Significance
This work represents a large, order-wide evolutionary analysis of the order Chiroptera (bats). Our pipeline for assembling sequence data and curating orthologous multiple sequence alignments includes methods for improving results when combining genomic and transcriptomic data sources. The resulting phylogenetic tree divides the order Chiroptera into Yinpterochiroptera and Yangochiroptera, in disagreement with the previous division into Megachiroptera and Microchiroptera and in agreement with some other recent molecular studies, and also provides evidence for other contested branch placements. We also performed a genome-wide analysis of positive selection and found 181 genes with signatures of positive selection. Enrichment analysis shows these positively selected genes to be primarily related to immune responses but also, surprisingly, collagen formation.
Keywords: Chiroptera, phylogenetics, transcriptome, gene alignment, orthologous genes
Abstract
Historically, the evolution of bats has been analyzed using a small number of genetic loci for many species or many genetic loci for a few species. Here we present a phylogeny of 18 bat species, each of which is represented in 1,107 orthologous gene alignments used to build the tree. We generated a transcriptome sequence of Hypsignathus monstrosus, the African hammer-headed bat, and additional transcriptome sequence for Rousettus aegyptiacus, the Egyptian fruit bat. We then combined these data with existing genomic and transcriptomic data from 16 other bat species. In the analysis of such datasets, there is no clear consensus on the most reliable computational methods for the curation of quality multiple sequence alignments since these public datasets represent multiple investigators and methods, including different source materials (chromosomal DNA or expressed RNA). Here we lay out a systematic analysis of parameters and produce an advanced pipeline for curating orthologous gene alignments from combined transcriptomic and genomic data, including a software package: the Mismatching Isoform eXon Remover (MIXR). Using this method, we created alignments of 11,677 bat genes, 1,107 of which contain orthologs from all 18 species. Using the orthologous gene alignments created, we assessed bat phylogeny and also performed a holistic analysis of positive selection acting in bat genomes. We found that 181 genes have been subject to positive natural selection. This list is dominated by genes involved in immune responses and genes involved in the production of collagens.
The bat order Chiroptera is one of the most common and diversely adapted orders of organisms on Earth. Estimates of the exact number of species vary, but all estimates show bats represent a large portion of the known mammalian species, representing ∼925 of 6,400 known species, about 15% (1, 2). Several characteristics of bats make them inherently interesting, most uniquely their ability to fly and echolocate. Bats are also notorious for harboring viruses that transmit to humans [called zoonotic viruses (3)]. For instance, bats are the established reservoir hosts for SARS coronavirus, Nipah virus, and Hendra virus (1, 4). Hypsignathus monstrosus, the hammer-headed bat, has been identified as a possible reservoir host of Ebola virus (5, 6). Marburg virus, a close relative of Ebola virus, has been isolated from Rousettus aegyptiacus (7, 8).
Unfortunately, the diversity that makes bats interesting also makes them difficult to study. Genetic information allows researchers to gain a deeper understanding of the evolutionary origins of such diverse taxa. There are several previous phylogenic analyses conducted with bats across the order Chiroptera (9–16). Given the difficulty of obtaining genetic data from a species group with such extreme diversity, there has historically been a tradeoff between number of loci analyzed and number of species analyzed (i.e., more loci can be reasonably analyzed only when a smaller number of species is considered). Some studies tackle hundreds of species at once. Most recently, Amador et al. constructed a tree containing 799 species by performing a supermatrix analysis with maximum likelihood and maximum parsimony methods using up to nine gene sequences per species (16). High-throughput sequencing is now lessening the burden of this tradeoff because a number of bats across the order Chiroptera have had their genomes or transcriptomes sequenced in bulk. Two studies have undertaken phylogenetic analyses of these large datasets (13, 15).
In most evolutionary studies, the unit of analysis is the orthologous multiple sequence alignment. Ideally, one creates a set of alignments representing each gene, each of which includes the correct ortholog from each species under analysis. However, accurately assembling short reads, identifying orthologs, and curating/vetting multiple sequence alignments from transcriptomic and genomic data are difficult tasks in an evolving field (17). In previous studies of bats that employed high-throughput sequencing data, these bioinformatic issues were partially addressed by collecting most of the data at the same time with similar methods: whole-genome sequencing in the case of Tsagkogeorga et al. (13) or transcriptomes in the case of Lei and Dong (15). However, the complexities of creating high-quality orthologous sequence alignments are compounded when public data from multiple sources are combined because these datasets have been collected with a variety of methods and from either chromosomal DNA or transcribed RNA. These issues may, in part, explain why the phylogenies of bats that have been created to date agree on much of the history of Chiroptera but differ in many ways as well.
The same data required to investigate phylogeny—namely, multiple sequence alignments of all available genes—is also highly valuable for guiding research into selective pressures that have influenced the genomes of bats. Of particular interest would be genes that have undergone positive natural selection in bats, possibly allowing them to better survive infection by viruses but also likely improving other processes that are specific to bat physiology, immunology, or ecology (18–20). Positive natural selection produces in gene alignments an unusually high ratio of DNA substitutions which change the protein sequence (dN) to those that do not (dS), measured by dN/dS > 1 (21, 22). Limited previous analysis of positive selection in bat genomes has been performed. Typically, selection has been analyzed only in key gene families for specific purposes (13, 23, 24). These studies yielded interesting results, but we still lack a holistic view of the categories of genes most affected by selection in bat genomes.
We set out to perform a metaanalysis of available Chiropteran annotated genomes and transcriptomes, with three principal goals: a carefully curated set of orthologous gene alignments, a high-confidence phylogeny, and positive selection measurements for each gene. In the process of obtaining these goals, we developed a curation and cleaning pipeline for producing high-quality orthologous multiple sequence alignments from datasets combining public data from multiple sources. We also produced transcriptomic data for R. aegyptiacus that complements previously published data (25, 26) and a transcriptomic dataset for H. monstrosus.
Results
Data Collection and Assembly.
The bat species analyzed in this study and the type of data associated with each are shown in Table 1. For a few bat species, genomic (chromosomal) sequences are available, and for others, transcriptomic sequences exist. For each bat species with an annotated genome project, we downloaded the relevant RefSeq database from the National Center for Biotechnology Information (NCBI) website and extracted the protein and coding sequence of the longest isoform of each gene for orthology search. After finding orthologous genes from each species, we selected the gene isoform which most closely matched the consensus sequence for further analysis (Methods). Genome assembly accession numbers, as well as basic assembly statistics, for each genome are given in SI Appendix, Table S1. Human, common shrew (Sorex araneus), and pig (Sus scrofa) genomes were also included as outgroups. Finally, we harvested mRNA and sequenced the transcriptomes of H. monstrosus and R. aegyptiacus on Illumina machines (Methods). For all other bats with available transcriptome data, we downloaded the raw sequencing reads from the Sequence Read Archive (SI Appendix, Table S2) (23, 27–38).
Table 1.
