Abstract
The hemagglutinin (HA) and neuraminidase (NA) of influenza A virus possess antagonistic activities on interaction with sialic acid (SA), which is the receptor for virus attachment. HA binds SA through its receptor-binding sites, while NA is a receptor-destroying enzyme by removing SAs. The function of HA during virus entry has been extensively investigated, however, examination of NA has long been focused to its role in the exit of progeny virus from infected cells, and the role of NA in the entry process is still under-appreciated. This review summarizes the current understanding of the roles of HA and NA in relation to each other during virus entry.
Keywords: influenza A virus, hemagglutinin, neuraminidase, virus entry
1. Introduction
Influenza viruses belong to the family Orthomyxoviridae of enveloped viruses and are classified into four types, A, B, C and the recently identified type D [1,2]. Three main types of influenza viruses (A, B and C) infect humans, with influenza A and B viruses usually cause annual epidemics, resulting in about 3 to 5 million cases of severe illness, and about 290,000 to 650,000 respiratory deaths annually worldwide [1]. Influenza A viruses from zoonotic sources can also result in occasional but devastating pandemics [3].
The influenza A virus (IAV) and type B virus (IBV) genomes each contain eight segmented, negative-sense, single-stranded viral RNA (vRNA) segments, coding for at least 11 proteins, of which hemagglutinin (HA) and neuraminidase (NA) are the major surface glycoproteins [4]. In contrast, influenza C virus (ICV) and type D virus (IDV) carry seven segments respectively, encoding only one glycoprotein, the haemagglutinin-esterase-fusion (HEF) protein, which combines both the function of HA and NA [5,6,7]. IAVs can be further classified into subtypes based on HA and NA antigenicity. To date, many combinations of 16 HA (H1-16) and 9 NA (N1-9) subtypes have been identified in wild birds, while two additional subtypes (H17N10 and H18N11) have recently been isolated in bats [8,9,10]. Two subtypes of IAVs (H1N1 and H3N2) and two antigenically distinct lineages of IBVs (Victoria and Yamagata) currently co-circulate in humans [11].
Both HA and NA interact with sialic acid (SA), which usually links to glycoproteins and glycolipids at the cell surface [12]. HA binds to SA moieties presented by cellular receptors, triggering virus entry by clathrin-mediated endocytosis, although other endocytic routes, including micropinocytosis and raft-dependent endocytosis, may also be used [13,14,15,16,17]. In contrast, NA removes SAs from cellular receptors and from newly synthesized HA and NA on nascent virions, enabling efficient release of progeny virus at the final stage of infection [18].
The antagonistic activities on SA binding suggest a close relationship between the functions of HA and NA. The role of the viral HA in attachment and infection has been well explored, however, examination of NA has largely focused on its role in the exit of progeny virus from infected cells [19]. Interestingly, increasing pieces of evidence strongly support an essential role of NA during virus entry process [19,20]. In this review, we summarize the current state of our understanding of how the interaction of HA and NA affects IAV entry.
2. Hemagglutinin
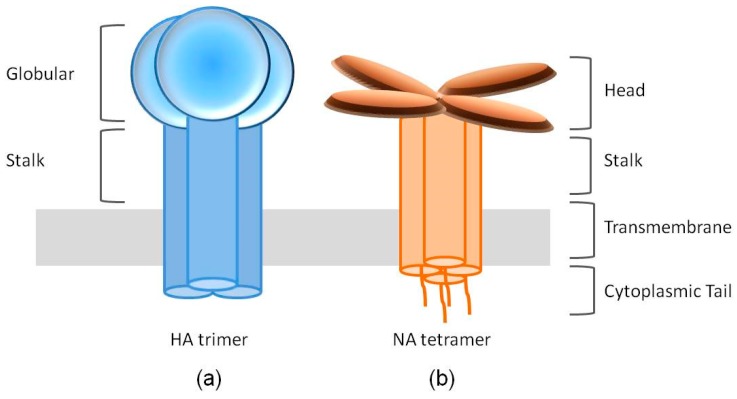
HA is translated as an uncleaved HA0 precursor protein, assembled as a homotrimer in the endoplasmic reticulum and transported to the plasma membrane via the secretory pathway. HA0 is further cleaved into an HA1-HA2 complex by a host protease. The mature HA1-HA2 complex consists of two domains: the membrane-proximal helix-rich stalk domain, primarily composed of HA2 with some HA1 residues, and the membrane-distal globular domain (also known as the “head”) comprised of HA1, containing a SA binding pocket [21,22] (Figure 1a).
Figure 1.
Cartoon structures of the spikes of hemagglutinin (HA) and neuraminidase (NA). (a) The HA spike is formed by an HA trimer. Each HA monomer contains two functional domains, the receptor binding globular domain and the helix-rich stalk domain. (b) NA exists as a tetramer of four identical monomers. Each NA monomer consists of four distinct structure domains, including the catalytic head, the stalk, the transmembrane region and the cytoplasmic tail.
Infection of IAV is initiated when HA binds SAs on the host cell surface and triggers internalization by endocytosis. The low pH condition of the maturing endosome triggers a series of pH-induced conformational changes in HA that ultimately result in the fusion of the viral and host endosomal membranes [23]. The linkage of terminal SA to the penultimate galactose occurs via carbon 3 or carbon 6, generating two isomers α-2,3 and α-2,6 SA, respectively [24]. The SA of α-2,3 isomer predominates in the avian gut, where influenza virus replicates in avian hosts, while the α-2,6 conformer is abundant in the human upper respiratory tract, the primary site of virus replication in the human host [25,26]. Correspondingly, avian and human influenza viruses are characterized by binding α-2,3 and α-2,6 isomers respectively [27,28,29,30,31,32,33,34,35,36,37]. This receptor specificity, which is partly directed by the structural features of the HA receptor binding pocket, is one of the most important determinants that influence host tropism, including interspecies adaptation and transmissibility [38,39].
