Abstract
Instability of transgene expression in plants is often associated with complex multicopy patterns of transgene integration at the same locus, as well as position effects due to random integration. Based on maize transposable elements Activator (Ac) and Dissociation (Ds), we developed a method to generate large numbers of transgenic barley (Hordeum vulgare var Golden Promise) plants, each carrying a single transgene copy at different locations. Plants expressing Ac transposase (AcTPase) were crossed with plants containing one or more copies of bar, a selectable herbicide (Basta) resistance gene, located between inverted-repeat Ds ends (Ds-bar). F1 plants were self-pollinated and the F2 generation was analyzed to identify plants segregating for transposed Ds-bar elements. Of Ds-bar transpositions, 25% were in unlinked sites that segregated from vector sequences, other Ds-bar copies, and the AcTPase gene, resulting in numerous single-copy Ds-bar plants carrying the transgene at different locations. Transgene expression in F2 plants with transposed Ds-bar was 100% stable, whereas only 23% of F2 plants carrying Ds-bar at the original site expressed the transgene product stably. In F3 and F4 populations, transgene expression in 81.5% of plants from progeny of F2 plants with single-copy, transposed Ds-bar remained completely stable. Analysis of the integration site in single-copy plants showed that transposed Ds-bar inserted into single- or low-copy regions of the genome, whereas silenced Ds-bar elements at their original location were inserted into redundant or highly repetitive genomic regions. Methylation of the non-transposed transgene and its promoter, as well as a higher condensation of the chromatin around the original integration site, was associated with plants exhibiting transgene silencing.
The successful introduction of transgenic crops into modern farming practices depends on maintaining and improving the agricultural performance of the modified plants and on the stable and predictable transmission and expression of the transgene in successive generations during seed production and commercial cultivation (McElroy, 1999). Inactivation of transgene expression has often been observed in plants and seems to be especially problematic in cereal crops. In these crop species the most frequently used biolistic transformation methods typically result in complex, multicopy transgene integration patterns, where it has been observed that more than 50% of T1 plants exhibit transgene silencing (Wan and Lemaux, 1994; Pawlowski et al., 1998; for review, see Iyer et al., 2000). Inactivation of expression in plants with single copies of the transgene can occur (Meyer and Heidmann, 1994), but it is not as frequent as the silencing observed when multiple copies of the transgene exist.
In addition to effects of copy number on transgene silencing (Assaad et al., 1993; Atkinson et al., 1998), this phenomenon is influenced by many other factors. These include the presence of inverted repeats in the complex integration patterns (Stam et al., 1997a, 1997b), the overexpression of the transgene (Que et al., 1997), the nature of the insertion site (Matzke and Matzke, 1998), the AT/CG composition of the transgene (Matzke and Matzke, 1998), and environmental factors (Meyer et al., 1992). These factors can trigger different mechanisms of transcriptional or post-transcriptional gene silencing.
Due to the exacerbated problems associated with transgene silencing, and especially silencing in multicopy insertion events, it is desirable to generate transgenic plants containing only a single copy of the transgene. With existing cereal transformation methods, however, the number of single-copy transgenic plants generated is usually low relative to the number of plants containing multicopy events. This is particularly problematic in crop species that are not readily transformed, since obtaining the large numbers of independent transformants to find the infrequent single insert is time consuming and costly. Furthermore, although it is possible with sufficient effort to identify a sufficient number of single-copy insertion events, this approach alone will not always overcome transgene expression instability problems. Transgene silencing has also been observed in events with single, simple-pattern transgene inserts associated with, for example, location of the insert, GC content of the region, and presence of vector DNA.
Attempts have been made to increase the number of single-copy transgenic plants. In general, Agrobacterium-mediated transformation leads to lower copy numbers of the transgene, and single-copy inserts in transgenic plants are observed more frequently. For some cereals, however, this method is not routine. Even though single-copy plants can be obtained more frequently in dicot and monocot (Cheng et al., 1997; Tingay et al., 1997) species, the use of Agrobacterium-mediated transformation does not circumvent the problem of transgene silencing (e.g. Ishida et al., 1996; Park et al., 1996). Other methods have been employed to increase the number of single-copy inserts, including the use of an Agrolistic method (Hansen and Chilton, 1996) and inhibitors of recombination (De Block et al., 1997). Another successful approach has been used in wheat and utilizes the bacteriophage cre/lox recombination system to resolve the complex integration patterns of multiple transgenes into single-copy integration patterns (Srivastava et al., 1999). However, these approaches do not always overcome transgene expression stability problems.
We developed a system in barley (Hordeum vulgare var Golden Promise) based on the maize transposons Activator (Ac) and Dissociation (Ds). In this system the transgene is inserted between the nonautonomous Ds-element inverted repeats and is translocated to different loci in the genome as a result of the action of Ac transposase (AcTPase). The Ds cassette carries bar (Ds-bar), which encodes phosphinothricin acetyl transferase, which confers resistance to the herbicide, Basta. This cassette can transpose to genetically unlinked sites, which segregate in the next generation from the remaining transgene copies, the other vector sequences, and the AcTPase gene. This method requires only a few original transformation events and results in high numbers of single-copy transgenic plants, each carrying a stabilized transgene at different genomic locations. Transposons are also subject to silencing (Fedoroff and Chandler, 1994), although at much reduced frequencies relative to most transgenes. Here we discuss the influence of transgene copy number and the nature of the integration site of the transposed Ds-bar element on transgene expression stability.
