Abstract
To understand antimicrobial resistance (AMR) patterns and mechanisms of horizontal gene transfer in human-associated environments is essential to AMR surveillance. Gram-negative bacteria (1122 isolates) from food-animal environments were characterized for antimicrobial susceptibility and AMR genes. Seventy five per cent of the isolates (837 of 1122) were resistant to at least one of the antibiotics tested. Resistance to more than three groups of antimicrobials (multidrug resistance) was observed in 43 isolates with most often encountered (12 of 43) resistance to β-lactams, tetracycline, quinolones and nitrofurantoin. The profile of frequently reported plasmid-mediated resistance gene in these isolates was determined. The mobility of these elements as plasmids or phages was examined. The blaCTX-M gene was present in the plasmid of 61 per cent and packed in induced phage fractions in 72 per cent of the isolates and blaTEM in 69 per cent phage fractions compared to 15 per cent presence in the plasmid.
Keywords: Antimicrobial resistance, environment, mobile genetic elements, prophage induction
Microbial infections are the second leading cause of death worldwide with infections due to Gram-negative bacteria increasing at an alarming rate1. The use of antimicrobials does not occur only with regard to human diseases and in hospital environment but is also a part of animal husbandry and agricultural practices2. Livestock and agricultural practices include the use of antibiotics for disease prevention and growth promotion of animals. The recurrent use of antibiotics in these environments makes these areas as potential reservoirs of drug-resistant bacteria which can spread to other animals, humans and the environment3.
The emergence of resistance has revealed mechanisms by which resistance genes spread across the bacterial kingdom, with apparent disregard for species barriers which is facilitated by horizontal gene transfer by mobile genetic elements2,3. The transmissibility and plasticity of antimicrobial resistance determinants (ARDs) imply an alarming potential to spread and diversify among bacterial populations with plasmids and phages playing a significant role in this4,5,6. ARDs blaTEM,, blaCTX-M, tetA, qnrA and qnrS have been reported in phage DNA fractions in the environment and separate studies have reported these in plasmids7,8,9,10,11. Bacterial genes can randomly get packed into phage heads by generalized transduction during infection with a lytic phage. We have reported blaCTX-M in lytic bacteriophages of Escherichia coli that transfer this ARD via generalized transduction12.
Thirty samples each of seafood, aquaculture farms (soil and water), poultry farms, piggeries, industrial effluents, domestic effluents and hospital effluents and 32 samples from livestock environments were collected at random (from July 2014 to 2016) from various locations in and around Mangalore, India. The study was conducted in the division of Infectious Diseases, Nitte Centre for Science Education & Research, Mangalore, India. Standard procedures for the isolation of Vibrio sps, Pseudomonas sps and Enterobacteriaceae members E. coli, Klebsiella sps, Enterobacter sps and Salmonella sps, were followed13,14,15,16.
Typical colonies were picked in each case and purified on tryptic soya agar. Isolated pure cultures were characterized by a battery of biochemical test13,14,15,16 and complemented with PCR-based methods for genotypic identification of certain species, namely E. coli17, Salmonella18 and Vibrio parahaemolyticus19. Antimicrobial susceptibility test was carried out by disk diffusion method for antibiotics representing the major groups such as penicillins [ampicillin (AMP) 10 μg, piperacillin/tazobactam 100/10 μg], cephalosporins [cefotaxime (CTX) 30 μg], aminoglycosides [gentamicin (GEN) 10 μg], quinolones [nalidixic acid (NA) 30 μg, ciprofloxacin (CIP) 5 μg], chloramphenicol (C) 30 μg, tetracycline (TET) 30 μg, nitrofurantoin (NIT) 300 μg, sulphonamides (co-trimoxazole 25 μg) and carbapenems [imipenem (IMP) 10 μg, meropenem (MRP)10 μg] as per Clinical Laboratory Standards International guidelines 201220. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains. Antimicrobial discs and control strains were procured from HiMedia Laboratories (Mumbai). PCR-based screening of selected ARDs was performed in a thermocycler (Applied Biosystems, USA).
The presence of ARDs in the plasmid and prophage DNA from Gram-negative isolates whose genotype confirmed the presence of resistance genes was tested. Plasmid extraction was performed by the alkaline lysis method21. Prophage induction was performed using mitomycin C at a final concentration of 1 μg/ml (Sigma Aldrich, India). Phage DNA was extracted from the lysate obtained after prophage induction22. PCR-based detected for ARDs in these fractions was performed.
