Abstract
Clustering is one of the most universal approaches for understanding complex data. A pivotal aspect of clustering analysis is quantitatively comparing clusterings; clustering comparison is the basis for many tasks such as clustering evaluation, consensus clustering, and tracking the temporal evolution of clusters. In particular, the extrinsic evaluation of clustering methods requires comparing the uncovered clusterings to planted clusterings or known metadata. Yet, as we demonstrate, existing clustering comparison measures have critical biases which undermine their usefulness, and no measure accommodates both overlapping and hierarchical clusterings. Here we unify the comparison of disjoint, overlapping, and hierarchically structured clusterings by proposing a new element-centric framework: elements are compared based on the relationships induced by the cluster structure, as opposed to the traditional cluster-centric philosophy. We demonstrate that, in contrast to standard clustering similarity measures, our framework does not suffer from critical biases and naturally provides unique insights into how the clusterings differ. We illustrate the strengths of our framework by revealing new insights into the organization of clusters in two applications: the improved classification of schizophrenia based on the overlapping and hierarchical community structure of fMRI brain networks, and the disentanglement of various social homophily factors in Facebook social networks. The universality of clustering suggests far-reaching impact of our framework throughout all areas of science.
Subject terms: Data mining, Network models, Modularity, Complex networks
Introduction
Clustering is one of the most basic and ubiquitous methods to analyze data1,2. Traditionally, clustering is viewed as separating data elements into disjoint clusters of comparable sizes. Complications to this simplistic picture are becoming more prevalent, particularly following the rise of network science and nuanced clustering methods that reveal heterogeneous cluster size distributions3,4, overlaps5–8, and hierarchical structure9–12. A growing consensus suggests that applying clustering is more about identifying appropriate techniques for the particular problem and properly interpreting the results, than developing a silver-bullet clustering method13,14.
The most fundamental step towards understanding, evaluating, and leveraging identified clusterings is to quantitatively compare them. Clustering comparison is the basis for clustering evaluation, consensus clustering, and tracking the temporal evolution of clusters, among many other tasks. The proliferation of nuanced clustering methods presents new challenges for clustering comparison3,15 and renders current methods susceptible to critical biases3,16–20. In addition to the consistent grouping of elements into clusters, similarity measures must account for many other aspects of clusterings, such as the number of clusters, the size distribution of those clusters, multiple element memberships when clusters overlap, and scaling relations between levels of hierarchical clusterings.
Despite the increasing prevalence of irregular cluster features, the effect of such structure on clustering similarity has received little attention. Here we illustrate that the most popular clustering similarity measures are vulnerable to critical biases, calling the appropriateness of their general usage into question. We also argue that these biases are maintained or exacerbated by extensions to accommodate overlapping or hierarchical clusterings21–24, suggesting that none of the existing frameworks for clustering similarity are adequate for comparing overlapping and hierarchically structured clusterings.
Here we propose a new element-centric framework for clustering similarity that naturally incorporates overlaps and hierarchy. In our approach, elements are compared based on the relationships induced by the cluster structure, in contrast to the traditional cluster-centric philosophy. As we will see, this change in perspective resolves many of the aforementioned difficulties and avoids the common biases induced by irregular cluster structure.
Bias in Clustering Comparisons
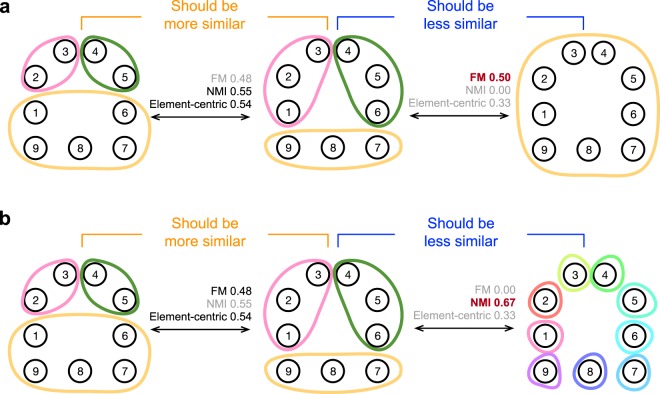
Every clustering similarity measure must trade-off between variation in three primary characteristics of clusterings: the grouping of elements into clusters, the number of clusters, and the size distribution of those clusters17,20,25–28. A failure to account for all three characteristics can result in a biased comparison in which clusterings with exaggerated features are favored over more intuitively similar clusterings. Before exploring these trade-offs further, we offer an illustrative example by the comparisons between clustering pairs shown in Fig. 1. Here we focus on three exemplary similarity measures—the normalized mutual information (NMI), Fowlkes-Mallows index (FM), and our element-centric similarity measure—and extend our discussion to a larger selection later. In the first set of comparisons (Fig. 1a), we demonstrate a bias towards clusterings with heterogeneous cluster sizes: NMI and the element-centric similarity determine the middle clustering is more similar to the left clustering than the right clustering, yet FM concludes the opposite—the middle clustering is more similar to the right clustering than the left—as it is biased by the large cluster in the right clustering. In the second set of comparisons (Fig. 1b), we illustrate a bias towards clusterings with more clusters: FM and the element-centric similarity determine the middle clustering is more similar to the left clustering than the right clustering, yet NMI concludes the opposite—the middle clustering is more similar to the right clustering than the left clustering—as it is biased by the number of clusters in the right clustering.
Figure 1.
Two examples of counter-intuitive bias in clustering comparisons. Four clusterings are considered over 9 elements, and compared using the Fowlkes-Mallows index (FM), normalized mutual information (NMI), and our element-centric similarity measure. We argue that the comparison between the clusterings on the left is more similar than the comparison between clusterings on the right. (a) Both NMI and the element-centric similarity follow this intuition, but FM is biased towards large clusters and suggests the comparison on the right is more similar. (b) Both FM and the element-centric similarity follow this intuition, but NMI is biased towards many clusters and suggests the comparison on the right is more similar.
One approach to correct biases in clustering comparison is to consider clustering similarity in the context of a random ensemble of clusterings18,26,29–32. Such a correction for chance uses the expected similarity of all pair-wise comparisons between clusterings specified by a random model to establish a baseline similarity value. However, the correction for chance approach has severe drawbacks20: (i) it is strongly dependent on the choice of random model assumed for the clusterings, which is often highly ambiguous, and (ii) no random model for overlapping or hierarchical clusterings has been suggested.
We introduce a simple set of synthetic clustering examples that illustrate the trade-offs between characteristics of clusterings. In each case, we outline the desired behavior for a measure of clustering similarity based on the extensive discussion in the literature16,17,20,21,25,27–30,33–35. Our intuition is based on the use of clustering similarity in practice: similar clusterings should have a similar number of clusters, of similar sizes, and elements should have similar memberships. Consider a typical case facing a practitioner of data science: we have three clustering methods M1, M2, and M3 such that M1 produces the clustering on the left of Fig. 1a,b, M2 produces the clustering on the top right of Fig. 1a, and M3 produces the clustering on the bottom right of Fig. 1b. Which method, M1, M2, or M3 performed best in recovering the ground-truth clustering in the middle of Fig. 1a,b? The answer depends on the clustering similarity measure used. Yet, to our best understanding, clustering M1 is the only clustering that reflects the number, sizes, and memberships of the ground-truth clustering. While other intuitions are possible (i.e. that offered by information theory or the correction for chance as discussed further in the SI, Section S2.11), we argue that the intuition adapted here most accurately captures the use of clustering comparisons in the literature36.
