Significance
The molecular-level cause of sleep is unknown. According to the fragment generation (FG) hypothesis discussed in the present paper, protease-generated protein fragments can be beneficial but toxic as well, and some fragments are eliminated slowly during wakefulness. We propose specific and experimentally verifiable ideas that allow a rigorous testing of the FG hypothesis. In particular, it is suggested that a major function of sleep involves a partial and reversible expansion of postsynaptic densities (PSDs) in excitatory synapses of the brain, thereby allowing an accelerated destruction of PSD-localized protein fragments by the ubiquitin/proteasome system. This and related suggestions, described in the paper, are compatible with current evidence about sleep and lead to testable predictions.
Keywords: fragment, extracellular, intracellular, proteolysis, sleep
Abstract
The molecular-level cause of sleep is unknown. In 2012, we suggested that the cause of sleep stems from cumulative effects of numerous intracellular and extracellular protein fragments. According to the fragment generation (FG) hypothesis, protein fragments (which are continually produced through nonprocessive cleavages by intracellular, intramembrane, and extracellular proteases) can be beneficial but toxic as well, and some fragments are eliminated slowly during wakefulness. We consider the FG hypothesis and propose that, during wakefulness, the degradation of accumulating fragments is delayed within natural protein aggregates such as postsynaptic densities (PSDs) in excitatory synapses and in other dense protein meshworks, owing to an impeded diffusion of the ∼3,000-kDa 26S proteasome. We also propose that a major function of sleep involves a partial and reversible expansion of PSDs, allowing an accelerated destruction of PSD-localized fragments by the ubiquitin/proteasome system. Expansion of PSDs would alter electrochemistry of synapses, thereby contributing to a decreased neuronal firing during sleep. If so, the loss of consciousness, a feature of sleep, would be the consequence of molecular processes (expansions of protein meshworks) that are required for degradation of protein fragments. We consider the concept of FG sentinels, which signal to sleep-regulating circuits that the levels of fragments are going up. Also discussed is the possibility that protein fragments, which are known to be overproduced during an epileptic seizure, may contribute to postictal sleep and termination of seizures. These and related suggestions, described in the paper, are compatible with current evidence about sleep and lead to testable predictions.
Sleep is universal among mammals and other vertebrates. Animals with much smaller nervous systems, such as insects, also sleep (1–8). Mechanisms that control sleep include circadian circuits, which underlie daily rhythms of sleep and other biological variables (9). However, circadian aspects of sleep do not define it entirely, because sleep has the homeostatic property of becoming longer after sleep deprivation (10, 11). In mammals, birds, and lizards, two alternating modes of sleep have been identified, the non-rapid eye movement (NREM) sleep and the rapid eye movement (REM) sleep (12, 13). The depth of NREM sleep is yet another sleep variable. In adult humans, NREM sleep occupies ∼80% of the total sleep time. For recent reviews of sleep, see refs. 14–20.
Despite advances in the understanding of neuronal circuits as well as genes, proteins, short peptides, and other compounds that regulate sleep, it is largely unknown why sleep exists and what it is for. Maladaptive aspects of sleep include elevated dangers of predation during sleep, the neglect of territorial defense and foraging for food, and the loss of parental care and mating opportunities. It is unknown what features of sleep did not allow these fitness costs to cause, through natural selection, a strong shortening or elimination of sleep during evolution.
Mammals, which contain ∼109 to ∼1012 neurons, sleep several hours per day. Insects, most of which contain significantly fewer than 106 neurons, also sleep on the order of hours per day. Bouts of sleep in insects vary, usually lasting about 5 min, close to durations of NREM sleep epochs in mammals such as mice, typically from ∼5 to ∼15 min. In adult humans, NREM sleep epochs usually range from ∼70 to ∼120 min. In sum, one clue about the cause of sleep is that its total duration per day is largely unrelated to the size of a nervous system, at least within the 1,000-fold range between mammals and insects.
Reversible transitions between wakefulness and sleep are controlled by neuronal circuits that reside in specific regions of the brain (21, 22). Sleep is also regulated by the skeletal muscle, possibly through sleep-promoting (somnogenic) cytokines such as interleukin-1 (IL-1) and tumor necrosis factor (TNFα) that can be secreted by the muscle (23, 24). Other findings indicate that a behaviorally overt sleep is an emergent state that coalesces from local sleep foci that might be, initially, single neurons (25–27). It is possible, therefore, that a cause that led to the advent of sleep during evolution may reside in individual neurons and other individual cells, as distinguished from higher-order settings such as networks of cells.
In what follows, we shall make a distinction between molecular-level hypotheses about the cause/function of sleep and propositions that are neither molecular nor overtly mechanistic. General notions include suggestions that the function of sleep is to optimize utilization of the organism’s energy flux (28) and/or to purge undesirable modes of interaction in neuronal networks (29). In 1995, it was proposed that the function of sleep may be a replenishment, during NREM sleep, of cerebral glycogen stores (30). This molecular-level guess about a major function of sleep is still an unsettled proposition (31, 32). Other suggested causes/functions of sleep tend to be higher-order concepts. They include the synaptic homeostasis hypothesis (33) and extensive evidence for a role of sleep in memory and learning (34, 35).
If the assumption (it remains an assumption) that the fundamental cause of sleep resides in individual neurons and other individual cells is correct, what is a molecular process(es) that comprises the cause of sleep? Homeostatic responses of animals to sleep deprivation that result in a longer “recovery” sleep suggest that sleep evolved to counteract a set of molecular processes, a form of stress that occurs during wakefulness and impairs both neural and other organismal functions. A hypothetical stressful process takes place in individual neurons, likely in other cells as well, and may also involve extracellular spaces. The stress keeps building up during wakefulness, impairs neural functions and other processes, and eventually causes sleep. Specific (largely obscure) molecular pathways act to reverse, during sleep, a stress-mediated impairment, bringing it down to a level at which the wakefulness can resume.
The Fragment Generation Hypothesis
In 2012, we suggested that the relevant stressful process, a specific molecular cause of sleep, is the production, during wakefulness, of numerous intracellular and extracellular protein fragments that can be transiently beneficial but can also perturb, through their cumulative and mechanistically diverse effects, the functioning of the brain and other organs (Fig. 1) (36). A part of the fragment generation (FG) hypothesis is the suggestion that some natural protein fragments are removed too slowly during wakefulness, and that the resulting accumulation of fragments gradually impairs cognition and other processes.
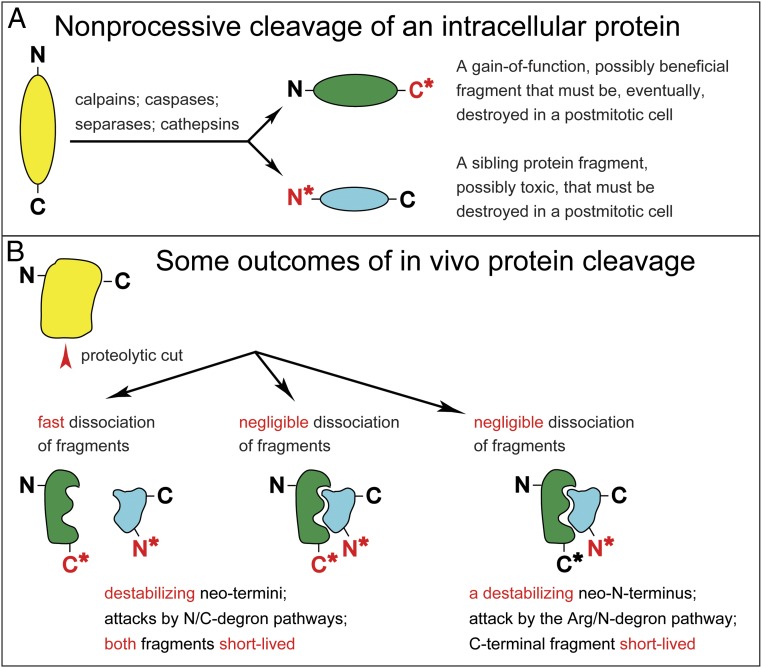
Fig. 1.
A nonprocessive cleavage of a protein and some of its outcomes. (A) A proteolytic cut generates a new N-terminus and a new C-terminus (marked by asterisks). (B) Two fragments resulting from a cut may either stay together indefinitely or dissociate, either rapidly or slowly. The rates of in vivo degradation of each fragment depend, in part, on the rate of their dissociation from each other, and also on whether the neo-N-terminus and/or the neo-C-terminus (marked by asterisks) bear destabilizing residues (marked in red), i.e., residues that can be recognized by N-degron and/or C-degron pathways (Fig. 2 and SI Appendix, Figs. S5 and S6). Depending on specific protein fragments and their Nt/Ct residues, these pathways can destroy not only dissociated fragments but also, selectively, either one or both of them even if these fragments are a part of a multisubunit protein complex. The diagram of two associated fragments on the Right illustrates the outcome (one of several possibilities) in which only the neo-N-terminus is destabilizing (denoted in red), while the neo-C-terminus (denoted in black) is not recognized by C-degron pathways.
In this 2012 concept, the production of intracellular and extracellular protein fragments, mediated by proteases, is decreased during sleep, specifically NREM sleep, while the removal or destruction of fragments is accelerated (36). It is unclear whether the FG hypothesis is also relevant to REM sleep, as REM and wakefulness appear to involve similar frequencies of Ca2+ transients. These influxes of calcium ions into the cytosol activate, in particular, calpain proteases, which generate a significant fraction of intracellular protein fragments (37).