Chiroptera data overview
| Species | Genome/transcriptome | Annotated genes | N50 | %GC | Assembly count |
| Artibeus jamaicensis | Transcriptome | 10,071 | 2,166 | 53.4 | 16 |
| Carollia brevicauda | Transcriptome | 3,954 | 1,284 | 51.3 | 12 |
| Cynopterus sphinx | Transcriptome | 6,232 | 1,653 | 49.8 | 12 |
| Desmodus rotundus | Transcriptome | 9,019 | 2,115 | 52.8 | 18 |
| Eptesicus fuscus | Genome | 13,248 | 2,235 | 54.2 | n/a |
| Hypsignathus monstrosus* | Transcriptome | 7,875 | 2,040 | 49.8 | 17 |
| Macrotus californicus | Transcriptome | 4,375 | 1,557 | 51.9 | 12 |
| Miniopterus schreibersii | Transcriptome | 11,089 | 2,202 | 53.4 | 19 |
| Murina leucogaster | Transcriptome | 9,267 | 2,055 | 53.6 | 14 |
| Myotis brandtii | Genome | 12,674 | 2,229 | 53.1 | n/a |
| Myotis davidii | Genome | 12,353 | 2,223 | 53.2 | n/a |
| Myotis lucifugus | Genome | 12,386 | 2,214 | 53.2 | n/a |
| Myotis ricketti | Transcriptome | 4,868 | 1,401 | 51.1 | 12 |
| Pteropus alecto | Genome | 13,295 | 2,235 | 52.4 | n/a |
| Pteropus vampyrus | Genome | 13,145 | 2,232 | 52.3 | n/a |
| Rhinolophus ferrumequinum | Transcriptome | 6,764 | 1,761 | 53.8 | 12 |
| Rousettus aegyptiacus* | Transcriptome | 9,714 | 2,235 | 52.8 | 18 |
| Tadarida brasiliensis | Transcriptome | 6,128 | 1,869 | 51.4 | 12 |
| Homo sapiens (outgroup) | Genome | 13,206 | 2,301 | 52.2 | n/a |
| Sorex araneus (outgroup) | Genome | 12,190 | 2,280 | 55.7 | n/a |
| Sus scrofa (outgroup) | Genome | 11,304 | 2,142 | 53.8 | n/a |
Each species analyzed in this study, along with basic information concerning data type (genomic or transcriptomic), the number of genes here placed in orthologous gene sets, the N50 statistic (50% of bases are in contigs of length at least N50), and guanine–cytosine content (%GC) of these genes. For bats with transcriptomic data, data were all assembled and annotated as part of this study, and the number of assemblies constructed and analyzed is listed. For the other species, genomic data and annotations were all downloaded from RefSeq. Transcriptome data for two species, shown with asterisks, were generated in this study.
For the sake of consistency, we constructed transcriptome assemblies using our own pipeline, even in the cases where authors made assemblies publicly available. Briefly, our pipeline consisted of removing adapter sequences with Trimmomatic (39), followed by using two of the most popular transcriptome assemblers, the De Bruijn graph-based Trinity (40) and Trans-ABySS (41) assemblers, with a range of input parameters. This resulted in multiple tentative assemblies per bat (Table 1 and Methods) (39–42).
Building Orthologous Gene Families.
The search for orthologous genes was performed in two primary steps. First, we searched for orthologs in the genomic datasets. With genomic data, one is able to use syntenic information to help predict orthology. We used all-v-all BLAST reciprocal best hits of the protein sequences, filtered using three sources of syntenic information: public orthology predictions from BioMart, proximity via whole genome alignment, and proximity of similar neighboring genes (43–45). Second, we searched for orthologs in transcriptomic datasets. We selected the best BLAST reciprocal best hits in transcriptomic data against genes found in the genomic orthologous gene sets, filtered by search using HMMER (46), a hidden Markov model-based homology search software package, and filtered by match length (Methods).
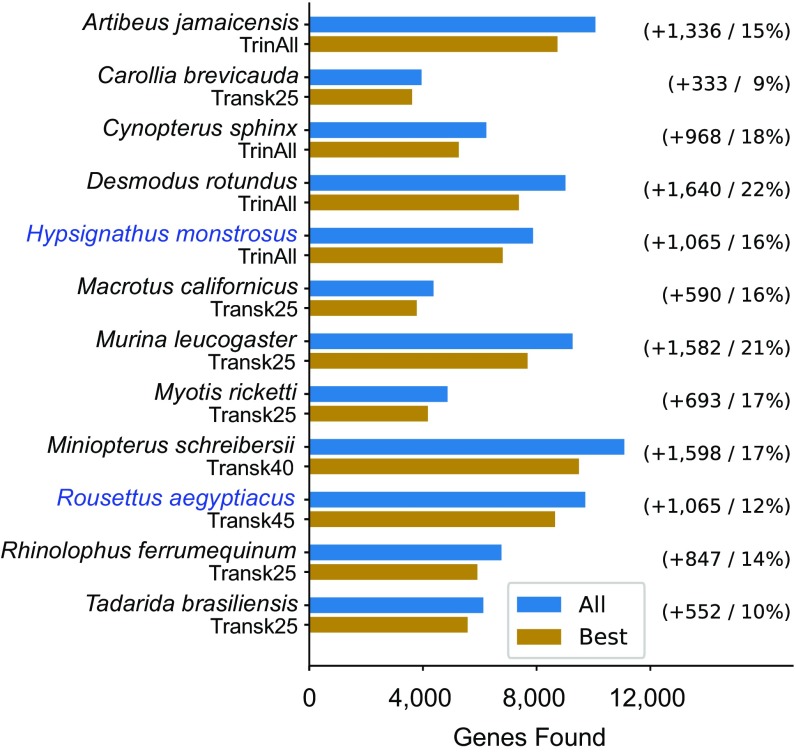
In all, we were able to place 192,686 transcripts into 11,677 orthologous gene sets, of which 1,107 contain genes from all species and outgroups. Fig. 1 shows the number of transcriptomic genes found by species. Also shown is the number of genes we would have found in each species had we known a priori which assembly would perform the best for each bat and only assembled that one. With the additional work of analyzing multiple assemblies per bat, we were able to identify 9–22% more genes per bat relative to the best single assembly, representing thousands of added transcripts and improvements to the completeness of the gene network.
Fig. 1.
Use of multiple assembly methods improves recovered gene counts. For each species where transcriptome data are being used, the number of genes placed in an orthologous gene family based on all assemblies is reported (blue), as well as the number of genes which could have been found from the best single assembly alone, had it been known a priori (gold). Which assembly was best for each species is shown as the labels for gold bars, where TrinAll indicates the Trinity assembly using all reads, and TranskXX indicates the Trans-ABySS assembly using the De Bruijn graph of k-mers of length XX. Number of genes added through use of multiple assemblies over the best assembly, and percentage increase, are shown to the right of the bars. Use of multiple assemblies added 9–22% more annotated orthologs per species relative to the best single assembly. Species with raw data generated for this paper are shown in blue type.
Multiple Sequence Alignment Cleaning.
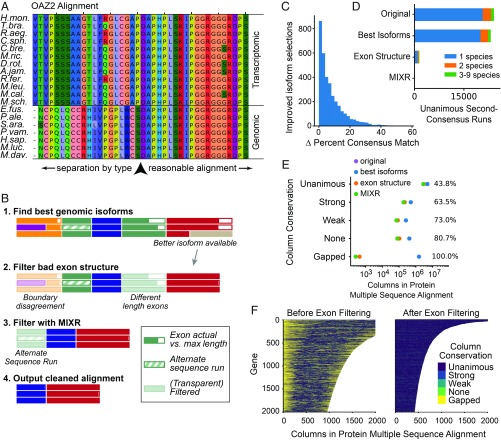
Manual inspection of many multiple sequence alignments of orthologous genes revealed a nonrandom source of error: the species were biased toward segregating by data type (i.e., genomic data vs. transcriptomic data). Some poorly aligned regions would tend to agree within data type but disagree between data types. An example is shown in Fig. 2A. Furthermore, the splits were observed to happen at sharp boundaries highly suggestive of exon boundaries. This effect can be largely explained by the fact that we chose the longest isoforms predicted from annotated genome sequences, even though the longest isoforms might not be expressed at high enough levels to appear in the transcriptomic datasets from a given tissue. Other artifacts in transcriptomic or genomic data assembly and annotation could also contribute to this effect. Quantification of this bias, based on the cleaning pipeline described below, is shown in SI Appendix, Fig. S2.
Fig. 2.
Multiple sequence alignment cleaning. (A) Some multiple sequence alignments were observed to demonstrate isoform selection biased toward separation of genomic and transcriptomic data, causing nonrandom, nongapped errors segregated by the artifact of data type. (B) The alignment cleaning pipeline. First, each gene derived from genomic data was revisited to choose the isoform which best matches the consensus alignment sequence. Second, exons were filtered by structure, where exons with disagreement about boundary positions in the alignment and exons with >1% length difference between species were filtered out. Last, exons were filtered using the MIXR algorithm described in the text. (C) A total of 3,444 improved genomic isoform selections were performed, shown as a function of the improvement in percent matching the consensus sequence, i.e., (percent matching after) – (percent matching before). (D) Counts of unanimous second-consensus runs after each step of the alignment cleaning pipeline (with steps progressing from top to bottom in the graph). Unanimous second-consensus runs are defined as runs of more than three columns in a row where some minority of bat sequences agree with each other but disagree with those of all other bats. The final output contains only seven such runs with two or more species, and these are only up to three species and five amino acids long. (E) In each step of the alignment cleaning pipeline, weakly conserved sites are filtered at higher rates, enriching the final alignments for conserved sequences. “Strong,” “Weak,” and “None” conservation categories are as defined by Clustal (47). Percent reduction in multiple sequence alignment column counts shown to the right. (F) Column conservation of the first 2,000 columns of the longest 2,000 alignments before and after exon filtering.