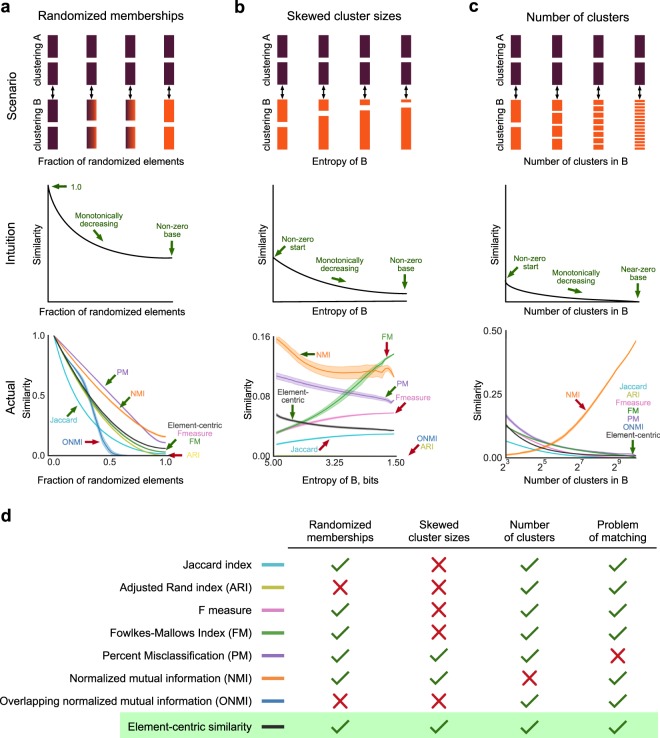
Since it is difficult to isolate changing cluster sizes or number of clusters from the grouping of elements into clusters, our examples consider the case of randomized element memberships; however, in practice, quantitative comparisons would make simultaneous trade-offs between all three aspects of clusterings. Here we expand our focus to seven exemplary similarity measures representing many of the most common measures from the literature—the Jaccard index, Adjusted Rand index (ARI)29, F measure, Fowlkes-Mallows index (FM)21, percentage matching (PM), the normalized mutual information (NMI), overlapping normalized mutual information (ONMI)23, and our element-centric similarity measure. We discuss another popular measure, the variation of information, in the SI, Section S2.10, due to its interpretation as a distance measure. These four examples suggest that the most common clustering similarity measures are subject to critical biases which render them inappropriate for comparing generalized clusterings—only our element-centric similarity measure displays the intuitive behavior in all examples and does not suffer from the problem of matching (Fig. 2d).
Figure 2.
Element-centric similarity behaves intuitively in three clustering similarity scenarios while common clustering similarity measures exhibit counter-intuitive behaviors. 1,024 elements are assigned to clusters according to the following scenarios (a–c) and compared using the Jaccard index, adjusted Rand index (ARI), the F measure, percentage matching (PM), normalized mutual information (NMI), overlapping normalized mutual information (ONMI), and our element-centric similarity. All results are averaged over 100 runs and error bars denote one standard deviation. (a) A clustering with 32 non-overlapping and equal-sized clusters is compared to a randomized version of itself where a fraction of the elements are shuffled. (b) A clustering with 32 non-overlapping and equal-sized clusters is compared against clusterings with increasing cluster size skewness. (c) A clustering with 8 non-overlapping and equal-sized clusters is compared against a clustering with c non-overlapping, equal-sized clusters and randomized element memberships for different values of c. (d) Only our element-centric similarity measure follows the intuitive behavior in all three scenarios and does not suffer from the problem of matching.
Bias in Randomized Membership
In the first example, the consistent grouping of elements is tested by comparing a clustering of 1,024 elements into 32 equally sized clusters against itself after a fraction of element memberships have been shuffled between clusters (Fig. 2a). Intuition suggests that as the randomization increases, the similarity between the original clustering and the shuffled clustering should decrease from the maximum value (1.0 in all cases) to some non-zero value, reflecting the fact that the number and sizes of clusters are still identical. However, two measures reach zero, ignoring the similarity of the cluster size sequences. The ONMI is particularly conservative, reporting no similarity at just over 50% randomization; ONMI’s surprising behavior highlights the difficulty of accommodating overlaps in a traditional similarity framework.
Bias in Skewed Cluster Sizes
The second example explores the bias favoring skewed cluster size sequences through a preferential attachment shuffling scheme (Fig. 2b). Starting from the same initial clustering of 1,024 elements into 32 equally sized clusters, we randomize all element memberships. The algorithm then proceeds to uniformly select a random element and reassign it to a new cluster based on the current sizes of those clusters. This procedure is run for a total of 5 × 106 steps, with a comparison to the original clustering performed every 500 steps. We argue that the desired clustering comparison behavior should reflect the cluster size differences, and that a decrease in the entropy of the cluster size sequence (reflecting an increase in cluster size heterogeneity) is reflected by the two clusterings becoming less similar. However, we now see three distinct types of behaviors exhibited by the clustering similarity measures. The NMI and our element-centric similarity measure exhibit the intuitive behavior and decrease as the clustering entropy decreases. The ONMI and ARI maintain a zero similarity for all comparisons regardless of the clustering entropy. Finally, the F measure and Jaccard index increase as the entropy decreases: They cannot account for the differences in the cluster size distribution. This increase is a consequence of their formulation in terms of the correctly co-assigned element pairs while disregarding the incorrectly co-assigned element pairs.
Bias in The Number of Clusters
Third, we investigate a scenario where the number and sizes of clusters in two clusterings diverge (Fig. 2c). Here we compare an initial clustering of 1,024 elements into 8 equally sized clusters against a second clustering generated by randomly assigning the elements to c regularly sized clusters, where c is the control parameter for the scenario. Hence, one clustering remains the same size, while the other has c regularly sized clusters. We see two distinctly different behaviors of the clustering similarity measures: the Jaccard index, F measure, ONMI, ARI and our element-centric similarity measure all follow our intuition and decrease with increasing c, while NMI increases with increasing c. The increasing behavior for NMI can be attributed to the aforementioned information-theoretic bias towards comparisons with more clusters16,19,20,34,37, and counters the large body of established literature controlling for the number of clusters in a clustering solution38,39. This bias makes NMI a particularly troubling measure for hierarchical clusterings where we expect the number of clusters to vary over several orders of magnitude.