In 2013, Nedergaard and colleagues (38) showed that some spaces between cells in the mouse brain become larger during NREM sleep, in part through decreases of astrocyte volumes. This change accelerates fluxes of interstitial fluid that transport substances from extracellular spaces in the brain to lymphatic vessels and eventually to the blood. These findings suggested that one function of sleep may be to facilitate the removal of harmful extracellular substances, including, possibly, β-amyloid (Aβ) peptides (38). In agreement with the FG hypothesis, the ∼40-residue Aβ peptides are protein fragments. They are produced by proteolytic cleavages of the amyloid precursor protein (APP), reside both extracellularly and intracellularly, and their levels in the brain increase during wakefulness and sleep deprivation (39–41).
The FG hypothesis posits accumulation, during wakefulness, of both intracellular and extracellular protein fragments (36). The latter are produced through cleavages by extracellular and membrane-embedded proteases. The removal of extracellular protein fragments from the brain may be selective inasmuch as fragments would often be soluble, in contrast to their full-length counterparts, many of which reside in insoluble or membrane-embedded extracellular meshworks.
Intracellular proteins, for example, the ones in the cytosol and the nucleus, can be cleaved primarily by two sets of nonprocessive proteases: calpains, which are activated by Ca2+ and/or phosphorylation (37), and caspases, which are activated by conditional dimerization and by cleavages of their precursors (42). Significant levels of activated caspases are present even in unstressed cells, including healthy neurons (43). A brief activation of calpains by a Ca2+ transient leads to irreversible cleavages of many intracellular proteins. Given the ubiquity of Ca2+ transients, these cleavages take place both in neurons and in other cell types (44–47). Activated calpains can, in turn, activate caspases (48, 49). Other intracellular proteases, e.g., separases, paracaspases, taspases, and cathepsins, or intramembrane proteases, e.g., secretases and rhomboid proteases, also generate intracellular protein fragments.
Cleavage sites for caspases and calpains are present in many intracellular proteins and are often conserved among at least vertebrates (SI Appendix, Figs. S1–S4). More than 1,500 mammalian proteins are substrates of caspases (42). The roughly 300 intracellular mammalian proteins that have been identified as substrates of calpains are, most likely, a small subset of the actual set of substrates, as calpain cleavage sites do not comprise a clear consensus sequence, and proteome-wide screens for calpain substrates are just beginning (37, 44–47, 50).
A part of the FG hypothesis is the concept of FG sentinels (36). They are defined, operationally, as somnogenic protein fragments that signal, to sleep-regulating neuronal circuits, that the levels of intracellular and/or extracellular protein fragments are going up.
The FG hypothesis suggests a link between fragments and epilepsy. Specifically, the known upsurges, during epileptic seizures, of extracellular and intracellular protein fragments (51–53) may directly cause the often observed postictal (postseizure) sleep.
We suggest that, during normal wakefulness, hundreds of different protein fragments may accumulate, at low fractional levels, in the brain and other sleep-relevant organs [such as the skeletal muscle (23, 24)] by the time sleep begins. The overall impact of protein fragments during wakefulness would stem from individually small but cumulative and mechanistically diverse effects of the sheer multitude of different fragments. If so, this aspect of the FG hypothesis would be analogous to the recently emerged understanding that many diseases as well as specific phenotypes (e.g., human height) are underlied by hundreds of variant genomic loci acting together, with each variant having a small effect (54, 55).
Degradation of Protein Fragments by N-Degron and C-Degron Pathways
Permanently postmitotic cells, such as, for example, mature neurons or myotubes (elongated multinucleated cells of the skeletal muscle), do not have the option of removing protein fragments through a dilution upon cell divisions. Consequently, either an ongoing or (transiently) delayed degradation of accumulating fragments is a necessity for such cells.
A cleavage of a protein generates a new N-terminus and a new C-terminus (Fig. 1). Thus, degradation signals (degrons) that are particularly relevant to protein fragments are N-degrons and C-degrons (Figs. 1 and 2 and SI Appendix, Figs. S5 and S6) (50, 56–64). The main determinant of N-degron is a destabilizing N-terminal (Nt)-residue of a protein. Other determinants of N-degron include a protein’s internal lysine (the site of polyubiquitylation). N-degron pathways (they were previously called “N-end rule pathways”) comprise proteolytic systems whose unifying feature is their ability to recognize N-degrons, thereby causing the degradation of targeted proteins by the 26S proteasome or autophagy in eukaryotes and by the ClpAP protease in bacteria (Figs. 1 and 2 and SI Appendix, Fig. S5) (57).
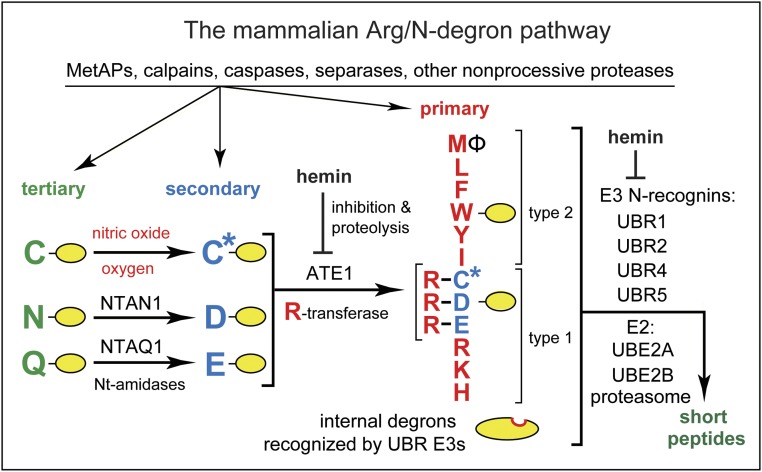
Fig. 2.
The mammalian Arg/N-degron pathway. Yellow ovals denote the rest of a protein substrate. Nt-residues are denoted by single-letter abbreviations. This pathway targets proteins for degradation through their specific unacetylated Nt-residues (50, 56–61). See SI Appendix, Fig. S5 for other N-degron pathways. “Primary,” “secondary,” and “tertiary” refer to mechanistically distinct classes of destabilizing Nt-residues. NTAN1 and NTAQ1 are Nt-amidases that convert, respectively, the tertiary destabilizing Nt-Asn and Nt-Gln to Nt-Asp and Nt-Glu. C* denotes oxidized Nt-Cys, either Cys-sulfinate or Cys-sulfonate, produced in vivo through reactions that require oxygen and nitric oxide. The ATE1 Arg-tRNA-protein transferase (R-transferase) conjugates Arg, a primary destabilizing residue, to Nt-Asp, Nt-Glu, and (oxidized) Nt-Cys. Hemin (Fe3+-heme) inhibits the enzymatic activity of R-transferase and accelerates its degradation in vivo. Hemin also binds to UBR1/UBR2 E3s and inhibits specific aspects of their activity (50, 56–61). “Type 1” and “type 2” refer, respectively, to two sets of primary destabilizing Nt-residues, basic (Arg, Lys, His) and bulky hydrophobic [Leu, Phe, Trp, Tyr, Ile, and also Met, if the latter is followed by a bulky hydrophobic residue (Ф)]. These Nt-residues are recognized by two substrate-binding sites (type 1 and type 2) of N-recognins, the pathway’s E3 Ub ligases UBR1, UBR2, UBR4, and UBR5. Besides recognizing Arg/N-degrons, these E3s contain, in addition, specific binding sites that are exposed conditionally and recognize internal (non-Nt) degrons of proteins that lack Arg/N-degrons (56–59).
Initially, most N-degrons are pro–N-degrons. They can be converted to N-degrons through a protease-mediated cleavage that exposes a destabilizing Nt-residue in the resulting C-terminal (Ct)-fragment. Proteases that include calpains, caspases, and separases have been shown to generate N-degrons in vivo through their cleavages of intracellular proteins (39, 50, 56, 57, 60). A different and mutually nonexclusive route to N-degrons is through Nt-modifications of proteins, including enzymatic Nt-acetylation, Nt-deamidation, Nt-arginylation, Nt-leucylation, and Nt-formylation of the α-amino groups of Nt-residues (Fig. 2 and SI Appendix, Fig. S5) (57, 65).
Although the identity of a destabilizing Nt-residue, in a natural Ct-fragment of a specific protein, can vary, in that Ct-fragment, from one animal species to another, the destabilizing nature of this (varying) residue is often strongly preserved (Fig. 2 and SI Appendix, Figs. S3–S5). Thus, remarkably, it is destabilizing activity of a neo-Nt-residue of a protein fragment (i.e., not the residue’s identity per se) that is under positive selection during evolution.
All 20 amino acids of the genetic code have been shown to act, in cognate sequence contexts, as destabilizing Nt-residues (Fig. 2 and SI Appendix, Fig. S5). Consequently, many cellular proteins and their natural fragments are short-lived N-degron substrates. The proteasome-mediated protein degradation by N-degron pathways is subunit selective, i.e., a targeted subunit (or its fragments) can be destroyed without damaging the rest of a protein complex (57). Conditional protein degradation by N-degron pathways has been shown to regulate many biological processes in all eukaryotes, from fungi and protists to animals and plants (56–59, 66).
