Abstract
Parasites of the genus Plasmodium infect a wide array of hosts, causing malaria in all major groups of terrestrial vertebrates including primates, reptiles, and birds. Molecular mechanisms explaining why some Plasmodium species are virulent, while other closely related malaria pathogens are relatively benign in the same hosts, remain unclear. Here, we present the RNA sequencing and subsequent transcriptome assembly of two avian Plasmodium parasites which can eventually be used to better understand the genetic mechanisms underlying Plasmodium species pathogenicity in an avian host. Plasmodium homocircumflexum, a cryptic, pathogenic species that often causes mortality and Plasmodium delichoni, a newly described, relatively benign malaria parasite that does not kill its hosts, were used to experimentally infect two Eurasian siskins (Carduelis spinus). RNA extractions were performed and RNA sequencing was completed using high throughput Illumina sequencing. Using established bioinformatics pipelines, the sequencing data from both species were used to generate transcriptomes using published Plasmodium species genomes as a scaffold. The finalized transcriptome of P. homocircumflexum contained 21,612 total contigs while that of P. delichoni contained 12,048 contigs. We were able to identify many genes implicated in erythrocyte invasion actively expressed in both P. homocircumflexum and P. delichoni, including the well described vaccine candidates Apical Membrane Antigen-1 (AMA-1) and Merozoite Surface Protein 1 (MSP1). This work acts as a stepping stone to increase available data on avian Plasmodium parasites, thus enabling future research into the evolution of pathogenicity in malaria.
Keywords: RNA sequencing, Transcriptome, Plasmodium, Avian malaria, Avian parasitology
1. Introduction
Plasmodium parasites, the causative agents of malaria, infect a diverse set of vertebrates, including birds, reptiles, amphibians, primates, and humans globally [1]. Being such widespread pathogens, they continue to pose a great threat to human health, infecting an estimated 212 million people in 2015 and resulting in approximately 429,000 deaths [2]. In addition to the human impact, bird populations are also affected by malaria [3–5]. Despite our knowledge of gene and protein level interactions in murine and primate Plasmodium species, less is known about the genetic mechanisms underlying pathogenicity in other animal models, such as birds.
In order to better elucidate these mechanisms, next generation sequencing techniques can be employed to obtain the full-length genomes or transcriptomes of the parasite and/or the host, allowing researchers to pinpoint conserved virulence genes and characterize gene expression [6–12]. However, transcriptomes and genomes of blood parasites in wild birds have not been well studied, largely due to the fact that bird erythrocytes are nucleated, unlike mammalian erythrocytes which are not. The presence of both host and parasite genetic material creates computational difficulties that prevent the use of standard bioinformatics pipelines. The problem is particularly compounded in the case of generating haemosporidian genomes, due to the presence of approximately 52 times more genomic material belonging to the avian host compared to that of the parasite [13]. However, when Plasmodium parasites undergo erythrocytic merogony, they reproduce asexually and invade many new red blood cells, generating a high level of genetic (transcriptional) activity that potentially increases the yield of sequence data from parasite RNA (rather than DNA) [7,8,13].
Recently, the RNA sequencing of avian and other malaria parasites has proven to be an effective technique to investigate conserved erythrocyte invasion genes, host-parasite interactions, and the expression of virulence genes. For example, the transcriptome sequencing of Plasmodium falciparum by Otto et al. [8] allowed the rectification of over 10% of gene models from the existing P. falciparum genome annotation, as well as the inclusion of 202 new splice sites and 107 novel transcripts [8]. In addition, Lauron et al. [14,15] sequenced the transcriptome of Plasmodium gallinaceum, revealing structurally conserved gene orthologues that are required for erythrocyte invasion, such as Apical Membrane Antigen-1 (AMA-1), Rhoptry Neck Protein 2 (RON2), and RH5 Interacting Protein (RIPR). Furthermore, they identified genes that are essential to the purine salvage pathway that show a high similarity to P. falciparum (the Plasmodium species that is responsible for the majority of human deaths) [14,15]. Moreover, Videvall et al. [12] sequenced the transcriptome of Plasmodium ashfordi, an avian Plasmodium parasite thought to be closely related to one of the parasites presented here, and examined gene expression profiles across different stages of infection, as well as across hosts [12].
Approximately 50 avian Plasmodium species have been described morphologically, but many more distinct genetic lineages are being identified using molecular methods, suggesting the presence of cryptic species [1,4,16–20]. New species descriptions, then, should ideally be based on multiple identification methods [4,21,22]. For example, Palinauskas et al. [4] described an example of a cryptic avian Plasmodium parasite using both morphological and molecular techniques. They were able to describe and differentiate Plasmodium homocircumflexum from Plasmodium circumflexum which are morphologically indistinguishable, but differ genetically and in their capacity to cause host mortality [4]. Here, using next generation sequencing of wild-caught avian parasites, we provide the transcriptome of P. homocircumflexum, as well as the newly described species, Plasmodium delichoni, both from a Eurasian siskin (Carduelis spinus) host.
