SUMMARY
Numerous classes of riboswitches have been found to regulate bacterial gene expression in response to physiological cues, offering new paths to anti-bacterial drugs. As common studies of isolated riboswitches lack the functional context of the transcription machinery, we here combine single-molecule, biochemical and simulation approaches to investigate the coupling between co-transcriptional folding of the pseudoknot-structured preQ1 riboswitch and RNA polymerase (RNAP) pausing. We show that pausing at a site immediately downstream of the riboswitch requires a ligand-free pseudoknot in the nascent RNA, a precisely spaced sequence resembling the pause consensus, and electrostatic and steric interactions with the RNAP exit channel. While interactions with RNAP stabilize the native fold of the riboswitch, binding of the ligand signals RNAP release from the pause. Our results demonstrate that the nascent riboswitch and its ligand actively modulate the function of RNAP and vice versa, a paradigm likely to apply to other cellular RNA transcripts.
Keywords: Riboswitch, single-molecule FRET, RNA polymerase, pausing, co-transcriptional folding
Graphical Abstract
eTOC blurb:
Proper folding of RNA during transcription is a critical prerequisite to the many functions of RNA. Widom et al. show that the co-transcriptional folding of a riboswitch is governed by interactions with RNA polymerase that concurrently affect the speed of transcription.
INTRODUCTION
To render bacteria responsive to rapidly changing environmental conditions, gene expression is regulated by a variety of means, including mechanisms in which riboswitches modulate messenger RNA (mRNA) synthesis, translation or stability in response to physiological cues. Riboswitches, structured non-coding RNA elements typically found in 5' untranslated regions, are thought to regulate the expression of up to 4% of genes in certain bacteria (Sherwood and Henkin, 2016). A typical riboswitch contains a ligand-binding aptamer domain upstream of an expression platform whose conformation changes in response to ligand binding by the aptamer domain, thereby regulating expression of the downstream gene. The at least 40 classes of riboswitches for which ligands have been identified thus far (Greenlee et al., 2018) sense diverse ligands including metabolites (Roth et al., 2007), nucleobases (Frieda and Block, 2012), other RNAs (Zhang and Ferré-D'Amaré, 2013), and more. Due to their critical importance in bacterial gene regulation, riboswitches present attractive targets for the development of new antibiotic therapies (Deigan and Ferré-D'Amaré, 2011).
Mechanistic studies have been performed on many isolated riboswitches (Duesterberg et al., 2015; Holmstrom et al., 2014; Liberman et al., 2015; Suddala et al., 2013); however, in the cell, riboswitch folding occurs in the context of the transcription machinery. The speed of transcription by RNA polymerase (RNAP) imposes kinetic constraints on the rate of riboswitch folding, while the nascent RNA itself has the potential to modulate transcription in a variety of different ways. Most studies of co-transcriptional RNA folding utilize readouts that result from transcription itself, including SHAPE probing to detect backbone flexibility (Watters et al., 2016), and single-molecule force and fluorescence techniques (Frieda and Block, 2012; Uhm et al., 2018). When naked RNA is compared to RNA folded during active transcription, the effects of interactions with the transcription machinery are convoluted with the kinetic effects of non-equilibrium folding. As a result, potential direct effects of the presence of the DNA template and RNAP on folding remain largely unexplored.
The preQ1 riboswitch from the Gram-positive bacterium Bacillus subtilis (Bsu) is thought to act through a transcriptional mechanism in which the binding of its ligand, the tRNA nucleotide precursor 7-methylamino-7-deazaguanine (preQ1), stabilizes a pseudoknot structure that favors the formation of a terminator hairpin, decreasing expression of enzymes involved in queuosine biosynthesis (Figure 1A) (Roth et al., 2007). With a minimal aptamer containing only 36 nucleotides, it is one of the smallest known riboswitches. By investigating this riboswitch in the context of an active transcription complex, we show that a network of interactions between the nascent RNA, DNA template and RNAP enable the transcription machinery to cross-couple regulation of nascent RNA folding and transcriptional pausing. Specifically, we used singlemolecule FRET (smFRET) to probe the conformation of the nascent riboswitch at and beyond a transcriptional pause observed immediately after the aptamer domain is synthesized (U46, Figure S1), finding that the presence of RNAP stabilizes the folded conformation of the riboswitch. Additionally, we found that the U46 pause itself is stabilized by interactions between a nascent RNA pseudoknot and RNAP in conjunction with a precisely spaced sequence that resembles the pause consensus identified in vivo (Figure S1D) (Larson et al., 2014; Vvedenskaya et al., 2014), and is destabilized (released) upon ligand binding. This pause, hereafter called the "que pause", exhibits features distinct from the two previously defined mechanisms for transcriptional pausing in bacteria (Artsimovitch and Landick, 2000), leading us to term it "Class III". Our observations demonstrate an unprecedented cross-coupling between riboswitch folding and RNAP pausing that is likely to govern the co-transcriptional folding of numerous RNAs.
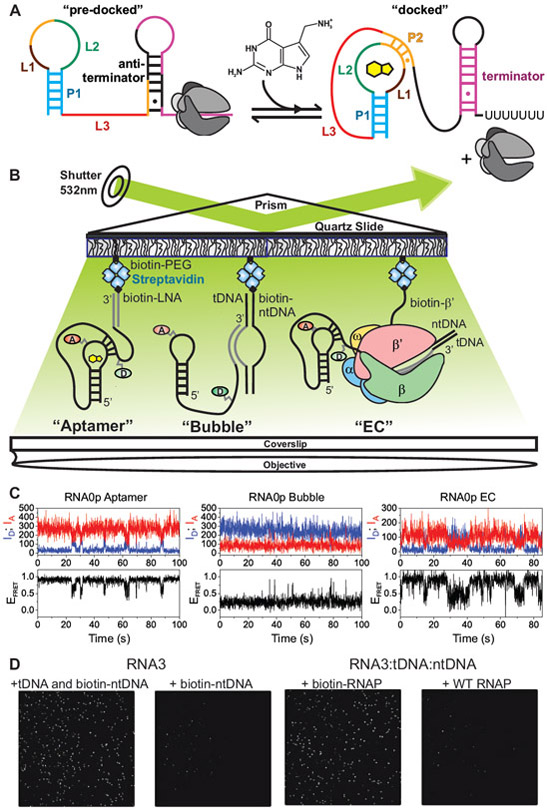
Figure 1. smFRET Investigation of the preQ1 Riboswitch in Active Transcription Complexes.
(A)Regulation of gene expression by the preQ1 riboswitch. Ligand-induced pseudoknot docking favors the formation of a terminator hairpin, leading to decreased expression of the downstream genes.
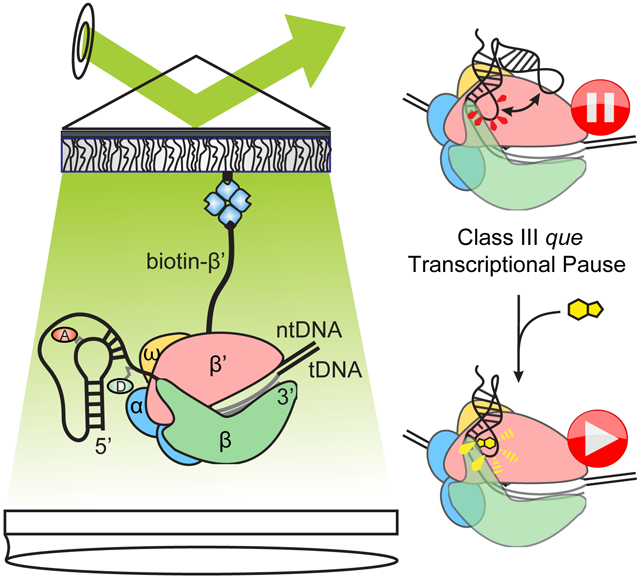
(B)smFRET experimental setup. “Aptamer” immobilization utilizes a biotinylated LNA capture probe. “Bubble” immobilization utilizes a partially complementary DNA bubble with a biotinylated nontemplate DNA. Elongation complex (“EC”) immobilization utilizes biotinylated Eco RNAP. Locations of donor (“D”, green) and acceptor (“A”, red) fluorophores are indicated.
(C)Representative smFRET traces for RNA0p under aptamer (left), bubble (middle) and EC (right) immobilization in the presence of 1 mM MgCl2 and 100 nM preQ1.
(D)iCCD images show that bubble immobilization is dependent on the tDNA in addition to the biotinylated ntDNA and the fluorophore-labeled RNA (left). Efficient EC immobilization requires biotinylated RNAP (right).
RESULTS
Direct Observation of Riboswitch Folding in Transcription Complexes
Due to the sensitivity of RNA folding to macromolecular crowding (Daher et al., 2018; Dupuis et al., 2014), ionic conditions (Suddala et al., 2015) and other factors, we hypothesized that the behavior of the preQ1 riboswitch would be significantly altered upon incorporation into a transcription elongation complex (EC) containing a DNA template and RNAP. We therefore used smFRET to investigate the effects of the transcription machinery on the folding of the nascent riboswitch. We immobilized the riboswitch on polyethylene glycol (PEG)-passivated, streptavidin-coated quartz slides for imaging via prism-based total internal reflection fluorescence (p-TIRF) microscopy. The donor fluorophore was placed at the 2' position of a guanosine residue within the L3 "tail" of the riboswitch, and the acceptor was placed on a 4-aminoallyl-uracil residue within the loop L2 (Figure 1). In an isolated RNA aptamer, the "docked" pseudoknot conformation in which the helix P2 is formed yields a high FRET efficiency (EFRET), while a mid-FRET state is observed for the "pre-docked" conformation in which P2 is not fully intact (Santner et al., 2012; Suddala et al., 2013; 2015) (Figure 1C).
We used a systematic series of immobilization strategies to study the riboswitch at varying degrees of biological complexity (Figure 1B). Immobilization through a biotinylated LNA capture probe (CP) allowed investigation of the isolated aptamer (henceforth called "aptamer immobilization"). A partially complementary DNA bubble with a biotinylated non-template (nt) strand enabled investigation of the nucleic acid framework of an EC ("bubble immobilization"). Finally, immobilization through biotinylated Escherichia coli (Eco) RNAP allowed observation of the riboswitch in a complete EC ("EC immobilization") (Daube and Hippel, 1992; Sidorenkov et al., 1998). While Eco RNAP has been found to exhibit more pronounced pausing than Bsu RNAP in some cases (Grundy and Henkin, 2004), many pauses are recognized by both RNAPs, including several that have been identified in riboswitches (Helmling et al., 2018; Lemay et al., 2011; Steinert et al., 2017). In our in vitro transcription assays, we found that while Eco and Bsu RNAPs did not yield identical results, they both recognized the que pause (Figure S1). As is frequently done, we therefore chose to use an experimental system based on the better studied Eco RNAP to identify not only specific features of preQ1 riboswitch folding in the context of a known transcription machinery, but also factors that may contribute to the folding of nascent RNA in general.
