Abstract
Duplication of the eukaryotic genome initiates from multiple origins of DNA replication whose activity is coordinated with the cell cycle. We have been studying the origins of DNA replication that control amplification of eggshell (chorion) genes during Drosophila oogenesis. Mutation of genes required for amplification results in a thin eggshell phenotype, allowing a genetic dissection of origin regulation. Herein, we show that one mutation corresponds to a subunit of the minichromosome maintenance (MCM) complex of proteins, MCM6. The binding of the MCM complex to origins in G1 as part of a prereplicative complex is critical for the cell cycle regulation of origin licensing. We find that MCM6 associates with other MCM subunits during amplification. These results suggest that chorion origins are bound by an amplification complex that contains MCM proteins and therefore resembles the prereplicative complex. Lethal alleles of MCM6 reveal it is essential for mitotic cycles and endocycles, and suggest that its function is mediated by ATP. We discuss the implications of these findings for the role of MCMs in the coordination of DNA replication during the cell cycle.
INTRODUCTION
Accurate duplication of the entire genome is essential for normal cell division. In the eukaryotic cell this awesome task is accomplished in a short period of time by initiating replication from multiple origins of DNA replication. To ensure that all of the genome is replicated exactly once, these origins must be regulated such that they initiate replication only once per cell cycle. The decision to initiate DNA replication and commit to a new round of cell division must also be coordinated with multicellular development.
In recent years much has been learned about how eukaryotic origins of DNA replication are regulated during the cell cycle. This has mainly come from pioneering work in the yeast Saccharomyces cerevisiae, but evidence indicates that this regulation is largely conserved in other eukaryotes. The essential feature is a two-step process in which a prereplication complex (pre-RC) of proteins assembles onto origin DNA early in G1, followed by activation of this complex upon entry into S phase (reviewed by Kelly and Brown, 2000; Diffley, 2001). This pre-RC is built sequentially, with the six subunit origin recognition complex (ORC) binding to origin DNA throughout the majority of the cell cycle (Diffley et al., 1994; Carpenter et al., 1996; Rowles et al., 1996). In G1, CDC6 and CDT1 proteins associate with the ORC, and are required for the loading of the hexameric minichromosome maintenance (MCM) complex into the pre-RC (Tanaka et al., 1997; Maiorano et al., 2000; Nishitani et al., 2000). Two kinase complexes, DBF4/CDC7 and Cyclin E/CDK2, are required for activation of the pre-RC and entry into S phase, in part through phosphorylation of pre-RC subunits directly (Lei et al., 1997; reviewed by Hengstschlager et al., 1999). With the rise in CDK activity, the CDC45 protein associates with the pre-RC, and is required for subsequent binding of proteins of the replication fork (Hopwood and Dalton, 1996; Hardy, 1997; Mimura and Takisawa, 1998; Zou and Stillman, 1998). During initiation, the pre-RC is remodeled with CDC6, CDT1, and MCM proteins leaving origin DNA (Liang et al., 1995; Cocker et al., 1996; Coleman et al., 1996; Romanowski et al., 1996). Evidence suggests that the MCM complex may act as the replicative helicase (Aparicio et al., 1997; reviewed by Lei and Tye, 2001). To ensure that origins initiate only once per cell cycle, continued CDK activity inhibits reassembly of a functional pre-RC during S, G2, and M phase, likely through phosphorylation of multiple pre-RC subunits (Dahmann et al., 1995; Nguyen et al., 2001). A negative regulator, geminin, also inhibits pre-RC assembly, in part by blocking the ability of CDT1 to load the MCM complex (Wohlschlegel et al., 2000; Tada et al., 2001). At the end of mitosis, cyclins and geminin are degraded permitting reassembly of the pre-RC in preparation for another S phase (McGarry and Kirschner, 1998; Noton and Diffley, 2000).
Although there have been great advances recently, the full biochemical mechanism for origin firing and rereplication control remains obscure. Moreover, although the cell cycle control of origin firing is largely conserved, there are distinctions between higher eukaryotes and S. cerevisiae. In this yeast, origins are ∼100–150 base pairs in size with an identifiable ARS consensus sequence, onto which the pre-RC assembles (Bell and Stillman, 1992; Diffley and Cocker, 1992; Marahrens and Stillman, 1994; reviewed by Bielinsky and Gerbi, 2001). In multicellular eukaryotes (the Metazoa), however, origins and pre-RC binding are less well defined. No DNA consensus has yet emerged for origins of DNA replication in Metazoa, and it is unclear what determines where the pre-RC binds and DNA replication initiates (reviewed by Bogan et al., 2000). Experiments that examine the regulated association of the pre-RC with bulk chromatin suggest that most of the yeast paradigm applies to Metazoa (Chong et al., 1995; Kubota et al., 1995; Carpenter et al., 1996; Coleman et al., 1996; Couéet al., 1996; Krude et al., 1996; Romanowski et al., 1996; Dimitrova et al., 1999). There are few reports, however, that examine the requirement of pre-RC proteins at defined origins (Li et al., 2000; Natale et al., 2000; Bielinsky et al., 2001). It is also possible that there are aspects of pre-RC structure and regulation in Metazoa that differ from yeast. During multicellular development the location where replication initiates on a metazoan chromosome can change, but what determines this modification of origin identity is also largely unknown (reviewed by Carminati and Orr-Weaver, 1996).
Some of the best defined origins of DNA replication in Metazoa control the developmental amplification of the eggshell (chorion) genes during Drosophila oogenesis (reviewed by Calvi and Spradling, 1999). This amplification represents a dramatic example of developmental reprogramming of DNA replication. At a precise time in oogenesis, somatic follicle cells that surround the developing oocyte switch from periodic genomic replication to continuous rereplication from origins resident at two chorion loci on the X and 3rd chromosome (Spradling and Mahowald, 1980; Calvi et al., 1998). This leads to amplification of the eggshell protein genes, and supports rapid synthesis of the eggshell later in oogenesis. Chorion DNA can amplify when transformed into ectopic genomic sites, which has led to the identification of subregions at the chorion loci that are required for replication (de Cicco and Spradling, 1984; reviewed by Orr-Weaver, 1991). Detection of replicating DNA by two-dimensional gel electrophoresis indicated that one of these subregions at the 3rd chorion locus is the preferred site at which replication initiates (Delidakis and Kafatos, 1989; Heck and Spradling, 1990; Lu et al., 2001).
Cyclin E is required for amplification, and therefore chorion origins are under cell cycle control similar to other origins (Calvi et al., 1998). This led to the model that chorion origins have a pre-RC–like complex that requires Cyclin E/CDK2, but escapes rereplication inhibition exerted by this kinase (Calvi et al., 1998). Amplification can be seen by immunofluorescence as subnuclear foci of bromodeoxyuridine (BrdU) incorporation and increased chorion copy number by fluorescence in situ hybridization in follicle cell nuclei (Calvi et al., 1998; Calvi and Spradling, 2001). Antibodies against Cyclin E, ORC2, ORC1, ORC5, CDT1, and CDC45 specifically label these subnuclear foci in follicle cells, implicating these proteins in amplification (Calvi et al., 1998; Asano and Wharton, 1999; Royzman et al., 1999; Loebel et al., 2000; Whittaker et al., 2000; reviewed by Spradling, 1999). Importantly, it has been shown that the two regions that are most critical for amplification at the 3rd chromosome locus are directly bound by Drosophila ORC2 (Austin et al., 1999).
Chorion gene amplification permits a genetic approach to dissect origin regulation. Flies homozygous for mild defects in essential S-phase genes live, but adult females have reduced amplification and lay eggs with thin eggshells. In the past few years, the molecular identification of genes with a thin eggshell phenotype has confirmed that proteins that are essential for genomic replication are also required for chorion gene amplification (Landis et al., 1997; Calvi et al., 1998; Landis and Tower, 1999; Royzman et al., 1999; Whittaker et al., 2000; Yamamoto et al., 2000; Bosco et al., 2001). This suggests that a thin eggshell is a sensitive and specific phenotype for a genetic dissection of origin function and regulation.
We have continued to take a genetic approach to identify the proteins that are required for chorion origin activity as a model for understanding the pre-RC and rereplication control. Herein, we show for the first time that MCMs, which are critical for licensing chromosomal origins, are also required for chorion gene amplification. This supports the idea that amplification requires assembly of a pre-RC–like complex onto chorion origins.