P. homocircumflexum is a virulent parasite originally isolated from a naturally infected Red-backed shrike (Lanius collurio). Its presence has been reported via sequencing data in the Collared flycatcher (Ficedula albicollis) [23], Red-rumped Warbling finch (Poospiza lateralis), and 5 other avian host species, although it cannot be confirmed if the parasite actually produces gametocytes and completes its lifecycle in each of these hosts [24]. It readily developed in all experimentally infected siskins, crossbills, and domestic canaries, killing 36.4% of experimentally infected canaries [4]. In experimental hosts with high parasitemia, P. homocircumflexum’s lethality was mainly due to a combination of two pathological effects. First, birds were dying because of anemia due to the loss of erythrocytes. Second, in both low and high parasitemia hosts, histological specimens revealed exoerythrocytic tissue stages developing in the endothelial cells of multiple organs, including the brain. These tissue stages can cause a blockage of capillaries in the brain resulting in severe cerebral paralysis and death [4,5].
Conversely, no deaths were observed in avian hosts infected with the newly described species P. delichoni, despite reaching up to 70% parasitemia in Eurasian siskins [25]. P. delichoni was discovered in the House martin Delichon urbicum, hence its name [25,26]. It has also been reported to naturally infect the Barn swallow (Hirundo rustica) [25] and the Collared flycatcher (Ficedula albicollis) [27,28], and it readily develops in siskins and canaries that are experimentally infected. High parasitemia levels were observed in siskins, while only light parasitemia (max 1%) developed in canaries [25].
This work offers insight into the gene expression of two newly described avian Plasmodium parasites, which can serve as the first step in identifying potential candidate genes that differ between deadly and benign parasites. The eventual goal being to elucidate novel evolutionarily conserved pathways involved in the pathogenicity of malaria. Additionally, by providing the complete transcriptome of two new Plasmodium species, these data serve as a public resource to future research in this area.
2. Methods
2.1. Experimental setup
The templates for molecular work were developed in the P. B. Šivickis Laboratory of Parasitology in Vilnius, Lithuania in 2015. Two wild juvenile (< 1-year-old) Eurasian siskins were caught in the beginning of summer with mist nets and maintained in a mosquito-free aviary. This bird species proved to be an excellent host for avian malaria experiments because it is easy to maintain in captivity and is highly susceptible to many Plasmodium species and their lineages [29]. Additionally, juvenile siskins caught early in the summer are uninfected with haemosporidian parasites at our study site [12]. Before experimental exposure, the wild-caught siskins were tested for possible presence of natural haemosporidian infections both by microscopic examination of blood films and PCR-based testing of their blood samples (for methods see [25]). Both birds were proven to be non-infected with blood parasites by both of these methods. Six control (non-infected) canaries were maintained in the same room with Eurasian siskins; the canaries were tested during this experiment the same way as the experimental siskins to ensure absence of malaria parasite transmission in the aviary. The controls remained non-infected for over 2 months (observation time). All birds were kept in a vector free room under controlled conditions (20 ± 1 °C, 50–60% relative humidity, natural light-dark photoperiod and were fed a standard diet for seed-eating bird species [25].
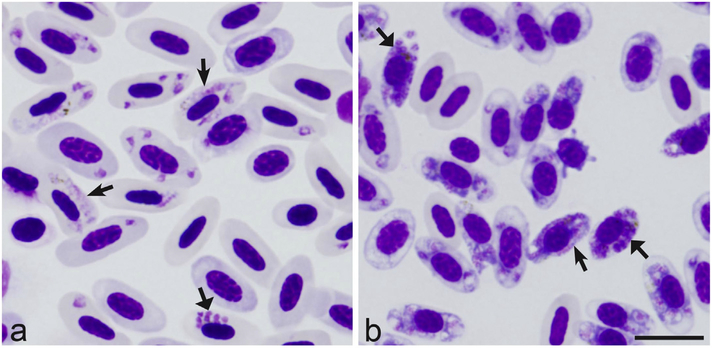
We used two inoculated siskins (one was exposed to P. homocircumflexum and one to P. delichoni) in this transcriptome study (bird A and B, respectively). Although the sample size is small, it provided enough variation to perform transcriptome analyses. Each bird was infected with a single injection of infected blood containing the erythrocytic stages of P. homocircumflexum (a mitochondrial lineage pCOLL4) and P. delichoni (a mitochondrial lineage pCOLL6). These parasite strains were obtained from one naturally infected House martin and one experimentally infected individual Domestic canary (Serinus canaria), respectively (for details see [4,25]). Infected blood of these donor birds was used to expose experimental siskins. A subinoculation of a freshly prepared mixture containing infected blood was made into the pectoral muscle of the recipient birds. The birds were exposed by subinoculation of about 250 μl of freshly prepared mixture, containing infected blood, 3.7% sodium citrate (anticoagulant) and 0.9% saline into their pectoral muscle (for details see [30]). The birds were observed continuously throughout the duration of the experiment. Blood for determining parasitemia intensity was sampled before and throughout the experiment in heparinized microcapillaries by puncturing the brachial vein once per 3–4 days. Blood films were prepared on glass slides for microscopic examinations and whole blood samples were also stored in SET-buffer for PCR-based analysis as described by Valkiūnas et al. [25]. Bird A had 85% of its red blood cells infected during peak parasitemia levels (determined via microscopy) before it died, whereas bird B reached a peak parasitemia level of 60% and survived. According to the original parasite descriptions and this study, the development of both P. delichoni [25] and P. homocircumflexum [4] is asynchronous in the blood, with all blood stages (meronts at all stages of their development and gametocytes) present simultaneously (Fig. 1). In the blood samples that were used for transcriptome analysis, meronts at various stages of their development predominated both in P. delichoni (82% per 100 infected red blood cells) and P. homocircumflexum (70%). Gametocytes represented 18% and 30% per 100 infected red blood cells during infection of P. delichoni and P. homocircumflexum, respectively. These blood samples were used in this study. About 75 μl of whole blood was collected from each bird and immediately stored in TRIzol® LS Reagent (Invitrogen, Grand Island, NY, USA) at a ratio of 1:10, before being taken back to San Francisco State University to be processed.