To investigate co-transcriptional folding wherein riboswitch properties and interactions with the transcription machinery may evolve as additional RNA is synthesized, we utilized RNAs containing varying amounts of the riboswitch expression platform. The shortest RNA species studied by smFRET is analogous to the species present at the que pause ("RNA0 pause", henceforth called "RNA0p"); we also investigated RNAs with an additional three (RNA3) or ten (RNA10) nucleotides of expression platform (Figure S2A). We first confirmed that our assembly procedures yielded the desired complexes and immobilization mechanisms. Efficient bubble immobilization required that the template DNA (tDNA) be present along with the fluorophore-labeled RNA and biotinylated ntDNA, indicating that the molecules we observe contain all three components. EC immobilization was efficient only with biotinylated RNAP, indicating the absence of nonspecific sticking to the slide (Figure 1D).
To determine the correct number of states with which to model our data, we fit the traces from several experimental conditions using 1-, 2- and 3-state kinetic models and computed the mean Bayesian information criterion (BIC) and modified Bayesian information criterion (BIC') (Lerner et al., 2018) across all of the traces (Table S3). Consistent with the interpretations made in earlier work on this riboswitch (Suddala et al., 2013; 2015), we found that the BIC indicated that the all of the data sets tested are best modeled with two states, and in no case did either criterion suggest more than two states. For a number of our data sets, however, the conformational fluctuations identified in the kinetic model exhibited bi-exponential kinetics. This indicates that, although a 2-state model may be the most parsimonious, those states likely include subpopulations with slight differences in structure that affect their propensity to interconvert between the global docked and pre-docked conformations, again similar to our prior observations in the isolated riboswitch (Suddala et al., 2013; 2015). In the current work, we focus on the effects of the transcription machinery on the equilibrium partitioning of population between the global docked and pre-docked states.
Addition of the DNA Template Electrostatically Disfavors Riboswitch Folding
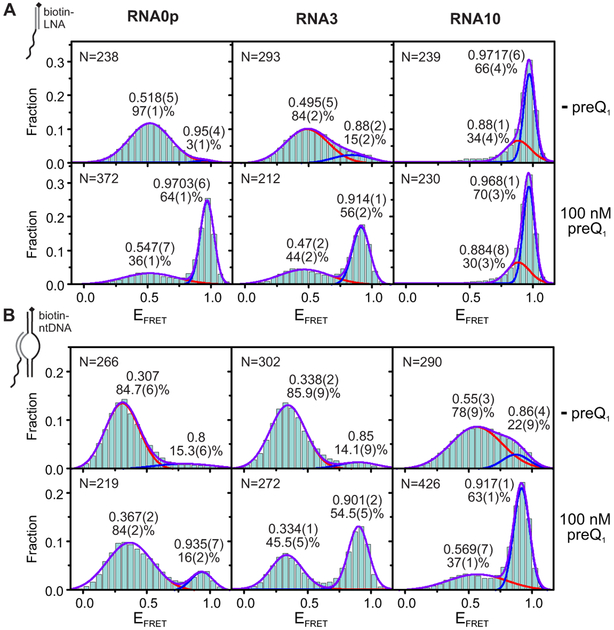
Consistent with previous studies of the isolated riboswitch aptamer (Suddala et al., 2013; 2015), we observed that under aptamer immobilization in the absence of Mg2+ or preQ1, RNA0p existed almost exclusively in the pre-docked mid-FRET state with EFRET ~ 0.5, while RNA3 also exhibited a minor fully-docked high-FRET population (15%, EFRET = 0.88) (Figure 2A). For both RNAs, addition of Mg2+ shifted the mid-FRET state to EFRET ~ 0.7, indicative of a more compact pre-docked conformation, while increasing the population of the high-FRET state (Figure 3A). Addition of preQ1 to either RNA (with or without Mg2+) shifted additional population to the high-FRET state while only slightly changing the EFRET of either state.
Figure 2. Riboswitch Folding Under Aptamer and Bubble Immobilization in the Absence of Mg2+; see also Figure S2.
Each histogram was fit with two Gaussians, and the resulting EFRET and population of each state are noted adjacent to the corresponding peak. The standard deviation of the last digit of each parameter (obtained via bootstrapping) is reported in parentheses. Values for which no standard deviation is reported were constrained during fitting. The number of traces included in each histogram ("N") is noted in the upper left corner.
(A)smFRET histograms of RNA0p (left), RNA3 (middle) and RNA10 (right) under aptamer immobilization in the absence of MgCl2 and the absence (top row) or presence (bottom row) of 100 nM preQ1.
(B)Corresponding smFRET histograms under bubble immobilization.
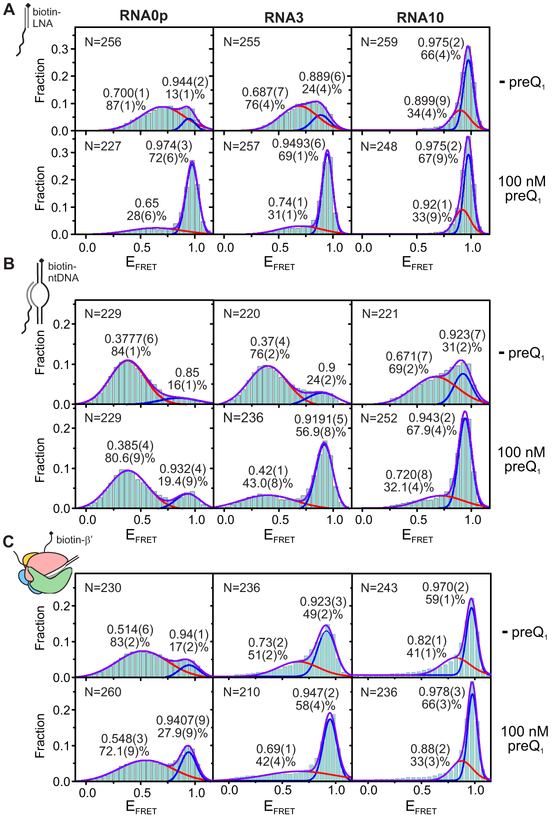
Figure 3. Riboswitch Folding Under Aptamer, Bubble and EC Immobilization in the Presence of 1 mM Mg2+; see also Figures S2 and S3.
Each histogram was fit with two Gaussians, and the resulting EFRET and population of each state are noted adjacent to the corresponding peak. The standard deviation of the last digit of each parameter (obtained via bootstrapping) is reported in parentheses. Values for which no standard deviation is reported were constrained during fitting. The number of traces included in each histogram ("N") is noted in the upper left corner.
(A)smFRET histograms of RNA0p (left), RNA3 (middle) and RNA10 (right) under aptamer immobilization in the presence of 1 mM MgCl2 and the absence (top row) or presence (bottom row) of 100 nM preQ1.
(B)Corresponding smFRET histograms under bubble immobilization.
(C)Corresponding smFRET histograms under EC immobilization.
In contrast, most RNA10 molecules occupied a very high-FRET state (EFRET ~ 0.97) regardless of Mg2+ or preQ1 concentration (Figures 2A and 3A), consistent with an alternate structure. Specifically, additional nucleotides in RNA10 are predicted to form Watson-Crick base pairs with nucleotides that make up P2 and L2 in the native conformation (Figure S2B). To test this notion, we used mutagenesis to generate a sequence whose most stable predicted fold was once again the native structure. This mutant responded to ligand, confirming that when wild-type (WT) RNA10 is heat-annealed with CP, the native aptamer fold is disfavored.
To isolate the effects of the DNA template from those of RNAP, we next probed the riboswitch as part of a bubble complex that mimics the nucleic acid scaffold of an EC. We found that addition of the DNA bubble led to a shift of population from the high-FRET to the mid-FRET state, and a shift of the mid-FRET state to lower EFRET (~ 0.35 for RNA0p and RNA3, Figure 2B). Furthermore, the Mg2+-dependent shift of the mid-FRET state to higher EFRET was also greatly diminished (Figure 3B). RNA0p is rendered nearly insensitive to preQ1 and Mg2+, but sensitivity is restored as additional bases are inserted to form RNA3 and RNA10. The effects of the bubble are relieved by increasing the K+ concentration (Figure S2C-D), indicating electrostatic repulsion by the DNA as the underlying cause for diminished pseudoknot docking. Bubble-immobilized RNA10 properly responds to Mg2+ and preQ1, in contrast to its misfolding under aptamer immobilization. These results show that the DNA template may play a role in shaping the folding free-energy landscape of nascent RNAs when in direct proximity.
Addition of RNAP to the Bubble Scaffold Restores Riboswitch Folding
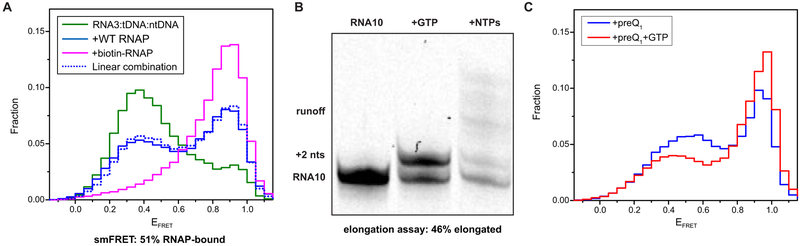
To probe the direct effects of RNAP on riboswitch folding, we assembled active ECs on a bubble scaffold identical to that described above. Immobilization of ECs through biotinylated RNAP proved to be critical to proper quantification of these effects, as experiments with the biotinylated ntDNA revealed that ~50% of bubble scaffolds were not bound by RNAP (Figure 4A). Specifically, the FRET histogram of RNA3-containing ECs immobilized through biotinylated ntDNA can be fit as a linear combination of the bubble histogram and the RNAP-immobilized EC histogram, with the EC histogram contributing 51%. Consistent with this result, addition of GTP to ECs under identical conditions resulted in an elongation efficiency of 46% (Figure 4B). On-slide addition of the substrate nucleotide (GTP) to RNA0p-containing ECs caused the histogram of RNA0p to evolve toward that of RNA3, indicating that the ECs remain active on the slide (Fig. 4C). Notably, obtaining ECs active under imaging conditions required optimization of the oxygen scavenging system (OSS) used to minimize photobleaching and blinking (Aitken et al., 2008) (Figure S3) to allow for stable binding of RNAP to the bubble scaffold and avoid nuclease degradation of the RNA.
Figure 4. Assembly and Activity of Riboswitch-Containing ECs.
(A)When RNA3-containing ECs are immobilized on a slide through biotinylated ntDNA, the FRET histogram (blue) corresponds to a linear combination (dashed blue) of the RNA3:tDNA:biotin-ntDNA histogram (green) and the RNA3:tDNA:ntDNA+biotin-RNAP histogram (purple), each contributing about 50%.
(B) A representative gel from a bubble-initiated elongation assay (the gel that was quantified is shown in Figure S3A).
(C)Incubation with GTP on the microscope slide causes the histogram of RNA0p ECs to evolve toward that of RNA3, indicating that nucleotides are being added.
Compared to bubble-immobilized complexes at the same low K+ concentration, the addition of RNAP shifted the mid-FRET state back to a higher EFRET value (~0.53 for RNA0p), and shifted a significant fraction of population back to the high-FRET state (Figures 3C and 4). Similar to bubble complexes, the closer the aptamer to the EC, the less stabilized the high-FRET state, which is evidence of steric hindrance. Surprisingly, however, the shift toward higher EFRET occurs even for ECs containing RNA0p, in which the 3' segment of the P2 helix is expected to be within the RNAP exit channel (Kang et al., 2017).
