Abstract
Background & objectives:
The temporal trends in the development of antimicrobial resistance (AMR) among Salmonella Typhi and Salmonella Paratyphi in India have not been systematically reported. We aimed to systematically review the temporal AMR trends (phenotypic and molecular mechanisms) in bacterial isolates from patients with enteric fever over two decades in India.
Methods:
To identify trends in AMR in India, resistance patterns among 4611 individual S. Typhi isolates and 800 S. Paratyphi A isolates, reported from 1992 to 2017 in 40 publications, were analysed. Molecular resistance determinants were extracted from 22 publications and also reviewed in accordance with the PRISMA guidelines. Articles were sourced using a predefined search strategy from different databases.
Results:
The analyses suggested that multidrug-resistant (MDR) enteric fever was declining in India and being replaced by fluoroquinolone (FQ) resistance. Mutations in gyrA and parC were key mechanisms responsible for FQ resistance, whereas MDR was largely driven by resistance determinants encoded on mobile genetic elements (plasmids, transposons).
Interpretation & conclusions:
The results reflect the effect of antimicrobial pressure which has been driving AMR in typhoidal Salmonella in India. Understanding these trends is important in planning future approaches to therapy, which serve as a baseline for assessment of the impact of new typhoid conjugate vaccines against these resistant organisms.
Keywords: Antimicrobial resistance, enteric fever, India, paratyphoid, prevention, typhoid
Enteric fever caused by serovars Typhi and Paratyphi A, B and C of the Salmonella enterica species accounts for over 25 million cases of febrile illness globally, with children being affected disproportionally1,2,3. India is endemic for enteric fever, where it is one of the main differential diagnoses for fever of unknown origin. In addition to the morbidity and mortality associated with enteric fever, the empiric and appropriate treatment of this disease continues to drive antimicrobial resistance (AMR). Multidrug-resistant (MDR) enteric fever isolates, defined as combined resistance to chloramphenicol, ampicillin and co-trimoxazole, were a common occurrence in the 1990s that necessitated the use of fluoroquinolones (FQs), subsequently cephalosporins and most recently azithromycin2.
The chronological trends in AMR among isolates of Salmonella Typhi and S. Paratyphi A in India have not been systematically reviewed. The WHO strategic group of experts committee, which makes global vaccine policy recommendations, emphasized the need for countries to strengthen the surveillance of typhoid fever and to monitor the occurrence of AMR strains before and after the programmatic implementation of the typhoid conjugate vaccines (TCVs)2,3. India has a unique advantage in that the tetanus-toxoid TCVs has already been licensed, and over five million doses have already been sold within the country4. It is, however, yet to be used programmatically, and one of the postulated uses of TCV is its direct and indirect effects in decreasing AMR.
This study was aimed to systematically review the temporal trends of antimicrobial resistance (AMR) in India. The objectives were two-fold: (i) to systematically delineate the historical trend of the proportion of expressed phenotypic resistance among typhoidal Salmonella to first-line antimicrobials, nalidixic acid, ciprofloxacin and cephalosporins; and (ii) to describe the molecular mechanisms of AMR in both serovars.
Material & Methods
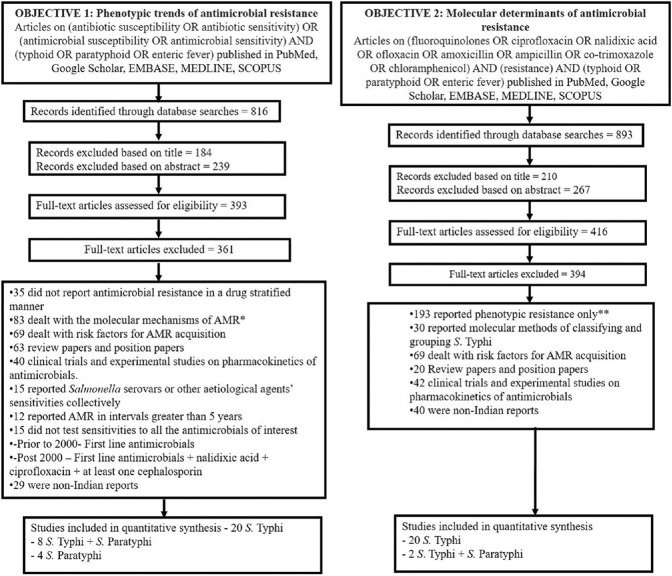
Search strategy: The key words and search strategy for objectives one and two included [(antibiotic susceptibility OR antibiotic sensitivity) OR (antimicrobial susceptibility OR antimicrobial sensitivity)] AND (typhoid OR paratyphoid OR enteric fever) and (fluoroquinolones OR ciprofloxacin OR nalidixic acid OR ofloxacin OR amoxicillin OR ampicillin OR co-trimoxazole OR chloramphenicol) AND (resistance) AND (typhoid OR paratyphoid OR enteric fever), respectively (Fig. 1). Databases searched included PubMed, Google Scholar, EMBASE, MEDLINE and SCOPUS. Filters such as time of publication, study design and language were not applied to ensure complete data collection.
Fig. 1.
Search Strategy and PRISMA flow diagram. *The eligibility of these excluded articles were screened for inclusion under objective 2, and non-duplicate articles were included. **The eligibility of these excluded articles were screened for inclusion under objective 1, and non-duplicate articles were included.
Phenotypic trends in antimicrobial resistance (AMR): For the purpose of this systematic review, an isolate was described as resistant to an antimicrobial if it was reported as ‘resistant’, ‘intermediately susceptible’, ‘intermediately resistant’ or ‘non-susceptible’ based on minimum inhibitory concentration (MIC) values or diameters of zones of inhibition via disc diffusion using customary interpretive criteria such as the Clinical and Laboratory Standards Institute (CLSI) or the European Committee on Antimicrobial Susceptibility Testing (EUCAST) standards5. For uniformity, studies prior to 2000 that reported sensitivities of at least the first-line antimicrobials were included, whereas studies conducted after 2000 which did not report antimicrobial sensitivities of chloramphenicol, co-trimoxazole, ampicillin/amoxicillin, nalidixic acid, ciprofloxacin or at least one cephalosporin were excluded. Studies that reported antibiograms collectively and had not stratified these into intervals shorter than five years were also excluded. These criteria were used to establish the validity of individual studies.
Isolates identified from reports were stratified based on year of isolation, geographic location and resistance phenotypes. Stratified isolates that were resistant to each antimicrobial were expressed as a proportion of all the isolates reported. The trends of antimicrobial resistance were expressed in five-year intervals as represented in Table I.
Table I.