The receptor binding profiles of 18 HA subtypes are different, moreover, subtle changes in the architecture of the SA-binding pocket of HA may result in altered SA-linkage preference [33,34]. For example, human H2 and H3 strains containing leucine at position 226 favor α-2,6-linked SA, whereas avian strains containing glutamine at this position display preferential binding to α-2,3 isomer [40,41]. In addition, amino acids at positions 189, 193, 194, 216, 198 211 and 222 may also influence the architecture of the pocket region, and have been shown to be important for SA binding and specificity [37,42,43,44].
3. Neuraminidase
NA is assembled as homotetramers when embedded in the envelope of the virus, with each of the monomers folding into four distinct structure domains: the cytoplasmic tail, the transmembrane region, the stalk and the catalytic head [19] (Figure 1b). The cytoplasmic tail, of which the sequence is nearly 100% conserved across all IAV subtypes, plays a critical role during NA transport and incorporation [45,46,47]. The N-terminal hydrophobic transmembrane domain, however, contains a variable sequence of amino acids spanning residues 7–29 and provides signals for translocation to the apical membrane and lipid raft association [48]. The stalk domains of different NA subtypes vary considerably in the number and sequence of amino acid residues, therefore, in the length, depending on which the NA tetramers may protrude slightly more or less above the viral envelope than the HA trimers. These differences can impact the NA enzymatic activity and virulence [45,49,50]. The catalytic head domain possesses the ability to cleave SA from nearby membrane glycoproteins to prevent virus trapping [51,52].
The function of NA the during release of the newly budded virions from host cells has long been well demonstrated, however, the roles of NA in IAV entry remain under-appreciated [20,53]. NA is involved in IAV entry likely in two ways: first, NA may play a role in direct receptor binding, complementing the receptor-binding function of HA; second, NA may help to release virions bound to the sialylated “decoy” receptors, facilitating virus movement on cell surface, therefore enabling HA access to SA expressed by entry receptors and subsequent efficient endocytosis [19,20].
3.1. Receptor Binding
In addition to the NA catalytic site, which binds SA and its analogues, various NA proteins, including N1, N2, N5, N6 and N9, have been reported to possess a second receptor binding site (SRBS), which is also referred to as the hemadsorption site [54,55,56,57,58]. Although these NA containing SRBSs are predominantly from avian sources, influenza A (H7N9) virus containing SRBS in the N9 NA emerged as a human infection in March 2013 [59]. Moreover, it has been demonstrated that a T401A substitution within the SRBS of N9 NA may be essential for the acquisition of altered HA receptor-binding properties and contributed to the spread of the novel H7N9 viruses [58].
The NAs of human H3N2 strains acquired an SA binding property [60,61,62]. However, in contrast to the SRBS, the receptor binding function of these N2 NAs resides in the same site with the catalytic site; mutations at threonine 148, histidine 150 or aspartate 151 near the catalytic site of these NAs contributes to the acquisition of receptor binding, and the NA mediated agglutination could be inhibited by NA inhibitors [60,61,62]. Yet the catalytic and receptor binding sites seem not to be identical since the relative sensitivity of the two functions to NA inhibitors varies [60]. The biological role of the NA catalytic site-associated receptor binding function for these H3N2 isolates remains unclear, but the fact that entry of MDCK cells with viruses having D151G substitution can be blocked by NA inhibitors suggests that it is an important function [63].
3.2. Catalytic Activity during Virus Entry
It has been well accepted that NA activity and cleavage of SA enables movement of the virion through airway mucus during infection [64]. The sialylated mucins within airway mucus may serve as a trap by presenting decoy receptors to which influenza virus binds, followed by mucociliary clearance [65]. Previous studies demonstrated that IAV interacts with the secreted mucus on human airway tissue, and bead-bound mucins inhibit the NA cleavage of the substrate [66]. Moreover, NA inhibitors have been shown to block IAV entry into cell cultures of human airway epithelium, while exogenous NA enhanced virus passage through the mucus layer [67,68]. This suggests that the catalytic activity of NA is essential to remove decoy SAs presented on mucins so that the virus can get access to functional receptors on the target cell surface.
When it comes to infection of target cells, not every binding attempt results in success infection. Most SA associated molecules bind to HA but could not mediate endocytosis of IAV. The infection of IAV, however, is initiated by HA binding to specific IAV entry receptor proteins, such as the killer-activating proteins NKp44 and NKp46 in natural killer cells, the epidermal growth factor receptor in lung epithelial cells, and the recently identified voltage-dependent Ca2+ channel Cav1.2 [69,70,71,72,73,74]. Therefore, most attached virions appear to require migration on the cell surface until meeting an entry receptor that subsequently mediates virus internalization.
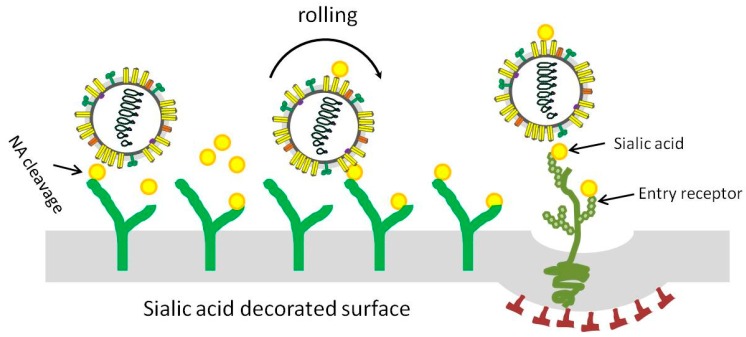
The catalytic activity of NA is crucial during virus migration, and HA and NA competitively cooperate as motile machinery to move IAV particles on the cell surface [75]. As shown in Figure 2, after binding of IAV to the SA decorated surface via HA, the NA may degrade SAs near the attachment site, creating a receptor density gradient that enables the rolling of virus particles on the surface. The recent development of a biolayer interferometry assay was used to study the dynamic and motile interaction of HA/NA-receptor during the initial infection of IAV and demonstrated the contribution of NA to virus-receptor binding and NA-dependent rolling on receptor-containing surfaces [76].
Figure 2.