Fig. 1.
P. delichoni and P. homocircumflexum display asynchronous development in blood. Parasitemia of Plasmodium delichoni (a) and Plasmodium homocircumflexum (b) in the Carduelis spinus blood samples, which were used for transcriptome research. Triangle wide short arrows – erythrocytic meronts. Short simple arrows – gametocytes. Scale bar = 10 μm. Note asynchronous development of parasites, with numerous young and mature meronts present in the same field of microscope.
Experimental procedures were approved by the Ethical Commission of the Baltic Laboratory Animal Science Association, Lithuania; Lithuanian State Food and Veterinary Office (permit 2015–05-07, no. G2–27); Environmental Protection Agency, Vilnius (permits 2015–04-08, no. 21 and 2015–04-27, no. 25); and the International Research Cooperation Agreement between the Zoological Institute of the Russian Academy of Sciences and Institute of Ecology of Nature Research Centre (25–05-2010). RNMS has permits to import avian blood to the USA (USDA veterinary permit number 114165).
2.2. RNA extraction and sequencing
Total RNA was extracted from the avian whole blood samples according to the TRIzol® LS Reagent (Invitrogen, Grand Island, NY, USA) protocol with the following modifications. Heavy phase lock gel tubes were used to localize RNA into an aqueous phase, while DNA and protein was left in an interphase and organic phase respectively, and stored for later analysis. RNA was precipitated using isopropyl alcohol and 250 μl of a high-salt solution (0.8 M sodium citrate, 0.2 M NaCl in RNAse free water). 1 μl of Pellet Paint NF (70,748 Millipore Sigma, Inc.) was used as a co-precipitant in order to visualize the pellet and aid in extraction. The RNA was then treated with Ambion® TurboDNase™ according to manufacturer’s instructions and re-extracted using phenol-chloroform with isoamyl and an ethanol precipitation [31]. The RNA pellet was re-suspended in RNAse free water. Bioanalyzer 2100 (Agilent Technologies Inc., USA) traces were taken at the Functional Genomics Laboratory at UC Berkeley (FGL) to determine the quality of the RNA before library preparation and sequencing, which took place at the FGL and the Vincent J. Coates Genomic Sequencing Laboratory at UC Berkeley respectively. While the P. homocircumflexum RNA sample received a sufficient RNA Integrity Number (RIN) [32] of 8.6, we were unable to extract RNA of this quality from P. delichoni after numerous attempts. The RIN value for the P. delichoni RNA sample was 3.9. Due to the differences in RNA quality, two different library construction methods were employed, each suited to maximize the yield for the respective sample. For Plasmodium delichoni, Illumina’s Ribo-Zero rRNA Removal Kit (Illumina Inc., USA) was used to deplete the rRNA. The library preparation was completed on the Apollo 324™ (WaferGen Biosystems, Fremont, Ca) with PrepX™ RNAseq Library Prep Kits, and 13 cycles of PCR amplification were used for index addition and library fragment enrichment. For P. homocircumflexum, mRNA enrichment was performed using polyA selection with the Invitrogen Dynabeads mRNA Direct kit. cDNA synthesis and SPIA amplification was then performed using the Ovation® RNAseq system by Nugen (San Carlos, Ca, USA). An S220 Focused-Ultrasonicator (Covaris®) was used to fragment the cDNA, and the fragmented DNA was cleaned & concentrated with the MinElute® PCR Purification kit (QIAGEN Inc.). The Library preparation was done on an Apollo 324TM with PrepX™ ILM 32i DNA Library Kits (WaferGen Biosystems, Fremont, CA), and 9 cycles of PCR amplification were used for library fragment enrichment. Sequencing was run in one lane as paired-end reads of 100 base pairs (bp) on the HiSeq4000 platform. Raw sequence reads (host and parasite) have been deposited in the NCBI Short Read Archive (SRA) with the following accession numbers: PRJNA343386 and PRJNA380974.