To better rationalize these observations, we explored the steric accessibility of P2 folding using MD simulations with an all-heavy-atom structure-based model built on the cryo-EM structure of an Eco RNAP EC (Kang et al., 2017), together with crystal (Klein et al., 2009) and NMR (Kang et al., 2009) structures of the preQ1 riboswitch (Figure 5). We found that while RNA0p is predicted to form fewer P2 base pairs than RNAs extended by 1-3 nucleotides (RNA1, RNA2 and RNA3), an average of 1-2 base pairs are intact (Table S4). In order for these base pairs to form, the 5' segment of P2 must "bend back" toward RNAP. This bent-back pose places the 5' leg of P2 near a cluster of basic residues in the β’ subunit and the flap-tip (FT) of the β subunit (Figures 5 and S4), conserved regions in the vicinity of the RNA exit channel (Guo et al., 2018; Kang et al., 2018).
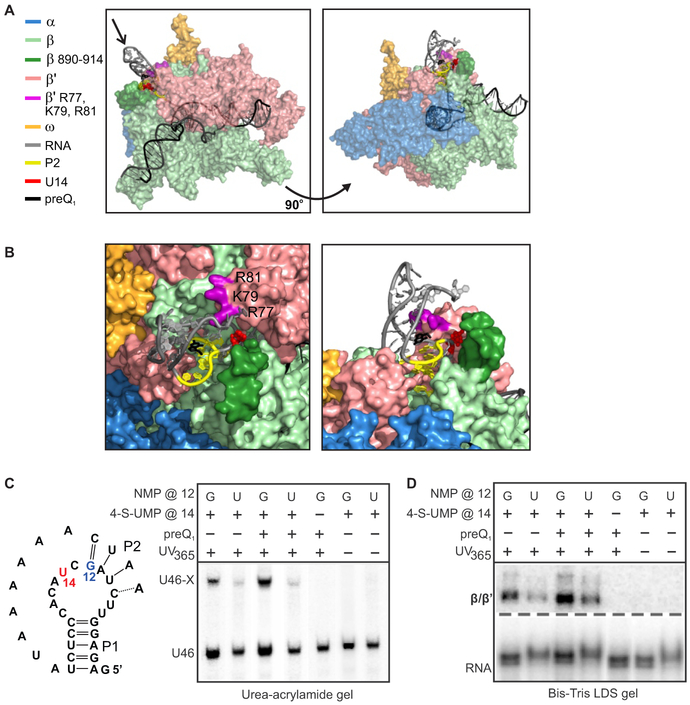
Figure 5. Simulations and Cross-Linking Experiments Show that P2 can Fold in the RNAP Exit Channel; see also Figure S4 and Table S4.
(A)An MD simulation snapshot from two angles with RNAP subunits colored as in Figure 1 and P2 colored in yellow. The β flap-tip (residues 890-914) and β' basic residues investigated later are shown in darker colors than the rest of their corresponding subunits. The model utilized a cryo-EM structure of an Eco EC (PDB ID: 6ALH), and crystal (PDB ID: 3FU2) and NMR (PDB ID: 2LIV) structures of the preQ1 riboswitch.
(B)Left: Close-up views of the simulation snapshot in (A) as seen in the direction along the arrow (left), and from the same angle as the right-hand snapshot (right).
(C)Left: schematic indicating position 12 within the aptamer, where a G-to-U mutation was investigated, and position 14, where a 4-thiouracil residue was incorporated for cross-linking. Right: Denaturing urea-acrylamide gel showing a shift of the RNA at the que pause upon cross-linking to RNAP, and controls lacking 4-thioU or UV illumination.
(D)Bis-Tris LDS gel containing the same samples as panel (C), indicating that the RNA cross-links to β or β'. Experiments with an RNAP deletion mutant confirmed that the observed cross-links are to β' (Figure S4E).
We probed for these predicted interactions through cross-linking experiments with 4-thio-UMP incorporated in or near P2. Indeed, when RNAP is stalled at the que pause site, both U10 and U14 efficiently cross-link to β', confirming that, as predicted, the 5' segment of P2 bends back to interact with RNAP (Figures 5 and S4). We found cross-linking to be dependent on the stability and accessibility of P2, being more efficient in the presence of preQ1 and less efficient in a G12-to-U mutant, in which P2 is weakened by disrupting its only G-C base pair. Interactions between the A-rich tail L3 and the minor groove of P1 may also contribute to stabilization of the cross-linking competent conformation (Feng et al., 2011; Klein et al., 2009). Together, our simulations and cross-linking results indicate that, at the que pause, residues near P2 interact with the β' subunit, potentially contributing to the stabilization of the docked conformation we observed through smFRET. Overall, our smFRET studies reveal that DNA and RNAP exert significant, opposing effects on nascent RNA folding, which are likely to play an important role in the regulation of transcription by other riboswitches.
PreQ1 Releases a Pseudoknot-Stabilized Transcriptional Pause
Our results so far are consistent with a model in which the balance between electrostatic repulsion by the DNA template and stabilizing interactions with RNAP plays a significant role in preQ1- and Mg2+-dependent nascent RNA folding. Because RNAP pausing plays a critical role in guiding the co-transcriptional folding of numerous RNAs (Pan et al., 1999; Perdrizet et al., 2012), we next used single-round transcription assays to further probe the relationship between riboswitch folding and pausing. In addition to the que pause at U46, we observed a terminator that the riboswitch is expected to regulate (U70) and a second U-tract terminator further downstream (U108) (Figures 6 and S1). Nearly 100% of complexes paused at U46, with a pause half-life of 42 seconds in the absence of preQ1 and a vast majority population (97%) with a significantly shortened half-life of 15 seconds in the presence of preQ1. In addition, we found that the raw data set for Bsu provided by Larson et al. (Larson et al., 2014) indicates that a transcriptional pause does, in fact, occur at this location in vivo (Figure S1E). Saturating concentrations of preQ1 increased termination efficiency modestly from 27% to 33%. Both effects can be fit globally with an apparent preQ1 binding affinity K1/2 of ~400 nM (Figure S5A), much weaker than the 5-10 nM for pseudoknot docking in the isolated aptamer, a reduction in apparent affinity also observed in other transcriptional riboswitches (Wickiser et al., 2005). The effect of preQ1 on termination remains modest upon the addition of transcription factors, cell extract, crowding agents, additional riboswitch RNA, and varied NTP concentrations (Figure S5B-D), suggesting that in vivo non-canonical factors may be involved in regulation of the downstream genes (Baird et al., 2010; Rinaldi et al., 2016).
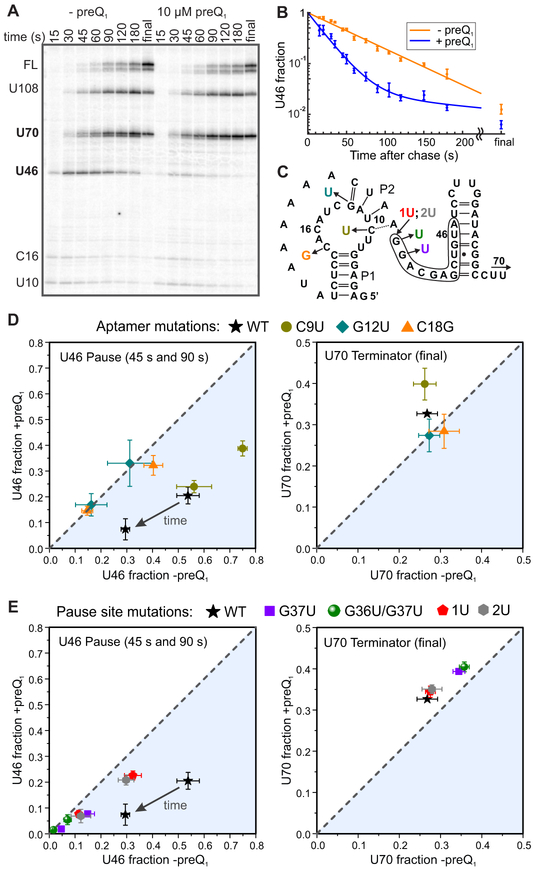
Figure 6. Effects of preQ1 on Transcriptional Pausing and Termination; see also Figures S5 and S6 and Tables S5 and S6.
All error bars represent the standard deviation of three independent replicates.
(A)Representative denaturing urea-acrylamide gel close-up showing two weak pauses within the aptamer domain (U12 and C16), the que pause immediately following the aptamer (U46), the terminator regulated by the riboswitch (U70), a U-tract arrest site (U108) and full-length RNA (FL). Here and elsewhere, all lanes and data points marked "final" were recorded after a 3-5 minute incubation with 200 μM NTPs.
(B)Fraction of complexes at the U46 site as a function of reaction time. A single-exponential fit is shown for the time course in the absence of preQ1 (orange), and a double-exponential fit is shown for the time course in the presence of 10 μM preQ1 (blue).
(C)Sequence of the aptamer and expression platform showing the final nucleotide added at the bands indicated in panel A, as well as the color-coded locations of mutations reported in panels D and E. U70 and U108 are beyond the region shown. The consensus-like pause sequence is outlined.
(D)Effects on pausing at U46 (left) and termination at U70 (right) of mutations within the aptamer domain. Data points above the dashed line indicate that addition of preQ1 increased the population of a particular species, while points below the dashed line indicate that preQ1 decreased the population of that species. PreQ1 had no effect on data points that fall on the dashed line.
(E)Effects on pausing (left) and termination (right) of mutations at the pause site.
The effect of preQ1 and the similarity of the sequence around U46 to the pause consensus suggested that the que pause has both sequence and structural determinants, so to further uncover its origins, several RNA mutations were analyzed (Figures 6C-E and S6). Altering the C18 base that forms a Watson-Crick pair with preQ1 (Roth et al., 2007) abolished the impact of preQ1 on pausing and termination, confirming that the riboswitch is responsible for the observed effects. A mutation that strengthens P2, stabilizing the docked state, (C9U, which converts a noncanonical C-A base pair to U-A) doubled the pause half-life (to 81 s), while the P2-weakening G12U mutation, which we also utilized in the cross-linking experiments presented above, shortened the pause (to 26 s) and eliminated the effect of preQ1. These aptamer mutations have smaller effects in the presence of preQ1, suggesting that the details of sequence- and structure-dependent interactions with RNAP are more critical in the less-folded apo state. As expected, mutation of key bases in the pause consensus sequence (G37U and G36U/G37U) greatly reduced pause half-life in the absence of preQ1 to 12 and 10 s, respectively. Addition of one U residue between the aptamer and the pause site (1U) also weakened pausing in the absence of preQ1 (23 s), with little effect in the presence of preQ1, and adding a second U (2U) had no significant further effect (22 s). The position of the pause is dictated by the consensus sequence rather than the riboswitch, as these insertions shift the pause site downstream by one and two nucleotides, respectively. These results indicate that the consensus sequence and strongly distance- and conformation-dependent interactions between the riboswitch and RNAP cooperate to stabilize the paused state, with ligand binding stabilizing the pseudoknot in a distinct way that facilitates pause release.