Representative model for influenza A virus moving on sialic acid (SA) decorated surface. In order to infect a target host cell, the virus might need to move either across airway mucus to reach the target cell or on the cell surface to access an entry receptor. After viral attachment, the NA could degrade SAs near the attachment site, resulting in reduced SA density; the SA density gradient promotes virus moving until successful infection occur.
3.3. Other Entry Steps
Using a cell-cell fusion assay, NA has also been shown to enhance the HA-mediated membrane fusion process, facilitating virus entry [77]. However, it seems that the enhancement of fusion and infectivity by NA is related to desialylation of the virion expressed HA [77].
When the NA genes from influenza H9N2, H5N1 or A(H1N1) pdm09 virus was respectively expressed on a PR8 background, the replication kinetics, both in vitro and in vivo, were not affected, but different effects on infection initiation, virus release and fusion of infected cells were observed, implicating a role for NA during the early stage of infection [78]. Quantitative proteomics analysis further demonstrated that many proteins postulated to be involved in cell-cell fusion were up-regulated in the cells infected with rPR8-H5N1NA or rPR8-H9N2NA viruses than the cells infected with wild-type virus, although the full mechanism remains to be explored [79].
4. Functional Balance between HA and NA
With respect to the complementary and opposite effects of HA and NA on SA binding, it is imperative that the relative activity of the two proteins is balanced, preventing the activity of one from overwhelming the role of the other; i.e., HA and NA competitive cooperation during the infection initiation and release of the newly formed virions. Three situations usually affect HA/NA balance: (i) adaptation to a new SA expression pattern after interspecies transmission occurs; (ii) antigenic drift, and (iii) presence of antivirals targeting either HA or NA [80]. Of note that the receptor-binding and receptor-destroying activities of HEF proteins of ICV and IDV should match accordingly. Since the recent study emphasized that ICV may infect the lower respiratory tract, and cause more severe disease and periodic outbreaks, while IDV also poses a potential threat to human health as an emerging pathogen, it is important to understand the characteristics of HEF proteins regarding the functional balance [81,82].
There is no straightforward method to determine the HA/NA balance of IAVs, and the most frequent method is to study each glycoprotein (HA or NA) separately. For example, the HA affinity against different substrates (α-2,3 or α-2,6 SA) can be estimated by glycan binding assay, and the NA activity can be assessed by different methods including chemiluminescence, fluorescence and glycan array assay [83,84,85]. However, not only the relative HA binding affinity and NA enzymatic activity to SA, but also several other critical factors comprehensively contribute to HA/NA balance, and the virus fitness is tuned in a much more complicated and finer way.
4.1. Relative HA Binding Affinity and NA Activity
All subtypes of H1-H16 and N1-N9 can be found amongst wild birds, however, only limited combinations of HA/NA occur frequently in nature whereas others rarely emerge, not to mention those infecting humans [86,87,88,89]. Laboratory attempts to produce viable reassortant viruses bearing particular subtype combinations also failed [90]. These results suggest some stringent requirements for the match between HA and NA.
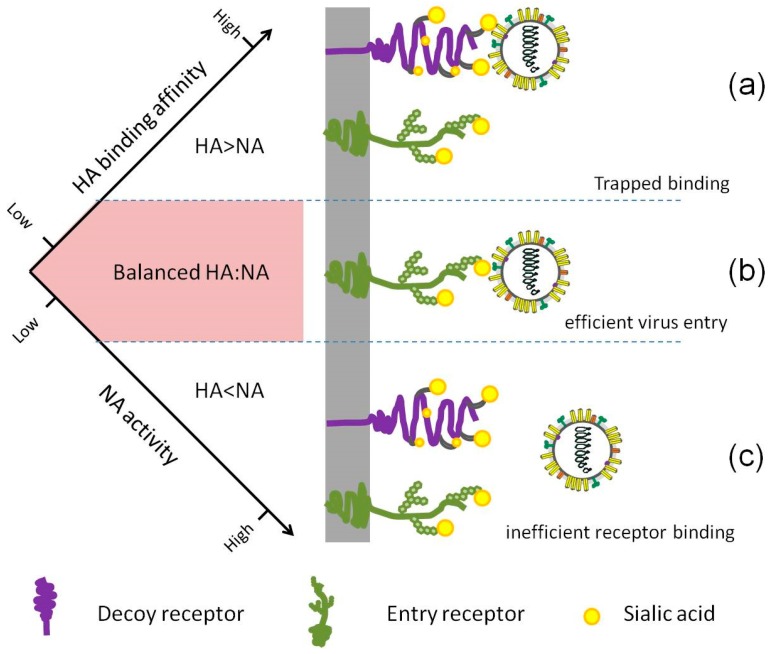
Neither HA binding affinity nor NA activity alone can guarantee a successful infection, replication or transmission, but a real stable balance between the HA and NA contributes good viral fitness (Figure 3). Interestingly, several extreme cases have been reported. Upon treatment with NA inhibitor drugs, several H3N2 virus isolates from the patients were shown to have little or no NA activity, in correspondence with which a weak binding HA was found [91,92]. Hooper and Bloom created a virus in the laboratory by introducing a G147R mutation in NA, paired with binding-deficient HA, the receptor-binding activity was entirely shifted from HA to NA [93].
Figure 3.
The relative HA binding affinity and NA activity need to be balanced for efficient entry. (a) If the HA and NA are mismatched and the NA activity is suboptimal, the virus may remain bound to decoy receptors, blocking virus movement and entry. (b) Efficient cleavage of SA from decoy receptors by NA enables HA access to the right entry receptors, followed by efficient endocytosis. (c) In the case that NA activity is too strong when compared to HA binding affinity, every binding attempt of the virus via HA will be disrupted by NA cleavage, resulting in failed binding.
4.2. HA:NA Ratio
The relative HA binding affinity and NA activity are the foremost determinants for virus infection into a cell. In addition, a number of physical characteristics of the virus that can influence the HA/NA balance are also important. For example, the ratio of HA-to-NA is one critical factor. On average, ~300–400 HA spikes and ~40–50 NA spikes are present on the surface of a virion, and the excess of HA over NA may compensate to the weak affinity of HA for SAs by forming more connections to form a stable interaction, and vice versa [49,94].
