Significance
The extensively studied rod-shaped bacteria Escherichia coli, Bacillus subtilis, and Caulobacter crescentus grow by dispersed insertion of new cell wall material along the entire length of the cell. An alternative prokaryotic growth mode—polar growth—is used by some Actinomycetales and Proteobacteria. The latter phylum includes the family Rhizobiaceae, of which Agrobacterium tumefaciens is a prominent member. The factors responsible for polar growth are largely unknown. Here we describe GROWTH POLE RING (GPR) protein from Agrobacterium with a striking localization as a ring of six foci at the growth pole. GPR is potentially the polar growth organizer in Agrobacterium, as its absence leads to severe morphological defects, including loss of cylindrical cell shape, while its overproduction leads to ectopic growth poles.
Keywords: Agrobacterium, bacterial polar growth, apolipoprotein, rod-shape morphology, GROWTH POLE RING protein
Abstract
Polar growth in Agrobacterium pirates and repurposes well-known bacterial cell cycle proteins, such as FtsZ, FtsA, PopZ, and PodJ. Here we identify a heretofore unknown protein that we name GROWTH POLE RING (GPR) due to its striking localization as a hexameric ring at the growth pole during polar growth. GPR also localizes at the midcell late in the cell cycle just before division, where it is then poised to be precisely localized at new growth poles in sibling cells. GPR is 2,115 aa long, with two N-terminal transmembrane domains placing the bulk of the protein in the cytoplasm, N- and C-terminal proline-rich disordered regions, and a large 1,700-aa central region of continuous α-helical domains. This latter region contains 12 predicted adjacent or overlapping apolipoprotein domains that may function to sequester lipids during polar growth. Stable genetic deletion or riboswitch-controlled depletion results in spherical cells that grow poorly; thus, GPR is essential for wild-type growth and morphology. As GPR has no predicted enzymatic domains and it forms a distinct 200-nm-diameter ring, we propose that GPR is a structural component of an organizing center for peptidoglycan and membrane syntheses critical for cell envelope formation during polar growth. GPR homologs are found in numerous Rhizobiales; thus, our results and proposed model are fundamental to understanding polar growth strategy in a variety of bacterial species.
How bacteria grow and divide is intensely investigated, given the importance of bacteria across all of biology, from microbes to man. Several model systems, including Escherichia coli, Bacillis subtilis, and Caulobacter crescentus, have provided a wealth of fundamental insights into lateral dispersed cell elongation (1). However, as more bacteria enter the ring of scrutiny, another mode of cell elongation, limited to one or both poles, has emerged (2–5). In gram-negative bacteria, a single growth pole is used by the Rhizobiales order of Alphaproteobacteria, which includes the plant pathogen/genetic engineer Agrobacterium and plant nitrogen-fixing symbiont Rhizobium, as well as the animal pathogens Brucella and Bartonella (2–14).
Little is known about the enzymes involved, their spatiotemporal expression, or what mechanisms regulate bacterial polar growth. To date, several molecular players for polar growth have been identified in Agrobacterium tumefaciens. First, Agrobacterium-specific homologs of the classic cell division factors FtsA and FtsZ localize to the growth pole during cell elongation and then migrate to the midcell during initiation and completion of septation (6, 9, 12, 13). At the division site, FtsA and FtsZ are then precisely located at the new growth poles in resulting sibling cells. Second, homologs of Caulobacter old and new pole-specific markers, PopZCc (15, 16) and PodJCc (17, 18), respectively, have opposite localizations in Agrobacterium. PopZAt exclusively marks the growing pole (13). PodJAt initially marks the old pole but then also accumulates at the growth pole later in the cell cycle; this relocation is correlated with the transition of the growth pole to an old pole, and PodJAt is proposed to facilitate this transition (13). It is likely that FtsZ also plays a role in the transition to an old pole, as cells depleted of FtsZ fail to divide but continue to grow (12). Genetic deletion or depletion of PodJAt or PopZAt causes severe growth defects (14, 19–21), attesting to their essential roles.
Notably, the amino acid sequences of PopZAt and PodJAt are distinct from their Caulobacter counterparts, each with only 23% identity (13). Nevertheless, as the search for Caulobacter homologs successfully identified critical pole-specific factors in Agrobacterium, we used this approach again to search for potential homologs of Caulobacter TipN, dubbed a landmark protein for establishing and perpetuating new pole polarity throughout most of its cell cycle (22, 23). We found an uncharacterized Agrobacterium protein with little amino acid identity to TipN but an overall similarity in secondary structural domains. Most remarkably, the Agrobacterium protein localizes into six subpolar foci arranged in a hexameric ring at the growth pole.
The lack of significant amino acid sequence similarity to TipN and its distinctive localization pattern led us to name the newly identified Agrobacterium protein GROWTH POLE RING (GPR). Here we describe the domain architecture of GPR, its localization during the Agrobacterium cell cycle, and genetic analyses revealing that GPR is essential for polar growth and rod shape morphology.
Results
Identification of GPR Protein.
As polar growth in Agrobacterium is not well understood, it is critical to identify factors that localize to the growth pole that may function to facilitate such growth. We used the C. crescentus new pole-localizing protein, TipN, as a query and identified an uncharacterized Agrobacterium protein, A9CJ72 (24), Atu1348 in the A. tumefaciens proteome, with low (∼20%) amino acid sequence identity to TipN. For reasons described below, we named Agrobacterium A9CJ72 the GPR protein.