MATERIALS AND METHODS
Plasmid Construction
All MCM6 plasmids were derived from the BDGP cDNA LD24958. The first step in construction of P{w+mC, Ub:FL:MCM6} (referred to as Ub:FL:MCM6) was the insertion of a polymerase chain reaction (PCR) product spanning the coding region from the cDNA into pBUF, a pBlueScript derivative that contains the Drosophila ubiquitin promoter and a FLAG epitope (a gift from J.J. Sekelsky, University of North Carolina, Chapel Hill, NC). This plasmid, pGS2, fuses a sequence encoding the FLAG epitope onto the amino terminus of MCM6. The 5-kb XbaI fragment from pGS2, containing the Ub promoter and FLAG-tagged MCM6 coding region, was then ligated into the P element vector pCasper 4 (Thummel et al., 1988).
For P{w+mC, UAS:FL:MCM6} (referred to as UAS:FL:MCM6), a PCR fragment containing the MCM6 coding region was ligated into the P vector pUAST (Brand and Perrimon, 1993), resulting in UAS:MCM6. The amino terminus of MCM6 was then replaced by digesting with BglII and ligating in a BglII PCR product from pGS2 that contained the FLAG:MCM6 fusion. Transformation of these P elements into a y w67c23 strain was by standard methods (Spradling and Rubin, 1982).
Genetics
Standard techniques were used for culture of Drosophila. Information about strains and genetic nomenclature can be found at http://www.flybase.harvard.edu. Initial deletion mapping used Df(1)JF5 (5E03-05;5E08), Df(1)5D (5D01;5E01-08), and Df(1)N73 (5C02;5D05-06). A y w fs(1)K1214 chromosome was constructed and crossed to the following mini-white P element strains for meiotic recombination mapping: EP(X)442 (5E4-5), EP(X)1402 (6A-B), EP(X)1364 (6C3-4), EP(X)1613 (6D1-2), EP(X)1388 (6D7-8) (Rorth et al., 1998). y w fs(1)K1214/w P{w+mC} females were crossed to the y w fs(1)K1214 test chromosome and female progeny with recombination in the yellow to P{w+mC} interval were tested for the thin eggshell phenotype. This allowed us to place fs(1)K1214 proximal or distal to the P{w+mC} element.
New deletions in 6C were created by mobilization of two different P elements by using standard methods (Spradling et al., 1995). These P elements, EP(X)1364 and EP(X)1445, contain the mini-white eye color gene (Rorth et al., 1998) (http://www.fruitfly.org). P element excision chromosomes were initially identified as a change in eye pigmentation when over an FM6 balancer that was mutant for white. In the next generation, these mutated X chromosomes were scored for lethality in males. For those strains that were male lethal, female siblings were crossed to fs(1)K1214 males and excision/fs(1)K1214 female progeny were scored for viability and noncomplementation of the thin eggshell phenotype.
Ethyl-methane sulfonate (EMS) alleles were created by feeding Drosophila males EMS by using standard techniques. These G0 males contained a y w67c23 X chromosome and were homozygous on the 2nd chromosome for the P{w+mC, Ub:FL:MCM6} transgene. These G0 males were crossed to C(1)DX y w f females that contained an attached X chromosome. This resulted in transmission of the mutagenized X chromosome to G1 sons in the next generation. These sons that also were hemizygous for P{w+mC, Ub:FL:MCM6} were crossed individually in vials to C(1)DX y w f females. In the next generation, those vials that contained predominantly red-eyed male offspring were saved as potential X-linked MCM6 lethals rescued by the MCM6 transgene. These putative MCM6 lethals were retested for rescue by MCM6 by crossing red-eyed males to C(1)DX y w f again, and for noncomplementation of MCM6 mutations by crossing to MCM6K1214, Df(1)6C-190, and Df(1)6C-310 alleles. Complementation and lethal phase data were obtained using standard methods and mutant larvae were identified using an FM7c balancer marked with green fluorescent protein (Casso et al., 2000). The strains that contain the deletions shown in Figure 2 and the four new EMS alleles have been deposited in the Bloomington Drosophila collection.
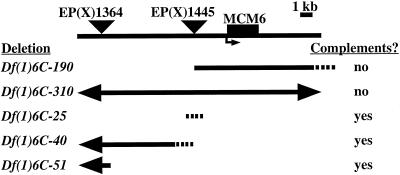
Figure 2.
Genetic and molecular mapping of fs(1)K1214. A molecular map showing the position of the MCM6 transcription unit (black box) relative to the two P elements (triangles) used to generate new deficiencies. The new deficiencies are indicated below. Solid lines represent deleted regions and dotted lines represent uncertainty in the extent of the deficiencies. Arrows indicate that the deficiency extends beyond the region shown (see MATERIALS AND METHODS). Whether the deficiencies complemented the fs(1)K1214 thin eggshell phenotype is indicated on the right. The two deficiencies that failed to complement deleted MCM6, whereas those that complemented did not delete the gene. Not shown are two lethal excision strains (6C-166 and 6C-157) that complemented, and in which we did not detect a deletion.
Southern Mapping of Deletions
Deletions created by P element excision were analyzed by Southern blotting. Genomic DNA was isolated from heterozygous adult females containing the deletion and balancer FM7c, and digested separately with BamHI, XhoI, or HindIII. Southern blots were probed with the MCM6 cDNA or PCR products from the 6C genomic interval. The PCR primers used to generate these probes were based on the BDGP genomic sequence AE003438. In this sequence the MCM6 transcription unit is between coordinates 134,362-137,127. The coordinates of the 5′ end of the PCR primer pairs are 121,330/121,958; 129,834/130,504. Signal intensity was quantified using a Storm PhosphorImager. Copy number was determined by comparison to the signal for genomic DNA from females homozygous for the parental, nondeleted P element chromosome and FM7c males on the same blot.
PCR Mapping of Deletions
Genomic DNA was prepared from male embryos containing the noncomplementing deletions Df(1)6C-310 and Df(1)6C-190. These embryos were identified by the absence of a green fluorescent protein-marked FM7c balancer chromosome. Genomic DNA from female siblings containing the FM7c balancer served as a control. PCR used the primers listed above for Southern mapping, and additional primer pairs with AE003438 coordinates: 123,962/124,477; 132,735/133,720; 137,851/138,537; 145,409/146,110; and 181,633/182,337. The absence of a PCR product in the mutant was evidence for the deletion extending into the region encompassed by that primer pair.
Sequencing of MCM6 Point Mutations
EMS induced alleles of MCM6 were amplified by PCR and subcloned into pBlueScript (Stratagene, La Jolla, CA), or directly sequenced. Sequencing was by the Taq FS Big Dye method (PerkinElmer, Boston, MA) on an ABI 377 sequencer. Both strands were sequenced at least once. For fs(1)K1214, the sequence of the wild-type MCM6 allele from the fs(1)K451 strain was used as a control. This strain was derived from the same isogenic X chromosome screen that yielded fs(1)K1214 (Komitopoulou et al., 1983). The M676K mutation in fs(1)K1214 destroys an Mlu I restriction site. We confirmed that this change is unique by digesting genomic PCR products with Mlu I from fs(1)K1214, fs(1)K451, and five other unrelated strains. The MCM6 lethal EMS alleles were induced on a y w67c23 X chromosome, which served as a control for their sequence.
Immunoprecipitation and Western Blotting
Standard methods were used for immunoprecipitation and analysis of FLAG:MCM6 (Harlow and Lane, 1999). Anti-FLAG antibodies and beads were purchased from Sigma (St. Louis, MO). Extracts were made from 20 pairs of ovaries in 500 μl of FLAG lysis buffer (Sigma) and incubated at 4°C for 3 h with anti-FLAG beads, or beads conjugated to mouse serum alone. Beads were washed several times in FLAG wash buffer (Sigma) and FLAG:MCM6 was eluted from the beads in 100 μl by addition of 3× FLAG peptide (Sigma). A Bradford assay was used to measure total protein in the input extract and equal amounts from the UAS:FLAG:MCM6 and y w control strain were loaded on 7.5% SDS-PAGE and electroblotted onto Hybond ECL membrane (Amersham Biosciences, Piscataway, NJ). Approximately 1/500 of the input and 1/10 of the pellet samples were loaded. Blots were incubated with anti-FLAG M5 antibody (1:2500) or antibodies to fly MCM2, MCM4, or MCM5 all at 1:1000 (Su et al., 1996). Proteins were detected using appropriate horseradish peroxidase-conjugated secondary antibodies and the ECL kit (Amersham Biosciences).
Immunolocalization and BrdU Labeling
BrdU labeling was as previously described (Calvi et al., 1998). Unless otherwise stated, tissues were fixed in 6% electron microscopy grade formaldehyde and labeling and microscopy was carried out essentially as described (Calvi et al., 1998). For localization of FLAG:MCM6, anti-FLAG M2 antibody (1:200) was used. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) or TOTO-3 (Molecular Probes, Eugene, OR) as described (Calvi and Spradling, 2001).