The noncoding regions (NCRs) of the eight segmented viral RNAs (vRNAs) of IAV consist of the highly conserved promoter region and the nonconserved segment-specific NCRs at both the 3′ and 5′ ends [95,96]. Nucleotides in the nonconserved segment-specific region are highly conserved both in the sequence and the length for the same segment of different IAV strains, except for the two subtype-determinant HA and NA segments, which are further subtype-specific [97]. Although the diversity in the subtype-specific NCRs of HA and NA segments has received little attention, they are proposed to affect vRNA synthesis, protein expression and genome packaging, which might subsequently influence the amounts of HA and NA on the virion surface, as well as HA:NA ratio [98,99,100,101]. This hypothesis is supported by the recent findings which show that HA NCR plays an important role in vRNA transcription and virus infectivity [102,103].
4.3. NA Accessibility
The ability of NA to access the cell surface SAs is also essential for the functional HA/NA balance. Variable lengths of the NA stalk region can have a significant impact on the accessibility, although the mechanism is yet to be fully understood. Using reverse genetic techniques, a series of NA mutations differing only in the stalk length were generated, and studies showed that altered stalk length did not interfere with NA activity to cleave fetuin or a small substrate in vitro, but the increased stalk length enhanced virus replication accordingly [104,105]. It is commonly accepted that reduced stalk length results in a diminished height of NA, and the towering HA may thus block the shorter NA from gaining access to the receptor SAs [106]. Oppositely, however, reduced NA accessibility may enhance virulence and transmission in some situations. It has been reported that NA stalk truncation has arisen from an evolutionary adaptation of avian IAVs from wild aquatic birds to domestic poultry [50,107,108,109,110]. The 2009 pandemic virus A(H1N1)pdm09 with truncated NA stalk showed greater lethality in mice and virulence in ferrets compared to the untruncated counterpart [111]. It is possible that in these situations, the reduced NA accessibility might reflect the reduced HA binding affinity or enhanced NA activity.
The distribution of NA on the virion may also affect the NA accessibility. Cryoelectron tomography studies have shown that IAV virions can be sorted into five classes based on their appearance in tomograms, and the NA tetramer exists mainly in two manners: the single NA spikes surrounded by HA, and local clusters of NA [49]. Altered virion morphology and NA distribution may potentially influence viral entry, as well as viral fitness [112].
5. Conclusions
The complementary and antagonistic effects of HA and NA on SA interaction suggest that HA and NA competitively cooperate with each other, instead of a simplistic view that HA is responsible for virus entry while NA is involved in viral particle release, in the viral life cycle. However, the effect of the functional HA/NA balance is still poorly understood. Current antiviral therapies mainly target NA, while recent approaches to new influenza therapy include monoclonal antibodies targeting the HA [113,114]. Caution should be exercised in these approaches since it is possible that HA binding inhibitors may re-balance HA to reduce NA activity mediated by NA inhibitors, and compensating mutations in the NA may allow escape from HA inhibition and vice versa. Thus further studies to investigate the interactive roles of HA and NA in the viral entry of influenza viruses have implications in the basic biology of influenza replication and pathogenesis and in the development of therapeutics and vaccines.
Author Contributions
Conceptualization, R.D. and L.R.; Writing—Original Draft preparation, R.D.; Writing—Review and Editing, L.R., R.D., and Q.C.; funding acquisition, R.D. and L.R.
Funding
The work was supported by (1) The Drug Innovation Major Project (Grant No. 2018ZX09711001); (2) the Key Research and Development Projects of Science and Technology Department of Shandong Province (Grant No. 2017CXGC1309); and (3) the Science Foundation of SDUTCM for outstanding youth (Grant No. 2018zk03).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.WHO . 2018 Influenza (Seasonal) Fact Sheet. WHO; Geneva, Switzerland: 2018. [(accessed on 20 May 2019)]. Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ [Google Scholar]
- 2.Hause B.M., Collin E.A., Liu R., Huang B., Sheng Z., Lu W., Wang D., Nelson E.A., Li F. Characterization of a novel influenza virus in cattle and swine: Proposal for a new genus in the orthomyxoviridae family. mBio. 2014;5 doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger J.K., Morens D.M. Pandemic influenza--including a risk assessment of H5N1. Rev. Sci. Tech. 2009;28:187–202. doi: 10.20506/rst.28.1.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resa-Infante P., Jorba N., Coloma R., Ortin J. The influenza virus RNA synthesis machine: Advances in its structure and function. RNA Biol. 2011;8:207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W., Zhang L., He W., Zhang X., Wen B., Wang C., Xu Q., Li G., Zhou J., Veit M., et al. Genetic evolution and molecular selection of the he gene of influenza C virus. Viruses. 2019;11:167. doi: 10.3390/v11020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su S., Fu X., Li G., Kerlin F., Veit M. Novel influenza D virus: Epidemiology, pathology, evolution and biological characteristics. Virulence. 2017;8:1580–1591. doi: 10.1080/21505594.2017.1365216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenthal P.B., Zhang X., Formanowski F., Fitz W., Wong C.H., Meier-Ewert H., Skehel J.J., Wiley D.C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houser K., Subbarao K. Influenza vaccines: Challenges and solutions. Cell Host Microbe. 2015;17:295–300. doi: 10.1016/j.chom.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong S., Li Y., Rivailler P., Conrardy C., Castillo D.A., Chen L.M., Recuenco S., Ellison J.A., Davis C.T., York I.A., et al. A distinct lineage of influenza a virus from bats. Proc. Natl. Acad. Sci. USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong S., Zhu X., Li Y., Shi M., Zhang J., Bourgeois M., Yang H., Chen X., Recuenco S., Gomez J., et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grohskopf L.A., Olsen S.J., Sokolow L.Z., Bresee J.S., Cox N.J., Broder K.R., Karron R.A., Walter E.B. Prevention and control of seasonal influenza with vaccines: Recommendations of the advisory committee on immunization practices (ACIP)--UNITED states, 2014-15 influenza season. MMWR Morb. Mortal. Wkly. Rep. 2014;63:691–697. [PMC free article] [PubMed] [Google Scholar]