Fig. 1 diagrams the predicted structural domains of GPR (25); SI Appendix, Fig. S1A provides more detailed predictions of secondary structure according to PHYRE2 (26). Two predicted N-terminal transmembrane domains suggest that the bulk of the protein resides in the cytoplasm (Fig. 1 and SI Appendix, Fig. S1A). The N (80 aa) and C (110 aa) termini are enriched in proline residues and are predicted to be disordered. Strikingly, 82% of GPR, encompassing 1,720 residues (160–1,890), is predicted to be comprised of contiguous α-helices (SI Appendix, Fig. S1A). This large central α-helical region (blue area in Fig. 1) contains numerous nonoverlapping and overlapping subdomains homologous to eukaryotic apolipoproteins (ApoLPs; Pfam P01442) (25) with solved structures (27–30). SI Appendix, Table S1 lists the locations of 12 predicted ApoLP domains in GPR, 8 of which have significant e-values and an average size of 204 aa. Caulobacter TipN and Agrobacterium GPR differ dramatically in length (882 aa vs. 2,115 aa), but have an overall similar placement of related structural domains, N-terminal membrane-spanning domains, N- and C-terminal disordered regions, and central regions of continuous alpha helices (26) (SI Appendix, Fig. S1 A and B). TipN is distinct from GPR, however, with a much shorter central α-helical region (520 aa) and no predicted ApoLP domains.
Fig. 1.
GPR domain structure. Predicted transmembrane regions are shown in green, disordered regions are in red, and overlapping and/or adjacent ApoLP domains (SI Appendix, Table S1 lists exact locations and lengths) are in blue. Other areas (white) are predominantly α-helical according to Pfam (25). SI Appendix, Fig. S1 provides more details.
ApoLPs have been extensively studied in eukaryotes, as they are essential for the formation of cardioprotective high-density lipoproteins (reviewed in ref. 31), where α-helical domains of ApoLPs sequester lipid droplets (reviewed in ref. 32). ApoLPs also have been identified in prokaryotes, primarily as structural components of lipid droplets (33–35). Due to data mining of bacterial genomes, their numbers are increasing; in 2015, prokaryotic ApoLPs represented approximately 16% of the total PF01442 family members, while as of October 2018, this proportion had more than doubled, to 41% (SI Appendix, Fig. S2). Interestingly, 272 of 326 (83%) bacterial sequences reported to date that contain predicted ApoLPs (Pfam P01442) (25) are from the Rhizobiales order, which includes Rhizobium and Agrobacterium species. Most ApoLPs in the Rhizobiales are large proteins and have an overall architecture similar to GPR, including two transmembrane domains near the N terminus.
To date, no ApoLPs have been identified in the Caulobacterales order, consistent with the absence of ApoLP domains in TipN. Furthermore, TipN and GPR likely have distinct functions, as they participate in distinctly different means of growth, dispersed in Caulobacter compared with polar in Agrobacterium. Potentially, the large central region of GPR carrying ApoLP domain in Agrobacterium represents an evolutionary diversification in the Rhizobiales order compared with the Caulobacterales order, which creates a new function (36–38).
GPR Localization.
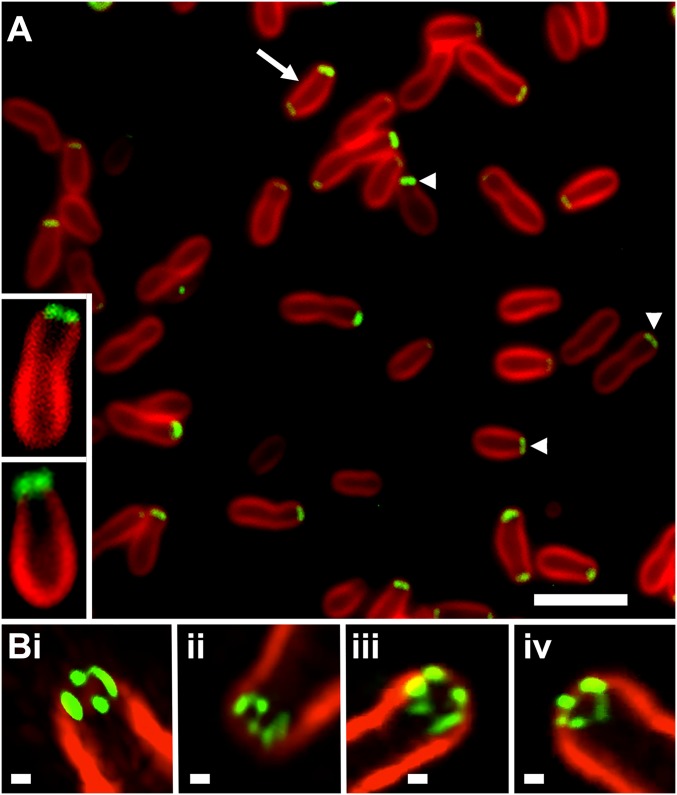
We created GFP fusions to the N and C termini of Agrobacterium GPR and visualized their localization. GFP fused to the N terminus reveals a broad, almost flat region of fluorescence near the growing pole (Fig. 2A; arrowheads) that differs from the tight focus of fluorescence observed with fusions to other growth pole- specific factors, such as FtsZ, FtsA, and PopZ (6, 13). On image enlargement, this broad band appears as paired foci very near, but not at, the tip of the growth pole (Figs. 2A, Insets and 3A).
Fig. 2.