C-degrons are degradation signals whose main determinant is a destabilizing Ct-motif (SI Appendix, Fig. S6) (57, 62–64). C-degrons and N-degrons are topologically analogous, can be cocreated by a single cut, and can be related functionally (Figs. 1 and 2 and SI Appendix, Figs. S5 and S6). For example, many calpain cleavage sites would yield, upon a cleavage of a protein, a putative (or confirmed) N-degron in the resulting Ct-fragment and a spatially adjacent putative C-degron in the sibling Nt-fragment (Figs. 1 and 2 and SI Appendix, Figs. S1, S3, S5, and S6). Thus, both fragments of a cleaved subunit in a protein complex can be destroyed through fragment-selective attacks by cognate N-degron and C-degron pathways, while preserving the rest of the complex (57).
In sum, the degradation of natural protein fragments, many of which, according to the FG hypothesis, are relevant to sleep physiology (36), would be carried out, to a large extent, by N-degron and C-degron pathways.
Reasons for Generating Protein Fragments, and Associated Costs
The evolutionary conservation of cleavage sites in intracellular and extracellular proteins (SI Appendix, Figs. S1–S4) suggests a positive selection for these sites. With caspases, the value of retaining, during evolution, their cleavage sites in individual proteins stems, in part, from the functions of caspases during apoptosis, a pathway of programmed cell death that sculpts both embryos and adults (42, 60). Cleavages by caspases mediate death-unrelated functions as well, including cell differentiation and long-term memory (43, 67).
As to calpains, cited below are examples of beneficial calpain-mediated cleavages. (i) Calpains regulate cell–cell interactions and intracellular cytoskeletons. For example, cortactin, an actin-binding protein, is cleaved by calpain-2 at a specific site. This (regulated) cleavage controls actin filaments, cell motility, and processes that include the inhibition of branching in axons (68, 69). (ii) In the marine snail Aplysia californica, type-C intracellular protein kinases are cleaved by calpains, yielding type-M (PKM) kinases that comprise unconditionally active catalytic domains and underlie the maintenance of memory in Aplysia (70). (iii) Pyroptosis, an immunostimulatory form of programmed cell death that can be physiologically beneficial, involves calpain-mediated cleavages of vimentin, a cytoskeletal protein (71). A pathologically high or prolonged calpain activity, which can occur in a disease, e.g., an intractable epilepsy, can be lethal for cells. However, both the first two examples above (68–70) and other known cases make clear that calpain-generated protein fragments are often not about cell death.
The FG hypothesis has emphasized a seemingly trivial but possibly significant dichotomy: A cleavage of a protein generates two fragments (36). Consequently, effects of a potentially beneficial fragment are linked to a possibly detrimental effect of the sibling fragment. This attribute of a proteolytic cleavage, i.e., a pervasive mechanistic link between a beneficial (at least in part) fragment and its often detrimental sibling fragment, might be relevant to the unexplained fact that sleep, despite its fitness costs (see Introduction), has not been strongly shortened or eliminated during evolution.
Gain-of-function features of protease-generated protein fragments may have become entrenched in early eukaryotes through being adaptive in some ways and despite being costly in other respects, given potentially detrimental sibling fragments and the necessity of destroying fragments in postmitotic cells. Early eukaryotes were, most likely, not obligatorily multicellular. If so, they were not postmitotic either and therefore could deal with less than beneficial fragments and other such proteins not only through degradation but also through cell divisions, either symmetric (dilution of fragments) or asymmetric ones, which could segregate some proteins (including aggregates) into one of two daughter cells. If so, an emergence, in early single-cell eukaryotes, of diverse proteases that generated protein fragments (some of which were beneficial) may have incurred relatively low fitness costs. As a result, molecular circuits based on partly beneficial protein fragments may have become entrenched. This disposition, which is difficult to reverse, would become a problem later, when some eukaryotes evolved to be obligatorily multicellular and developed a division of labor among cells of different types, including terminally differentiated, postmitotic cells.
In this FG-based scenario, the advent of sleep during evolution may have been caused, at least in part, by the emergence of postmitotic cells, such as neurons and myotubes, in multicellular eukaryotes. If so, some functions of modern sleep as well as complexities of its regulation may be later additions to simpler versions of sleep that coevolved with nonprocessive proteases that generated protein fragments and with processive proteasome-mediated pathways that recognized and destroyed these fragments.
On a Possible Delay in Degradation of Fragments During Wakefulness
The FG hypothesis posits that sleep would be up-regulated by processes that increase the levels of protein fragments (36). Epileptic seizures are known to up-regulate fragments (51–53) and sleep deprivation is predicted to do so (36). These processes involve excitatory (glutamatergic) synapses, which contain postsynaptic densities (PSDs). A PSD is a disk-shaped aggregate (meshwork) of roughly 500 different proteins. PSDs are located underneath synapse-encompassing patches of the plasma membrane in dendritic spines of postsynaptic neurons. PSDs are rich in scaffold/cytoskeletal proteins (72–81). Specific scaffold proteins bind to cytosolic domains of glutamate receptors such as AMPAR, NMDAR, and mGluR1, transiently trapping these and other transmembrane proteins within a PSD (80). PSDs form, grow, adopt a range of shapes, and disappear in ways that depend on fates of dendritic spines in which PSDs reside (81).
PSDs contain both calpains and caspases (37, 82–84). Once activated, these proteases can cleave cytoskeletal proteins as well as cytosolic domains of PSD-localized transmembrane receptors. Owing to their location, immediately beneath the plasma membrane and within regions of Ca2+ transients, PSDs encounter highest local Ca2+ concentrations in postsynaptic neurons and are, therefore, major sites at which Ca2+-activated calpains generate protein fragments.
Studies of oligomeric proteins described a number of cases in which a cleaved protein subunit stays embedded (as one or both of its fragments) within a complex. Some oligomeric proteins continue to be active, at least for a while, even after cleavages of their essential subunits, which remain embedded in a complex as fragments (61, 85–87).
We suggest that a delay in destroying a fragment that is formed and retained in a protein complex within a PSD may become particularly pronounced owing to a difficulty that is unique to dense protein meshworks: In addition to an often slow or negligible dissociation of subunit’s fragments from a (cleaved) oligomeric complex, these fragments may also be shielded from the bulky polyubiquitylation–degradation machinery, inasmuch as both fragments and protein complexes that contain them would reside in diffusion-impeding meshworks such as PSDs (Fig. 3).
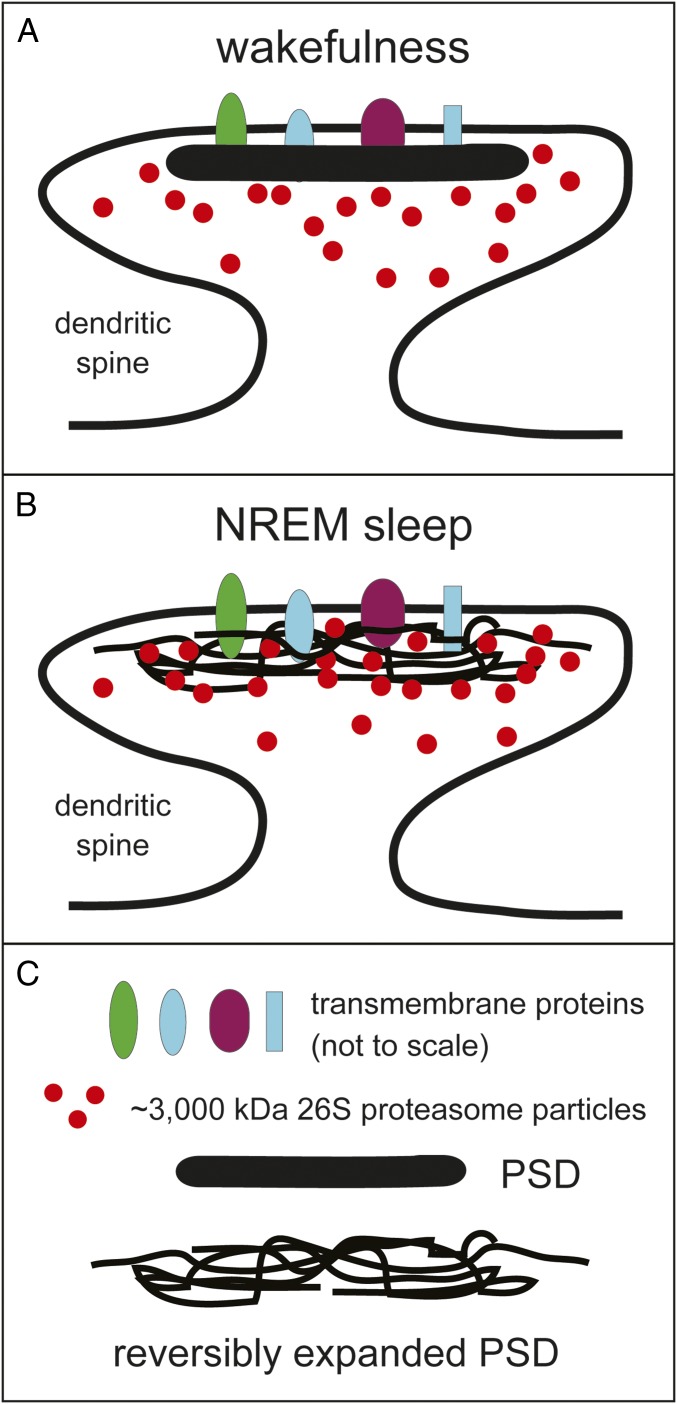
Fig. 3.
