Abstract
Background
The mechanisms whereby neuroinflammation negatively affects neuronal function in the hypothalamus are not clear. Our previous study determined that obesity-mediated chronic inflammation elicits sex-specific impairment in reproductive function via reduction in spine density in gonadotropin-releasing hormone (GnRH) neurons. Neuroinflammation and subsequent decrease in GnRH neuron spine density was specific for male mice, while protection in females was independent of ovarian estrogens.
Methods
To examine if neuroinflammation-induced cytokines can directly regulate GnRH gene expression, herein we examined signaling pathways and mechanisms in males in vivo and in GnRH-expressing cell line, GT1–7.
Results
GnRH neurons express cytokine receptors, and chronic or acute neuroinflammation represses GnRH gene expression in vivo. Leukemia inhibitory factor (LIF) in particular represses GnRH expression in GT1–7 cells, while other cytokines do not. STAT3 and MAPK pathways are activated following LIF treatment, but only MAPK pathway, specifically p38α, is sufficient to repress the GnRH gene. LIF induces cFOS that represses the GnRH gene via the −1,793 site in the enhancer region. In vivo, following high-fat diet, cFOS is induced in GnRH neurons and neurons juxtaposed to the leaky blood brain barrier of the organum vasculosum of the lamina terminalis, but not in the neurons further away.
Conclusion
Our results indicate that the increase in LIF due to neuroinflammation induces cFOS and represses the GnRH gene. Therefore, in addition to synaptic changes in GnRH neurons, neuroinflammatory cytokines directly regulate gene expression and reproductive function, and the specificity for neuronal targets may stem from the proximity to the fenestrated capillaries.
Keywords: Leukemia inhibitory factor, Gonadotropin-releasing hormone, Neuroinflammation, Hypothalamus, High-fat diet
Introduction
Gonadotropin-releasing hormone (GnRH) is the final brain output for the regulation of reproduction. GnRH neurons, which are scattered in the hypothalamus, synthesize and secrete GnRH that acts on the anterior pituitary to stimulate the synthesis and secretion of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) from gonadotrope cells [1, 2]. LH and FSH act on the gonads to promote steroidogenesis and gametogenesis. GnRH neuronal processes, named “dendrons,” by Herbison group [3], form an interwoven network that receives direct synaptic and neuropeptide input from upstream regulatory neurons, most notably kisspeptin [4, 5]. This GnRH network integrates other signals that impinge on reproduction, such as stress [6, 7], endocrine disruptors [8], circadian rhythms [9, 10], metabolism [11, 12], and acute inflammation during infection [13–15].
Previous studies have implicated acute inflammation, elicited with an injection of lipopolysaccharide (LPS), in the impairment of reproductive function [14, 16, 17]. LPS-challenged rodents exhibited reduced levels of LH and GnRH mRNA, diminished release of LH and GnRH, and increased levels of proinflammatory cytokines, such as tumor necrosis factor (TNF)-α, interleukin (IL)-1β, and IL-6 in the circulation [14, 18–20]. Centrally administered cytokines also provoked reduced LH and GnRH levels, but the mechanism whereby these cytokines mediate their effects is unknown [16, 17, 21–23]. More recently, our group determined that low-grade, chronic inflammation caused by high-fat diet (HFD)-induced obesity may also directly affect GnRH neurons, resulting in reduced levels of LH in circulation and diminished GnRH mRNA levels in the hypothalamus, specifically in male mice [24]. We, and others, have reported that diet-induced obese mice and people exhibited increased levels of proinflammatory cytokines, TNF-α, IL-1β, and IL-6, in circulation [24, 25] and in the hypothalamus, at the mRNA level and protein level [24, 26]. Additionally, we also identified that leukemia inhibitory factor (LIF), a member of the IL-6 family, is increased in the circulation and locally produced in male mice hypothalami in obesity [24]. Interestingly, LIF is increased in a sex-specific manner only in males that exhibit reduction in GnRH mRNA and gonadotropin hormones, but not in females that lack changes in GnRH or gonadotropin hormones. IL-6, a prototypical member of the family, on the other hand, is increased in both sexes. Our previous study postulated that impairment of GnRH neurons stems from reduction in spine density and consequently the connectivity of the GnRH network [24]. However, GnRH neurons express several cytokine receptors [27] and inflammation-induced cytokines may directly regulate intracellular signaling pathways in the GnRH neurons.
Herein, we focus on delineating the mechanisms by which inflammatory cytokines influence GnRH gene expression to provide insight into the etiology of neuroinflammation-induced impairment of reproductive function. TNF-α, IL-1β, and IL-6 are key players in the regulation of immune response and inflammatory processes during infection [28]. In the central nervous system, TNF-α and IL-1β regulate synaptic plasticity, neurodegeneration, learning, and memory [29–32]. During infection, both of these cytokines mediate the physiological and behavioral responses in sickness such as, inducing fever, inhibiting food intake, causing nausea and fatigue [33–35]. TNF-α and IL-1β mediate their effects through activation of downstream signaling molecules: nuclear factor-κB (NF-κB), Janus kinase and signal transducers and activators of transcription pathway (JAK-STAT), and mitogen activated protein kinases (MAPK) [36–38]. Similarly, IL-6 is produced in response to infection and stress, and in turn stimulates various cell populations, also through the JAK-STAT and MAPK pathways [39]. In the brain, IL-6 is involved in degenerative responses [40, 41]. However, IL-6 is also induced following TNF-α or IL-1β treatment and is involved in the negative feedback that ultimately contributes to the dampening of the immune response and activating tissue repair [42].
LIF is a member of IL-6 family that is induced during inflammatory response [43]. However, its functions are not limited to inflammation: LIF has been demonstrated to play a crucial, non-redundant role in embryo implantation in both mice and humans [44–46]. LIF also maintains stem cells and regulates differentiation of germ cells [47, 48]. In the brain, LIF regulates neuronal function and neuronal response to injury [49–51]. With respect to GnRH neurons, LIF has been shown to regulate the migration of GN11 immature GnRH neuron cell line and regulate the release of GnRH in GT1–7 cells [52–54]. LIF binds its specific receptor, which, similarly to the other members of the IL-6 family, recruits and signals through the GP130 signals transducer, activating the JAK-STAT pathway [45].
Signaling pathways involved in the regulation of GnRH gene transcription by any of these cytokines have not been elucidated. GnRH neurons in the rodent hypothalamus are located in the preoptic area surrounding organum vasculosum laminae terminalis (OVLT) and send long processes to the median eminence (ME) where secretion occurs from the terminals. Both OVLT and ME are areas that contain fenestrated capillaries and a leaky blood-brain barrier [55]. A subpopulation of GnRH neurons extends their processes into the OVLT and across the blood-brain barrier, where they may be able to directly respond to circulating molecules, including cytokines [56]. OVLT and surrounding thermoregulatory neurons are involved in changes in body temperature and inducing fever in response to systemic inflammation. Pyrogenic, proinflammatory cytokines, TNF-α, IL-1β, and IL-6, produced locally in the hypothalamus, or from the circulation via fenestrated capillaries in the OVLT, stimulate thermoregulatory neurons to increase the body temperature [57–60]. We postulate that these cytokines directly regulate GnRH neurons in the proximity to OVLT.
About 800–1,200 GnRH neurons are scattered throughout the forebrain of a mouse [61]. This poses a challenge for molecular studies of GnRH neurons in vivo. GT1–7 cells are the only model of mature, terminally differentiated, GnRH-producing neurons, and have been used to identify regulatory elements and transcription factors important for GnRH transcription [62, 63]. GT1–7 cells allow for the investigation of molecular mechanisms and direct effects on GnRH gene expression without confounding variables that may be present in in vivo studies. Here, we combine in vivo mouse studies and investigate the direct effects of cytokines on GnRH gene expression in GT1–7 cells. Our results delineate the molecular mechanisms and signaling pathways that LIF activates and strongly suggest that LIF directly affects GnRH neurons to regulate GnRH gene expression in infection or obesity induced inflammation of the hypothalamus.
Materials and Methods
Animals
C57BL/6J mice were maintained under a 12-h light, 12-h dark cycle and received food and water ad libitum. All experiments were performed with approval from the University of California (Riverside, CA, USA) Animal Care and Use Committee and in accordance with the National Institutes of Health Animal care and Use Guidelines. C57BL/6J male mice were placed on either an HFD (D12492, 60% kcal from fat; 5.21 kcal/g; protein 20% kcal; fat 60% kcal [lard 0.32 g/g diet, soybean oil 0.03 g/g]; carbohydrate 20% kcal; Research Diet, New Brunswick, NJ, USA) or control diet (CTR, D12450J, 10% kcal from fat; matching sucrose levels to HFD; 3.82 kcal/g; protein 20% kcal; fat 10% kcal [lard 0.02 g/g diet, soybean oil 0.025 g/g]; carbohydrate 70% kcal; Research Diet, New Brunswick, NJ, USA) from weaning age for 12 weeks. Mice treated with vehicle or LPS were fed standard food pellets (STD, 5,053, 4.07 kcal/g; protein 24% kcal; fat 13%; carbohydrates 63%; St. Louis, MO, USA) from weaning. LPS (2.5 mg/kg body weight) from Escherichia coli (catalog # L4391; Sigma, USA) was administered by intraperitoneal injection. Mice were sacrificed 24 h after LPS treatment in parallel with the corresponding control groups. At least 10 animals/diet or treatment were analyzed unless otherwise indicated, and differences from corresponding controls were compared by Student’s t test. GnRH-GFP mice were kindly provided by Dr. Suzanne Moenter [64] to facilitate investigation of fluorescently labeled GnRH neurons.
Histological Analysis and Immunohistochemistry
For brain collection, animals were perfused with ice cold phosphate buffer saline (PBS) solution followed by 4% paraformaldehyde solution. Brains were postfixed in 4% paraformaldehyde for 2 h at room temperature and cryopreserved in 30% sucrose/PBS solution for 3 days at 4 ° C before freezing in OTC. Frozen brains from GnRH-GFP mice were sectioned to 30 μm sections and stained for GFP to visualize GnRH neurons and for GP130 or cFOS. Slides were blocked with 20% goat serum and incubated with primary antibodies against GFP (1: 5,000 raised in chicken, Table 1) at 4 ° C for 48 h. After PBS washes, slides were incubated with FITC/Alexa 488 goat anti-chicken IgG (1: 300, Molecular Probes, Eugene, OR) for 1 h. Slides were then incubated with primary antibodies against GP130 (1: 500, MAB4681; R&D Systems, Minneapolis, MN, USA) or cFOS (1: 300, SC-52; Santa Cruz Biotechnology Inc. Dallas, TX, USA) for 48 h at 4 ° C followed by Alexa 594 goat-anti-rat IgG (1: 300; Molecular Probes) or biotinylated goat anti-rabbit IgG (1: 300; Vector Laboratories, USA), and Cy5-streptavidin (1: 300, 434,316, Life Tech Corp., Eugene, OR, USA) for 1 h each at room temperature, respectively. Sections were mounted, and slides covered using VectaShield mounting media with DAPI (H-1500, Vector Laboratories). Secondary antibody-only controls were performed to determine antibody specificity. Images were obtained using a Leica microscope system.
