Abstract
A major catabolic pathway for gibberellin (GA) is initiated by 2β-hydroxylation, a reaction catalyzed by GA 2-oxidase. We have isolated and characterized a cDNA, designated Oryza sativa GA 2-oxidase 1 (OsGA2ox1) from rice (Oryza sativa L. cv Nipponbare) that encodes a GA 2-oxidase. The encoded protein, produced by heterologous expression in Escherichia coli, converted GA1, GA4, GA9, GA20, and GA44 to the corresponding 2β-hydroxylated products GA8, GA34, GA51, GA29, and GA98, respectively. Ectopic expression of the OsGA2ox1 cDNA in transgenic rice inhibited stem elongation and the development of reproductive organs. These transgenic plants were deficient in endogenous GA1. These results indicate that OsGA2ox1 encodes a GA 2-oxidase, which is functional not only in vitro but also in vivo. OsGA2ox1 was expressed in shoot apex and roots but not in leaves and stems. In situ hybridization analysis revealed that OsGA2ox1 mRNA was localized in a ring at the basal region of leaf primordia and young leaves. This ring-shaped expression around the shoot apex was drastically decreased after the phase transition from vegetative to reproductive growth. It was absent in the floral meristem, but it was still present in the lateral meristem that remained in the vegetative phase. These observations suggest that OsGA2ox1 controls the level of bioactive GAs in the shoot apical meristem; therefore, reduction in its expression may contribute to the early development of the inflorescence meristem.
Gibberellins (GAs) are endogenous phytohormones that are involved in the regulation of the life cycle of plants. Therefore, biosynthesis of GAs has been intensively studied. Bioactive GAs, such as GA1 and GA4, are synthesized from trans-geranylgeranyl diphosphate by the sequential action of cyclases in plastids, membrane-associated mono-oxygenases in the endoplasmic reticulum, and soluble 2-oxoglutarate-dependent dioxygenases (2ODDs) in the cytosol (Hedden and Kamiya, 1997; Lange, 1998). During the last 10 years, genes for GA 20-oxidase and GA 3β-hydroxylase have been cloned. They encode 2ODDs that catalyze the later steps in GA biosynthesis, namely, the oxidation of the C-20 group and the introduction of the 3β-hydroxyl group, respectively. In Arabidopsis, the transcript levels of GA 20-oxidase genes and a GA 3β-hydroxylase gene (GA4) are subject to feedback regulation (Chang et al., 1995; Phillips et al., 1995). These results indicate that the endogenous levels of bioactive GAs are maintained at proper levels through the regulation of transcript levels of GA 20-oxidase and GA 3β-hydroxylase.
Catabolism of GAs is another important factor that regulates the endogenous levels of bioactive GAs. In many plant species, bioactive GAs are 2β-hydroxylated to produce biologically inactive GAs. This step is catalyzed by a third 2ODD, GA 2-oxidase. This enzyme also inactivates immediate precursors of bioactive GAs such as GA20 and GA9 (Ross et al., 1995). Thus, GA 2-oxidase has been considered to play an important role in the regulation of plant growth through the reduction of endogenous levels of bioactive GAs.
The first GA 2-oxidase cDNA was cloned from runner bean (Phaseolus coccineus) by functional screening, and then three GA 2-oxidase cDNAs were cloned from Arabidopsis by database screening (Thomas et al., 1999). Two cDNAs for GA 2-oxidase were also isolated from garden pea (Pisum sativum) by functional screening and reverse transcription (RT)-PCR (Lester et al., 1999; Martin et al., 1999). A loss-of-function mutation in the GA 2-oxidase gene of garden pea results in the slender phenotype. This indicates that GA 2-oxidase has an important role in the regulation of elongation growth (Lester et al., 1999; Martin et al., 1999).
We describe the cloning and characterization of a GA 2-oxidase gene, OsGA2ox1, from rice (Oryza sativa L. cv Nipponbare). Based on detailed expression analysis of OsGA2ox1, we suggest that the GA 2-oxidase encoded by this gene participates in the phase transition from vegetative to reproductive growth.
RESULTS
Isolation of a GA 2-Oxidase Gene from Rice
The predicted amino acid sequences of conserved regions among 2ODDs, including GA 20-oxidase and GA 3β-hydroxylase (Prescott, 1993), were used to search for putative GA 2-oxidase genes in databases. Arabidopsis genomic sequence T31E10.11 (accession no. AC004077) has relatively high homology with the sequences of GA 3β-hydroxylase genes, although its deduced amino acid sequence did not have M-G-L-A-A/P-H-T-D motif that is conserved in the GA 3β-hydroxylases reported. The sequence of Marah macrocarpa dioxygenase mRNA M7–3 (accession no. Y09113; MacMillan et al., 1997) had high homology with T31E10.11. The full-length cDNA corresponding to T31E10.11 was isolated from Arabidopsis inflorescences by RT-PCR, and the 2β-hydroxylation activity of the recombinant protein was confirmed in vitro. This cDNA was identical to the cDNA clone AtGA2ox3 reported by Thomas et al. (1999).
The predicted amino acid sequences of AtGA2ox3, M7-3, and two rice GA 3β-hydroxylases (H. Itoh, M. Ueguchi-Tanaka, N. Sentoku, H. Kitano, M. Matsuoka, and M. Kobayashi, unpublished data) were compared to design degenerate oligonucleotide primers. Using total RNA from rice shoot as a template, RT-PCR with degenerate primers produced one sequence of the expected length with significant homology to GA 2-oxidases from Arabidopsis. This clone was used for further screening to isolate corresponding full-length cDNA and genomic clones.