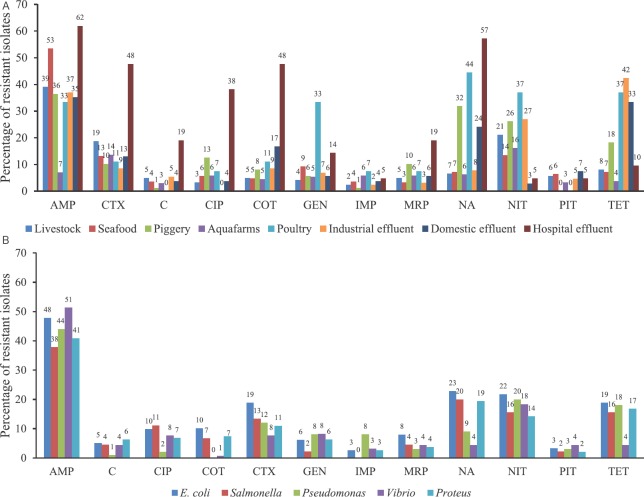
In this study, 1122 Gram-negative bacteria were isolated from 242 samples and the antimicrobial susceptibility patterns were studied (Figs. A and B). 74.6 per cent of the isolates (837 of 1122) were resistant to at least one of the antibiotics tested. Of the 1122 isolates, 498 were resistant to AMP (44.4%), 220 to NIT (19.6%), 156 to TET (13.9%), 158 to CTX (13.2%), 120 to NA (10.7%), 73 to GEN (6.5%), 66 to co-trimethoxazole (5.9%), 59 to CIP (5.3%), 43 each (3.8%) to chloramphenicol and IMP, 58 to meropenem (5.2%) and 48 to piperacillin/tazobactam (4.3%). A total of 342 isolates were resistant to a single antibiotic tested while 243 of the resistant isolates (21%) were concomitantly resistant to more than one class of antimicrobials with resistance to AMP with NIT encountered more frequently (26 isolates) followed by resistance to TET with NIT (17 isolates) and AMP and NA (14 isolates). Resistance to more than three groups of antimicrobials (multidrug resistance) was observed in 43 isolates (~4%) with resistance to β-lactams, TET, quinolones and NIT most often encountered (12 isolates). The resistance patterns in multidrug-resistant isolates are depicted in Table. Chloramphenicol has been suggested as a drug of choice against multi-drug resistant Gram-negative bacteria23. However, in our study, of the 43 isolates resistant to chloramphenicol, 41 were concomitantly resistant to other groups of antimicrobials including quinolones and cephalosporin.
Figure 1.
Resistance profile of Gram-negative bacteria: (A) Resistance pattern observed in different sources; (B) Resistance pattern observed in different genera. AMP, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; COT, co-trimoxazole; CTX, cefotaxime; GEN, gentamicin; IMP, imipenem; MRP, meropenem; NA, nalidixic; NIT, nitrofurantoin; PIT, piperacillin/tazobactam; TET, tetracycline.
Table.
Resistance patterns observed in multidrug-resistant isolates
| Number of isolates | AMP and/CTX | C | NA and/CIP | TET | GEN | COT | NIT |
|---|---|---|---|---|---|---|---|
| 12 | R | R | R | R | |||
| 3 | R | R | R | R | |||
| 3 | R | R | R | R | |||
| 1 | R | R | R | R | |||
| 4 | R | R | R | R | |||
| 2 | R | R | R | R | |||
| 1 | R | R | R | R | |||
| 2 | R | R | R | R | |||
| 1 | R | R | R | R | |||
| 2 | R | R | R | R | |||
| 2 | R | R | R | R | |||
| 1 | R | R | R | R | |||
| 1 | R | R | R | R | |||
| 1 | R | R | R | R | |||
| 1 | R | R | R | R | R | ||
| 2 | R | R | R | R | R | ||
| 3 | R | R | R | R | R | R | |
| 1 | R | R | R | R | R | R | R |
AMP, ampicillin; CTX, cefotaxime; C, chloramphenicol; NA, nalidixic; CIP, ciprofloxacin; TET, tetracycline; GEN, gentamicin; COT, co-trimoxazole; NIT, nitrofurantoin
AMR determinants for genes frequently reported to be mobile through elements such as plasmids and phages were detected in a small fraction (72 isolates) of the resistant isolates. Nearly 56 isolates harboured genes coding for β-lactamases and the determinants blaTEM and blaCTX-M were concomitantly detected in 10 of these isolates; 27 isolates harboured genes coding for TET resistance and multiple variants of the tet gene in the same culture were observed in 11 isolates. Sulphonamide resistance encoded by the sul genes was observed in 20 isolates with multiple variants of the sul genes observed in eight isolates. Determinants for fluoroquinolone resistance, qnrA, qnrB, qnrS and qepA were present in one, three, one and two isolates, respectively. Extended spectrum β-lactamase (ESBL) in combination with sul genes were detected in six isolates and ESBL with plasmid-mediated quinolone resistance (PMQR) genes in four. ESBL, sul and PMQR genes together were observed in four isolates while ESBLs with tet genes were detected in only one isolate.
The property of the ARD being plasmid encoded and/or inducible in the temperate phage fraction was studied. The blaCTX-M genes were observed to be extremely mobile being present in the plasmid of 61 per cent (11 of 18) and packed in prophage fractions in 72 per cent (13 of 18) of the isolates. The blaTEM variants were more mobile in phage fractions (69%) compared to being plasmid encoded (15%). Of the TET resistance determinants, tetA was plasmid encoded in about 80 per cent of the isolates and in the inducible phage fraction in 33 per cent of the isolates. The sul genes were completely chromosomal and were not present in either the plasmid or prophage.