Enteric fever pathogen isolates derived from reports systematically reviewed in this study
| Year | Total number | Proportion of Salmonella Typhi-resistant isolates | |||||
|---|---|---|---|---|---|---|---|
| CH | AM | TMX | NA | FQ | CEPH | ||
| Pre-2001 | 854 | 0.51 | 0.56 | 0.58 | - | - | - |
| 2001-2005 | 1259 | 0.28 | 0.44 | 0.41 | 0.63 | 0.08 | 0.03 |
| 2006-2010 | 902 | 0.09 | 0.35 | 0.06 | 0.76 | 0.15 | 0.01 |
| 2011-2015 | 1596 | 0.07 | 0.17 | 0.13 | 0.82 | 0.63 | 0.04 |
| Proportion of Salmonella Paratyphi A-resistant isolates | |||||||
| Pre-2001 | 179 | 0.22 | 0.21 | 0.26 | - | - | - |
| 2001-2005 | 261 | 0.29 | 0.43 | 0.21 | 0.59 | 0.03 | 0.00 |
| 2006-2010 | 26 | 0.00 | 0.04 | 0.00 | 0.77 | 0.58 | 0.04 |
| 2011-2015 | 329 | 0.01 | 0.05 | 0.01 | 0.91 | 0.60 | 0.05 |
CH, chloramphenicol; AM, ampicillin; TXM, co-trimoxazole; NA, nalidixic acid; FQ, fluoroquinolone; CEPH, cephalosporin
Molecular determinants of AMR: For the second objective, molecular mechanisms of AMR of isolates reported in studies either collectively or individually were included. These were only stratified based on the country of isolation and type of mechanism reported as methods used to study these mechanisms were heterogeneous over the years and techniques employed were also changed, thus making temporal comparisons challenging.
Data extraction & risk of bias (RoB): Data from the respective studies were extracted under the following: (i) study identifier including first author, year of publication, year of study commencement, duration of study, country, study design and sampling population (hospital-based/community and travel-associated/endemic or outbreak); (ii) methodology: sample size, site of isolation and antimicrobial susceptibility testing and interpretive criteria. For the studies included to evaluate molecular determinants, the technique of molecular detection was also recorded; and (iii) results: number of S. Typhi and S. Paratyphi A isolates, frequency of MDR, nalidixic acid-resistant, FQ-resistant and cephalosporin-resistant strains. In addition, data pertaining to the molecular mechanisms of MDR, FQ and cephalosporin resistance were also extracted. Study-specific data extraction was done twice - overall for objectives 1 and 2 separately.
Risk of bias (RoB) was assessed using two tools (Table II). The first classifies studies based on low-, moderate- or high-RoB and is known as the Quality in Prognosis Studies tool6. The second is known as the Joanna Briggs Institute (JBI) tool7 and reports RoB dichotomously. The JBI was adapted for use in this study similar to the adaptations used by Tadesse et al8. These RoB analyses were performed separately on studies selected to meet the first and second objectives. The isolates derived from these studies were used for the frequency analysis. Parameters assessed for bias across the two tools included (i) population description, i.e. whether community or hospital setting; (ii) study design, sample size and sampling techniques; (iii) use of appropriate performance standards and quality control in microbiologic techniques such as bacteriologic culture and antimicrobial sensitivity; and (iv) the statistical analysis used for reporting summary measures.
Table II.
Studies included in the systematic review in which phenotypic AMR trends of S. Typhi isolates were analysed
| Year of study | Year of publication | Author & Reference | No. of isolates | Study Design | Risk of Bias | |
|---|---|---|---|---|---|---|
| QUIPS | JBI | |||||
| 2012 | 2017 | Harichandran & Dinesh17 | 79 | Retrospective | Low | No |
| 2016 | 2016 | Sharvani et al33 | 167 | Retrospective | Low | No |
| 2013-2014 | 2015 | Misra et al26 | 50 | Retrospective | Low | No |
| 2015 | 2015 | Narain & Gupta29 | 220 | Prospective | Low | No |
| 2012 | 2014 | Srirangaraj et al34 | 16 | Retrospective | Low | No |
| 2014 | 2017 | Dahiya et al13 | 380 | Retrospective | Low | No |
| 2010 | 2013 | Choudhary et al12 | 322 | Retrospective | Low | No |
| 2012 | 2013 | Venkatesh et al35 | 251 | Retrospective | Low | No |
| 2008-2010 | 2013 | Gupta et al16 | 257 | Retrospective | Low | No |
| 2010-2012 | 2013 | Jain & Chugh18 | 266 | Retrospective | Low | No |
| 2008 | 2011 | Kumar et al22 | 128 | Retrospective | Low | No |
| 2011 | 2011 | Adhikary et al9 | 2 | Case Report | Low | Yes |
| 2000-2006 | 2010 | Verma et al36 | 159 | Retrospective | Low | No |
| 2008 | 2009 | Kumar et al21 | 50 | Retrospective | Low | No |
| 1990 | 1992 | Rodrigues et al31 | 74 | Retrospective | Low | No |
| 2004 | 2007 | Joshi & Amarnath19 | 25 | Retrospective | Low | No |
| 2002 | 2007 | Capoor et al11 | 178 | Retrospective | Low | No |
| 2003 | 2007 | Banerjee et al10 | 60 | Retrospective | Low | No |
| 2004-2005 | 2006 | Manchanda et al25 | 56 | Retrospective | Low | No |
| 2006 | 2006 | Ray et al30 | 70 | Cross-sectional | Low | No |
| 1999-2004 | 2006 | Mohanty et al27 | 629 | Retrospective | Low | No |
| 2001-2004 | 2006 | Lakshmi et al23 | 60 | Retrospective | Low | No |
| 2003-2004 | 2005 | Dutta et al14 | 379 | Retrospective | Low | No |
| 2004 | 2005 | Senthilkumar et al32 | 6 | Retrospective | Low | No |
| 2002 | 2004 | Madhulika et al24 | 157 | Cross-sectional | Low | No |
| 1997-2001 | 2002 | Gautam et al15 | 436 | Retrospective | Low | No |
| 2001-2003 | 2005 | Kadhiravan et al20 | 50 | Retrospective | Low | No |
| 2006-2007 | 2010 | Nagshetty et al28 | 84 | Retrospective | Low | No |
Results
Phenotypic trends of AMR
Thirty two (Fig. 1)studies (Table II)9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36 satisfied the inclusion criteria from which 49 yr-stratified summaries of S. Typhi antimicrobial-resistant isolates were obtained. For instance, Gautam et al15 reported the isolates of their study in a year-stratified manner for five years, therefore providing five serial year-stratified summaries. Of these 49 yr-stratified summaries, 27 were undertaken prior to the year 2005 and over 80 per cent were retrospective in study design. The summaries obtained from each report were pooled into the following temporal intervals: pre-2001, 2001-2005, 2006-2010 and 2011-2015 and expressed as a proportion of resistant isolates for each antimicrobial (Table I). 19 yr-stratified summaries of antimicrobial-resistant S. Paratyphi A were obtained, of which 11 were prior to the year 2005. The various studies included in this systematic review were found in the medium-to-low spectrum in the RoB assessment (Table III)37,38,39.
Table III.