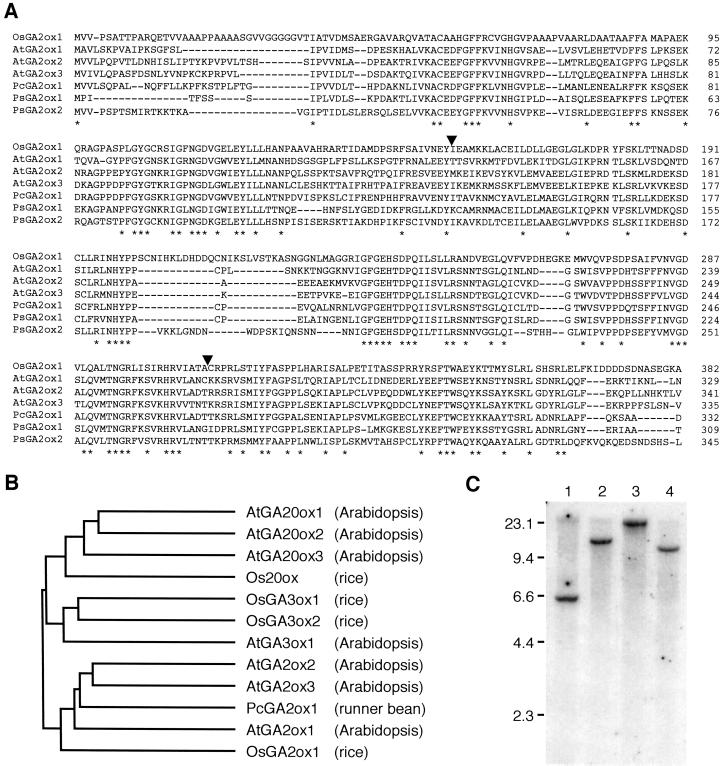
The isolated full-length cDNA contained an open reading frame of 1,146 bp encoding a protein of 382 amino acids, and was designated OsGA2ox1 (Oryza sativa GA 2-oxidase 1). The predicted amino acid sequence of OsGA2ox1 contained the amino acids that are conserved within the 2ODDs for GA biosynthesis, including His-241, Asp-243, and His-302 (Fig. 1A, numbers refer to OsGA2ox1 sequence), the amino acid residues that are supposed to associate at the catalytic site and bind with Fe2+ (Valegard et al., 1998). When compared with other 2ODDs for GA biosynthesis, the deduced amino acid sequence of OsGA2ox1 showed highest homology with GA 2-oxidases (Fig. 1B). However, the phylogenetic relationship revealed that GA 2-oxidases from dicot plants share relatively high (47%–69%) amino acid identity with each other but significantly lower (< 44%) identity with OsGA2ox1 (Fig. 1B; Table I).
Figure 1.
Sequence analysis and genomic DNA gel-blot analysis of OsGA2ox1. A, Alignment of deduced amino acid sequences of GA 2-oxidases. Asterisks indicate identical amino acids among the enzymes from various plants. Positions of introns are marked by arrowheads. AtGA2ox1, AtGA2ox2, and AtGA2ox3, GA 2-oxidases from Arabidopsis (accession nos. AJ132435, AJ132436, and AJ132437); PcGA2ox1, GA 2-oxidase from runner bean (accession no. At132438); PsGA2ox1 and PsGA2ox2, GA 2-oxidases from garden pea (accession nos. AF100954 and AF100955). B, Phylogenetic relationships among GA 2-oxidases. AtGA20ox1, AtGA20ox2, and AtGA20ox3, GA 20-oxidases from Arabidopsis (accession nos. X83379, X83380, X83381); Os20ox, GA 20-oxidase from rice (accession no. U50333); AtGA3ox1, GA 3β-hydroxylase from Arabidopsis (accession no. L37126); OsGA3ox1 and OsGA3ox2, 3β-hydroxylase from rice (H. Itoh, M. Ueguchi-Tanaka, N. Sentoku, H. Kitano, M. Matsuoka, and M. Kobayashi, unpublished data). C, Genomic DNA gel-blot analysis of OsGA2ox1. Five micrograms of genomic DNA was digested with ApaI (lane 1), BamHI (lane 2), EcoRI (lane 3), or HindIII (lane 4). Mr markers are indicated at the left in kilobases.
Table I.
Amino acid sequence homology between GA 2-oxidases
| No. | OsGA2ox1 No. 1 | AtGA2ox1 No. 2 | AtGA2ox2 No. 3 | AtGA2ox3 No. 4 | PcGA2ox1 No. 5 | PsGA2ox1 No. 6 | PsGA2ox2 No. 7 |
|---|---|---|---|---|---|---|---|
| 1 | — | 37 | 36 | 36 | 36 | 44 | 38 |
| 2 | 51 | — | 56 | 53 | 57 | 48 | 57 |
| 3 | 51 | 75 | — | 69 | 62 | 47 | 55 |
| 4 | 49 | 71 | 84 | — | 55 | 47 | 53 |
| 5 | 49 | 74 | 75 | 67 | — | 50 | 56 |
| 6 | 58 | 65 | 65 | 64 | 67 | — | 50 |
| 7 | 52 | 73 | 74 | 68 | 72 | 67 | — |
Values are percentage of amino acid identity (above the diagonal) or similarity (below the diagonal).