2.3. De Novo transcriptome assembly and filtering
Raw reads were quality assessed with FastQC (v. 0.10.1) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), followed by quality and adapter trimming using the Trinity utility Trimmomatic (v. 2.3.2) [33], with the following parameters: LEADING:5 TRAILING:5 SLIDINGWINDOW:4:15 MINLEN:36. At this point, the data now contained both Plasmodium sequences as well as bird sequences. The initial bulk of bird reads were removed by mapping the trimmed reads to the zebra finch (Taeniopygia guttata) genome (the most closely related species to the Eurasia siskin with a sequenced genome at the time this research was conducted) using Bowtie2 with default parameters (v. 2.2.8) [34]. Anything that matched was removed and the remaining reads for each sample were separately passed into Trinity for transcriptome assembly using default parameters (v. 2.3.2) [33]. Due to the fact that many bird contigs were still present, each preliminary transcriptome was then BLASTx queried for protein amino acid sequence similarity (e-value < 1e-5) against known Plasmodium species in order to differentiate between Plasmodium contigs and bird contigs. The BLASTx query was performed against 2 custom databases thereby creating 2 versions of each transcriptome. The “Initial Database” contained the 15 available (at the time) Plasmodium species downloaded from NCBI (National Center for Biotechnology Information, 2017). The “Updated Database” contained the now 16 Plasmodium species available at NCBI, in addition to the newly released, first available avian Plasmodium genomes of P. gallinaceum and P. relictum [35] as well as species from 7 additional genera within the Apicomplexa phylum (Eimeria, Babesia, Toxoplasma, Cryptosporidium, Neospora, Hammondia, and Theileria). From henceforth, anytime the final transcriptome is referenced, it is referring to the version created with the Updated Database. The Initial Database Version was used for 2 purposes: 1) as a comparison to examine what differences there would be once the newly released genomes and additional apicomplexan parasite genera were added, and 2) for the gene ontology and KEGG pathway mapping because more results were able to be obtained (see Analysis). The positive BLASTx matches were then separated from non-matches (most likely bird sequences) using a custom BLAST parser script developed by Samantha Taffner and Elvin Lauron [15]. Similar contigs were clustered with CDHIT-EST (v. 4.6) using a similarity threshold of 97% [36]. Lastly, the remaining contigs were BLASTn searched using a 90% nucleotide sequence similarity threshold against Taeniopygia guttata to remove any remaining bird sequences that may have been highly conserved and therefore mistakenly matched to any of the genes within the Initial or Updated Plasmodium databases. The matching bird sequences were removed using the custom BLAST parser script and the remaining contigs represent the final transcriptome of P. delichoni and P. homocircumflexum respectively (Supplementary Dataset S1-S2, Supporting information).
2.4. Analysis
The assembled contigs in the Initial Database Version of the transcriptomes were fed into Blast2Go [37] where they were mapped for GO terms and annotated. Furthermore, KEGG pathway maps were drawn to visualize the enzymatic pathways of the putative genes in the two Plasmodium spp. transcriptomes. The Initial Database Versions were used to generate gene ontology/KEGG pathway data because more information was able to be retrieved due to the fact that more contigs had best BLAST hits to P. falciparum before the newly available avian Plasmodium spp. genomes were added to the database, and the gene ontology information is based largely on P. falciparum.
Gene expression levels were calculated using the Trinity RSEM utility, while transcriptome statistics were calculated using the Trinity script Trinitystats.pl (v. 2.3.2) [33]. We searched for putative erythrocyte invasion gene orthologues in the finalized transcriptome of each species using the following parameters: an e-value ≤1e-10, an FPKM ≥ 1, and a subject coverage ≥ 60%.
3. Results
3.1. Transcriptome characterization
The final transcriptome using the Updated Database (see Methods) of P. homocircumflexum contains 15,742 contigs not including isoforms (21,612 with isoforms) containing 17,175,763 total assembled bases (Table 1). The average contig length is 795 nucleotides (nt) long, with a contig N50 value of 1081. The transcriptome is extremely AT biased with a GC content of 21.64%. Somewhat comparably, the final transcriptome of P. delichoni has 11,305 contigs (12,048 including isoforms) containing 5,680,962 total nts, and a GC content of 23.93%. However, its contig N50 value is much lower at 516 with an average contig length of 472 nt.
Table 1.
Transcriptome summary statistics table.
| Transcriptome statistics table | ||
|---|---|---|
| P. homocircumflexum | P. delichoni | |
| Raw Paired End Reads | 80.5 M | 128 M |
| Total Contigs (without Isoforms) | 15,742 | 11,305 |
| Total Contigs including Isoforms | 21,612 | 12,048 |
| Average Contig Length (nt) | 795 | 472 |
| Contig N50 Value (nt) | 1081 | 516 |
| GC Content (%) | 21.64 | 23.93 |
| Total Assembled Bases (nt) | 17,175,763 | 5,680,962 |
The transcriptomes of P. homocircumflexum and P. delichoni have a comparable total contig number, GC content, and average contig length to other recently published avian Plasmodium transcriptomes [12,14].