Studies included in the systematic review in which phenotypic AMR trends of S. Paratyphi isolates were analysed
| Year of study | Year of publication | Author & Reference | No. of isolates | Study design | Risk of Bias | |
|---|---|---|---|---|---|---|
| QUIPS | JBI | |||||
| 1996-2001 | 2000 | Chandel et al37 | 83 | Retrospective | Low | No |
| 1997-2001 | 2002 | Gautam et al15 | 94 | Retrospective | Low | No |
| 2012-2014 | 2017 | Harichandran & Dinesh17 | 22 | Retrospective | Low | No |
| 2004 | 2004 | Harish et al38 | 1 | NA | Low | No |
| 2010-2011 | 2013 | Jain & Chugh18 | 75 | Retrospective | Low | No |
| 2012 | 2013 | Venkatesh et al35 | 92 | Cross-sectional | Low | No |
| 2004 | 2007 | Joshi19 | 25 | Cross-sectional | Low | No |
| 2014-2015 | 2015 | Misra et al26 | 14 | Case Report | Low | No |
| 1999-2000 | 2006 | Mohanty et al27 | 198 | Retrospective | Low | No |
| 2014 | 2015 | Narain & Gupta29 | 5 | unknown | Low | No |
| 2013 | 2016 | Sharvani et al33 | 152 | Cross-sectional | Low | No |
| 2001-2002 | 2003 | Tankhiwale et al39 | 39 | Retrospective | Low | No |
Of the 4611 S. Typhi isolates obtained from the various studies, 41 per cent (1936 S. Typhi isolates) were from the 2011-2015 time period, although the time period between 2000 and 2004 had 21 yr-stratified summaries, making up 43 per cent of the total year-wise summaries in this study. Nalidixic acid, ciprofloxacin and cephalosporin trends were only analysed from the year 2000 as these drugs were not routinely tested as part of antimicrobial sensitivity studies prior to this period, although preliminary reports of ciprofloxacin resistance surfaced as early as 199240. Fig. 2 summarises the pan-Indian AMR trends, which indicate a decline in MDR and a high level of FQ resistance.
Fig. 2.
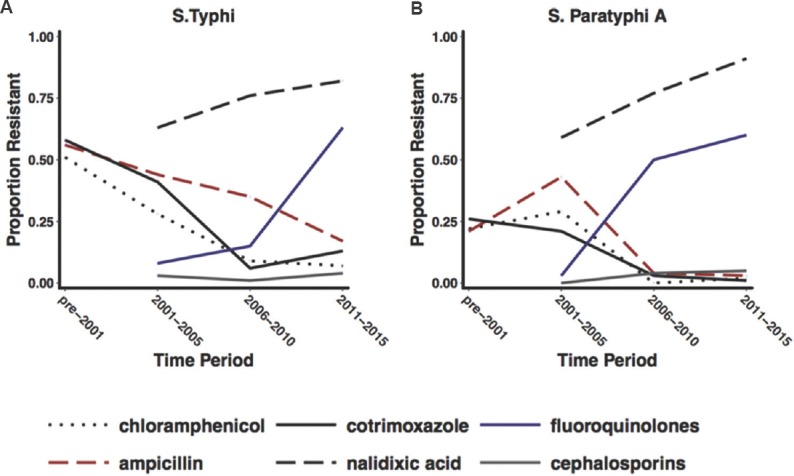
Temporal representation of AMR trends of enteric fever isolates from Indian reports. (A and B) graphical representations of the proportion of Salmonella Typhi and Salmonella Paratyphi A isolates obtained from various Indian reports that were resistant to antimicrobials (indicated by coloured lines). Isolates represented in this graph were consolidated from published reports between the 1990s and 2017 from endemic and epidemic sources, assembled systematically. Source: Refs 9-39.
The temporal trends of AMR showed a steady decline in the proportion of MDR isolates and accounted for less than 20 per cent of isolates obtained between 2011 and 2015, whereas resistance to FQs continued to increase during this period (from 10% in 2001-2005 to 66% in 2011-2015), necessitating the use of third-generation cephalosporins in the treatment of enteric fever. Third-generation cephalosporin resistance remained constant across all time periods (Table I and Fig. 2). Azithromycin is often used for the treatment of enteric fever, but the number of reports on the susceptibility was too few to be presented in this study although there are sporadic reports of phenotypic resistance41,42,43. The scenario was similar with the S. Paratyphi A isolates (Table I).
Molecular determinants of AMR
A total of 880 S. Typhi and 11 S. Paratyphi A isolates spanning 22 studies (Table IV)11,18,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63 were included for the analysis of molecular mechanisms. Most studies (76%) incorporated the polymerase chain reaction method using specific probes of interest to study the molecular determinants of AMR. There was only one study46 which looked at the mechanisms of FQ resistance other than single-nucleotide polymorphisms (SNPs) in quinolone resistance-determining region (QRDR) (qnr genes, the aac(6’)-lb-cr gene, oqxAB and qepA genes). All other studies only looked at QRDR SNPs.
Table IV.
Studies included in the systematic review in which molecular characteristics of AMR in S. Typhi and Paratyphi A were analysed
| Author & Reference | Year of publication | No. of S. Typhi analysed | No. of S. Paratyphi analysed | Risk of Bias | |
|---|---|---|---|---|---|
| QUIPS | JBI | ||||
| Capoor et al44 | 2009 | 14 | - | ||
| Capoor11 | 2007 | 12 | - | Low | No |
| Chau et al54 | 2007 | 23 | - | Low | No |
| Dahiya et al57 | 2014 | 18 | - | Low | No |
| Das et al58 | 2017 | 165 | - | Low | No |
| Devanga Ragupathi et al59 | 2016 | 1 | - | Low | No |
| Dutta et al60 | 2008 | 2 | - | Low | Yes |
| Dutta et al61 | 2014 | 18 | - | Low | No |
| Elumalai et al62 | 2016 | 1 | - | Low | Yes |
| Gaind et al63 | 2006 | 8 | 7 | Low | No |
| Geetha et al45 | 2014 | 36 | - | Low | No |
| Gopal et al46 | 2016 | 131 | - | Low | No |
| Jain and Chugh18 | 2013 | 266 | - | Low | No |
| Kumarasamy et al47 | 2012 | 1 | - | Low | No |
| Misra et al48 | 2016 | 100 | - | Low | No |
| Mohanty et al49 | 2010 | 1 | - | Low | Yes |
| Nath & Maurya50 | 2010 | 1 | - | Low | No |
| Ramachandran et al51 | 2017 | 2 | - | Low | No |
| Renuka et al52 | 2004 | 52 | 4 | Low | No |
| Shanahan et al55 | 2000 | 2 | - | Low | No |
| Shanahan et al53 | 1998 | 20 | - | Low | No |
| Thamizhmani et al56 | 2012 | 6 | - | Low | No |
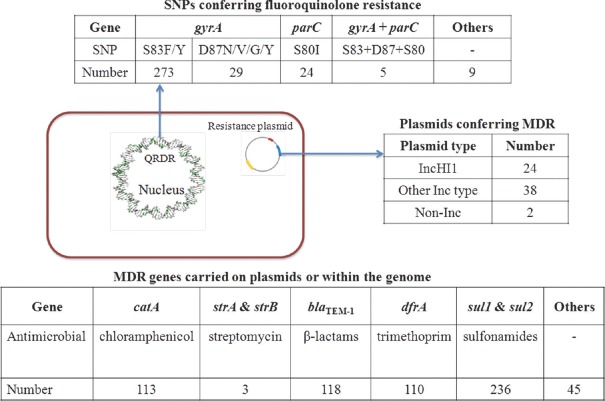
Genetic signatures implicated in FQ resistance were very distinct amongst the identified Indian isolates. SNPs in gyrA, gyrB, parC and parE, which include the QRDR in the S. Typhi genome, as well as FQ resistance conferring plasmids containing qnrB2, qnrB4 and qnrS1 genes, were reported64. It was apparent that FQ resistance in S. Typhi was frequently linked to mutations with gyrA (Fig. 3). A frequent position for SNPs in gyrA is codon 83, with the S83F being the most common occurring in 244 isolates. S80I was the most common SNP in the parC gene, detected in 24 isolates, together with a concordant SNP in S83F. The S83Y mutation was detected in 29 isolates, while 18 isolates harboured the mutation gyrA D87N, further underpinning the importance of gyrA-associated SNPs, likely in response to antimicrobial selection pressure. Isolates harbouring combinations of three SNPs in gyrA, at codons 83 and 87 as well as mutations at codon 80 in parC, are associated with a high level of ciprofloxacin resistance and designated as ‘triple mutants’64. SNPs in parE and gyrB were also observed but to a much lower extent (three and seven isolates, respectively). The qnrB2, qnrB4 and qnrS1 resistance determinants were found in S. Typhi but are still rare when compared with QRDR mutations.