The Problem of Matching
Finally, we recount one of the oldest biases discussed in the literature, the problem of matching15,40,41. The problem of matching is a symptom of all set-matching methods which identify a “best match” for each cluster. As a result, the measures completely ignore what happens to elements in the “unmatched” part of each cluster. For example, suppose is a clustering with K equal-sized clusters over N elements, with , and clustering is obtained from by moving a small fraction of the elements in each cluster to the cluster . Likewise, the clustering is obtained from by reassigning the same fraction of the elements in each evenly between the other clusters. In this case, measures suffering from the problem of matching would say the similarity between and is equal to the similarity between and , contradicting the intuition that is more similar to than . For the measures considered here, only the percentage matching similarity measure suffers from the problem of matching. Despite this issue, it is important to note that the percentage matching has been used both in practice and in theory, typically when the clusterings are assumed to be relatively similar.
Consequence for Extensions to Overlapping and Hierarchical Structure
The three examples discussed in this section illustrate biases in the case of disjoint clusterings without hierarchy. Despite the increasing prevalence of overlapping and hierarchical structured clusterings, there is a lack of intuition for the trade-offs encountered by clustering similarity measures in the presence of such structure. However, exploring the behavior of similarity measures when comparing partitions reveals useful insights into how these measures behave when comparing other clustering structures. The presence of overlaps can exaggerate the heterogeneity in cluster sizes8, especially if one considers each overlap region as a separate cluster (i.e. as considered by the Omega index). Since hierarchical clusterings reflect cluster structure over many scales, the sizes of these clusters typically vary by orders of magnitude; for example, the benchmark models typically used to capture hierarchical structure in networks are full k-ary trees, and thus the number of clusters grows exponentially in the number of levels9,23.
All but one of the similarity measures for overlapping or hierarchical clusterings simplifies to one of the cases we have studied: the Omega index is equivalent to the adjusted Rand index for partitions22, hierarchical mutual information reduces to NMI on partitions24, and the Fowlkes-Mallows analysis of dendrograms considers each cut of the dendrogram independently, thus producing a curve of comparisons between partitions21. The overlapping normalized mutual information (ONMI) is the only measure which does not reduce to another measure on partitions23, yet we have demonstrated that it has particularly unintuitive behavior in our examples. In sum, all existing measures for overlapping or hierarchical clusterings either inherit critical biases from their simpler counterparts on flat-partitions, or are inadequate for handling overlapping and hierarchical clusterings.
Element-Centric Clustering Comparisons
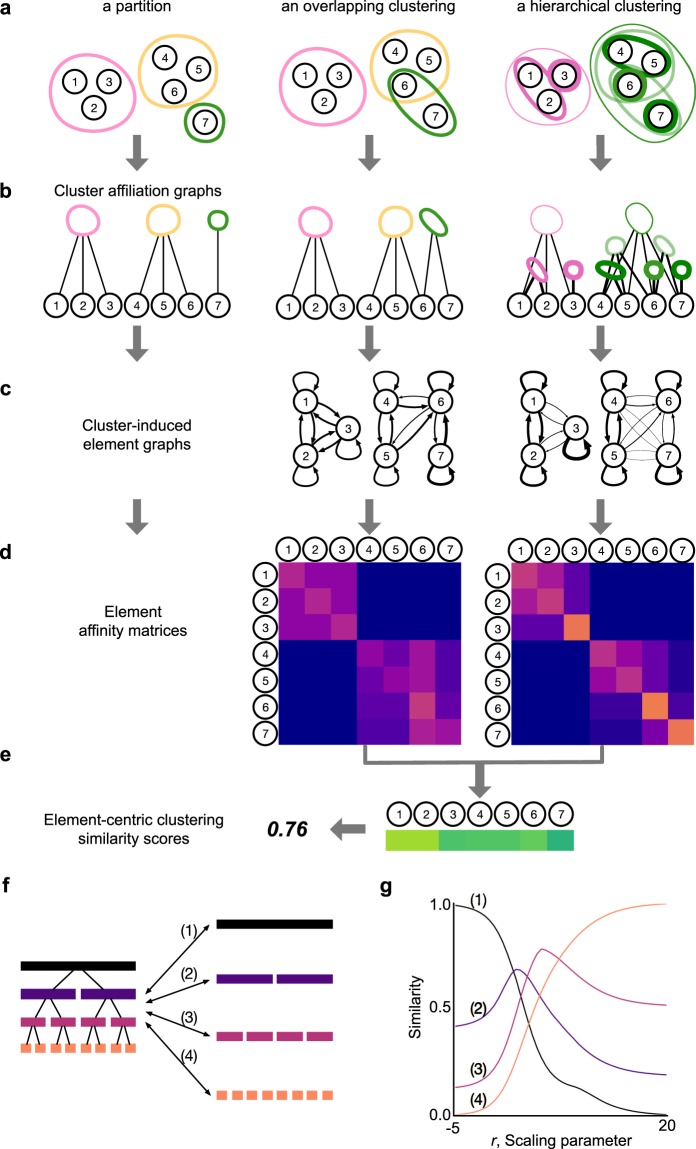
Our element-centric clustering similarity approach captures cluster-induced relationships between the elements through the cluster affiliation graph, a bipartite graph where one vertex set corresponds to the original elements and the other corresponds to the clusters. Specifically, a cluster affiliation graph is constructed for a clustering of labeled elements as a bipartite graph where one vertex set corresponds to the original elements V and the other vertex set corresponds to the cluster set C. An undirected edge is placed between element and cluster if , i.e. the element is a member of the cluster. Notice that an element’s membership in multiple overlapping clusters can be directly incorporated with multiple edges in the cluster affiliation graph. For hierarchically structured clusterings, each cluster is assigned a hierarchical level by re-scaling the hierarchy’s acyclic graph (dendrogram) according to the maximum path length from the roots42. The weight of the cluster affiliation edge is given by the hierarchy weighting function h(lβ):
| 1 |
where r is a scaling parameter that determines the relative importance of membership at different levels of the hierarchy (further discussed below).
The cluster affiliation graph is then projected onto the element vertices to produce the cluster-induced element graph, which is a weighted, directed graph that summarizes the inter-element relationships induced by common cluster memberships43 (see Fig. 3c). In the cluster-induced element graph, with weighted adjacency matrix W, each edge wij between elements vi and vj has weight:
| 2 |
where aiγ are the entries of the N × K bipartite adjacency matrix for the cluster affiliation graph.
Figure 3.
The element-centric perspective naturally incorporates overlaps and hierarchy. (a) Three examples of clusterings: a partition, a clustering with overlap, and a clustering with both overlapping and hierarchical structure. (b) Cluster affiliation graphs derived from the overlapping and hierarchical clusterings. (c) Cluster-induced element graphs found by projecting the cluster affiliation graphs in (b) to the element vertices. (d) The element-affinity matrices found as the personalized pagerank equilibrium distribution. (e) The corrected L1 metric distance between each affinity distribution in (d) gives an element-wise similarity between clusterings, the average element-wise similarity provides the final element-centric clustering similarity score. (f) A binary hierarchical clustering is compared to each of its individual levels. (g) The hierarchical scaling parameter for element-centric similarity acts as a “zooming lens”, refocusing the similarity to different levels (1–4) of the hierarchical comparison in (f).