Table 1.
List of antibodies
| Antibody | Species | Dilution | Provider, cat. # |
|---|---|---|---|
| GFP | chicken | 1:5,000 | Abcam, ab1397 |
| GP130 | rat | 1:300 | R&D Systems, MAB4681; clone#125623 |
| STAT3 | mouse | 1:1,000 | Cell Signaling, 9139 |
| p-STAT3 | rabbit | 1:1,000 | Cell Signaling, 9145 |
| p38 | rabbit | 1:1,000 | Cell Signaling, 9212 |
| p-p38 | rabbit | 1:1,000 | Cell Signaling, 9211 |
| ERK 1/2 | rabbit | 1:1,000 | Cell Signaling, 9102 |
| p-ERK 1/2 | rabbit | 1:1,000 | Cell Signaling, 9101 |
| JNK | rabbit | 1:1,000 | Cell Signaling, 9252 |
| p-JNK | mouse | 1:1,000 | Cell Signaling, 9255 |
| OCT-1 | rabbit | 1:500 | Abcam, ab66132 |
| cFOS | rabbit | 1:300, 1:1,000 | Santa Cruz Biotechnology, sc-52 |
| β-Tubulin | rabbit | 1:1,000 | Santa Cruz Biotechnology, sc-9104 |
To quantify the number of cFOS-expressing GnRH neurons, coronal sections of the preoptic area in the hypothalamus of GnRH-GFP mice were stained for GFP (green) and cFOS (red). Three hundred GnRH-GFP neurons from each of the 4 male mice from control and HFD group were counted for the co-labeling with cFOS, and results represented as a percent of total GFP labeled neurons. To assess expression of cFOS in other cells, two 100 × 100 μm areas, one area proximal to the OVLT and one more dorsal, in the same section were counted to quantify the number of cFOS-expressing cells, where DAPI staining was used to identify cell nuclei. Statistical differences (p < 0.05) were determined by Student’s t test.
qPCR Analysis
Hypothalami were dissected, total RNA extracted using MicroRNA kit from Ambion and reverse transcribed using Superscript III (Invitrogen, CA, USA). qPCR was performed using an iQ SYBR Green supermix and an IQ5 real-time PCR machine (BioRad Laboratories, Hercules, CA, USA), with primers listed in Table 2, under the following conditions: 95 ° C for 15 min, followed by 40 cycles at 95 ° C for 20 s, 56 ° C for 30 s, and 72 ° C for 30 s. The amount of the gene of interest was calculated by comparing the threshold cycle obtained for each sample with the standard curve generated in the same run and normalized to the β2-microglobulin housekeeping gene in the same sample using the ΔΔCt method. Replicates were averaged. After each run, a melting curve analysis was performed to confirm that a single amplicon was generated in each reaction. Statistical differences (p < 0.05) in expression were determined by Student’s t test using JMP software (SAS Institute, Cary, NC, USA).
Table 2.
Primers used in qRT-PCR
| Primers | Forward | Reverse |
|---|---|---|
| Gnrh (GnRH) | CTACTGCTGACTGTGTGTTTG | CATCTTCTTCTGCCTGGCTTC |
| Il6 (IL-6) | TTCTCTGGGAAATCGTGGAAAT | TCCAGTTTGGTAGCATCCATCA |
| Tnfa (TNF-α) | ATGTCTCAGCCTCTTCTCATTCC | GCTTGTCACTCGAATTTTGAGAA |
| Il1β (IL-1β) | GCAACTGTTCCTGAACTCAACTG | CACAGCCACAATGAGTGATACTG |
| Lif (LIF) | ATGTGCGCCTAACATGACAG | TATGCGACCATCCGATACAG |
| B2m (β2-microglobulin) | TGACCGGCCTGTATGCTATCCA | CAGTGTGAGCCAGGATATAGAAAGAC |
| LIFR | TCAGTTTCAGCCAGGAGTAA | GCAATAATCAATCCCACAGA |
| IL-6R | AAGCAGCAGGCAATGTTACC | CATAAATAGTCCCCAGTGTCG |
| GP130 | GCGTACACAGATGAAGGTGGGAAAGA | GCTGACTGCAGTTCTGCTTGA |
Cell Culture
GT1–7 cells, kindly provided by Pamela Mellon (University of California, La Jolla, CA, USA), were cultured in DMEM (Cellgro, Mediatech Inc., Herndon, VA, USA) with 10% FBS. LIF (34–8521; eBioscience, USA) and IL-6 (216–16; PeproTech, Rocky Hill, NJ, USA) were reconstituted in PBS containing 0.1% BSA and stored in aliquots at −80 ° C until use. RNA was isolated with TRIzol (Life Tech, Carlsbad, CA, USA) and RT-PCR performed as previously described [65, 66].
Western Blot Analysis
Whole cell lysates were obtained after treatment with 10 ng/mL LIF or vehicle for times indicated, using lysis buffer (20 mM Tris-HCl, pH 7.4, 140 mM NaCl, 0.5% Nonidet P-40, 0.5 mM EDTA) with 1% protease inhibitor cocktail (P8340; Sigma, USA) and 1 mM PMSF. Protein content was determined using Bradford reagent (Bio-Rad Laboratories). An equal amount of protein from each sample was resolved on a 10% SDS-PAGE and electrotransferred to a nitrocellulose membrane. Membranes were blocked in TBST (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Tween-20) with 10% non-fat milk for 2 h at room temperature and incubated overnight with specific antibodies for p-STAT3 (1: 1,000; 9134; Cell Signaling, Danvers, MA, USA), STAT3 (1: 1,000, 9139; Cell Signaling), p-p38 (1: 1,000, 9211; Cell Signaling), p38 (1: 1,000, 9212; Cell Signaling), p-ERK½ (1: 1,000, 9101; Cell Signaling), ERK½ (1: 1,000, 9102; Cell Signaling), p-JNK (1: 1,000, 9255; Cell Signaling), JNK (1: 1,000, 9252; Cell Signaling), OCT-1 (1: 500, ab66132, Abcam, Cambridge, UK), cFOS (1: 1,000, SC-52; Santa Cruz Biotechnology), and β-tubulin (1: 1,000, SC-9104; Santa Cruz Biotechnology). The bands were visualized with horseradish peroxidase (HRP)-linked secondary antibodies and enhanced chemiluminescence reagent (Amersham Bioscience), as described by the manufacturer. Blots were exposed to autoradiography film (Bioexpress, USA). Each experiment was performed 3 times, and representative images are presented.
Transfections
Cells were plated into 12-well plates and transfected using Fugene 6 reagent (Roche Applied Science), as described previously [66–69]. Wells were transfected with 500 ng of reporter plasmid, 100 ng of β-galactosidase reporter plasmid driven by the Herpesvirus thymidine kinase promoter, as an internal control for the efficiency of the transfection, and 200 ng of expression vectors or empty vector control, as indicated in the figure legends. 24 h after transfection, cells were switched to serum-free media (DMEM with 0.1% BSA) and treated with either 10 ng/mL LIF, 20 ng/mL IL-6, or vehicle for 24 h. Following treatment, cells were lysed in 0.1 M potassium phosphate buffer, pH 7.8, with 0.2% Triton X-100. Luciferase activity in the lysates was measured with a Veritas Microplate luminometer (Turner Biosystems, Sunnyvale, CA, USA) after injection with 100 μL of luciferase assay buffer (100 mM Tris-HCl, pH 7.8, 15 mM MgSO4, 10 mM ATP, and 65 μM luciferin). β-Galactosidase activity was measured using the Tropix Galacto-light β-galactosidase assay (Applied Biosystems, Foster City, CA, USA). All experiments were performed three independent times and in triplicates within each experiment. Luciferase values were normalized to β-galactosidase values for each sample. Results are presented as an average of three experiments. Statistical significance, p < 0.05, was determined with ANOVA followed by Tukey’s post hoc test using JMP software (SAS Institute).
Plasmids
The reporter plasmids were kindly provided by Pamela Mellon (University of California, La Jolla, CA, USA). The −5-kb rat GnRH (−4,984 to +22 relative to the transcription start site), GnRH E/P, which contains GnRH-E1 (−1,863 to −1,571)/GnRH-P (−173 to +112), and GnRH-P (−173 to +112) luciferase reporters have previously been described [70–73]. Luciferase reporter plasmids containing mutations of AP1 binding site in GnRH enhancer and GnRH promoter, and reporter plasmids with RSV promoter fused to GnRH enhancer (GnRHe/RSVp), have also been previously described [70–73]. The expression vectors for wild-type STAT3 (Stat3 Flag pRC/CMV, #8707, dominant negative (DN) STAT3 (Stat3 Y705F Flag pRC/CMV, #8704), constitutively active (CA) STAT3 (Stat3-C Flag pRC/CMV, #8722), and luciferase reporter containing STAT3 response element (SBR, 4xM67 pTATA TK-Luc, #8688) were purchased from Addgene (Cambridge, MA, USA) and have previously been described [74]. Expression vectors for CA RAS, MEK1, MEK2, MKK4, MKK7, MKK3, MKK6, and CA isoforms of p38 (p38α, p38β, p38λ, p38δ) were a gift from Peiqing Sun (The Scripps Research Institute, La Jolla, CA, USA) [75]. Expression vector for cFOS has been previously described [65, 76–78].
Results
Inflammation in the Hypothalamus Induces Inflammatory Cytokines and Represses GnRH
Inflammation, either acute, caused by an injection of LPS, or chronic, elicited by HFD, negatively affects hypothalamic neurons and in particular, reproductive function [21, 79]. We and others postulated this impairment is due to repression of the GnRH gene, which is the final brain signal in the control of reproduction. Inflammatory cytokines have previously been proposed to negatively regulate GnRH neurons [16, 18, 80]. We initiated our studies with an analysis of cytokine mRNA levels in the hypothalami of C57BL/6J male mice, induced by LPS injection, a bacterial endotoxin known to elicit an inflammatory response. Our studies revealed that TNF-α (Tnf) mRNA was induced 22-fold, IL-1β (Il1b) was induced 7-fold, IL-6 (Il6) 1.6-fold, and LIF (Lif) 2.2-fold after LPS injection (Fig. 1a). Gnrh mRNA level in mice treated with LPS was reduced by 72% (Fig. 1a).
Fig. 1.
GnRH expression is repressed when cytokine levels are increased. a Hypothalami from male mice 24 h after injection with either vehicle control (CTR, white bars) or lipopolysaccharide (LPS, gray bars). b Hypothalami from male mice following 12-week feeding with control (CTR, white bars) or high-fat diet (HFD, black bars). Cytokine and GnRH (Gnrh1) expression was assayed with RT-qPCR. TNF-α, (Tnf) tumor necrosis factor alpha; IL-1β, (Il1b) interleukin-1 beta; IL-6, (Il6) interleukin-6; LIF, (Lif) leukemia inhibitory factor. Asterisk indicates significant difference (p < 0.05) determined with the t test.