The corresponding genomic DNA that completely covers the OsGA2ox1 coding region was also cloned. By comparing the genomic DNA and cDNA sequences, we revealed that OsGA2ox1 consists of three exons and two introns (Fig. 1A). This exon/intron structure is also conserved in the AtGA2ox3 coding sequence.
To investigate the existence of a related sequence for OsGA2ox1 in the rice genome, we digested rice genomic DNA with several restriction enzymes and subjected it to DNA gel-blot analysis at low stringency, using OsGA2ox1 cDNA fragment as a probe. As shown in Figure 1C, only one band was obtained; this indicates that the OsGA2ox1 protein is encoded by a single-copy gene in the rice genome.
Function of Recombinant OsGA2ox1 Protein
Recombinant OsGA2ox1 protein was prepared by expressing the OsGA2ox1 cDNA in Escherichia coli. The catalytic properties of the recombinant protein were examined by incubating cell lysate with GAs identified from rice (Kobayashi et al., 1988, 1989). GAs without a free carboxyl at C-19 were converted to the corresponding 2β-hydroxylated product by the recombinant OsGA2ox1 protein (Table II). On the other hand, no metabolite was identified for GA19 and GA53; both GAs have a free carboxyl at C-19. This indicates that OsGA2ox1 is a GA 2-oxidase.
Table II.
Metabolism of recombinant OsGA2ox1 protein
| Substrate | Producta | KRI | Characteristic Ions at m/z (% Relative Intensity of Base Peak)b |
|---|---|---|---|
| 2H2-GA44 | GA98 | 2,962 | 522 (47), 507 (5), 373 (4), 345 (4), 240 (21), 209 (100) |
| 2H2-GA20 | GA29 | 2,688 | 509 (100), 494 (6), 450 (2), 379 (6), 210 (25) |
| 2H3-GA1 | GA8 | 2,818 | 597 (100), 582 (7), 538 (8), 451 (5), 210 (20) |
| 2H3-GA9 | GA51 | 2,539 | 389 (22), 331 (30), 287 (78), 271 (76), 227 (100) |
| GA4 | GA34 | 2,678 | 506 (100), 459 (1), 416 (3), 313 (7), 288 (3) |
Substrate GAs were incubated with recombinant OsGA2ox1 protein, and the product GAs were identified by full-scan GC-MS and KRI.
Identification of metabolic products by GC-MS on the basis of KRI and full-scan mass spectra of the methyl ester trimethylsilylether derivatives.
Based on ions with a mass-to-charge ratio (m/z) of >200.
Ectopic Expression of the OsGA2ox1 cDNA in Transgenic Rice
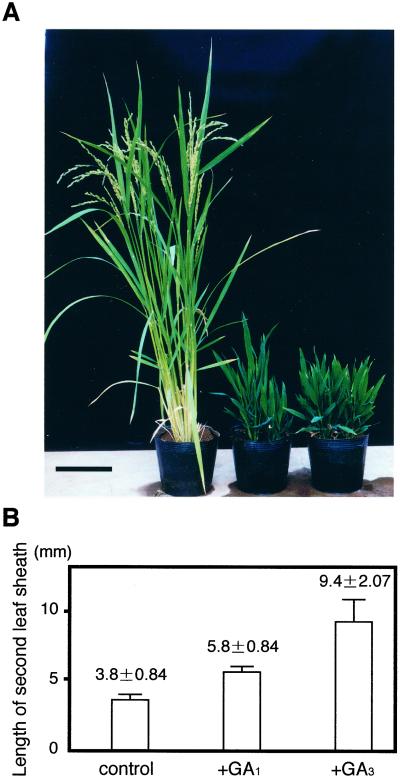
To assess the activity of the OsGA2ox1 gene product in vivo, we fused the full-length OsGA2ox1 cDNA to the rice actin promoter in the sense orientation and introduced it into wild-type rice by Agrobacterium-mediated gene transfer. All primary transformants (46 independent lines) showed dwarf phenotype. The final plant height of extremely dwarfed transformants was less than 15 cm, whereas that of wild type was 90 cm. Their leaf blades were dark green and shorter and wider than those of wild-type plants, a typical phenotype for GA-deficient dwarf rice (Fig. 2A). Although wild-type plants flowered approximately 90 d after sowing, the formation of floral organs and internode elongation in the extremely dwarfed transformants were not observed even at 120 d after sowing. The transformants did not bear any seeds, but exogenous application of GA3 could rescue this phenotype (data not shown).
Figure 2.
Ectopic expression of OsGA2ox1 in transgenic rice plants. A, Typical phenotype of transgenic rice plants carrying the Act::OsGA2ox1 gene approximately 120 d after germination. Left, wild-type (cv Nipponbare); center and right, Act::OsGA2ox1 transformants. In contrast to wild-type plants, flowering of transformants was impeded. Bar = 10 cm. B, Effect of exogenous GA treatment on elongation of the second leaf sheath of transgenic seedlings. Ten nanograms of GA1 or GA3 was dropped on the seedlings, which were then grown for 3 d. Each data point is the average for five plants.
We have previously reported that major endogenous GAs in vegetative tissues of rice are biosynthesized via the early 13-hydroxylation pathway, and GA1 is the major bioactive GA that regulates shoot elongation (Kobayashi et al., 1988, 1989, 1994). Therefore, we compared the levels of 13-hydroxylated GAs in the leaves of wild-type and extremely dwarfed transgenic plants (Table III). In the transformants, the level of GA1 decreased to less than one-fourth that of wild-type plants. In contrast, the levels of GA8 and GA29, the 2β-hydroxylated metabolites of GA1 and GA20, were elevated approximately 2.5- and 6.8-fold, respectively, in the leaves of transformants.