RESULTS
Single-Copy Transgenic Plants
From crosses between plants expressing functional AcTPase and plants from three independent transformants that contain one to six copies of Ds-bar we obtained numerous families of F1 plants carrying both elements. Progeny of four self-pollinated F1 plants were analyzed (200 F2 plants derived from line A8-1 and 100 F2 plants from A8-5, A1-5, and A18-5) to identify F2 plants containing a single copy of transposed Ds-bar that had segregated away from the gene encoding AcTPase (AcTPase) and other copies of Ds-bar. These plants were designated “TNP” (transposed) in contrast to segregating plants designated “nTNP” (non-transposed) carrying Ds-bar at the original integration sites and also not containing AcTPase.
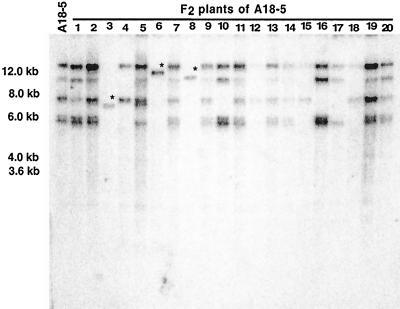
Among 200 F2 progeny (designated a “family”) from F1 plant A8-1, which contained a single Ds-bar element at its original integration site after microparticle bombardment, we identified 21 plants (designated “lines”) with a single Ds-bar element transposed to a new position, as confirmed by PCR and DNA-blot hybridization analysis. From F1 plant A8-5, a sibling of A8-1 also carrying a single Ds-bar element at its original integration site, we obtained 17 F2 TNP plants (hereafter referred to as lines). Among progeny of F1 plants A1-5 and A18-5, which have different and independently transformed parents from the A8-1 and A8-5 families and which contain six and five copies of Ds-bar, respectively, we identified in the F2 generation five TNP lines from each family. Figure 1 shows a DNA-blot hybridization of samples from different F2 lines derived from A18-5. In this blot the hybridization pattern of DNA from three plants containing a single copy of Ds-bar is seen as different-sized HindIII fragments. Each of the HindIII sites is unique and is predicted to be in the genomic DNA flanking the site of Ds-bar integration. The presence or absence of AcTPase was, in all cases, analyzed by PCR using AcTPase-specific primers (data not shown). Other single-copy events were analyzed in an identical manner (data not shown).
Figure 1.
Transposon-mediated generation of single-copy transgenic plants. DNA-blot hybridization shows the integration pattern of Ds-bar after HindIII digest of genomic DNA from F1 parent, A18-5 (far left lane), and 20 of its F2 progeny. In three plants (i.e. nos. 3, 6, and 8), a single Ds-bar cassette has transposed to an unlinked site, segregating away from other copies. The asterisk above the band marks the position in single-copy Ds-bar plants that differs from the parental bands. Mr markers in kilobases are indicated on the left.
Transgene and Transgene Expression Stability
The physical transmission of bar from the single-copy TNP lines followed Mendelian segregation in the F3 generation in the newly created single-copy, Ds-bar plants, as evidenced by DNA-blot hybridization analysis of genomic DNA. In the TNP and nTNP lines from families A8-1, A8-5, A1-5, and A18-5, functional expression of the bar gene product was analyzed in later generations by monitoring herbicide resistance. While screening the F2 populations from the four families to identify single-copy Ds-bar individuals, plants were painted with Basta. All TNP lines in the F2 generation were completely resistant to the herbicide. In contrast, up to 40% of all nTNP lines derived from the four families showed reduced numbers of Basta-resistant plants, consistent with transgene silencing.
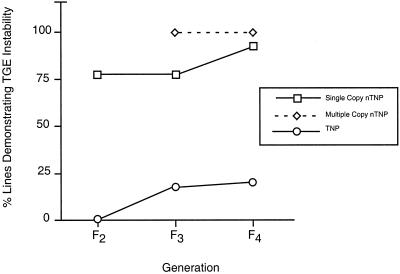
Transgene-expression stability of lines derived from the four families was monitored in advanced generations in 17 nTNP lines and 35 TNP lines, all of which were Basta-resistant in the F2 generation. Between 40 and 50 F3 and F4 plants, derived from each of the 52 F2 lines, were tested for herbicide sensitivity. To avoid damage or death of the entire plant in the case of herbicide sensitivity and to ensure seed set, only one leaf of each plant was painted with Basta. We observed no expression of the bar gene product in F3 progeny of 14 of the 17 nTNP lines (82.4%), as evidenced by sensitivity of the plants in the line to Basta (Table I). In the F3 generation 100% of all nTNP lines derived from multicopy families A1-5 and A18-5 contained plants in which Basta sensitivity was evident. In contrast, only six of the 35 TNP lines (17.1%) produced F3 progeny showing transgene silencing (Table I). The number of plants within nTNP lines that showed evidence of transgene silencing was significantly higher statistically (P < 0.1%; chi-square analysis) than within TNP lines (Table I). Even though only seeds from Basta-resistant F3 plants were collected, transgene silencing continued to persist into the F4 generation. Only one of 17 nTNP lines produced completely resistant F4 plants; 94% of all nTNP lines contained between 1% and 48% of plants with silencing of the bar gene product (Table I; Fig. 2). It is striking that all 29 TNP lines, which were completely Basta-resistant in F3, also produced completely resistant F4 plants. Among F4 progeny from the six TNP lines that exhibited transgene silencing in the F3 generation, plants were again Basta-sensitive in numbers comparable with those in the nTNP lines (data not shown). Progeny of one TNP plant line (A1-5-54) derived from family A1-5 were completely Basta-resistant in the F3 generation, but showed signs of transgene expression instability in F4.