Obesity is considered a state of chronic inflammation as opposed to acute inflammation elicited by LPS injection [24, 25, 81]. Thus, we exposed male C57BL/6J mice to HFD or to CTR diet for 12 weeks. Diet-induced obesity induced TNF-α 1.6-fold and IL-1β 1.7-fold, although due to variability, it did not reach significance (Fig. 1b). IL-6 was significantly induced 3.1-fold and LIF 1.6-fold, compared to mice fed CTR diet (Fig. 1b). Further analysis revealed significantly reduced Gnrh mRNA levels by 46%, compared to the controls (Fig. 1b). Thus, increase in locally produced cytokines, acute and chronic, correlates with repression of GnRH mRNA expression.
LIF Represses GnRH
We then determined whether these locally produced, inflammatory cytokines can regulate GnRH gene expression directly. Due to the scarcity and scattered location of GnRH neurons in mice, in vivo analysis of molecular mechanisms whereby cytokines regulate the GnRH gene is not possible. To analyze direct effects of cytokines on gene expression we employed GT1–7 cells that are an established Gnrh expressing and GnRH secreting cell model [62]. We expressed GnRH 5-kb luciferase reporter (5-kb GnRH luc) in GT1–7 cells and treated with inflammatory cytokines. LIF repressed GnRH reporter expression by 45.7% compared to CTR (Fig. 2a, black bar), similar to what was observed in vivo with LPS injection and HFD. TNF-α, IL-1β, and IL-6 had no effect on GnRH expression. To further analyze the effect of LIF treatment on the GnRH gene, RNA was isolated from GT1–7 cells treated with either vehicle or LIF, reverse transcribed and quantitative RT-PCR performed. LIF treatment reduced endogenous GnRH mRNA levels by 39.1% (Fig. 2b). Taken together, these results indicate that LIF represses the Gnrh gene.
Fig. 2.
GnRH expression is suppressed by LIF. a GT1–7 cells transiently transfected with the 5-kb GnRH reporter (5-kb GnRH Luc) treated with vehicle (CTR, white bar), or TNF-α, IL-1β, IL-6, and LIF, for 24 h. Luciferase values were normalized to β-galactosidase values for each sample. Results are presented as an average of three experiments performed in triplicate. Statistical significance (p < 0.05) was determined with ANOVA followed by Tukey’s post hoc test and indicated with an asterisk. b RT-qPCR using total RNA from GT1–7 cells treated with LIF, 10 ng/mL for 24 h, demonstrates repression of endogenous GnRH mRNA (Gnrh1). Asterisk indicates significant difference determined with the t test (p < 0.05).
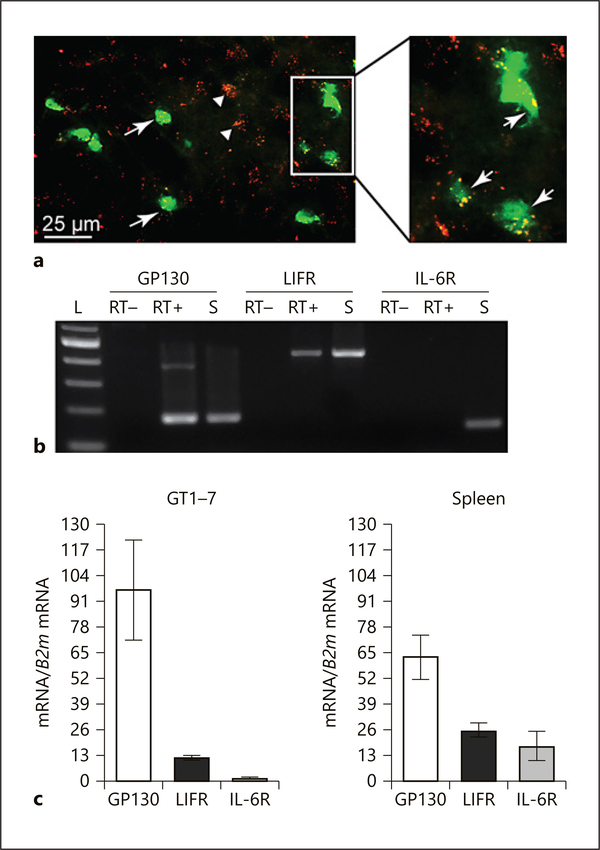
Since LIF and IL-6 belong to the same cytokine family, it was surprising that LIF repressed GnRH while IL-6 did not. These two cytokines share GP130 signaling receptor, but each has a specific receptor for ligand binding. We analyzed the expression of their specific receptors on GnRH neurons to explain why IL-6 had no effect on GnRH gene expression. We first demonstrated the expression of GP130 cytokine signaling receptor (red) on GnRH neurons (green) in vivo using immunocytochemistry of hypothalamic slices (Fig. 3a). This suggests that LIF and IL-6 cytokines can act on GnRH neurons in the mouse. Since antibodies for specific LIF receptor (LIFR) and IL-6 receptor (IL-6R) were not effective in immunohistochemistry, we analyzed mRNA expression of these receptors using RNA isolated from GT1–7 cells. Products of expected size for GP130 (177 bp), and LIFR (452 bp), were present in both GT1–7 cells and spleen, which served as a positive control, but absent in negative control samples lacking reverse transcriptase (Fig. 3b). IL-6R expression (156 bp) was absent in GT1–7 cells, but present in spleen (Fig. 3b). qRT-PCR analysis reveals that the expression of IL-6R is limited in GT1–7 cells, compared to the expression of GP130 and LIFR, while expression levels of these receptors was similar in spleen (Fig. 3c). These results indicate that LIF can directly bind GnRH neurons and affect GnRH gene expression.
Fig. 3.
GP130 is expressed in GnRH neurons in vivo and in GT1–7 cells. a Immunohistochemistry of GnRH neurons genetically labeled with GFP (green), stained for GP130 receptor (red). White arrows indicate GnRH neurons that express GP130, while arrowheads indicate GP130 staining in non-GnRH cells. b RT-PCR demonstrates expression of GP130 (177 bp) and LIFR (452 bp), but not IL-6R (156 bp) in GT1–7 cells (RT+), while all three receptors are expressed in the spleen (S). RT–, GT1–7 cells mRNA used without reverse transcriptase serves as negative controls; L, size ladder. c RT-qPCR demonstrates relative expression of GP130, LIFR, and IL-6R in GT1–7 cells and the spleen.
LIF Functions via GnRH Enhancer to Repress the GnRH Gene
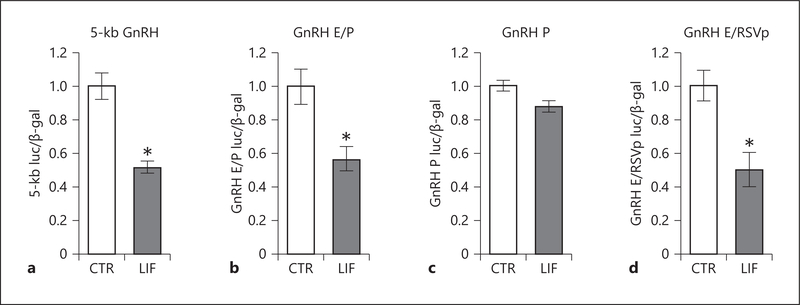
The 5-kb GnRH reporter contained two upstream regulatory elements: the 300-bp enhancer (−1,863 to −1,571) and the evolutionarily conserved promoter (−173 to +1), that confer neuron-specific activation of the GnRH, in culture and in vivo [70, 82, 83]. To map the elements necessary for LIF-mediated repression of GnRH expression, GT1–7 cells were transiently transfected with a luciferase reporter containing the 5 kb regulatory region upstream of the GnRH transcription start site (5-kb GnRH), a reporter containing the enhancer and promoter without intervening sequences (GnRH E/P) or a reporter containing the promoter (GnRH P). LIF repressed luciferase activity of the 5-kb GnRH reporter and of the reporter containing the enhancer and promoter, GnRH E/P luc, by 45.7 and 42.6%, respectively (Fig. 4a, b). Luciferase activity of the reporter containing only the promoter, GnRH P luc, did not change in response to LIF (Fig. 4c). Next, to examine if the enhancer is sufficient for repression, the reporter containing enhancer fused to heterologous RSV promoter was examined (GnRH E/RSVp luc). LIF treatment significantly repressed GnRH E/RSVp luc reporter by 45.2% (Fig. 4d). Given that the GnRH E/P and GnRH E/RSVp reporters were repressed to a similar degree by LIF as the full-length 5-kb GnRH reporter, these results indicate that the enhancer is sufficient and necessary for repression by LIF.
Fig. 4.
LIF represses GnRH gene expression through the enhancer region. The 5-kb GnRH reporter containing 5 kb of the GnRH gene regulatory sequence from the transcriptional start site (a), reporter containing GnRH enhancer (E, −1,863/−1,571) and promoter (P, −173/+1) (b), reporter containing GnRH promoter −173/+1 (P) (c), and reporter containing GnRH enhancer (E, −1,863/−1,571) linked to the heterologous RSV (Raus Sarcoma Virus) promoter (GnRH E/RSVp luc) (d) were transfected in GT1–7 cells and cells treated with LIF for 24 h. Results demonstrate sufficiency of the enhancer for GnRH repression by LIF. Statistical significance (p < 0.05), indicated with an asterisk, was determined with a t test.
