned, and polymorphisms specific to each member can be identified. For other purposes, better positional information is required. One means of achieving this is through radiation hybrid maps (Cox et al., 1990). Seed of addition lines monosomic for a single maize chromosome have been irradiated. The radiation treatment breaks chromosomes resulting in the apparently random loss of sequences (whether oat or corn). The hexaploid oat background of the addition lines tolerates the loss of sequences, facilitating the development of a set of radiation hybrid lines carrying a portion of a maize chromosome (Riera-Lizarazu et al., 2000). The position of a maize sequence on a chromosome is deduced from the presence versus absence of that sequence in the different radiation hybrid lines. Radiation hybrid mapping has the dual advantage of allowing high-throughput applications and having high resolution. The human radiation hybrid map contains over 30,000 markers (Deloukas et al., 1998). We are pursuing a two-track strategy for developing maize radiation hybrid mapping. Low-resolution maps will use a few well-characterized lines; these lines will be used to map large numbers of sequences to large regions on a chromosome. Several lines that may prove useful have been recovered from chromosome 9. For example, lines M9RH035 and M9RH0872 carry the distal ends of maize chromosome 9S and 9L, respectively (Riera-Lizarazu et al., 2000). High-resolution radiation hybrid maps will use larger numbers of lines to achieve increased resolution. Sequences of particular interest can be mapped to very small regions, perhaps one to two megabases. These positional data combined with expression data from EST studies and other sources will provide researchers with a valuable tool for associating sequences with phenotypes and identifying important genes.
MATERIALS AND METHODS
Plant Materials
Detailed procedures for the production of OMA plants, the establishment of fertile OMA lines, and their characterization were described by Riera-Lizarazu et al. (1996) and Kynast et al. (2001). OMA lines used in this study were monosomic (OMAm) or disomic (OMAd) for the maize chromosome (Table I). The first number following OMAm or OMAd indicates which maize (Zea mays) chromosome is carried, and the second number traces the chromosome back to the ancestral oat- (Avena sativa) maize F1 plant. Thus OMAm1.1 is monosomic for maize chromosome 1, and it originated from the first OMA plant carrying maize chromosome 1. Leaf tissue was harvested from OMA lines along with tissue from the Seneca 60 maize parent and the Starter-1 oat parent for most of the OMA lines. DNA was isolated using a cetyl-trimethyl-ammonium bromide (CTAB) procedure (Saghai-Maroof et al., 1984), and the concentration was determined using PicoGreen according to the supplier's directions (Molecular Probes, Eugene, OR).
Table I.
Oat-maize addition materials
| Chromosome | Designation | Oat Parent | Status | Reference |
|---|---|---|---|---|
| 1 | OMAm1.1 | Preakness 1 | Haploid F1 plant | Kynast et al. (2001) |
| 2 | OMAd2.1 | Starter 1 | Fertile diploid line | Riera-Lizarazu et al. (1996) |
| 3 | OMAd3.1 | Sun II | Fertile diploid line | Riera-Lizarazu et al. (1996) |
| 4 | OMAd4.1 | Starter 1 | Fertile diploid line | Riera-Lizarazu et al. (1996) |
| 5 | OMAm5.17 | Hybrida | Haploid F1 plant | Kynast et al. (2001) |
| 6 | OMAd6.1 | Starter 1 | Fertile diploid line | Maquieira (1997) |
| 7 | OMAd7.1 | Gaf Park | Fertile diploid line | Riera-Lizarazu et al. (1996) |
| 8 | OMAm8.1 | Gaf Park | Fertile monosomic line | Riera-Lizarazu et al. (1996) |
| 9 | OMAd9.1 | Starter 1 | Fertile diploid line | Riera-Lizarazu et al. (1996) |
| 10 | OMAm10.1 | Gaf Park | Haploid F1 plant | Kynast et al. (2001) |
Hybrid is an oat F1 hybrid (MN97201-1 × MN841801-1).