To ameliorate nonrandom assembly and isoform-selection artifacts, we developed a three-step cleaning algorithm for the multiple sequence alignments (Fig. 2B). First, we revisited each gene derived from genomic data and replaced it, if necessary, with the isoform closest to the consensus sequence. This resulted in improvements to 3,444 transcripts, with transcripts improving their match to the consensus sequence by an average of 8%, although there was a wide range of percent improvements (Fig. 2C).
Second, we removed exons if the species did not all agree on the exon structure. Specifically, we removed exons if all species did not agree on the aligned exon boundaries or if exon sequences differed too much in length. For phylogenetic analysis, where all genes are used in aggregate to fit a single tree, we required exact agreement in exon lengths. For gene-level positive selection analysis we required exons differ by no more than 1%. This cutoff was chosen because we observed it to be a transition point to high-gap exons in our data (SI Appendix, Fig. S1). Fig. 2 D–F are based on the latter threshold, and SI Appendix, Fig. S4, shows the equivalent figures for the former.
Third, we developed the Mismatching Isoform eXon Remover (MIXR) software package to detect and remove exons with alternate consensus runs directly (available at http://github.com/hawkjo/mixr). By alternate consensus run, we mean the situation as shown in Fig. 2A, where some subset of species has a long run of amino acids which are internally consistent but which disagree from the majority of the sequences. While no substitution pattern in a single column of an alignment can distinguish real biological mutations from artifactual mismatching of isoforms, runs of an alternate consensus sequence are exponentially unlikely as a function of run length according to all site substitution models, so we want to remove them before analysis. Briefly, the MIXR algorithm works as follows. First, we define the alternate-consensus-run score, designed to give high scores to runs where some subset of species is internally consistent but differs from the majority of species. We then find the maximum alternate-consensus-run score for all species bipartitions (divisions of the species into two subsets) in a given alignment, such as the bipartition into transcriptomic and genomic species in Fig. 2A. Finally, exons with significant scores are removed. See Methods for additional details.
Note that in all three of our cleaning steps, we have tried to avoid filtering on sequence divergence as much as possible. This is because our intention is to perform positive selection analysis on the cleaned alignments, an analysis which measures the ratio between nonsynonymous and synonymous mutations. Filtering strategies based on whether the protein sequences agree will directly bias the data in favor of synonymous mutations, and the exon-length-matching and MIXR steps are expected to have this effect to some degree. However, to whatever degree this is true in our data, it will have a conservative effect, biasing the data to fewer rather than more significant hits in a positive selection analysis.
To measure the efficacy of each of our filtering steps, we looked at two different measures of the filtering process. First, we looked at unanimous second-consensus runs. We define unanimous second-consensus runs to be runs in an alignment where some subset of species has unanimous agreement of the amino acid sequence but disagrees with the majority of species. This is correlated with but distinct from the alternate-consensus runs defined for the MIXR algorithm as it is independent of the substitution model used for the MIXR scoring function. The results after each step of the cleaning process are shown in Fig. 2D. We see that the most significant reduction in second-consensus runs is due to the exon structure filtering step, and MIXR cleaned up essentially all of the runs which passed the previous two steps. After cleaning, only seven runs of more than three columns were detected, and those were up to only five columns in length. Furthermore, all but one of these alignments contained only species with genome data available, removing the concern of separation by data type.
As a second measure of alignment improvement, we checked that the filtered sequences had increased overall sequence conservation. Our strategy, as expected, preferentially discriminates against more weakly conserved sites [as defined by CLUSTAL (47)], filtering >80% of nonconserved sites vs. 44% for unanimous sites, and almost all gapped sites (Fig. 2E). Furthermore, both the exon-structure filtering step and the MIXR algorithm improve sequence conservation. Fig. 2F shows the positions and distribution of conserved sites in the first 2,000 multiple sequence alignments. Many of the poor quality exons are at the ends of the alignments, which is to be expected. The ortholog finding process scores sequences based on length of agreement, which naturally tends to include matching sections in the center. The ends are then more free to vary, with variability expected due to differences in isoform selection, as well as due to incomplete or incorrect transcript assembly. Transcripts tend to have less coverage near the ends and correspondingly poorer assembly. This also helps explain why so many unanimous sites end up being filtered: correctly matched exons will still be filtered if partial assembly results in exons of different lengths. From these results, as well as manual inspection, the alignments have significantly fewer erroneous columns after exon filtering.
Phylogenetic Analysis.
Phylogenetic trees were constructed from these multiple sequence alignments, using multiple strategies and software packages. First, using the 1,107 genes found in all species, we constructed the species tree with a partitioned nucleotide analysis in Mr. Bayes (48), which fits for one species tree while allowing the model parameters to vary for each gene. Next, using the same genes, we constructed the species tree with concatenated data using RAxML (49). This we did for the full coding sequence (CDS), the amino acid sequence, and each codon position separately. We additionally fit the phylogenetic tree using RAxML with the CDS sequence of the longest 100 alignments after subjecting the corresponding amino acid alignments to manual inspection. The full length of all 100 alignments looked highly credible to manual inspection. Finally, we constructed the 1,107 gene trees with all species using Mr. Bayes and determined the species tree via coalescent analysis as implemented in ASTRAL (50). This was done for the full CDS sequence and each codon position separately.
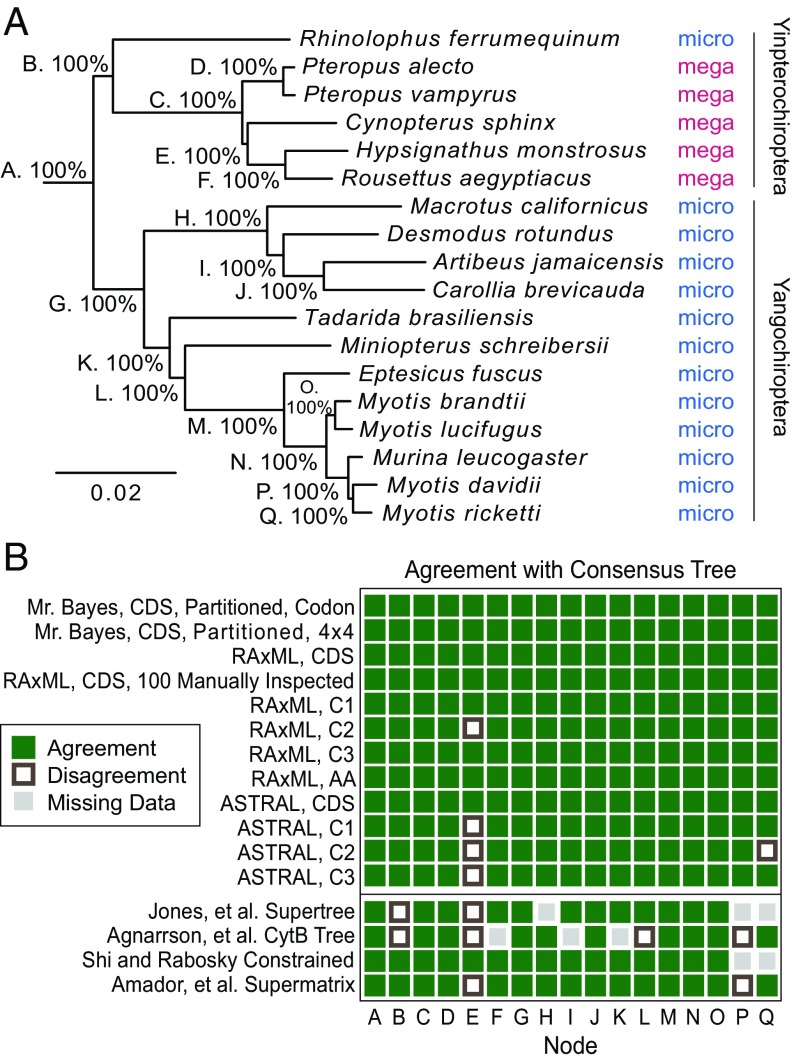
The final tree is shown in Fig. 3A, with reported posterior probabilities given from the Mr. Bayes partitioned analysis. We refer to the final tree instead of a specific version of the final tree due to the strong consensus between methods. A summary of how the methods agreed or disagreed is shown in Fig. 3B. All methods converged on nearly the same species tree. In fact, due to the large amount of data, all nodes resolved with 100% reported posterior probability in both Mr. Bayes analyses. The only species not consistently placed in every analysis were Cynopterus sphinx and Murina leucogaster. Their alternative placements are shown in SI Appendix, Fig. S5.