RESULTS
To identify genes that regulate origins of DNA replication, we have been searching for mutations that impair chorion gene amplification. Females homozygous for these mutations produce inviable embryos with thin eggshells. One such female sterile mutation on the X chromosome is fs(1)K1214 (Komitopoulou et al., 1983). Southern blotting had shown that females homozygous for fs(1)K1214 amplify the chorion genes on the X and 3rd chromosomes to only 14 and 6% of wild-type levels, respectively (Orr et al., 1984). Eggs laid by homozygous fs(1)K1214 females were flaccid with thin, fragile eggshells (Figure 1, A-D). The two chorion dorsal appendages that protrude prominently from the anterior of the wild-type eggshell were noticeably thinner and less rigid in the mutant (Figure 1, A and B).
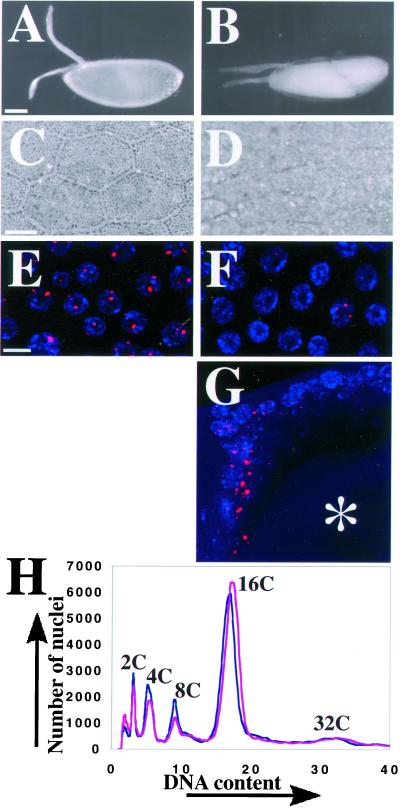
Figure 1.
The eggshell and cellular phenotype of fs(1)K1214. (A) An egg from a wild-type mother is turgid with a thick eggshell that includes two prominent, rigid dorsal appendages. (B) An egg from a homozygous fs(1)K1214 mother is flaccid with a thin eggshell and flimsy dorsal appendages. Anterior is to the left and dorsal is up. Bar, 100 μm (A and B). (C) A 40× phase contrast image of a wild-type eggshell has a hexagonal pattern. Each hexagon represents a “footprint” of the follicle cell that formed that unit of the eggshell late in oogenesis before it died and was sloughed off. (D) Dorsal view of an eggshell produced by an fs(1)K1214 mother shows that it is less phase dense than wild-type, although some follicle cell footprints are evident in the dorsal/anterior on the left. Bar (C and D), 10 μm. (E) BrdU labeling (red) in wild-type stage 10B follicle cell nuclei (blue) reveals four spots of incorporation. The two most prominent spots are the amplifying chorion loci on the X and 3rd chromosome, whereas the faint spots represent unknown loci. (F) Most mutant fs(1)K1214 follicle cells in stage 10B have undetectable BrdU incorporation, whereas a few have faint or nearly wild-type incorporation at amplifying loci. Images in E and F represent a composite stack of eight, 1-μm confocal sections. Bar, 10 μm. (G) Lateral confocal image of a stage 12 egg chamber in an fs(1)K1214 ovary. Shown are the dorsal anterior follicle cells that are mostclosely apposed to the oocyte nucleus (asterisk), and which have the most robust BrdU labeling at chorion loci and elsewhere in the nucleus. Image represents a composite stack of 16, 1 μm confocal sections. Dorsal is up and anterior is to the left. (H) Flow sorting of DAPI stained nuclei from wild-type (blue) and fs(1)K1214 (magenta) ovaries indicates there is no significant difference in DNA content between them. The smaller peaks from the less abundant, but higher ploidy, nurse cells also gave no evidence for endocycle defects in fs(1)K1214 (our unpublished results).
Cellular Phenotype of fs(1)K1214 Reveals Altered DNA Replication during Late Oogenesis
In wild-type females the chorion genes amplify during late stages of oogenesis in the somatic follicle cells that surround the Drosophila egg chamber (reviewed by Calvi and Spradling, 1999). These follicle cells undergo several modifications to their cell cycle during oogenesis (Calvi et al., 1998). They first proliferate in a canonical mitotic cycle, and then, at stage 6 of oogenesis, they enter an endocycle characterized by alternating G and S phases. Most cells achieve a ploidy of 16C and arrest by stage 10A. A small amount of amplification of the chorion genes on the 3rd chromosome occurs during these endocycles. Later, at the onset of stage 10B, chorion genes on the X and 3rd chromosome begin a period of continuous rereplication, whereas the majority of the genome does not replicate. This can be visualized in the microscope as subnuclear foci of BrdU incorporation from stage 10B until stage 14, close to the end of oogenesis and the demise of follicle cells (Calvi et al., 1998; Calvi and Spradling, 2001). Follicle cell nuclei have four BrdU foci; the two largest are the amplifying chorion genes on the 3rd and X chromosome, and the two small foci represent unidentified amplifying genes (Figure 1E). This period of continuous amplification can be thought of as an extended S phase during which only some origins can refire.
To characterize the cellular phenotype of fs(1)K1214, we examined ovaries from homozygous mutant females by BrdU and DAPI labeling. The follicle cells in the mitotic and endocycles appeared normal (our unpublished results). Beginning in stage 10B, however, fs(1)K1214 egg chambers displayed two characteristics that differed from wild type. First, consistent with the known amplification defect, they had reduced incorporation of BrdU into the four foci in follicle cell nuclei (Figure 1F). Surprisingly, incorporation was mosaic among cells within an egg chamber. Although most cells had undetectable incorporation of BrdU, some cells (∼0–50/800 total follicle cells in an egg chamber) had incorporation close to 50% of wild-type levels. Second, most nuclei with detectable labeling at chorion had incorporated BrdU in other parts of the nucleus, indicating that there was inappropriate genomic replication in stage 10B (Figure 1G). There was a spatial bias within the egg chamber for detectable BrdU incorporation. The dorsal-anterior cells that are most closely apposed to the underlying oocyte nucleus most often had intense BrdU labeling during stages 10B-12 (Figure 1G). These included the cells that are destined to form the dorsal appendages and those that migrate to centripetal positions of the egg chamber to define the anterior of the eggshell. Cells in the posterior of the egg chamber, which are close to the oocyte nucleus earlier in oogenesis, also frequently had detectable BrdU incorporation at chorion loci and elsewhere in the nucleus. Both populations of cells receive signals from the underlying oocyte nucleus for anterior-posterior and dorsal-ventral patterning. The BrdU labeling confirms that fs(1)K1214 has a severe defect in amplification, but reveals that the severity of this defect is variable among cells. Moreover, this labeling also indicates that fs(1)K1214 causes inappropriate replication of genomic regions late in oogenesis.
The inappropriate genomic BrdU incorporation in stage 10B may represent additional replication beyond the normal 16C follicle cell arrest. Alternatively, fs(1)K1214 may have defects in replication during earlier endocycles, and the BrdU labeling may represent delayed replication that should have occurred before stage 10B. To address this question, we analyzed the DNA content of fs(1)K1214 follicle cell nuclei by nuclear flow sorting (Lilly and Spradling, 1996; Calvi et al., 1998). This indicated that fs(1)K1214 follicle cells in the mitotic (2C and 4C) and endocycle (8C and 16C) had similar DNA contents to those of wild-type (Figure 1H). Given that dorsal-anterior cells most often had extra BrdU labeling, we measured the DNA content of these nuclei in the microscope by quantifying DAPI fluorescence (n = 100). This also did not reveal a significant difference in DNA content between fs(1)K1214 and wild-type (our unpublished results). Thus, the evidence suggests that fs(1)K1214 follicle cells do not have appreciable extra genomic replication beyond the final 16C DNA content. It is possible that this labeling in stage 10B represents defects in earlier endocycles and completion of the final 8C-16C S phase that normally occurs in stage 9–10A. The absence of evidence for earlier defects indicates that, if endocycle S phases are abnormal in fs(1)K1214, the impairment is subtle.