Localization of GFP-GPR at the growth pole. (A) On WFM, most cells show a single broad polar focus of GFP (arrowheads), and a few cells (arrow) show additional weak localization at the opposite pole. (Insets) Cells with paired polar foci. (Scale bar: 3 μm.) (B) SIM 3D reconstructions show subpolar localization and multiple foci. Movies S1–S4 correspond to Fig. 2B, i–iv, respectively. FM4-64 stains the older nongrowing pole; weak or no FM4-64 staining occurs at the growth pole. (Scale bar: 100 nm.)
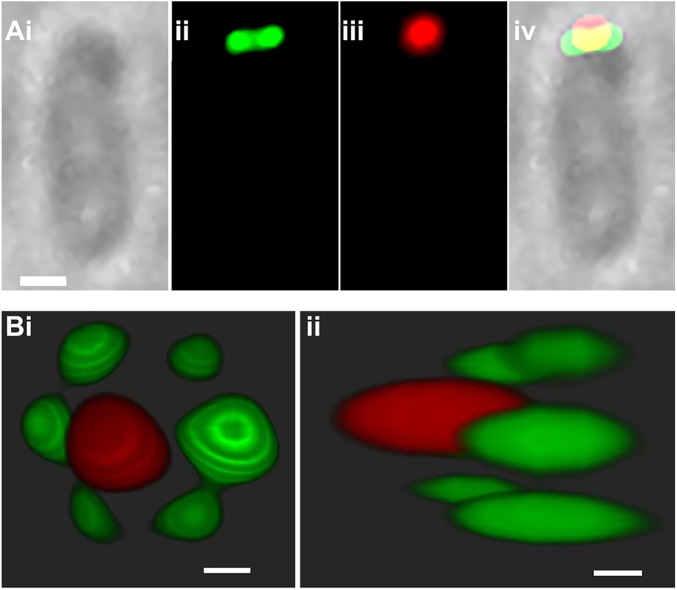
Fig. 3.
Localization of PopZ-GFP and GFP-GPR. (A) PopZ-RFP fluorescence overlaps with GFP-GPR. Some PopZ-RFP extends beyond GFP-GPR toward the tip of the growth pole, while the bulk of PopZ overlaps with GPR. Compare bright field (i), green (ii), and red (iii) fluorescence with both green and red fluorescence in the merged image (iv). (Scale bar: 500 nm.) (B) SIM confirms that PopZ localizes at the tip of growth pole, while GFP-GPR is subpolar and peripheral. (i) growth-pole view; (ii) lateral view. Movie S5 shows an animation of the 3D reconstruction of PopZ and GPR localization from SIM images. BF, bright field. (Scale bar: 50 nm.)
As paired foci in single optical sections are indicative of a potential ring structure, we used high-resolution structured illumination microscopy (SIM) to better resolve these polar foci. Indeed, GPR resolves into a ring of four to six foci subpolar to the growth pole (Fig. 2B). Some foci are round and likely represent single foci, while other foci likely represent overlapping fluorescence from two adjacent round foci, as exemplified by the cell shown in Fig. 2B, i. Movies S1–S4 show different rotations of the four cells shown in Fig. 2B. These multiple foci with a unique ring-like localization at the growth pole are the basis for naming Agrobacterium protein A9CJ72 the GPR protein.
GFP-GPR occasionally exhibits bipolar localization (Fig. 2A, arrow), and we performed time-lapse studies to further analyze this observation (see below). C-terminal GFP fusions to GPR resulted in abnormal growth patterns with multiple ectopic growth poles (SI Appendix, Fig. S3) suggesting that GFP blocks an essential function at the C terminus of GPR.
GPR Function Is Sensitive to Levels of Ectopic GFP-GPR.
For the images presented in Fig. 2 and throughout, we used very low levels of induction of GFP-GPR expression. Here, as in previous work, we cloned genes of interest (as fusions to GFP) into a low-copy number plasmid carrying a tightly regulated lac promoter (39), and induced protein expression by adding 2.5–10 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (6, 9, 13, 14, 19). For example, 10 mM and 5 mM IPTG resulted in expression of FtsZ-GFP or FtsA-GFP at approximately 10% of the levels of endogenous FtsZ or FtsA, respectively; these levels of ectopic induction did not alter cell morphology or cell cycle timing (6, 9, 13, 14, 19). However, for cells to remain viable during ectopic expression of GFP-GPR, it was essential to reduce the IPTG concentration to only 0.25 mM. SI Appendix, Fig. S4 shows cells carrying GFP-GPR induced for expression with 0.25 mM, 0.5 mM, 1.0 mM, and 2 mM IPTG. As the concentration of IPTG increased, the fluorescence of GFP-GPR increased, as expected; however, with more GFP-GPR expression, cells produced more ectopic growth poles. Ectopic growth poles were first observed with 0.5 mM IPTG induction, but not with 0.25 mM IPTG induction, and 0.25 mM induction was sufficient to allow observation of GFP-GPR by fluorescence microscopy.
The foregoing data suggest that WT GPR function is very sensitive to ectopic expression of GFP-GPR, potentially because GFP-GPR was induced to higher levels than expected. We tested the relative protein levels of GFP-GPR compared with WT GPR under different IPTG induction conditions. SI Appendix, Fig. S5 shows that 0.1 mM IPTG was not sufficient to induce GFP-GPR expression, 0.25 mM IPTG resulted in GFP-GPR expression at approximately 20% of WT levels, and 0.5 mM or 1.0 mM IPTG resulted in GFP-GPR expression at 50% or higher compared with WT. Thus, higher levels of IPTG significantly increased the ratio of the ectopic GFP fusion protein compared with endogenous WT GPR, and this may account for the altered cell shape and ectopic poles observed.