Interactions with the β Flap-Tip of RNAP Stabilize Pausing
We next tested the effects of RNAP-riboswitch contacts revealed by MD simulations. We first substituted basic β' residues (R77A, K79A and R81A, Figure 5B) that lie near the stem of the his pause hairpin in E. coli ECs (Guo et al., 2018; Kang et al., 2018) and have been implicated in the function of put antiterminator RNA structures (Sen et al., 2002). Arg77 is conserved between Eco and Bsu RNAPs, and Lys79 and Arg81 are also positively charged in Bsu RNAP. We found that individual point mutations led to modest decreases in pause half-life in the absence of preQ1, whereas a triple mutant had the most significant effect (~2-fold). In contrast, effects on pausing were negligible in the presence of preQ1 (Figure S6B). This suggests that the pause is partially stabilized by electrostatic interactions between the exit channel and the pause-promoting, ligand-free docked conformation of the riboswitch. All variants showed decreased termination at U70, with the R77A and triple mutants showing the most extreme effects, though the termination-enhancing activity of preQ1 was retained (Figure S5E and I).
We next tested the effect of the β FT, which is required for the pause-stabilizing effects of RNA duplexes (Hein et al., 2014). In contrast to the point mutants, which had effects only in the absence of preQ1, deleting the FT by removing residues K890-K914 of β (ΔFT) decreased the pause half-life by about 30% in both the presence and absence of preQ1 (Figure S6C). The effect of preQ1-dependent conformational changes on pausing was therefore maintained for ΔFT RNAP, despite an overall decrease in pause half-life. In contrast, we observed no effect of the G12U mutation on pausing by ΔFT RNAP, indicating that pausing by ΔFT is not stabilized by the pseudoknot. Together, our experiments on RNAP variants suggest that both electrostatic interactions with exit channel residues and steric interactions with the FT contribute to pseudoknot-dependent pause stabilization in the absence of preQ1. By contrast, in the preQ1-bound docked state, only steric interactions with the FT contribute to pause destabilization. These observations support the notion that the docked states in the presence and absence of preQ1 are distinct, and involve different interactions with RNAP.
DISCUSSION
We used smFRET, biochemical transcription assays and MD simulations to study the co-transcriptional folding of the Bsu preQ1 riboswitch. Isolated aptamers exhibited the previously observed pre-docked (mid-FRET) and docked (high-FRET) conformations, with the docked conformation being stabilized upon addition of preQ1 and Mg2+, and the pre-docked conformation becoming more compact upon addition of Mg2+. Similar Mg2+-dependent changes in the absence of ligand have previously been observed by NMR in the isolated Bsu and Fusobacterium nucleatum (Fnu) preQ1 riboswitches. In the Bsu riboswitch, addition of Mg2+ leads to changes in the ligand-binding pocket that make it resemble but not perfectly mimic the ligand-bound state (Suddala et al., 2013). In the Fnu riboswitch, addition of Mg2+ leads to signals suggestive of P2 base pairing and interactions between P1 and the 3' tail (Santner et al., 2012). These structural insights may explain some of the variation in EFRET that we observe upon addition of Mg2+ and ligand. For example, we found that addition of preQ1 shifts the EFRET of the docked conformation to slightly higher values, a change that accompanies the concomitant rearrangement of the ligand-binding pocket. The significant changes in the EFRET of the pre-docked conformation upon addition of Mg2+ are consistent with formation of the P1-3' tail interactions observed in both the Bsu (Suddala et al., 2013) and Fnu (Santner et al., 2012) preQ1 riboswitches.
We also found that the DNA template disfavors nascent RNA folding in a distance-dependent manner. The EFRET of the pre-docked conformation was significantly lowered and its dependence on Mg2+ weakened by addition of the DNA. We found that addition of KCl increased the EFRET of the pre-docked conformation in a continuous fashion (Figure S3C-D), suggesting that there is no one distinct pre-docked conformation or that its dynamics remain unresolved. The addition of RNAP reverses this effect, restoring preQ1-dependent docking and compaction of the pre-docked state despite steric hindrance of folding in the exit channel (Figure 3). Our observations provide concrete evidence for a previously-hypothesized chaperone activity of RNAP, which has proven difficult to investigate by other means (Pan and Sosnick, 2006; Zhang and Landick, 2016).
Few studies exist in which naked RNA was directly compared to RNA in bubble complexes or ECs, with the only changes being the addition of those components of the transcription machinery. A study utilizing the fluorescent adenine analogue 2-aminopurine showed that addition of the DNA template had little effect on base stacking in an RNA hairpin, while addition of bacteriophage T7 RNAP to the bubble decreased stacking significantly (Datta and Hippel, 2008). A similar pattern was observed with single-stranded nascent RNA (Datta et al., 2006). Our investigation reveals very different behavior for a pseudoknot, with the DNA template having a significant effect and RNAP favoring RNA folding. Addition of Eco RNAP to a bubble scaffold decreased the rate of annealing of an antisense oligonucleotide (designed to mimic folding of the his pause hairpin) to the nascent RNA, but to a lesser extent than might be expected, indirectly implying a chaperoning function of RNAP (Hein et al., 2014). However, Hein et al. investigated a bimolecular annealing process, and their results therefore cannot be directly compared to our measurements of intramolecular pseudoknot folding.
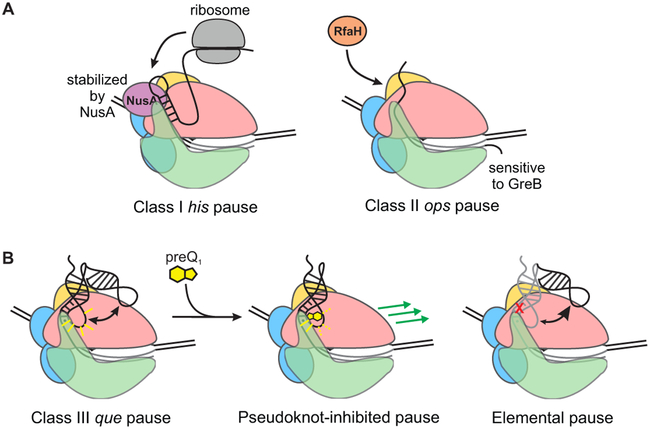
Conversely, using transcription assays, we showed that riboswitch folding modulates RNAP pausing (Figure 6). Current evidence points to the existence of an "elemental" paused state, in which sequences similar to the consensus trigger conformational changes that interfere with nucleotide addition. Elemental pauses have so far been found to be stabilized by two primary mechanisms (Artsimovitch and Landick, 2000; Zhang and Landick, 2016) (Figure 7A). In "hairpin-stabilized" or Class I pausing, a hairpin in the nascent RNA causes conformational rearrangements in the exit channel that stabilize the pause by allosterically promoting a "swiveled" conformation of RNAP (Kang et al., 2018). Class I pauses are further stabilized by the transcription factor NusA through interactions with the nascent RNA and the β flap domain (Hein et al., 2014; Toulokhonov et al., 2001), and are resolved by the trailing ribosome (Landick et al., 1985). In a second mechanism (Class II), the elemental pause enables backtracking by RNAP, leading to disengagement of the 3' end of the RNA from the active site. Class II pauses are sensitive to GreB, which enhances RNA cleavage by RNAP, and NusG, which antagonizes backtracking (Herbert et al., 2010). The Class II ops pause provides the opportunity for the antitermination factor RfaH to bind, which temporarily stabilizes the pause before promoting elongation and suppressing pausing elsewhere (Artsimovitch and Landick, 2002).
Figure 7. Mechanisms for Pause Stabilization and Control of Gene Expression; see also Figure S7.
(A)At a Class I pause, the trailing ribosome promotes RNAP escape and couples transcription to translation to regulate attenuation in amino acid biosynthesis operons. At a Class II pause, RfaH binds to the EC and antiterminates transcription of long operons.
(B)At the Class III que pause, the preQ1 ligand chases RNAP from the pause and rearranges the riboswitch structure to promote termination, converting a pseudoknot-stabilized pause (left) into a pseudoknot-inhibited pause (middle). Disrupting the pseudoknot yields an elemental pause (right).
The que pause we studied similarly requires an initial elemental pause, evidenced by the decrease in pause lifetime upon mutation of the consensus sequence (G37U and G36U/G37U mutants; Figures 6E and S7). The que pause is destabilized by moving the riboswitch further from RNAP (1U and 2U insertions), destabilizing P2 (G12U mutant), or deleting the β FT. We propose that these variants represent elemental or only weakly pseudoknot-stabilized pauses, lasting 22-25 seconds under our experimental conditions (Figure 7). In the WT and C9U variants, in contrast, the elemental pause is further stabilized by interactions between the pseudoknot and RNAP, resulting in a 2- to 4-fold increase in the pause half-life over that of the elemental pause.
These results suggest an analogy to the hairpin-stabilized Class I pause, and the que pause described here indeed shares intriguing characteristics with other pauses that are dependent on nascent RNA structure. Notably, the first base-pair of P2 is the same distance from the 3' end of the nascent RNA as is the first base-pair of the his pause hairpin (Toulokhonov et al., 2001). This is also identical to the distance at which initial folding of a pseudoknot was observed in the fluoride riboswitch (Watters et al., 2016). Differences in RNAP translocation register yield a 1-2 nucleotide variation in the position of the paired base relative to RNAP; nevertheless, these similarities indicate a common length-scale for nucleation of RNA folding. Insertion of two nucleotides between the hairpin and the his pause site reduces the pause lifetime and eliminates the ability of an oligonucleotide that disrupts the hairpin to inhibit pausing (Toulokhonov et al., 2001), suggesting that the hairpin no longer affects pausing at this distance. Our 1U and 2U variants exhibit the same effect, with potential modulation of nascent RNA structure through preQ1 binding having little effect on pausing. Similar to our results, it has previously been found that deletion of the β FT has no effect on the duration of an elemental pause, while it destabilizes a his pause-complex mimic in which the hairpin is replaced with a duplex (Hein et al., 2014). The que pause therefore shares certain characteristics with the Class I his pause despite being stabilized by a pseudoknot rather than a hairpin.
However, there are several significant differences between the properties of the que pause and canonical Class I and Class II pauses. Notably, Bsu and Eco RNAPs both respond to the que pause signal, whereas Bsu RNAP does not recognize the his pause at all (Artsimovitch et al., 2000). The FT has a much larger effect on Class I pauses than on the que pause, and the presence of a pseudoknot rather than a hairpin also renders the que pause insensitive to NusA (Figure S5G and S6D), which requires either a simple duplex of at least 10 base-pairs (Kolb et al., 2014) or a loop of greater than 4 nucleotides (Toulokhonov et al., 2001) for maximum stimulation of pausing. The folded riboswitch instead presents a 4 base-pair helix at the same position as the his pause hairpin, and instead of a loop presents a triplex-like structure in which L3 forms Hoogsteen-face interactions with the helix P1 (Klein et al., 2009). preQ1 binding has the effect of chasing RNAP away from the que pause, a function performed by the ribosome in the case of Class I pauses (Landick et al., 1985). Unlike Class II pauses, the RNA at the que pause does not undergo cleavage in response to GreB (Figure S5H), indicating that RNAP is not in a backtracked position. Our results suggest that the que pause represents a new mechanism in which an elemental pause is stabilized by a pseudoknot in the nascent RNA next to the RNAP exit channel, which we term "pseudoknot-stabilized" or "Class III" pausing (Figure 7B).