- 12.Matrosovich M., Herrler G., Klenk H.D. Sialic acid receptors of viruses. Top. Curr. Chem. 2015;367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verma D.K., Gupta D., Lal S.K. Host lipid rafts play a major role in binding and endocytosis of influenza A virus. Viruses. 2018;10:650. doi: 10.3390/v10110650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edinger T.O., Pohl M.O., Stertz S. Entry of influenza A virus: Host factors and antiviral targets. J. Gen. Virol. 2014;95:263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 15.Rust M.J., Lakadamyali M., Zhang F., Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 2004;11:567–573. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sieczkarski S.B., Whittaker G.R. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J. Virol. 2002;76:10455–10464. doi: 10.1128/JVI.76.20.10455-10464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Vries E., Tscherne D.M., Wienholts M.J., Cobos-Jimenez V., Scholte F., Garcia-Sastre A., Rottier P.J., de Haan C.A. Dissection of the influenza a virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLOS Pathog. 2011;7:e1001329. doi: 10.1371/journal.ppat.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichelberger M.C., Wan H. Influenza neuraminidase as a vaccine antigen. Curr. Top. Microbiol. Immunol. 2015;386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- 19.McAuley J.L., Gilbertson B.P., Trifkovic S., Brown L.E., McKimm-Breschkin J.L. Influenza virus neuraminidase structure and functions. Front. Microbiol. 2019;10:39. doi: 10.3389/fmicb.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen F., Wan X.F. Influenza neuraminidase: Underrated role in receptor binding. Trends Microbiol. 2019 doi: 10.1016/j.tim.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 a resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 22.Weis W., Brown J.H., Cusack S., Paulson J.C., Skehel J.J., Wiley D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 23.Luo M. Influenza virus entry. Adv. Exp. Med. Biol. 2012;726:201–221. doi: 10.1007/978-1-4614-0980-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traving C., Schauer R. Structure, function and metabolism of sialic acids. Cell. Mol. Life Sci. CMLS. 1998;54:1330–1349. doi: 10.1007/s000180050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. Avian flu: Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 26.Wan H., Perez D.R. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology. 2006;346:278–286. doi: 10.1016/j.virol.2005.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradley K.C., Jones C.A., Tompkins S.M., Tripp R.A., Russell R.J., Gramer M.R., Heimburg-Molinaro J., Smith D.F., Cummings R.D., Steinhauer D.A. Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic h1n1 isolates (novel 2009 H1N1) Virology. 2011;413:169–182. doi: 10.1016/j.virol.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 28.Connor R.J., Kawaoka Y., Webster R.G., Paulson J.C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 29.Ibricevic A., Pekosz A., Walter M.J., Newby C., Battaile J.T., Brown E.G., Holtzman M.J., Brody S.L. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 2006;80:7469–7480. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamblin S.J., Haire L.F., Russell R.J., Stevens D.J., Xiao B., Ha Y., Vasisht N., Steinhauer D.A., Daniels R.S., Elliot A., et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 31.Maines T.R., Chen L.M., Van Hoeven N., Tumpey T.M., Blixt O., Belser J.A., Gustin K.M., Pearce M.B., Pappas C., Stevens J., et al. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology. 2011;413:139–147. doi: 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell R.J., Stevens D.J., Haire L.F., Gamblin S.J., Skehel J.J. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj. J. 2006;23:85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- 33.Stevens J., Blixt O., Glaser L., Taubenberger J.K., Palese P., Paulson J.C., Wilson I.A. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Shelton H., Ayora-Talavera G., Ren J., Loureiro S., Pickles R.J., Barclay W.S., Jones I.M. Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J. Virol. 2011;85:1875–1880. doi: 10.1128/JVI.01822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada S., Suzuki Y., Suzuki T., Le M.Q., Nidom C.A., Sakai-Tagawa Y., Muramoto Y., Ito M., Kiso M., Horimoto T., et al. Haemagglutinin mutations responsible for the binding of h5n1 influenza A viruses to human-type receptors. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z.Y., Wei C.J., Kong W.P., Wu L., Xu L., Smith D.F., Nabel G.J. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science. 2007;317:825–828. doi: 10.1126/science.1135165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens J., Blixt O., Tumpey T.M., Taubenberger J.K., Paulson J.C., Wilson I.A. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 38.Mair C.M., Ludwig K., Herrmann A., Sieben C. Receptor binding and ph stability—ow influenza A virus hemagglutinin affects host-specific virus infection. Biochim. Biophys. Acta (BBA)-Biomembr. 2014;1838:1153–1168. doi: 10.1016/j.bbamem.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Russell C.J., Hu M., Okda F.A. Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol. 2018;26:841–853. doi: 10.1016/j.tim.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu R., McBride R., Paulson J.C., Basler C.F., Wilson I.A. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J. Virol. 2010;84:1715–1721. doi: 10.1128/JVI.02162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ha Y., Stevens D.J., Skehel J.J., Wiley D.C. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 hong kong pandemic influenza virus. Virology. 