GPR Forms a Large Hexameric Ring at the Growth Pole.
Wide-field fluorescence microscopy (WFM) revealed GFP-GPR as paired foci near the growth pole tip (Fig. 2). Similar WFM during coexpression of GFP-GPR with PopZ-RFP showed PopZ-RFP closer to the tip of one end of the cell and GFP-GPR just below and overlapping PopZ-RFP (Fig. 3A). SIM imaging confirmed this localization and further revealed that GFP-GPR forms six distinct foci (Fig. 3B), as suggested by images in Fig. 2B; this notable localization is further evidenced in Movie S5. Each GFP-GPR focus is approximately 50 nm in diameter, similar in size to the PopZ-RFP focus. As the space between each focus is also ∼50 nm, the ring has a circumference of ∼600 nm with a diameter of ∼200 nm.
Localization of GPR During the Agrobacterium Cell Cycle.
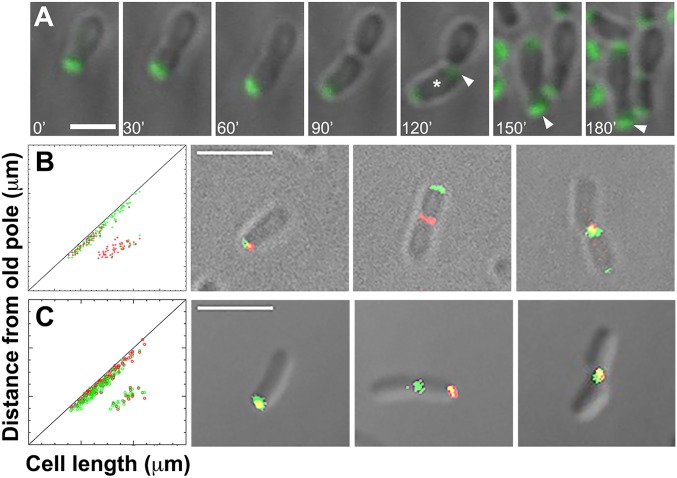
Fig. 4A presents images of a single cell expressing GFP-GPR following time-lapse observation. GFP-GPR is at the growing pole until cell division (at 90 min) and then is located at the new growth poles, resulting from midcell constriction in sibling cells (at 120 min). The previous new pole has become an old pole (4), yet GFP-GPR remains at this site, reflecting that GPR is membrane-anchored. That GPR remains at this old growth pole even until the next cell division (at 180 min) may suggest the time it takes for its removal and/or degradation. These results imply that GFP-GPR observed in the sibling new cell poles is likely to be newly synthesized; that is, it is unlikely that a membrane-anchored protein would easily migrate to the new poles. Movie S6 shows the same cell shown in Fig. 4.
Fig. 4.
GPR localization during the cell cycle and quantification of GPR, FtsZ, and PopZ localization to the growth pole or the midcell. (A) Time-lapse localization of GFP-GPR. Note that between 120 and 150 min, the lower cell (white asterisk) rotates (Movie S7) approximately 120° (clockwise), so that its growth pole faces downward at 150 min. The old pole is indicated by an arrowhead. (B and C) Coexpression of GFP-GPR and FtsZ-RFP (B) and GFP-GPR and PopZ-RFP (C). Red and green foci at the growth pole and the midcell were plotted along the diagonal or in the region between the diagonal and the x-axis, respectively, in 127 cells (B) and 100 cells (C). The tick marks on the x- and y-axes correspond to 0.5 μm. Representative cell images are shown to the right of each graph. While red and green foci occur together at the growth pole, FtsZ-RFP can occur at the midcell without GFP-GPR (but not vice versa), and GFP-GPR can occur at the midcell without PopZ-RFP (but not vice versa). (Scale bar: A, 2 μm; B and C, 3 μm.)
Notably, how long GFP-GPR remains at the new old pole varies. Movie S7 shows two different cells, one cell in which GFP-GPR remains at the new old pole and another cell in which it disappears relatively soon after the first cell division. The daughter that contains the original old pole does not contain GFP-GPR at its (very) old pole; in contrast, the daughter that originates from polar growth at the new pole contains GFP-GPR at its (new) old pole. These results also underscore that daughter cells are not equivalent.
To independently assess GPR localization during the cell cycle, we performed a quantitative assessment of the localization of GFP-GPR in cells coexpressing FtsZ-RFP or PopZ-RFP that each exhibit distinct time frames for their relocation from the growth pole to the midcell. FtsZ marks the new growth pole in Agrobacterium and then relocates to the midcell region approximately 60–70 min before septation (13), while PopZ remains at the growth pole until septation (13). Instead of elapsed time, we used cell length as a measure of progression through the cell cycle, and more than 100 green or red fluorescent foci were plotted relative to cell length (Fig. 4 B and C); foci that localize at the growth pole are plotted on the diagonal, and foci that occur at the midcell are plotted between the diagonal and the x-axis. GPR and FtsZ colocalize at the growth pole in shorter cells, corresponding to early stages of the cell cycle, and GPR remains at the growth pole as cells elongate up to 4 μm. In contrast, in cells ranging from 2.6 to 4 μm, FtsZ is rarely found at the growth pole and instead occurs at the midcell.