Table III.
Concentration (ng g−1 fresh weight) of GAs in leaves of wild-type and transgenic rice plant overexpressing OsGA2ox1 cDNA
| Plant | GA53 | GA44 | GA19 | GA20 | GA1 | GA8 | GA29 |
|---|---|---|---|---|---|---|---|
| Wild type | 0.37 | 0.41 | 2.72 | 0.66 | 0.21 | 0.39 | 0.17 |
| Transformant | 0.20 | 0.31 | 5.97 | 0.74 | 0.05 | 0.97 | 1.15 |
The level of endogenous GAs in the shoots of wild-type (cv Nipponbare) and transgenic plants were analyzed by GC-SIM using internal standards.
We also determined the effects of exogenous application of GA1 and GA3 on the elongation of the second leaf sheath in the transgenic seedlings. GA1 can be inactivated by GA 2-oxidase, but GA3 is not. GA1 and GA3 showed an almost equivalent effect on the elongation of dwarf rice, cv Tan-ginbozu (Murakami, 1972). In our results, however, GA1 was much less active than GA3 on the elongation of the transformants (Fig. 2B).
Expression of OsGA2ox1 Gene in Rice
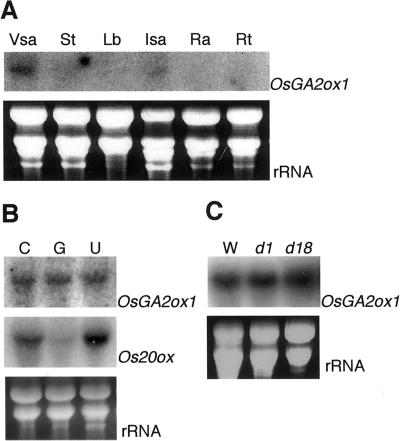
We performed RNA gel-blot analysis to determine the expression pattern of OsGA2ox1 in various organs of rice. A single strong band was detected in RNA from vegetative shoot apices (Fig. 3A). A relatively weak band was also observed in RNA from inflorescence shoot apices, but not from stems, leaf blades, or rachises.
Figure 3.
RNA gel-blot analysis of OsGA2ox1. Ten micrograms of total RNA was loaded per lane. The blot was probed with 32P-labeled OsGA2ox1 or Os20ox cDNA. The lower part of A and B shows ethidium bromide-stained RNAs corresponding to the above lanes for loading control. A, OsGA2ox1 expression in various organs of wild-type rice. Total RNA was extracted from vegetative shoot apices (Vsa), stems (St), leaf blades (Lb), inflorescence shoot apices (Isa), rachises (Ra), and roots (Rt). B, Effect of GA3 and uniconazole treatment on the levels of OsGA2ox1 and Os20ox transcripts. Total RNA was extracted from wild-type seedlings treated with 10 μm GA3 (G) or 10 μm uniconazole (U) or from untreated control plants (C). C, OsGA2ox1 expression in rice dwarf mutants. Total RNA was extracted from wild-type (W), GA-insensitive dwarf mutant (d1), and GA-responsive dwarf mutant (d18) seedlings.
The expression of AtGA2ox1 and AtGA2ox2 is up-regulated by the application of GA3 (Thomas et al., 1999). We examined the effects of GA3 and uniconazole (an inhibitor of GA biosynthesis) on the level of OsGA2ox1 mRNA. We also determined the expression of the rice GA 20-oxidase gene, Os20ox (Toyomasu et al., 1997), as a control. The expression of Os20ox was up-regulated by uniconazole and down-regulated by GA3 (Fig. 3B), but the levels of OsGA2ox1 transcripts were not affected (Fig. 3B). Moreover, expression levels of OsGA2ox1 in the dwarf mutants d1 (GA-insensitive dwarf; Ueguchi-Tanaka et al., 2000) and d18 (loss-of-function mutation in the rice GA 3β-hydroxylase gene; Murakami, 1972) were similar to those in the wild-type plants (Fig. 3C).
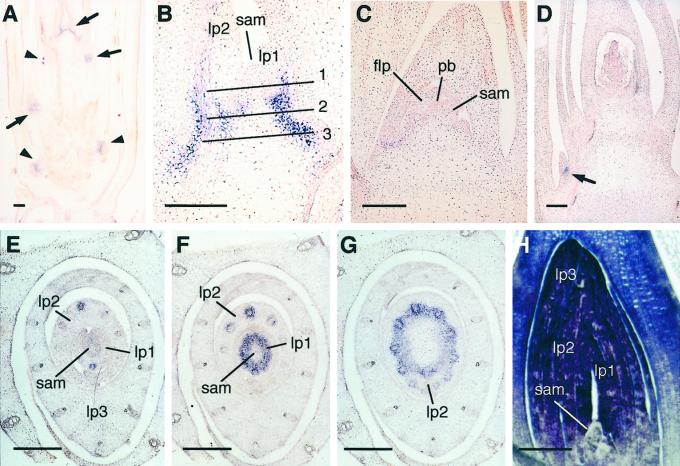
To determine more precisely the spatial pattern of OsGA2ox1 expression in rice, we conducted in situ hybridization with a digoxygenin-labeled OsGA2ox1 antisense-strand RNA probe. Figure 4A shows the localization of OsGA2ox1 mRNA in seedlings. Purple staining, which indicates the presence of OsGA2ox1 mRNA, occurred around the shoot apical and lateral meristems (arrows) and around the meristems of crown roots in culms (arrowheads). Control sections, hybridized with a sense-strand RNA probe, showed no signal above background staining (data not shown).