Table I.
Transgene expression stability in nTNP and TNP plant lines
| F1 Plant | No. of Copies | No. of Lines with Transgene Expression Instability/Total Lines Analyzed
|
|||||
|---|---|---|---|---|---|---|---|
| F2
|
F3
|
F4
|
|||||
| nTNPa | TNPb | nTNP | TNP | nTNP | TNP | ||
| A8-1 | 1 | 6/8 (75%) | 0/15 (0%) | 6/8 (75%) | 2/15 (13%) | 7/8 (88%) | 2/15 (13%) |
| A8-5 | 1 | 4/5 (80%) | 0/10 (0%) | 4/5 (90%) | 2/10 (20%) | 5/5 (100%) | 2/10 (20%) |
| A1-5 | 6 | N.D. | N.D. | 2/2 (100%) | 1/5 (20%) | 2/2 (100%) | 2/5 (40%) |
| A18-5 | 5 | N.D. | N.D. | 2/2 (100%) | 1/5 (20%) | 2/2 (100%) | 1/5 (20%) |
nTNP lines are “non-transposed,” containing Ds-bar at the original integration site and do not contain AcTPase.
TNP lines are “transposed” and are progeny of F1 plants selected for a single copy of Ds-bar transposed to a new location.
Figure 2.
Average frequency of transgene expression instability in F2, F3, and F4 populations. Lines from F2, F3, and F4 populations were assessed for functional expression of the product of bar by testing for Basta sensitivity. Percentages represent the number of lines in which at least one plant exhibited Basta sensitivity divided by the total number of lines per family tested. TNP, Transposed Ds-bar element, no ActPase; nTNP, Ds-bar element remaining at original site of integration following bombardment, no AcTPase; TGE, transgene expression.
Analysis of the Integration Sites of Ds-bar
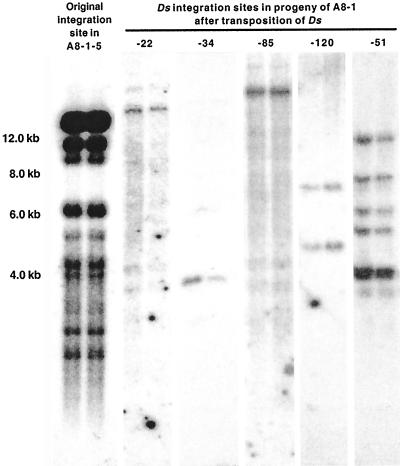
To analyze the redundancy of the genomic region into which the Ds-bar element from single-copy nTNP and TNP plants had inserted, DNA blots of HindIII-digested DNA from non-transgenic barley plants (var Golden Promise) were hybridized with probes from genomic regions flanking the Ds element. Flanking regions were isolated from the single-copy nTNP plant (A8-1-5), three silenced TNP plants (e.g. see Fig. 3, A8-1-51), and six stably expressing TNP plants (e.g. see Fig. 3, A8-1-22, -34, -85, and -120) that have Ds-bar in different locations. Analysis of the DNA blots showed that the single-copy Ds-bar element from the silenced nTNP plant A8-1-5 had integrated into a highly repetitive genomic region. Two of the three TNP plants with silenced transgene expression (e.g. see Fig. 3, A8-1-51) had transposed into highly or moderately repetitive regions of the barley genome. The Ds-bar elements from all stably expressing TNP plants had transposed to single-copy or low-copy regions (up to three hybridizing fragments) of the barley genome (e.g. Fig. 3, A8-1-22, -34, -85, and -120 and data not shown). Only one line, A8-1-25, in which Ds-bar had inserted into a non-repetitive genomic region (data not shown), exhibited signs of transgene silencing and the silencing was observed in only one of 31 (3%) bar-containing F4 plants.
Figure 3.
Analysis of Ds-bar integration sites. Genomic DNA flanking single-copy Ds-bar elements was isolated from six different transgenic plants by thermal asymmetric interlaced-PCR. PCR fragments were then used as hybridization probes with DNA blots of HindIII-digested genomic DNA from non-transgenic plants. The two lanes on left are results after hybridization with a probe specific to the genomic DNA flanking the original Ds-bar insertion site of line A8-1-5. Other sets of lanes show results after hybridization with probes flanking the insertion sites from five lines in which Ds-bar has transposed to a new location: four lines that stably express the transgene, e.g. nos. 22, 34, 85, 120, and one line that does not, 51. Mr markers in kilobases are indicated on left.