STAT3 Is Not Necessary for GnRH Repression by LIF
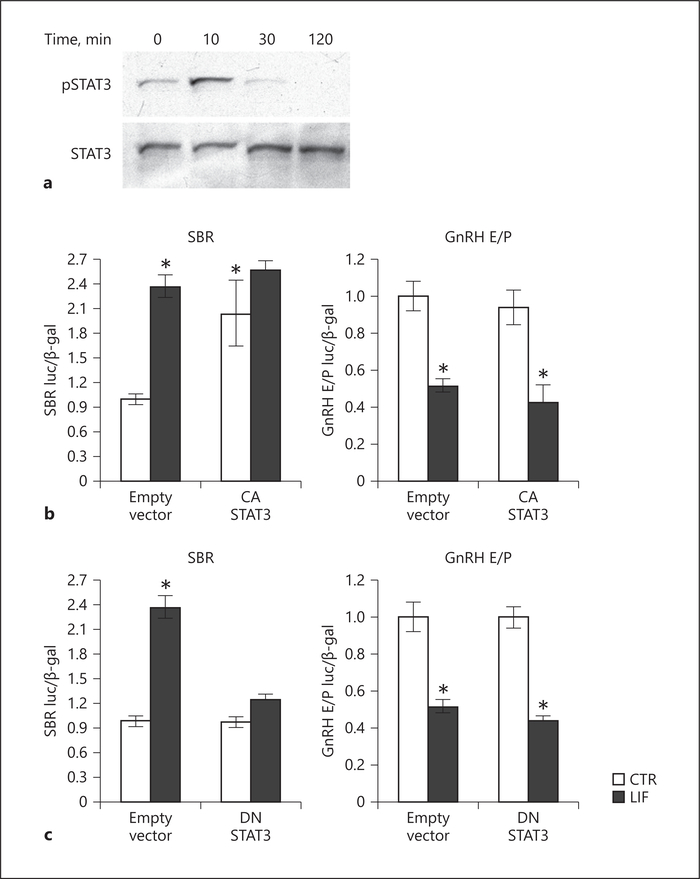
LIF signals through GP130 to activate the STAT pathway. To analyze signaling pathways activated by LIF, cells were treated with LIF and Western blots performed using whole-cell lysate. LIF treatment resulted in increased levels of STAT3 phosphorylation (Fig. 5a). To determine whether STAT3 is sufficient to repress GnRH gene expression, GT1–7 cells were co-transfected with a CA STAT3 mutant (CA STAT3) and STAT binding region (SBR), a reporter containing 6 copies of the STAT3 response element that serves as a positive control; or with the GnRH E/P reporter. CA STAT3 overexpression induced STAT3-SBR to the similar level as induction by LIF. LIF treatment together with CA STAT3 overexpression did not further increase induction over CA STAT3 alone, indicating that CA STAT3 maximally induces the SBR reporter. On the other hand, CA STAT3 overexpression did not affect the expression of the GnRH E/P reporter or its repression by LIF (Fig. 5b). We then analyzed necessity of STAT3 by co-transfecting DN mutant STAT3 (DN STAT3) with SBR or with GnRH E/P. While transfection with DN STAT3 inhibited LIF induction of SBR positive control, DN STAT3 did not prevent repression of GnRH E/P by LIF (Fig. 5c). Taken together, these results demonstrate that STAT3 is not necessary for GnRH repression by LIF.
Fig. 5.
STAT3 is not sufficient or necessary for GnRH repression by LIF. a Analysis of STAT3 phosphorylation by Western blot of GT1–7 cell lysates after 10 ng/mL LIF treatment, for 10, 30, and 120 min. b Co-transfection of a reporter containing 6 copies of the STAT3 response element (SBR) or GnRH E/P with a constitutively active (CA) STAT3 demonstrates that CA-STAT3 is sufficient to induce SBR, but not GnRH E/P. c Co-transfection of a dominant negative (DN) STAT3 inhibits LIF induction of SBR-luciferase, but does not prevent LIF repression of the GnRH E/P reporter. Statistical significance (asterisk, p < 0.05) was determined with ANOVA followed by Tukey’s post hoc test.
LIF Activates p38 to Repress GnRH
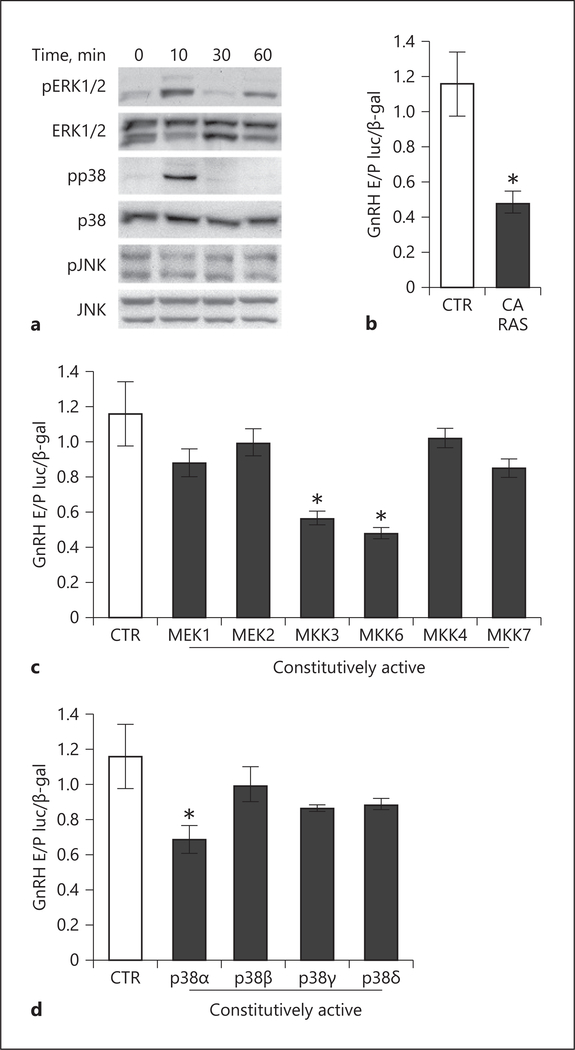
LIF signaling has been shown to activate the mitogen-activated protein kinase (MAPK) pathway, which includes extracellular signal-regulated protein kinases (ERK½), p38, and the c-Jun N-terminal kinases (JNK) in a variety of other cells, including mouse embryonic stem cells, 3T3-L1, and AtT20 pituitary corticotrope cells [84, 85]. LIF treatment of GT1–7 cells resulted in phosphorylation of ERK½ and p38, but no changes in JNK phosphorylation were observed (Fig. 6a). To further delineate the necessity of MAPK signaling pathway, co-transfection assays with expression plasmid containing CA RAS (CA RAS), that is upstream to the MAPK pathway, resulted in repression of GnRH expression by 51.4% (Fig. 6b). To identify the MAPK kinase pathway sufficient for repression, co-transfection assays were conducted using CA forms of MEK1 and MEK2 that lead to ERK½ activation, MKK3 and MKK6 that that activate p38, or MKK4 and MKK7 that activate JNK. CA MEK1, MEK2, MKK4, and MKK7 had no effect on GnRH expression. GnRH expression was repressed with CA MKK3 and MKK6, which lead to activation of p38, by 40.9 and 48.2%, respectively (Fig. 6c).
Fig. 6.
p38α is sufficient to repress GnRH gene expression. a LIF treatment activates ERK½, p38, but not JNK, demonstrated by Western blots using whole-cell lysate of GT1–7 cells treated with LIF for times indicated above each lane. The GnRH reporter is repressed by co-transfection with constitutively active (CA) RAS (b), CA MKK3 and CA MKK6, upstream activators of p38 (c), and CA p38α (d). Statistical significance (p < 0.05), indicated by an asterisk, was determined with ANOVA followed by Tukey’s post hoc test.
The p38 kinases have four isoforms, p38α, p38β, p38γ, and p38σ [86]. To explore the role of these isoforms on GnRH, CA mutants of p38 isoforms were analyzed in co-transfection assays. Reporter activity was repressed by 31% with CA p38α, while overexpression of CA p38β, p38γ, and p38σ had no effect on luciferase activity (Fig. 6d). These results demonstrate that LIF represses GnRH transcriptional activity via the MAPK pathway, specifically p38, and highlights the important role that p38α plays in this repression.
cFOS Is Induced by LIF in GT1–7 Cells and Represses GnRH
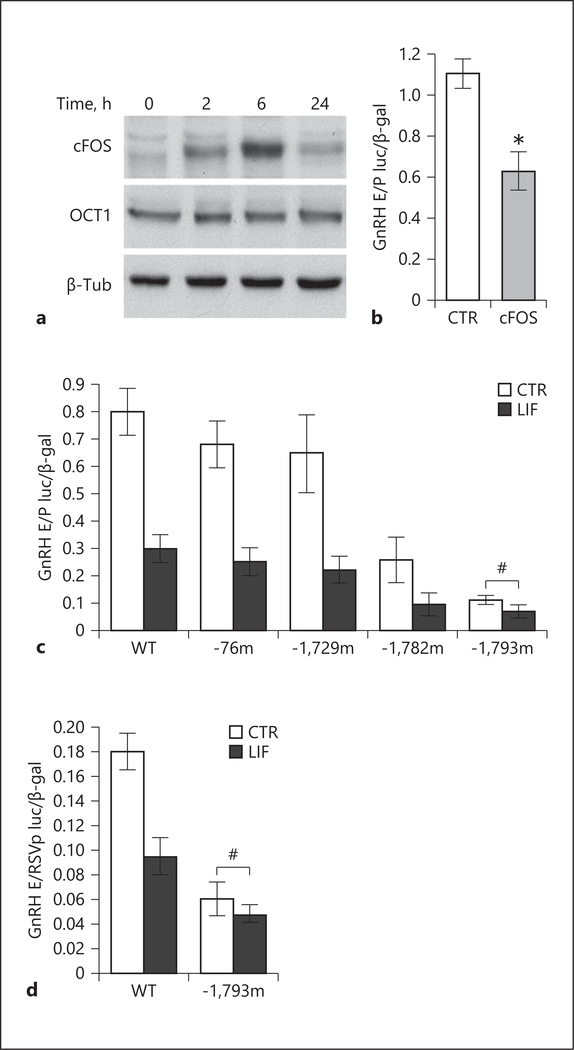
OCT-1 has been previously identified as an essential regulator of GnRH gene transcription [87]. Western blot analysis revealed that OCT-1 protein level was not affected by LIF treatment in GT1–7 cells, suggesting that OCT-1 may not mediate LIF repression of the GnRH gene. GnRH gene repression by cFOS has previously been shown [72], and thus we next examined cFOS protein levels. Indeed, Western blots demonstrated that cFOS was induced by LIF (Fig. 7a). The GnRH E/P reporter was repressed by 43.4% with overexpression of cFOS (Fig. 7b). This indicates that induction of cFOS is likely mediating LIF repression of GnRH.
Fig. 7.
cFOS mediates LIF repression of the GnRH gene. a LIF treatment of GT1–7 cells induces cFOS, but not OCT-1, demonstrated by Western blots of nuclear extracts. b Overexpression of cFOS, by co-transfection of cFOS expression vector, is sufficient to repress the GnRH E/P reporter. Asterisk indicates significant difference (p < 0.05) determined with the t test. c Transient transfections of reporters containing the mutations of the putative cFOS binding sites in GT1–7 cells demonstrate that the mutation of the cFOS binding site at −1,793 abolishes LIF repression of the GnRH E/P reporter, indicated by the pound sign #. d The same mutation was created in the GnRH E/RSVp reporter and also abrogates the repression by LIF. Statistical significance (p < 0.05), indicated by an asterisk, was determined with ANOVA followed by Tukey’s post hoc test. # indicates lack of repression.