On the possibility of a reversible expansion of natural dense meshworks during sleep. A partial and reversible expansion of postsynaptic densities (PSDs) and analogous protein aggregates is the hypothetical reason for a proposed delay in degradation of PSD-localized protein fragments during wakefulness. (A) Schematic of a dendritic spine that contains a PSD (solid black shape) during wakefulness. (B) Same as in A but during NREM sleep, with an expansion of PSD indicated by interweaved black curves, and with 26S proteasome particles (red circles) having access to the interior of PSD. (C) Notations. Transmembrane proteins, denoted by colored ovals and rectangles, are depicted not to scale (all of these proteins are smaller than the 26S proteasome). Red circles denote the ∼3,000-kDa 26S proteasome particles, located, according to this model, largely outside PSDs during wakefulness but partly within PSDs during NREM sleep. See the main text for details.
PSDs are not the only natural aggregates in which a diffusion of large complexes (particularly the ∼3,000-kDa 26S proteasome) may be slow or negligible. For example, analogous meshworks of cytoskeletal and transmembrane proteins are also present on the postsynaptic sides of inhibitory synapses. Such synapses usually do not involve dendritic spines but form directly on dendrites (88).
Gephyrin (GPHN), a postsynaptic scaffold protein in inhibitory synapses, can be cleaved, at a specific site, by activated calpains (88). As to PSDs of excitatory synapses, they contain many calpain substrates, including PSD95, PSD93, PSD97, GRIP1, spectrin (SPTBN1), ezrin (EZR), stargazin (CACGN2), p35 (CDK5R1), and cytosolic domains of PSD-entrapped transmembrane proteins such as NMDAR, AMPAR, mGluR1, and N-cadherin (37, 73, 74, 77). For most of these proteins, specific physiological functions of cleavages by calpains are still unknown. The PSD-localized calpain substrates cited above are but a subset of a future comprehensive list.
Many calpain/caspase-mediated cleavages of PSD proteins produce N-degrons and/or C-degrons in newly generated protein fragments (Figs. 1 and 2 and SI Appendix, Figs. S1–S6) (50, 60). Therefore, a delay in degradation of protein fragments within, e.g., a PSD would be caused by the conjectured steric inaccessibility of at least some PSD-localized protein fragments, rather than by a scarcity of degrons in these fragments (Fig. 2 and SI Appendix, Figs. S1 and S2). In contrast, “soluble” degron-bearing intracellular fragments, produced through cleavages of proteins outside of meshworks, would be efficaciously destroyed, during either wakefulness or sleep, by the ubiquitin (Ub)–proteasome system (UPS) that includes N-degron and C-degron pathways.
The proposed impediment (steric hindrance) to the destruction of fragments that are generated within PSDs and other dense meshworks may stem from electrochemical constraints on the design of postsynaptic PSDs. These meshworks may have to be sufficiently compact (dense) for allowing neurons to fire frequently enough during wakefulness. If so, the necessity of dense PSDs during wakefulness, and the cost of that necessity—an impairment of protein degradation within PSDs—may be a fundamental molecular-level reason for the existence of NREM sleep. A transition from wakefulness to a different regimen, referred to as sleep, may deal with the problem of steric hindrance through a process proposed below.
Might PSDs Reversibly Expand During NREM Sleep?
During wakefulness, the meshwork of a PSD is suggested to be dense enough to be a significant obstacle to entry and diffusion of large protein complexes that include, particularly, the ∼3,000-kDa 26S proteasome (Fig. 3). What follows is a specific hypothesis, and its ramifications, about sleep–wake transitions in PSDs and analogous protein meshworks.
1) It is proposed that the onset of NREM sleep involves, among other things, a reversible physical expansion (loosening) of the PSD meshwork (Fig. 3). The envisioned expansion may be caused by any number of specific protein modifications by PSD-localized enzymes that carry out, for example, phosphorylation–dephosphorylation and/or acetylation–deacetylation of specific PSD proteins. As a result, some (but not all) interprotein distances in PSDs would become larger, specifically large enough to allow a relatively unimpeded diffusion of the ∼3,000-kDa 26S proteasome and Ub ligase complexes within an expanded PSD (Fig. 3). This alteration would accelerate degradation of fragments that had been generated during wakefulness within PSDs and analogous meshworks. The nature of expansion-regulating enzymatic modifications of PSD proteins remains to be determined. It has been shown, for example, that phosphorylation of PSD95 and other PSD proteins can influence their intermolecular and intramolecular interactions (89).
In sum, it is proposed that protein fragments are generated within PSDs and related natural meshworks during wakefulness (owing to Ca2+ transients and activation of at least calpains), and that PSDs, in that state, are too dense for an entry and diffusion of the ∼3,000-kDa 26S proteasome. A reversible PSD expansion, by allowing a relatively unimpeded diffusion of the 26S proteasome within PSDs, makes possible the destruction of PSD-localized fragments that formed during wakefulness. By down-regulating excitability of postsynaptic neurons, expansions of PSDs would lead to an overall decrease of excitatory neuronal activity, and thereby would shift the system to a different regimen, sleep. In this model, at least NREM sleep is the “price” of being able to efficaciously destroy PSD-localized protein fragments that accumulated during wakefulness (Fig. 3).
It should be noted that a compact PSD, during wakefulness, is not necessarily expected to lack 26S proteasome particles altogether. The key idea is that the 26S proteasome can neither efficaciously diffuse within a compact PSD nor readily enter it.
2) The notion of a reversible PSD expansion during NREM sleep (Fig. 3) presumes that transmembrane receptors, which are dynamically (transiently) trapped, within PSDs, by scaffold proteins would continue to be largely retained in expanded PSDs as well. Thus, the envisioned expansion of PSDs would be not only moderate but also selective in regard to specific contacts, as the proposed expansion would preserve interactions of scaffold proteins with cytosolic domains of PSD-localized transmembrane proteins.
3) Firing, by a postsynaptic neuron, of an action potential results from multiple inputs, including Ca2+ transients at neuron’s dendrites. The known decrease of the frequency of Ca2+ transients and action potentials during NREM sleep (21) would be caused, at least in part, by the envisioned physical expansion of PSDs during sleep.
4) If a decrease of firing by neurons during NREM sleep would be found to be actually caused, in part, by a physical expansion of PSDs, such an understanding would also explain why PSDs cannot stay expanded all of the time, inasmuch as the mean frequency of firing must be higher during wakefulness. In sum, a lower overall activity of neurons during NREM sleep is suggested to be caused, in part, by an expansion of PSDs and analogous protein meshworks.
5) It is unknown why sleep is accompanied by the loss of consciousness. A quiet wakefulness would seemingly suffice, but this is not what actually happens. Consciousness requires activity of many neuronal circuits, particularly of the cortico-thalamic system (90). If the proposed expansions of PSDs and analogous meshworks (Fig. 3) actually take place during NREM sleep, the necessity of PSD expansion for destroying accumulated protein fragments and the (presumed) incompatibility of expanded PSDs with frequent firing by neurons would suffice to account for the loss of consciousness during at least NREM sleep. In this model, the loss of consciousness is caused by expansion of PSD meshworks and is an outcome of molecular constraints (a delayed destruction of protein fragments) that underlie the fundamental cause/function of sleep. We are not aware of other suggestions about a specific molecular-level cause of losing consciousness during sleep. A nonmechanistic explanation, proposed a century ago, is that the loss of consciousness prevents new experiences and thereby assists memory consolidation during sleep (91).
6) One prediction of the PSD-expansion model is that postsynaptic neurons would contain, during wakefulness, the 26S proteasome in dendritic spines, i.e., spatially close to PSDs, but largely not within PSDs (Fig. 3). Current evidence is not inconsistent with this prediction. The autophosphorylated Ca2+/calmodulin-dependent protein kinase IIα (CamKIIα), a component of spines, has been shown to bind to the 26S proteasome (92). Up-regulation of neuronal activity causes the movement of CamKIIα from dendritic shafts to nearby spines, in which CamKIIα acts as a “sink” for the 26S proteasome, increasing its entry into spines (92). These results were obtained using light microscopy, which does not distinguish between locations of specific proteins within a PSD vs. the rest of a spine. An electron microscopy study suggested that the movement of CamKIIα into spines concentrates this kinase (and also, by inference, the associated 26S proteasome particles) immediately beneath the dense PSD meshwork (79). In sum, it remains to be determined exactly where the 26S proteasome resides within a spine, and whether the distribution of proteasome particles changes during wakefulness vs. NREM sleep.
7) Varying in size from ∼100 to ∼1,000 kDa, Ub ligase complexes are smaller than the ∼3,000-kDa 26S proteasome. Therefore, a variant of the PSD-expansion model is that at least some Ub ligases, as well as ∼100-kDa Ub-activating enzymes (let alone the 9-kDa Ub) should be able to diffuse within compact PSDs and analogous protein meshworks during wakefulness, in contrast to the 26S proteasome. If so, these Ub ligases could target and polyubiquitylate their PSD-localized substrates, including protein fragments, even during wakefulness. In contrast, the 26S proteasome is envisioned to gain an efficacious access to the interior of PSDs either largely or solely during NREM sleep (Fig. 3).