Methylation Analysis of the Promoter and Coding Region of Ds-bar
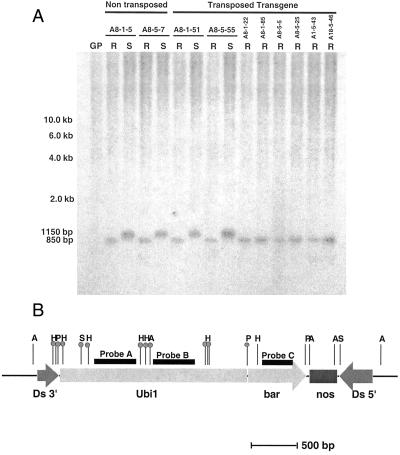
To assess the methylation status of the Ds-bar element a series of DNA hybridization blots were constructed to analyze the ubiquitin promoter region driving bar gene expression and the bar-coding region itself. DNAs from different silenced and non-silenced nTNP and TNP plants were digested with six different methylation-sensitive enzymes, and hybridization patterns with multiple probes were analyzed to provide information about the methylation status at specific restriction sites. The data in Figure 4A shows the results of digestion with HpaII; the positions of restriction sites located within the ubiquitin promoter and first intron region are indicated in Figure 4B. All DNA samples from Basta-sensitive plants showed evidence of methylation at certain HpaII sites in the ubiquitin promoter region. In contrast, DNA samples from Basta-resistant plants showed no evidence of methylation at these HpaII sites. Based on the map position of HpaII sites (Fig. 4B) and predicted fragment sizes after HpaII digestion, it appears that certain sites are more prone to becoming methylated than are others. For example, HpaII sites I and III were non-methylated in all plants shown in Figure 4A, resulting in fragment sizes of 850 or 1,150 bp after hybridization with probe A (Fig. 4B). Analysis of additional plants, however, showed that in a few cases HpaII sites I and III were hypermethylated (data not shown). In contrast to that situation, HpaII sites II and V were always methylated in silenced plants, as confirmed by hybridization with probes B and C (data not shown).
Figure 4.
A, Methylation analysis of Basta-sensitive and Basta-resistant plants from lines with transposed and non-transposed Ds-bar cassette. DNA blots of HpaII-digested genomic DNA from Basta-sensitive and Basta-resistant plants were hybridized with probe A (B) from the 5′ end of the ubiquitin promoter of the Ds-bar cassette. Lanes containing DNA from Basta-resistant plants have an 850-bp hybridization fragment, as expected for a non-methylated HpaII site in the ubiquitin promoter region. Digests of DNA from Basta-sensitive plants have fragments of 1,150 bp, indicating methylation of an HpaII site in the immediate 5′ region of probe A (B). R, Basta-resistant; S, Basta-sensitive; GP, Golden Promise non-transgenic control. B, Methylation analysis of promoter and coding region of Ds-bar cassette in Basta-sensitive lines. Diagram summarizes results of digests with different methylation-sensitive enzymes (A, AvaI; H, HpaII/MspI; P, PstI; Sc, SacII; and S1, SalI) and hybridization with three different probes (A, B, and C). HpaII/MspI sites are numbered I through VII from left to right. A total of 20 different Basta-sensitive plants from eight different lines were analyzed. Based on the size of hybridization fragments and information regarding the restriction sites in the Ds-bar cassette, the position of specific methylated sites was determined. In some cases different HpaII/MspI sites are not sufficiently separated to allow differentiation between individual sites. In accordance with this, closely connected sites were marked as methylated if at least one of the sites was methylated. A circle on top of the lines symbolizes methylation was detected at that site in at least some plants. Size bar indicates 500 bp.
Restriction digests with other methylation-sensitive enzymes showed an increased level of methylation in the promoter region in all silenced plants (data not shown). The methylation pattern within the analyzed promoter/intron region varied from plant to plant, but, in general, was higher in Basta-sensitive F4 plants than in F3 plants. Figure 4B shows a diagram of the analyzed promoter/intron region with the sites indicated that were found to be methylated in at least some plants. In most silenced plants only a few of the sites shown in Figure 4B were methylated or partially methylated; however, in non-silenced plants none of these sites were methylated. The highest degree of methylation was found in silenced single-copy nTNP plants and in silenced TNP plants in which the transposed Ds-bar element had reinserted into highly repetitive genomic regions (e.g. Fig. 3, plant A8-1-51).
DNase I Sensitivity Tests
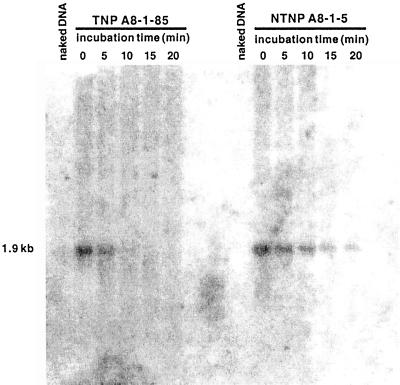
To determine if the silencing observed in nTNP and TNP plants is correlated with changes in the contiguous chromatin structure, the DNase I sensitivity of the transgene region was determined using isolated nuclei from several silenced and non-silenced plants. Chromatin of two silenced nTNP plants, four silenced TNP plants, and five non-silenced plants was analyzed. Figure 5 shows the results of a representative experiment. The hybridization signal of the expected 1.9-kb fragment in nTNP plant A8-1-5, which shows unstable expression and has a single Ds-bar element, decreases slowly over a 20-min period, indicative of low accessibility of the transgene region to DNase I. In contrast, in TNP line A8-1-85, which exhibits stable expression of the transgene, the hybridization signal decreases rapidly and disappears completely after a 15-min incubation period with DNase I. The transgene of a silenced TNP plant (A8-1-51) exhibited a similar low accessibility to DNase I as that observed in silenced nTNP plants. Analysis of the comparison of the DNase I sensitivity of different silenced and non-silenced TNP plants showed that silencing was associated with a compact chromatin structure in the vicinity of the transgene in 10 of 11 analyzed plants. The chromatin of one silenced TNP plant showed a DNase I sensitivity that was comparable with the sensitivity of chromatin in non-silenced TNP plants. The accessibility to DNase I varied between different non-silenced TNP lines, but was very similar between individual plants from the same TNP line (data not shown).
Figure 5.