Several cFOS binding sites were previously identified in the GnRH regulatory region [71–73]. To identify the site involved in LIF repression of GnRH, GT1–7 cells were transfected with luciferase reporters containing mutation of the putative cFOS sites at −76 in the promoter region (−76m), −1,729 (−1,729m), −1,782 (−1,782m), and −1,793 (−1,793m) in the enhancer and their expression was compared to wild-type GnRH E/P. LIF repressed luciferase activity of all reporters used by 61.0, 61.8, 65.9, and 61.6% respectively, except the −1,793 mutant, indicating that the mutation of this site prevents LIF repression of GnRH (Fig. 7c). Thus, the −1,793 site is involved in LIF repression of GnRH. We confirmed the role of this site by transfecting the mutation of the −1,793 site in the GnRH E/RSVp reporter and comparing the repression of the mutant to the wild-type reporter containing the GnRH enhancer linked to the RSV promoter (GnRH E/RSVp). Similar to what was observed in Figure 7c, LIF repression of GnRH was inhibited with a mutation of the −1,793 site (Fig. 7d). Therefore, LIF induces cFOS which represses GnRH gene expression via the −1,793 site.
cFOS Expression Is Induced with HFD in Mice
Since we determined that cFOS mediates LIF repression of the GnRH gene and we observed reduced GnRH mRNA levels in the hypothalami of mice fed HFD, we explored cFOS expression in the hypothalamus. We concentrated specifically on the sections containing the OVLT where the largest numbers of GnRH neuron soma are found. In the hypothalamus of GnRH-GFP mice fed HFD, a significant increase in the percent of double-labeled green GnRH neurons with cFOS (red) was observed compared to CTR (Fig. 8a, b.1). In CTR mice, 17.9% of GnRH neurons expressed cFOS, while in the HFD mice, 36.3% of GnRH neurons expressed cFOS, suggesting that HFD induces cFOS expression in GnRH neurons. To examine the specificity of this increase, we also counted cells that express cFOS in two other areas, one proximal to OVLT and one more dorsal from OVLT, using DAPI to identify cell nuclei. We determined that there is an increase in the percent of cFOS-positive neurons in the area proximal to the OVLT of mice fed HFD (45.4%) compared to CTR (30.5%) (Fig. 8a.2, b.2). However, in the area dorsal to OVLT, we did not observe a difference in the number of neurons that express cFOS (Fig. 8b.3). Thus, obesity induces cFOS expression in neurons in the proximity to OVLT and in GnRH neurons. Given that cFOS mediates LIF repression of the GnRH gene in GT1–7 cells, and the reduced levels of GnRH mRNA observed in vivo coupled with an increased cFOS expression in GnRH neurons following LIF induction by inflammatory stimuli, our data together demonstrate that LIF represses Gnrh mRNA in vivo through cFOS. Furthermore, LIF may mediate repression of the Gnrh mRNA and reproductive function caused by inflammation.
Fig. 8.
More GnRH neurons express cFOS following high-fat diet (HFD). a Coronal sections of the preoptic area in the hypothalamus of GnRH-GFP mice following HFD stained for GFP (green) and cFOS (red). White arrows indicate GnRH neurons labeled with GFP and cFOS. Numbered squares correspond to enlarged areas below; 1, GnRH neurons green, cFOS red; arrows indicate GnRH neurons that express cFOS, an arrowhead points to the GnRH neuron without cFOS; 2–3, DAPI channel is included to facilitate cell count; nuclei blue, cFOS magenta. Arrows indicate cells labeled with cFOS and DAPI. b Quantification of neurons expressing cFOS in control (CTR, white bars) and HFD male mice (black bars): 1, increase in the percent of GnRH neurons with cFOS is observed in HFD compared to control. 2, quantification of the neurons that express cFOS, proximal to the OVLT delineated with #2 square; 3, quantification of the neurons that express cFOS, dorsally from the OVLT delineated with #3 square following control and HFD. Three hundred GnRH neurons were counted in each mouse. Statistical significance (p < 0.05), indicated by an asterisk, was determined with the t test.
Discussion
Both infection and obesity negatively affect hypothalamic function, GnRH neuronal network and reproduction, but the mechanisms are unknown. We postulated that neuroinflammation is a common characteristic between these conditions and that inflammatory cytokines may mediate impairment of reproductive function by both infection and obesity. Classical pro-inflammatory cytokines, TNF-α, IL-1β, and IL-6, are induced in the brain during inflammation, when GnRH and LH expression and secretion are diminished, but direct regulation of GnRH neuron function by cytokines remains inconclusive. In our recently published report [24] and herein, we identified a novel cytokine, LIF, which is induced during these inflammatory states that directly affects GnRH gene expression. In this report, we also determined the precise mechanisms of GnRH gene repression and signaling pathways that are necessary for LIF-mediated effects on GnRH neurons. Finally, we demonstrated that the transcription factor induced by LIF to repress the GnRH gene is increased in a location-specific manner following HFD in vivo.
Repression of Gnrh mRNA expression is consistently observed during inflammatory states. In agreement with previous reports, acute inflammation, elicited with an injection of LPS and chronic inflammation, caused by HFD-induced obesity, suppresses GnRH mRNA in our studies. LPS treatment, resulting in inflammation, represses Gnrh in ewes [88], birds [89], and rats [90]. Infusion of the pro-inflammatory IL-1β cytokine into the rodent hypothalamus also represses Gnrh expression [16]. We and others reported that obese mice fed HFD, with a low-grade chronic inflammation, exhibit lower Gnrh mRNA [24, 91, 92]. These reports suggested that repression of Gnrh mRNA during inflammation may be a result of increased cytokine concentration. Alternatively, microglia, brain-resident immune cells, are involved in the regulation of synaptic transmission and activity-dependent structural remodeling [93]. In neuroinflammation, in response to injury, infection, or disease, microglia engulf damaged synapses [93]. We demonstrated decreased synaptic spine density in GnRH neurons following obesity-mediated neuroinflammation and microglia activation, indicating reduced connectivity [24]. This reduced synaptic activity may regulate Gnrh expression in an activity-dependent manner [94, 95]. Some synaptic proteins are regulated in an activity-dependent manner at the transcriptional level [96]. This may be the case with GnRH gene expression as well, since several hypothalamic factors, such as RFamide-related peptide 3 (RFRP −3) – a mammalian gonadotropin-inhibitory hormone ortholog, senktide – a neurokinin B receptor agonist, and oxytocin alter both GnRH secretion and Gnrh transcription [97–99]. However, since GnRH neurons express cytokine receptors [27], herein, we investigated if the Gnrh gene is repressed via activation of cytokine receptor signaling pathways.
We identified LIF as a critical player in the neuroinflammation-induced impairment of GnRH gene expression. In the brain, LIF is involved in neural stem cell maintenance and axonal growth, and in modulation of gene expression [100]. During embryonic development, LIF is expressed in the olfactory placode, which led to the hypothesis that LIF affects GnRH neuron migration [54]. Our results demonstrate that LIF represses GnRH gene expression, which is in agreement with findings that GnRH mRNA is repressed during embryonic GnRH neuron migration [101], in addition to repression during inflammation. To analyze the effects of infection on GnRH, previous studies analyzed classical inflammatory markers, TNF-α, IL-1β, and IL-6, but not LIF. Although LPS, or TNF-α or IL-1β administration in the ventricle consistently reduces GnRH secretion in vivo [18, 20, 21, 23], some have found that IL-6 treatment of the hypothalamic slices stimulates GnRH secretion [19, 102]. While IL-1β and TNF-α are prototypical pro-inflammatory cytokines, IL-6 may function as either a pro- or anti-inflammatory cytokine depending on timing and stimulus. IL-6 synthesis in the hypothalamus is induced by IL-1β [103], and IL-6 may be engaged in the negative feedback to dampen inflammation [42]. Thus, results may vary due to the differences between in vivo and in vitro treatments, since initially induced cytokines may cause a cascade of other cytokines in vivo. For this reason, we concentrated on analyzing the effects of LIF in GT1–7 cells. LIF alone repressed Gnrh mRNA and GnRH reporter, similarly to HFD-induced and LPS-elicited repression of GnRH mRNA in vivo. LIF is induced by IL-1β in vivo. Therefore, IL-1β repression of GnRH mRNA, demonstrated in previous studies, may occur via induction of LIF.
We determined that LIF induces cFOS which represses GnRH gene expression. GnRH repression by cFOS has been demonstrated previously and several cFOS binding sites have been identified in the GnRH regulatory region [72, 73]. However, a specific site necessary for GnRH repression has not been identified, until now. The GnRH gene is inhibited by PKC signaling pathway, which induces cFOS [70, 104]; and PKC activation by TPA treatment represses the GnRH gene via the −1,793 site [71]. Here, we demonstrate that this same site at −1,793 is involved in LIF repression of the GnRH gene. This site is juxtaposed to the OCT-1 binding site, and OCT-1 is the critical transcription factor that regulates GnRH gene expression [71, 87, 105, 106]. Thus, although OCT-1 protein levels do not change with LIF treatment, its interaction with cFOS [107], likely modulates that level of GnRH gene expression.
Signaling pathways involved in the regulation of GnRH gene transcription by cytokines have not been elucidated. GP130, the common signal transducer for IL-6 and LIF family, is expressed in GnRH neurons in vivo. LIF treatment activates STAT3 in GT1–7 cells, a model of mature GnRH neurons, as we show herein, and in GN11 cells, a model of immature, migrating GnRH neurons [54]. However, STAT3 does not play a role in GnRH gene repression. To exert its effect on GnRH gene expression, LIF activates MAPK-p38 pathway, and in particular p38α is critical for repression. This is an important finding, since while other p38 isoforms have normal physiological functions, p38α is involved in inflammatory processes [108]. Moreover, p38α is the target for several potential therapeutics for chronic inflammatory disorders.
Leptin is necessary for pubertal transition and normal reproductive function [109–111]. Since leptin is elevated in obese and HFD-fed mice, leptin was previously analyzed as a mediator of obesity-induced impairment of reproductive function [10, 112–114]. Additionally, a mouse model with leptin overexpression exhibits early puberty and lower GnRH at 21 weeks of age [115], which may correlate with precocious puberty in obese girls and lower Gnrh mRNA in obese mice in our study. However, previous studies determined that leptin does not directly affect GnRH neurons in vivo, since deletion of the leptin receptor specifically in GnRH neurons had no effect on fertility [116]. STAT3 signaling molecule that is activated by leptin, is required for leptin’s role in energy balance, but is not required for leptin’s effect on fertility [117, 118]. STAT3 studies demonstrate separate roles for leptin signaling pathways, involved in metabolic function that require STAT3, and reproductive function that do not. Although leptin receptor is similar to the IL-6 and LIF signaling receptor GP130 and can activate the same signaling pathways, leptin does not engage GP130 [119]. Given that in vivo studies determined that leptin influences reproductive function via its actions in the brain, but indirectly, via GnRH neuron afferents, it is unlikely that elevated leptin levels in our studies had direct effects on GnRH gene expression.