Standards on recommended antimicrobial use for animals and training of personnel involved in farm practices are not readily available resulting in inappropriate and indiscriminate use of antimicrobials in this sector. Our study highlights the need to control resistance in human-associated environments by systematic surveillance and the institution of antimicrobial stewardship practices in these sectors.
Acknowledgment:
The authors acknowledge Nitte (Deemed to be University), Mangaluru, for providing the necessary facilities to carry out this study.
Footnotes
Financial support & sponsorship: The authors acknowledge the Indian Council of Medical Research, New Delhi, for the financial support rendered through the research grant number AMR/37/2011-ECD-1..
Conflicts of Interest: None.
References
- 1.Bockstael K, van Aerschot A. Antimicrobial resistance in bacteria. Cent Eur J Med. 2009;4:141–55. [Google Scholar]
- 2.Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, et al. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950-2002. Emerg Infect Dis. 2012;18:741–9. doi: 10.3201/eid1805.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andremont A. Commensal flora may play key role in spreading antibiotic resistance. ASM News. 2003;69:601–7. [Google Scholar]
- 4.Harbarth S, Samore MH. Antimicrobial resistance determinants and future control. Emerg Infect Dis. 2005;11:794–801. doi: 10.3201/eid1106.050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbosa TM, Levy SB. The impact of antibiotic use on resistance development and persistence. Drug Resist Updat. 2000;3:303–11. doi: 10.1054/drup.2000.0167. [DOI] [PubMed] [Google Scholar]
- 6.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsley LC, Consuegra EJ, Kakirde KS, Land AM, Harper WF, Jr, Liles MR. Identification of diverse antimicrobial resistance determinants carried on bacterial, plasmid, or viral metagenomes from an activated sludge microbial assemblage. Appl Environ Microbiol. 2010;76:3753–7. doi: 10.1128/AEM.03080-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colomer-Lluch M, Imamovic L, Jofre J, Muniesa M. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob Agents Chemother. 2011;55:4908–11. doi: 10.1128/AAC.00535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colomer-Lluch M, Jofre J, Muniesa M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One. 2011;6:e17549. doi: 10.1371/journal.pone.0017549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colomer-Lluch M, Jofre J, Muniesa M. Quinolone resistance genes (qnrA and qnrS) in bacteriophage particles from wastewater samples and the effect of inducing agents on packaged antibiotic resistance genes. J Antimicrob Chemother. 2014;69:1265–74. doi: 10.1093/jac/dkt528. [DOI] [PubMed] [Google Scholar]
- 11.Shousha A, Awaiwanont N, Sofka D, Smulders FJ, Paulsen P, Szostak MP, et al. Bacteriophages isolated from chicken meat and the horizontal transfer of antimicrobial resistance genes. Appl Environ Microbiol. 2015;81:4600–6. doi: 10.1128/AEM.00872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohan Raj JR, Vittal R, Huilgol P, Bhat U, Karunasagar I. T4-like Escherichia coli phages from the environment carry blaCTX-M. Lett Appl Microbiol. 2018;67:9–14. doi: 10.1111/lam.12994. [DOI] [PubMed] [Google Scholar]
- 13.MacFaddin JF. 3rd ed. Philadelphia: Lippincott Williams and Wilkins; 2000. Biochemical tests for identification of medical bacteria. [Google Scholar]
- 14.Bacteriological Analytical Manual. Maryland: U.S. Food and Drug Administration. [accessed on March 29, 2018]. updated 7 March, 2018. Available from: https://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm064948.htm .
- 15.Bacteriological Analytical Manual. Maryland: U.S. Food and Drug Administration. [accessed on March 29, 2018]. updated 7 March, 2018. Available from: https://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070149.htm .
- 16.Bacteriological Analytical Manual. Maryland: U.S. Food and Drug Administration; updated 7 March, 2018. [accessed on March 29, 2018]. Available from: https://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070830.htm .
- 17.Bej AK, DiCesare JL, Haff L, Atlas RM. Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl Environ Microbiol. 1991;57:1013–7. doi: 10.1128/aem.57.4.1013-1017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galán JE, Ginocchio C, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–9. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 19.Taniguchi H, Ohta H, Ogawa M, Mizuguchi Y. Cloning and expression in Escherichia coli of Vibrio parahaemolyticus thermostable direct hemolysin and thermolabile hemolysin genes. J Bacteriol. 1985;162:510–5. doi: 10.1128/jb.162.2.510-515.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI Document M100-S22. Wayne, PA: CLSI; 2012. Clinical and Laboratory Standards Institute. Performance standard for antimicrobial susceptibility testing; 22nd informational supplement. [Google Scholar]
- 21.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–23. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su MT, Venkatesh TV, Bodmer R. Large and small-scale preparation of bacteriophage λ lysate and DNA. Biotechniques. 1998;25:44–6. doi: 10.2144/98251bm08. [DOI] [PubMed] [Google Scholar]
- 23.Sood S. Chloramphenicol - A potent armament against multi-drug resistant (MDR) gram negative bacilli? J Clin Diagn Res. 2016;10:DC01–3. doi: 10.7860/JCDR/2016/14989.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]