Analysis of chromatin structure near Ds-bar cassettes from lines with a transposed or non-transposed Ds-bar cassette. Intact nuclei from plants with (i.e. TNP A8-1-85) or without (i.e. nTNP A8-1-5) transposed Ds-bar were treated with DNaseI for 0 to 20 min. DNA was isolated, digested with BglI to obtain fragments of a specific reference size (1.9 kb), electrophoretically separated, blotted, and hybridized with probe C (Fig. 4B). Naked DNA (no intact chromatin) from both plants, treated for 2 min with DNase I, served as a control.
DISCUSSION
Transgene expression instability is a common phenomenon in genetically transformed plants from monocot species (Iyer et al., 2000). The majority of cases of transgene silencing can be correlated with the existence of multicopy inserts, sequence rearrangements, and the random nature of integration into the genome, all of which occur frequently when methods of direct gene transfer are used for transformation (Hiei et al., 1994). Transgene silencing occurs in single-copy transgenic plants due to other factors such as insertion position, but the silencing is much less frequent than in multicopy plants. With most present methods of transformation, it is often necessary to screen large numbers of independent transformants to identify single-copy transgenic plants. This makes the process labor intensive, expensive, and, in recalcitrant species, essentially not feasible. These facts make the development of a single-copy gene delivery system that inserts genes into transcriptionally active regions of potential value.
Agrobacterium-mediated gene delivery in cereals leads to lower copy-number insertions, and single-copy transgenic plants are more frequent (Cheng et al., 1997; Tingay et al., 1997). This method, however, does not eliminate the problem of gene silencing in cereals (Ishida et al., 1996; Park et al., 1996) because even Agrobacterium-generated transgenic plants containing a single-copy of the transgene showed evidence of silencing (Waterhouse et al., 1998).
In the present study we describe a different method of generating large numbers of single-copy transgenic plants in barley by crossing AcTPase-expressing plants with plants containing one or multiple Ds-elements into which an herbicide resistance gene was inserted. We show that a single Ds-bar element can transpose from a multicopy locus to linked and unlinked sites. In the latter case the element can segregate away from the other copies of the Ds element and from the AcTPase gene (Koprek et al., 2000), physically stabilizing the element in its new location. The resulting single-copy element can be re-activated by providing AcTPase in trans. In this way, hundreds of independent, single-copy transgenic plants can readily be generated and the transgene of interest can be separated away from the rest of the vector sequences, e.g. plasmid backbone and selectable marker genes.
Analysis of transgene expression in plants carrying bar in new locations following transposition showed that plants from most TNP lines were completely Basta-resistant in the F3 and F4 generation, exhibiting no signs of necrosis after herbicide treatment. In contrast, a large proportion of lines with one or multiple copies in their original integration site (82% in F3 and 94% in F4) showed signs of transgene inactivation. This striking difference occurred in the F4 generation despite the fact that only Basta-resistant F3 plants were chosen for generation-advance. In all cases studied, gene-silenced plants exhibited increased methylation of cytosine residues in the ubiquitin promoter region linked to bar and a more condensed chromatin structure in the immediate vicinity of the transgene or the transposed silenced Ds-bar element. These observations are consistent with the observations of others that transcriptional gene silencing often correlates with changes in the methylation status (Stam et al., 1997a, 1997b) and with a locally condensed chromatin structure (Ye and Signer, 1996; van Blokland et al., 1997).
Despite the dramatic improvement in transgene expression stability in the TNP plants, silencing was observed in a few TNP lines in our experiments, indicating that additional factors play a role in transgene expression stability. The Ds-bar construct contains short, inverted repeats and inverted repeats are known to induce transcriptional and post-transcriptional gene silencing (for review, see Muskens et al., 2000). In addition, methylation-induced, heritable, and reversible transposon inactivation has been described for all major maize transposons (Fedoroff and Chandler, 1994). This phenomenon can be triggered by the terminal inverted repeats of the transposon, as well as by the transposon copy number. Although copy number is not a factor, the inverted repeats are present on the cassette. This mechanism could account for some of the silencing in TNP and nTNP lines in our experiments. However, since all TNP plants, whether silenced or not, contain the same terminal inverted repeats and subterminal repeats of Ds and in the same structural context in the single intact Ds copy, this would not seem to explain the differences in expression stability among plants from the same line. The analysis of transgene expression in advanced generations will show if the Ds-bar elements go through cycles of silencing that are typical for maize transposable elements. To date, tracking transgene expression stability through the F6 generation in numerous lines has turned up little evidence of transgene expression instability (T. Koprek and S. Rangel, unpublished data).
Another factor that might play a decisive role in transgene expression stability is the nature of the chromosomal location of the transgene insertion, which cannot be controlled and varies from line to line. In our experiments most transposed Ds-bar elements tested (80%) re-inserted into single-copy genomic DNA regions and all but one of these lines produced stably expressing progeny. The existence of the one unstable single-copy line indicates that apart from insertion into genomic DNA of low redundancy, other factors like vicinity to matrix attachment regions (Spiker and Thompson, 1996; Holmes-Davis and Comai, 1998), isochore compatibility of the transgene with the flanking genomic DNA (Kumpatla et al., 1998; Matzke and Matzke, 1998), and higher-order chromatin structure (Sun and Elgin, 1999) might influence transgene expression, but these other factors do not appear to affect transgene expression stability in many Ds-delivered lines.