3.2. Species similarities
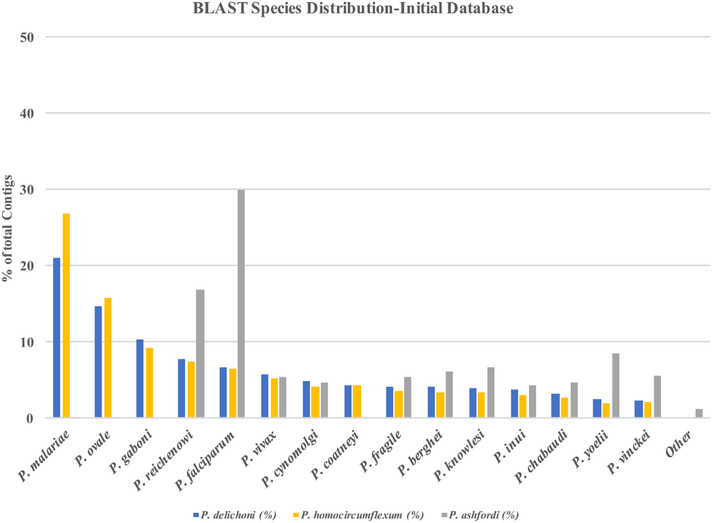
The first draft of the P. homocircumflexum and P. delichoni transcriptomes were created prior to the release of the first available avian Plasmodium genomes of P. gallinaceum and P. relictum [35]. After their release, the transcriptomes were re-assembled (see Methods) and differences were assessed. Using the BLASTx program (e-value < 1e-5), we examined the distribution of species to which P. homocircumflexum and P. delichoni had the highest level of amino acid sequence homology (e.g. 30% of P. homocircumflexum’s contigs displayed the best homology match to P. malariae, compared to all other species within the given database, with an e-value < 1e-5) (Supplementary Dataset S3-S4, Supporting information). This was done to identify the closest known relatives of P. homocircumflexum and P. delichoni. Furthermore, it enabled us to identify the potential limitations posed by the current lack of data available on avian Plasmodium parasites, and how that affects researchers’ abilities to assemble transcriptomes using the exclusionary bioinformatics methods described here. The Initial Database Version of both P. homocircumflexum and P. delichoni had a similar best BLAST hit species distribution, with the majority of contigs receiving hits from P. malariae (26.94% and 21.06% of contigs respectively), and then P. ovale (15.74% and 14.74%), and P. gaboni (9.21% and 10.33%). P. homocircumflexum and P. delichoni had 54.41% and 48.20% of contigs respectively result in hits to Plasmodium parasites that infect humans, 35.36% and 39.34% of contigs matching to Plasmodium primate parasites, and 10.24% and 12.46% of contigs matching to Plasmodium rodent parasites (Fig. 2). Rather than a custom database, Videvall et al. [12] BLASTed their recently published transcriptome of P. ashfordi against the entire NCBI non-redundant protein database. Only 0.92% of P. ashfordi contigs matched to parasites of birds (72 out of 7860 total contigs) [12]. Interestingly, P. falciparum received only the 5th most BLAST hits in both P. homocircumflexum and P. delichoni (6.49% and 6.68% respectively) compared to nearly a third (29.91%) of P. ashfordi contigs which had best BLAST matches to P. falciparum. The differences seen between the work presented here and that of Videvall et al. [12] is explained by the availability of additional Plasmodium sequence data such as that of P. malariae which was published in 2017, as well as the fact that P. falciparum is the best characterized Plasmodium genome available and is likely overrepresented in the NCBI non redundant protein database.
Fig. 2.
BLAST species distribution prior to the inclusion of the two newly available avian Plasmodium genomes.
Plasmodium delichoni and Plasmodium homocircumflexum were queried for protein amino acid sequence similarity (e-value <1e-5) against the custom Initial Database, compared to the data from [12] for Plasmodium ashfordi which was similarly queried against the NCBI non-redundant database. This demonstrates different results that can occur as new Plasmodium sequence data becomes available.