Genetic and Molecular Mapping of fs(1)K1214
The phenotype of fs(1)K1214 suggested that the molecular identification of the gene would provide insight into the cell cycle regulation of chorion gene amplification. The genetic location of fs(1)K1214 had been previously mapped to the large cytogenetic interval 5D5–6C12 (Orr et al., 1984). We found that three deficiency strains, Df(1)JF5, Df(1)5D, and Df(1)N73, that collectively delete 5C2–5E8 complemented the fs(1)K1214 thin eggshell phenotype. We further refined the location of fs(1)K1214 by meiotic recombination relative to single genetically marked P elements of known location (Rorth et al., 1998; Spradling et al., 1999). This indicated that fs(1)K1214 mapped to cytogenetic interval 6C (see MATERIALS AND METHODS).
During this analysis, we were using in situ hybridization to polytene chromosomes to map the genomic location of cDNAs from BDGP. We found that a cDNA similar to the yeast pre-RC component MCM6 hybridized to cytogenetic interval 6C3-4 on the X chromosome. This location has also been reported recently by two other laboratories (Ohno et al., 1998; Feger, 1999), and has been confirmed subsequently by the genomic sequence of D. melanogaster (Adams, 2000). Given that fs(1)K1214 mapped genetically to this interval, and that the MCM complex is essential for origin function, we deemed it likely that fs(1)K1214 is a mutation in the fly homolog of MCM6. To test this, we created deletions in the 6C cytogenetic interval by imprecise P element excision, and asked whether the failure to complement fs(1)K1214 corresponded to deletion of the MCM6 gene. We recovered seven X-linked lethal excisions. All excisions were viable in females in trans to fs(1)K1214, but two failed to complement the female sterility and resulted in thin eggshells (Figure 2). Molecular characterization of the P excisions by Southern blotting and PCR revealed detectable deletions in five of the seven lethal chromosomes (Figure 2). The two deletions that failed to genetically complement fs(1)K1214 deleted the genomic region corresponding to the MCM6 cDNA, whereas all those that complemented did not remove this region. These results, therefore, were consistent with fs(1)K1214 being a lesion in MCM6.
MCM6 Rescues fs(1)K1214 Phenotype
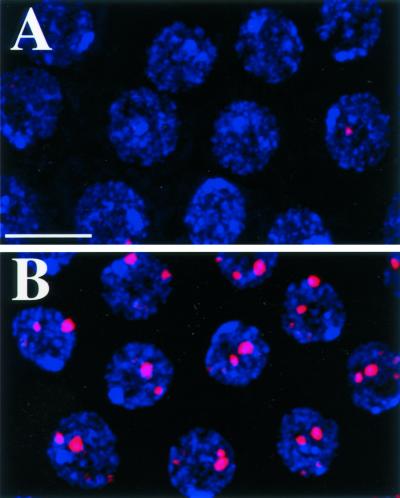
To confirm that fs(1)K1214 is MCM6, we asked whether transgenes containing the MCM6 cDNA could rescue the fs(1)K1214 phenotype. We transformed flies with a FLAG-tagged MCM6 cDNA under control of the fly ubiquitin promoter Ub:FL:MCM6, which is expressed in nearly all cells. One copy of Ub:FL:MCM6 reverted the eggshells produced by homozygous fs(1)K1214 mothers to virtually wild-type appearance. This transgene also rescued the fs(1)K1214 cellular BrdU phenotype, restoring wild-type incorporation at chorion loci and eliminating the inappropriate genomic replication seen after stage 10A in the mutant (Figure 3, A and B). None of the fs(1)K1214 stage 10B egg chambers had normal BrdU incorporation, whereas 92% of fs(1)K1214; Ub:FL:MCM6/+ egg chambers and 97% of wild-type controls had normal BrdU incorporation (n = 61).
Figure 3.
MCM6 rescues fs(1)K1214. (A and B) BrdU labeling (red) in stage 10B follicle cell nuclei (blue) from females homozygous for fs(1)K1214 without (A) and with (B) 1 copy of Ub:FL:MCM6. Images represent a composite stack of eight, 1-μm confocal sections. Bar, 10 μm.
Sequence of the MCM6 allele in fs(1)K1214 indicated that it contains a missense mutation that changes a methionine to a lysine at amino acid position 676 (M676K), in a region of relatively low conservation between fly and MCM6 genes from other organisms (Figure 7, A and D) (see below). The combined genetic and molecular results strongly suggest that fs(1)K1214 is a mutation in MCM6, and therefore, that this MCM family member is required for chorion gene amplification. This mutation will hereafter be referred to as MCM6K1214.
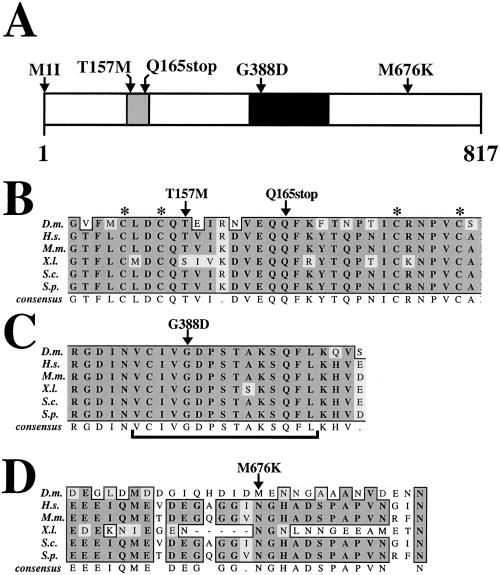
Figure 7.
Sequence of MCM6 mutant alleles. (A) Amino acid changes in the MCM6 mutant alleles are indicated above a schematic representation of the protein. The fs(1)K1214 mutation is homozygous viable and changes a methionine at 676 to a lysine (M676K). The four other mutations are homozygous lethal. The black shaded portion represents the highly conserved MCM box. Gray shading indicates the putative C4 Zinc finger. See Table 1 for allele numbers. M, methionine; I, isoleucine; T, threonine; Q, glutamine; K, lysine. (B-D) ClustalW alignment of selected regions of Drosophila MCM6 protein with MCM6 proteins from other species is shown to indicate the degree of conservation of the amino acid residues that are changed in the mutants (arrows above). (B) Lethal T157M mutation changes a highly conserved threonine that lies between the cysteine pairs (asterisks above) of a putative C4 Zinc finger motif. The Q165stop mutation predicts a translation stop within this motif. (C) Lethal G388D mutation lies within a sequence similar to the Walker A motif (bracket below) that is conserved among proteins that bind and hydrolyze ATP. (D) Viable but amplification defective M676K mutation lies within a region of low conservation between Drosophila and other MCM6 proteins. Not shown is M1I, which mutates the putative initiator methionine. Conserved regions are boxed. Dark shading indicates identical residues. Light shading indicates conservative substitutions. No shading indicates residues that are not conserved. D.m., Drosophila melanogaster; H.s., Homo sapiens; M.m., Mus musculus; X.l., Xenopus laevis; S.p., Schizosaccharomyces pombe; S.c., S. cerevisiae.
MCM6 Associates with Other MCM Subunits in Follicle Cells during Amplification
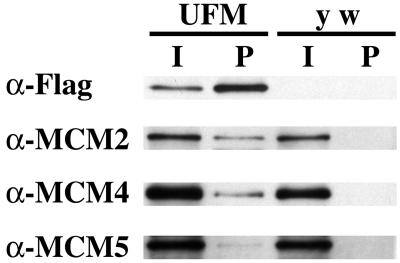
MCM proteins assemble into a hexameric complex, and all six subunits are essential for DNA replication in yeast (reviewed by Tye, 1999b). The requirement for MCM6, therefore, suggested that other MCM proteins may participate in amplification. To address this, we used a second rescue construct based on the two part GAL4/UAS system to ask whether MCM6 associates with other MCM proteins during amplification (Brand and Perrimon, 1993). We transformed flies with an MCM6 cDNA tagged with the FLAG epitope and under control of the GAL4-responsive UAS promoter (UAS:FL:MCM6). When combined with the c323 GAL4 enhancer trap line, FLAG:MCM6 was produced only in stage 10A-14 follicle cells of the ovary, the postendocycle period of continuous amplification (see below; Figure 5F) (Calvi et al., 1998). Extracts made from these ovaries, and those lacking the transgene, were immunoprecipitated with anti-FLAG antibodies, and Western blots were incubated with antibodies available for three other MCM subunits, MCM2, 4, and 5 (Su et al., 1996). This gave evidence for enrichment of these other MCM proteins in the pellet from ovaries expressing FLAG:MCM6, but not in the y w strain, which lacks the transgene (Figure 4). This indicates that precipitation of MCM 2, 4, and 5 is dependent on FLAG:MCM6, and not due to nonspecific associations with the FLAG antibody or beads. Although the ratio of pellet/input is low for MCM 2, 4, and 5, it should be noted that the input represents protein from all cells in the ovary, whereas the pellet represents the protein precipitated by FLAG:MCM6 from only a minority of those cells undergoing amplification. Moreover, these cells also contained untagged, wild-type MCM6, which competes with FLAG:MCM6 for binding to the other subunits. It is also important to note that the ratio of pellet/input for MCM 2, 4, and 5 is relatively lower than that for FLAG:MCM6, in part, because this epitope-tagged protein was produced in stoichiometric excess over the other MCM subunits. These results suggest, therefore, that MCM6 physically associates with at least three other subunits of the MCM complex at a time when chorion origins are rereplicating.