Our cross-linking results would seem to indicate that addition of preQ1 strengthens interactions with RNAP (Figure 5), and on the basis of our transcription assays in the absence of preQ1, this would be expected to promote pausing. However, in our cross-linking experiments, pause release is impossible due to the absence of NTPs. In this situation, preQ1 has the sole effect of stabilizing a bent-back, cross-linking competent conformation in which P2 is intact. When the presence of NTPs makes pause release possible, this conformation, which clearly differs from the pause-promoting bent-back conformation, appears to instead yield "pseudoknot-inhibited" analogue of the hairpin-inhibited pause escape mechanism (Zhang and Landick, 2016). The G12U mutant does not transition into a pseudoknot-inhibited pause because it binds preQ1 too weakly, leading to minimal effects of preQ1 on pausing and termination. The 1U and 2U variants do not transition into a pseudoknot-inhibited state for one or both of two reasons: 1) The increased distance between the pseudoknot and RNAP means that RNAP does not "feel" the conformational change upon preQ1 binding; and/or 2) preQ1 binding does not cause an extensive structural rearrangement, as seen in our MD and smFRET results.
In summary, we have demonstrated an intimate cross-coupling between bacterial riboswitch folding and transcriptional pausing that is likely to govern the co-transcriptional folding of numerous RNAs. Using a combination of approaches that are readily applicable to a wide variety of RNA species, we found that the presence of stalled RNAP provides a favorable free energy landscape for folding of the nascent preQ1 riboswitch. In turn, the ligand-free but pre-folded riboswitch pseudoknot stabilizes the paused state of RNAP via interactions with the β FT and with positively charged residues on the β’ subunit, while preQ1 binding stabilizes a distinct docked conformation that counteracts pausing. As pseudoknots are a common RNA structural motif, we expect that numerous examples of Class III pausing will be found among the thousands of transcriptional pauses that have been identified (Larson et al., 2014; Vvedenskaya et al., 2014). Our results reveal an additional, so far underappreciated layer to bacterial gene regulation that can be exploited for antibiotic drug intervention (Blount and Breaker, 2006; Deigan and Ferré-D'Amaré, 2011; Howe et al., 2015).
STAR METHODS TEXT
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nils Walter (nwalter@umich.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains, Plasmids and Growth Conditions
E. coli JM109 competent cells were used for maintenance of all transcription template plasmids. E. coli XL10 gold ultracompetent cells were used for maintenance of biotinylated RNAP expression plasmids. E. coli BLR (DE3) competent cells were used for overexpression of biotinylation-tagged RNAP, and XJb (DE3) cells were used for overexpression of RNAP variants. All strains were grown at 37 °C in Luria-Bertani media or on Luria-Bertani media-agar plates supplemented with 100 μg/mL carbenicillin. Plasmids used in this study are listed in the Key Resources Table.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli: JM109 competent cells | Promega | Cat#L2001 |
| E. coli: BLR (DE3) competent cells | EMD Millipore | Cat#69053 |
| E. coli: XJb (DE3) competent cells | Zymo Research | Cat#T3051 |
| E. coli: XL10 gold ultracompetent cells | Stratagene | Cat#200314 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cy5 mono-reactive dye pack | GE Healthcare Life Sciences | Cat#PA25001 |
| T4 RNA ligase 2 | New England Biolabs | Cat#M0239S |
| E. coli RNA polymerase, core enzyme | New England Biolabs | Cat#M0550S |
| E. coli RNA polymerase, holoenzyme | New England Biolabs | Cat#M0551S |
| E. coli RNA polymerase, core enzyme with biotinylation tag | This study | N/A |
| E. coli RNA polymerase, holoenzyme with R77A mutation on β' subunit | This study | N/A |
| E. coli RNA polymerase, holoenzyme with K79A mutation on β' subunit | This study | N/A |
| E. coli RNA polymerase, holoenzyme with R81A mutation on β' subunit | This study | N/A |
| E. coli RNA polymerase, holoenzyme with R77A/K79A/R81A triple mutation on β' subunit | This study | N/A |
| E. coli RNA polymerase, holoenzyme with β flap tip deleted | Irina Artsimovitch | N/A |
| ApC RNA dinucleotide | TriLink Biotechnologies | Cat#O-31002 |
| High purity rNTP set | GE Healthcare Life Sciences | Cat#27-2025-01 |
| α-32P GTP, 3000 Ci/mmol | Perkin Elmer | Cat#BLU006H250UC |
| Trolox | Fisher Scientific | Cat#218940050 |
| Protocatechuic acid | Soltec Ventures | Cat#CL107 |
| Protocatechuate 3,4-dioxygenase | Sigma Aldrich | Cat#P8279 |
| Glucose | Sigma Aldrich | Cat#G8270 |
| Glucose oxidase from Aspergillus Niger | Sigma Aldrich | Cat#G7141 |
| Catalase from bovine liver | Sigma Aldrich | Cat#C40 |
| Catalase from Corynebacterium glutamicum | Sigma Aldrich | Cat#02071 |
| Deposited Data | ||
| NET-Seq data from B. subtilis | Gene Expression Omnibus | Accession#GSE56720 |
| Oligonucleotides | ||
| Sequences provided in Tables S1 and S2 | Dharmacon/Integrated DNA Technologies | |
| Recombinant DNA | ||
| pVR: E. coli RecA promoter followed by 5'UTR of QueC gene from B. subtilis | This study | N/A |
| pVR2: pVR but with a 12-nucleotide stretch lacking A residues inserted after the promoter | This study | N/A |
| pVR2-m1: C9U mutant | This study | N/A |
| pVR2-m2: C12U mutant | This study | N/A |
| pVR2-m3: C18G mutant | This study | N/A |
| pVR2-m5: 1U mutant | This study | N/A |
| pVR2-m6: 2U mutant | This study | N/A |
| pVR2-m7: G37U mutant | This study | N/A |
| pVR2-m8: G36U/G37U mutant | This study | N/A |
| pIA999: overexpression plasmid for E. coli RNAP with biotinylation tag | Irina Artsimovitch | N/A |
| pIA1291: overexpression plasmid for E. coli RNAP with β' R77A mutation | This study | N/A |
| pIA1292: overexpression plasmid for E. coli RNAP with β' K79A mutation | This study | N/A |
| pIA1293: overexpression plasmid for E. coli RNAP with β' R81A mutation | This study | N/A |
| pIA1294: overexpression plasmid for E. coli RNAP with β' R77A/K79A/R81A triple mutation | This study | N/A |
| Software and Algorithms | ||
| ImageQuant | Molecular Dynamics | |
| Custom smFRET analysis code in MATLAB and IDL | This study | N/A |
METHOD DETAILS
Single-molecule FRET
Design and preparation of RNA constructs
It is widely known that transcription initiation can be bypassed by incubating RNAP with a pre-assembled scaffold consisting of a partially complementary DNA bubble and an RNA primer (Daube and Hippel, 1992; Hein et al., 2014; Kolb et al., 2014). Our DNA bubble was designed based on a sequence from (Kolb et al., 2014), with minor changes so that it was predicted to not interfere with formation of the native riboswitch structure (Table S1). “EC RNAs” 0, 3 and 10 were then designed to contain 10 nucleotides that were complementary to the template strand of the bubble, as well as the desired number of bases from the expression platform. The sequence of the capture probe (CP) was then designed to sequester in a duplex the same nucleotides that form the RNA:DNA hybrid in the bubble complex, and LNA bases were included to achieve a Tm of ~80 °C. The RNA:LNA or RNA:DNA hybrid sequence was not based on the expression platform, and was identical in the three RNA constructs with the exception of two bases that had to be changed to accommodate the portion of the aptamer oligonucleotide that became part of the hybrid in the shortest construct, RNA0p. The riboswitch aptamer sequence included a G-to-C mutation at the second position to suppress formation of alternate structures containing helix P1a (Kang et al., 2009). Aptamer labeling locations were chosen to match those used in previous work on the isolated aptamer (Suddala et al., 2013; 2015). Ligation enabled smFRET constructs of multiple lengths to be prepared from one aptamer construct.
All oligonucleotides used in smFRET experiments were purchased HPLC-purified from Dharmacon (in the case of fluorophore-labeled RNA) or Integrated DNA Technologies (in the case of DNA and unlabeled RNA). The riboswitch aptamer construct contained a 2’ Dy547 label at G37 and a 5-aminoallyl-uridine (5-N-U) at position 14. To label the 5-N-U, 4 nmol RNA was incubated for 4 hours at room temperature in a 50 μL reaction containing 30 μL of DMSO, one Cy5 mono-reactive dye pack (GE Healthcare Life Sciences) and 0.1 M sodium bicarbonate (pH 9). The RNA was isolated by ethanol precipitation. The labeled aptamer (200 pmol) and 5’ phosphorylated EC RNA (400 pmol) were annealed with a DNA or LNA splint (400 pmol-1 nmol) in a 100 μL mixture containing 150 mM NaCl, 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA. This annealing mixture was diluted to 500 μL in T4 RNA ligase 2 buffer, and 40 units of T4 RNA ligase 2 (New England Biolabs) were added. The reaction mixture was incubated at 37 °C for 2 hours. The reaction mixture was extracted with an equal volume of 25:24:1 phenol:chloroform:isoamyl alcohol, then concentrated down to ~20 μL in a vacuum centrifuge at 45° C and purified by denaturing, 8 M urea, 8% polyacrylamide gel electrophoresis (PAGE). The product band was extracted and the RNA isolated by ethanol precipitation.
Protein preparation
E. coli core RNAP bearing an “AviTag” biotinylation tag (GLNDIFEAQKIEWH) on the C-terminus of the β’ subunit was expressed from plasmid pIA999 in BLR(DE3) cells (Kay et al., 2009). The cells were grown in LB media supplemented with 20 μM biotin and 100 μg/mL carbenicillin, induced with 1 mM IPTG at an OD600 of 0.7 and harvested 3 hours post-induction. The protein was purified essentially as described (Svetlov and Artsimovitch, 2015). Briefly, the his-tagged protein was purified on a nickel-NTA agarose column, followed by a heparin column, and finally by a Mono Q ion-exchange column. The purified protein was dialyzed into storage buffer (10 mM Tris-HCl, pH 7.5, 50% glycerol, 100 mM NaCl, 0.1 mM EDTA, 0.1 mM DTT) and stored at −80° C. Biotinylation of the β’ subunit by endogenous BirA was verified by incubating biotin-tagged and wild-type proteins with streptavidin, followed by native PAGE analysis. Further in vitro biotinylation was not performed. RNAP mutants were prepared by the same protocol, except that a Resource Q ion-exchange column was used instead of Mono Q.