2003;309:209–218. doi: 10.1016/S0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 42.Stevens J., Chen L.M., Carney P.J., Garten R., Foust A., Le J., Pokorny B.A., Manojkumar R., Silverman J., Devis R., et al. Receptor specificity of influenza a H3N2 viruses isolated in mammalian cells and embryonated chicken eggs. J. Virol. 2010;84:8287–8299. doi: 10.1128/JVI.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maines T.R., Jayaraman A., Belser J.A., Wadford D.A., Pappas C., Zeng H., Gustin K.M., Pearce M.B., Viswanathan K., Shriver Z.H., et al. Transmission and pathogenesis of swine-origin 2009 a(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradley K.C., Galloway S.E., Lasanajak Y., Song X., Heimburg-Molinaro J., Yu H., Chen X., Talekar G.R., Smith D.F., Cummings R.D., et al. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J. Virol. 2011;85:12387–12398. doi: 10.1128/JVI.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blok J., Air G.M. Variation in the membrane-insertion and "stalk" sequences in eight subtypes of influenza type a virus neuraminidase. Biochemistry. 1982;21:4001–4007. doi: 10.1021/bi00260a015. [DOI] [PubMed] [Google Scholar]
- 46.Barman S., Adhikary L., Chakrabarti A.K., Bernas C., Kawaoka Y., Nayak D.P. Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza A virus neuraminidase in raft association and virus budding. J. Virol. 2004;78:5258–5269. doi: 10.1128/JVI.78.10.5258-5269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mintaev R.R., Alexeevski A.V., Kordyukova L.V. Co-evolution analysis to predict protein-protein interactions within influenza virus envelope. J. Bioinform. Comput. Biol. 2014;12:1441008. doi: 10.1142/S021972001441008X. [DOI] [PubMed] [Google Scholar]
- 48.Barman S., Nayak D.P. Analysis of the transmembrane domain of influenza virus neuraminidase, a type ii transmembrane glycoprotein, for apical sorting and raft association. J. Virol. 2000;74:6538–6545. doi: 10.1128/JVI.74.14.6538-6545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris A., Cardone G., Winkler D.C., Heymann J.B., Brecher M., White J.M., Steven A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuoka Y., Swayne D.E., Thomas C., Rameix-Welti M.A., Naffakh N., Warnes C., Altholtz M., Donis R., Subbarao K. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J. Virol. 2009;83:4704–4708. doi: 10.1128/JVI.01987-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colman P.M., Varghese J.N., Laver W.G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 52.Burmeister W.P., Ruigrok R.W., Cusack S. The 2.2 A resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. Embo J. 1992;11:49–56. doi: 10.1002/j.1460-2075.1992.tb05026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong J., Xu W., Zhang J. Structure and functions of influenza virus neuraminidase. Curr. Med. Chem. 2007;14:113–122. doi: 10.2174/092986707779313444. [DOI] [PubMed] [Google Scholar]
- 54.Varghese J.N., Colman P.M., van Donkelaar A., Blick T.J., Sahasrabudhe A., McKimm-Breschkin J.L. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc. Natl. Acad. Sci. USA. 1997;94:11808–11812. doi: 10.1073/pnas.94.22.11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Air G.M. Influenza neuraminidase. Influenza Other Respir. Viruses. 2012;6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun X., Li Q., Wu Y., Wang M., Liu Y., Qi J., Vavricka C.J., Gao G.F. Structure of influenza virus N7: The last piece of the neuraminidase “jigsaw” puzzle. J. Virol. 2014;88:9197–9207. doi: 10.1128/JVI.00805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uhlendorff J., Matrosovich T., Klenk H.D., Matrosovich M. Functional significance of the hemadsorption activity of influenza virus neuraminidase and its alteration in pandemic viruses. Arch. Virol. 2009;154:945–957. doi: 10.1007/s00705-009-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai M., McBride R., Dortmans J., Peng W., Bakkers M.J.G., de Groot R.J., van Kuppeveld F.J.M., Paulson J.C., de Vries E., de Haan C.A.M. Mutation of the second sialic acid-binding site, resulting in reduced neuraminidase activity, preceded the emergence of H7N9 influenza A virus. J. Virol. 2017;91 doi: 10.1128/JVI.00049-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benton D.J., Wharton S.A., Martin S.R., McCauley J.W. Role of neuraminidase in influenza a(H7N9) virus receptor binding. J. Virol. 2017;91 doi: 10.1128/JVI.02293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohr P.G., Deng Y.M., McKimm-Breschkin J.L. The neuraminidases of mdck grown human influenza A (H3N2) viruses isolated since 1994 can demonstrate receptor binding. Virol. J. 2015;12:67. doi: 10.1186/s12985-015-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin Y.P., Gregory V., Collins P., Kloess J., Wharton S., Cattle N., Lackenby A., Daniels R., Hay A. Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: A role in virus attachment? J. Virol. 2010;84:6769–6781. doi: 10.1128/JVI.00458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mogling R., Richard M.J., Vliet S.V., Beek R.V., Schrauwen E.J.A., Spronken M.I., Rimmelzwaan G.F., Fouchier R.A.M. Neuraminidase-mediated haemagglutination of recent human influenza A(H3N2) viruses is determined by arginine 150 flanking the neuraminidase catalytic site. J. Gen. Virol. 2017;98:1274–1281. doi: 10.1099/jgv.0.000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gulati S., Smith D.F., Cummings R.D., Couch R.B., Griesemer S.B., St George K., Webster R.G., Air G.M. Human H3N2 influenza viruses isolated from 1968 to 2012 show varying preference for receptor substructures with no apparent consequences for disease or spread. PLoS ONE. 2013;8:e66325. doi: 10.1371/journal.pone.0066325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burnet F.M. Mucins and mucoids in relation to influenza virus action; inhibition of virus haemagglutination by glandular mucins. Aust. J. Exp. Biol. Med. Sci. 1948;26:371–379. doi: 10.1038/icb.1948.38. [DOI] [PubMed] [Google Scholar]