Figure 5.
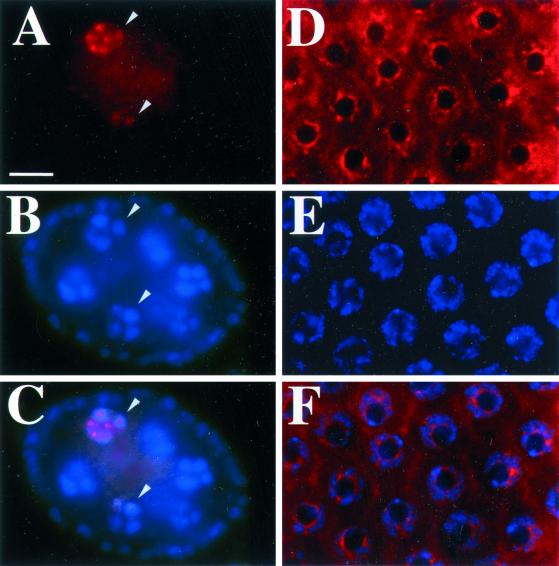
MCM6 is chromatin associated but not visibly concentrated at chorion loci. (A-C) Stage 4 egg chamber from the Ub:FL:MCM6 strain stained with DAPI (B) and anti-FLAG antibody (A). DAPI staining in B shows four of the 15 nurse cells in the central part of the egg chamber that have condensed polytene chromosomes at this stage. The smaller nuclei on the periphery are within follicle cells. Anti-FLAG staining in A and merged image in C shows that MCM6 is associated with chromosomes in only two of these nuclei (arrowheads) consistent with asynchronous DNA replication in these egg chambers. (D and F) Anti-FLAG staining of stage 10B follicle cells from the UAS:FL:MCM6 strain does not reveal focal staining corresponding to chorion loci. (D) Anti-FLAG (E) DAPI staining (F) merged image. The intense DAPI spots in E correspond to the heterochromatic chromocenter. Bar, 10 μm.
Figure 4.
MCM6 associates with MCM2, 4, and 5 during amplification. Western blot of immunoprecipitation with anti-FLAG antibody by using extracts from ovaries of c323GAL4; UAS:FL:MCM6 (UFM), which expresses FLAG:MCM6 specifically in follicle cells during amplification, or the transformation host lacking the transgene (y w). I = 1/500 of input from total extract, p = 1/10 pellet. Duplicate blots were probed with antibodies against FLAG, MCM2, MCM4, and MCM5 giving evidence for coimmunoprecipitation of FLAG:MCM6 with MCM2 and 4, and, minimally, with MCM5. Note that the input for MCM2, 4, and 5 is from all cells in the ovary, whereas the pellet represents protein precipitated with FLAG:MCM6 expressed only in amplifying follicle cells. Equal amounts of y w and UFM were loaded based on measurement of total protein in the input extract.
MCM6 Association with Chromatin Is Cell Cycle Regulated but Is not Visibly Concentrated at Chorion Loci
With immunofluorescence, a number of replication proteins are visibly concentrated at chorion loci during amplification. We therefore asked whether MCM6 is concentrated at chorion loci by labeling follicle cells from the c323GAL4; UAS:FL:MCM6 strain with anti-FLAG antibody. Nuclear labeling appeared beginning in stage 10A/B and continued until stage 13. Although this expression rescues amplification in MCM6K1214, staining with anti-FLAG antibody did not reveal subnuclear foci corresponding to amplifying chorion genes (Figure 5, D-F). In fact, most of the labeling did not coincide with chromatin as indicated by DAPI counterstaining (Figure 5E). To ask whether MCM6 associates with chromatin at other times, we labeled ovaries containing the Ub:FL:MCM6 transgene, which expresses in all germline and somatic cells of the ovary. The 15 germline nurse cells in a stage 4 egg chamber have pseudopolytene chromosomes, but undergo replication at different times relative to one another (Dej and Spradling, 1999). Some nurse cell nuclei in a stage 4 chamber had FLAG:MCM6 associated with chromatin, whereas others did not (Figure 5, A-C). This suggests that MCM6, like other MCMs, cycles on and off chromatin (Su and O'Farrell, 1997, 1998; reviewed by Tye, 1999a). Treatment of nuclei with Triton-X or high salt reduced nucleoplasmic staining and enhanced detection of periodic association of MCM6 with chromatin, but did not reveal focal staining at chorion loci during amplification in stage 10B (our unpublished results). The absence of focal staining was not unique to FLAG:MCM6, because labeling with antibodies against MCM2, 4, and 5 also appeared distributed throughout follicle cell nuclei and not concentrated at chorion foci (our unpublished results) (Royzman et al., 1999). These results indicate that MCM6, like other members of the MCM complex, associates with chromatin periodically during cell cycles but is not visibly concentrated at chorion loci during amplification.
MCM6 Is Required for Cell Cycles during Development
Because of their central role in origin function, all MCM genes are essential for viability in yeast. Mutations in two other MCM family members in the fly, MCM2 and discs proliferation abnormal (MCM4), result in lethality before adulthood (Feger et al., 1995; Treisman et al., 1995). The MCM6K1214/MCM6K1214 and MCM6K1214/Df females had a severe defect in amplification, but were otherwise viable and normal. To ask whether MCM6 is required during development, we screened for new lethal mutations in the gene after treating flies with EMS (see MATERIALS AND METHODS). We recovered four mutations that were fully viable in trans to MCM6K1214, but failed to complement the thin eggshell phenotype, indicating that they were alleles of MCM6 (Table 1). These new alleles failed to complement each other and resulted in complete lethality before adulthood, which was rescued by MCM6 transgenes (Table 1). Analysis of the lethal phase for different allele combinations indicated that most mutant offspring survived through embryogenesis. Similar to mutations in MCM2, MCM4, and other cell cycle genes, up to 50% of the mutant class survived until metamorphosis but died as pupae that lacked any sign of adult structures (n > 200 expected mutant class). The lethal phase was similar when these EMS alleles were in trans to MCM6 deletions or the Y chromosome, indicating that they are close to null.
Table 1.
Complementation phenotypes of MCM6 alleles
| Allelea | MCM6K1214 | MCM62 | MCM63 | MCM64 | MCM65 | Df(6C)c |
|---|---|---|---|---|---|---|
| MCM6K1214 (M676K) | fs-teb | fs-te | fs-te | fs-te | fs-te | fs-te |
| MCM62 (M1I) | l | l | l | l | l | |
| MCM63 (Q165stop) | l | l | l | l | ||
| MCM64 (T157M) | l | l | l | |||
| MCM65 (G388D) | l | l | ||||
| Df(6C) | l |
Allele names are shown with amino acid position and change shown in parentheses (see Figure 5).
fs-te, females sterile and laid eggs with thin eggshells; l, lethal before adulthood.
Similar results were obtained when the mutations were in trans to either noncomplementing deletion or the Y chromosome in males.
Examination of 3rd instar larvae with lethal mutations in MCM6 indicated that they were 50% the size of their wild-type siblings. Because most of larval growth is due to an increase in cell size associated with endoreplication, this suggests that MCM6 is required for the endocycle. The cells of the imaginal disk and brain, however, proliferate during larval life by a standard mitotic division cycle. MCM6 mutant larvae had no identifiable imaginal discs, and brain lobes that were reduced to 50% of the diameter of wild type (Figure 6, A and B). This suggests that MCM6 is required during mitotic division cycles. Like MCM2 and MCM4, abundant MCM6 mRNA is loaded into the early embryo from the mother (Feger et al., 1995; Ohno et al., 1998). It is likely, therefore, that MCM6 is required early in development, but that it is only after these maternal stores become depleted that an essential function in cell proliferation becomes evident as a defect in metamorphosis.
Figure 6.