The traditional notion of pair-wise co-occurrence in a cluster is now captured by the (binary) presence of an edge in the cluster-induced element graph. However, the focus on element pairs misses high-order relations (triplets, quadruplets, etc.), which are useful for characterizing cluster structure29. Such high-order co-occurrences can be captured through the presence of paths in the cluster-induced element graph. The weight of the path accounts for the relative importance of elements in the presence of overlapping and hierarchical cluster structures. Here, we incorporate every possible path between elements obtaining the equilibrium distribution for a personalized diffusion process on the graph (often called “personalized pagerank” or “random walk with restart”)44–46. Given a cluster-induced element graph with weighted adjacency matrix W, the personalized PageRank (PPR) affinity from element vi to all elements vj is found as the stationary distribution of a diffusion process with restart probability 1.0 − α to vi which takes the form:
| 3 |
where vi is an N-vector with 1 in the ith entry, and 0 otherwise. The value of α controls the influence of overlapping clusters and hierarchical clusters with shared lineages; here we use α = 0.90.
In general, for large data sets and clusterings with many overlapping and hierarchical clusters, the calculation of personalized pagerank can be a computationally expensive process. However, there are some computational simplifications that can be made. First, the personalized PageRank affinity of partitions (disjoint clusterings) can be analytically solved—the affinity value for each co-clustered element pair is a linear function of the inverse cluster size, 1/|cβ|, and 0 otherwise:
| 4 |
where δ is the Kronecker delta function, element vi is in cluster cγ, and element vj is in cluster cβ. Second, when several elements share exactly the same cluster memberships, their resulting personalized pagerank affinity vectors are related by simple permutations; therefore, the personalized pagerank affinity vector need only be calculated once for each common cluster membership set. Third, due to the utility of personalized pagerank for recommendation systems, there have been many algorithms for the approximation of personalized pagerank47,48. The worst-case computational complexity of element-centric similarity will only occur for highly overlapping and deeply hierarchical clusterings, which were previously incomparable using traditional clustering similarity methods.
The element-wise similarity of an element vi in two clusterings and is found by comparing the stationary probability distributions and induced by the PPR processes on the two cluster-induced element graphs. Here, we use the normalized L1 metric for probability distributions corrected to account for the PPR process:
| 5 |
The L1 metric was chosen because it is invariant to the magnitude of the probability values, i.e. it treats all cluster sizes equally. Other popular probability metrics (Hellinger, Euclidean, etc.) extenuate the differences in small or large probability values, thereby disproportionately favoring clusters based on their sizes. The final element-centric similarity score of two clusterings , is the average of the element-wise similarities:
| 6 |
A full implementation of the element-centric clustering similarity, and all other clustering similarity measures discussed here, is provided in the CluSim python package49. As illustrated in Fig. 3, our element-centric framework unifies disjoint, overlapping, and hierarchical clustering comparison in a single framework.
Interpretations of Element-Centric Similarity
Cluster affiliation graph and cluster-induced element graph
The cluster affiliation graph provides a convenient representation of element membership in multiple clusters at different scales of the hierarchy. Unweighted variants of the affiliation graph are common approaches to study the relationship between labels and data in network science43,50. Our weighted extension reflects the varying importance of membership at different scales of the hierarchy.
The element-centric philosophy suggests a focus on common memberships between data elements induced by the cluster structure, rather than overlaps between clusters induced by elements (as suggested by the cluster-centric philosophy). The cluster-induced element graph captures these relationships by integrating over all shared cluster memberships through the projection of the cluster affiliation graph onto the element nodes. This projection has three important features. First, the induced relationship between two elements is normalized by the size of the cluster capturing the fact that co-occurrence in larger clusters implies less direct influence between elements than co-occurrence in smaller clusters. Second, the weight for each element is normalized by the sum over all of its cluster memberships reflecting the idea that membership in many clusters reduces the relative influence from any one of the clusters. Third, in the presence of overlap or hierarchy, the weights in the cluster-induced element graph can be asymmetric (i.e. ) arising from the fact that multiple cluster affiliations will change the respective local neighborhoods of individual elements. Note that our normalization for the edge-weights in the cluster-induced element graph is equivalent to the landing probability of a two-step random walk on the cluster affiliation graph from element vi to element vi.
Element-wise scores
Beyond naturally accommodating generalized clusterings, our element-centric similarity can provide detailed insights into how two clusterings differ because the similarity is calculated at the level of individual elements. Specifically, the individual element-wise scores directly measure how similar the clusterings appear from the perspective of each element. The distribution of element-wise similarity scores can also provide insight into how the clusterings differ. For example, the ranked-distribution of element-wise scores reflects the differences in cluster structure: a flat distribution occurs when all elements have the same similarity score, suggesting that the clusterings differ equally across all elements; a skewed distribution occurs when some elements have much higher or lower similarity than the rest, suggesting that the clusterings are distinguished by a subset of elements.
Average agreement and frustration
Our element-centric similarity measure also reveals the consistency of element groupings within an arbitrary set of clusterings. The average agreement between a reference clustering and a set of clusterings measures the regular grouping of elements with respect to a reference clustering. Specifically, given a clustering and a set of clusterings , the element-wise average agreement for element vi is evaluated as:
| 7 |
The frustration within a set of clusterings reflects the consistency with which elements are grouped by the clusterings. For the set of clusterings , the element-wise frustration for element vi is given by:
| 8 |
Interpretation of overlap
The element-centric framework naturally incorporates the multiple memberships that occur in overlapping clusterings. First, as discussed above, element membership in multiple clusters is directly captured by multiple edges in the cluster affiliation graph, and is propagated into asymmetric weights in the cluster-induced element graph. Second, the integration over local paths through the personalized PageRank process means that the presence of multiple memberships for elements is not isolated to the overlapping elements, but propagates throughout the clusters which overlap. This is because shared elements introduce additional information into the system, namely that the clusters share common features. Essentially, in the absence of any other information, if two clusters overlap (share some elements), then their elements should be more similar compared with the case where the clusters are disjoint.
For example, let us simplify our discussion to talk about counting element triplets in the overlapping clustering from Fig. 3a. When no overlaps are present, it is simple to declare whether all three elements co-occur within the same cluster or not; so elements 1, 2, 3 all co-occur in the pink cluster, but elements 1, 2, 4 do not. However, in the presence of overlap, additional decisions must be made. Consider elements 4,6,7. Elements 4 and 6 co-occur in the same yellow cluster, and elements 6 and 7 co-occur in the same green cluster, but how should one determine if the elements 4 and 7 co-occur? Clearly, this triplet has important information. Indeed, it specifically defines what the “overlap” means for this clustering. Thus, the element-centric similarity measure does not disregard this triplet, but retains it with a reduced weight determined by the α parameter. Namely, the triplet 4,6,7 co-occurs less strongly than the triplet without overlap 1, 2, 3.