Fig. 3.
Chiroptera phylogeny. (A) The consensus phylogeny for the bats considered in this study, plotted with FigTree (102). Branch lengths and posterior probability for nodes are from the Mr. Bayes partitioned analysis with full codon model, which agrees at all nodes with the consensus tree. Historical designations of microbat and megabat are listed, as well as the newer divisions of Yinpterochiroptera and Yangochiroptera. (B) Comparison of trees constructed by various methods with the consensus tree. For each tree method, the rectangle at each node indicates agreement or disagreement with the consensus tree on the implied split at the node. Comparison with previously published trees is shown below. For nodes B, E, L, and P, the previously published trees agree with either our consensus phylogeny or a single alternative hypothesis: R. ferrumequinum above node G, C. sphinx above node C, M. schreibersii above node H, or M. leucogaster above node N, respectively. AA, amino acid sequence; C1–C3, the coding sequence restricted to those bases in the first, second, or third codon position, respectively; 100 manually inspected, phylogeny constructed using the longest 100 alignments after manual inspection for artifacts (none observed).
Also included in Fig. 3B are comparisons with previously published trees, in particular, those that included most of the species in our analysis. All species placements in our final tree agree with these trees, with the exception of two previously controversial species, Rhinolophus ferrumequinum and Miniopterus schreibersii; one particularly close node involving C. sphinx; and one surprising placement, M. leucogaster. Other order-wide phylogenetic analyses were omitted from Fig. 3B due to minimal overlap with the species considered here.
The placement of R. ferrumequinum addresses the first branching of the order Chiroptera. The traditional division of order Chiroptera into Megachiroptera and Microchiroptera, the large and small bats, respectively, has been challenged in recent years as molecular phylogenetic analyses have gained prominence. An alternative history has been proposed, dividing bats into two suborders named Yinpterochiroptera and Yangochiroptera (51). In the proposal, the microbat families Rhinopomatidae, Rhinolophidae, Hipposideridae, and Megadermatidae are joined with the megabats to form the new clade Yinpterochiroptera, while the rest of the microbats form Yangochiroptera. Our phylogeny agrees with this model, with R. ferrumequinum falling into Yinpterochiroptera (Fig. 3A). This restructuring has gained additional recent support (9, 11, 13, 15, 52), although it must be noted that the accurate rooting of ancient species groups is a notoriously difficult aspect of phylogenetics (44).
The placement of M. schreibersii has also been unclear. Agnarsson’s cytochrome B-based phylogeny places M. schreibersii just outside node H on our phylogeny. On the other hand, our placement of M. schreibersii agrees with Hoofer et al. (53), who argued that due to this placement and the large divergence, Miniopteridae deserves to be its own family, and this agrees with the other phylogenies shown in Fig. 3B as well.
The most surprising placement in our tree is that of M. leucogaster. In all of our calculated trees shown in Fig. 3B, with 100% reported posterior probability where calculated, M. leucogaster disrupts the monophyly of the genus Myotis (Fig. 3B and SI Appendix, Fig. S5). One must keep in mind, however, that the quantity of data here considered is so large it will tend to produce results with 100% reported posterior probability at each node, pushing the question of credibility further upstream to the quality of the data. No data assembly and cleaning strategy, including our own, is perfect, and it is possible that sufficient errors remain in our data as to result in an erroneous topology of such closely related species. It at first even seems suggestive, given our previous observations of bias by data type, that M. leucogaster has moved closer than expected to Myotis ricketti, the Myotis species represented by transcriptomic data. SI Appendix, Fig. S6, takes a focused look at the data for these five bats in the 100 manually inspected alignments. Strikingly, the bipartition of M. leucogaster as outgroup to Myotis has less than or equal the support of any other Myotis bat being outgroup to Myotis + M. leucogaster. This includes the hypothesis of M. ricketti as outgroup, which should have near-zero support under the data-type bias hypothesis. Meanwhile, the bipartition of M. leucogaster, M. ricketti, and Myotis davidii vs. the other two Myotis bats is the bipartition most clearly supported by the data. Furthermore, the relationships between the ranges of the bats in this clade roughly match our consensus phylogeny: M. ricketti lives in southeast Asia, M. davidii lives in central China, M. leucogaster lives in southeast Asia and central China, Myotis brandtii lives across Europe and parts of northern Asia, and Myotis lucifugus lives in North America (54–58). We recognize that this placement is surprising given previously published work, but our results suggest the placement of M. leucogaster may merit further consideration.
One final species placement of note is Desmodus rotundus. The family Phyllostomidae (New World leaf-nosed bats) here consists of the bats within the clade defined by node H. Wetterer, et al. (59) proposed that the genus Desmodus be placed sister to the rest of the phyllostomids, which were to have formed the subfamily Phyllostominae. Our placement of Macrotus californicus, however, disrupts Phyllostominae, in agreement with the tree proposed by Rojas et al. (60).
Positive Selection Analysis.
Finally, we looked within bat genomes for signatures of positive selection acting on protein-coding genes. We used the PAML software package (61) to identify dN/dS > 1 codons within each of the orthologous gene alignments that we created. We used codon models M8 and M8a, which estimate the evolutionary pressures that have acted at specific codon sites over the entire phylogeny of species included. These models do not consider unique evolutionary scenarios that have affected gene evolution along different branches over the phylogenetic tree like some other models. In M8, the data in each multiple sequence alignment is fit to a model where one group of codons from within the alignment is allowed to evolve with dN/dS > 1. M8a is the null version of this model, where that same category of codons is allowed but is constrained to dN/dS = 1. Because there is one additional degree of freedom in M8, the data will usually fit better to the M8 model (as estimated by a likelihood value). A gene was deemed to be evolving under positive selection when the data are a statistically better fit to M8 than to M8a (P < 0.05 after correction for multiple tests; Dataset S1). We performed this analysis on 10,650 genes that met criteria of having at least six species and 30 codons represented in the alignment. Out of these, 181 genes met the statistical criteria for being subject to positive natural selection, and these genes were hand-curated for function (Dataset S1). Table 2 shows a subset of the genes. Nineteen percent are known to be involved in immunity or the replication cycles of pathogens. Surprisingly, 8% are known to be involved in collagen formation, including 10 out of the 27 top scoring genes for positive selection (Dataset S1). We also looked at the gene ontology (GO) classifications which are overrepresented in the list of genes under positive selection and present this data in Dataset S2. The GO categories most enriched for positively selected genes are dominated by immune responses and collagen formation.
Table 2.