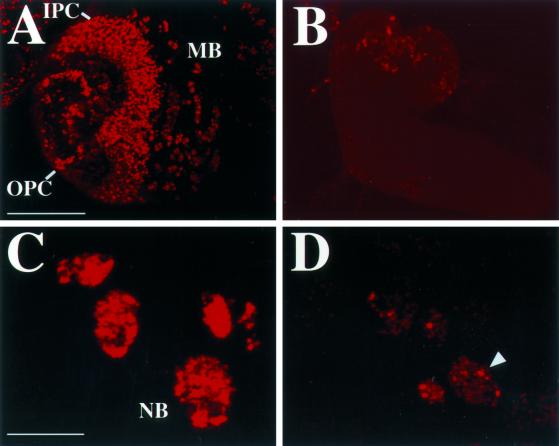
MCM6 mutant brains have a severe reduction in DNA replication and cell proliferation. (A) Brain hemisphere from a wild-type late 3rd instar larva labeled with BrdU (red) to detect proliferating cells in S phase. Labeling reveals numerous cells in S phase in the outer proliferation center (OPC), inner proliferation center (IPC), and midbrain (MB). (B) Brain from an MCM6 mutant larva of the same stage is greatly reduced in size and has few cells that detectably incorporate BrdU. A and B are the same magnification and exposure. Bar (A and B), 100 μm. (C) Cluster of cells from the midbrain of a wild-type larva that are in S phase has robust BrdU incorporation in the nucleus. These represent the nuclei of large neuroblast stem cells (NB) and its smaller descendants. (D) Nuclei from MCM6 mutant cells have greatly reduced incorporation of BrdU into small foci. The larger foci (one indicated by arrowhead) represent heterochromatin that replicates late in S phase. Bar (C and D), 10 μm.
To analyze cell proliferation directly, we incubated brains from wandering 3rd instar larvae in BrdU and detected incorporation with anti-BrdU antibodies. In wild-type brains, the inner and outer proliferation centers contained several hundred cells that were in S phase and stained positively for BrdU (Figure 6A). In the midbrain and ventral ganglion, isolated groups of cells were positive for BrdU, which represent neuroblast stem cells and their descendants (Figure 6C). In contrast, for all four MCM6 lethal alleles, BrdU incorporation was greatly reduced. Mutant brains had <100 cells that labeled weakly with BrdU, and distinct proliferation centers were not apparent (n = 20 brains) (Figure 6B). Nuclear BrdU incorporation in the mutant was punctate, appearing as 100–300 small foci (Figure 6D). One focus corresponded to the heterochromatic chromocenter as evidenced by colocalization with bright DAPI counterstaining, a pattern similar to that normally seen late in S phase (our unpublished results) (Figure 6D). These results indicate that these mutations in MCM6 greatly diminish DNA replication and cause severe defects in cell proliferation.
To gain insight into the nature of the mutations, we sequenced the coding region of MCM6 from the mutants and compared it with the parental strain used in the mutagenesis. The wild-type sequence was identical to other sequences that have been reported for fly MCM6, except for silent polymorphisms in the 3rd position of some codons (Feger, 1999). This predicts a protein of 817 amino acids (Figure 7A). The central part of the protein contains the MCM box (amino acids 379–531), which is the most highly conserved region among different MCM proteins in D. melanogaster and other species. Within this region are sequences highly similar to the Walker A and B motifs, predicting a role for MCM6 in ATP binding and hydrolysis (Walker et al., 1982). In the amino terminus of the MCM6 protein (amino acids 152–179) there is a putative noncanonical C4 Zinc finger motif that is conserved among MCM2, 4, 6, and 7 family members (Ohno et al., 1998; Feger, 1999; reviewed by Tye, 1999a).
Sequence of two of the lethal alleles predicts that they severely hamper MCM translation. MCM62 mutates the start methionine to an isoleucine (M1I) (Figure 7A). Translation beginning at the next ATG seven nucleotides downstream would result in an aberrant eight amino acid peptide that is out of frame with MCM6. MCM63 changes a glutamine to a stop codon before the MCM box at position 165 (Q165stop) (Figure 7, A and B). The other two lethal alleles contained missense mutations that are potentially more informative about MCM6 protein function. MCM64 substitutes a methionine at position 157 for the normal threonine (T157M) (Figure 7, A and B). This residue is two amino acids carboxy-terminal to the first C pair of the putative C4 Zinc finger. MCM65 contains a missense mutation within the Walker A box that changes a glycine at position 388 to an aspartate (G388D) (Figure 7, A-C). The mutated glycine is perfectly conserved among MCM6 homologs and other proteins that contain this subtype of Walker A box, including proteins as distant as prokaryotic transcription factors that are known to hydrolyze ATP and promote opening of DNA at promoters (Koonin, 1993). The recovery of this lethal mutation in a random mutagenesis of MCM6 strongly suggests that ATP binding, and perhaps hydrolysis, is essential for MCM6 function.
DISCUSSION
Chorion gene amplification requires proteins that are essential for G1/S progression and origin firing and permits a genetic and molecular dissection of origin regulation in vivo. We have shown that fly MCM6 is required for amplification and is essential for cell cycles in earlier development. Based on our evidence it is likely that MCM6 and additional MCM family members associate with chorion origins to form an amplification complex (AC) that resembles the pre-RC. This is significant because it suggests that a continued investigation into the regulation and binding of this complex to chorion origins should reveal cell cycle mechanisms that control chromosome duplication in Metazoa.
Role for MCMs in Chromosome Duplication and Amplification
Our evidence indicates that MCM6 is required for mitotic cycles, endocycles, and the special S phase associated with the amplification of chorion genes. The full biochemical picture for DNA unwinding at chromosomal origins and replication forks is far from complete (reviewed by Lei and Tye, 2001). The evidence to date, however, indicates that the MCM complex has a role in both of these processes. Mutations in MCM5 that bypass the essential function of CDC7 kinase have premature origin unwinding suggesting a role for the MCM complex in this activity (Hardy et al., 1997; Geraghty et al., 2000). Consistent with a role in elongation, ChIP experiments in yeast suggest that MCMs are found at origins in G1, and travel bidirectionally outward from origins during S phase (Aparicio et al., 1997; Tanaka et al., 1997). Moreover, all six MCMs are required continuously during S phase (Labib et al., 2000). There have been numerous reports of MCM subcomplexes in vivo, and some of these have been shown to have helicase, single-stranded DNA binding, and ATPase activity in vitro (Su et al., 1996; Ishimi, 1997; Kelman et al., 1999; You et al., 1999; Chong et al., 2000; Lee and Hurwitz, 2000). An MCM 4/6/7 subcomplex with these activities appears to be particularly stable in vivo and has robust helicase activity in vitro (Ishimi, 1997; Lee and Hurwitz, 2001). We found that MCM6 associates with at least MCM 2, 4, and 5 in follicle cells during amplification, suggesting that multiple MCM subunits may have a role in origin unwinding or fork elongation at chorion loci. The mutation of the conserved glycine in the Walker A box of MCM6 resulted in lethality early in development, implicating binding, and perhaps hydrolysis, of ATP as an essential function of this subunit. This glycine is perfectly conserved among all MCM genes in flies and other organisms, and in proteins as distant as the NtrC class of prokaryotic transcription factors that are known to have ATPase activity and melt DNA at promoters (Koonin, 1993). This supports our suggestion that an essential function of MCM6 in DNA replication is mediated by ATP. Because we did not examine the mutant protein directly, however, it is also possible that the mutation has other effects on protein structure or stability.
The T157M mutation lies within the paired cysteines of the putative C4 Zinc finger that is conserved in MCM2, 4, 6, and 7. It is currently unclear whether the putative Zinc fingers mediate DNA binding or, as has been shown for other Zinc fingers, protein–protein association (reviewed by Wolfe et al., 2000). Like other MCM family members, we found that FLAG:MCM6 cycles on and off chromatin, suggesting that the critical function of MCM6 involves chromatin association during chromosome duplication and amplification. Given the abundance of MCM subunits and subcomplexes in the cell, it is possible that MCM6 has other functions that have yet to be revealed. Recently, MCM3 and MCM5 have been implicated in transcriptional control in mammalian cells (DaFonseca et al., 2001).