- 65.Button B., Cai L.H., Ehre C., Kesimer M., Hill D.B., Sheehan J.K., Boucher R.C., Rubinstein M. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science. 2012;337:937–941. doi: 10.1126/science.1223012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen M., Zhang X.Q., Senaati H.P., Chen H.W., Varki N.M., Schooley R.T., Gagneux P. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol. J. 2013;10:321. doi: 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matrosovich M.N., Matrosovich T.Y., Gray T., Roberts N.A., Klenk H.D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang X., Steukers L., Forier K., Xiong R., Braeckmans K., Van Reeth K., Nauwynck H. A beneficiary role for neuraminidase in influenza virus penetration through the respiratory mucus. PLoS ONE. 2014;9:e110026. doi: 10.1371/journal.pone.0110026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ho J.W., Hershkovitz O., Peiris M., Zilka A., Bar-Ilan A., Nal B., Chu K., Kudelko M., Kam Y.W., Achdout H., et al. H5-type influenza virus hemagglutinin is functionally recognized by the natural killer-activating receptor NKP44. J. Virol. 2008;82:2028–2032. doi: 10.1128/JVI.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Achdout H., Meningher T., Hirsh S., Glasner A., Bar-On Y., Gur C., Porgador A., Mendelson M., Mandelboim M., Mandelboim O. Killing of avian and swine influenza virus by natural killer cells. J. Virol. 2010;84:3993–4001. doi: 10.1128/JVI.02289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arnon T.I., Achdout H., Lieberman N., Gazit R., Gonen-Gross T., Katz G., Bar-Ilan A., Bloushtain N., Lev M., Joseph A., et al. The mechanisms controlling the recognition of tumor- and virus-infected cells by NKP46. Blood. 2004;103:664–672. doi: 10.1182/blood-2003-05-1716. [DOI] [PubMed] [Google Scholar]
- 72.Mandelboim O., Lieberman N., Lev M., Paul L., Arnon T.I., Bushkin Y., Davis D.M., Strominger J.L., Yewdell J.W., Porgador A. Recognition of haemagglutinins on virus-infected cells by NKP46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 73.Eierhoff T., Hrincius E.R., Rescher U., Ludwig S., Ehrhardt C. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 2010;6:e1001099. doi: 10.1371/journal.ppat.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujioka Y., Nishide S., Ose T., Suzuki T., Kato I., Fukuhara H., Fujioka M., Horiuchi K., Satoh A.O., Nepal P., et al. A sialylated voltage-dependent ca(2+) channel binds hemagglutinin and mediates influenza A virus entry into mammalian cells. Cell Host Microbe. 2018;23:809–818.e5. doi: 10.1016/j.chom.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 75.Sakai T., Nishimura S.I., Naito T., Saito M. Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery. Sci. Rep. 2017;7:45043. doi: 10.1038/srep45043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo H., Rabouw H., Slomp A., Dai M., van der Vegt F., van Lent J.W.M., McBride R., Paulson J.C., de Groot R.J., van Kuppeveld F.J.M., et al. Kinetic analysis of the influenza A virus ha/na balance reveals contribution of na to virus-receptor binding and na-dependent rolling on receptor-containing surfaces. PLoS Pathog. 2018;14:e1007233. doi: 10.1371/journal.ppat.1007233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Su B., Wurtzer S., Rameix-Welti M.A., Dwyer D., van der Werf S., Naffakh N., Clavel F., Labrosse B. Enhancement of the influenza a hemagglutinin (HA)-mediated cell-cell fusion and virus entry by the viral neuraminidase (NA) PLoS ONE. 2009;4:e8495. doi: 10.1371/journal.pone.0008495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Q., Huang S., Chen J., Zhang S., Chen Z. Na proteins of influenza A viruses H1N1/2009, H5N1, and H9N2 show differential effects on infection initiation, virus release, and cell-cell fusion. PLoS ONE. 2013;8:e54334. doi: 10.1371/journal.pone.0054334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sui Z., Wen B., Gao Z., Chen Q. Fusion-related host proteins are actively regulated by na during influenza infection as revealed by quantitative proteomics analysis. PLoS ONE. 2014;9:e105947. doi: 10.1371/journal.pone.0105947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaymard A., Le Briand N., Frobert E., Lina B., Escuret V. Functional balance between neuraminidase and haemagglutinin in influenza viruses. Clin. Microbiol. Infect. 2016;22:975–983. doi: 10.1016/j.cmi.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 81.Thielen B.K., Friedlander H., Bistodeau S., Shu B., Lynch B., Martin K., Bye E., Como-Sabetti K., Boxrud D., Strain A.K., et al. Detection of influenza C viruses among outpatients and patients hospitalized for severe acute respiratory infection, minnesota, 2013–2016. Clin. Infect. Dis. 2018;66:1092–1098. doi: 10.1093/cid/cix931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Asha K., Kumar B. Emerging influenza D virus threat: What we know so far! J. Clin. Med. 2019;8 doi: 10.3390/jcm8020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stevens J., Blixt O., Paulson J.C., Wilson I.A. Glycan microarray technologies: Tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol. 2006;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wetherall N.T., Trivedi T., Zeller J., Hodges-Savola C., McKimm-Breschkin J.L., Zambon M., Hayden F.G. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: Report of the neuraminidase inhibitor susceptibility network. J. Clin. Microbiol. 2003;41:742–750. doi: 10.1128/JCM.41.2.742-750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cohen M., Fisher C.J., Huang M.L., Lindsay L.L., Plancarte M., Boyce W.M., Godula K., Gagneux P. Capture and characterization of influenza A virus from primary samples using glycan bead arrays. Virology. 2016;493:128–135. doi: 10.1016/j.virol.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Venkatesh D., Poen M.J., Bestebroer T.M., Scheuer R.D., Vuong O., Chkhaidze M., Machablishvili A., Mamuchadze J., Ninua L., Fedorova N.B., et al. Avian influenza viruses in wild birds: Virus evolution in a multihost ecosystem. J. Virol. 2018;92 doi: 10.1128/JVI.00433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaverin N.V., Matrosovich M.N., Gambaryan A.S., Rudneva I.A., Shilov A.A., Varich N.L., Makarova N.V., Kropotkina E.A., Sinitsin B.V. Intergenic HA-NA interactions in influenza A virus: Postreassortment substitutions of charged amino acid in the hemagglutinin of different subtypes. Virus Res. 2000;66:123–129. doi: 10.1016/S0168-1702(99)00131-8. [DOI] [PubMed] [Google Scholar]
- 88.Alexander D.J. Report on avian influenza in the eastern hemisphere during 1997–2002. Avian Dis. 2003;47:792–797. doi: 10.1637/0005-2086-47.s3.792. [DOI] [PubMed] [Google Scholar]