Some of the 181 genes under positive selection in bats
| Genes | |
| Immunity/pathogens (n = 35, 19% of total) | ANPEP/CD13 |
| C4BPB/C4BP | |
| C5/complement C5 | |
| CALCOCO2/NDP52 | |
| CCL21 | |
| CCL25/TECK | |
| CD36* | |
| CD53 | |
| CD63 | |
| CD83 | |
| CFP/properdin | |
| CRISP3* | |
| CTSW/cathepsin W | |
| CYBB/cytochrome b-245 beta chain | |
| DPP4/CD26 | |
| ENPP3/E-NPP3/CD203c | |
| GYPA/CD235a/glycophorin A | |
| HLA-DMB | |
| ICAM3/CD50 | |
| IL7/interleukin 7 | |
| IQGAP2 | |
| LTF/lactotransferrin* | |
| LY49 | |
| LY6G5C | |
| MMP12 | |
| MX1/MxA | |
| ORC2 | |
| PON3/paraoxonase 3 | |
| PRF1/perforin 1 | |
| SAMHD1 | |
| SEMA7A/semaphorin 7A | |
| SIGLEC1/CD169 | |
| SRGN/serglycin | |
| TF/transferrin | |
| ZC3HAV1/ZAP | |
| Sexual reproduction (n = 15, 8% of total) | CATSPER3 |
| CCDC136 | |
| CCDC146 | |
| CRISP3* | |
| FAM217A† | |
| FETUB/fetuin B | |
| HbE1/HbE/embryonic hemoglobin subunit | |
| IQUB† | |
| LTF/lactotransferrin* | |
| MSMB/IgBF | |
| PRR9† | |
| PZP | |
| RBM44† | |
| SLC26A8 | |
| TMEM62 | |
| Collagen formation (n = 9, 5% of total) | ANO5 |
| CD36* | |
| COL11A2 | |
| COL16A1 | |
| COL18A1 | |
| COL1A2 | |
| COL28A1 | |
| COL3A1 | |
| COL4A1 | |
| COL4A2 | |
| COL4A3 | |
| COL4A5 | |
| COL4A6 | |
| COL5A3 | |
| COL7A1 | |
| COL9A2 | |
| MMP12 | |
| TMEM38B/TRIC-B | |
| TSC1 | |
| Peroxisome (n = 9, 5% of total) | ACAA1 |
| ACOX2 | |
| ACSL5 | |
| AGPS | |
| DHRS4 | |
| EPHX2 | |
| PECR | |
| PHYH | |
| PXMP4 |
These genes are placed in two categories on this table.
Relatively little is known about these genes, but they are expressed solely in the testis.
Discussion
Understanding the evolutionary history of bats is important not just for the study of Chiropteran zoology but also for the study of bats as reservoirs of deadly human viruses. Knowledge of an accurate phylogeny improves analysis of positive selection in bat genomes because dN/dS analysis requires both gene alignments and a phylogenetic relationship of the orthologs being analyzed. Also, the reliable identification of gene orthologs will allow molecular biologists to functionally test differences in these genes from one species to the next (62). Functional studies such as these will allow us to understand whether some bats have unique features of their immunity that allow them to harbor viruses that are dangerous to humans (63). Herein, we have curated multiple sequence alignments of thousands of bat genes. Using both genomic and transcriptomic data, we were able to find 11,677 orthologous gene families. To enhance these alignments, we provided transcriptome data for two of these bats, H. monstrosus and R. aegyptiacus, from which we annotated 7,858 and 9,682 genes, respectively. We furthermore developed a general data cleaning method for filtering exons with nonrandom structural errors, in this case observed to result from genomic vs. transcriptomic data. For this method, we developed the MIXR software package which directly detects and removes alternate consensus run artifacts, and is available at http://github.com/hawkjo/mixr. The multiple sequence alignments that we have created for bat genes, both before and after exon filtering, are available for use by the wider bat and virology communities (http://numerical.recipes/chiroptera/). Using these alignments, we examined the history both of speciation and of positive selection in 18 species of bats. This study hopefully sets the stage for continued and more in-depth study of the evolution and functional differentiation of bat genes relevant to immunity and beyond.
Using these orthologous gene families, we were able to reconstruct the phylogeny of the order Chiroptera using multiple methods. Due to the sheer scale of the data, we resolved each node in the tree with 100% reported posterior probability, although the topology differed slightly depending on the analysis method. Our results support the division of Chiroptera into the two suborders Yinpterochiroptera and Yangochiroptera, in disagreement with the traditional division into Megachiroptera and Microchiroptera. However, we acknowledge that rooting ancient clades continues to be a difficult phylogenetic problem, and further data may shed more light on this issue. We furthermore provide evidence for the placement of M. schreibersii, in which we agree with Hoofer and Bussche, supporting their proposal for the separation of Miniopteridae into its own family (53). We also provide evidence for the disruption of proposed subfamily Phyllostominae by D. rotundus. Most intriguingly, we saw M. leucogaster placed in the Myotis genus, which will require further investigation.
Finally, we have analyzed positive selection of genes during the speciation of bats, on a genome-wide basis. Previously, work aimed at describing adaptive evolution in bats primarily focused on their unique traits, selecting families of genes to study for selection. These studies can broadly be divided into two categories, those that dealt with specific life traits, such as echolocation or metabolism related to frugivory (24, 64–73), and those that were related to pathogens or immunity (74–80). There are three studies that used larger datasets, two of which used whole-genome data (13, 23, 24). Unlike our study, Tsagkogeorga et al. (13) used genome-wide data to ask which genes might explain the alternative subordinal topologies supported by their data. Similarly, the Shen et al. (24) study was specifically interested in energy metabolism in bats due to their energy-expensive mode of locomotion, flight. Finally, Zhang et al. (23) used whole-genome data available from two distantly related bats, along with orthologs from a number of mammals, and inferred adaptive evolution in the innate immune pathway and DNA damage checkpoint pathway. Our work, in comparison, includes more Chiropteran species and a holistic analysis of selection in the bat genome. The addition of more species, and the generation of a high-confidence tree for these species, gives us better resolution for detecting adaptive evolution specifically within Chiroptera (81).
We find that the positively selected genes in bats are dominated by genes involved with immunity. This could have something to do with the high pathogen load that bats are thought to carry (3), but on the other hand, this finding is not unusual. Bats now join many other species groups in the finding that immune processes stand out for the strength of positive natural selection that has shaped them. The same has been found in many diverse species groups including primates (82, 83), fish (84), and insects (85). It has been noted that the bats have some usual aspects of their immune systems (86–88), which could be consistent with the evolutionary signatures of rapid sequence evolution that we observe in many genes involved in immunity.
Less clear to us is why so many genes involved in collagen formation seem to be under positive selection. Collagens are a family of structural proteins that form connective tissues in the body, including tendons, ligaments, and skin (89). The walls of veins, arteries, and capillaries also contain collagen (90). Collectively, the different forms of collagen constitute the most abundant protein in mammalian bodies (89). Bat wings consist of a network of collagen (91), and bats often have injuries on their wings which need to heal quickly (92). Recently, Pseudogymnoascus destructans, a fungal pathogen that has killed more than 6 million bats, has been reported to damage collagen (93). It is less clear how pathogens would have placed pressure on collagen-formation pathways in the past, across many species. Alternatively, it seems possible that the demanding physical and physiological constraints inherent in muscle-powered flight put bats at an edge of what is evolutionarily achievable. In that case, one might expect to see, after speciation events, comparatively more positive selection in wing- and flight-related genes compared with genes involved in other adaptations, with collagen an indicator of the former set.
In summary, we have provided a way to combine genomic and transcriptomic data to build reliable multiple sequence alignments. We have created multiple sequence alignments for bat genes and made them publicly available. We have used these to produce a phylogeny and to assess positive selection in bat genes. Despite this progress, bats will continue to present challenges that push the limits of genomics and phylogenetics because of their high levels of sequence divergence.
Methods
Transcriptome Sequencing of H. monstrosus and R. aegyptiacus.