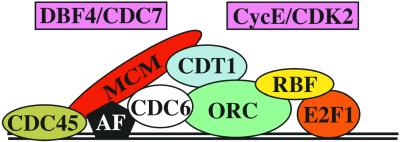
The phenotype of MCM6K1214 indicates that at least this MCM family member is required for amplification of chorion genes. Previous experiments indicated that ORC2 binds to the regions at the 3rd chromosome chorion locus that are essential for amplification (Austin et al., 1999). Although we have not shown binding of the MCM complex directly to chorion origins, the requirement for MCM6, together with previous evidence for involvement of other pre-RC subunits in amplification (Asano and Wharton, 1999; Austin et al., 1999; Royzman et al., 1999; Loebel et al., 2000; Whittaker et al., 2000), suggests that MCM6 is part of an AC that is similar to the pre-RC at other origins (Figure 8). Given that a heteromeric MCM complex is essential for replication, and our finding that MCM6 coimmunoprecipitates with other MCMs during amplification, it seems likely that the AC will contain a complex of MCM subunits. A further analogy with the pre-RC is suggested by recent evidence that E2F1 and RBF may contact ORC at chorion origins, and the finding that Rb is localized to replication foci in mammalian cells (Kennedy et al., 2000; Bosco et al., 2001). The regulation of this proposed AC is similar to that of the pre-RC in that there is evidence that two kinase complexes required for normal S phase, CDK2/Cyclin E and DBF4/CDC7, are also required for the activation of chorion origins (Calvi et al., 1998; Landis and Tower, 1999).
Figure 8.
Schematic model for the composition and regulation of the Amplification Complex (AC). Based on our results and those of previous investigations, it is likely that an AC resembling the pre-RC assembles onto chorion origins. Current evidence suggests that the AC contains most of the components that comprise pre-RCs at other origins (colored ovals), and responds to S-phase kinases (boxes). CDC6 (white oval) is the only known pre-RC component that has not been linked to amplification. Unlike the pre-RC, the AC may contain one or more amplification factors (AF) (black pentagon) that permit rereplication in the presence of constitutively high CDK activity. Recent evidence suggests that E2F1 and RBF1 participate directly in the regulation of chorion and other origins. See text for references.
There are few well-characterized origins of DNA replication. The potential to genetically and molecularly characterize the assembly of the AC onto specific DNA sequences at chorion origins provides an opportunity to explore origin structure and regulation in Metazoa. The obvious distinction from most origins is that chorion origins rereplicate. The previous finding that Cyclin E is constitutively high in follicle cell nuclei during amplification led us to suggest that the AC may contain an amplification factor (AF) (Figure 8) (Calvi et al., 1998). This proposed AF would allow the AC to locally escape rereplication inhibition that is exerted by Cyclin E/CDK2 on other origins in follicle cell nuclei. During chromosome duplication, the assembly of the pre-RC culminates with the binding of the MCM complex, which is essential for origin licensing. Uncovering the regulation of MCM association with chorion origins may be key to understanding this rereplication phenomenon, and should provide insight into how origins normally initiate replication only once per cell cycle.
Why does the MCM6K1214 allele have severe defects in amplification but no detectable impairment in earlier cell cycles? Sequence of this allele revealed a missense mutation that changes the methionine at position 676 to a lysine (M676K), in a region of low conservation that is carboxy terminal to the MCM box. One copy of this mutant allele is sufficient to support normal development, yet two copies are insufficient for full chorion gene amplification. It may be that M676K is revealing a special role for this region of the MCM6 protein in amplification. Alternatively, this mutation may slightly reduce function in all cell cycles, and chorion gene amplification may require an overall higher level of MCM6 function than does earlier development. In support of this latter suggestion, there are missense mutations in many other essential cell cycle genes that result in severe amplification defects, but no apparent defect earlier in development (reviewed by Calvi and Spradling, 1999). We therefore favor the interpretation that the restricted temporal window for repeated replication of chorion genes makes defects in amplification an extremely sensitive phenotype for slight reductions in function of genes essential for S phase. Thus, the thin eggshell phenotype is the fly analogue of the mini-chromosome maintenance assay in yeast that led to the initial identification of MCM genes (Maine et al., 1984).
Differential Replication during Oogenesis
BrdU labeling of MCM6K1214 revealed that dorsal-anterior and posterior cells most often had detectable incorporation of BrdU at chorion and elsewhere in the genome in stages 10B-13. These two groups of cells receive the highest levels of patterning signals from the underlying oocyte, suggesting that pathways that determine dorsal-ventral and anterior-posterior polarity intersect with DNA replication activity. This remains an untested hypothesis because we did not examine BrdU labeling in double mutants for patterning genes and MCM6K1214. The integration of axis patterning with amplification makes sense, however, given that follicle cells in the dorsal-anterior and posterior make specialized chorion structures. Follicle cells in the dorsal anterior of the egg chamber express high levels of several Zinc finger transcription factors from the Broad-Complex (Tzolovsky et al., 1999). These genes are required for dorsal appendage formation, and misexpression alters the normal transition from endocycles to chorion gene amplification during stage 10. It is possible that these transcription factors play a role in augmenting amplification in response to signals from the oocyte.
In MCM6K1214 we observed extra BrdU incorporation in stage 10B, but measurement of DNA content indicates that this does not represent substantial replication beyond the normal 16C arrest. It is therefore unclear whether mutation of MCM6 results in a small amount of extra replication, or a minor delay in completion of the last endocycle. Little is known regarding what controls the arrest of genomic replication and onset of continuous amplification in stage 10. Recent evidence suggests that fly RBF1, a gene related to human retinoblastoma protein, is required for amplification and to inhibit replication of other genomic regions in stage 10B (Bosco et al., 2001). Earlier during stage 6, it appears that the Notch pathway, which is required for A/P and D/V patterning, is involved in the transition of follicle cells from mitotic cycles to endocycles (Lopez-Schier and St Johnston, 2001). Interpreting what the extra BrdU labeling in MCM6K1214 indicates about the developmental and cell cycle control of DNA replication awaits further insights into the mechanisms that control origin activity during endocycles and amplification.
ACKNOWLEDGMENTS
We thank J. Sekelsky for FLAG epitope plasmid pBUF, T.T. Su for MCM antibodies, and the Bloomington Drosophila Stock Center for providing fly strains. Thanks to Jennifer Bandura for help with embryo injections. Thanks also to S. Dinardo, A.-K. Bielinsky, and anonymous reviewers for helpful comments on the manuscript. This research was supported by an ACS-IRG pilot grant 78-002-22 and Public Health Service grant R01GM61290-01 to B.R.C.
Footnotes
DOI:10.1091/mbc.01–08-0400.