Figure 4.
In situ mRNA localization of OsGA2ox1 and OsGA3ox2. Purple staining indicates the presence of OsGA2ox1 (A–G) and OsGA3ox2 (H) mRNA. A, Longitudinal section of seedling at 7 d after germination. Arrows and arrowheads indicate the shoot and root meristem, respectively. B, Median longitudinal section of the vegetative shoot apex at higher magnification than (A). Lines 1, 2, and 3 indicate the approximate planes of bisection shown in E, F, and G. C, Median longitudinal section of the inflorescence meristem. D, Median longitudinal section of the floral meristem at the primary rachis branch stage. Arrow indicates the lateral meristem. E through G, Serial cross sections around the vegetative shoot apex. H, Longitudinal section of seedling at 3 d after sowing. sam, Shoot apical meristem; lp1 through 3, leaf primordium 1 through 3; flp, flag leaf primordium; pb, primary rachis branch.
Figure 4B shows a near-median longitudinal section of a rice vegetative shoot apex at a higher magnification than Figure 4A. Lines 1, 2, and 3 indicate approximate planes of the cross-sections shown in Figure 4, E, F, and G, respectively. The OsGA2ox1 expression appeared as pairs of signals on opposite flanks of the shoot apical meristem (SAM), which was located in the basal region of leaf primordia (Fig. 4B). Serial cross-sections (Fig. 4, E–G) better show the spotted expression of OsGA2ox1 localized as a doughnut-ring shape around the boundary of leaf primordia (Fig. 4B).
The ring-shaped expression of OsGA2ox1 around the SAM was drastically decreased just after the phase transition from vegetative to inflorescence stage (Fig. 4C). At the stage of primary rachis-branch primordia differentiation, OsGA2ox1 was barely expressed around the SAM but was strongly expressed around the lateral meristem, which was still in the vegetative phase (arrow in Fig. 4D).
We also examined the in situ mRNA localization of a rice GA 3β-hydroxylase gene, OsGA3ox2, around the vegetative shoot apex. As shown in Figure 4H, the OsGA3ox2 mRNA was strongly expressed in young leaves, but it was not expressed in the SAM or in the basal regions of young leaves throughout the vegetative phase.
DISCUSSION
The concentration of bioactive GAs is tightly maintained by the balance between their synthesis and catabolism, as indicated by the fact that GAs reduce the expression of GA20ox genes and stimulate the expression of GA2ox genes. GA 2-oxidase catalyzes the catabolism, namely, the conversion of bioactive GAs and their immediate precursors into biologically inactive GAs by 2β-hydroxylation (Ross et al., 1995). The expression of GA 2-oxidase genes from Arabidopsis was stimulated by the application of GA3 (Thomas et al., 1999). This indicates that the expression of these genes is regulated through feedback to maintain endogenous levels of bioactive GAs.
We cloned OsGA2ox1, the first GA 2-oxidase gene from rice. Four of six GA 2-oxidases from runner bean, Arabidopsis, and garden pea catalyze not only 2β-hydroxylation but also the further oxidation at C-2. On the other hand, the recombinant OsGA2ox1 protein catalyzed a single step of the 2β-hydroxylation. These results demonstrate that OsGA2ox1 inactivates the bioactive GAs and their precursors. The physiological and biochemical phenotypes of transgenic rice plants that ectopically express the OsGA2ox1 cDNA further support this conclusion.
Two or more GA 2-oxidase genes have been cloned from Arabidopsis and garden pea, and their expression was observed in various organs (Lester et al., 1999; Martin et al., 1999; Thomas et al., 1999). Because OsGA2ox1 was expressed only in limited tissues and was not induced by the application of GA3, there may be another GA 2-oxidase gene that maintains bioactive GA levels in various organs of rice. We have tried unsuccessfully so far to clone another GA 2-oxidase gene from rice. Only one band appeared in the DNA gel-blot analysis (Fig. 2C). Another GA 2-oxidase gene may have relatively less sequence similarity with those genes already reported. Our recent results have revealed that one of the rice 3β-hydroxylases, OsGA3ox1, had activity for 2β-hydroxylation of bioactive GAs (H. Itoh, M. Ueguchi-Tanaka, N. Sentoku, H. Kitano, M. Matsuoka, and M. Kobayashi, unpublished data). Thus, there are at least two genes in the rice genome whose products have 2β-hydroxylation activity.
OsGA2ox1 was expressed in a limited region around the shoot apex and was not affected by the levels of bioactive GAs. Tissue-specific expression has been reported for GA4H, a GA 3β-hydroxylase gene expressed in germinating seeds of Arabidopsis. The expression of GA4H was induced by the phytochrome signal to promote germination and was not affected by the exogenous application of GA3 or uniconazole (Yamaguchi et al., 1999). OsGA2ox1, similarly, may have a specific role in the regulation of developmental processes in the shoot apex, rather than in the maintenance of bioactive GA levels in leaf tissues.