PCR Assays for Detecting Maize Sequences in OMA Material
PCR assays were used to detect maize sequences in OMA material. Three groups of sequences were tested. First, 50 probes that had been mapped previously by Southern-blot hybridization were tested (Davis et al., 1999). These probes have been sequenced and mapped to chromosomes (Fig. 1). Remapping these probes provided a comparison between Southern blot and PCR assays. Second, 50 maize genomic sequences were mapped. These STSs originated from the hypomethylated fraction of the maize genome that is believed to be enriched for genes (Rabinowicz et al., 1999). Third, 300 EST sequences from the maize database, ZmDB, were mapped (Gai et al., 2000).
PCR primers were designed using the program Primer 3 (Rozen and Skaletsky, 1998). Default values in the primer design program were used with the following exceptions: the optimum product size was set to 250 nucleotides (nts), the optimum primer size was 23 nts, maximum self-complementarity value was 5.00, and maximum 3′-self-complementarity value was 2.00. Oligonucleotide primers were ordered either from Integrated DNA Technologies (Coralville, IA) or from MWG-Biotech (High Point, NC).
The standard PCR reaction used for this work contained 1× PCR buffer supplied with the Taq polymerase, 1.67 mm of each dNTP, 0.5 μm forward oligonucleotide primer, 0.5 μm reverse oligonucleotide primer, approximately 100 ng of genomic DNA, and 0.15 units of enzyme in a 15-μL reaction. HotStarTaq from Qiagen USA (Valencia, CA) was used. Cycling conditions were based on a touchdown program previously described for use with maize (Chin et al., 1996). A 15-min incubation at 95°C activated the modified Taq polymerase. This was followed by two cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min. Nine cycles followed in which the annealing temperature was reduced from 64°C to 56°C in 1-degree increments; other conditions remained the same. This was followed by 29 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. After the last cycle, a 1-min incubation at 72°C was added in some cases. Samples were duplicated and were retested if there was a discrepancy between the duplicates. After amplification, the entire PCR reaction was fractionated on an agarose gel. One percent agarose gels were run in Tris-borate EDTA buffer unless higher percentage agarose gels were required to resolve products. Sets of reactions also included an oat Starter-1 DNA control, a maize Seneca 60 DNA control, and TE (10 mm Tris, pH 8.0, 1 mm EDTA) for a “no DNA” control.
A few PCR products were cloned and sequenced for further study. Products from three standard reactions were pooled, ethanol precipitated, and resuspended in 10 μL of TE. One microliter of the resuspended PCR product was ligated into the vector pGEM-T Easy (Promega, Madison, WI) according to manufacturer's directions, and a portion of the ligation reaction was used to transform frozen competent DH5α cells (Life Technologies, Rockville, MD). Transformed cells were plated on selective media and incubated overnight. Colonies were inoculated into Luria-Bertani media containing ampicillin and grown overnight. Plasmid DNA was isolated using Wizard DNA miniprep kits from Promega. Plasmid templates were sequenced using m13-forward or m13-reverse sequencing primers by the University of Minnesota Advanced Genetic Analysis Center.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rex Bernardo and Paula Olhoft for critically reading this manuscript, Adrian Stec and Tera Secker for technical help, and suggestions from two reviewers, which improved the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. 9872650).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
LITERATURE CITED
- Ahn SN, Tanksley SD. Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr B, Burr FA, Thompson KH, Albertsen MC, Stuber CW. Gene mapping with recombinant inbreds in maize. Genetics. 1988;118:519–526. doi: 10.1093/genetics/118.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SP. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- Chin ECL, Senior ML, Shu H, Smith JSC. Maize simple repetitive DNA sequences: abundance and allele variation. Genome. 1996;39:866–873. doi: 10.1139/g96-109. [DOI] [PubMed] [Google Scholar]