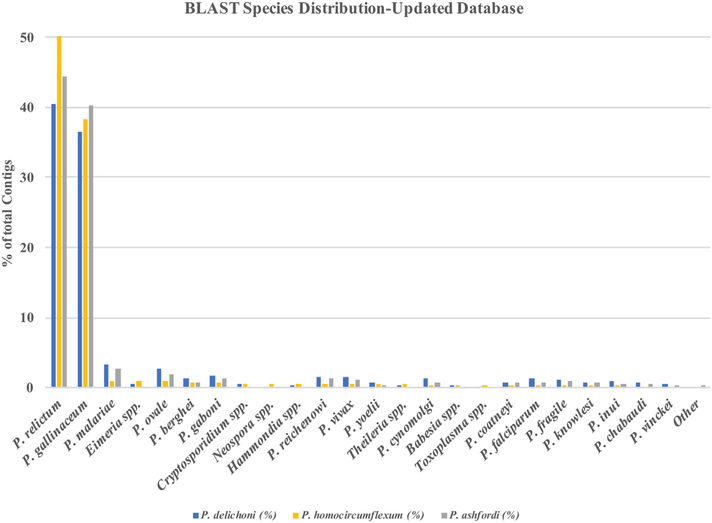
After adding the additional species and using the Updated Database in the annotation, as well as including the transcriptome of P. ashfordi in the analysis, 89.02% of P. homocircumflexum’s contigs, 77.02% of P. delichoni’s contigs, and 84.62% of P. ashfordi’s contigs had a best BLASTx match to either P. gallinaceum or P. relictum (Fig. 3). Additionally, the amount of contigs that matched to P. falciparum decreased to only 0.34%, 1.34%, and 0.83% respectively. Overall, these results show an obvious but previously unseen result, that these species have the highest sequence similarity to other avian Plasmodium parasites, rather than to the previously shown primate parasites, due to the overrepresentation of Plasmodium parasites that infect primates in databases, and the lack of available avian Plasmodium genomes. This demonstrates the importance of these newly available genomes in the assembly of avian Plasmodium transcriptomes, and how previous work without it may have been skewed [35].
Fig. 3.
BLAST species distribution after the inclusion of the two newly available avian Plasmodium genomes.
Plasmodium delichoni, P. homocircumflexum, and P. ashfordi were queried for protein amino acid sequence similarity (e-value < 1e-5) against the custom Updated Database which includes the two newly available avian Plasmodium genomes of P. gallinaceum and P. relictum. The distribution is heavily skewed towards the newly available avian Plasmodium species once included.
3.3. Gene ontology and KEGG pathway mapping
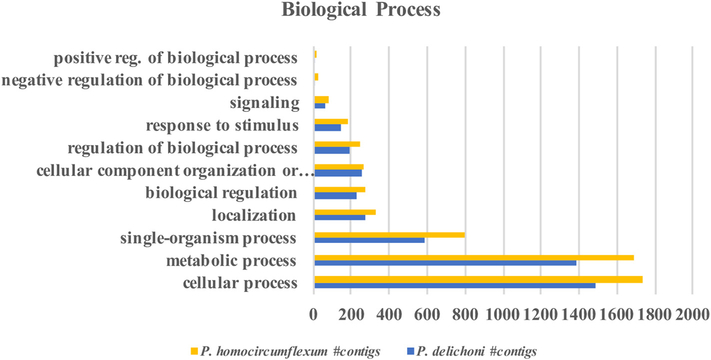
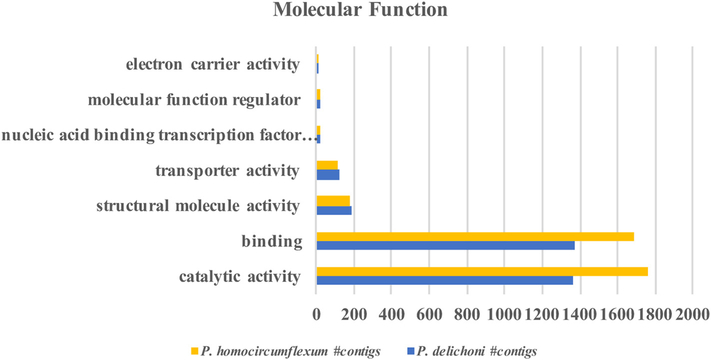
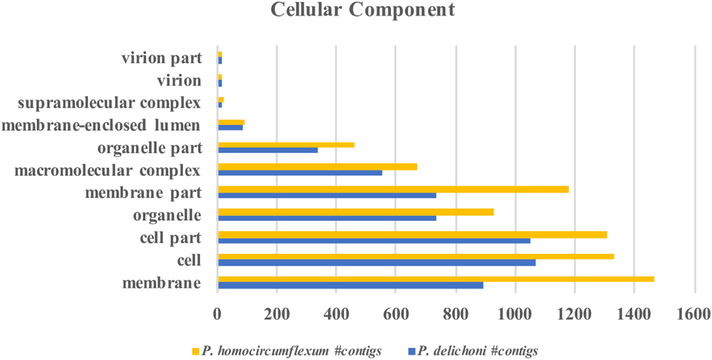
Functional analysis information was derived using the Initial Database Version of the two transcriptomes. This is because the gene ontology information is derived from the BLAST results, and since the majority of contigs in the Updated Database Version have best BLAST hits to the recently published avian Plasmodium genomes, whilst the majority of gene ontology information available is based on P. falciparum, more information was retrievable when using the earlier version. After BLAST results were uploaded into Blast2Go, sequences were mapped to Gene Ontology (GO) terms and subsequently annotated (Supplementary Dataset S5, Supporting information). A total of 3960 sequences were annotated with GO terms in the P. homocircumflexum transcriptome, while 2936 were annotated in the P. delichoni transcriptome. GO terms were grouped according to Biological Processes, Molecular Functions, and Cellular Components (Fig. 4, 5, 6). P. homocircumflexum had more contigs than P. delichoni in all categories except ‘virion’ and ‘virion part’ in Cellular Components, and ‘molecular function regulator’ in Molecular functions. However, it is important to note this could just be due to a differing number of contigs among the two annotations.