In contrast, the omega index counts element co-occurrences very conservatively and states that elements 4 and 7 do not co-occur. It continues to make the distinction that 4 and 6 didn’t co-occur either because 4 doesn’t have the exact same memberships as 6. Thus it throws away valuable information about the cluster structure.
Interpretation of hierarchy
Our element-centric framework is flexible and allows natural choices to accommodate alternative interpretations of hierarchy. For example, our choice of hierarchical weighting function and the scaling parameter, r, reflects a continuum in the hierarchy (Fig. 3g): lower r emphasizes higher levels and reflects a divisive hierarchy, in which lower levels of the dendrogram are treated as refinements of the higher levels, while larger r puts emphasis on lower levels and reflects an agglomerative hierarchy, in which higher levels of the dendrogram are seen as a coarsening of the lower level cluster structure. Other interpretations of hierarchy can be implemented by changing the specific hierarchical weighting function; for example, constant function ( above) collapses the hierarchy into an overlapping clustering with each cluster weighted equally.
Relation to other similarity measures
Our choice of L1 comparisons between personalized pagerank distributions was based on a principled extension of element co-occurrence. This choice can be replaced by another measure of graph similarity or probability metric with an alternative intuition of the trade-offs associated with clustering similarity51. Indeed, several common clustering similarity measures can be recovered by adapting other choices of graph similarity; all pair-counting measures can be recovered from graph set operations between cluster-induced element graphs from disjoint clusterings. The Rand index, in particular, is recovered by applying the graph-edit distance between the two cluster-induced element graphs from disjoint clusterings.
Applications
Element-centric comparisons reveal insights into how K-means clusterings differ
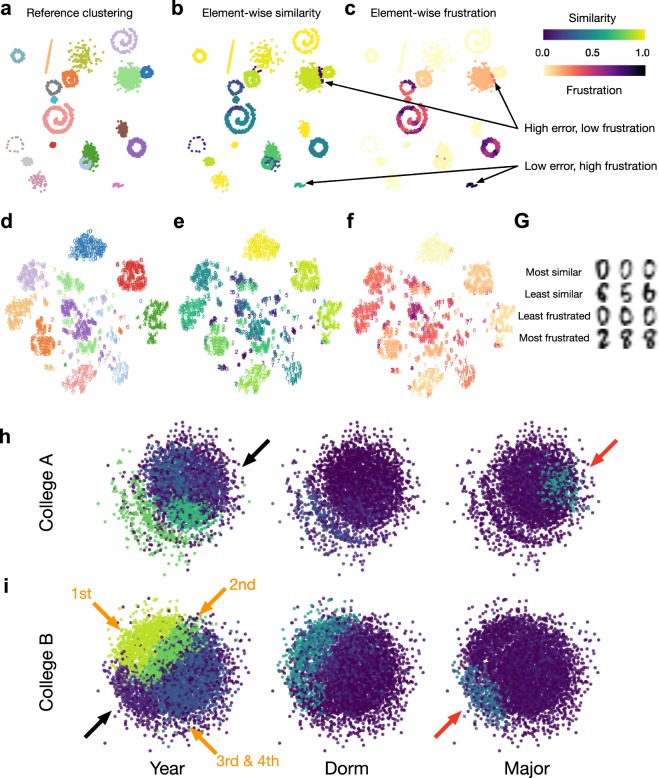
Beyond serving as a global measure of clustering similarity, our element-centric similarity also provides detailed insights into how clusterings differ, in contrast to other measures. Consider an illustrative example from K-means clustering shown in Fig. 4a and detailed in the SI, Section S3.1; 19 clusters were randomly placed in a square with a randomly selected arrangement (Gaussian blob, anisotropic blob, circle, or spiral) and size. K-means has difficulty when the predefined clusters overlap or are circularly arranged52. This difficulty can be explicitly quantified by calculating the average element-wise similarity between the predefined clustering and 100 uncovered clusterings (Fig. 4b). The element-wise frustration, found by averaging over all pair-wise comparisons between the 100 uncovered clusterings, reveals data points that are consistently grouped into similar clusters or are assigned to drastically different clusters (Fig. 4c). The combination of similarity and frustration identifies specific elements which are consistently grouped into an incorrect cluster (Fig. 4b,c: high error, low frustration), or those elements which K-means cannot consistently decide on a grouping (Fig. 4b,c: low error, high frustration).
Figure 4.
Element-wise clustering similarity reveals insights into how clusterings differ. (a–c) A K-means clustering example. (a) The planted clustering. (b) The average element-wise similarity between the planted clustering and 100 K-means clusterings. (c) The average element-wise frustration between 100 K-means clusterings. (d–g) A handwriting classification example. (d) The labeled handwritten digit data projected using t-SNE dimensionality reduction for visualization. (e) The average element-wise similarity between the labels and 100 K-means clusterings. (f) The average element-wise frustration between 100 K-means clusterings. (g) Exemplar digits that are 1) consistently grouped as in the ground-truth clustering, 2) consistently clustered differently from the ground-truth clustering, 3) least frustrated, and 4) most frustrated. (h,i) Facebook friendship networks for (h) College A and (i) College B. The element-wise similarity between user affiliation to school year, dorm, and major compared to Newman’s modularity optimized by the Louvain method demonstrates that social networks can be organized by a convolution of different attributes (black vs red arrows). The similarity to school year attenuates with student’s status (1st year–4th year, orange arrows).
We also present a real-world example of handwriting recognition53 (Fig. 4d and SI, Section S3.2). The same procedure reveals that some clusters of digits are correctly and consistently identified (“0”), while the error mostly results from incorrect grouping of other digit clusters (“9”, “8”, and “1”; Fig. 4e). Element-wise frustration shows that there are some digits that cannot be consistently classified (“3” and “8”, Fig. 4f), while some errors are regularly made (“1” and “9”). The extreme examples of these two types of error are shown in Fig. 4g.
The convolution of meta-data in social networks
We now use our framework to explore the community structure of Facebook college friendship networks. Previous research has suggested that friendship networks at major universities are organized into clusters which reflect the graduation year, dormitory, or student major54,55. However, the details of the organizing principles underlying this similarity are unknown. Here we demonstrate and visualize how multiple attributes interact and contribute to community structure.
The Facebook friendship networks analyzed here were originally released as part of the Facebook 100 data set54,55. This dataset contains a snapshot of all friendships at each of 100 schools in the fall of 2005. Additionally, the data includes several categorical variables shared by the users on their individual pages: gender, class year, high school, major, and dormitory residence. Here, we analyze the networks in two schools: the Oberlin (College A) and Rochester networks (College B). For each school we took the largest connected component and uncovered clusterings using the Louvain method56. The categorical data for year, dorm and major were used to create three non-overlapping clusterings. Every student with missing categorical data was placed into an individual singleton cluster.