Fig. 3.
Molecular determinants of AMR in enteric fever isolates from India. Fluoroquinolone resistance occurs through mutational DNA gyrase enzyme of the bacteria which is encoded by gyrA, gyrB, parC and parE genes (quinolone resistance-determining region). Number refers to the number of isolates harbouring the respective determinant of antimicrobial resistance as identified through the review. Amino acids: S, serine; F, phenylalanine; Y, tyrosine; D, asparagine; N, aspartic acid; I, isoleucine.
The recent decline in MDR S. Typhi across South and South-East Asia has been accompanied by a decrease in the proportion of isolates carrying IncHI1 plasmids64,65, which often harbour the resistance genes responsible for MDR typhoid
(Fig. 3). Such resistance genes are clustered on composite transposons and include catA, sul1, sul2, dfrA, blaTEM-1, strA, strB, tetA, tetB, tetC and tetD. These MDR-associated genes can also be found integrated on the chromosome of H58 S. Typhi in isolates from countries including India and Bangladesh64,65 Other plasmids identified in S. Typhi included R27-like, B7-like and those falling into IncH and IncN, but these are relatively uncommon. Extended-spectrum β-lactamase (ESBL)-producing S. Typhi isolates, which confer resistance to third-generation cephalosporins, have been reported in India and Pakistan66,67. The Indian isolates carried IncX3 and IncA plasmids which encoded blaSHV-12 and blaCMY-2 determinants66, as well as blaTEM-1B and blaDHA-1, probably on an IncN plasmid59.
Discussion
The rapidly changing antimicrobial pressure in India has selected certain clones of S. Typhi which continue to adapt to changing pressures. The dominant clone currently circulating is known as H58 and has constantly evolved over the last 15 yr as evidenced by Bayesian estimates64. These H58 strains comprise two main lineages namely lineage I and lineage II68. Analysis of enteric fever isolates from Nepal suggested that lineage I strains were dominant in the 1990s and were gradually replaced by lineage II strains which are now the most prevalent. The distinction of lineages is important due to their varying capacities in carrying AMR-determining genes. While lineage I is more strongly associated with MDR, lineage II strains favour FQ resistance68 with a rapidly expanding highly FQ-resistant sub-population known as ‘triple mutants’64. These triple mutations are most commonly identified in S. Typhi isolates from South Asia, often as a distinct sub-group within the main H58 clonal population64.
The decline in MDR typhoid as seen in the results is likely due to the infrequent use of chloramphenicol and co-trimoxazole in India and in the Indian subcontinent in general. The first-line antimicrobials namely chloramphenicol, co-trimoxazole and ampicillin were widely used in the 1990s which prompted both S. Typhi and S. Paratyphi A to adapt to this antimicrobial pressure. Both organisms subsequently acquired resistance to these antimicrobials via acquisition of the full suite of seven acquired AMR genes that are typically located within a composite transposon, comprising Tn6029 (sul2, strA, strAB and blaTEM-1) and Tn21 (dfrA7, sul1) inserted within Tn9 (catA), which is often carried on the IncHI1 group of plasmids64. This plasmid possesses genes which confer resistance to sulphonamides (sul1, sul2), ampicillin (blaTEM-1), trimethoprim (dfrA7), chloramphenicol (catA) and streptomycin (strAB). The horizontal transfer of these plasmids to S. Typhi and S. Paratyphi A also meant that these plasmids could be lost in the absence of such antimicrobial pressure, as was seen at the turn of the century when FQs became the drug of choice and the first-line antimicrobials fell out of favour among clinicians due to widespread resistance.
Ciprofloxacin and ofloxacin were the choices for both empirical therapy and treatment of culture-proven enteric fever, resulting in FQ-associated antimicrobial pressure. The spread of FQ resistance across India was enhanced by the emergence of the H58 clade, which dominated circulating S. Typhi populations in India by the late 1990s, with an apparent increased fitness advantage and enhanced transmission success69,70,71. These clones of S. Typhi and S. Paratyphi A accumulated non-synonymous SNPs in the genome inducing conformational changes in DNA gyrase and topoisomerase IV, the main sites of FQ action64,72. The genes in which SNPs occur include gyrA, parC, parE and gyrB, with gyrA SNPs correlating strongly with treatment failure69. Accumulating mutations in the QRDR cause S. Typhi to gradually increase the MIC values of ciprofloxacin. Ciprofloxacin-susceptible strains (MIC - 0.06 μg ml) are known to acquire a gyrA S83F single mutation with a subsequent increase in MIC values (0.12-0.5 μg ml), and additional gyrA and parC mutations continue to cause an increase in MICs up to 4 μg ml71.
The standard method of antimicrobial sensitivity testing, i.e. disc diffusion, suggested that S. Typhi was still relatively sensitive to ciprofloxacin despite ongoing treatment failure and relapse73,74. A WHO report comprising an antimicrobial surveillance study of enteric fever isolates from 15 sites across India between 2008 and 2010 revealed that sensitivity of nalidixic acid was a good indicator of FQ sensitivity, but nalidixic acid resistance correlated poorly with ciprofloxacin resistance74. The fact that nalidixic acid breakpoints on disc diffusion correlated more accurately with ciprofloxacin-related treatment outcomes prompted a revision in the CLSI-recommended breakpoints for ciprofloxacin. A report from Veeraraghavan et al75 compared breakpoints for ciprofloxacin using the CLSI guidelines before and after the 2012 revision and also with the EUCAST guidelines and found that only three per cent of isolates were sensitive using the revised guidelines versus 95 per cent of isolates that were sensitive using the older guidelines. The sensitivities of isolates reported using EUCAST breakpoints were comparable to the revised CLSI breakpoints75. In our analysis, the trend lines of changing nalidixic acid and ciprofloxacin resistance over time seem to converge from 2011, which may in large part be due to revisions in the CLSI guidelines.
In the face of FQ resistance, third-generation cephalosporins and azithromycin have become the preferred treatment choices for enteric fever. However, the most contemporary concern stems from the emergence of ESBLs produced by various Gram-negative species, which has originated as a result of the widespread cephalosporin use which has subsequently led to treatment failure with third-generation cephalosporins in India59,66. More worryingly, reports from Pakistan67,76 detailing an extensively drug-resistant typhoid outbreak in populous parts of the Sindh province76 are a cause for concern. These isolates had a composite transposon as described above and an additional IncY plasmid containing blaCTX-M15 and qnrS genes77, conferring resistance to the first-line antimicrobials, FQs and third-generation cephalosporins. Cephalosporins were the most commonly used antimicrobial in India followed by broad-spectrum penicillins, FQs and macrolides as per a 2014 report78 and more recently by a 2018 report79. This indirectly portrays the antimicrobial pressure exerted by the use of cephalosporins, which has consequently led to the production of ESBLs by Gram-negative bacteria, including S. Typhi59,66,67.