- Coe EH, Hoisington DA, Neuffer MG. The genetics of corn. In: Sprague GF, Dudley JW, editors. Corn and Corn Improvement. Madison, WI: American Society of Agron; 1988. pp. 81–259. [Google Scholar]
- Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: a somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- Davis GL, McMullen MD, Baysdorf C, Musket T, Grant D, Staebell M, Xua G, Polacco M, Kosterd L, Melia-Hancock S. A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics. 1999;152:1137–1172. doi: 10.1093/genetics/152.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloukas P, Shuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckmann JS. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Emerson RA, Beadle GW, Fraser AC (1935) A summary of linkage studies in maize. Cornell Univ Agric Exp Sta Memoir 180
- Ewing RM, Kahla AB, Poirot O, Lopez F, Audic S, Claverie JM. Large-scale statistical analysis of rice ESTs reveal correlated patterns of gene expression. Genome Res. 1999;9:950–959. doi: 10.1101/gr.9.10.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Frary A, Grandillo E, van der Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Freeling M, Fowler J. A nine-step way to characterize a morphological mutant. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 209–211. [Google Scholar]
- Gai X, Lai S, Xing L, Brendel V, Walbot V. Gene discovery using the maize genome database ZmDB. Nucleic Acids Res. 2000;28:94–96. doi: 10.1093/nar/28.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JG, Eis PS, Law SM, Reynaldo LP, J. R Prudent JR, Marshall DJ, Allawi HT, Mast AL, Dahlberg JE, Kwiatkowski RW. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc Natl Acad Sci USA. 2000;97:8272–8277. doi: 10.1073/pnas.140225597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris T, Slocum M, Wright S, Schaeffer A, Nienhuis J. Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theor Appl Genet. 1986;72:761–769. doi: 10.1007/BF00266542. [DOI] [PubMed] [Google Scholar]
- Helentjaris T, Weber D, Wright S. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics. 1988;118:353–363. doi: 10.1093/genetics/118.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L. The use of a genetic map of biallelic markers in linkage studies. Nat Genet. 1997;17:21–24. doi: 10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- Kynast RG, Riera-Lizarazu O, Vales MI, Okagaki RJ, Maquieira S, Chen G, Ananiev EV, Odland WE, Russell CD, Stec AO. A complete set of maize individual chromosome additions to the oat genome. Plant Physiol. 2001;125:1216–1227. doi: 10.1104/pp.125.3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquieira S. Production and characterization of plants from oat × maize and oat × pearl millet. MS thesis. St. Paul, MN: University of Minnesota; 1997. [Google Scholar]
- Okubo K, Hori N, Matoba R, Niiyama T, Fukushima A, Kojima Y, Matsubara K. Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene expression. Nat Genet. 1992;2:173–179. doi: 10.1038/ng1192-173. [DOI] [PubMed] [Google Scholar]
- Rabinowicz PD, Schutz K, Dedhia N, Yordan C, Parnell LD, Stein L, McCombie WR, Martienssen RA. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat Genet. 1999;23:305–308. doi: 10.1038/15479. [DOI] [PubMed] [Google Scholar]
- Riera-Lizarazu O, Rines HW, Phillips RL. Cytological and molecular characterization of oat × maize partial hybrids. Theor Appl Genet. 1996;93:123–135. doi: 10.1007/BF00225737. [DOI] [PubMed] [Google Scholar]
- Riera-Lizarazu O, Vales MI, Ananiev EV, Rines HW, Phillips RL. Production and characterization of maize chromosome 9 radiation hybrids derived from an oat-maize addition line. Genetics. 2000;156:327–339. doi: 10.1093/genetics/156.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (1998) Primer3. version 0.2, available through the Whitehead Institute for Biomedical Research/MIT center for Genome Research website. http://www-genome.wi.mit.edu/genome_software/other/primer3.html. January 1999–July 2000
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–818. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature. 1995;378:199–206. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Wilson WA, Harrington SE, Woodman WL, Lee M, Sorrells ME, McCouch SR. Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics. 1999;153:453–473. doi: 10.1093/genetics/153.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.