Coexpression of GFP-GPR and PopZ-RFP confirms that PopZ remains at the growth pole throughout the cell cycle (13) and in the longest cells, as expected. However, GPR relocates to the midcell just slightly before PopZ, as (i) there are a few more green foci in cells 2–3.5 cm long, (ii) we detect green foci at the midcell without red foci, and (iii) we rarely detect red PopZ foci at the midcell without green GPR foci.
Deletion or Depletion of GPR Causes Severe Growth Defects.
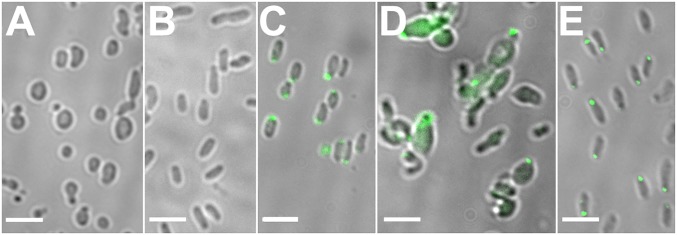
To test whether GPR is essential for polar growth, we created a genetic deletion of gpr (SI Appendix, Fig. S6 shows deletion strategy). Δgpr cells grow slowly, with a doubling time of 4–5 h in liquid cultures (vs. 90 min for WT); however, cell cycle length varies between cells (see below). Strikingly, Δgpr cells are coccoid and vary in diameter and overall shape (Fig. 5A). Few cells are elongated but misshapen (Fig. 5A, arrows). There is little evidence of normal polar growth, except some cells produce round projections (Fig. 5A, encircled). A few cells produce two small “buds” (Fig. 5A, arrowheads) that may lead to mini cells.
Fig. 5.
Deletion of GPR results in spherical cells. (A) Δgpr cells are mostly coccoid. Some cells appear to be making new round cells (encircled), a few cells produce two buds (arrowheads), and a few cells are elongated but misshapen (arrows). (B) Time-lapse imaging of two dividing cells, labeled 1 and 2 (0 min), giving rise to cells numbered 3, 4, and 5 (images derived from Movie S8). The times shown were chosen to illustrate newly arising or dividing cells and do not represent a constant time difference between each panel. (Scale bar: 3 μm.)
Nevertheless, at least some Δgpr cells grow in liquid culture and form small colonies on agar plates. Time-lapse imaging (Fig. 5B and Movie S8) shows that round cell 1 produces cell 2, which produces cell 3; after a delay cell 2 produces cell 4, and then cell 5 forms. Thus, Δgpr daughter cells are not synchronous in their cell cycle progression. Normally, two cells should produce four cells and then eight cells; instead, we observe cell number 3 before cell number 4, and cell number 5 before cell number 6.
Δgpr daughter cells have an additional phenotype, failure to separate following division, so one can easily distinguish the order of formation of cells 3, 4, and 5 (Fig. 5B); such images are representative of 20% of Δgpr cells with round protrusions (Fig. 5A). In addition, at least 40% of the cells are unusually large and spherical; such cells grow in all directions over a long period (14 h) and produce a quartet of daughter cells that remain attached (Movie S9).
The coccoid phenotype of Δgpr disappeared following ectopic expression of WT GPR under control of the lac promoter (SI Appendix, Fig. S7). Surprisingly, rescue occurred following extremely low levels (0.025 mM) of IPTG (10-fold less IPTG than used previously to induce GFP-GPR expression), suggesting that cells do not require high amounts of GPR to survive. In support, higher levels of IPTG resulted in abnormal branched cells with ectopic poles. Thus, GPR levels must be tightly controlled in WT cells.
Since stable genetic deletion of gpr caused such a strong phenotype, we next used a riboswitch strategy (SI Appendix, Fig. S8) to make RS::gpr. Without theophylline, RS::gpr exhibits a coccoid phenotype identical to Δgpr, and WT cell shape is restored on addition of theophylline (Fig. 6 A and B). Notably, inducible expression of GFP-GPR with low levels of IPTG (0.025 mM) also rescues RS::gpr grown in the absence of theophylline (Fig. 6C).
Fig. 6.
Riboswitch-controlled depletion of GPR mimics stable genetic deletion of GPR and alters PopZ-RFP localization. RS::gpr cells were imaged at 24 h after growth without theophylline (A) or at 24 h after growth in 0.5 mM theophylline (B). Expression of GFP-GPR was induced with 0.025 mM IPTG for 24 h in the RS::gpr strain grown in the absence of theophylline (C). PopZ-RFP was introduced into RS::gpr and grown in the presence of 0.25 mM IPTG without theophylline (D) or 0.25 mM IPTG plus 0.5 mM theophylline (E).
Loss of GPR Impacts Localization of PopZ.
PopZ localizes very close to GPR (Fig. 3), and GPR relocates to the midcell just before PopZ. To test whether PopZ position is dependent on GPR, we monitored PopZ localization following depletion of GPR in RS::GPR (Fig. 6 D and E). In GPR-depleted cells, PopZ-GFP appears mostly diffuse, with a few random foci in the misshapen cells (Fig. 6D). WT PopZ-GFP localization is rescued when this strain is grown in the presence of theophylline (Fig. 6E). Thus, PopZ can still form foci on its own, but that proper localization to the cell pole requires rod shape morphology, dependent on GPR. These data demonstrate the utility of RS::gpr to assay phenotypic effects following depletion of GPR.