REFERENCES
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Aparicio O, Weinstein D, Bell S. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Asano M, Wharton RP. E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J. 1999;18:2435–2448. doi: 10.1093/emboj/18.9.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RJ, Orr-Weaver TL, Bell SP. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bielinsky A-K, Blitzblau H, Beall EL, Ezrokhi M, Smith HS, Botchan MR, Gerbi SA. Origin recognition complex binding to a metazoan replication origin. Curr Biol. 2001;11:1427–1431. doi: 10.1016/s0960-9822(01)00444-4. [DOI] [PubMed] [Google Scholar]
- Bielinsky AK, Gerbi SA. Where it all starts: eukaryotic origins of DNA replication. J Cell Sci. 2001;114:643–651. doi: 10.1242/jcs.114.4.643. [DOI] [PubMed] [Google Scholar]
- Bogan JA, Natale DA, DePamphilis ML. Initiation of eukaryotic DNA replication: conservative or liberal? J Cell Physiol. 2000;184:139–150. doi: 10.1002/1097-4652(200008)184:2<139::AID-JCP1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Bosco G, Du W, Orr-Weaver TL. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat Cell Biol. 2001;3:289–295. doi: 10.1038/35060086. [DOI] [PubMed] [Google Scholar]
- Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes Dev. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi BR, Spradling AC. Chorion gene amplification in Drosophila: a model for metazoan origins of DNA replication and S-phase control. Methods. 1999;18:407–417. doi: 10.1006/meth.1999.0799. [DOI] [PubMed] [Google Scholar]
- Calvi BR, Spradling AC. The nuclear location and chromatin organization of active chorion amplification origins. Chromosoma. 2001;110:159–172. doi: 10.1007/s004120100131. [DOI] [PubMed] [Google Scholar]
- Carminati JL, Orr-Weaver TL. Changes in DNA replication during animal development. In: DePamphilis ML, editor. DNA Replication in Eukaryotic Cells. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1996. pp. 409–434. [Google Scholar]
- Carpenter P, Mueller P, Dunphy W. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- Casso D, Ramirez-Weber F, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech Dev. 2000;91:451–454. doi: 10.1016/s0925-4773(00)00248-3. [DOI] [PubMed] [Google Scholar]
- Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc Natl Acad Sci USA. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Mahbubani H, Khoo C, Blow J. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Cocker J, Piatti S, Santocanale C, Nasmyth K, Diffley J. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Coleman T, Carpenter P, Dunphy W. The Xenopus Cdc6 protein is essential for the intiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Coué M, Kearsey S, Méchali M. Chromatin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J. 1996;15:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- DaFonseca CJ, Shu F, Zhang JJ. Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-gamma. Proc Natl Acad Sci USA. 2001;98:3034–3039. doi: 10.1073/pnas.061487598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann C, Diffley JFX, Nasmyth KA. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- de Cicco D, Spradling A. Localization of a cis-acting element responsible for the developmentally regulated amplification of Drosophila chorion genes. Cell. 1984;38:45–54. doi: 10.1016/0092-8674(84)90525-7. [DOI] [PubMed] [Google Scholar]
- Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126:293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- Delidakis C, Kafatos F. Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J. 1989;8:891–901. doi: 10.1002/j.1460-2075.1989.tb03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF. DNA replication: building the perfect switch. Curr Biol. 2001;11:367–370. doi: 10.1016/s0960-9822(01)00196-8. [DOI] [PubMed] [Google Scholar]
- Diffley J, Cocker J. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Diffley J, Cocker J, Dowell S, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feger G. Identification and complete cDNA sequence of the missing Drosophila MCMs: DmMCM3, DmMCM6 and DmMCM7. Gene. 1999;227:149–155. doi: 10.1016/s0378-1119(98)00596-4. [DOI] [PubMed] [Google Scholar]
- Feger G, Vaessin H, Su T, Wolff E, Jan L, Jan Y. dpa, a member of the MCM family, is required for mitotic DNA replication but not endoreplication in Drosophila. EMBO J. 1995;14:5387–5398. doi: 10.1002/j.1460-2075.1995.tb00223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty DS, Ding M, Heintz NH, Pederson DS. Premature structural changes at replication origins in a yeast minichromosome maintenance (MCM) mutant. J Biol Chem. 2000;275:18011–18021. doi: 10.1074/jbc.M909787199. [DOI] [PubMed] [Google Scholar]
- Hardy CF. Identification of Cdc45p, an essential factor required for DNA replication. Gene. 1997;187:239–246. doi: 10.1016/s0378-1119(96)00761-5. [DOI] [PubMed] [Google Scholar]
- Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1999. [Google Scholar]
- Heck M, Spradling A. Multiple replication origins are used during Drosophila chorion gene amplification. J Cell Biol. 1990;110:903–914. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengstschlager M, Braun K, Soucek T, Miloloza A, Hengstschlager-Ottnad E. Cyclin-dependent kinases at the G1-S transition of the mammalian cell cycle. Mutat Res. 1999;436:1–9. doi: 10.1016/s1383-5742(98)00022-2. [DOI] [PubMed] [Google Scholar]
- Hopwood B, Dalton S. Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc Natl Acad Sci USA. 1996;93:12309–12314. doi: 10.1073/pnas.93.22.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Brown GW. Regulation of chromosome replication. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- Kelman Z, Lee JK, Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc Natl Acad Sci USA. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Barbie DA, Classon M, Dyson N, Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komitopoulou K, Gans M, Margaritis L, Kafatos F, Masson M. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster with special attention to eggshell mutants. Genetics. 1983;105:897–920. doi: 10.1093/genetics/105.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T, Musahl C, Laskey RA, Knippers R. Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J Cell Sci. 1996;109:309–318. doi: 10.1242/jcs.109.2.309. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Landis G, Kelley R, Spradling A, Tower J. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc Natl Acad Sci USA. 1997;94:3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G, Tower J. The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development. 1999;126:4281–4293. doi: 10.1242/dev.126.19.4281. [DOI] [PubMed] [Google Scholar]
- Lee JK, Hurwitz J. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J Biol Chem. 2000;275:18871–18878. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- Lee JK, Hurwitz J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc Natl Acad Sci USA. 2001;98:54–59. doi: 10.1073/pnas.98.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Tye BK. Initiating DNA synthesis. from recruiting to activating the MCM complex. J Cell Sci. 2001;114:1447–1454. doi: 10.1242/jcs.114.8.1447. [DOI] [PubMed] [Google Scholar]
- Li CJ, Bogan JA, Natale DA, DePamphilis ML. Selective activation of pre-replication complexes in vitro at specific sites in mammalian nuclei. J Cell Sci. 2000;113:887–898. doi: 10.1242/jcs.113.5.887. [DOI] [PubMed] [Google Scholar]
- Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Lilly M, Spradling A. The Drosophila endocycle is controlled by cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 1996;10:2514–2526. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- Loebel D, Huikeshoven H, Cotterill S. Localization of the DmCdc45 DNA replication factor in the mitotic cycle and during chorion gene amplification. Nucleic Acids Res. 2000;28:3897–3903. doi: 10.1093/nar/28.20.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Schier H, St Johnston D. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 2001;15:1393–1405. doi: 10.1101/gad.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zhang H, Tower J. Functionally distinct, sequence-specific replicator and origin elements are required for Drosophila chorion gene amplification. Genes Dev. 2001;15:134–146. doi: 10.1101/gad.822101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GT, Sinha P, Tye BK. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B. Replicator dominance in a eukaryotic chromosome. EMBO J. 1994;13:3395–3400. doi: 10.1002/j.1460-2075.1994.tb06642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale DA, Li CJ, Sun WH, DePamphilis ML. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J. 2000;19:2728–2738. doi: 10.1093/emboj/19.11.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Noton E, Diffley JF. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- Ohno K, Hirose F, Inoue YH, Takisawa H, Mimura S, Hashimoto Y, Kiyono T, Nishida Y, Matsukage A. cDNA cloning and expression during development of Drosophila melanogaster MCM3, MCM6 and MCM7. Gene. 1998;217:177–185. doi: 10.1016/s0378-1119(98)00358-8. [DOI] [PubMed] [Google Scholar]
- Orr W, Komitopoulou K, Kafatos F. Mutants suppressing in trans chorion gene amplification in Drosophila. Proc Natl Acad Sci USA. 1984;81:3773–3777. doi: 10.1073/pnas.81.12.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. Drosophila chorion genes: cracking the eggshell's secrets. Bioessays. 1991;13:97–105. doi: 10.1002/bies.950130302. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine M, Rowles A, Blow J, Laskey R. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rorth P, et al. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong J, Brown L, Howell M, Evan G, Blow J. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Royzman I, Austin RJ, Bosco G, Bell SP, Orr-Weaver TL. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 1999;13:827–840. doi: 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. ORC binding, gene amplification, and the nature of metazoan replication origins. Genes Dev. 1999;13:2619–2623. doi: 10.1101/gad.13.20.2619. [DOI] [PubMed] [Google Scholar]
- Spradling A, Mahowald A. Amplification of genes for chorion proteins during oogenesis in Drosophila melanogaster. Proc Natl Acad Sci USA. 1980;77:1096–1100. doi: 10.1073/pnas.77.2.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Feger G, O'Farrell P. Drosophila MCM protein complexes. Mol Biol Cell. 1996;7:319–329. doi: 10.1091/mbc.7.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, O'Farrell P. Chromosome association of minichromosome maintenance proteins in Drosophila mitotic cycles. J Cell Biol. 1997;139:13–21. doi: 10.1083/jcb.139.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, O'Farrell PH. Chromosome association of minichromosome maintenance proteins in Drosophila endoreplication cycles. J Cell Biol. 1998;140:451–460. doi: 10.1083/jcb.140.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Boulet AM, Lipshitz HD. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- Treisman J, Follette P, O'Farrell P, Rubin G. Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant. Genes Dev. 1995;9:1709–1715. doi: 10.1101/gad.9.14.1709. [DOI] [PubMed] [Google Scholar]
- Tye BK. MCM proteins in DNA replication. Annu Rev Biochem. 1999a;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- Tye BK. Minichromosome maintenance as a genetic assay for defects in DNA replication. Methods. 1999b;18:329–334. doi: 10.1006/meth.1999.0793. [DOI] [PubMed] [Google Scholar]
- Tzolovsky G, Deng WM, Schlitt T, Bownes M. The function of the broad-complex during Drosophila melanogaster oogenesis. Genetics. 1999;153:1371–1383. doi: 10.1093/genetics/153.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker AJ, Royzman I, Orr-Weaver TL. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev. 2000;14:1765–1776. [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Yamamoto RR, Axton JM, Yamamoto Y, Saunders RD, Glover DM, Henderson DS. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics. 2000;156:711–721. doi: 10.1093/genetics/156.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Komamura Y, Ishimi Y. Biochemical analysis of the intrinsic Mcm4-Mcm6-Mcm7 DNA helicase activity. Mol Cell Biol. 1999;19:8003–8015. doi: 10.1128/mcb.19.12.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]