- 89.Munster V.J., Baas C., Lexmond P., Waldenstrom J., Wallensten A., Fransson T., Rimmelzwaan G.F., Beyer W.E., Schutten M., Olsen B., et al. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 2007;3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wagner R., Matrosovich M., Klenk H.D. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 91.Ferraris O., Kessler N., Valette M., Lina B. Evolution of the susceptibility to antiviral drugs of a/H3N2 influenza viruses isolated in france from 2002 to 2005. Vaccine. 2006;24:6656–6659. doi: 10.1016/j.vaccine.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 92.Richard M., Erny A., Care B., Traversier A., Barthelemy M., Hay A., Lin Y.P., Ferraris O., Lina B. Rescue of a H3N2 influenza virus containing a deficient neuraminidase protein by a hemagglutinin with a low receptor-binding affinity. PLoS ONE. 2012;7:e33880. doi: 10.1371/journal.pone.0033880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hooper K.A., Bloom J.D. A mutant influenza virus that uses an N1 neuraminidase as the receptor-binding protein. J. Virol. 2013;87:12531–12540. doi: 10.1128/JVI.01889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sauter N.K., Bednarski M.D., Wurzburg B.A., Hanson J.E., Whitesides G.M., Skehel J.J., Wiley D.C. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: A 500-MHZ proton nuclear magnetic resonance study. Biochemistry. 1989;28:8388–8396. doi: 10.1021/bi00447a018. [DOI] [PubMed] [Google Scholar]
- 95.Robertson J.S. 5′ and 3′ terminal nucleotide sequences of the rna genome segments of influenza virus. Nucleic Acids Res. 1979;6:3745–3757. doi: 10.1093/nar/6.12.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Desselberger U., Racaniello V.R., Zazra J.J., Palese P. The 3′ and 5′-terminal sequences of influenza A, B and C virus rna segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 97.Wang L., Lee C.W. Sequencing and mutational analysis of the non-coding regions of influenza A virus. Vet. Microbiol. 2009;135:239–247. doi: 10.1016/j.vetmic.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 98.Bergmann M., Muster T. Mutations in the nonconserved noncoding sequences of the influenza A virus segments affect viral vRNA formation. Virus Res. 1996;44:23–31. doi: 10.1016/0168-1702(96)01335-4. [DOI] [PubMed] [Google Scholar]
- 99.Zheng H., Palese P., Garcia-Sastre A. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication. Virology. 1996;217:242–251. doi: 10.1006/viro.1996.0111. [DOI] [PubMed] [Google Scholar]
- 100.Goto H., Muramoto Y., Noda T., Kawaoka Y. The genome-packaging signal of the influenza A virus genome comprises a genome incorporation signal and a genome-bundling signal. J. Virol. 2013;87:11316–11322. doi: 10.1128/JVI.01301-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benkaroun J., Robertson G.J., Whitney H., Lang A.S. Analysis of the variability in the non-coding regions of influenza A viruses. Vet. Sci. 2018;5 doi: 10.3390/vetsci5030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang J., Li J., Zhao L., Cao M., Deng T. Dual roles of the hemagglutinin segment-specific noncoding nucleotides in the extended duplex region of the influenza A virus RNA promoter. J. Virol. 2017;91 doi: 10.1128/JVI.01931-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao L., Peng Y., Zhou K., Cao M., Wang J., Wang X., Jiang T., Deng T. New insights into the nonconserved noncoding region of the subtype-determinant hemagglutinin and neuraminidase segments of influenza A viruses. J. Virol. 2014;88:11493–11503. doi: 10.1128/JVI.01337-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Els M.C., Air G.M., Murti K.G., Webster R.G., Laver W.G. An 18-amino acid deletion in an influenza neuraminidase. Virology. 1985;142:241–247. doi: 10.1016/0042-6822(85)90332-0. [DOI] [PubMed] [Google Scholar]
- 105.Castrucci M.R., Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J. Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baigent S.J., McCauley J.W. Glycosylation of haemagglutinin and stalk-length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus Res. 2001;79:177–185. doi: 10.1016/S0168-1702(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 107.Hoffmann T.W., Munier S., Larcher T., Soubieux D., Ledevin M., Esnault E., Tourdes A., Croville G., Guerin J.L., Quere P., et al. Length variations in the NA stalk of an H7N1 influenza virus have opposite effects on viral excretion in chickens and ducks. J. Virol. 2012;86:584–588. doi: 10.1128/JVI.05474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Blumenkrantz D., Roberts K.L., Shelton H., Lycett S., Barclay W.S. The short stalk length of highly pathogenic avian influenza H5N1 virus neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J. Virol. 2013;87:10539–10551. doi: 10.1128/JVI.00967-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun Y., Tan Y., Wei K., Sun H., Shi Y., Pu J., Yang H., Gao G.F., Yin Y., Feng W., et al. Amino acid 316 of hemagglutinin and the neuraminidase stalk length influence virulence of H9N2 influenza virus in chickens and mice. J. Virol. 2013;87:2963–2968. doi: 10.1128/JVI.02688-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bi Y., Xiao H., Chen Q., Wu Y., Fu L., Quan C., Wong G., Liu J., Haywood J., Liu Y., et al. Changes in the length of the neuraminidase stalk region impact H7N9 virulence in mice. J. Virol. 2016;90:2142–2149. doi: 10.1128/JVI.02553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park S., Il Kim J., Lee I., Bae J.Y., Yoo K., Nam M., Kim J., Sook Park M., Song K.J., Song J.W., et al. Adaptive mutations of neuraminidase stalk truncation and deglycosylation confer enhanced pathogenicity of influenza A viruses. Sci. Rep. 2017;7:10928. doi: 10.1038/s41598-017-11348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wasilewski S., Calder L.J., Grant T., Rosenthal P.B. Distribution of surface glycoproteins on influenza A virus determined by electron cryotomography. Vaccine. 2012;30:7368–7373. doi: 10.1016/j.vaccine.2012.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ison M.G. Optimizing antiviral therapy for influenza: Understanding the evidence. Expert Rev. Anti-Infect. Ther. 2015;13:417–425. doi: 10.1586/14787210.2015.1018183. [DOI] [PubMed] [Google Scholar]
- 114.Nachbagauer R., Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin. Microbiol. Infect. 2017;23:222–228. doi: 10.1016/j.cmi.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]