We demonstrate that cFOS expression is increased specifically in GnRH neurons and in cells located close to the OVLT, but not in cells located more dorsally, following HFD. Others reported increased cFOS following HFD in specific areas and postulated that the location of cFOS induction is dependent on the function of the neuronal population in which it is induced. For example, cFOS was increased in the reward circuitry following HFD [120] and in the dorsomedial and lateral hypothalamus [121]. cFOS is similarly induced following LPS treatment in the hypothalamus [122] and this induction is location specific [123]. These studies also postulated that the location is dependent on the neuron function and suggested that cFOS was induced following neuron activation, since cFOS is the most commonly used marker of neuronal activity. As an immediate early gene, cFOS is induced via activity-dependent transcription in neurons. cFOS expression is increased in activated GnRH neurons, during the preovulatory LH surge and after kisspeptin treatment [124, 125]. We, on the other hand, posit that location of the cFOS induction is dependent on the proximity to the vasculature. The roles of cFOS as a transcription factor, involved in the GnRH gene regulation, and as a marker of neuronal activity, are not mutually exclusive. Thus, we postulate that increased LIF concentration in the proximity to GnRH neurons induces cFOS in GnRH neurons to repress GnRH gene expression. Specificity for GnRH neurons may be due to either their proximity to the OVLT or because GnRH processes extend beyond the blood brain barrier into the OVLT.
Our studies delineate the mechanisms of neuroinflammation-mediated impairment of GnRH gene expression, and may explain repression of reproductive function during infection and in obesity. We identified LIF as the cytokine that mediates these effects and p38α as a critical signaling molecule. We also postulate that the proximal location of GnRH neurons to the leaky blood brain barrier, where the local concentration of this cytokine may be the highest, permits for repression of GnRH gene expression via cFOS induction. Future studies will determine the in vivo role of p38α in neuroinflammation-mediated GnRH repression.
Acknowledgement
The authors thank Suzanne Moenter for GnRH-GFP mouse line and Pamela Mellon for GT1–7 cell line. We also thank Pamela Mellon, Hanne Hoffmann, Kellie Breen, and Peiqing Sun for plasmids. We are very grateful to Mark Lawson for his help with statistical analyses.
Funding Sources
This study was supported by R01 HD091167 from NIH NICHD to D. Coss. Nancy Lainez was supported in part by Biomedical Sciences graduate program.
Footnotes
Disclosure Statement
The authors have no conflicts of interest to declare
Statement of Ethics
All experiments were performed with approval from the University of California (Riverside, CA, USA) Animal Care and Use Committee and in accordance with the National Institutes of Health Animal care and Use Guidelines.
References
- 1.Coss D Regulation of reproduction via tight control of gonadotropin hormone levels. Mol Cell Endocrinol. 2018. March; 463: 116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thackray VG, Mellon PL, Coss D. Hormones in synergy: regulation of the pituitary gonadotropin genes. Mol Cell Endocrinol. 2010. January; 314(2): 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herde MK, Iremonger KJ, Constantin S, Herbison AE. GnRH neurons elaborate a longrange projection with shared axonal and dendritic functions. J Neurosci. 2013. July; 33(31): 12689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarkson J, Han SY, Piet R, McLennan T, Kane GM, Ng J, et al. Definition of the hypothalamic GnRH pulse generator in mice. Proc Natl Acad Sci USA. 2017. November; 114(47):E10216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman RL, Lehman MN. Kisspeptin neurons from mice to men: similarities and differences. Endocrinology. 2012. November; 153(11): 5105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breen KM, Karsch FJ. New insights regarding glucocorticoids, stress and gonadotropin suppression. Front Neuroendocrinol. 2006. July; 27(2): 233–45. [DOI] [PubMed] [Google Scholar]
- 7.Acevedo-Rodriguez A, Kauffman AS, Cher-rington BD, Borges CS, Roepke TA, Laconi M. Emerging insights into hypothalamic-pituitary-gonadal axis regulation and interaction with stress signalling. J Neuroendocrinol. 2018. October; 30(10):e12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gore AC, Patisaul HB. Neuroendocrine disruption: historical roots, current progress, questions for the future. Front Neuroendocrinol. 2010. October; 31(4): 395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo KA, La JL, Stephens SB, Poling MC, Padgaonkar NA, Jennings KJ, et al. Circadian Control of the Female Reproductive Axis Through Gated Responsiveness of the RFRP-3 System to VIP Signaling. Endocrinology. 2015. July; 156(7): 2608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans MC, Anderson GM. Integration of circadian and metabolic control of reproductive function. Endocrinology. 2018. November; 159(11): 3661–73. [DOI] [PubMed] [Google Scholar]
- 11.Manfredi-Lozano M, Roa J, Tena-Sempere M. Connecting metabolism and gonadal function: novel central neuropeptide pathways involved in the metabolic control of puberty and fertility. Front Neuroendocrinol. 2018. January; 48: 37–49. [DOI] [PubMed] [Google Scholar]
- 12.Evans MC, Anderson GM. Neuroendocrine integration of nutritional signals on reproduction. J Mol Endocrinol. 2017. February; 58(2):R107–28. [DOI] [PubMed] [Google Scholar]
- 13.Rivest S, Rivier C. The role of corticotropin-releasing factor and interleukin-1 in the regulation of neurons controlling reproductive functions. Endocr Rev. 1995. April; 16(2): 177–99. [DOI] [PubMed] [Google Scholar]
- 14.Kalra PS, Edwards TG, Xu B, Jain M, Kalra SP. The anti-gonadotropic effects of cytokines: the role of neuropeptides. Domest Anim Endocrinol. 1998. September; 15(5): 321–32. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y, Cai D. Hypothalamic inflammation and GnRH in aging development. Cell Cycle. 2013. September; 12(17): 2711–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivest S, Lee S, Attardi B, Rivier C. The chronic intracerebroventricular infusion of interleukin-1 beta alters the activity of the hypothalamic-pituitary-gonadal axis of cycling rats. I. Effect on LHRH and gonadotropin biosynthesis and secretion. Endocrinology. 1993. December; 133(6): 2424–30. [DOI] [PubMed] [Google Scholar]
- 17.Rivest S, Rivier C. Centrally injected interleukin-1 beta inhibits the hypothalamic LHRH secretion and circulating LH levels via prostaglandins in rats. J Neuroendocrinol. 1993. August; 5(4): 445–50. [DOI] [PubMed] [Google Scholar]
- 18.Dondi D, Limonta P, Montagnani Marelli M, Piva F. Mechanism of action of interleukin-1 in modulating gonadotropin secretion. In vivo and in vitro studies. Biol Signals Recept. 1998. Jan-Feb; 7(1): 55–60. [DOI] [PubMed] [Google Scholar]
- 19.Feleder C, Wuttke W, Moguilevsky JA. Hypothalamic relationships between interleukin-6 and LHRH release affected by bacterial endotoxin in adult male rats. Involvement of the inhibitory amino acid system. Biol Signals. 1998. January; 7(1): 7–14. [PubMed] [Google Scholar]
- 20.Watanobe H, Hayakawa Y. Hypothalamic interleukin-1 beta and tumor necrosis factoralpha, but not interleukin-6, mediate the endotoxin-induced suppression of the reproductive axis in rats. Endocrinology. 2003. November; 144(11): 4868–75. [DOI] [PubMed] [Google Scholar]
- 21.Rivier C, Vale W. Cytokines act within the brain to inhibit luteinizing hormone secretion and ovulation in the rat. Endocrinology. 1990. August; 127(2): 849–56. [DOI] [PubMed] [Google Scholar]
- 22.Feng YJ, Shalts E, Xia LN, Rivier J, Rivier C, Vale W, et al. An inhibitory effects of interleukin-1a on basal gonadotropin release in the ovariectomized rhesus monkey: reversal by a corticotropin-releasing factor antagonist. Endocrinology. 1991. April; 128(4): 2077–82. [DOI] [PubMed] [Google Scholar]
- 23.Yoo MJ, Nishihara M, Takahashi M. Tumor necrosis factor-alpha mediates endotoxin induced suppression of gonadotropin-releasing hormone pulse generator activity in the rat. Endocr J. 1997. February; 44(1): 141–8. [DOI] [PubMed] [Google Scholar]
- 24.Lainez NM, Jonak CR, Nair MG, Ethell IM, Wilson EH, Carson MJ, et al. Diet-Induced Obesity Elicits Macrophage Infiltration and Reduction in Spine Density in the Hypothalami of Male but Not Female Mice. Front Immunol. 2018. September; 9: 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010; 72(1): 219–46. [DOI] [PubMed] [Google Scholar]
- 26.Thaler JP, Yi CX, Schur EA, Guyenet SJ, Hwang BH, Dietrich MO, et al. Obesity is associated with hypothalamic injury in rodents and humans. J Clin Invest. 2012. January; 122(1): 153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jasoni CL, Todman MG, Han SK, Herbison AE. Expression of mRNAs encoding receptors that mediate stress signals in gonadotropin-releasing hormone neurons of the mouse. Neuroendocrinology. 2005; 82(5–6): 320–8. [DOI] [PubMed] [Google Scholar]
- 28.Drutskaya MS, Efimov GA, Astrakhantseva IV, Kruglov AA, Nedospasov SA. Making anti-cytokine therapy more selective: studies in mice. Cytokine. 2018. January; 101: 33–8. [DOI] [PubMed] [Google Scholar]
- 29.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003. September; 3(9): 745–56. [DOI] [PubMed] [Google Scholar]
- 30.Rizzo FR, Musella A, De Vito F, Fresegna D, Bullitta S, Vanni V, et al. Tumor Necrosis Factor and Interleukin-1β Modulate Synaptic Plasticity during Neuroinflammation. Neural Plast. 2018. May; 2018: 8430123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kempuraj D, Thangavel R, Natteru PA, Selvakumar GP, Saeed D, Zahoor H, et al. Neuroinflammation Induces Neurodegeneration. J Neurol Neurosurg Spine. 2016; 1(1): 1003. [PMC free article] [PubMed] [Google Scholar]
- 32.Allan SM, Tyrrell PJ, Rothwell NJ. Interleukin-1 and neuronal injury. Nat Rev Immunol. 2005. August; 5(8): 629–40. [DOI] [PubMed] [Google Scholar]
- 33.Olmos G, Lladó J. Tumor necrosis factor alpha: a link between neuroinflammation and excitotoxicity. Mediators Inflamm. 2014; 2014: 861231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sankowski R, Mader S, Valdés-Ferrer SI. Systemic inflammation and the brain: novel roles of genetic, molecular, and environmental cues as drivers of neurodegeneration. Front Cell Neurosci. 2015. February; 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yirmiya R, Pollak Y, Morag M, Reichenberg A, Barak O, Avitsur R, et al. Illness, cytokines, and depression. Ann N Y Acad Sci. 2000; 917(1): 478–87. [DOI] [PubMed] [Google Scholar]