Analysis of chromosomal DNA from TNP lines A8-1-51 (Fig. 3) and A8-5-55 (data not shown) in which Ds-bar reinserted into repetitive DNA sequences showed a high degree of methylation in the ubiquitin promoter region of the transgene (Fig. 4, A and B) and plants derived from these lines showed signs of transgene silencing. Insertion of transgenes in or near repetitive DNA or heterochromatic regions has been shown to correlate with inactivation of the transgene (Pröls and Meyer, 1992). The results of gene-tagging experiments with Ac/Ds in Arabidopsis and tomato show that Ds elements frequently transpose into transcriptionally active regions (Parinov et al., 1999; Meissner et al., 2000), and this predisposition could be an explanation for the high percentage of TNP plants that express bar stably over several generations.
Our results show that the use of transposable elements as a transporter for transgenes leads to the generation of large numbers of independent single-copy transgenic plants from only two original transformants, one with Ds and one with transposase—a situation that has positive implications for recalcitrant species and varieties within a species. In addition, the increased stability of transgene expression in most TNP plants, as well as the ability to eliminate selectable marker genes and other vector sequences, gives rise to a system of gene delivery that has great utility for generating crop plants with improved agronomic characteristics. The system described here can also be used as a valuable tool to gain further insight into mechanisms of gene silencing.
MATERIALS AND METHODS
Plasmids
Plasmid pSP-Ds-Ubi-bar containing the Ds-bar element consists of the Streptomyces hygroscopicus phosphinothricin acetyl transferase gene (bar) under control of the Ubi1 promoter and first intron from maize and the nos terminator as a 0.9-kbp ClaI-NotI restriction fragment. The UbiI-bar-nos cassette is flanked by a 254-bp 5′ sequence and a 340-bp 3′ sequence from Ds, derived from pDs7 (Wirtz et al., 1997) as a 0.59-kbp SalI-BamHI restriction fragment. The construction of the entire plasmid has been described in detail previously (McElroy et al., 1997; Koprek et al., 2000).
Plant Material
Transgenic barley (Hordeum vulgare var Golden Promise) plants containing the Ds element with a ubiquitin-bar expression cassette as an insert or the AcTPase gene under transcriptional control of the ubiquitin promoter from maize or the putative AcTPase promoter region were generated by particle bombardment of immature embryos (Wan and Lemaux, 1994). AcTPase-expressing plants were crossed with plants that contained one, five, or six Ds-bar copies. Resulting F1 plants were self-pollinated; F2 plants in which Ds had transposed to an unlinked site and segregated away from linked sequences were identified by DNA-blot hybridization and PCR analysis (Koprek et al., 2000).
Transgene Expression Test
Plants negative for the AcTPase gene and containing single or multiple Ds-bar copies in their original integration site (non-transposed, nTNP) or a single transposed Ds-bar in a new location (transposed, TNP) were treated with the herbicide Basta to monitor expression of bar (Wan and Lemaux, 1994). A total of 17 nTNP lines (eight lines derived from A8-1, five lines from A8-5, and two lines each from A1-5 and A18-5) and 35 TNP lines (15 lines derived from A8-1, 10 lines from A8-5, and five lines each from A1-5 and A18-5) were tested. When plants were in the three-leaf stage, 2.5 cm-long leaf segments were painted with an aqueous solution containing 0.5% (v/v) Basta and 0.1% (v/v) Triton X-100 using a cotton swap. Plants showing any sign of necrosis due to the herbicide were scored as sensitive. In F3 generation, plants with a transposed Ds-bar cassette were expected to segregate the Ds-bar element in a 3:1 ratio. Any plants scored as sensitive were analyzed by PCR to confirm the presence or absence of the transgene; sensitive plants containing the transgene were scored as silenced. Resistant plants were self-pollinated and progeny of these plants (F4 generation) were analyzed in an identical manner.
DNA-Blot Hybridization
Genomic DNA was isolated from leaf tissue (Cone, 1989) when plants were in the four-leaf stage. To identify plants containing a single copy of Ds-bar, 10 μg of DNA was digested with HindIII, which does not cut within the 3.6-kb Ds-bar fragment. The digested DNA was separated in 0.8% (w/v) agarose gels, transferred to nylon membranes (Bio-Rad, Hercules, CA), and hybridized with radioactively labeled (Promega, Madison, WI) Ds-specific probe. Analysis of the methylation status of bar and the mapping of its location in non-silenced and silenced plants was performed. Genomic DNA was digested to completion with AvaI, HpaII, MspI, PstI, SacII, and SalI (HpaII was purchased from New Enlgand Biolabs, Beverly, MA], all other enzymes were from Promega) alone or in combination with the non-methylation sensitive enzymes DraI or BglI. Digested DNA was treated as above. Hybridizations were carried out consecutively with bar expression cassette probes A, B, and C (Fig. 4B). Blots were stripped after exposure to x-ray film and were re-probed according to manufacturer's instructions.
PCR Analysis
Genomic DNA (0.5 μg) was subjected to PCR amplification in a thermocycler (model PTC-100, MJ Research, Waltham, MA). PCR reactions (50 μL) contained 1× PCR buffer (Promega), 200 μm of each dNTP, 1.5 mm MgCl2, 1 μm primer, 1% (w/v) dimethyl sulfoxide, and 2.5 units of Taq DNA polymerase (Promega). The primer pairs used for the bar-coding region were BAR5′: (5′-TGC ACC ATC GTC AAC CAC TA-3′) and BAR3′: (5′-ACA GCG ACC ACG CTC TTG AA-3′) and were used to produce a PCR product of 311 bp. PCR reactions were performed with an initial denaturation step at 94°C for 2 min followed by 10 cycles of a touch-down program with decreasing annealing temperatures from 65°C to 60°C in increments of −0.5°C per cycle for 30 s, an extension at 72°C for 30 s, a denaturation at 94°C for 45 s, a subsequent 25 amplification cycles for 30 s at 60°C, 30 s at 72°C, and 45 s at 94°C, and a final extension at 72°C for 5 min. PCR products were analyzed by gel electrophoresis in 1.1% (w/v) agarose gels.