Element-centric similarity reveals that school year closely captures the modular structure for most of the network, confirming previous results54,55. However, our element-centric similarity further illustrates that this similarity is particularly high for the students in their 1st or 2nd years, and fails to capture the clustering structure of other students (Fig. 4h,i black arrows). In these cases, the students’ major gradually takes over the cohort-based connections (Fig. 4h,i red arrows). This result, which has only become straight-forward through our framework, supports the intuition that network structure results from the convolution of multiple attributes14.
Element-centric comparisons of overlapping and hierarchical clustering in brain networks
Finally, to further illustrate the utility of our element-centric similarity measure, we demonstrate its ability to capture meaningful differences in overlapping and hierarchical clustering structure by classifying schizophrenic individuals based on the community structure of resting-state fMRI brain networks. There are several known distinctive and interpretable properties of resting-state fMRI brain networks in schizophrenia57–61. Network communities, in particular, are hypothesized to capture functionally integrated modules in the brain that reflect key properties of schizophrenia57. Our goal for this example is not to introduce a superior classification of schizophrenic subjects, rather, upon controlling the clustering method and data set, we demonstrate that our measure can extract more useful information than the other state-of-the-art clustering comparison methods for overlapping clusterings (ONMI, Omega index). We extract communities with overlapping and hierarchical structure using OSLOM community detection62 from the functional brain networks of 48 subjects (29 healthy controls and 19 individuals diagnosed with schizophrenia) analyzed in a previous study58 (see SI, Section S3.3 for details). The similarity between each pair of the subjects’ hierarchical and overlapping clusterings was found using our element-centric similarity measure, producing a 48 × 48 similarity matrix (Fig. 5a).
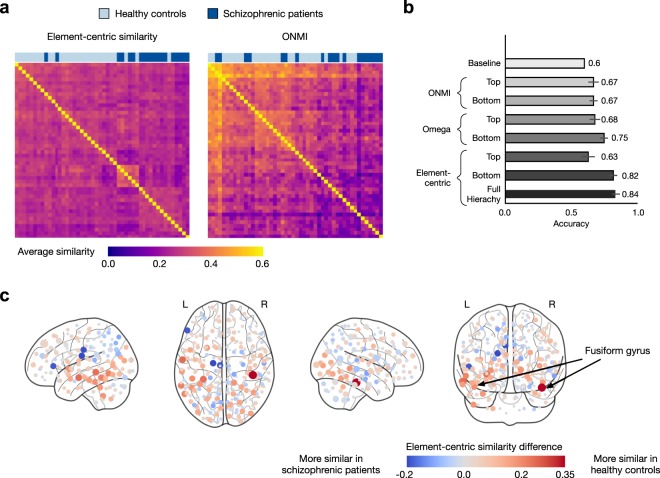
Figure 5.
Our element-centric similarity better differentiates the overlapping and hierarchical community structure of functional brain networks in healthy and schizophrenic individuals. (a) Hierarchical clustering of average pair-wise element-centric similarity using the entire OSLOM hierarchy closely reflects the true classification of participants as healthy (light blue) or schizophrenic (dark blue), while hierarchical clustering of the average pair-wise similarity using ONMI on the bottom level of the OSLOM hierarchy fails to uncover patient classification. (b) Classification accuracy using different clustering similarity measures averaged over 100 instances of 10-fold cross-validation, error bars denote one standard deviation. (c) The difference in element-centric similarity for each brain region when comparing amongst the healthy controls minus the similarity when comparing amongst the schizophrenic individuals; ROIs within the Fusiform gyrus are more consistently clustered in the healthy controls than in the schizophrenic individuals.
The subject-subject similarity matrix was then used in conjunction with a weighted k-nearest neighbors classifier to perform a binary classification of subjects as either schizophrenic or healthy controls. Evaluated by a nested 10-fold cross-validation procedure, our approach achieves an average accuracy of 84%, outperforming other measures (ONMI, the Omega index, Fig. 5b). Note that, classification based on individual levels from the hierarchy does not perform as well as the method using the full hierarchy. Even when limited only to the overlapping clustering at the bottom of the OSLOM hierarchy, our element-centric clustering similarity outperforms both ONMI and the Omega Index.
Our element-centric clustering similarity measure also provides insights into which brain regions are consistently clustered within groups. To find such group differences, we consider the element-centric similarity between all healthy controls, and the element-centric similarity between all schizophrenic patients. As seen in Fig. 5c, the difference between the means of these two groups highlights several regions which are consistently clustered into similar functional modules in the healthy controls or schizophrenic patients. In particular, regions of interest (ROIs) located in the Fusiform gyrus (Brodmann Area 37) were consistently clustered in the healthy controls but displayed great variability in cluster structure for the schizophrenic patients. This result is corroborated by the fact that the Fusiform gyrus has previously been associated with abnormal activation in schizophrenia during semantic tasks63,64.
Summary and Discussion
In summary, we present an element-centric framework that intuitively unifies the comparison of disjoint, overlapping, and hierarchically structured clusterings. We argue that our element-centric similarity does not suffer from the common counter-intuitive biases of existing measures, and that it also provides insights into how clusterings differ at the level of individual elements.
Our framework suggests straight-forward extensions to more complex scenarios, such as soft or fuzzy clusterings, hierarchical clusterings specified by dendrograms with merge distance information, and hyper-graph similarity. The framework also provides a measure of pair-wise similarity between elements, akin to the nodal association matrix of Bassett et al.65, and an element-wise clustering similarity which summarizes the difference in relationships induced by overlapping and hierarchically structured clusterings from the perspective of individual elements. Both of these objects hold promise for use in clustering ensemble methods66,67.
As clustering methods advance to uncover more nuanced and accurate organizational structure of complex systems, so too should clustering similarity measures facilitate meaningful comparisons of these organizations. The element-centric framework proposed here provides an intuitive quantification of clustering similarity that holds great promise for uncovering the relationships amongst all types of clusters, such as network communities, ontogenies, and dendrograms. The ubiquity of clustering in all areas of science suggests extensive potential impact of our framework.
Supplementary information
Acknowledgements
We would like to thank Dae-Jin Kim for assistance with the interpretation of the schizophrenia classifications, and Hu Cheng for assistance processing the fMRI timeseries. We thank Randall D. Beer, Luis M. Rocha, Filippo Radicchi, Sune Lehmann, Olaf Sporns, Alessio Cardillo, and Artemy Kolchensiky for helpful discussions. Supported in part by National Institute for Mental Health Grant 2R01MH074983 to W.P.H. Y.Y.A. thanks Microsoft Research for support through a Microsoft Research Faculty Fellowship. We thank the anonymous reviewers for their insightful comments.
Author Contributions
A.J.G. and Y.Y.A. developed the method, A.J.G. and I.B.W. performed the analysis, W.P.H. contributed the fMRI data, A.J.G., I.B.W., W.P.H. and Y.Y.A. participated in interpreting results, A.J.G. and Y.Y.A. wrote the manuscript. All authors reviewed and edited the manuscript.
Data Availability
All data used in this work is available upon request. A full implementation of the element-centric similarity measure is available in the open-source python package: CluSim49.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alexander J. Gates, Email: a.gates@northeastern.edu
Yong-Yeol Ahn, Email: yyahn@iu.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44892-y.