Fig. 4.
Gene ontology mapping results for “Biological Process”.
Gene ontology information was assessed for both Plasmodium species using the Blast2Go program. Using sequence homology to closely related species, gene function was inferred, grouped into one of three main categories, and then further grouped into multiple, more specific categories. The gene ontology patterns under “Biological Process” for Plasmodium homocircumflexum and Plasmodium delichoni are distributed similarly, with differences mostly attributed to a different number of total contigs in each respective transcriptome.
Fig. 5.
Gene ontology mapping results for “Molecular Function”.
The gene ontology patterns under “Molecular Function” for Plasmodium homocircumflexum and Plasmodium delichoni are distributed similarly.
Fig. 6.
Gene ontology mapping results for “Cellular Component”.
The gene ontology patterns under “Cellular Component” for Plasmodium homocircumflexum and Plasmodium delichoni are distributed similarly.
KEGG pathway analysis was also performed resulting in 827 sequences in the P. homocircumflexum transcriptome mapping to 157 unique enzymes in 81 KEGG pathways, with the top 4 pathways represented being: 491 contigs;21 enzymes to purine metabolism, 372;3 to thiamine metabolism, 108;13 to pyrimidine metabolism, 93;41 to biosynthesis of antibiotics. Similarly, 683 contigs in the P. delichoni transcriptome mapped to 146 unique enzymes in 76 KEGG pathways. The same 4 pathways were represented with the most sequences in P. delichoni with 452 contigs;23 enzymes to purine metabolism, 348;5 to thiamine metabolism, 95;12 to pyrimidine metabolism, and 60;43 to biosynthesis of antibiotics.
3.4. Invasion genes
Among the successfully assembled erythrocyte invasion genes found, two are among a handful of potential candidates for a malaria vaccine: Apical Membrane Antigen-1 (AMA-1) which was assembled in both Plasmodium species, and Merozoite surface protein 1 (MSP1) which was assembled only in P. homocircumflexum [7]. MSP1 aids in the initial binding of the merozoite to the erythrocyte as well as mediating rupture [38,39], while AMA-1, an integral membrane bound protein, has been shown to be critical in erythrocyte invasion in P. falciparum by helping to form the tight junction between the parasite and red blood cell [39–41].
Putative erythrocyte invasion gene orthologues were searched for in both transcriptomes using an e-value ≤1e-10, FPKM ≥ 1, and a subject coverage ≥ 60%, as cutoff parameters. While FPKM was examined to confirm a gene was being actively expressed, individual FPKM expression values for invasion genes could not be compared across species (see Discussion). The following invasion genes had a contig in the P. delichoni transcriptome that met these parameters: AMA-1, bromodomain protein 1 (BDP1), glideosome-associated protein 50, myosin A tail domain interacting protein, reticulocyte binding protein, and rhomboid protease 4 (ROM4) (Supplementary Dataset S7, Supporting information).
In P. homocircumflexum, substantially more contigs matched to known erythrocyte invasion genes using these parameters. Subtracting for multiple isoforms of the same contig, the following genes had 1 or more contigs match (number of matching contigs separated by a semicolon if more than one): AMA-1, BDP1; 3, fructose-bisphosphate aldolase, glideosome-associated protein 45, glideosome-associated protein 50, MSP1, Merozoite TRAP-like protein, myosin A tail domain interacting protein; 2, reticulocyte binding protein; 5, ROM1, ROM4, rhoptry-associated membrane antigen, rhoptry associated leucine zipper-like protein 1, rhoptry neck protein-5 (Supplementary Dataset S6, Supporting information).
4. Discussion
A fundamental question in biomedical research remains insufficiently understood. Why are some pathogens virulent, while other closely related species belonging to the same genus are benign in the same host species? This issue is still difficult to analyze in wildlife haemosporidian parasites, particularly because genomes and transcriptomes have only been studied in a few species, resulting in little information about the evolution of virulence in wildlife malaria parasites and other haemosporidians [11–15,35]. It remains unclear how virulence of Plasmodium parasites can vary in natural systems. The avian malaria model provides excellent opportunities to address these questions experimentally [5,12]. These studies are of particular importance, given that virulent malaria parasites can threaten naive populations and even lead to the extinction of some endemic avifaunas [3]. This study helps to address the first step in solving this problem by providing the transcriptomes of two avian Plasmodium parasites that can later be further analyzed. Increasing the pool of genomic and transcriptomic data on avian Plasmodium parasites is important so that future studies can make genus wide comparisons.