As with most community-acquired infections, single-drug therapy (monotherapy) has been a common practice in the management of enteric fever. Monotherapy with the former first-line antimicrobials may not be an unreasonable option in India as evidenced by the results from this systematic review. A case report from Nepal suggests that treatment with co-trimoxazole results in complete remission of H58-related typhoid which was FQ-resistant but not MDR79. However, a more judicious approach might involve combination therapy with a first- line antimicrobial and perhaps azithromycin. This approach could potentially facilitate the conservation of cephalosporins and reduce the antimicrobial pressure currently exerted by the widespread use of this class of drugs. The decrease in MDR as highlighted in these data following the scant use of first- line antibiotics (amoxicillin, chloramphenicol and co-trimoxazole) suggests that an additional option of cycling these antimicrobials potentially exists, on the condition that close monitoring of antimicrobial susceptibility is feasible.
India is not only one of the largest global consumers of antibiotics, but also one of the countries with the highest rates of AMR80. Between 2000 and 2015, antimicrobial consumption expressed in defined daily doses increased by 103 per cent (3.2 billion in 2000-6.5 billion in 2015), making it the number one consumer of antimicrobials in low- and middle-income countries78. The strongest factor attributed to this trend was an increase in the use of cephalosporins81, due to changing prescribing practices for enteric fever and other infections including those of the respiratory tract, skin and soft tissue as well as gonococcal infections81. Cephalosporins replaced penicillins and quinolones for infection management in both empirical and definitive treatment78. Antimicrobials available to the community from both private and public sector pharmacies included FQs, cephalosporins, macrolides and co-trimoxazole82, and more recently carbapenems, with chloramphenicol being rarely prescribed or used over the counter. The excessive use of third-generation cephalosporins for acute febrile illnesses83 as well as respiratory tract infections84 and the inappropriate usage of FQs for diarrhoea82,85 all contribute to antimicrobial pressure which impacts treatment options for bloodstream infections such as enteric fever. Fixed-drug combinations that are available for use include combinations of FQs with antiprotozoal drugs, FQs with azithromycin or cefixime and cefixime with azithromycin, often licensed for use by State Drug Licensing Authorities without documented central regulatory approval86. Social factors that contribute to rising AMR include access to antimicrobials without prescription and the use of pharmacies and informal providers as sources of healthcare by the general public, exposure to antimicrobial residues in animal husbandry (such as ciprofloxacin used for growth promotion in poultry) leading to a general increase in antimicrobial pressure in the environment, plus the lack of established monitored standards for antimicrobial residues in pharmaceutical industry effluents87.
This study was limited by the fact that these isolates did not represent the antibiogram of Indian isolates in its entirety. Most isolates in this study were obtained from tertiary care settings, with almost no representation from community settings although it is plausible that the antibiogram of isolates would not be very different between community and hospital settings as far as enteric fever is concerned. Finally, the CLSI breakpoints were significantly revised in 2011, and it was not possible to ascertain how quickly laboratories transitioned to the new breakpoint guidelines which might have a bearing on the estimation of ciprofloxacin resistance around the 2011-2012 period.
The problem of AMR in the pathogens which cause enteric fever underscores the importance of controlling the spread of typhoid through the deployment of vaccines and prudent antimicrobial use in the short term. Immunization could theoretically reduce the number of circulating MDR, FQ- and cephalosporin-resistant strains and, furthermore, decrease the incidence of undifferentiated febrile illness, thereby reducing the need for empirical antimicrobial therapy.
Footnotes
Financial support & sponsorship: The authors acknowledge the Bill & Melinda Gates Foundation for their support in ongoing enteric fever related studies by our respective groups. The first author (CDB) is a Rhodes scholar funded by the Rhodes trust. The last author (AJP) received grants from Bill & Melinda Gates Foundation, during the conduct of the study; grants from Okairos, grants from Pfizer, outside the submitted work.
Conflicts of Interest: The last author (AJP) chairs the UK Department of Health's (DH) Joint Committee on Vaccination and Immunisation (JCVI) and is a member of the World Health Organization's (WHO) Strategic Advisory Group of Experts. The views expressed in this manuscript do not necessarily reflect the views of JCVI, DH, or WHO. Other authors have no competing interests to declare.
References
- 1.Buckle GC, Walker CL, Black RE. Typhoid fever and paratyphoid fever: Systematic review to estimate global morbidity and mortality for 2010. J Glob Health. 2012;2:010401. doi: 10.7189/jogh.02.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geneva, Switzerland: 2017. Oct 17-19, Summary of the October 2017 Meeting of the Strategic Advisory Group of Experts on Immunization. [Google Scholar]
- 3.SAGE Working Group on Typhoid Vaccines. Background Paper to SAGE on typhoid Vaccine Policy recommendations. 2017 Sep 24; [Google Scholar]
- 4.World Health Organization. Safety of Typhoid Vaccines. [accessed on April 3, 2018]. Available from: http://www.who.int/vaccine_safety/committee/topics/typhoid/Dec_2016/en/
- 5.Britto CD, Wong VK, Dougan G, Pollard AJ. A systematic review of antimicrobial resistance in Salmonella enterica serovar Typhi, the etiological agent of typhoid. PLoS Negl Trop Di. 2018;12:e0006779. doi: 10.1371/journal.pntd.0006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayden JA, van der Windt DA, Cartwright JL, Côté P, Bombardier C. Assessing bias in studies of prognostic factors. Ann Intern Med. 2013;158:280–6. doi: 10.7326/0003-4819-158-4-201302190-00009. [DOI] [PubMed] [Google Scholar]
- 7.Aromataris E, Fernandez R, Godfrey C, Holly C, Khalil H. Australia: The Joanna Briggs Institute; 2014. Methodology for JBI umbrella reviews. Joanna Briggs Institute Reviewers' Manual: 2014 edition / Supplement; pp. 1–34. [Google Scholar]