References
- 1.Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Computing Surveys (CSUR) 1999;31:264–323. doi: 10.1145/331499.331504. [DOI] [Google Scholar]
- 2.Fortunato S. Community detection in graphs. Physics Reports. 2010;486:75–174. doi: 10.1016/j.physrep.2009.11.002. [DOI] [Google Scholar]
- 3.He H, Garcia EA. Learning from imbalanced data. IEEE Transactions on Knowledge and Data Engineering. 2009;21:1263–1284. doi: 10.1109/TKDE.2008.239. [DOI] [Google Scholar]
- 4.Leskovec J, Lang KJ, Dasgupta A, Mahoney MW. Community structure in large networks: Natural cluster sizes and the absence of large well-defined clusters. Internet Mathematics. 2009;6:29–123. doi: 10.1080/15427951.2009.10129177. [DOI] [Google Scholar]
- 5.Palla G, Derenyi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 6.Ahn Y-Y, Bagrow JP, Lehmann S. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–764. doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- 7.Gopalan PK, Blei DM. Efficient discovery of overlapping communities in massive networks. PNAS. 2013;110:14534–14539. doi: 10.1073/pnas.1221839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Leskovec J. Structure and overlaps of ground-truth communities in networks. ACM Transactions on Intelligent Systems and Technology (TIST) 2014;5:26. [Google Scholar]
- 9.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási A-L. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 10.Sales-Pardo M, Guimera R, Moreira AA, Amaral LAN. Extracting the hierarchical organization of complex systems. PNAS. 2007;104:15224–15229. doi: 10.1073/pnas.0703740104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delvenne J-CC, Yaliraki SN, Barahona M. Stability of graph communities across time scales. PNAS. 2010;107:12755–12760. doi: 10.1073/pnas.0903215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang P, Moore C. Scalable detection of statistically significant communities and hierarchies, using message passing for modularity. PNAS. 2014;111:18144–18149. doi: 10.1073/pnas.1409770111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinberg, J. An Impossibility Theorem for Clustering, 463–470, NIPS’02 (MIT Press, 2002).
- 14.Peel L, Larremore DB, Clauset A. The ground truth about metadata and community detection in networks. Science Advances. 2017;3:e1602548. doi: 10.1126/sciadv.1602548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meila M. Comparing clusterings—an information based distance. Journal of Multivariate Analysis. 2007;98:873–895. doi: 10.1016/j.jmva.2006.11.013. [DOI] [Google Scholar]
- 16.White AP, Liu WZ. Technical note: Bias in information-based measures in decision tree induction. Machine Learning. 1994;15:321–329. [Google Scholar]
- 17.Pfitzner D, Leibbrandt R, Powers D. Characterization and evaluation of similarity measures for pairs of clusterings. Knowledge and Information Systems. 2009;19:361–394. doi: 10.1007/s10115-008-0150-6. [DOI] [Google Scholar]
- 18.Vinh NX, Epps J, Bailey J. Information Theoretic Measures for Clusterings Comparison: Variants, Properties, Normalization and Correction for Chance. Journal of Machine Learning Research. 2010;11:2837–2854. [Google Scholar]
- 19.Zhang P. Evaluating accuracy of community detection using the relative normalized mutual information. Journal of Statistical Mechanics: Theory and Experiment. 2015;2015:P11006. doi: 10.1088/1742-5468/2015/11/P11006. [DOI] [Google Scholar]
- 20.Gates AJ, Ahn Y-Y. The impact of random models on clustering similarity. Journal of Machine Learning Research. 2017;18:1–28. [Google Scholar]
- 21.Fowlkes EB, Mallows CL. A method for comparing two hierarchical clusterings. Journal of the American Statistical Association. 1983;78:553–569. doi: 10.1080/01621459.1983.10478008. [DOI] [Google Scholar]
- 22.Collins LM, Dent CW. Omega: A general formulation of the rand index of cluster recovery suitable for non-disjoint solutions. Multivariate Behavioral Research. 1988;23:231–242. doi: 10.1207/s15327906mbr2302_6. [DOI] [PubMed] [Google Scholar]
- 23.Lancichinetti A, Fortunato S, Kertész J. Detecting the overlapping and hierarchical community structure in complex networks. New Journal of Physics. 2009;11:033015. doi: 10.1088/1367-2630/11/3/033015. [DOI] [Google Scholar]
- 24.Perotti JI, Tessone CJ, Caldarelli G. Hierarchical mutual information for the comparison of hierarchical community structures in complex networks. Physical Review E. 2015;92:062825. doi: 10.1103/PhysRevE.92.062825. [DOI] [PubMed] [Google Scholar]
- 25.Meila, M. Comparing clusterings: an axiomatic view. In Proceedings of the 22nd International Conference on Machine Learning, 577–584 (ACM, New York, NY, USA, 2005).
- 26.Albatineh AN, Niewiadomska-Bugaj M, Mihalko D. On similarity indices and correction for chance agreement. Journal of Classification. 2006;23:301–313. doi: 10.1007/s00357-006-0017-z. [DOI] [Google Scholar]
- 27.Amigó E, Gonzalo J, Artiles J, Verdejo F. A comparison of extrinsic clustering evaluation metrics based on formal constraints. Information Retrieval. 2009;12:461–486. doi: 10.1007/s10791-008-9066-8. [DOI] [Google Scholar]
- 28.Souto, M. C. P. D. et al. A comparison of external clustering evaluation indices in the context of imbalanced data sets. Brazilian Symposium on Neural Networks 49–54 (2012).
- 29.Hubert L, Arabie P. Comparing partitions. Journal of Classification. 1985;2:193–218. doi: 10.1007/BF01908075. [DOI] [Google Scholar]
- 30.Vinh, N. X., Epps, J. & Bailey, J. Information theoretic measures for clusterings comparison: is a correction for chance necessary? In Proceedings of the 26th Annual International Conference on Machine Learning, 1073–1080 (ACM, 2009).
- 31.Albatineh AN, Niewiadomska-Bugaj M. Correcting jaccard and other similarity indices for chance agreement in cluster analysis. Advances in Data Analysis and Classification. 2011;5:179–200. doi: 10.1007/s11634-011-0090-y. [DOI] [Google Scholar]
- 32.Romano, S., Bailey, J., Nguyen, V. & Verspoor, K. Standardized mutual information for clustering comparisons: one step further in adjustment for chance. In Proceedings of the 31st International Conference on Machine Learning (ICML-14), 1143–1151 (2014).
- 33.Rosenberg, A. & Hirschberg, J. V-Measure: A Conditional Entropy-Based External Cluster Evaluation Measure. In EMNLP-CoNLL, vol. 7, 410–420 (2007).
- 34.Amelio, A. & Pizzuti, C. Is normalized mutual information a fair measure for comparing community detection methods? In Proceedings of the 2015 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining 2015, 1584–1585 (ACM, 2015).