Isolation and Analysis of Regions Flanking Ds Inserts
The redundancy of the DNA flanking the integration site of transposed and non-transposed Ds-elements was analyzed by hybridizing DNA blots of HindIII-digested genomic DNA from non-transgenic var Golden Promise plants with isolated Ds-flanking genomic DNA regions. Genomic DNA flanking Ds-element inserts from 10 different transposition events was isolated using thermal asymmetric interlaced-PCR (Liu et al., 1995) with random primers as described previously (Koprek et al., 2000). Specific amplified tertiary PCR products were analyzed on 1.5% (w/v) agarose gels, isolated using the QIAquick gel extraction kit (Qiagen, Valencia, CA), and used as probes for DNA-blot hybridizations as described above.
Nuclei Isolation and DNase I Sensitivity Test
Five grams of young leaves were ground in liquid N2; the powder was resuspended in 25 mL of Hamilton buffer (10 mm Tris-HCl, pH 7.6, 1.14 m Suc, 5 mm MgCl2, and 5 mm β-mercaptoethanol; Hamilton et al., 1972) and the slurry was gently stirred and filtered through three layers of Miracloth (Calbiochem, La Jolla, CA) in 10 mL of the same buffer. The final pellet was resuspended, washed in 5 mL of DNase I digestion buffer (50 mm Tris-HCl, pH 8.0, 0.3 m Suc, 5 mm MgCl2, 1.5 mm NaCl, 0.1 mm CaCl2, and 5 mm β-mercaptoethanol), and centrifuged at 1,000g for 10 min. The pellet was resuspended in 1,550 μL of DNase I digestion buffer and 50 μL of the nuclei suspension was stained with 4′,6-diamino-phenylindole. Nuclei were counted under a fluorescence microscope. All manipulations were carried out at 4°C.
For DNase I digestion, aliquots containing 5 × 105 nuclei (usually in 100–150 μL) were transferred into separate reaction tubes and DNase I digestion buffer was added to a final volume of 200 μL per reaction. Nuclei were treated with 1 unit/mL DNase I (Promega) at 25°C for 0, 5, 10, 15, and 20 min. The reaction was stopped by adding 40 μL of stop buffer (0.25 m EGTA, 0.25 m EDTA). Chromatin-free (naked) DNA was separated from the isolated nuclei and treated with 1 unit of DNase I for 2 min. DNA was extracted and purified as described above, resuspended in 20 μL of Tris-EDTA, and digested with BglI to create reference-sized DNA fragments. The DNA was separated on 0.8% (w/v) agarose gels, blotted, and hybridized as described above with a probe specific for the bar-coding region (probe C; Fig. 4B).
Footnotes
This work was supported by the Deutscheforschungsgemeinschaft, by the U.S. Department of Agriculture Cooperative Extension Service through the University of California, and by the Novartis Agricultural Discovery Institute.
LITERATURE CITED
- Assaad FF, Tucker KL, Signer ER. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol Biol. 1993;22:1067–1085. doi: 10.1007/BF00028978. [DOI] [PubMed] [Google Scholar]
- Atkinson RG, Bieleski LRF, Gleave AP, Janssen B-J, Morris BAM. Post-transcriptional silencing of chalcone synthase in petunia using a geminivirus-based episomal vector. Plant J. 1998;15:593–604. doi: 10.1046/j.1365-313x.1998.00211.x. [DOI] [PubMed] [Google Scholar]
- Cheng M, Fry JE, Shengzhi P, Huaping Z, Hironaka CM, Duncan DR, Conner TW, Wan Y. Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 1997;115:971–980. doi: 10.1104/pp.115.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K. Yet another rapid plant DNA prep. Maize Gen Coop Newslett. 1989;63:68. [Google Scholar]
- De Block M, Debrouwer D, Moens T. The development of a nuclear male sterility system in wheat: expression of the barnase gene under the control of tapetum specific promoters. Theor Appl Genet. 1997;95:125–131. [Google Scholar]
- Fedoroff NV, Chandler V. Inactivation of maize transposable elements. In: Paszkowski J, editor. Homologous Recombination and Gene Silencing in Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 349–385. [Google Scholar]
- Hamilton RH, Kunsch U, Temperli A. Simple rapid procedures for the isolation of tobacco leaf nuclei. Anal Biochem. 1972;49:48–57. doi: 10.1016/0003-2697(72)90241-2. [DOI] [PubMed] [Google Scholar]
- Hansen G, Chilton M-D. “Agrolistic” transformation of plant cells: integration of T-strands generated in planta. Proc Natl Acad Sci USA. 1996;93:14978–14983. doi: 10.1073/pnas.93.25.14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Holmes-Davis R, Comai L. Nuclear matrix attachment regions and plant gene expression. Trends Plant Sci. 1998;3:91–97. [Google Scholar]
- Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat Biotechnol. 1996;14:745–750. doi: 10.1038/nbt0696-745. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Kumpatla SP, Chandrasekharan MB, Hall TC. Transgene silencing in monocots. Plant Mol Biol. 2000;43:323–346. doi: 10.1023/a:1006412318311. [DOI] [PubMed] [Google Scholar]
- Koprek T, McElroy D, Louwerse J, Williams-Carrier R, Lemaux PG. An efficient method for dispersing Ds elements in the barley genome as a tool for determining gene function. Plant J. 2000;24:253–264. doi: 10.1046/j.1365-313x.2000.00865.x. [DOI] [PubMed] [Google Scholar]