- 8.Tadesse G, Tessema TS, Beyene G, Aseffa A. Molecular epidemiology of fluoroquinolone resistant Salmonella in Africa: A systematic review and meta-analysis. PLoS One. 2018;13:e0192575. doi: 10.1371/journal.pone.0192575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adhikary R, Joshi S. Dual Salmonella typhi Typhi infection. Indian J Pathol Microbiol. 2011;54:849–50. doi: 10.4103/0377-4929.91522. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee A, Kalghatgi AT, Singh P, Nagendra A, Singh Z, Handa SK. Epidemiological investigation of an outbreak of enteric fever. Med J Armed Forces India. 2007;63:322–4. doi: 10.1016/S0377-1237(07)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capoor MR, Nair D, Aggarwal P, Mathys V, Dehem M, Bifani PJ. Salmonella enterica serovar Typhi: Molecular analysis of strains with decreased susceptibility and resistant to ciprofloxacin in India from 2001-2003. Braz J Infect Dis. 2007;11:423–5. doi: 10.1590/s1413-86702007000400011. [DOI] [PubMed] [Google Scholar]
- 12.Choudhary A, Gopalakrishnan R, Nambi PS, Ramasubramanian V, Ghafur KA, Thirunarayan MA. Antimicrobial susceptibility of Salmonella enterica serovars in a tertiary care hospital in southern India. Indian J Med Res. 2013;137:800–2. [PMC free article] [PubMed] [Google Scholar]
- 13.Dahiya S, Sharma P, Kumari B, Pandey S, Malik R, Manral N, et al. Characterisation of antimicrobial resistance in Salmonellae during 2014-2015 from four centres across India: An ICMR antimicrobial resistance surveillance network report. Indian J Med Microbiol. 2017;35:61–8. doi: 10.4103/ijmm.IJMM_16_382. [DOI] [PubMed] [Google Scholar]
- 14.Dutta S, Sur D, Manna B, Bhattacharya SK, Deen JL, Clemens JD. Rollback of Salmonella enterica serotype Typhi resistance to chloramphenicol and other antimicrobials in Kolkata, India. Antimicrob Agents Chemother. 2005;49:1662–3. doi: 10.1128/AAC.49.4.1662-1663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautam V, Gupta NK, Chaudhary U, Arora DR. Sensitivity pattern of Salmonella serotypes in Northern India. Braz J Infect Dis. 2002;6:281–7. doi: 10.1590/s1413-86702002000600003. [DOI] [PubMed] [Google Scholar]
- 16.Gupta V, Singla N, Bansal N, Kaistha N, Chander J. Trends in the antibiotic resistance patterns of enteric fever isolates - a three year report from a tertiary care centre. Malays J Med Sci. 2013;20:71–5. [PMC free article] [PubMed] [Google Scholar]
- 17.Harichandran D, Dinesh KR. Antimicrobial susceptibility profile, treatment outcome and serotype distribution of clinical isolates of Salmonella enterica subspecies enterica: A 2-year study from Kerala, South India. Infect Drug Resist. 2017;10:97–101. doi: 10.2147/IDR.S126209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain S, Chugh TD. Antimicrobial resistance among blood culture isolates of Salmonella enterica in New Delhi. J Infect Dev Ctries. 2013;7:788–95. doi: 10.3855/jidc.3030. [DOI] [PubMed] [Google Scholar]
- 19.Joshi S, Amarnath SK. Fluoroquinolone resistance in Salmonella Typhi and S. Paratyphi A in Bangalore, India. Trans R Soc Trop Med Hyg. 2007;101:308–10. doi: 10.1016/j.trstmh.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Kadhiravan T, Wig N, Kapil A, Kabra SK, Renuka K, Misra A. Clinical outcomes in typhoid fever: adverse impact of infection with nalidixic acid-resistant Salmonella Typhi. BMC Infect Dis. 2005;5:37. doi: 10.1186/1471-2334-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar Y, Sharma A, Mani KR. High level of resistance to nalidixic acid in Salmonella enterica serovar Typhi in Central India. J Infect Dev Ctries. 2009;3:467–9. doi: 10.3855/jidc.419. [DOI] [PubMed] [Google Scholar]
- 22.Kumar Y, Sharma A, Mani KR. Re-emergence of susceptibility to conventionally used drugs among strains of Salmonella Typhi in central west India. J Infect Dev Ctries. 2011;5:227–30. doi: 10.3855/jidc.1310. [DOI] [PubMed] [Google Scholar]
- 23.Lakshmi V, Ashok R, Susmita J, Shailaja VV. Changing trends in the antibiograms of Salmonella isolates at a tertiary care hospital in Hyderabad. Indian J Med Microbiol. 2006;24:45–8. doi: 10.4103/0255-0857.19894. [DOI] [PubMed] [Google Scholar]
- 24.Madhulika U, Harish BN, Parija SC. Current pattern in antimicrobial susceptibility of Salmonella Typhi isolates in Pondicherry. Indian J Med Res. 2004;120:111–4. [PubMed] [Google Scholar]
- 25.Manchanda V, Bhalla P, Sethi M, Sharma VK. Treatment of enteric fever in children on the basis of current trends of antimicrobial susceptibility of Salmonella enterica serovar typhi and paratyphi A. Indian J Med Microbiol. 2006;24:101–6. doi: 10.4103/0255-0857.25182. [DOI] [PubMed] [Google Scholar]
- 26.Misra R, Prasad KN, Amrin N, Kapoor P, Singh S, Ghar M. Absence of multidrug resistance in Salmonella enterica serotypes Typhi and Paratyphi A isolates with intermediate susceptibility to ciprofloxacin. Trans R Soc Trop Med Hyg. 2015;109:538–40. doi: 10.1093/trstmh/trv036. [DOI] [PubMed] [Google Scholar]
- 27.Mohanty S, Renuka K, Sood S, DAS BK, Kapil A. Antibiogram pattern and seasonality of Salmonella serotypes in a North Indian tertiary care hospital. Epidemiol Infect. 2006;134:961–6. doi: 10.1017/S0950268805005844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagshetty K, Channappa ST, Gaddad SM. Antimicrobial susceptibility of Salmonella Typhi in India. J Infect Dev Ctries. 2010;4:70–3. doi: 10.3855/jidc.109. [DOI] [PubMed] [Google Scholar]
- 29.Narain U, Gupta R. Emergence of resistance in community-acquired enteric fever. Indian Pediatr. 2015;52:709. doi: 10.1007/s13312-015-0704-0. [DOI] [PubMed] [Google Scholar]
- 30.Ray P, Sharma J, Marak RSK, Garg RK. Predictive efficacy of nalidixic acid resistance as a marker of fluoroquinolone resistance in Salmonella enterica var Typhi. Indian J Med Res. 2006;124:105–8. [PubMed] [Google Scholar]
- 31.Rodrigues C, Mehta A, Mehtar S, Blackmore PH, Hakimiyan A, Fazalbhoy N, et al. Chloramphenicol resistance in Salmonella typhi. Report from Bombay. J Assoc Physicians India. 1992;40:729–32. [PubMed] [Google Scholar]
- 32.Senthilkumar B, Prabakaran G. Multidrug resistant Salmonella typhi in asymptomatic typhoid carriers among food handlers in Namakkal district, Tamil Nadu. Indian J Med Microbiol. 2005;23:92–4. doi: 10.4103/0255-0857.16046. [DOI] [PubMed] [Google Scholar]
- 33.Sharvani R, Hemavathi, Dayanand DK, Shenoy P, Sarmah P. Antibiogram of Salmonella Isolates: Time to consider antibiotic salvage. J Clin Diagn Res. 2016;10:DC06–8. doi: 10.7860/JCDR/2016/18102.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srirangaraj S, Kali A, Charles MV. A study of antibiogram of Salmonella enterica serovar Typhi isolates from Pondicherry, India. Australas Med J. 2014;7:185–90. doi: 10.4066/AMJ.2014.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatesh BM, Joshi S, Adhikary R, Bhaskar BH. Antibiogram of Salmonella typhii and Salmonella paratyphi A in a tertiary care hospital in 2012. Indian J Pathol Microbiol. 2013;56:484–5. doi: 10.4103/0377-4929.125418. [DOI] [PubMed] [Google Scholar]