It is noteworthy that OsGA3ox2, the major GA 3β-hydroxylase in the vegetative tissue of rice (H. Itoh, M. Ueguchi-Tanaka, N. Sentoku, H. Kitano, M. Matsuoka, and M. Kobayashi, unpublished data), was expressed in young leaves but not in the SAM itself (Fig. 4H). This result is consistent with the previous report that the young leaves are the source of bioactive GAs (Choi et al., 1995). In contrast, the OsGA2ox1 mRNA was localized in a ring at the basal region of young leaves. This pattern of gene expression leads us to speculate that OsGA2ox1 inactivates GAs synthesized in young leaves to keep the levels of bioactive GAs low in the SAM. This may be one of the mechanisms for repressing internode elongation during the vegetative phase.
It is interesting that the expression of OsGA2ox1 around the shoot apex disappeared after flower induction. It has been suggested that GA is involved in the phase transition in various plant species (Evans and Poethig, 1995; Chien and Sussex, 1996; McDaniel and Hartnett, 1996; Telfer et al., 1997; Scott et al., 1999). In a long-day plant, Arabidopsis, GA has been proposed to promote flowering by activating the LEAFY promoter (Okamuro et al., 1996; Blazquez et al., 1997, 1998; Blazquez and Weigel, 1999, 2000). GA also activates SaMADS A, a gene involved in the regulation of floral transition in the long-day plant Sinapis alba (Bonhomme et al., 2000). In a short-day plant, Pharbitis nil, application of uniconazole inhibited the flowering response induced by short-day treatment, and the inhibition by uniconazole was canceled by further application of GA1 (Wijayanti et al., 1996, 1997). These studies indicate that GA is required for the differentiation and/or development of the shoot apex during the phase transition. This hypothesis is consistent with the fact that, in transgenic rice plants ectopically expressing the OsGA2ox1 cDNA, flowering time was significantly delayed and the development of reproductive organs was impeded. From this point of view, the down-regulation of OsGA2ox1 in the shoot apex may be one of the initial events required for the development of inflorescence meristem.
MATERIALS AND METHODS
Plant Materials
Seeds of tall rice (Oryza sativa L. cv Nipponbare), two GA-insensitive dwarf cultivars (Daikoku-dwarf, T65-d1, and Hosetsu-dwarf, d18-ID18h), and OsGA2ox1 transformants were sterilized in 1% (v/v) NaClO for 1 h and sown on an agar medium. Seedlings were grown in a growth chamber with a cycle of 16-h light/8-h dark at 30°C. To investigate the effects of GA3 and uniconazole on the expression of OsGA2ox1, seeds of wild-type rice were sown on an agar medium containing 10 μm GA3 or 10 μm uniconazole and grown for 3 d.
Isolation of Partial Fragment Encoding Rice GA 2-Oxidase
To amplify a GA 2-oxidase from rice, total RNA was extracted from whole seedlings. RT-PCR was performed by using two degenerate oligonucleotide primers (forward primer, 5′-GGNTTYGGNGARCAYWCNGAYCC-3′; reverse primer, 5′-ACNCKRTGYYTAYISWIWYVARICKICCRTTI- GT-3′). Amplified fragments (approximately 200 bp) were cloned into pCR II (Invitrogen, Carlsbad, CA), and their sequences were determined. One of the 64 independent clones showed homology with 2ODDs and was predicted to encode a rice GA 2-oxidase gene. The deduced amino acid sequence for this clone was used to search the DDBJ nucleotide sequence database, and one rice expressed sequence tag clone (accession no. C72618) was obtained. Oligonucleotide primers were designed based on the sequence of this clone (forward primer, 5′-GCGGCGTTCTTCGCG-3′; reverse primer, 5′-ATGCTAGCCTTGGTGCTAA-3′) and used in RT-RCR. An amplified fragment (230 bp) was cloned into pCR II (Invitrogen), and the sequence was confirmed. This DNA fragment was used to screen the cDNA and genomic libraries.
Screening of Rice cDNA and Genomic Libraries
A cDNA library was constructed with mRNA prepared from immature rice seed, and a genomic library was constructed from rice genomic DNA partly digested with Sau3AI. The libraries were screened with a 32P-labeled probe prepared as described above. Hybridization was performed in 5× SSC (1× SSC contains 0.15 m NaCl, 15 mm sodium citrate), 5× Denhardt's solution (1× Denhardt's solution contains 0.02% [w/v] Ficoll, 0.02% [w/v] polyvinylpyrrolidone, 0.02% [w/v] bovine serum albumin), 0.5% (w/v) SDS, and 20 mg L−1 salmon sperm DNA at 65°C for 14 h. Then filters were washed in 2× SSC, 0.1% (w/v) SDS, at room temperature.
Sequence Analysis
Nucleotide sequences were determined by a dideoxy-nucleotide chain-termination method using an automated sequencing system (ABI377). The cDNA and genomic clones were completely sequenced on both strands, including a large intron. The cDNA and amino acid sequences were analyzed by Lasergene computer software (DNAStar, Madison, WI).
DNA Gel-Blot Analysis
Five micrograms of the genomic DNA isolated from rice leaves was digested with ApaI, BamHI, EcoRI, and HindIII, electrophoresed on a 0.7% (w/v) agarose gel, then transferred on to nylon membranes (Hybond N+ membrane Amersham, Buckinghamshire, England) and hybridized with a 32P-labeled probe prepared as described above. Hybridization was performed in 5× SSC, 5× Denhardt's solution, 0.5% (w/v) SDS, 10% (w/v) dextran sulfate, and 20 mg L−1 salmon sperm DNA at 65°C for 14 h. The filter was washed in 2× SSC, 0.1% (w/v) SDS, at 65°C, and then further washed in 0.2× SSC, 0.1% (w/v) SDS, at 65°C.