- Kumpatla SP, Chandrasekharan MB, Iyer LM, Li G, Hall TC. Genome intruder scanning and modulation systems and transgene silencing. Trends Plant Sci. 1998;3:97–104. [Google Scholar]
- Liu Y-G, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Matzke AJ, Matzke MA. Position effects and epigenetic silencing of plant transgenes. Curr Opin Plant Biol. 1998;1:142–148. doi: 10.1016/s1369-5266(98)80016-2. [DOI] [PubMed] [Google Scholar]
- McElroy D. Moving agbiotech downstream. Nat Biotechnol. 1999;17:1071–1074. doi: 10.1038/15054. [DOI] [PubMed] [Google Scholar]
- McElroy D, Louwerse JD, McElroy SM, Lemaux PG. Development of a simple transient assay for Ac/Ds activity in cells of intact barley tissue. Plant J. 1997;11:157–165. doi: 10.1046/j.1365-313x.1997.11010157.x. [DOI] [PubMed] [Google Scholar]
- Meissner R, Chague V, Zhu Q, Emmanuel E, Elkind Y, Levy AA. A high throughput system for transposon tagging and promoter trapping in tomato. Plant J. 2000;22:265–274. doi: 10.1046/j.1365-313x.2000.00735.x. [DOI] [PubMed] [Google Scholar]
- Meyer P, Heidmann I. Epigenetic variants of a transgenic petunia line show hypermethylation in transgene DNA: an indication for specific recognition of foreign DNA in transgenic plants. Mol Gen Genet. 1994;243:390–399. doi: 10.1007/BF00280469. [DOI] [PubMed] [Google Scholar]
- Meyer P, Linn F, Heidmann I, Meyer H, Niedenhof I, Saedler H. Endogenous and environmental factors influence 35S promoter methylation of a maize A1 gene construct in transgenic petunia and its color phenotype. Mol Gen Genet. 1992;231:345–352. doi: 10.1007/BF00292701. [DOI] [PubMed] [Google Scholar]
- Muskens MWM, Vissers APA, Mol JNM, Kooter JM. Role of inverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Mol Biol. 2000;43:243–260. doi: 10.1023/a:1006491613768. [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang W-C, Kumaran M, Sundaresan V. Analysis of flanking sequences from Dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell. 1999;11:2263–2270. doi: 10.1105/tpc.11.12.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Pinson SR, Smith RH. T-DNA integration into genomic DNA of rice following Agrobacterium inoculation of isolated shoot apices. Plant Mol Biol. 1996;32:1135–1148. doi: 10.1007/BF00041397. [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Torbert KA, Rines HW, Somers DA. Irregular patterns of transgene silencing in allohexaploid oat. Plant Mol Biol. 1998;38:597–607. doi: 10.1023/a:1006090731414. [DOI] [PubMed] [Google Scholar]
- Pröls F, Meyer P. The methylation patterns of chromosomal integration regions influence gene activity of transferred DNA in Petunia hybrida. Plant J. 1992;2:465–475. doi: 10.1046/j.1365-313x.1992.t01-20-00999.x. [DOI] [PubMed] [Google Scholar]
- Que Q, Wang H-Y, English JJ, Jorgensen RA. The frequency and degree of co-suppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell. 1997;9:1357–1368. doi: 10.1105/tpc.9.8.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiker S, Thompson WF. Nuclear matrix attachment regions and transgene expression in plants. Plant Physiol. 1996;110:15–21. doi: 10.1104/pp.110.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Anderson OD, Ow DW. Single-copy transgenic wheat generated through the resolution of complex integration patterns. Proc Natl Acad Sci USA. 1999;96:11117–11121. doi: 10.1073/pnas.96.20.11117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M, De Bruin R, Kenter S, Van Der Hoorn RAL, Van Blokland R, Mol JNM, Kooter JM. Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J. 1997a;12:63–82. [Google Scholar]
- Stam M, Mol JNM, Kooter JM. The silence of genes in transgenic plants. Ann Bot. 1997b;79:3–12. [Google Scholar]
- Sun FL, Elgin SC. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/s0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- Tingay S, McElroy D, Kalla R, Fieg S, Wang M, Thornton S, Brettell R. Agrobacterium tumefaciens-mediated barley transformation. Plant J. 1997;11:1369–1376. [Google Scholar]
- van Blokland R, Ten Lohuis M, Meyer P. Condensation of chromatin in transcriptional regions of an inactivated plant transgene: evidence for an active role of transcription in gene silencing. Mol Gen Genet. 1997;257:1–13. doi: 10.1007/s004380050617. [DOI] [PubMed] [Google Scholar]
- Wan Y, Lemaux PG. Generation of large numbers of independently transformed fertile barley plants. Plant Physiol. 1994;104:37–48. doi: 10.1104/pp.104.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz U, Osborne B, Baker B. Ds excision from extrachromosomal geminivirus vector DNA is coupled to vector DNA replication in maize. Plant J. 1997;11:125–135. doi: 10.1046/j.1365-313x.1997.11010125.x. [DOI] [PubMed] [Google Scholar]
- Ye F, Signer ER. RIGS (repeat-induced gene silencing) in Arabidopsis is transcriptional and alters chromatin configuration. Proc Natl Acad Sci USA. 1996;93:10881–10886. doi: 10.1073/pnas.93.20.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]