- 36.Verma S, Thakur S, Kanga A, Singh G, Gupta P. Emerging Salmonella Paratyphi A enteric fever and changing trends in antimicrobial resistance pattern of Salmonella in Shimla. Indian J Med Microbiol. 2010;28:51–3. doi: 10.4103/0255-0857.58730. [DOI] [PubMed] [Google Scholar]
- 37.Chandel DS, Chaudhry R, Dhawan B, Pandey A, Dey AB. Drug-resistant Salmonella enterica serotype paratyphi A in India. Emerg Infect Dis. 2000;6:420–1. doi: 10.3201/eid0604.000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harish BN, Madhulika U, Parija SC. Isolated high-level ciprofloxacin resistance in Salmonella enterica subsp. enterica serotype Paratyphi A. J Med Microbiol. 2004;53(Pt 8):819. doi: 10.1099/jmm.0.05451-0. [DOI] [PubMed] [Google Scholar]
- 39.Tankhiwale SS, Agrawal G, Jalgaonkar SV. An unusually high occurrence of Salmonella enterica serotype Paratyphi A in patients with enteric fever. Indian J Med Res. 2003;117:10–2. [PubMed] [Google Scholar]
- 40.Ugboko H, De N. Mechanisms of antibiotic resistance in Salmonella typhi. Int J Curr Microbiol App Sci. 2014;3:461–76. [Google Scholar]
- 41.Rai S, Jain S, Prasad KN, Ghoshal U, Dhole TN. Rationale of azithromycin prescribing practices for enteric fever in India. Indian J Med Microbiol. 2012;30:30–3. doi: 10.4103/0255-0857.93017. [DOI] [PubMed] [Google Scholar]
- 42.Kumar VA, Kumar A, Khan S, Dinesh KR, Karim S. Revised ciprofloxacin breakpoints for Salmonella: Is it time to write an obituary? J Clin Diagn Res. 2013;7:2467–9. doi: 10.7860/JCDR/2013/7312.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel SR, Bharti S, Pratap CB, Nath G. Drug resistance pattern in the recent isolates of Salmonella Typhi with special reference to cephalosporins and azithromycin in the gangetic plain. J Clin Diagn Res. 2017;11:DM01–DM03. doi: 10.7860/JCDR/2017/23330.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capoor MR, Nair D, Walia NS, Routela RS, Grover SS, Deb M, et al. Molecular analysis of high-level ciprofloxacin resistance in Salmonella enterica serovar Typhi and S. Paratyphi A: need to expand the QRDR regio. Epidemiol Infect. 2009;137:871–8. doi: 10.1017/S0950268808001076. [DOI] [PubMed] [Google Scholar]
- 45.Geetha VK, Yugendran T, Srinivasan R, Harish BN. Plasmid-mediated quinolone resistance in typhoidal Salmonellae: a preliminary report from South India. Indian J Med Microbiol. 2014;32:31–4. doi: 10.4103/0255-0857.124292. [DOI] [PubMed] [Google Scholar]
- 46.Gopal M, Elumalai S, Arumugam S, Durairajpandian V, Kannan MA, Selvam E, et al. GyrA ser83 and ParC trp106 mutations in Salmonella enterica serovar Typhi isolated from typhoid fever patients in tertiary care hospital. J Clin Diagn Res. 2016;10:DC14–8. doi: 10.7860/JCDR/2016/17677.8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumarasamy K, Krishnan P. Report of a Salmonella enterica serovar Typhi isolate from India producing CMY-2 AmpC beta-lactamase. J Antimicrob Chemother. 2012;67:775–6. doi: 10.1093/jac/dkr514. [DOI] [PubMed] [Google Scholar]
- 48.Misra R, Thakare R, Amrin N, Prasad KN, Chopra S, Dhole TN. Antimicrobial susceptibility pattern and sequence analysis of DNA gyrase and DNA topoisomerase IV in Salmonella enterica serovars Typhi and Paratyphi A isolates with decreased susceptibility to ciprofloxacin. Trans R Soc Trop Med Hyg. 2016;110:472–9. doi: 10.1093/trstmh/trw051. [DOI] [PubMed] [Google Scholar]
- 49.Mohanty S, Gaind R, Paglietti B, Paul P, Rubino S, Deb M. Bacteraemia with pleural effusions complicating typhoid fever caused by high-level ciprofloxacin-resistant Salmonella enterica serotype Typhi. Ann Trop Paediatr. 2010;30:233–40. doi: 10.1179/146532810X12786388978760. [DOI] [PubMed] [Google Scholar]
- 50.Nath G, Maurya P. Drug resistance patterns in Salmonella enterica subspecies enterica serotype Typhi strains isolated over a period of two decades, with special reference to ciprofloxacin and ceftriaxone. Int J Antimicrob Agents. 2010;35:482–5. doi: 10.1016/j.ijantimicag.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 51.Ramachandran A, Shanthi M, Sekar U. Detection of blaCTX-M extended spectrum beta-lactamase producing Salmonella enterica Serotype Typhi in a tertiary care centre. J Clin Diagnostic Res. 2017;11:DC21–DC24. doi: 10.7860/JCDR/2017/30150.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renuka K, Kapil A, Kabra SK, Wig N, Das BK, Prasad VV, et al. Reduced susceptibility to ciprofloxacin and gyra gene mutation in North Indian strains of Salmonella enterica serotype Typhi and serotype Paratyphi A. Microb Drug Resist. 2004;10:146–53. doi: 10.1089/1076629041310028. [DOI] [PubMed] [Google Scholar]
- 53.Shanahan PM, Jesudason MV, Thomson CJ, Amyes SG. Molecular analysis of and identification of antibiotic resistance genes in clinical isolates of Salmonella Typhi from India. J Clin Microbiol. 1998;36:1595–600. doi: 10.1128/jcm.36.6.1595-1600.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chau TT, Campbell JI, Galindo CM, Van Minh Hoang N, Diep TS, Nga TT, et al. Antimicrobial drug resistance of Salmonella enterica serovar typhi in asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother. 2007;51:4315–23. doi: 10.1128/AAC.00294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shanahan PM, Karamat KA, Thomson CJ, Amyes SG. Characterization of multi-drug resistant Salmonella Typhi isolated from Pakistan. Epidemiol Infect. 2000;124:9–16. doi: 10.1017/s0950268899003453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thamizhmani R, Bhattacharya D, Sayi DS, Bhattacharjee H, Muruganandam N, Ghosal SR, et al. Emergence of fluoroquinolone resistance in Salmonella enterica serovar Typhi in Andaman and Nicobar Islands, India. Indian J Med Res. 2012;136:98–101. [PMC free article] [PubMed] [Google Scholar]
- 57.Dahiya S, Kapil A, Lodha R, Kumar R, Das BK, Sood S, et al. Induction of resistant mutants of Salmonella enterica serotype Typhi under ciprofloxacin selective pressure. Indian J Med Res. 2014;139:746–53. [PMC free article] [PubMed] [Google Scholar]
- 58.Das S, Samajpati S, Ray U, Roy I, Dutta S. Antimicrobial resistance and molecular subtypes of Salmonella enterica serovar Typhi isolates from Kolkata, India over a 15 years period 1998-2012. Int J Med Microbiol. 2017;307:28–36. doi: 10.1016/j.ijmm.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 59.Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Shankar BA, Munusamy E, Anandan S, Veeraraghavan B. Draft genome sequence of blaTEM-1-mediated cephalosporin-resistant Salmonella enterica serovar Typhi from bloodstream infection. J Glob Antimicrob Resist. 2016;7:11–2. doi: 10.1016/j.jgar.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Dutta S, Sur D, Manna B, Sen B, Bhattacharya M, Bhattacharya SK, et al. Emergence of highly fluoroquinolone-resistant Salmonella enterica serovar Typhi in a community-based fever surveillance from Kolkata, India. Int J Antimicrob Agents. 2008;31:387–9. doi: 10.1016/j.ijantimicag.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 61.Dutta S, Das S, Mitra U, Jain P, Roy I, Ganguly SS, et al. Antimicrobial resistance, virulence profiles and molecular subtypes of Salmonella enterica serovars Typhi and Paratyphi A blood isolates from Kolkata, India during 2009-2013. PLoS One. 2014;9:e101347. doi: 10.1371/journal.pone.0101347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elumalai S, Seetharaman S. Molecular analysis of fluoroquinolone resistance in Salmonella enterica serovar Typhi from a breast abscess case. Indian J Pathol. Microbiol. 2016;59:261. doi: 10.4103/0377-4929.182030. [DOI] [PubMed] [Google Scholar]