Discussion
We have identified a previously unknown protein, which we named GROWTH POLE RING (GPR), with a striking localization as a ring of six foci at the growth pole in A. tumefaciens. GPR plays a major role in polar growth, as its absence leads to severe morphological defects, including loss of cylindrical cell shape. In corollary, its overproduction leads to ectopic growth poles; the finding that overproduction only slightly above WT levels leads to multiple growth poles implies that GPR expression must be tightly regulated. While prokaryotic proteins with single ApoLP domains (associated with lipid droplets) have been reported (33), to date, GPR is the first prokaryotic protein identified with an extended multiple ApoLP domain structure anchored in the inner membrane. GPR potentially represents a new class of proteins with a critical role in polar growth and cell morphogenesis in the Rhizobiales.
We propose that GPR functions as a scaffold for assembly of polar growth machinery essential for peptidoglycan (PG) and/or phospholipid (PL)-dependent membrane biogenesis (Fig. 7A). Polar growth necessitates a mechanism to ensure that the growing pole is not amorphous or unstructured. This need is compounded by the fact that A. tumefaciens lacks an underlying cytoskeleton, such as provided by the actin homolog MreB in model systems like E. coli and B. subtilis, where MreB serves as a scaffold for circumferential PG synthesis during cell elongation by interspersed growth (40–42). Indeed, the numerous ApoLP domains in GPR support the hypothesis that GPR provides a scaffold for membrane assembly, as ApoLPs are well known to sequester lipids in eukaryotes (32). The dramatically abundant α-helical regions of GPR, including a continuous stretch of more than 1,700 amino acids, may provide a hydrophobic environment for lipid assembly.
Fig. 7.
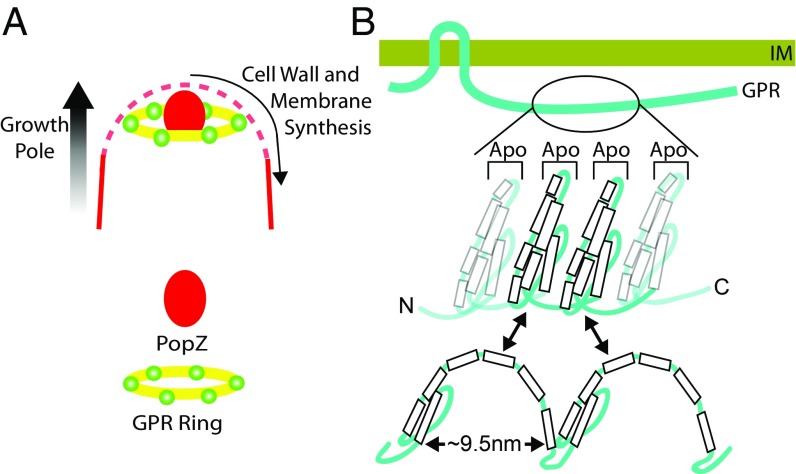
Models of the GPR hexamer ring (A) and monomer secondary structure (B). (A) Six GPR complexes (green spheres) form a (yellow) ring-shaped scaffold ∼100 nm from the tip of the growth pole to organize membrane and PG synthesis. The solid red line indicates outer membrane (OM) staining with red lipophilic dye FM4-64; the dashed red line indicates OM stains less intensively at the growth pole (6). (B) Flexible/reversible structure (based on a model for ApoLP A1; figure 5B in ref. 44) of GPR ApoLP domains (extended vs. compact) may facilitate a variety of configurations between adjacent GPR monomers that maintain the diameter of the GPR scaffold. C, C terminus; IM, inner membrane; N, N terminus.
Mycobacterium also grows by polar growth and produces a polar complex that contains at least three PG synthetic enzymes (MurG, GlfT2, and Pks13) (43). This complex resides in a band called the subpolar space, 0.5 μm from the tip of the growth pole, and the diameter of the complex corresponds to the diameter of the mature cylindrical cell. In striking contrast, the GPR ring complex localizes approximately 100 μm below the tip of the new pole, and the diameter of the GPR ring is only approximately 200 nm. These latter data reveal that Agrobacterium polar growth initially occurs in a much narrower ring of activity. Indeed, independent measurements of new pole cell diameter in different-sized new cells demonstrated that approximately one-third of the upper part of the new cell is of significantly narrower diameter during the early stages of polar growth, and later, PG synthesis increases the new cell diameter until it corresponds to the diameter of the old cell compartment just before division (9). Mycobacterium and Agrobacterium present surprisingly different examples of what might comprise the geometry of cell wall synthesis complexes during polar growth. Nevertheless, they both suggest a general mechanism for polar growth via a ring structure near the growth pole(s).
How might GPR foci be organized to be ∼50 nm apart in a GPR hexamer of ∼200 nm diameter? An extended linear string of 1,700 helically arranged amino acids would be ∼600 nm long. More likely, GPR ApoLP domains fold into specific 3D shapes. The ApoLP structure is very dynamic, and monomers fold as bundles of helices or extended curved structures in the presence of lipids (44, 45). ApoLPs form two types of complexes of ∼9–10 nm diameter when associated with lipids: (i) three ApoLPs arranged in a trefoil with their alpha helical regions on the surface of lipid spheres or (ii) two stacked ApoLPs around a lipid disk (reviewed in ref. 32).