Interestingly, behavioral tests that retrieved a specific long-term memory in mice were found to increase the in vivo polyubiquitylation of hippocampal proteins (93). Polyubiquitylated proteins are recognized and destroyed by the 26S proteasome. Increased in vivo levels of polyubiquitylated hippocampal proteins suggest that their proteasome-mediated degradation was the rate-limiting step under those conditions. (If polyubiquitylation, rather than degradation, were the rate-limiting step, polyubiquitylated proteins would not be expected to accumulate.) The authors (93) did not consider this interpretation, but their data are consistent with the possibility that proteins in their preparations (a significant fraction of those proteins resided in PSDs) were polyubiquitylated in vivo but not degraded, as yet, under conditions of memory retrieval experiments with awake mice.
8) Regulation of sleep involves neurons that fire frequently during wakefulness but are largely silent during, e.g., NREM sleep. Conversely, neurons that maintain NREM sleep are active during NREM but relatively silent during wakefulness (15). The proposed PSD-expansion model may also be relevant to neurons that maintain NREM sleep, except that this circuit would operate “in reverse”: Neurons that fire frequently during NREM sleep but tend not to fire during wakefulness may accumulate protein fragments during NREM sleep and destroy them later, during wakefulness.
9) A particularly deep NREM sleep, called a “slow-wave” sleep and defined, in part, by its electroencephalographic features, is up-regulated in cortical regions of the brain that have been recently hyperactive (35). Molecular/mechanistic aspects of a slow-wave NREM sleep are largely unknown. Given the PSD-expansion model (Fig. 3), one possibility is that a deeper NREM sleep may involve a stronger PSD expansion. This would lead to a less impeded diffusion of the 26S proteasome within PSDs and, consequently, to a faster destruction of PSD-localized fragments and other proteins. According to the FG hypothesis (36), protein fragments would be overproduced in a hyperactive region of the brain, in comparison with a less exercised region. If so, a PSD-expansion model in which a greater depth of NREM sleep signifies a stronger PSD expansion (and, consequently, a faster degradation of fragments; Fig. 3) may account for the observed connection between hyperactivity of a cortical region and the depth of its subsequent NREM sleep.
10) Durations of NREM sleep epochs vary widely during sleep and vary even more among different species. For example, human NREM epochs, during a single night, range from ∼70 to ∼120 min, and alternate with (typically) shorter REM sleep epochs. In contrast, while the total durations of mouse and human sleep are comparable (several hours per day), a mouse sleeps in much shorter NREM epochs, usually between ∼5 and ∼15 min (1).
What is the benefit of a NREM epoch as brief as 5 min? Enzymatic modifications that mediate the proposed expansion of PSDs upon a NREM sleep (Fig. 3) would be expected to occur rapidly, on the scale of seconds. The ensuing proteasome-mediated processive destruction of protein fragments within an expanded PSD would be irreversible. Consequently, even a brief NREM epoch would attain, by its end, a ratchet-like “improvement,” i.e., a decrease in the load of protein fragments. If so, specific durations of NREM epochs would be relatively unimportant (in contrast to the total duration of NREM sleep), in agreement with the observed scatter of NREM sleep epochs.
11) The FG hypothesis is relevant to intracellular and extracellular settings (including ones outside the brain) that involve accumulation of protease-generated protein fragments whose destruction entails a significant delay. At least for intracellular proteins, this delay would happen either because one or both fragments resulting from a cut would still reside in a cleaved but otherwise intact protein complex, or because the ∼3,000-kDa 26S proteasome cannot get access to fragments (owing to their location within a dense meshwork), or for both of these (very different) reasons together. One example of meshworks outside the brain are natural cytoskeletal aggregates that underlie the architecture of skeletal muscle and its myotubes (94). Consequently, a delayed destruction of protein fragments (accumulated during wakefulness) within protein meshworks of muscle cells may be relevant to the known role of skeletal muscle in sleep regulation (24) and may also play a role in muscle fatigue.
Verification of the PSD expansion/compaction model (Fig. 3) and other aspects of the FG hypothesis is a tractable challenge, since all proposed conjectures are concrete enough to be falsifiable. If the FG hypothesis proves correct at least in outline, it would imply that the dynamics of intracellular and extracellular protein fragments, in the brain and in other organs, may underlie not only molecular-level roles of sleep but its higher-order functions as well, including the extensively documented role of sleep in learning and memory (34, 35). Several lines of evidence indicate that memory and learning involve natural protein fragments, both in mammals and in the Aplysia marine snail (67, 70). Protein fragments are also likely to play a role in the complex dynamics of dendritic spines (33, 95).
Up-Regulation of Protein Fragments in Epilepsy and Their Possible Links to Sleep
For most extracellular or intracellular proteins, the percentages of their cleavages during normal wakefulness are envisioned, in the FG hypothesis, to be low (<10%, and often <<10%). In addition, not all fragments are expected to accumulate during wakefulness, as some of them would be destroyed efficaciously.
Generation of fragments can be accelerated in a disease, for example, during epileptic seizures, which involve higher frequencies of Ca2+ transients and therefore an increased activation of at least calpain proteases. Seizures have been shown to up-regulate cleavages of intracellular and extracellular proteins strongly enough to make specific protein fragments in the brain readily detectable by immunoblotting (51–53).
Descriptions of human and rodent epilepsy mention a frequent occurrence of sleep, either immediately or soon after a seizure (96–98). In agreement with this evidence, our own surveys of Internet chat rooms that facilitate correspondence among epilepsy patients indicated that postictal sleep is a recurrent subject of their discussions. “I slept it off” and other remarks to that effect are frequent descriptions of postictal experiences that one encounters at these websites.
Electroconvulsive therapy (ECT), a version of induced seizure, is still used to treat an otherwise intractable depression. ECT is also associated with an enhanced sleep of a patient soon after treatment, as remarked upon, repeatedly, by ECT patients and their physicians (99). For example: “People often found they were very sleepy immediately after their ECT treatment and wanted to go to bed.” “Her daughter would sleep for up to twelve to fourteen hours afterwards” (after ECT treatments) (www.healthtalk.org/peoples-experiences/mental-health/electroconvulsive-treatment/side-effects-having-ect).
Two more descriptions of a postictal sleep: “If the convulsion became more pronounced his orderlies quickly brought someone to him whose presence he found relaxing. … ‘Peter Alexeevich, here is the person to whom you wished to speak,’ his worried orderly would say and then withdraw. The Tsar would lie down and place his shaking head on the woman’s lap and she would stroke his forehead and temples, speaking to him softly and reassuringly. Peter would fall asleep, and when he awoke an hour or two later, he was always refreshed and in far better humor than he had been before” (100).
“A boy had an epileptic seizure. I’d never seen one. I knew something was going on behind me but didn’t turn around to look until the boy was asleep on the floor. He was snoring and his mother stood over his body while his sister ran to use the pay phone” (101).
The FG hypothesis predicts postictal sleep, given the demonstrated upsurges of protein fragments in the brain that are caused by seizures (51–53). For the same reason, a prolonged but reversible comatose state, upon a relatively mild physical brain trauma, may also be caused, in part, by protein fragments that are produced by up-regulated Ca2+ transients (which activate calpains) in injured cells, and by upswings of extracellular proteases. A reversible coma of this kind, while not nearly as reversible as a NREM sleep, may be mechanistically analogous to sleep through their common attribute of protein fragments, with much higher levels of fragments in the case of a temporary coma.
Most seizures in humans are spatially localized and self-terminating. (Seizures lasting significantly longer than 10 min are often fatal.) How a seizure manages to stop is not well understood. Processes that would counteract a seizure include a Na+-dependent K+ efflux that hyperpolarizes the plasma membrane and thereby reduces its excitability; a hyperpolarizing influx of chloride ions, via γ-aminobutyric acid (GABA) ionotropic receptors; a decrease of excitatory neurotransmitter glutamate in hyperactive presynaptic neurons; and up-regulation of neuromodulators, such as adenosine and endocannabinoids (102, 103).
Protein fragments that are overproduced during a seizure may cause not only a postictal sleep but may also act to reduce excitability of neurons during a seizure. The verifiable possibilities, below, are neutral in regard to identities of fragments, which remain to be identified. (i) An extracellular fragment that is relevant to seizure termination would be overproduced locally, via a seizure-enhanced proteolytic cleavage of an extracellular (or partly extracellular) full-length protein. Such a fragment would act through its binding, largely within the region of seizure, to a cognate neuronal surface-exposed protein, e.g., an ion channel. The fragment would act as an antagonist of neuronal excitability. (A full-length precursor of this fragment would be inactive as an antagonist.) (ii) A mutually nonexclusive possibility involves upsurges of Ca2+ transients and increases of seizure-suppressing intracellular protein fragments (51–53).