- 63.Gaind R, Paglietti B, Murgia M, Dawar R, Uzzau S, Cappuccinelli P, et al. Molecular characterization of ciprofloxacin-resistant Salmonella enterica serovar Typhi and Paratyphi A causing enteric fever in India. J Antimicrob Chemother. 2006;58:1139–44. doi: 10.1093/jac/dkl391. [DOI] [PubMed] [Google Scholar]
- 64.Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47:632–9. doi: 10.1038/ng.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong VK, Holt KE, Okoro C, Baker S, Pickard DJ, et al. International Typhoid Consortium. Molecular surveillance identifies multiple transmissions of typhoid in west Africa. PLoS Negl Trop Dis. 2016;10:e0004781. doi: 10.1371/journal.pntd.0004781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodrigues C, Kapil A, Sharma A, Devanga Ragupathi NK, Inbanathan FY, Veeraraghavan B, et al. Whole-Genome Shotgun Sequencing of cephalosporin-resistant Salmonella enterica serovar Typhi. Genome Announc. 2017;5 doi: 10.1128/genomeA.01639-16. pii: e01639-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Munir T, Lodhi M, Ansari JK, Andleeb S, Ahmed M. Extended spectrum beta lactamase producing cephalosporin resistant Salmonella Typhi, reported from Rawalpindi, Pakistan. J Pak Med Assoc. 2016;66:1035–6. [PubMed] [Google Scholar]
- 68.Britto CD, Dyson ZA, Duchene S, Carter MJ, Gurung M, Kelly DF, et al. Laboratory and molecular surveillance of paediatric typhoidal Salmonella in Nepal: Antimicrobial resistance and implications for vaccine policy. PLoS Negl Trop Dis. 2018;12:e0006408. doi: 10.1371/journal.pntd.0006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pham Thanh D, Karkey A, Dongol S, Ho Thi N, Thompson CN, Rabaa MA, et al. A novel ciprofloxacin-resistant subclade of H58 Salmonella Typhi is associated with fluoroquinolone treatment failure. Elife. 2016;5:e14003. doi: 10.7554/eLife.14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saad NJ, Bowles CC, Grenfell BT, Basnyat B, Arjyal A, Dongol S, et al. The impact of migration and antimicrobial resistance on the transmission dynamics of typhoid fever in Kathmandu, Nepal: A mathematical modelling study. PLoS Negl Trop Dis. 2017;11:e0005547. doi: 10.1371/journal.pntd.0005547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cuypers WL, Jacobs J, Wong V, Klemm EJ, Deborggraeve S, Van Puyvelde S. Fluoroquinolone resistance in Salmonella: insights by whole-genome sequencing. Microb genomics. 2018;4 doi: 10.1099/mgen.0.000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holt KE, Phan MD, Baker S, Duy PT, Nga TV, Nair S, et al. Emergence of a globally dominant inchi1 plasmid type associated with multiple drug resistant typhoid. PLoS Negl Trop Dis. 2011;5:e1245. doi: 10.1371/journal.pntd.0001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parry CM. The treatment of multidrug-resistant and nalidixic acid-resistant typhoid fever in Vietnam. Trans R Soc Trop Med Hyg. 2004;98:413–22. doi: 10.1016/j.trstmh.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 74.Rupali P, Abraham OC, Jesudason MV, John TJ, Zachariah A, Sivaram S, et al. Treatment failure in typhoid fever with ciprofloxacin susceptible Salmonella enterica serotype Typhi. Diagn Microbiol Infect Dis. 2004;49:1–3. doi: 10.1016/j.diagmicrobio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 75.Veeraraghavan B, Sharma A, Ranjan P, Kapil A. Revised ciprofloxacin breakpoints for Salmonella Typhi: Its implications in India. Indian J Med Microbiol. 2014;32:161–3. doi: 10.4103/0255-0857.129804. [DOI] [PubMed] [Google Scholar]
- 76.Qamar FN, Saleem K, Shakoor S, Yousufzai T, Kazi M, Lohana H, et al. Kampala, Uganda: 2017. Proceedings of 10th International Conference on Typhoid and other Invasive Salmonelloses; p. 40. [Google Scholar]
- 77.Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, et al. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio. 2018;9:e00105–18. doi: 10.1128/mBio.00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, et al. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14:742–50. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 79.Karki M, Pandit S, Baker S, Basnyat B. Cotrimoxazole treats fluoroquinolone-resistant Salmonella Typhi H58 infection. BMJ Case Rep 2016. 2016;2016 doi: 10.1136/bcr-2016-217223. pii: bcr2016217223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. 2016. [accessed on April 11, 2018]. Available from: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf .
- 81.Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, et al. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A. 2018;115:E3463–E3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kotwani A, Holloway K. Trends in antibiotic use among outpatients in New Delhi, India. BMC Infect Dis. 2011;11:99. doi: 10.1186/1471-2334-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson ML, Kadam D, Kagal A, Khadse S, Kinikar A, Valvi C, et al. Antibiotic utilization and the role of suspected and diagnosed mosquito-borne illness among adults and children with acute Febrile illness in Pune, India. Clin Infect Dis. 2017;66:1602–9. doi: 10.1093/cid/cix1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gandra S, Singh SK, Jinka DR, Kanithi R, Chikkappa AK, Sharma A, et al. Point prevalence surveys of antimicrobial use among hospitalized children in six hospitals in India in 2016. Antibiotics (Basel) 2017;6 doi: 10.3390/antibiotics6030019. pii: E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kotwani A, Chaudhury RR, Holloway K. Antibiotic-prescribing practices of primary care prescribers for acute diarrhea in New Delhi, India. Value Health. 2012;15:S116–9. doi: 10.1016/j.jval.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 86.McGettigan P, Roderick P, Kadam A, Pollock A. Threats to global antimicrobial resistance control: Centrally approved and unapproved antibiotic formulations sold in India. Br J Clin Pharmacol. 2019;85:59–70. doi: 10.1111/bcp.13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gandra S. Scoping Report on Antimicrobial Resistance in India. [accessed April 3, 2018]. Available from: https://cddep.org/publications/scopingreport-antimicrobial-resistance-india/