RNA was isolated from the immortalized cell lines HypLu/45.1 and HypNi/1.1 (H. monstrosus) and RoNi/7.1 (R. aegyptiacus) (94) using the AllPrep DNA/RNA mini kit (Qiagen- 80204). mRNA isolation, clean up, and library prep was done by the Genome Sequencing and Analysis Facility at University of Texas at Austin. Total RNA was enriched for mRNA using the Poly(A) Purist Magnetic kit (Life Technologies; AM1922). The mRNA was fragmented using the NEBNext Mg Fragmentation module (NEB; E6150S) and column purified using RNeasy MinElute kit (Qiagen; 74204). mRNA ends were repaired using T4 Polynucleotide kinase (NEB; M0201L) and ATP (Ambion; AM8110G). A final concentration step was run using the RiboMinus Concentration Module (ThermoFisher; K155005). Adaptors were ligated (NEB; E6120L), and mRNA was converted to cDNA (Invitrogen; 18080093). The library was then PCR amplified and size selected. H. monstrosus cDNA was sequenced on Illumina HiSeqs 2000 and 2500, both generating 2 × 101 bp paired end reads, and R. aegyptiacus cDNA was sequenced on Illumina HiSeq 2000 and NextSeq, with 2 × 101 bp and 2 × 151 bp reads, respectively. Reads are available on the Sequence Read Archive under project numbers SRP158567 and SRP158571. Assembled transcripts are available on GenBank at accession numbers GHDN00000000 and GHDO00000000.
Data Cleaning and Assembly.
Sequencing data were first cleaned to remove sequencing adaptors and low-quality bases with Trimmomatic (39), with appropriate settings for each dataset. Trinity (40) was run with all reads, with all reads with in silico normalization, and with ∼35 million read subsamples for those bats with large datasets as recommended in Francis et al. (95). Trans-ABySS (41) was run with k-mer lengths of 32, 64, and all multiples of 5 from 25 to 60.
For genomic data, loci annotated as alternative loci were ignored, as well as readthrough genes. A few instances appeared with isoforms labeled as separate genes; these were manually reduced to one longest isoform.
Ortholog Search.
First, we found orthologs between all species using only the genomic datasets, where syntenic evidence helps confirm orthology rather than paralogy (see below). The first step was all-vs.-all BLAST-ing, filtered for e values no higher than 10−5. Reciprocal BLAST hits (for limitations, see ref. 96) were considered as tentative ortholog predictions (43). Tentative predictions were further filtered by including evidence from public ortholog annotation in Biomart and, where available, syntenic evidence (see below). The resulting ortholog prediction graphs for each gene were filtered to remove any connected components with paths connecting two genes from the same species.
Next, we added orthologs from the transcriptomes. First, genomic transcripts were translated to amino acid sequence, and blastx-tblastn reciprocal BLAST hits were found between all genomic high-quality orthologous genes and all transcriptome assemblies, again using an e-value cutoff of 10−5. We then created HMMER models of each of the genomic orthologous gene sets and searched within the reciprocal BLAST hit contigs for the best HMMER hit for each gene, filtering for those hits with e value below 10−10, extracting only the portion of each contig specified in the HMMER hit. We then filtered all transcripts which differed in length from the median genomic sequence length by more than 25%.
Syntenic Evidence.
Whole-genome alignments were performed following a procedure described on the University of California, Santa Cruz, Genome Browser wiki, which for our purposes only required aligning, chaining, and netting. Alignment was performed using Lastz (97), while chaining and netting were performed with kentUtils (45). Tentative orthologs are considered to have evidence of synteny if they are in syntenic regions, as defined by the top-level nodes of the net.
The algorithm used for determining gene-proximity evidence of synteny is a simple extension of the algorithm in Jarvis et al. (44). Let species A and species B have genes a1 and b1, respectively, which are tentatively orthologs. Then let a2 be the nearest gene to a1 on the same chromosome which has a tentative ortholog in species B, b2. If the number of genes between a1 and a2 and the number of genes between b1 and b2 are both less than 5, then we consider the ortholog pair a1–b1 to have syntenic evidence. If there are at least five genes in each direction, but the above is not true, then there is evidence against synteny. In the case of not enough genes to either side, it is undetermined.
Multiple Sequence Alignments and Best Genomic Isoforms.
Multiple sequence alignments were generated on amino acid sequences with MAFFT L-INS-i, the slower but more accurate version of the popular MAFFT alignment software (98). Manual inspection revealed that while the HMMER models had quite consistently found the same isoform from all of the transcriptomic datasets, the genomic data were slightly less consistent. This is not surprising, as different species can have different annotated longest isoforms. So, for each gene in each species with genomic data, we went back and found the isoform most similar to the consensus isoform.
The algorithm for finding best splice variant for a gene g in a given orthologous set S is as follows. First, find the consensus sequence of S from the MAFFT L-INS-i alignment. The consensus sequence is the identity in each column of the amino acid which is found in a majority of transcripts, or X if no single value is the majority. Next, align all splice variants of gene g against the consensus sequence, again using MAFFT L-INS-i, and score each by the number of nongap, non-X positions in agreement with the consensus. Select the splice variant with the highest score. This resulted in improved selection of 3,444 splice variants.
Finally, we realigned for final gene alignments. Corresponding CDS alignments were created using pal2nal (99).
MIXR.
We developed the MIXR software to detect and remove alternate consensus runs (available at http://github.com/hawkjo/mixr). The MIXR algorithm finds the maximum alternate-consensus-run score for each hypothesis bipartition of species and removes exons which overlap runs with significant scores—or species if removing exons would eliminate the entire alignment (see flowchart in SI Appendix, Fig. S3). To implement this algorithm outline, we needed to define three things: hypothesis bipartitions of species, the alternate-consensus-run scoring function, and the method used to determine significance.
First, we define hypothesis bipartitions of species. Scoring all bipartitions is computationally intractable because for n species, there are 2n bipartitions. We use as a heuristic the set of all bipartitions in the alignment, by which we mean subsets of species A and B for which there exists a column i in the protein alignment where the sets of amino acids in Ai and Bi are disjoint, i.e., no amino acid in Ai is in Bi and vice versa. Note that this step does not require internal agreement within Ai or Bi, just disjointness between them. On average, this required looking at fewer than 23 bipartitions per alignment.
Second, we define the scoring function. Let A and B be a bipartition of species such that A is the larger (or equal) subset, and let Ai and Bi be the corresponding sets of amino acids in the ith column. We wish to reward internal agreement within B and penalize agreement between A and B. Let M be a log-likelihood amino acid transition scoring matrix. We here use PAM30 from the NCBI toolkit, the scoring matrix used by BLAST recommended for detecting shorter sequences (100). Then we define the score of the ith alignment column, Si, to be
if Si so calculated is positive and zero otherwise, where S0 = 0. That is, the minimum agreement in B must be high and the maximum agreement between A and B low for several columns in a row to gain a high score. The factor of 2 in the second term reflects penalizing both agreement from A to B and B to A and is necessary to penalize complete agreement between A and B.
Finally, to determine significance, we build and score negative control sequences. The negative control sequences are constructed by shuffling all columns from all alignments which contain all species to construct sequences at least 100× as long as the longest alignment. For each alignment, we select the negative control sequences corresponding to the species in the alignment, truncate them to 100× the length of the alignment in question, and find the maximum score for each hypothesis bipartition. Alternate-consensus-run scores larger than their corresponding negative control score thus have P values ≲ 0.01, and are considered significant, to be removed as shown in flowchart SI Appendix, Fig. S3. No multiple hypothesis correction is performed, conservatively removing borderline cases.
Phylogenetic Analyses.
Partitioned analyses in Mr. Bayes (48) used two runs with four chains, the 4 × 4 model run for 1,000,000 steps and the full codon model for 30,000 steps. Gamma models were also subsampled, resulting in a >99% reported posterior probability for the gtrsubmodel[123456] model under the 4 × 4 model and six models with >5% posterior probability for the full codon model: gtrsubmodels 123425 (24%), 123454 (17%), 123456 (17%), 123424 (16%), 123121 (10%), and 123124 (10%). RAxML (49) was run with the rapid bootstrapping and ML algorithm, the GTRGAMMA and PROTGAMMAGTR models, and with 100 different starting trees. Gene trees were created using Mr. Bayes using 100,000 steps sampled every 10 steps with 20,000 step burn-in and an inverse gamma model.
Positive Selection Analysis.