smFRET experiments
The RNA of interest was combined at a final concentration of 0.5 μM with either capture probe (CP) or template DNA (tDNA) and nontemplate DNA (ntDNA) in buffer A (50 mM Tris-HCl, pH 7.5, 100 mM KCl). When CP was used, its concentration was 5 μM; DNA concentrations were 10 μM when immobilizing through 3’-biotinylated ntDNA and 1 μM when immobilizing through biotinylated RNAP. The mixture was annealed by incubating at 90 °C for 2 min, then 37 °C for 10 min, then RT for 10 min. For experiments in the absence of RNAP, the annealing mixture was diluted in buffer A to a concentration of 15-50 μM RNA and flowed onto a slide that had previously been passivated with a 10:1 ratio of PEG to biotinylated PEG, then incubated for 10-15 min with a 0.2 mg/mL solution of streptavidin. For experiments performed in the presence of RNAP, the annealed nucleic acid scaffold was diluted to 50 nM RNA in buffer B (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 1 mM MgCl2), and E. coli RNAP was added to 250 nM. This mixture was incubated for 15 min at 37 °C, then diluted in buffer B to 50 pM RNA for immobilization. Imaging was performed at 62 ms time resolution in buffer A or B (where indicated, supplemented with preQ1 or additional KCl), containing 44 mM glucose, 165 U/mL glucose oxidase from Aspergillus niger, 2170 U/mL catalase from Corynebacterium glutamicum and 5 mM trolox as an oxygen scavenger (Aitken et al., 2008). This oxygen scavenger was chosen based on measurements of RNAP elongation activity in buffers containing various OSS components (Figure S4). The slide was incubated in each imaging buffer for 10 min prior to data collection. A laser power of 10-15 mW at 532 nm was used to excite the sample in a prism-based total internal reflection geometry, and emission from DY547 and Cy5 were recorded simultaneously using an intensified CCD camera (I-Pentamax, Princeton Instruments). The sample was additionally excited by a 640 nm laser at the beginning and end of each data set to verify the presence of Cy5. For on-slide elongation assays, ECs were prepared, immobilized and imaged as described above. Imaging buffer containing 1 mM GTP was then added, and imaging was repeated after a 10-minute incubation.
smFRET data analysis
Locations of molecules and fluorophore intensity traces for each molecule were extracted from raw movie files using IDL (Research Systems) and analyzed using Matlab (The Math Works). Traces were manually selected for further analysis using the following criteria: single-step photobleaching, a Cy5 fluorescence intensity of >200 (arbitrary units) when excited at 640 nm, a total (DY547+Cy5) intensity of >200 when excited at 532 nm, and a fluorescence duration (prior to photobleaching) of >3 s and, if transitions were observed in the trace, anticorrelation between donor and acceptor signals. The FRET efficiency (EFRET) was calculated as IA/(IA+ID), where IA and ID represent the background corrected fluorescence intensities of the acceptor (Cy5) and donor (DY547) fluorophores, respectively. FRET histograms were created by sorting the EFRET observed for the first 100 frames of a large number of traces (>200 for main text data, >100 for supplementary data) into 22 bins with centers ranging from EFRET = −0.05 to EFRET = 1.05. Plotting and fitting of FRET efficiency histograms was performed in OriginLab 8.1. Uncertainties in the histogram fit parameters were computed by bootstrapping, splitting the entire set of data points into three batches and fitting them independently.
Hidden Markov Modeling (HMM) analysis was performed using the segmental k-means algorithm in the QuB software suite as previously described (Blanco and Walter, 2010). To select the number of states to model our data with, HMM was performed on a selection of data sets using 1-, 2-, and 3-state models. We used fits to the corresponding histograms to select the initial values for optimization of the average EFRET and relative populations of the states. For each trace, we computed the Bayesian information criterion (BIC) and modified Bayesian information criterion (BIC'), as defined by Lerner et al. (Lerner et al., 2018), according to Equations 1-3:
| (1) |
| (2) |
| (3) |
where q is the number of states in the model being tested, K is the number of parameters in that model, L is the log-likelihood of the HMM fit, n is the number of data points in a given trace, and the number of states p varies over all of the models being considered. We then computed the mean BIC or BIC' over all of the traces in that data set. We found that for our data, BIC overvalued long traces while BIC' overvalued short traces. Unsurprisingly, then, BIC' suggested models with less than or equal to the number of states suggested by BIC. In none of the cases we tested did either metric suggest more than two states, and BIC suggested 2 states in all cases tested. We therefore used a 2-state model for the fitting and interpretation of all of our smFRET data.
Transcription Assays
Bubble-initiated transcription assays
Annealed nucleic acid scaffolds were prepared using RNA10 as described above for smFRET. Additional buffer components such as oxygen scavengers were added either immediately after annealing the nucleic acid scaffold, or after incubating the scaffold with RNAP (noted as "after RNAP" in Figure S4). Scaffolds were diluted to 50 nM in buffer B, E. coli RNAP core enzyme (New England Biolabs) was added to 200 nM, and the mixture was incubated at 37 °C for 15 min. The reaction mixture was then treated with 200 μM GTP and incubated for 10 min, after which an aliquot was removed and quenched into loading buffer (1x = 4 M urea, 25 mM EDTA, 45 mM Tris-borate, pH 8.3). The remaining reaction was treated with 200 μM ATP, CTP and UTP and incubated for an additional 10 min, after which another aliquot was removed and quenched. The reaction aliquots were analyzed by denaturing, 8 M urea, 15% PAGE and visualized by detecting the Cy5 fluorescence using a Typhoon 9410 Variable Mode Imager (GE Healthcare Life Sciences). Gel images for these and all following experiments were analyzed in ImageQuant (Molecular Dynamics). For RNA degradation assays (Figure S4), a 10 nM solution of RNA10 was prepared in buffer B, including the specified OSS components and RNase inhibitors at the same concentrations used for smFRET. The mixture was incubated at 37 °C for 30 min and analyzed by denaturing, 8 M urea, 15% PAGE.
We found through these assays that a number of common OSS components interfered with transcription in various ways. The OSS substrate protocatechuic acid (PCA) was found to interfere with transcription, as was the combined glucose/glucose oxidase/catalase (GOD/CAT) OSS, indicating inhibition by its product, D-gluconic acid. PCA and D-gluconic acid are both carboxylic acids, as are many known inhibitors of bacterial RNAP (Elgaher et al., 2014), and are therefore negatively charged under our imaging conditions. Inhibition was relieved by adding the OSS after incubating the bubble scaffold with RNAP, suggesting that PCA and D-gluconic acid interfere with binding rather than elongation. It is therefore likely that at the millimolar concentrations used for oxygen scavenging, they bind to and neutralize the electropositive cleft of RNAP where the transcription bubble resides. The maximum elongation efficiency was obtained by adding the GOD/CAT OSS after incubation with RNAP; therefore, this protocol was used for all experiments reported herein. Certain OSS enzymes, particularly catalase from bovine liver, were found to have RNase contamination and were therefore avoided, echoing reports of DNase contamination in OSS enzymes (Senavirathne et al., 2015).
Promoter-initiated transcription assays
A 151-nucleotide portion of the 5’ UTR of the queCDEF operon from B. subtilis, including the preQ1 riboswitch, was cloned into the pUC18 plasmid between the KpnI and EcoRI restriction sites. The E. coli RecA promoter was inserted immediately upstream of the riboswitch between the BamHI and KpnI sites. In addition, 12 nucleotides not found in the wild-type sequence were inserted after the promoter in order to generate a 19-nucleotide stretch in which the RNA transcript lacks any adenosine residues. Mutant plasmids were generated by site-directed mutagenesis using the primers in Table S2. Transcription templates for all experiments except those utilizing ΔFT RNAP were prepared by PCR using the "Forward PCR primer" and "Reverse PCR primer" indicated in Table S2. Transcription templates for ΔFT RNAP and associated WT controls contained the Gal1 promoter instead of RecA, and were generated in two PCR steps using first "Forward Gal1 primer 1" and "Reverse PCR primer", then "Forward Gal1 primer 2" and "Reverse PCR primer".
For transcription assays, halted complexes (HCs) were prepared in buffer C (20 mM Tris-acetate, pH 8.0, 20 mM sodium acetate, 2 mM magnesium acetate, 5% glycerol, 14 mM 2-mercaptoethanol, 0.1 mM EDTA) containing 1 μM labeling NTP, 5 μM of two other NTPs (omitting ATP), 56 nM α32P-GTP or CTP (3000 Ci/mmol), 100 μM ApC dinucleotide primer, and 40 nM DNA template. E. coli RNAP holoenzyme (New England Biolabs) was added to 80 nM, and the mixture was incubated for 15 min at 37 °C. 0.9 reaction volumes of HC was treated with 0.1 reaction volumes of pre-warmed chase solution in buffer C, which contained 200 μg/mL rifampicin and 10x the desired final concentrations of all four NTPs and preQ1. The mixture was incubated at 37 °C, and reaction aliquots were quenched at the desired times into an equal volume of loading buffer (8 M urea, 0.8× TBE, 0.2% bromophenol blue, 0.2% xylene cyanol, 1 mM EDTA). For most experiments, additional NTPs were then added to 200 μM, and a final aliquot was removed 3-5 minutes later. For experiments with RNAP variants (point mutants in the β' subunit and ΔFT RNAP), the concentration of RNAP was 50 nM instead of 80 nM. Sequencing ladders were prepared by combining 9 μL of HC with 1 μL of chase solution containing 250 μM of each NTP, in addition to one 3’-OMe NTP (at 25 μM for 3'-OMe GTP and 15 μM for 3’-OMe ATP, UTP and CTP). Reactions were incubated for 15 min at 37 °C before being quenched with 10 μL of loading buffer. Reaction aliquots were denatured before loading 1.5-2.5 μL of each onto a denaturing 8 M urea, 10% polyacrylamide sequencing gel. The gel was dried and exposed to a phosphor screen (typically overnight), which was then scanned on a Typhoon 9410 Variable Mode Imager.
For reactions including transcription factors, cell extract or RNA hairpin and the associated control reactions, the template was purified by spermine precipitation (Hoopes and McClure, 1981). These reactions were performed by preparing HCs at twice the concentration described above, adding an equal volume of 2x the desired final concentration of factor, extract or hairpin, and 30 seconds later adding the chase solution. The final concentration of all transcription factors was 100 nM. In the case of Eco NusA, it was verified that concentrations as high as 500 nM had no effect beyond that seen at 100 nM.
Transcription data analysis
Volumes of the pause band following the aptamer domain (U46), the terminator in the expression platform (U70), the terminator following the expression platform (U108), and the two bands resulting from runoff were determined using ImageQuant (Molecular Dynamics). For each lane, these volumes were background corrected by determining the volume above background of all points that had a higher intensity than the average intensity of a background box positioned in the same lane. Two background boxes were used in each lane: one to correct band U46 (positioned just below that band), and one to correct bands U70, U108 and the two runoff bands (positioned between U70 and U108). The relative intensity of each band in a given lane was determined by dividing the background-corrected volume of that band by the sum of the volumes of all five bands mentioned above. Two pause bands were observed within the aptamer domain, but these were omitted from the analysis due to their low intensity and lack of variation in response to preQ1 and mutations. Error bars in transcription quantifications represent the standard deviation of the intensity fraction of each band across three independent experiments. To determine the K1/2 for regulation of pausing and termination by preQ1, the relative intensities of U46 and U70 at 45 seconds and 90 seconds after chasing, and 5 min after adding a further chase to 200 μM of each NTP were plotted as a function of preQ1 concentration. These six data sets were globally fit with equation 4:
| (4) |
where F is the intensity fraction in a given band, y0 is the intensity fraction at zero preQ1, and A is the amplitude of the change that occurs upon addition of preQ1. A global fit was performed in which y0 and A were allowed to vary between data sets, but all were required to have the same value of K1/2.