- 36.Pegoretti V, Baron W, Laman JD, Eisel UL. Selective Modulation of TNF-TNFRs Signaling: Insights for Multiple Sclerosis Treatment. Front Immunol. 2018. April; 9: 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grebe A, Hoss F, Latz E. NLRP3 Inflammasome and the IL-1 Pathway in Atherosclerosis. Circ Res. 2018. June; 122(12): 1722–40. [DOI] [PubMed] [Google Scholar]
- 38.Ballak DB, Stienstra R, Tack CJ, Dinarello CA, van Diepen JA. IL-1 family members in the pathogenesis and treatment of metabolic disease: focus on adipose tissue inflammation and insulin resistance. Cytokine. 2015. October; 75(2): 280–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinrich PC, Behrmann I, Müller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998. September; 334(Pt 2): 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Covarrubias AJ, Horng T. IL-6 strikes a balance in metabolic inflammation. Cell Metab. 2014. June; 19(6): 898–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauer J, Denson JL, Brüning JC. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 2015. February; 36(2): 92–101. [DOI] [PubMed] [Google Scholar]
- 42.Del Giudice M, Gangestad SW. Rethinking IL-6 and CRP: why they are more than inflammatory biomarkers, and why it matters. Brain Behav Immun. 2018. May; 70: 61–75. [DOI] [PubMed] [Google Scholar]
- 43.Villiger PM, Geng Y, Lotz M. Induction of cytokine expression by leukemia inhibitory factor. J Clin Invest. 1993. April; 91(4): 1575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suman P, Malhotra SS, Gupta SK. LIF-STAT signaling and trophoblast biology. JAK-STAT. 2013. October; 2(4):e25155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicola NA, Babon JJ. Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev. 2015. October; 26(5): 533–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosario GX, Stewart CL: The Multifaceted Actions of Leukaemia Inhibitory Factor in Mediating Uterine Receptivity and Embryo Implantation. Am J Reprod Immunol. 2016; 75: 246–255. [DOI] [PubMed] [Google Scholar]
- 47.Ohtsuka S, Nakai-Futatsugi Y, Niwa H. LIF signal in mouse embryonic stem cells. JAK-STAT. 2015. September; 4(2):e1086520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onishi K, Zandstra PW. LIF signaling in stem cells and development. Development. 2015. July; 142(13): 2230–6. [DOI] [PubMed] [Google Scholar]
- 49.Patterson PH. Leukemia inhibitory factor, a cytokine at the interface between neurobiology and immunology. Proc Natl Acad Sci USA. 1994. August; 91(17): 7833–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holmberg KH, Patterson PH. Leukemia inhibitory factor is a key regulator of astrocytic, microglial and neuronal responses in a low-dose pilocarpine injury model. Brain Res. 2006. February; 1075(1): 26–35. [DOI] [PubMed] [Google Scholar]
- 51.Bauer S, Patterson PH. Leukemia inhibitory factor promotes neural stem cell self-renewal in the adult brain. J Neurosci. 2006. November; 26(46): 12089–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dozio E, Ruscica M, Galliera E, Corsi MM, Magni P. Leptin, ciliary neurotrophic factor, leukemia inhibitory factor and interleukin-6: class-I cytokines involved in the neuroendocrine regulation of the reproductive function. Curr Protein Pept Sci. 2009. December; 10(6): 577–84. [DOI] [PubMed] [Google Scholar]
- 53.Dozio E, Watanobe H, Ruscica M, Maggi R, Motta M, Magni P. Expression of functional ciliary neurotrophic factor receptors in immortalized gonadotrophin-releasing hormone-secreting neurones. J Neuroendocrinol. 2005. May; 17(5): 286–91. [DOI] [PubMed] [Google Scholar]
- 54.Magni P, Dozio E, Ruscica M, Watanobe H, Cariboni A, Zaninetti R, et al. Leukemia inhibitory factor induces the chemomigration of immortalized gonadotropin-releasing hormone neurons through the independent activation of the Janus kinase/signal transducer and activator of transcription 3, mitogen-activated protein kinase/extracellularly regulated kinase ½, and phosphatidylinositol 3-kinase/Akt signaling pathways. Mol Endocrinol. 2007. May; 21(5): 1163–74. [DOI] [PubMed] [Google Scholar]
- 55.Miyata S New aspects in fenestrated capillary and tissue dynamics in the sensory circumventricular organs of adult brains. Front Neurosci. 2015. October; 9: 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herde MK, Geist K, Campbell RE, Herbison AE. Gonadotropin-releasing hormone neurons extend complex highly branched dendritic trees outside the blood-brain barrier. Endocrinology. 2011. October; 152(10): 3832–41. [DOI] [PubMed] [Google Scholar]
- 57.Romanovsky AA, Almeida MC, Aronoff DM, Ivanov AI, Konsman JP, Steiner AA, et al. : Fever and hypothermia in systemic inflammation: recent discoveries and revisions. Front Biosci 2005; 10: 2193–2216. [DOI] [PubMed] [Google Scholar]
- 58.Shibata M, Blatteis CM. Human recombinant tumor necrosis factor and interferon affect the activity of neurons in the organum vasculosum laminae terminalis. Brain Res. 1991. October; 562(2): 323–6. [DOI] [PubMed] [Google Scholar]
- 59.Dinarello CA. Infection, fever, and exogenous and endogenous pyrogens: some concepts have changed. J Endotoxin Res. 2004; 10(4): 201–22. [DOI] [PubMed] [Google Scholar]
- 60.Ott D, Murgott J, Rafalzik S, Wuchert F, Schmalenbeck B, Roth J, et al. Neurons and glial cells of the rat organum vasculosum laminae terminalis directly respond to lipopolysaccharide and pyrogenic cytokines. Brain Res. 2010. December; 1363: 93–106. [DOI] [PubMed] [Google Scholar]
- 61.Forni PE, Wray S. GnRH, anosmia and hypogonadotropic hypogonadism—where are we? Front Neuroendocrinol. 2015. January; 36: 165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiner RI, Wetsel W, Goldsmith P, Martinez de la Escalera G, Windle J, Padula C, et al. Gonadotropin-releasing hormone neuronal cell lines. Front Neuroendocrinol. 1992. April; 13(2): 95–119. [PubMed] [Google Scholar]
- 63.Mellon PL, Eraly SA, Belsham DD, Lawson MA, Clark ME, Whyte DB, et al. An immortal cell culture model of hypothalamic gonadotropin-releasing hormone neurons. Methods 1995; 7: 303–310. [Google Scholar]
- 64.Suter KJ, Song WJ, Sampson TL, Wuarin JP, Saunders JT, Dudek FE, et al. Genetic targeting of green fluorescent protein to gonadotropin-releasing hormone neurons: characterization of whole-cell electrophysiological properties and morphology. Endocrinology. 2000. January; 141(1): 412–9. [DOI] [PubMed] [Google Scholar]
- 65.Jonak CR, Lainez NM, Boehm U, Coss D. GnRH Receptor Expression and Reproductive Function Depend on JUN in GnRH Receptor‒Expressing Cells. Endocrinology. 2018. March; 159(3): 1496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jonak CR, Lainez NM, Roybal LL, Williamson AD, Coss D. c-JUN Dimerization Protein 2 (JDP2) Is a Transcriptional Repressor of Follicle-stimulating Hormone β (FSHβ) and Is Required for Preventing Premature Reproductive Senescence in Female Mice. J Biol Chem. 2017. February; 292(7): 2646–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roybal LL, Hambarchyan A, Meadows JD, Barakat NH, Pepa PA, Breen KM, et al. Roles of binding elements, FOXL2 domains, and interactions with cJUN and SMADs in regulation of FSHβ. Mol Endocrinol. 2014. October; 28(10): 1640–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lindaman LL, Yeh DM, Xie C, Breen KM, Coss D. Phosphorylation of ATF2 and interaction with NFY induces c-Jun in the gonadotrope. Mol Cell Endocrinol. 2013. January; 365(2): 316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coss D, Hand CM, Yaphockun KK, Ely HA, Mellon PL. p38 mitogen-activated protein kinase is critical for synergistic induction of the FSH(beta) gene by gonadotropin-releasing hormone and activin through augmentation of c-Fos induction and Smad phosphorylation. Mol Endocrinol. 2007. December; 21(12): 3071–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eraly SA, Mellon PL. Regulation of gonadotropin-releasing hormone transcription by protein kinase C is mediated by evolutionarily conserved, promoter-proximal elements. Mol Endocrinol. 1995; 9(7): 848–59. [DOI] [PubMed] [Google Scholar]
- 71.Tang Q, Mazur M, Mellon PL. The protein kinase C pathway acts through multiple transcription factors to repress gonadotropin-releasing hormone gene expression in hypothalamic GT1–7 neuronal cells. Mol Endocrinol. 2005. November; 19(11): 2769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glidewell-Kenney CA, Shao PP, Iyer AK, Grove AM, Meadows JD, Mellon PL. Neurokinin B causes acute GnRH secretion and repression of GnRH transcription in GT1–7 GnRH neurons. Mol Endocrinol. 2013. March; 27(3): 437–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoffmann HM, Gong P, Tamrazian A, Mellon PL. Transcriptional interaction between cFOS and the homeodomain-binding transcription factor VAX1 on the GnRH promoter controls Gnrh1 expression levels in a GnRH neuron maturation specific manner. Mol Cell Endocrinol. 2018. February; 461: 143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999. August; 98(3): 295–303. [DOI] [PubMed] [Google Scholar]
- 75.Kwong J, Chen M, Lv D, Luo N, Su W, Xiang R, et al. Induction of p38δ expression plays an essential role in oncogenic ras-induced senescence. Mol Cell Biol. 2013. October; 33(19): 3780–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reddy GR, Xie C, Lindaman LL, Coss D. GnRH increases c-Fos half-life contributing to higher FSHβ induction. Mol Endocrinol. 2013. February; 27(2): 253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ely HA, Mellon PL, Coss D. GnRH induces the c-Fos gene via phosphorylation of SRF by the calcium/calmodulin kinase II pathway. Mol Endocrinol. 2011. April; 25(4): 669–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coss D, Jacobs SB, Bender CE, Mellon PL. A novel AP-1 site is critical for maximal induction of the follicle-stimulating hormone beta gene by gonadotropin-releasing hormone. J Biol Chem. 2004. January; 279(1): 152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nelson SM, Fleming R. Obesity and reproduction: impact and interventions. Curr Opin Obstet Gynecol. 2007. August; 19(4): 384–9. [DOI] [PubMed] [Google Scholar]
- 80.Feleder C, Refojo D, Jarry H, Wuttke W, Moguilevsky JA. Bacterial endotoxin inhibits LHRH secretion following the increased release of hypothalamic GABA levels. Different effects on amino acid neurotransmitter release. Neuroimmunomodulation. 1996. Nov-Dec; 3(6): 342–51. [DOI] [PubMed] [Google Scholar]