Positive selection analysis was performed on the 10,650 orthologous genes alignments that had at least six species and 30 amino acids represented in the alignment. For each analysis, we used the species tree generated in this study. Using PAML (61), each alignment was fit to the M8 and M8a codon models, both performed with the f61 codon model. Both the M8 and M8a models assume the dN/dS value for a given aligned codon is either a draw from a binned beta distribution (with fit parameters a and b) or a draw from a floating bin with parameter dN/dS = ω. The parameter ω can float to values >1 in M8 and is set to ω = 1 for M8a. P values were calculated via χ2 test on twice the difference in reported log likelihoods of the two models, a likelihood-ratio test of nested models. GO category enrichment among positively selected genes was performed with the STRING database online software (101).
Software Versions.
Software versions used in this project were Trimmomatic 0.32, Trinity 20140717, Trans-ABySS 1.5.1, BLAST 2.2.29+, HMMER 3.1b2, MAFFT 7.221beta, Lastz 1.02.00, kentUtils 302, Mr. Bayes 3.2.6, RAxML 8.2.6, ASTRAL 4.7.8, FigTree v1.4.2, PAML 4.8, and STRING 11.0.
Supplementary Material
Acknowledgments
We thank the Texas Advanced Computing Center for hundreds of thousands of hours of compute time. Thanks to Claire Hemingway, Kathleen Lyons, Siavash Mirarab, Susan Tsang, and Qing Yang for valuable insights into the project and manuscript. This work was supported by NIH grant R01-AI-137011 (to S.L.S.), DFG grant SPP 1596 (DR 772/10-2; to C.D.), and the RAPID consortium of the Bundesministerium für Bildung und Forschung (01KI1723A; to C.D.). M.E.K. is supported by a National Science Foundation Graduate Research Fellowship, and S.L.S. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease.
Footnotes
The authors declare no conflict of interest.
Data deposition: MIXR software package is available on GitHub at http://github.com/hawkjo/mixr. Multiple sequence alignments are available at http://numerical.recipes/chiroptera/. Assembled transcripomic sequences for H. monstrosus and R. aegyptiacus are available on GenBank (accession nos. GHDN00000000 and GHDO00000000). Raw sequenced reads are available at the Sequence Read Archive (accession nos. SRP158567 and SRP158571).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814995116/-/DCSupplemental.
References
- 1.Calisher C. H., Childs J. E., Field H. E., Holmes K. V., Schountz T., Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19, 531–545 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Society of Mammalogists, Mammal Diversity Database. https://mammaldiversity.org/. Accessed 18 June 2018.
- 3.Olival K. J., et al. , Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith I., Wang L.-F., Bats and their virome: An important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 3, 84–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leroy E. M., et al. , Fruit bats as reservoirs of Ebola virus. Nature 438, 575–576 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Leroy E. M., et al. , A serological survey of Ebola virus infection in central African nonhuman primates. J. Infect. Dis. 190, 1895–1899 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Towner J. S., et al. , Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 5, e1000536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Towner J. S., et al. , Marburg virus infection detected in a common African bat. PLoS One 2, e764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones K. E., Purvis A., MacLarnon A., Bininda-Emonds O. R. P., Simmons N. B., A phylogenetic supertree of the bats (Mammalia: Chiroptera). Biol. Rev. Camb. Philos. Soc. 77, 223–259 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Eick G. N., Jacobs D. S., Matthee C. A., A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera). Mol. Biol. Evol. 22, 1869–1886 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Teeling E. C., et al. , A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307, 580–584 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Agnarsson I., Zambrana-Torrelio C. M., Flores-Saldana N. P., May-Collado L. J., A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia). PLoS Curr. 3, RRN1212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsagkogeorga G., Parker J., Stupka E., Cotton J. A., Rossiter S. J., Phylogenomic analyses elucidate the evolutionary relationships of bats. Curr. Biol. 23, 2262–2267 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Shi J. J., Rabosky D. L., Speciation dynamics during the global radiation of extant bats. Evolution 69, 1528–1545 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Lei M., Dong D., Phylogenomic analyses of bat subordinal relationships based on transcriptome data. Sci. Rep. 6, 27726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amador L. I., Moyers Arévalo R. L., Almeida F. C., Catalano S. A., Giannini N. P., Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. J. Mamm. Evol. 25, 37–70 (2018). [Google Scholar]
- 17.Yang Z., Rannala B., Molecular phylogenetics: Principles and practice. Nat. Rev. Genet. 13, 303–314 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Daugherty M. D., Malik H. S., Rules of engagement: Molecular insights from host-virus arms races. Annu. Rev. Genet. 46, 677–700 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Meyerson N. R., Sawyer S. L., Two-stepping through time: Mammals and viruses. Trends Microbiol. 19, 286–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandl J. N., Schneider C., Schneider D. S., Baker M. L., Going to bat(s) for studies of Disease tolerance. Front. Immunol. 9, 2112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura M., Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 267, 275–276 (1977). [DOI] [PubMed] [Google Scholar]
- 22.Yang Z., Bielawski J. P., Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. (Amst.) 15, 496–503 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G., et al. , Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339, 456–460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y.-Y., et al. , Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. U.S.A. 107, 8666–8671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavlovich S. S., et al. , The Egyptian Rousette genome reveals unexpected features of bat antiviral immunity. Cell 173, 1098–1110.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee A. K., et al. , De novo transcriptome reconstruction and annotation of the Egyptian rousette bat. BMC Genomics 16, 1033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw T. I., et al. , Transcriptome sequencing and annotation for the Jamaican fruit bat (Artibeus jamaicensis). PLoS One 7, e48472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong D., Lei M., Liu Y., Zhang S., Comparative inner ear transcriptome analysis between the Rickett’s big-footed bats (Myotis ricketti) and the greater short-nosed fruit bats (Cynopterus sphinx). BMC Genomics 14, 916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Low D. H. W., et al. , Dracula’s children: Molecular evolution of vampire bat venom. J. Proteomics 89, 95–111 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Gracheva E. O., et al. , Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats. Nature 476, 88–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francischetti I. M. B., et al. , The “Vampirome”: Transcriptome and proteome analysis of the principal and accessory submaxillary glands of the vampire bat Desmodus rotundus, a vector of human rabies. J. Proteomics 82, 288–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z., et al. , Unique expression patterns of multiple key genes associated with the evolution of mammalian flight. Proc. Biol. Sci. 281, 20133133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fushan A. A., et al. , Gene expression defines natural changes in mammalian lifespan. Aging Cell 14, 352–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L., et al. , Deep RNA sequencing reveals complex transcriptional landscape of a bat adenovirus. J. Virol. 87, 503–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips C. J., et al. , Dietary and flight energetic adaptations in a salivary gland transcriptome of an insectivorous bat. PLoS One 9, e83512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papenfuss A. T., et al. , The immune gene repertoire of an important viral reservoir, the Australian black flying fox. BMC Genomics 13, 261 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei M., Dong D., Mu S., Pan Y.-H., Zhang S., Comparison of brain transcriptome of the greater horseshoe bats (Rhinolophus ferrumequinum) in active and torpid episodes. PLoS One 9, e107746 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips C. D., Baker R. J., Secretory gene recruitments in vampire bat salivary adaptation and potential convergences with sanguivorous leeches. Front. Ecol. Evol. 3, 122 (2015). [Google Scholar]
- 39.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabherr M. G., et al. , Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson G., et al. , De novo assembly and analysis of RNA-seq data. Nat. Methods 7, 909–912 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Martin J. A., Wang Z., Next-generation transcriptome assembly. Nat. Rev. Genet. 12, 671–682 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jarvis E. D., et al. , Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kent J., kentUtils. https://github.com/ENCODE-DCC/kentUtils. Accessed 15 April 2016.