To determine the pause efficiency and lifetime from detailed time courses, the natural logarithm of the fraction paused was calculated at each time point t (seconds) and the following equation was fit to the resulting data:
| (5) |
Fitting to the logarithm of the data rather than the raw data allowed the time points with low paused fraction to still contribute to the fit. a1 and a2 correspond to the fraction of elongation complexes that pause with half-lives -Ln[2]/m1 and -Ln[2]/m2, respectively. The constraint of a1+a2 ≤ 1 was used in all fits.
Cross-linking Assays
100 nM DNA template and 20 nM RNAP holoenzyme were incubated in buffer D (20 mM Tris-HCl, 120 mM KCl, 5 mM MgCl2, 5% glycerol (w/v), 1 mM 2-mercaptoethanol, pH 7.9) at 37°C for 15 min. ApU (100 μM) and starting NTPs (30 μM CTP, GTP, and ATP) were added to the promoter bound RNAP, and the reaction was incubated for 30 min at 37 °C. The EC at position 6 was immobilized on 10 μL Ni2+-NTA agarose beads at 37°C for 15 min. Each following walk (indicated in Figure S5) was completed with 5 intermediate washes with buffer D to eliminate unbound NTPs. Each wash involved addition of 1 mL of buffer D followed by a 5 second spin on a tabletop centrifuge to pellet the Ni2+ beads with attached ECs. The supernatant was then removed by pipette. The cross-linkable analogue 4-thio-UTP or 6-thio-GTP (10 μM) was introduced into the RNA at the desired position. Upon reaching A36, 10 μCi [α-32P\-GTP, 3000 Ci/mmol was added for 2 min. 5 μM ATP, CTP, and GTP were then added to reach C43, then 5 μM UTP and GTP to extend to U46. ECs C43 and U46 were incubated with 40 μM preQ1 at 37°C for 5 min and exposed to 365 nm UV light (UVP, 8W Model UVLMS-38, Upland, CA) for 10 min on ice. The reactions were either quenched with stop buffer (9M Urea, 20 mM EDTA, 1XTBE, 0.5 % Brilliant Blue R and 0.5% Xylene Cyanol FF) and the sample resolved on denaturing 7M urea 15% polyacrylamide-gel, or in LDS loading dye and separated on a 4-12% Bis-Tris Novex gel (Invitrogen). The gels were dried and exposed to phosphor screens.
Simulations
Molecular dynamics simulations were performed to explore the folding landscape of the preQ1 riboswitch bound to the RNA polymerase elongation complex. Structure-based or Gō-type simulations are particularly useful for exploring slow processes like folding that are computationally expensive to probe with more precise explicit solvent simulations. Structure-based models were originally used in protein folding (Shea et al., 1998) and are based on energy landscape theory and the principle of minimal frustration (Onuchic and Wolynes, 2004). In order to fold on biological time scales, native interactions must be more stabilizing than nonnative interactions, so structure-based models only include interactions between native contacts. We chose to use an all heavy atom structure-based model that has been extensively calibrated, and has been used for RNA folding (Lutz et al., 2014; Whitford et al., 2009) and functional studies of the ribosome, a mixed protein and RNA system (Whitford et al., 2010).
To generate a structure-based model, a native structure is needed to specify the interactions that ought to be stabilized. Atoms in close proximity within a known structure are chosen to be contact pairs that interact within the model; the shadow algorithm was used to calculate the contact map (Noel et al., 2012). Since no structure of the preQ1 riboswitch docked to RNAP existed, a model was created from structures of various components. Run input files for various structural elements were prepared using SMOG 2 (Noel et al., 2010; 2016), and then stitched together as described. The preQ1 riboswitch was modeled using contacts present in its crystal structure (PDB ID: 3FU2) (Klein et al., 2009). Its NMR structure (PDB ID: 2L1V) (Kang et al., 2009) was used to fill in 5’ and L2 gaps from the crystal structure: any contacts and interactions with missing residues were used to supplement the interactions defined by the crystal structure. An E. coli RNAP model was adopted from a cryo-EM structure of an EC (PDB ID: 6ALH) (Kang et al., 2017). The transcription bubble was modeled by creating single- and double-stranded sections of RNA and DNA using the nucleic acid builder (http://structure.usc.edu/make-na/server.html), and then stitching the resulting topologies together appropriately. Interactions of residues in either the riboswitch or the polymerase overrode those interactions in the DNA bubble model, so the bubble was only used to supplement interactions for missing residues. Since several topologies were being combined, and contact and dihedral interactions are scaled slightly differently in each, the dihedral and contact energies were both set to typical values of 0.7 energy units. We refer to the supplemental information of (Hayes et al., 2014) for a discussion of units in the model. Models with 0 to 3 nucleotides inserted between the riboswitch and RNAP (RNA0p, RNA1, RNA2 and RNA3) were made by modifying the sequences generated with the nucleic acid builder (sequences are reported in Table S1).
The resulting model essentially stitches together models of the riboswitch and the RNAP. Since the riboswitch and RNAP fold independently, the model is valid, but only accounts for steric interactions between the riboswitch and RNAP, and neglects any specific interactions that could be obtained from a structure of the complex. Consequently, some questions are beyond the scope of the model, while others can be robustly answered. The average number of base pairs formed in the model should be fairly robust, as the model identifies structures of the riboswitch that lead to a minimal disruption of RNAP structure. Likewise, the model should be able to elucidate the sterically allowed binding poses for the folded riboswitch, whereas identifying the dominant pose is beyond the scope of the model, as the populations of competing poses can be shifted by specific interactions between the riboswitch and RNAP.
Equilibrium simulations of riboswitch folding were run using umbrella sampling on the Q (number of formed contacts) reaction coordinate with replica exchange between umbrellas to allow trapped high-energy folded poses to relax. 11 replicas were used with an exchange attempts every 5000 steps, an umbrella k constant of 0.005 energy units per contact squared, and umbrella equilibria Q0 evenly spaced among replicas from 0 to 282 contacts. The weighted histogram analysis method (WHAM) (Kumar et al., 1992) was then used to reweight each frame of each replica to an unbiased simulation. Simulations started from an unfolded preQ1 riboswitch structure, and allowed the riboswitch to fold at a temperature of T = 85 units. (The folding temperature of the riboswitch is approximately 90 units in the presence of the polymerase.) Simulations were run for 50 million steps with the first 12.5 million steps discarded for equilibration. Simulations with the preQ1 ligand included 5 ligands in a cubic box of 20 nm side length. The binding affinity of preQ1 was not calibrated.
Simulations were run with and without preQ1 and varying numbers of inserted nucleotides to determine whether steric constraints allowed P2 to fold (Table S3). A base pair was considered formed if the central hydrogen bond between purine N1 and pyrimidine N3 was within a factor of 1.5 of its native distance. (In the A•C mismatch, the only hydrogen bond was used instead.) The number of formed base pairs was counted for each frame and averaged using the weight given by WHAM over all replicas in the simulation. The average number of base pairs was largely unchanged in the absence or presence of preQ1 for RNA1, RNA2 and RNA3 (1.8-2.4 versus 2.0-2.4, respectively), and depends weakly on number of inserts, but changes substantially in the zero insert case (RNA0p), where it is lower by about half a base pair with preQ1 and a whole base pair without preQ1. To check the robustness of the results, the contacts in the RNA linker (all residues between the 3’ end of P2 and the 5’ end of the RNA-DNA duplex) were removed. This allows the RNA to pull slightly out of the exit tunnel, and makes the RNA more flexible so it can twist more. While about half an additional base pair could form with the more flexible model, the trends remained the same: one to three inserts formed roughly the same number of base pairs as each other, and no inserts formed half a base pair less than them. Finally, a model of the flap tip deletion mutant was made in which residues K890-K914 of the beta subunit were removed. Simulations of the flap tip deletion mutant achieved results intermediate between the other two models, but showed less sensitivity to preQ1 in the RNA0p system.
To connect with cross-linking studies and to elucidate the folded pose, the frequency with which the cross-linking residue U14 was within 8 Å of each protein residue was calculated. Two poses of the riboswitch were identified that involved interactions primarily between the riboswitch and β’ (collectively called "mode 1"), along with a third pose where U14 is wedged between the clamp and flap tip ("mode 2"). Within mode 1, simulations with the default parameters (rigid linker) yielded a pose where U14 was positioned near K76-K79, while with the floppy linker, an additional pose was observed in which it is positioned near T393. These results are in agreement with cross-linking results that show that U14 interacts with the β’ subunit.
DATA AND SOFTWARE AVAILABILITY
Custom scripts used for this work are available upon request.