Hypersomnolence of Patients with Calpain-Activating Wolfram Syndrome
The FG hypothesis predicts that sleep would be enhanced by an up-regulation of calpain activity (36). Wolfram syndrome involves early-onset diabetes and neurodegeneration, including optic nerve atrophy (104). This recessive birth defect is caused by mutations in WFS1 or WFS2, which encode proteins embedded in the endoplasmic reticulum membrane (105). One function of WFS1 and WFS2 is to down-regulate the activity of calpain-2. Cells of Wolfram syndrome patients and other human or mouse cells with decreased (or absent) WFS1 or WFS2 exhibit abnormally high calpain activity and increased levels of calpain-generated fragments of (at least) spectrin, a natural substrate of calpains (105).
In agreement with the FG hypothesis, a significant but underexplored feature of the Wolfram syndrome is hypersomnolence, an excessive daytime sleepiness often accompanied by a prolonged nighttime sleep (104). It would be, therefore, informative to investigate the hypersomnolence of Wolfram syndrome patients and to analyze sleep in WFS1/WFS2-lacking mice (105) vis-à-vis protein fragments in these settings.
Other human sleep disorders that involve hypersomnolence [but no narcolepsy (106)] occur in 1 out of ∼10,000 births and are referred to as idiopathic (cause unknown) conditions (https://www.orpha.net/consor/cgi-bin/OC_Exp.php?lng=en&Expert=33208). Both the FG hypothesis and hypersomnolence of Wolfram syndrome patients suggest a screening approach in which cells from patients with idiopathic hypersomnolence disorders would be examined for an up-regulation of calpain activity and increased generation of, e.g., spectrin fragments. This can be done using white blood cells as well as primary fibroblasts from skin biopsies.
FG Sentinels
An FG sentinel is defined as a protein that signals, to sleep-regulating circuits, that the levels of some protein fragments, the ones that the sentinel reports about, are going up (36). This would couple the levels of fragments to outputs of sleep-regulating circuits in the brain.
A precursor of an FG-sentinel that reports, for example, about levels of calpain-generated protein fragments would be a protein that is inactive as a sentinel and contains one or more calpain cleavage sites. A cleavage of this protein by a calpain would confer, on at least one of resulting fragments, the ability to act as an FG sentinel. Analogous designs can underlie a sentinel specific for caspases or a sentinel for, e.g., extracellular metalloproteases. Such sentinels would react not to fragments themselves but to an up-regulation of a protease(s) that generates specific classes of fragments. The concept of FG sentinels, which remains to be verified, does not place a priori constraints on mechanisms through which an FG sentinel would actually function. Cited below are a few (out of many) plausible but far from certain candidates for FG sentinels.
The Panx1 Pannexin Channel.
ATP can be released from cells by vesicular exocytosis and also through transmembrane channels such as Panx1, which can be activated by effectors that include Ca2+ transients (107). Extracellular ATP can be converted, by plasma membrane-embedded nucleotidases, to extracellular adenosine, a natural somnogen (18). Extracellular ATP can also increase proteolytic processing and secretion of inflammatory cytokines, including IL-1 and TNFα. They, too, can act as somnogens (23). It has been shown that a caspase-mediated cleavage of Panx1 (at a cleavage site conserved during vertebrate evolution) activates Panx1 as an ATP release channel (108). Thus, a caspase-generated Nt-fragment of Panx1 is a potential FG sentinel that can up-regulate extracellular ATP and thereby signal, through both adenosine increases and upsurges of inflammatory cytokines, about the levels of fragment-producing caspase activities in a cell. (There is no evidence, so far, about whether or not Panx1 might also be cleavable by calpains.)
Calcineurin.
Calcineurin (CaN) is a ubiquitously expressed Ca2+/calmodulin-dependent Ser/Thr phosphatase. In Drosophila, both strong increases and strong decreases of CaN activity have been shown to inhibit sleep (109, 110). CaN is a heterodimer of the catalytic A-subunit and regulatory B-subunit. In the absence of Ca2+, the A-subunit of CaN is autoinhibited by a domain of the same subunit. The A-subunit can be cleaved by a calpain or caspase at two different but close by sites. Remarkably, either one of those cleavages would separate the autoinhibitory and catalytic domains of the A-subunit, thereby converting a regulated CaN phosphatase into an unconditionally active one (111, 112). Since a moderate increase of the phosphatase activity of CaN is somnogenic (109), a physiologically relevant activation of CaN through a cleavage of some of its molecules by a calpain or caspase may underlie the function of CaN as an FG sentinel, a verifiable proposition.
Aβ Peptides.
The ∼40-residue Aβ peptides are produced by proteolytic cleavages of the APP protein. Short-term physiological effects of Aβ peptides are many and include, for example, a suppression of specific aspects of cannabinoid receptor activity (113). Extracellular/intracellular levels of Aβ peptides in the brain increase during both wakefulness and sleep deprivation (40, 41). Might Aβ peptides function as FG sentinels? Available evidence neither supports nor contradicts this idea. Other potential FG sentinels can be cited as well, but these examples suffice to define the concept.
Concluding Remarks
This paper considers the 2012 FG hypothesis about the fundamental cause of sleep (36), making FG concepts more detailed through several ideas.
One of them is the possibility of an impeded diffusional access, of the UPS proteolytic machinery (particularly the ∼3,000-kDa 26S proteasome), to dense protein meshworks such as PSDs of excitatory synapses and other natural protein aggregates during wakefulness (Fig. 3). An impeded diffusion of the 26S proteasome in these aggregates is proposed to be the reason for a (presumed) slow or negligible destruction of protein fragments that are produced during wakefulness within PSDs and analogous meshworks.
Fragments that form within dense natural aggregates during wakefulness and are relevant to sleep may be present not only in the brain but also in quasipermanent cytoskeletal meshworks that underlie the architectures of other organs, such as the skeletal muscle and its myotubes. If so, a slow or negligible degradation of protein fragments within cytoskeletal aggregates of the muscle during wakefulness might be relevant to the demonstrated involvement of muscle in the regulation of sleep (24).
Protein fragments that accumulate during wakefulness are proposed to be destroyed faster during NREM sleep, owing to a (hypothetical) enzymatically controlled and reversible expansion of PSDs and analogous natural meshworks (Fig. 3). The envisioned PSD expansion during NREM sleep would be moderate but significant enough to allow access, within PSDs, by particles such as the ∼3,000-kDa 26S proteasome.
Extracellular cytoskeletal protein meshworks, including, possibly, perineural nets, may also undergo reversible cycles of expansion (during NREM sleep) and contraction (during wakefulness). An expansion would facilitate elimination of extracellular protein fragments that accumulate during wakefulness.
FG sentinels would convey to sleep-regulating circuits that the levels of protein fragments are going up. Potential FG sentinels include fragments of cytoskeletal proteins, transmembrane channels, kinases, and phosphatases. Somnogenic cytokines such as TNFα and IL-1 (23) might also function as FG sentinels, if the proteolytic processing and secretion of these cytokines by, e.g., the skeletal muscle during wakefulness could be shown to be increased in response to higher levels of protein fragments in the muscle.
It is also proposed that the known overproduction of extracellular and intracellular protein fragments during epileptic seizures may be a cause of postictal sleep and may also contribute to seizure termination.
The notion that a sufficiently dense meshwork would impede, with physiological consequences, a diffusion of large particles within the meshwork may be relevant not only to PSDs. Analogous settings may include the sarcomeres of skeletal and cardiac muscles, and also nucleosomal fibers of chromatin, some of which are associated with the 26S proteasome and other UPS components (114). Sleep up-regulates chromosome dynamics (a measure of three-dimensional mobility of chromatin domains in the nucleus) and facilitates repair of double-strand breaks in DNA (115, 116). At least the latter process involves the 26S proteasome (117). One possibility is that a decreased or halted diffusion of the 26S proteasome within (reversibly) compacted chromosomal regions may be a part of circuits that regulate the proteasome-mediated destruction of histone fragments and other proteins in the vicinity of DNA. [Subsets of nucleosomal histones are cleaved by nonprocessive proteases during cell differentiation, stresses, and other transitions (118, 119).]
The set of testable conjectures that comprise the FG hypothesis is compatible, to our knowledge, with data about functions of sleep in the regulation of dendritic spines, memory, and the immune system, given the already existing evidence that these structures and processes involve protein fragments. Extracellular and intracellular fragments, possibly hundreds of them, produced at low but physiologically significant levels, are conjectured to be a problem for systems that comprise fragment-generating proteases and fragment-destroying proteasomal pathways (36). The emergence of sleep as a solution of this problem, and proposed mechanistic features of that solution might account for the inability of natural selection to strongly shorten or eliminate sleep during evolution, despite fitness costs of sleep. Specific properties of extant sleep, including its different kinds, its intricate regulation, and its roles in memory, immunity, and other aspects of modern organisms may be adaptations that emerged since the early setting of a simpler primordial sleep. In sum, the FG hypothesis suggests a verifiable, molecular, and possibly chief reason because of which sleep evolved in the first place.
Supplementary Material
Acknowledgments
I am most grateful to D. Tsao, W. Tansey, D. Anderson, U. Hartl, A. Hershko, J. Raskatov, and O. Sundin for their comments on the manuscript. Studies in the author’s laboratory are supported by the NIH Grants DK039520 and GM031530.
Footnotes
The author declares no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904709116/-/DCSupplemental.