Enzyme Assays
Full-length OsGA2ox1 cDNA was inserted in the sense orientation as a translational fusion into the pMAL-c2 expression vector (New England Biolabs, Beverly, MA), and expressed in Escherichia coli strain JM109. Cell lysate was prepared as described by Rebers et al. (1999). The presence of OsGA2ox1 recombinant protein in the cell lysate was confirmed by SDS-PAGE. After the addition of 2-ketogluta-rate (final concentration, 5 mm), ascorbate (5 mm), and FeSO4 (0.5 mm), the cell lysate (total volume, 200 μL) was incubated with substrate GA at 30°C for 1 h. Then the reaction was adjusted to pH 2, and the product was extracted with ethyl acetate. The extract was analyzed by full-scan gas chromatography-mass spectrometry with an Automass mass spectrometer (JEOL, Akishima, Japan) connected to a Hewlett-Packard 5890 series II gas chromatograph (Kobayashi et al., 1996). Product GAs were identified by their mass spectrum and Kovats retention index.
Plasmid Constructs and Plant Transformation
The full-length cDNA of OsGA2ox1 was excised as an XbaI-EcoRV fragment and inserted between the rice actin promoter and the nopaline synthase polyadenylation signal of hygromycin-resistant binary vector pAct-Hm2. This vector is modified from pBI-H1 (Ohta et al., 1990) and contains a rice actin promoter. The resulting fusion construct was introduced into Agrobacterium tumefaciens strain EHA101 by electroporation. Agrobacterium-mediated transformation of rice was performed as described by Hiei et al. (1994). Transgenic plants were selected on media containing 50 mg L−1 hygromycin.
Analysis of Endogenous GA levels
Endogenous GA levels in the transgenic plants were analyzed by a modification of the procedure of Choi et al. (1995). Plant tissues were homogenized in liquid nitrogen and extracted twice with an excess volume of 80% (v/v) methanol. The extract was amended with 5 ng of deuterated GAs (2H2-GA53, 44, 19, 20, 29, 8 and 2H5-GA1) as internal standards. After concentration in vacuo, the aqueous residue was submitted to a solvent fractionation procedure to give the acidic ethyl acetate-soluble fraction. After two steps of purification with Bond Elut C18 and Bond Elut DEA cartridge columns (Varian, Harbor City, CA; Kobayashi et al., 1993), the fraction was submitted to HPLC on a column of Nucleosil 5 C18 (i.d., 6 mm; length, 100 mm; Macherey-Nagel, Düren, Germany). The column was eluted with a mixture of solvents, A (10% [v/v] methanol, 0.05% [v/v] acetic acid) and B (0.05% [v/v] acetic acid in methanol). After injection, the concentration of solvent B was increased from 20% to 75% over 32 min. The elute was collected according to the retention time of each GA, concentrated, and derivatized to methyl ester trimethylsilyl ether or trimethylsilyl ester trimethylsilyl ether. It was then analyzed by gas chromatography-selected ion monitoring (GC-SIM). The conditions for GC-SIM were as described by Kobayashi et al. (1996).
Biological Activity of GA1 and GA3 on the Elongation of OsGA2ox1 Transgenic Plants
The biological activity was evaluated by micro-drop assay (Murakami, 1972).
RNA Gel-Blot Analysis
Total RNA was separately prepared from various organs of rice for analysis. Ten micrograms of each RNA preparation was electrophoresed on a 1.2% (w/v) agarose gel, then transferred on to Hybond N+ membrane and hybridized with a 32P-labeled probe prepared as described above. The hybridization and washing conditions were as for the DNA gel-blot analysis.
In Situ Hybridization Analysis
Plant materials were fixed in 4% (w/v) paraformaldehyde and 0.25% (w/v) glutaraldehyde in 100 mm sodium phosphate buffer (pH 7.4) overnight at 4°C, dehydrated through a graded ethanol series and t-butanol series (Sass, 1958), and finally embedded in Paraplast Plus (Sherwood Medical, St. Louis). Microtome sections (7–10 μm thick) were applied to glass slides treated with silane. Hybridization with a digoxygenin-labeled RNA probe and immunological detection were conducted according to the methods of Kouchi and Hata (1993).
Footnotes
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (grant no. 11306003 to S.I.); by the Program for Promotion of Basic Research Activities for Innovation Biosciences (grant to H.T. and M.M.); and by a research fellowship from the Japan Society for the Promotion of Science (to T.S.).