- 46.Eddy S., HMMER User’s Guide (Biological Sequence Analysis Using Profile Hidden Markov Models, 2003). [Google Scholar]
- 47.Larkin M. A., et al. , Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Ronquist F., Huelsenbeck J. P., MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirarab S., et al. , ASTRAL: Genome-scale coalescent-based species tree estimation. Bioinformatics 30, i541–i548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Springer M. S., Teeling E. C., Madsen O., Stanhope M. J., de Jong W. W., Integrated fossil and molecular data reconstruct bat echolocation. Proc. Natl. Acad. Sci. U.S.A. 98, 6241–6246 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoofer S. R., Reeder S. A., Hansen E. W., Bussche R. A. V. D., Molecular phylogenetics and taxonomic review of noctilionoid and vespertilionoid bats (Chiroptera: Yangochiroptera). J. Mammal. 84, 809–821 (2003). [Google Scholar]
- 53.Hoofer S. R., Bussche R. A. V. D., Molecular phylogenetics of the chiropteran family vespertilionidae. Acta Chiropt. 5 (Suppl), 1–63 (2003). [Google Scholar]
- 54.Stubbe M., et al. , Murina leucogaster. The IUCN Red List of Threatened Species 2016 (International Union for Conservation of Nature, 2016), p e.T13943A22093328. [Google Scholar]
- 55.Hutson A.M., et al. , Myotis brandtii: The IUCN Red List of Threatened Species 2008 (International Union for Conservation of Nature, 2008), p e.T14125A4397500. [Google Scholar]
- 56.Smith A. T., Johnston C. H., Jones G., S. Rossiter, Myotis davidii: The IUCN Red List of Threatened Species 2008 (International Union for Conservation of Nature, 2008), p e.T136250A4265409. [Google Scholar]
- 57.Arroyo-Cabrales J., Álvarez-Castañeda S. T., Myotis lucifugus: The IUCN Red List of Threatened Species 2008 (International Union for Conservation of Nature, 2008), p e.T14176A4415629. [Google Scholar]
- 58.Csorba G., Bates P., Myotis pilosus: The IUCN Red List of Threatened Species 2008 (International Union for Conservation of Nature, 2008), p e.T14193A4418772. [Google Scholar]
- 59.Wetterer A. L., Rockman M. V., Simmons N. B., Phylogeny of phyllostomid bats (mammalia: Chiroptera): Data from diverse morphological systems, sex chromosomes, and restriction sites. Bull. Am. Mus. Nat. Hist. 248, 1–200 (2000). [Google Scholar]
- 60.Rojas D., Warsi O. M., Dávalos L. M., Bats (chiroptera: Noctilionoidea) challenge a recent origin of extant neotropical diversity. Syst. Biol. 65, 432–448 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Banerjee A., Misra V., Schountz T., Baker M. L., Tools to study pathogen-host interactions in bats. Virus Res. 248, 5–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker M. L., Schountz T., Wang L.-F., Antiviral immune responses of bats: A review. Zoonoses Public Health 60, 104–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan L., et al. , Adaptive evolution of Leptin in heterothermic bats. PLoS One 6, e27189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin Q., et al. , Molecular evolution of the nuclear factor (Erythroid-Derived 2)-like 2 gene Nrf2 in old world fruit bats (Chiroptera: Pteropodidae). PLoS One 11, e0146274 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen B., Han X., Zhang J., Rossiter S. J., Zhang S., Adaptive evolution in the glucose transporter 4 gene Slc2a4 in Old World fruit bats (family: Pteropodidae). PLoS One 7, e33197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen B., Han X., Jones G., Rossiter S. J., Zhang S., Adaptive evolution of the myo6 gene in old world fruit bats (family: Pteropodidae). PLoS One 8, e62307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang L., et al. , Adaptive evolution of the Hox gene family for development in bats and dolphins. PLoS One 8, e65944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li G., Wang J., Rossiter S. J., Jones G., Zhang S., Accelerated FoxP2 evolution in echolocating bats. PLoS One 2, e900 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li G., et al. , The hearing gene Prestin reunites echolocating bats. Proc. Natl. Acad. Sci. U.S.A. 105, 13959–13964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang L., Shen B., Irwin D. M., Zhang S., Parallel evolution of the glycogen synthase 1 (muscle) gene Gys1 between Old World and New World fruit bats (order: Chiroptera). Biochem. Genet. 52, 443–458 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Chen Y., Shen B., Zhang J., Jones G., He G., Cloning and molecular evolution of the aldehyde dehydrogenase 2 gene (Aldh2) in bats (Chiroptera). Biochem. Genet. 51, 7–19 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Zhou Y., Dong D., Zhang S., Zhao H., Positive selection drives the evolution of bat bitter taste receptor genes. Biochem. Genet. 47, 207–215 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Demogines A., Farzan M., Sawyer S. L., Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J. Virol. 86, 6350–6353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Escalera-Zamudio M., et al. , The evolution of bat nucleic acid-sensing Toll-like receptors. Mol. Ecol. 24, 5899–5909 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Fuchs J., et al. , Evolution and antiviral specificities of interferon-induced Mx proteins of bats against Ebola, influenza, and other RNA viruses. J. Virol. 91, e00361-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He G., He B., Racey P. A., Cui J., Positive selection of the bat interferon alpha gene family. Biochem. Genet. 48, 840–846 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang H., et al. , Selective evolution of Toll-like receptors 3, 7, 8, and 9 in bats. Immunogenetics 69, 271–285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kammerer R., et al. , Recent expansion and adaptive evolution of the carcinoembryonic antigen family in bats of the Yangochiroptera subgroup. BMC Genomics 18, 717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schad J., Voigt C. C., Adaptive evolution of virus-sensing toll-like receptor 8 in bats. Immunogenetics 68, 783–795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McBee R. M., Rozmiarek S. A., Meyerson N. R., Rowley P. A., Sawyer S. L., The effect of species representation on the detection of positive selection in primate gene data sets. Mol. Biol. Evol. 32, 1091–1096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Lee R., Wiel L., van Dam T. J. P., Huynen M. A., Genome-scale detection of positive selection in nine primates predicts human-virus evolutionary conflicts. Nucleic Acids Res. 45, 10634–10648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cagan A., et al. , Natural selection in the great apes. Mol. Biol. Evol. 33, 3268–3283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiao J., et al. , Transcriptome analysis revealed positive selection of immune-related genes in tilapia. Fish Shellfish Immunol. 44, 60–65 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Roux J., et al. , Patterns of positive selection in seven ant genomes. Mol. Biol. Evol. 31, 1661–1685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou P., et al. , Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc. Natl. Acad. Sci. U.S.A. 113, 2696–2701 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hayward J. A., et al. , Differential evolution of antiretroviral restriction factors in pteropid bats as revealed by APOBEC3 gene complexity. Mol. Biol. Evol. 35, 1626–1637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schountz T., Baker M. L., Butler J., Munster V., Immunological control of viral infections in bats and the emergence of viruses highly pathogenic to humans. Front. Immunol. 8, 1098 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ricard-Blum S., The collagen family. Cold Spring Harb. Perspect. Biol. 3, a004978 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Townsley M. I., Structure and composition of pulmonary arteries, capillaries, and veins. Compr. Physiol. 2, 675–709 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheney J. A., Allen J. J., Swartz S. M., Diversity in the organization of elastin bundles and intramembranous muscles in bat wings. J. Anat. 230, 510–523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ceballos-Vasquez A., Caldwell J. R., Faure P. A., Seasonal and reproductive effects on wound healing in the flight membranes of captive big brown bats. Biol. Open 4, 95–103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.O’Donoghue A. J., et al. , Destructin-1 is a collagen-degrading endopeptidase secreted by Pseudogymnoascus destructans, the causative agent of white-nose syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, 7478–7483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kühl A., et al. , Comparative analysis of Ebola virus glycoprotein interactions with human and bat cells. J. Infect. Dis. 204 (Suppl_3), S840–S849 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Francis W. R., et al. , A comparison across non-model animals suggests an optimal sequencing depth for de novo transcriptome assembly. BMC Genomics 14, 167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dalquen D. A., Dessimoz C., Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals. Genome Biol. Evol. 5, 1800–1806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harris R. S., “Improved pairwise alignment of genomic DNA,” Dissertation, The Pennsylvania State University, (2007).
- 98.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suyama M., Torrents D., Bork P., PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic. Acids. Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pearson W. R., Selecting the right similarity‐scoring matrix. Curr. Protoc. Bioinforma. 43, 3.5.1–3.5.9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.von Mering C., et al. , STRING: Known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic. Acids. Res. 33 (Suppl_1), D433–D437 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rambaut A., FigTree, a graphical viewer of phylogenetic trees. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 19 June 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.