References
- 1.Siegel JM. (2005) Clues to the functions of mammalian sleep. Nature 437:1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oikonomou G, Prober DA (2017) Attacking sleep from a new angle: Contributions from zebrafish. Curr Opin Neurobiol 44:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nath RD, et al. (2017) The jellyfish Cassiopea exhibits a sleep-like state. Curr Biol 27:2984–2990.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joiner WJ. (2016) Unraveling the evolutionary determinants of sleep. Curr Biol 26:R1073–R1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anafi RC, Kayser MS, Raizen DM (2019) Exploring phylogeny to find the function of sleep. Nat Rev Neurosci 20:109–116. [DOI] [PubMed] [Google Scholar]
- 6.Geissmann Q, Beckwith EJ, Gilestro GF (2019) Most sleep does not serve a vital function: Evidence from Drosophila melanogaster. Sci Adv 5:eaau9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honda T, et al. (2018) A single phosphorylation site of SIK3 regulates daily sleep amounts and sleep need in mice. Proc Natl Acad Sci USA 115:10458–10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kempf A, Song SM, Talbot CB, Miesenböck G (2019) A potassium channel β-subunit couples mitochondrial electron transport to sleep. Nature 568:230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum ID, Bell B, Wu MN (2018) Time for bed: Genetic mechanisms mediating the circadian regulation of sleep. Trends Genet 34:379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pimentel D, et al. (2016) Operation of a homeostatic sleep switch. Nature 536:333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allada R, Cirelli C, Sehgal A (2017) Molecular mechanisms of sleep homeostasis in flies and mammals. Cold Spring Harb Perspect Biol 9:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shein-Idelson M, Ondracek JM, Liaw HP, Reiter S, Laurent G (2016) Slow waves, sharp waves, ripples, and REM in sleeping dragons. Science 352:590–595. [DOI] [PubMed] [Google Scholar]
- 13.Peever J, Fuller PM (2017) The biology of REM sleep. Curr Biol 27:R1237–R1248. [DOI] [PubMed] [Google Scholar]
- 14.Artiushin G, Sehgal A (2017) The Drosophila circuitry of sleep-wake regulation. Curr Opin Neurobiol 44:243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bringmann H. (2018) Sleep-active neurons: Conserved motors of sleep. Genetics 208:1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiNuzzo M, Nedergaard M (2017) Brain energetics during the sleep-wake cycle. Curr Opin Neurobiol 47:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirelli C. (2017) Sleep, synaptic homeostasis and neuronal firing rates. Curr Opin Neurobiol 44:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wigren HK, Porkka-Heiskanen T (2018) Novel concepts in sleep regulation. Acta Physiol (Oxf) 222:e13017. [DOI] [PubMed] [Google Scholar]
- 19.Rattenborg NC, Martinez-Gonzalez D (2015) Avian versus mammalian sleep: The fruits of comparing apples and organges. Curr Sleep Med Rep 1:55–63. [Google Scholar]
- 20.Krueger JM, Frank MG, Wisor JP, Roy S (2016) Sleep function: Toward elucidating an enigma. Sleep Med Rev 28:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scammell TE, Arrigoni E, Lipton JO (2017) Neural circuitry of wakefulness and sleep. Neuron 93:747–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber F, Dan Y (2016) Circuit-based interrogation of sleep control. Nature 538:51–59. [DOI] [PubMed] [Google Scholar]
- 23.Rockstrom MD, et al. (2018) Tumor necrosis factor alpha in sleep regulation. Sleep Med Rev 40:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehlen JC, et al. (2017) Bmal1 function in skeletal muscle regulates sleep. eLife 6:e26557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siclari F, Tononi G (2017) Local aspects of sleep and wakefulness. Curr Opin Neurobiol 44:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vyazovskiy VV, et al. (2011) Local sleep in awake rats. Nature 472:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krueger JM, Nguyen JT, Dykstra-Aiello CJ, Taishi P (2019) Local sleep. Sleep Med Rev 43:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt MH. (2014) The energy allocation function of sleep: A unifying theory of sleep, torpor, and continuous wakefulness. Neurosci Biobehav Rev 47:122–153. [DOI] [PubMed] [Google Scholar]
- 29.Crick F, Mitchison G (1983) The function of dream sleep. Nature 304:111–114. [DOI] [PubMed] [Google Scholar]
- 30.Benington JH, Heller HC (1995) Restoration of brain energy metabolism as the function of sleep. Prog Neurobiol 45:347–360. [DOI] [PubMed] [Google Scholar]
- 31.Petit JM, Burlet-Godinot S, Magistretti PJ, Allaman I (2015) Glycogen metabolism and the homeostatic regulation of sleep. Metab Brain Dis 30:263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bellesi M, de Vivo L, Koebe S, Tononi G, Cirelli C (2018) Sleep and wake affect glycogen content and turnover at perisynaptic astrocytic processes. Front Cell Neurosci 12:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tononi G, Cirelli C (2014) Sleep and the price of plasticity: From synaptic and cellular homeostasis to memory consolidation and integration. Neuron 81:12–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasch B, Born J (2013) About sleep’s role in memory. Physiol Rev 93:681–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donlea JM. (2019) Roles for sleep in memory: Insights from the fly. Curr Opin Neurobiol 54:120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varshavsky A. (2012) Augmented generation of protein fragments during wakefulness as the molecular cause of sleep: A hypothesis. Protein Sci 21:1634–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briz V, Baudry M (2017) Calpains: Master regulators of synaptic plasticity. Neuroscientist 23:221–231. [DOI] [PubMed] [Google Scholar]
- 38.Xie L, et al. (2013) Sleep drives metabolite clearance from the adult brain. Science 342:373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brower CS, Piatkov KI, Varshavsky A (2013) Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol Cell 50:161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shokri-Kojori E, et al. (2018) β-Amyloid accumulation in the human brain after one night of sleep deprivation. Proc Natl Acad Sci USA 115:4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ju YS, et al. (2017) Slow wave sleep disruption increases cerebrospinal fluid amyloid-β levels. Brain 140:2104–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Julien O, Wells JA (2017) Caspases and their substrates. Cell Death Differ 24:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee A, Williams DW (2017) More alive than dead: Non-apoptotic roles for caspases in neuronal development, plasticity and disease. Cell Death Differ 24:1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell RL, Davies PL (2012) Structure-function relationships in calpains. Biochem J 447:335–351. [DOI] [PubMed] [Google Scholar]
- 45.Croall DE, Ersfeld K (2007) The calpains: Modular designs and functional diversity. Genome Biol 8:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Liu MC, Wang KKW (2008) Calpain in the CNS: From synaptic function to neurotoxicity. Sci Signal 1:re1. [DOI] [PubMed] [Google Scholar]
- 47.Ono Y, Sorimachi H (2012) Calpains: An elaborate proteolytic system. Biochim Biophys Acta 1824:224–236. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz-Vela A, González de Buitrago G, Martínez-A C (1999) Implication of calpain in caspase activation during B cell clonal deletion. EMBO J 18:4988–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakagawa T, Yuan J (2000) Cross-talk between two cysteine protease families. Activation of caspase-12 by calpain in apoptosis. J Cell Biol 150:887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piatkov KI, Oh J-H, Liu Y, Varshavsky A (2014) Calpain-generated natural protein fragments as short-lived substrates of the N-end rule pathway. Proc Natl Acad Sci USA 111:E817–E826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gascón S, Sobrado M, Roda JM, Rodríguez-Peña A, Díaz-Guerra M (2008) Excitotoxicity and focal cerebral ischemia induce truncation of the NR2A and NR2B subunits of the NMDA receptor and cleavage of the scaffolding protein PSD-95. Mol Psychiatry 13:99–114. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, et al. (2007) Calpain-mediated collapsin response mediator protein-1, -2, and -4 proteolysis after neurotoxic and traumatic brain injury. J Neurotrauma 24:460–472. [DOI] [PubMed] [Google Scholar]
- 53.Xu J, et al. (2009) Extrasynaptic NMDA receptors couple preferentially to excitotoxicity via calpain-mediated cleavage of STEP. J Neurosci 29:9330–9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood AR, et al. ; Electronic Medical Records and Genomics (eMEMERGEGE) Consortium; MIGen Consortium; PAGEGE Consortium; LifeLines Cohort Study (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Visscher PM, et al. (2017) 10 years of GWAS discovery: Biology, function, and translation. Am J Hum Genet 101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varshavsky A. (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20:1298–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varshavsky A. (2019) N-degron and C-degron pathways of protein degradation. Proc Natl Acad Sci USA 116:358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tasaki T, Sriram SM, Park KS, Kwon YT (2012) The N-end rule pathway. Annu Rev Biochem 81:261–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibbs DJ, Bacardit J, Bachmair A, Holdsworth MJ (2014) The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol 24:603–611. [DOI] [PubMed] [Google Scholar]