LITERATURE CITED
- Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell. 1998;10:791–800. doi: 10.1105/tpc.10.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D. Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol. 1999;120:1025–1032. doi: 10.1104/pp.120.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D. Integration of floral inductive signals in Arabidopsis. Nature. 2000;404:889–892. doi: 10.1038/35009125. [DOI] [PubMed] [Google Scholar]
- Bonhomme F, Kurz B, Melzer S, Bernier G, Jacqmard A. Cytokinin and gibberellin activate SaMADS A, a gene apparently involved in regulation of the floral transition in Sinapis alba. Plant J. 2000;24:103–111. doi: 10.1046/j.1365-313x.2000.00859.x. [DOI] [PubMed] [Google Scholar]
- Chang H-H, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 Locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien JC, Sussex IM. Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996;111:1321–1328. doi: 10.1104/pp.111.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y-H, Yoshizawa K, Kobayashi M, Sakurai A. Distribution of endogenous gibberellin in vegetative shoots of rice. Plant Cell Physiol. 1995;36:997–1001. [Google Scholar]
- Evans MM, Poethig RS. Gibberellins promote vegetative phase change and reproductive maturity in maize. Plant Physiol. 1995;108:475–487. doi: 10.1104/pp.108.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Gaskin P, Spray CR, Phinney BO, MacMillan J. The metabolism of gibberellin A20 to gibberellin A1 by tall and dwarf mutants of Oryza sativa and Arabidopsis thaliana. Plant Physiol. 1994;106:1367–1372. doi: 10.1104/pp.106.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Gaskin P, Spray CR, Suzuki Y, Phinney BO, MacMillan J. Metabolism and biological activity of gibberellin A4 in vegetative shoots of Zea mays, Oryza sativa, and Arabidopsis thaliana. Plant Physiol. 1993;102:379–386. doi: 10.1104/pp.102.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Sakurai A, Saka H, Takahashi N. Quantitative analysis of endogenous gibberellins in normal and dwarf cultivars of rice. Plant Cell Physiol. 1989;30:963–969. [Google Scholar]
- Kobayashi M, Yamaguchi I, Murofushi N, Ota Y, Takahashi N. Fluctuation and localization of endogenous gibberellins in rice. Agric Biol Chem. 1988;52:1189–1194. [Google Scholar]
- Kobayashi M, Yoshizawa K, Sakurai A, Nakamura T. Analysis of endogenous gibberellins and abscisic acid in vegetative shoots of normal and weeping Japanese cherry (Prunus spachiana) Biosci Biotech Biochem. 1996;60:159–160. [Google Scholar]
- Kouchi H, Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- Lange T. Molecular biology of gibberellin synthesis. Planta. 1998;204:409–419. doi: 10.1007/s004250050274. [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB. Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 1999;19:65–73. doi: 10.1046/j.1365-313x.1999.00501.x. [DOI] [PubMed] [Google Scholar]
- MacMillan J, Ward DA, Phillips AL, Sanchez-Beltran MJ, Gaskin P, Lange T, Hedden P. Gibberellin biosynthesis from gibberellin A12-aldehyde in endosperm and embryos of Marah macrocarpa. Plant Physiol. 1997;113:1369–1377. doi: 10.1104/pp.113.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P. The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol. 1999;121:775–781. doi: 10.1104/pp.121.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel CN, Hartnett LK. Flowering as metamorphosis: two sequential signals regulate floral initiation in Lolium temulentum. Development. 1996;122:3661–3668. doi: 10.1242/dev.122.11.3661. [DOI] [PubMed] [Google Scholar]
- Murakami Y. Dwarfing genes in rice and their relation to gibberellin biosynthesis. In: Carr DJ, editor. Plant Growth Substances. Berlin: Springer-Verlag; 1972. pp. 166–174. [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K. Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol. 1990;31:805–813. [Google Scholar]
- Okamuro JK, den Boer BG, Lotys-Prass C, Szeto W, Jofuku KD. Flowers into shoots: photo and hormonal control of a meristem identity switch in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:13831–13836. doi: 10.1073/pnas.93.24.13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NJE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott AG. A dilemma of dioxygenases (or where biochemistry and molecular biology fail to meet) J Exp Bot. 1993;44:849–861. [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang Y-Y, Imai R, Sekimoto H, Kamiya Y. Regulation of gibberellin biosynthesis genes during flower and early development of tomato. Plant J. 1999;17:241–250. doi: 10.1046/j.1365-313x.1999.00366.x. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Reid JB, Swain SM, Hasan O, Poole AT, Hedden P, Willis CL. Genetic regulation of gibberellin deactivation in Pisum. Plant J. 1995;7:513–523. [Google Scholar]
- Sass AE. Botanical Micro Technique. Ed 3. Ames: Iowa State University Press; 1958. [Google Scholar]
- Scott DB, Jin W, Ledford HK, Jung H-S, Honma MA. EAF1 regulates vegetative-phase change and flowering time in Arabidopsis. Plant Physiol. 1999;120:675–684. doi: 10.1104/pp.120.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Sekimoto H, Numers C, Phillips AL, Hedden P, Kamiya Y. Cloning and characterization of a cDNA encoding gibberellin 20-oxidase from rice (Oryza sativa) seedlings. Physiol Plant. 1997;99:111–118. [Google Scholar]
- Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M. Rice dwarf mutant, d1, which is defective in the α subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA. 2000;97:11638–11643. doi: 10.1073/pnas.97.21.11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegard K, van Scheltinga AC, Lloyd MD, Hara T, Ramaswamy S, Perrakis A, Thompson A, Lee HJ, Baldwin JE, Schofield CJ. Structure of a cephalosporin synthase. Nature. 1998;394:805–809. doi: 10.1038/29575. [DOI] [PubMed] [Google Scholar]
- Wijayanti L, Fujioka S, Kobayashi M, Sakurai A. Effect of uniconazole and gibberellin on the flowering of Pharbitis nil. Biosci Biotech Biochem. 1996;60:852–855. doi: 10.1271/bbb.60.852. [DOI] [PubMed] [Google Scholar]
- Wijayanti L, Kobayashi M, Fujioka S, Sakurai A. Gibberellin transport from the cotyledon to plumule in the flowering of Pharbitis nil. Biosci Biotech Biochem. 1997;61:1384–1385. doi: 10.1271/bbb.60.852. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun TP. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1999;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]