- 35.van der Hoef, H. & Warrens, M. J. Understanding information theoretic measures for comparing clusterings. Behaviormetrika 1–18 (2018).
- 36.Lancichinetti, A., Fortunato, S. & Radicchi, F. Benchmark graphs for testing community detection algorithms. Physical Review E78 (2008). [DOI] [PubMed]
- 37.Amelio A, Pizzuti C. Correction for closeness: Adjusting normalized mutual information measure for clustering comparison. Computational Intelligence. 2016;33:579–601. doi: 10.1111/coin.12100. [DOI] [Google Scholar]
- 38.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2001;63:411–423. doi: 10.1111/1467-9868.00293. [DOI] [Google Scholar]
- 39.Newman M, Reinert G. Estimating the Number of Communities in a Network. Physical Review Letters. 2016;117:078301. doi: 10.1103/PhysRevLett.117.078301. [DOI] [PubMed] [Google Scholar]
- 40.Meila, M. Comparing clusterings by the variation of information. In Learning Theory and Kernel Machines, 173–187 (Springer, 2003).
- 41.Rezaei M, Franti P. Set matching measures for external cluster validity. IEEE Trans Knowl Data Eng. 2016;28:2173–2186. doi: 10.1109/TKDE.2016.2551240. [DOI] [Google Scholar]
- 42.Czégel D, Palla G. Random walk hierarchy measure: what is more hierarchical, a chain, a tree or a star. Scientific Reports. 2015;5:17994. doi: 10.1038/srep17994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou T, Ren J, Medo M, Zhang Y-C. Bipartite network projection and personal recommendation. Physical Review E. 2007;76:046115. doi: 10.1103/PhysRevE.76.046115. [DOI] [PubMed] [Google Scholar]
- 44.Haveliwala TH. Topic-sensitive pagerank: A context-sensitive ranking algorithm for web search. IEEE Transactions on Knowledge and Data Engineering. 2003;15:784–796. doi: 10.1109/TKDE.2003.1208999. [DOI] [Google Scholar]
- 45.Tong, H., Faloutsos, C. & Pan, J. Y. Fast random walk with restart and its applications. In ICDM’06. Sixth International Conference on Data Mining, 613–622 (IEEE Computer Society, 2006).
- 46.Kloumann IM, Ugander J, Kleinberg J. Block models and personalized pagerank. PNAS. 2017;114:33–38. doi: 10.1073/pnas.1611275114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lofgren, P. A., Banerjee, S., Goel, A. & Seshadhri, C. Fast-ppr: scaling personalized pagerank estimation for large graphs. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 1436–1445 (ACM, 2014).
- 48.Gleich DF, Kloster K. Seeded pagerank solution paths. European Journal of Applied Mathematics. 2016;27:812–845. doi: 10.1017/S0956792516000280. [DOI] [Google Scholar]
- 49.Gates AJ, Ahn Y-Y. CluSim: a Python package for the comparison of clusterings. Journal of Open Source Software. 2019;4:1264. doi: 10.21105/joss.01264. [DOI] [Google Scholar]
- 50.Yang, J. & Leskovec, J. Community-Affiliation Graph Model for Overlapping Network Community Detection. In 2012 IEEE 12th International Conference on Data Mining (ICDM), 1170–1175 (IEEE, 2012).
- 51.Blondel VD, Gajardo A, Heymans M, Senellart P, Van Dooren P. A Measure of similarity between graph vertices: applications to synonym extraction and web searching. SIAM Review. 2004;46:647–666. doi: 10.1137/S0036144502415960. [DOI] [Google Scholar]
- 52.Jain AK. Data clustering: 50 years beyond k-means. Pattern Recognition Letters. 2010;31:651–666. doi: 10.1016/j.patrec.2009.09.011. [DOI] [Google Scholar]
- 53.Alimoglu, F. & Alpaydin, E. Methods of combining multiple classifiers based on different representations for pen-based handwritten digit recognition. In Proceedings of the Fifth Turkish Artificial Intelligence and Artificial Neural Networks Symposium (1996).
- 54.Traud AL, Kelsic ED, Mucha PJ, Porter MA. Comparing community structure to characteristics in online collegiate social networks. SIAM Review. 2011;53:526–543. doi: 10.1137/080734315. [DOI] [Google Scholar]
- 55.Traud AL, Mucha PJ, Porter MA. Social structure of facebook networks. Physica A: Statistical Mechanics and its Applications. 2012;391:4165–4180. doi: 10.1016/j.physa.2011.12.021. [DOI] [Google Scholar]
- 56.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment. 2008;2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 57.Alexander-Bloch A, et al. The discovery of population differences in network community structure: new methods and applications to brain functional networks in schizophrenia. Neuroimage. 2012;59:3889–3900. doi: 10.1016/j.neuroimage.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng H, et al. Nodal centrality of functional network in the differentiation of schizophrenia. Schizophrenia Research. 2015;168:345–352. doi: 10.1016/j.schres.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fornito A, Zalesky A, Pantelis C, Bullmore ET. Schizophrenia, neuroimaging and connectomics. Neuroimage. 2012;62:2296–2314. doi: 10.1016/j.neuroimage.2011.12.090. [DOI] [PubMed] [Google Scholar]
- 60.Du, W. et al. High classification accuracy for schizophrenia with rest and task fmri data. Frontiers in Human Neuroscience6 (2012). [DOI] [PMC free article] [PubMed]
- 61.Arbabshirani, M. R., Kiehl, K., Pearlson, G. & Calhoun, V. D. Classification of schizophrenia patients based on resting-state functional network connectivity. Frontiers in Neuroscience7 (2013). [DOI] [PMC free article] [PubMed]
- 62.Lancichinetti A, Radicchi F, Ramasco JJ, Fortunato S. Finding Statistically Significant Communities in Networks. PLoS One. 2011;6:e18961. doi: 10.1371/journal.pone.0018961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kircher TT, et al. Differential activation of temporal cortex during sentence completion in schizophrenic patients with and without formal thought disorder. Schizophrenia research. 2001;50:27–40. doi: 10.1016/S0920-9964(00)00042-6. [DOI] [PubMed] [Google Scholar]
- 64.Kuperberg GR, Deckersbach T, Holt DJ, Goff D, West WC. Increased temporal and prefrontal activity in response to semantic associations in schizophrenia. Archives of General Psychiatry. 2007;64:138–151. doi: 10.1001/archpsyc.64.2.138. [DOI] [PubMed] [Google Scholar]
- 65.Bassett DS, et al. Robust detection of dynamic community structure in networks. Chaos: An Interdisciplinary Journal of Nonlinear Science. 2013;23:013142. doi: 10.1063/1.4790830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003;52:91–118. doi: 10.1023/A:1023949509487. [DOI] [Google Scholar]
- 67.Lancichinetti A, Fortunato S. Consensus clustering in complex networks. Scientific Reports. 2012;2:00336. doi: 10.1038/srep00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this work is available upon request. A full implementation of the element-centric similarity measure is available in the open-source python package: CluSim49.