Our data suggest that the 1,700-aa central region of GPR contains approximately eight ApoLPs, each ∼200 aa long. These eight ApoLPs may provide an extremely flexible region that may undergo significant rearrangements from bundled to extended configurations during the assembly of membranes and PG. Based on the structure of ApoLP A4 (44), Fig. 7B graphically depicts two ApoLP domains in GPR, alternating between helix bundles and extended conformations. It is highly likely that additional, as-yet unidentified proteins may be structural and functional components of the GPR hexamer. Future investigations using a variety of approaches likely will lead to abundant and informative discoveries regarding GPR function during polar growth in the Rhizobiales.
Materials and Methods
Strains and Cell Growth Conditions.
The strains and plasmids used in this study are listed in SI Appendix, Table S2. WT A. tumefaciens C58 was transformed with the relevant plasmids and grown at 28 °C in Luria Broth (LB) medium containing appropriate antibiotics.
Bioinformatics.
We used TipN protein from C. crescentus (CC_1485 from strain CB15) as the query in a BLAST search of the Uniprot database Microbial Proteomes with default search parameters (24). GPR (Atu1348) was identified from a manual inspection of the list of hits generated by this search. GPR domains were identified using Pfam (25) and Phyre2 (26).
Molecular Cloning and Strain Construction.
Standard molecular cloning techniques were used (46). To construct pJZ251 (Plac::gpr-gfp), gpr was amplified by PCR from A. tumefaciens genomic DNA with NdeI sites at each end and subcloned into pCR2.1-Topo (Thermo Fisher Scientific), and then gpr was isolated from NdeI digestion and ligated to similarly digested pJZ210. A similar strategy was used to construct pJZ253 (Plac::gfp-gpr), except that gpr was PCR-amplified with 5′ AvrII and 3′ KpnI sites.
Δgpr and RS::gprAt were created as described previously (14, 19). In brief, C58 was transformed with pJZ298 (Δgpr) or pJZ274 (RS::gpr) (SI Appendix, Table S2), followed by selecting for a single cross-over into the genome by growth on carbenicillin and then selecting for a second recombination by growth on sucrose. Δgpr and RS::gprAt were verified by PCR amplification of the relevant genomic region and sequencing.
Time-Lapse and Fluorescence Microscopy.
Unless noted otherwise, lactose-inducible expression was achieved by diluting overnight cultures to 108 cells/mL and adding 0.25 mM IPTG for 4–5 h before single-time or time-lapse imaging by deconvolution fluorescence microscopy as described previously (13). Long-term time-lapse imaging of Δgpr was done using the CellASIC ONIX system (EMD Millipore) on a Zeiss Axio Observer Z1 microscope. For quantitative analyses, C58 cells were cotransformed with pJZ253 (GFP-GPR) and pJZ269 (PopZ-RFP) or with pJZ253 and pTC077 (FtsZ-RFP); cell length and position of the fluorescent foci were determined using ObjectJ (47). All images were processed using Fiji software (48).
Superresolution Microscopy.
Superresolution images were captured using a Zeiss Elyra PS.1 structured illumination microscope (SIM) equipped with a Zeiss Plan-Apochromat 100×/1.46 oil immersion objective lens and a pco.edge scientific complementary metal-oxide semiconductor camera with a 1.6× tube lens. GFP-GPR fluorescence was detected with 488-nm laser excitation. FM4-64 or RFP fluorescence was detected with 561-nm laser excitation. The lateral pixel size was 41 nm × 41 nm in the recorded images. Z-stacks were acquired by capturing 20 slices with a 0.1-μm step size. The 3D SIM images were reconstructed using ZEN 2012 (black edition) (Zeiss) and processed with Imaris 8.1 (Bitplane).
Protein Analysis.
Preparation of Agrobacterium cell lysates, gel electrophoresis, and Western blot analysis were performed as described previously (49). The primary antibody was commercially prepared rabbit polyclonal anti-GPR antibody (Genscript) generated against the peptide FEEKPETRKTAKPK (amino acids 27–41) and used at a dilution of 1:1,000.
Supplementary Material
Acknowledgments
We thank our colleagues John-Marc Chandonia and Steven Brenner for discussions on ApoLP. We also thank Steven Ruzin and Denise Schichnes (College of Natural Resources Biological Imaging Facility at University of California, Berkeley) for assistance with fluorescent imaging and SIM. The Biological Imaging Facility is supported in part by National Institutes of Health Program S10 (Award 1S1OD018136-01). Research in the P.Z. laboratory is supported by National Science Foundation Grant MCB-0923840. J.S.R.-E. received fellowship support from the Secretaria Nacional de Educacion Superior, Ciencia y Tecnologia, Ecuador. N.G. received support from the College of Natural Resources Sponsored Projects for Undergraduate Research program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905900116/-/DCSupplemental.