Supplementary Material
Highlights:
smFRET, biochemistry and simulations probe co-transcriptional riboswitch folding
Nascent RNA folding is disfavored by the DNA template and aided by RNAP
Transcriptional pausing is stabilized by RNA pseudoknot and destabilized by ligand
ACKNOWLEDGEMENTS
We thank Dr. Catherine Eichhorn and Dr. Hashim Al-Hashimi for generous gifts of plasmids containing the preQ1 riboswitch, Dr. Paul Lund for help mining previously published deep sequencing data, and Dr. Tina Henkin for a gift of Bsu RNAP. This work was supported by National Institutes of Health (NIH) R01 grants GM062357 and GM118524 to N.G.W., R01 grant GM67153 to I.A., R01 grant GM037554 to C.L.B. III, and NIH NSRA F32 postdoctoral fellowship GM113297 and K99 pathway to independence award GM120457 to J.R.W. V.R. acknowledges the Undergraduate Research Opportunity Program and Department of Biophysics for additional funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Aitken CE, Marshall RA, and Puglisi JD (2008). An Oxygen Scavenging System for Improvement of Dye Stability in Single-Molecule Fluorescence Experiments. Biophys. J 94, 1826–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Svetlov V, Anthony L, Burgess RR, and Landick R (2000). RNA Polymerases from Bacillus subtilis and Escherichia coli Differ in Recognition of Regulatory Signals In Vitro. J. Bacteriol 182, 6027–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, and Landick R (2000). Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. USA 97, 7090–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, and Landick R (2002). The Transcriptional Regulator RfaH Stimulates RNA Chain Synthesis after Recruitment to Elongation Complexes by the Exposed Nontemplate DNA Strand. Cell 109, 193–203. [DOI] [PubMed] [Google Scholar]
- Baird NJ, Kulshina N, and Ferré-D'Amaré AR (2010). Riboswitch function:Flipping the switch or tuning the dimmer? RNA Biol. 7, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M, and Walter NG (2010). Analysis of Complex Single-Molecule FRET Time Trajectories. Methods Enzymol. 472, 153–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, and Breaker RR (2006). Riboswitches as antibacterial drug targets. Nat. Biotechnol 24, 1558–1564. [DOI] [PubMed] [Google Scholar]
- Daher M, Widom JR, Tay W, and Walter NG (2018). Soft Interactions with Model Crowders and Non-canonical Interactions with Cellular Proteins Stabilize RNA Folding. J. Mol. Biol 430, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, and Hippel, von P (2008). Direct Spectroscopic Study of Reconstituted Transcription Complexes Reveals That Intrinsic Termination Is Driven Primarily by Thermodynamic Destabilization of the Nucleic Acid Framework. J. Biol. Chem 283, 3537–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Johnson NP, and Hippel, von PH (2006). Mapping the Conformation of the Nucleic Acid Framework of the T7 RNA Polymerase Elongation Complex in Solution Using Low-energy CD and Fluorescence Spectroscopy. J. Mol. Biol 360, 800–813. [DOI] [PubMed] [Google Scholar]
- Daube SS, and Hippel VP (1992). Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science 258, 1320–1324. [DOI] [PubMed] [Google Scholar]
- Deigan KE, and Ferré-D'Amaré AR (2011). Riboswitches: Discovery of Drugs That Target Bacterial Gene-Regulatory RNAs. Acc. Chem. Res 44, 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesterberg VK, Fischer-Hwang IT, Perez CF, Hogan DW, and Block SM (2015). Observation of long-range tertiary interactions during ligand binding by theTPP riboswitch aptamer. eLIFE 4, e12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis NF, Holmstrom ED, and Nesbitt DJ (2014). Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc. Natl. Acad. Sci. USA 111, 8464–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgaher WAM, Fruth M, Groh M, Haupenthal J, and Hartmann RW (2014). Expanding the scaffold for bacterial RNA polymerase inhibitors: design, synthesis and structure–activity relationships of ureido-heterocyclic-carboxylic acids. RSC Adv. 4, 2177–2194. [Google Scholar]
- Feng J, Walter NG, and Brooks CL III (2011). Cooperative and Directional Folding of the preQ1 Riboswitch Aptamer Domain. J. Am. Chem. Soc 133, 4196–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieda KL, and Block SM (2012). Direct Observation of Cotranscriptional Folding in an Adenine Riboswitch. Science 338, 397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee EB, Stav S, Atilho RM, Brewer KI, Harris KA, Malkowski SN, Arachchilage GM, Perkins KR, Sherlock ME, and Breaker RR (2018). Challenges of ligand identification for the second wave of orphan riboswitch candidates. RNA Biol. 0, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, and Henkin TM (2004). Kinetic Analysis of tRNA-Directed Transcription Antitermination of the Bacillus subtilis glyQS Gene In Vitro. J. Bacteriol 186, 5392–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, Schultz P, and Weixlbaumer A (2018). Structural Basis for NusA Stabilized Transcriptional Pausing. Mol. Cell 69, 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes RL, Noel JK, Whitford PC, Mohanty U, Sanbonmatsu KY, and Onuchic JN (2014). Reduced Model Captures Mg2+ -RNA Interaction Free Energy of Riboswitches. Biophys. J 106, 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein PP, Kolb KE, Windgassen T, Bellecourt MJ, Darst SA, Mooney RA, and Landick R (2014). RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat. Struct. Mol. Biol 21, 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmling C, Klötzner D-P, Sochor F, Mooney RA, Wacker A, Landick R, Furtig B, Heckel A, and Schwalbe H (2018). Life times of metastable states guide regulatory signaling in transcriptional riboswitches. Nat. Commun 944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Zhou J, Mooney RA, La Porta A, Landick R, and Block SM (2010). E. coli NusG Inhibits Backtracking and Accelerates Pause-Free Transcription by Promoting Forward Translocation of RNA Polymerase. J. Mol. Biol 399, 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom ED, Polaski JT, Batey RT, and Nesbitt DJ (2014). Single-Molecule Conformational Dynamics of a Biologically Functional Hydroxocobalamin Riboswitch. J. Am. Chem. Soc 136, 16832–16843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes BC, and McClure WR (1981). Studies on the Selectivity of DNA Precipitation by Spermine. Nucleic Acids Res. 9, 5493–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, et al. (2015). Selective small-molecule inhibition of an RNA structural element. Nature 526, 672–677. [DOI] [PubMed] [Google Scholar]
- Kang JY, Mishanina TV, Bellecourt MJ, Mooney RA, Darst SA, and Landick R (2018). RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing. Mol. Cell 69, 802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Olinares PDB, Chen J, Campbell E, Mustaev A, Chait BT, Gottesman ME, and Darst SA (2017). Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. eLIFE 6, e25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Peterson R, and Feigon J (2009). Structural Insights into Riboswitch Control of the Biosynthesis of Queuosine, a Modified Nucleotide Found in the Anticodon of tRNA. Mol. Cell 33, 784–790. [DOI] [PubMed] [Google Scholar]
- Kay BK, Thai S, and Volgina VV (2009). High-Throughput Biotinylation of Proteins. Methods Mol Biol 498, 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Edwards TE, and Ferré-D'Amaré AR (2009). Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat. Struct. Mol. Biol 16, 343–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb KE, Hein PP, and Landick R (2014). Antisense Oligonucleotide-stimulated Transcriptional Pausing Reveals RNA Exit Channel Specificity of RNA Polymerase and Mechanistic Contributions of NusA and RfaH. J. Biol. Chem 289, 1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Bouzida D, Swendson R, Kollman PA, and Rosenberg JM (1992). The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. J. Comput. Chem 13, 1011–1021. [Google Scholar]
- Landick R, Carey J, and Yanofsky C (1985). Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc. Natl. Acad. Sci. USA 82, 4663–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, and Weissman JS (2014). A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344, 1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J-F, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Massé E, and Lafontaine DA (2011). Comparative Study between Transcriptionally- and Translationally-Acting Adenine Riboswitches Reveals Key Differences in Riboswitch Regulatory Mechanisms. PLoS Genet 7, e1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E, Ingargiola A, and Weiss S (2018). Characterizing highly dynamic conformational states: The transcription bubble in RNAP-promoter open complex as an example. J. Chem. Phys 148, 123315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman JA, Suddala KC, Aytenfisu A, Chan D, Belashov IA, Salim M, Mathews DH, Spitale RC, Walter NG, and Wedekind JE (2015). Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics. Proc. Natl. Acad. Sci. USA 112, E3485–E3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz B, Faber M, Verma A, Klumpp S, and Schug A (2014). Differences between cotranscriptional and free riboswitch folding. Nucleic Acids Res 42, 2687–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel JK, Whitford PC, Sanbonmatsu KY, and Onuchic JN (2010). SMOG@ctbp: simplified deployment of structure-based models in GROMACS. Nucleic Acids Res. 38, W657–W661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel JK, Levi M, Raghunathan M, Lammert H, Hayes RL, Onuchic JN, and Whitford PC (2016). SMOG 2: A Versatile Software Package for Generating Structure-Based Models. PLOS Comput Biol 12, e1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel JK, Whitford PC, and Onuchic JN (2012). The Shadow Map: A General Contact Definition for Capturing the Dynamics of Biomolecular Folding and Function. J Phys Chem B 116, 8692–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuchic JN, and Wolynes PG (2004). Theory of protein folding. Curr. Opin. Struc. Biol 14, 70–75. [DOI] [PubMed] [Google Scholar]
- Pan T, and Sosnick T (2006). RNA Folding During Transcription. Annu. Rev. Biophys 35, 161–175. [DOI] [PubMed] [Google Scholar]
- Pan T, Artsimovitch I, Fang X-W, Landick R, and Sosnick TR (1999). Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. USA 96, 9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdrizet GA II, Artsimovitch I, Furman R, Sosnick TR, and Pan T (2012). Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Natl. Acad. Sci. USA 109, 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi AJ, Lund PE, Blanco MR, and Walter NG (2016). The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts. Nat. Commun [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Winkler WC, Regulski EE, Lee BWK, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, Welz R, et al. (2007). A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol 14, 308–317. [DOI] [PubMed] [Google Scholar]
- Santner T, Rieder U, Kreutz C, and Micura R (2012). Pseudoknot Preorganization of the preQ1 Class I Riboswitch. J. Am. Chem. Soc 134, 11928–11931. [DOI] [PubMed] [Google Scholar]
- Sen R, King RA, Mzhavia N, Madsen PL, and Weisberg RA (2002). Sequence-specific interaction of nascent antiterminator RNA with the zinc-finger motif of Escherichia coli RNA polymerase. Mol. Microbiol 46, 215–222. [DOI] [PubMed] [Google Scholar]
- Senavirathne G, Liu J, Lopez MA, Hanne J, Martin-Lopez J, Lee J-B, Yoder KE, and Fishel R (2015). Widespread nuclease contamination in commonly used oxygen-scavenging systems. Nat. Meth 12, 901–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea J-E, Nochomovitz YD, Guo Z, and Brooks CL III, (1998). Exploring the space of protein folding Hamiltonians: The balance of forces in a minimalist β-barrel model. J. Chem. Phys 109, 2895–2903. [Google Scholar]
- Sherwood AV, and Henkin TM (2016). Riboswitch-Mediated Gene Regulation: Novel RNA Architectures Dictate Gene Expression Responses. Annu. Rev. Microbiol 70, 361–374. [DOI] [PubMed] [Google Scholar]
- Sidorenkov I, Komissarova N, and Kashlev M (1998). Crucial Role of the RNA:DNA Hybrid in the Processivity of Transcription. Mol. Cell 2, 55–64. [DOI] [PubMed] [Google Scholar]
- Steinert H, Sochor F, Wacker A, Buck J, Helmling C, Hiller F, Keyhani S, Noeske J, Grimm S, Rudolph MM, et al. (2017). Pausing guides RNA folding to populate transiently stable RNA structures for riboswitch-based transcription regulation. eLIFE 6, e21297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suddala KC, Rinaldi AJ, Feng J, Mustoe AM, Eichhorn CD, Liberman JA, Wedekind JE, Al-Hashimi HM, Brooks CL III, and Walter NG (2013). Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic Acids Res. 41, 10462–10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suddala KC, Wang J, Hou Q, and Walter NG (2015). Mg 2+Shifts Ligand-Mediated Folding of a Riboswitch from Induced-Fit to Conformational Selection. J. Am. Chem. Soc 137, 14075–14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlov V, and Artsimovitch I (2015). Purification of Bacterial RNA Polymerase: Tools and Protocols In Methods in Molecular Biology, (New York, NY: Springer New York; ), pp. 13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, and Landick R (2001). Allosteric Control of RNA Polymerase by a Site That Contacts Nascent RNA Hairpins. Science 292, 730–733. [DOI] [PubMed] [Google Scholar]
- Uhm H, Kang W, Ha KS, Kang C, and Hohng S (2018). Single-molecule FRET studies on the cotranscriptional folding of a thiamine pyrophosphate riboswitch. Proc. Natl. Acad. Sci. USA 115, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, and Nickels BE (2014). Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 3344, 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Strobel EJ, Yu AM, Lis JT, and Lucks JB (2016). Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat. Struct. Mol. Biol 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, and Sanbonmatsu KY (2010). Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA 16, 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, and Sanbonmatsu KY (2009). Nonlocal Helix Formation Is Key to Understanding S-Adenosylmethionine-1 Riboswitch Function. Biophys. J 96, L7–L9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, and Crothers DM (2005). The Speed of RNA Transcription and Metabolite Binding Kinetics Operate an FMN Riboswitch. Mol. Cell 18, 49–60. [DOI] [PubMed] [Google Scholar]
- Zhang J, and Ferré-D'Amaré AR (2013). Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 500, 363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, and Landick R (2016). A Two-Way Street: Regulatory Interplay between RNA Polymerase and Nascent RNA Structure. Trends Biochem. Sci 41, 293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Custom scripts used for this work are available upon request.