- 60.Piatkov KI, Brower CS, Varshavsky A (2012) The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc Natl Acad Sci USA 109:E1839–E1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Piatkov KI, Colnaghi L, Békés M, Varshavsky A, Huang TT (2012) The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol Cell 48:926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koren I, et al. (2018) The eukaryotic proteome is shaped by E3 ubiquitin ligases targeting C-terminal degrons. Cell 173:1622–1635.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin HC, et al. (2018) C-terminal end-directed protein elimination by CRL2 ubiquitin ligases. Mol Cell 70:602–613.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rusnac DV, et al. (2018) Recognition of the diglycine C-end degron by CRL2(KLHDC2) ubiquitin ligase. Mol Cell 72:813–822.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim JM, et al. (2018) Formyl-methionine as an N-degron of a eukaryotic N-end rule pathway. Science 362:eaat0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen SJ, Wu X, Wadas B, Oh J-H, Varshavsky A (2017) An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 355:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Z, Sheng M (2012) Caspases in synaptic plasticity. Mol Brain 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perrin BJ, Amann KJ, Huttenlocher A (2006) Proteolysis of cortactin by calpain regulates membrane protrusion during cell migration. Mol Biol Cell 17:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mingorance-Le Meur A, O’Connor TP (2009) Neurite consolidation is an active process requiring constant repression of protrusive activity. EMBO J 28:248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farah CA, et al. (2016) A PKM generated by calpain cleavage of a classical PKC is required for activity-dependent intermediate-term facilitation in the presynaptic sensory neuron of Aplysia. Learn Mem 24:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis MA, et al. (2019) Calpain drives pyroptotic vimentin cleavage, intermediate filament loss, and cell rupture that mediates immunostimulation. Proc Natl Acad Sci USA 116:5061–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tao CL, et al. (2018) Differentiation and characterization of excitatory and inhibitory synapses by cryo-electron tomography and correlative microscopy. J Neurosci 38:1493–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frank RA, Grant SG (2017) Supramolecular organization of NMDA receptors and the postsynaptic density. Curr Opin Neurobiol 45:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen H, Tang AH, Blanpied TA (2018) Subsynaptic spatial organization as a regulator of synaptic strength and plasticity. Curr Opin Neurobiol 51:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen X, et al. (2015) PSD-95 family MAGUKs are essential for anchoring AMPA and NMDA receptor complexes at the postsynaptic density. Proc Natl Acad Sci USA 112:E6983–E6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bosch M, et al. (2014) Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron 82:444–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huganir RL, Nicoll RA (2013) AMPARs and synaptic plasticity: The last 25 years. Neuron 80:704–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buonarati OR, Hammes EA, Watson JF, Greger IH, Hell JW (2019) Mechanisms of postsynaptic localization of AMPA-type glutamate receptors and their regulation during long-term potentiation. Sci Signal 12:eaar6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dosemeci A, Weinberg RJ, Reese TS, Tao-Cheng JH (2016) The postsynaptic density: There is more than meets the eye. Front Synaptic Neurosci 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borgdorff AJ, Choquet D (2002) Regulation of AMPA receptor lateral movements. Nature 417:649–653. [DOI] [PubMed] [Google Scholar]
- 81.Yuste R. (2010) Dendritic Spines (MIT, Cambridge, MA: ). [Google Scholar]
- 82.Averna M, et al. (2015) Physiological roles of calpain 1 associated to multiprotein NMDA receptor complex. PLoS One 10:e0139750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Louneva N, et al. (2008) Caspase-3 is enriched in postsynaptic densities and increased in Alzheimer’s disease. Am J Pathol 173:1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Westphal D, Sytnyk V, Schachner M, Leshchyns’ka I (2010) Clustering of the neural cell adhesion molecule (NCAM) at the neuronal cell surface induces caspase-8- and -3-dependent changes of the spectrin meshwork required for NCAM-mediated neurite outgrowth. J Biol Chem 285:42046–42057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cohn MA, et al. (2007) A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 28:786–797. [DOI] [PubMed] [Google Scholar]
- 86.Shoshan-Barmatz V, Weil S, Meyer H, Varsanyi M, Heilmeyer LM (1994) Endogenous, Ca2+-dependent cysteine-protease cleaves specifically the ryanodine receptor/Ca2+ release channel in skeletal muscle. J Membr Biol 142:281–288. [DOI] [PubMed] [Google Scholar]
- 87.Wang L, Alzayady KJ, Yule DI (2016) Proteolytic fragmentation of inositol 1,4,5-trisphosphate receptors: A novel mechanism regulating channel activity? J Physiol 594:2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Specht CG, et al. (2013) Quantitative nanoscopy of inhibitory synapses: Counting gephyrin molecules and receptor binding sites. Neuron 79:308–321. [DOI] [PubMed] [Google Scholar]
- 89.Wu Q, Sun M, Bernard LP, Zhang H (2017) Postsynaptic density 95 (PSD-95) serine 561 phosphorylation regulates a conformational switch and bidirectional dendritic spine structural plasticity. J Biol Chem 292:16150–16160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tononi G, Boly M, Massimini M, Koch C (2016) Integrated information theory: From consciousness to its physical substrate. Nat Rev Neurosci 17:450–461. [DOI] [PubMed] [Google Scholar]
- 91.Jenkins HG, Dallenbach KM (1924) Obliviscence during sleep and waking. Am J Psychol 35:605–612. [Google Scholar]
- 92.Bingol B, et al. (2010) Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell 140:567–578. [DOI] [PubMed] [Google Scholar]
- 93.Lee SH, et al. (2008) Synaptic protein degradation underlies destabilization of retrieved fear memory. Science 319:1253–1256. [DOI] [PubMed] [Google Scholar]
- 94.Berthier C, Blaineau S (1997) Supramolecular organization of the subsarcolemmal cytoskeleton of adult skeletal muscle fibers. A review. Biol Cell 89:413–434. [DOI] [PubMed] [Google Scholar]
- 95.Turrigiano G. (2012) Homeostatic synaptic plasticity: Local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol 4:a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Malow BA. (2007) The interaction between sleep and epilepsy. Epilepsia 48:36–38. [DOI] [PubMed] [Google Scholar]
- 97.Gast H, et al. (2014) Epileptic seizures as condensed sleep: An analysis of network dynamics from electroencephalogram signals. J Sleep Res 23:268–273. [DOI] [PubMed] [Google Scholar]
- 98.Lamberts RJ, Sander JW, Thijs RD (2011) Postictal sleep: Syncope or seizure? Seizure 20:350–351. [DOI] [PubMed] [Google Scholar]
- 99.McCall WV, Rosenquist PB (2015) Letter to the editor: The effect of ECT on sleep–A comment to Winkler et al. J Psychiatr Res 61:239–240. [DOI] [PubMed] [Google Scholar]
- 100.Massie R. (1980) Peter the Great (Alfred A. Knopf, New York: ), p 136. [Google Scholar]
- 101.Sedaris D. (April 15, 2017) The IHOP Years. New Yorker. Available at https://www.newyorker.com/culture/personal-history/david-sedaris-the-ihop-years. Accessed April 30, 2019.
- 102.Lado FA, Moshé SL (2008) How do seizures stop? Epilepsia 49:1651–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zubler F, Steimer A, Gast H, Schindler KA (2014) Seizure termination. Int Rev Neurobiol 114:187–207. [DOI] [PubMed] [Google Scholar]
- 104.Urano F. (2016) Wolfram syndrome: Diagnosis, management, and treatment. Curr Diab Rep 16:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu S, et al. (2014) A calcium-dependent protease as a potential therapeutic target for Wolfram syndrome. Proc Natl Acad Sci USA 111:E5292–E5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mahoney CE, Cogswell A, Koralnik IJ, Scammell TE (2019) The neurobiological basis of narcolepsy. Nat Rev Neurosci 20:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Penuela S, Gehi R, Laird DW (2013) The biochemistry and function of pannexin channels. Biochim Biophys Acta 1828:15–22. [DOI] [PubMed] [Google Scholar]
- 108.Sandilos JK, et al. (2012) Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J Biol Chem 287:11303–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakai Y, et al. (2011) Calcineurin and its regulator sra/DSCR1 are essential for sleep in Drosophila. J Neurosci 31:12759–12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tomita J, et al. (2011) Pan-neuronal knockdown of calcineurin reduces sleep in the fruit fly, Drosophila melanogaster. J Neurosci 31:13137–13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu HY, et al. (2004) Critical role of calpain-mediated cleavage of calcineurin in excitotoxic neurodegeneration. J Biol Chem 279:4929–4940. [DOI] [PubMed] [Google Scholar]
- 112.Mukerjee N, McGinnis KM, Park YH, Gnegy ME, Wang KK (2000) Caspase-mediated proteolytic activation of calcineurin in thapsigargin-mediated apoptosis in SH-SY5Y neuroblastoma cells. Arch Biochem Biophys 379:337–343. [DOI] [PubMed] [Google Scholar]
- 113.Orr AL, et al. (2014) β-Amyloid inhibits E-S potentiation through suppression of cannabinoid receptor 1-dependent synaptic disinhibition. Neuron 82:1334–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Geng F, Wenzel S, Tansey WP (2012) Ubiquitin and proteasomes in transcription. Annu Rev Biochem 81:177–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zada D, Bronshtein I, Lerer-Goldshtein T, Garini Y, Appelbaum L (2019) Sleep increases chromosome dynamics to enable reduction of accumulating DNA damage in single neurons. Nat Commun 10:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bellesi M, Bushey D, Chini M, Tononi G, Cirelli C (2016) Contribution of sleep to the repair of neuronal DNA double-strand breaks: Evidence from flies and mice. Sci Rep 6:36804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Butler LR, et al. (2012) The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J 31:3918–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou P, Wu E, Alam HB, Li Y (2014) Histone cleavage as a mechanism for epigenetic regulation: Current insights and perspectives. Curr Mol Med 14:1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Duncan EM, et al. (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135:284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.