- 81.Odegaard JI, Chawla A. Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science. 2013. January; 339(6116): 172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Whyte DB, Lawson MA, Belsham DD, Eraly SA, Bond CT, Adelman JP, et al. A neuronspecific enhancer targets expression of the gonadotropin-releasing hormone gene to hypothalamic neurosecretory neurons. Mol Endocrinol. 1995. April; 9(4): 467–77. [DOI] [PubMed] [Google Scholar]
- 83.Lawson MA, MacConell LA, Kim J, Powl BT, Nelson SB, Mellon PL. Neuron-specific expression in vivo by defined transcription regulatory elements of the GnRH gene. Endocrinology. 2002; 143: 1404–12. [DOI] [PubMed] [Google Scholar]
- 84.Schiemann WP, Nathanson NM. Raf-1 independent stimulation of mitogen-activated protein kinase by leukemia inhibitory factor in 3T3-L1 cells. Oncogene. 1998. May; 16(20): 2671–9. [DOI] [PubMed] [Google Scholar]
- 85.Ernst M, Oates A, Dunn AR. Gp130-mediated signal transduction in embryonic stem cells involves activation of Jak and Ras/mitogenactivated protein kinase pathways. J Biol Chem. 1996. November; 271(47): 30136–43. [DOI] [PubMed] [Google Scholar]
- 86.Askari N, Diskin R, Avitzour M, Capone R, Livnah O, Engelberg D. Hyperactive variants of p38alpha induce, whereas hyperactive variants of p38gamma suppress, activating protein 1-mediated transcription. J Biol Chem. January; 282(1): 91–9. [DOI] [PubMed] [Google Scholar]
- 87.Clark ME, Mellon PL. The POU homeodomain transcription factor Oct-1 is essential for activity of the gonadotropin-releasing hormone neuron-specific enhancer. Mol Cell Biol. 1995. November; 15(11): 6169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haziak K, Herman AP, Tomaszewska-Zaremba D. Effects of central injection of anti-LPS antibody and blockade of TLR4 on GnRH/LH secretion during immunological stress in anestrous ewes. Mediators Inflamm. 2014; 2014: 867170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lopes PC, Wingfield JC, Bentley GE. Lipopolysaccharide injection induces rapid decrease of hypothalamic GnRH mRNA and peptide, but does not affect GnIH in zebra finches. Horm Behav. 2012. July; 62(2): 173–9. [DOI] [PubMed] [Google Scholar]
- 90.Nappi RE, Rivest S. Effect of immune and metabolic challenges on the luteinizing hormone-releasing hormone neuronal system in cycling female rats: an evaluation at the transcriptional level. Endocrinology. 1997. April; 138(4): 1374–84. [DOI] [PubMed] [Google Scholar]
- 91.Tortoriello DV, McMinn J, Chua SC. Dietary-induced obesity and hypothalamic infertility in female DBA/2J mice. Endocrinology. 2004. March; 145(3): 1238–47. [DOI] [PubMed] [Google Scholar]
- 92.Nam KN, Mounier A, Wolfe CM, Fitz NF, Carter AY, Castranio EL, et al. Effect of high fat diet on phenotype, brain transcriptome and lipidome in Alzheimer’s model mice. Sci Rep. 2017. June; 7(1): 4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schafer DP, Lehrman EK, Stevens B. The “quad-partite” synapse: microglia-synapse interactions in the developing and mature CNS. Glia. 2013. January; 61(1): 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bao J, Lin H, Ouyang Y, Lei D, Osman A, Kim TW, et al. Activity-dependent transcription regulation of PSD-95 by neuregulin-1 and Eos. Nat Neurosci. 2004. November; 7(11): 1250–8. [DOI] [PubMed] [Google Scholar]
- 95.Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013. January; 493(7432): 327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jordan BA, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat Neurosci. April; 10(4): 427–35. [DOI] [PubMed] [Google Scholar]
- 97.Grachev P, Li XF, Kinsey-Jones JS, di Domenico AL, Millar RP, Lightman SL, et al. Suppression of the GnRH pulse generator by neurokinin B involves a κ-opioid receptor-dependent mechanism. Endocrinology. 2012. October; 153(10): 4894–904. [DOI] [PubMed] [Google Scholar]
- 98.Salehi MS, Khazali H, Mahmoudi F, Janahmadi M: Oxytocin Intranasal Administration Affects Neural Networks Upstream of GNRH Neurons. J Mol Neurosci 2017; 62(3–4): 356–362. [DOI] [PubMed] [Google Scholar]
- 99.Xiang W, Zhang B, Lv F, Ma Y, Chen H, Chen L, et al. The Inhibitory Effects of RFamide-Related Peptide 3 on Luteinizing Hormone Release Involves an Estradiol-Dependent Manner in Prepubertal but Not in Adult Female Mice. Biol Reprod. 2015. August; 93(2): 30. [DOI] [PubMed] [Google Scholar]
- 100.Zigmond RE. gp130 cytokines are positive signals triggering changes in gene expression and axon outgrowth in peripheral neurons following injury. Front Mol Neurosci. 2012. January; 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wierman ME, Kiseljak-Vassiliades K, To- bet S. Gonadotropin-releasing hormone (GnRH) neuron migration: initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front Neuroendocrinol. 2011. January; 32(1): 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamaguchi M, Koike K, Yoshimoto Y, Matsuzaki N, Miyake A, Tanizawa O. Interleukin-6 stimulates gonadotropin-releasing hormone secretion from rat hypothalamic cells. Horm Res. 1991; 35(6): 252–6. [DOI] [PubMed] [Google Scholar]
- 103.Moro JA, Carretero J, Alonso MI, Martín C, Gato A, Mano AL. Prenatal expression of interleukin 1beta and interleukin 6 in the rat pituitary gland. Cytokine. 2008. December; 44(3): 315–22. [DOI] [PubMed] [Google Scholar]
- 104.Wetsel WC, Eraly SA, Whyte DB, Mellon PL. Regulation of gonadotropin-releasing hormone by protein kinase-A and -C in immortalized hypothalamic neurons. Endocrinology. 1993. June; 132(6): 2360–70. [DOI] [PubMed] [Google Scholar]
- 105.Rave-Harel N, Miller NL, Givens ML, Mellon PL. The Groucho-related gene family regulates the gonadotropin-releasing hormone gene through interaction with the homeodomain proteins MSX1 and OCT1. J Biol Chem. 2005. September; 280(35): 30975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eraly SA, Nelson SB, Huang KM, Mellon PL. Oct-1 binds promoter elements required for transcription of the GnRH gene. Mol Endocrinol. 1998. April; 12(4): 469–81. [DOI] [PubMed] [Google Scholar]
- 107.Hafezi F, Marti A, Grimm C, Wenzel A, Remé CE. Differential DNA binding activities of the transcription factors AP-1 and Oct-1 during light-induced apoptosis of photoreceptors. Vision Res. 1999. July; 39(15): 2511–8. [DOI] [PubMed] [Google Scholar]
- 108.Schieven GL. The p38alpha kinase plays a central role in inflammation. Curr Top Med Chem. 2009; 9(11): 1038–48. [DOI] [PubMed] [Google Scholar]
- 109.Elias CF, Purohit D. Leptin signaling and circuits in puberty and fertility. Cell Mol Life Sci. 2013. March; 70(5): 841–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bellefontaine N, Elias CF. Minireview: Metabolic control of the reproductive physiology: insights from genetic mouse models. Horm Behav. 2014. June; 66(1): 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roa J, Tena-Sempere M. Connecting metabolism and reproduction: roles of central energy sensors and key molecular mediators. Mol Cell Endocrinol. 2014. November; 397(1–2): 4–14. [DOI] [PubMed] [Google Scholar]
- 112.Donato J Jr, Cravo RM, Frazão R, Elias CF. Hypothalamic sites of leptin action linking metabolism and reproduction. Neuroendocrinology. 2011; 93(1): 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fernandez MO, Sharma S, Kim S, Rickert E, Hsueh K, Hwang V, et al. Obese Neuronal PPARγ Knockout Mice Are Leptin Sensitive but Show Impaired Glucose Tolerance and Fertility. Endocrinology. 2017. January; 158(1): 121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Quennell JH, Howell CS, Roa J, Augustine RA, Grattan DR, Anderson GM. Leptin deficiency and diet-induced obesity reduce hypothalamic kisspeptin expression in mice. Endocrinology. 2011. April; 152(4): 1541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yura S, Ogawa Y, Sagawa N, Masuzaki H, Itoh H, Ebihara K, et al. Accelerated puberty and late-onset hypothalamic hypogonadism in female transgenic skinny mice overexpressing leptin. J Clin Invest. 2000. March; 105(6): 749–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Quennell JH, Mulligan AC, Tups A, Liu X, Phipps SJ, Kemp CJ, et al. Leptin indirectly regulates gonadotropin-releasing hormone neuronal function. Endocrinology. 2009. June; 150(6): 2805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singireddy AV, Inglis MA, Zuure WA, Kim JS, Anderson GM. Neither signal transducer and activator of transcription 3 (STAT3) or STAT5 signaling pathways are required for leptin’s effects on fertility in mice. Endocrinology. 2013. July; 154(7): 2434–45. [DOI] [PubMed] [Google Scholar]
- 118.Bates SH, Stearns WH, Dundon TA, Schubert M, Tso AW, Wang Y, et al. STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature. 2003. February; 421(6925): 856–9. [DOI] [PubMed] [Google Scholar]
- 119.Baumann H, Morella KK, White DW, Dembski M, Bailon PS, Kim H, et al. The full-length leptin receptor has signaling capabilities of interleukin 6-type cytokine receptors. Proc Natl Acad Sci USA. 1996. August; 93(16): 8374–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Valdivia S, Patrone A, Reynaldo M, Perello M. Acute high fat diet consumption activates the mesolimbic circuit and requires orexin signaling in a mouse model. PLoS One. 2014. January; 9(1):e87478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xin X, Storlien LH, Huang XF. Hypothalamic c-fos-like immunoreactivity in high-fat diet-induced obese and resistant mice. Brain Res Bull. 2000. July; 52(4): 235–42. [DOI] [PubMed] [Google Scholar]
- 122.Beynon AL, Coogan AN. Diurnal, age, and immune regulation of interleukin-1β and interleukin-1 type 1 receptor in the mouse suprachiasmatic nucleus. Chronobiol Int. 2010. September; 27(8): 1546–63. [DOI] [PubMed] [Google Scholar]
- 123.Belevych N, Buchanan K, Chen Q, Bailey M, Quan N. Location-specific activation of the paraventricular nucleus of the hypothalamus by localized inflammation. Brain Behav Immun. 2010. October; 24(7): 1137–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee WS, Smith MS, Hoffman GE. cFos Activity Identifies Recruitment of Luteinizing Hormone-Releasing Hormone Neurons During the Ascending Phase of the Proestrous Luteinizing Hormone Surge. J Neuroendocrinol. 1992. April; 4(2): 161–6. [DOI] [PubMed] [Google Scholar]
- 125.Kauffman AS, Clifton DK, Steiner RA. Emerging ideas about kisspeptin-GPR54 signaling in the neuroendocrine regulation of reproduction. Trends Neurosci. 2007. October; 30(10): 504–11. [DOI] [PubMed] [Google Scholar]