References
- 1.den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA (2008) Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev 32:321–344. [DOI] [PubMed] [Google Scholar]
- 2.Howell M, Brown PJ (2016) Building the bacterial cell wall at the pole. Curr Opin Microbiol 34:53–59. [DOI] [PubMed] [Google Scholar]
- 3.Brown PJB, Kysela DT, Brun YV (2011) Polarity and the diversity of growth mechanisms in bacteria. Semin Cell Dev Biol 22:790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameron TA, Zupan JR, Zambryski PC (2015) The essential features and modes of bacterial polar growth. Trends Microbiol 23:347–353. [DOI] [PubMed] [Google Scholar]
- 5.Kysela DT, Brown PJB, Huang KC, Brun YV (2013) Biological consequences and advantages of asymmetric bacterial growth. Annu Rev Microbiol 67:417–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zupan JR, Cameron TA, Anderson-Furgeson J, Zambryski PC (2013) Dynamic FtsA and FtsZ localization and outer membrane alterations during polar growth and cell division in Agrobacterium tumefaciens. Proc Natl Acad Sci USA 110:9060–9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueroa-Cuilan WM, Brown PJB (2018) Cell wall biogenesis during elongation and division in the plant pathogen Agrobacterium tumefaciens. Curr Top Microbiol Immunol 418:87–110. [DOI] [PubMed] [Google Scholar]
- 8.Brown PJB, et al. (2012) Polar growth in the alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA 109:1697–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron TA, Anderson-Furgeson J, Zupan JR, Zik JJ, Zambryski PC (2014) Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. MBio 5:e01219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eswara PJ, Ramamurthi KS (2017) Bacterial cell division: Nonmodels poised to take the spotlight. Annu Rev Microbiol 71:393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuru E, et al. (2012) In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl 51:12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howell ML, et al. (2019) Agrobacterium tumefaciens divisome proteins regulate the transition from polar growth to cell division. Mol Microbiol 111:1074–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grangeon R, Zupan JR, Anderson-Furgeson J, Zambryski PC (2015) PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens. Proc Natl Acad Sci USA 112:11666–11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson-Furgeson JC, Zupan JR, Grangeon R, Zambryski PC (2016) Loss of PodJ in Agrobacterium tumefaciens leads to ectopic polar growth, branching, and reduced cell division. J Bacteriol 198:1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C (2008) A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134:956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowman GR, et al. (2008) A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viollier PH, Sternheim N, Shapiro L (2002) Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA 99:13831–13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinz AJ, Larson DE, Smith CS, Brun YV (2003) The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol 47:929–941. [DOI] [PubMed] [Google Scholar]
- 19.Grangeon R, Zupan J, Jeon Y, Zambryski PC (2017) Loss of PopZAt activity in Agrobacterium tumefaciens by deletion or depletion leads to multiple growth poles, minicells, and growth defects. MBio 8:e01881-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howell M, et al. (2017) Absence of the polar organizing protein PopZ causes aberrant cell division in Agrobacterium tumefaciens. J Bacteriol 199:e00101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehrle HM, et al. (2017) Polar organizing protein PopZ is required for chromosome segregation in Agrobacterium tumefaciens. J Bacteriol 199:e00111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam H, Schofield WB, Jacobs-Wagner C (2006) A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124:1011–1023. [DOI] [PubMed] [Google Scholar]
- 23.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH (2006) Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025–1037. [DOI] [PubMed] [Google Scholar]
- 24.The UniProt Consortium (2017) UniProt: The universal protein knowledge base. Nucleic Acids Res 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Gebali S, et al. (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson WS, Thompson TB (2007) The structure of apolipoprotein A-I in high-density lipoproteins. J Biol Chem 282:22249–22253. [DOI] [PubMed] [Google Scholar]
- 28.Huang R, et al. (2011) Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat Struct Mol Biol 18:416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng X, et al. (2012) The structure of dimeric apolipoprotein A-IV and its mechanism of self-association. Structure 20:767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker RG, et al. (2014) The structure of human apolipoprotein A-IV as revealed by stable isotope-assisted cross-linking, molecular dynamics, and small-angle X-ray scattering. J Biol Chem 289:5596–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothblat GH, Phillips MC (2010) High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr Opin Lipidol 21:229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips MC. (2013) New insights into the determination of HDL structure by apolipoproteins: Thematic review series: High density lipoprotein structure, function, and metabolism. J Lipid Res 54:2034–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, et al. (2012) The proteomics of lipid droplets: Structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res 53:1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Y, et al. (2012) Identification of the major functional proteins of prokaryotic lipid droplets. J Lipid Res 53:399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, et al. (2014) Integrated omics study delineates the dynamics of lipid droplets in Rhodococcus opacus PD630. Nucleic Acids Res 42:1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vetsigian K, Goldenfeld N (2005) Global divergence of microbial genome sequences mediated by propagating fronts. Proc Natl Acad Sci USA 102:7332–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang P, Riley M (2001) A comparative genomics approach for studying ancestral proteins and evolution. Adv Appl Microbiol 50:39–72. [DOI] [PubMed] [Google Scholar]
- 38.Barton HJ, Zeng K (2018) New methods for inferring the distribution of fitness effects for INDELs and SNPs. Mol Biol Evol 35:1536–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan SR, Gaines J, Roop RM 2nd, Farrand SK (2008) Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bisson-Filho AW, et al. (2017) Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355:739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, et al. (2017) GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355:744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Teeffelen S, et al. (2011) The bacterial actin MreB rotates, and rotation depends on cell wall assembly. Proc Natl Acad Sci USA 108:15822–15827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meniche X, et al. (2014) Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA 111:E3243–E3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mei X, Atkinson D (2011) Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high-density lipoprotein (HDL) by dimerization. J Biol Chem 286:38570–38582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mei X, Liu M, Herscovitz H, Atkinson D (2016) Probing the C-terminal domain of lipid-free apoA-I demonstrates the vital role of the H10B sequence repeat in HDL formation. J Lipid Res 57:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green MR, Sambrook J (2012) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, New York: ), 4th Ed. [Google Scholar]
- 47.Vischer NOE, et al. (2015) Cell age-dependent concentration of Escherichia coli divisome proteins analyzed with ImageJ and ObjectJ. Front Microbiol 6:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindelin J, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baron C, Llosa M, Zhou S, Zambryski PC (1997) VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1. J Bacteriol 179:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.