The transcriptomes of P. homocircumflexum and P. delichoni provided here are comparable to the recently published avian Plasmodium transcriptomes of P. ashfordi [12] and P. gallinaceum [14,15]. Plasmodium parasites are known to have very AT-biased genomes, and even more recently it was shown that to date, avian malaria parasites in particular have the most AT-biased genes of all known eukaryotes [42]. As such, GC content can be used as a parameter to compare Plasmodium genomes and transcriptomes [42]. The transcriptome of Plasmodium falciparum, perhaps the most notable and best characterized Plasmodium species for its ability to cause infection and death in humans, has a GC content of 23.79%. The transcriptomes of Plasmodium homocircumflexum and P. delichoni similarly have a GC content of 21.64% and 23.93%, also in line with the recently published P. ashfordi transcriptome at 21.22%. Both P. homocircumflexum and P. delichoni have vastly more total contigs (including isoforms) than P. falciparum, at 21,612 and 12,048 compared to 5800, likely due to multiple isoforms, fragmented contigs, and potential non-coding RNA [12]. This can be attributed in part, to the de novo transcriptome assembly approach taken here which will unavoidably create more contigs than there are genes in the genome [33]. The de novo approach was necessary, however, due to the lack of a genome available to serve as a scaffold for either of the Plasmodium species studied here. Furthermore, with 128 million and 80.5 million raw sequence reads for P. homocircumflexum and P. delichoni respectively, reduced sequencing depth can also partially explain the amount of fragmented contigs. The same is true for the average length of contigs, at almost 2200 nts in P. falciparum, compared to 794 nt in P. homocircumflexum and 471 nt in P. delichoni [12]. However, these metrics are much more similar to the avian parasite transcriptomes of P. ashfordi and P. gallinaceum, which have 11,954 and 17,832 total contigs respectively, as well as 753 nt and 893 nt average contig lengths respectively. P. delichoni stands out here as having a particularly low average contig length, likely due to the lower quality of RNA that was used in sequencing. Additionally, the different library construction methods employed can affect the number of contigs as well. (see Methods). RNA of a sufficient RIN value was particularly difficult to isolate from these samples, something we speculate may have been due to their intercontinental transport. Despite this, many genes of interest were found common to all species of Plasmodium.
By creating two versions of each transcriptome with either the Initial Database or Updated Database, we were able to show the substantially different BLAST matches that resulted, and consequently the differences in the transcriptome assembly that followed. This is especially important because due to the nucleated avian erythrocytes, it is common to use (as we have done here) an exclusionary procedure of nucleotide and/or protein sequence similarity matching, and then sorting contigs based on matches to host or parasite. Therefore, the more closely related the species in the queried database are to the sample, the more likely bioinformatics processing is able to accurately remove all host contigs and retain all parasite contigs. This may perhaps be demonstrated by the difference in contigs retained in the transcriptome version using the Initial Database as opposed to the Updated Database. Including isoforms, and after accounting for contigs that matched to one of the 7 additional genera added to the Updated Database, 1752 additional contigs were identified in the transcriptome of P. homocircumflexum, presumably due to the addition of the two newly released avian Plasmodium genomes. Similarly, 233 additional contigs were retained in the transcriptome of P. delichoni. It is also important to note, however, that the BLAST algorithm can have some variation from run to run. Moreover, while these additionally captured contigs are helpful in ensuring that the transcriptomes presented here are as complete as possible in representing all of the genes being expressed at the time of blood isolation, due to this work’s reliance on previously published Plasmodium data, we were likely unable to identify novel genes that do not contain orthologues in related Plasmodium species.
It should also be noted for comparison purposes that although Videvall et al. [12]’s approach differed slightly from this research in that they performed protein sequence similarity queries against the entire NCBI non-redundant protein database, only 1.22% of P. ashfordi contigs had matches to species outside of the 15 Plasmodium species included in the custom Initial Database. Furthermore, after performing a protein sequence similarity search of the completed transcriptome of P. ashfordi against the Updated Database, the number of contigs that had matches to species outside of our database was reduced to 0.41%. Therefore, we concluded that performing a protein sequence similarity search against the entire NCBI non-redundant protein database was not necessary in our case as it would not have changed the results dramatically.
We identified many genes implicated in erythrocyte invasion being actively expressed in both P. homocircumflexum and P. delichoni, including the well described vaccine candidates, AMA-1 and MSP1. Additional invasion genes of note (full list in Results) include BDP1, which encodes a protein that regulates other invasion genes’ expression by binding to chromatin at transcriptional start sites [12,43], myosin A tail interacting protein and glidesome-associated proteins 45 and 50 (GAP 45/GAP50), which are implicated in aiding parasite gliding motion at the parasite motor complex [12,44,45], and ROM1/ROM4 which have been associated with the cleavage of transmembrane adhesins during host invasion in P. falciparum [12,46,47]. Furthermore, we discovered transcript orthologues of reticulocyte binding protein in both P. homocircumflexum and P. delichoni, helping to elucidate and confirm the discrepancies between the work of Videvall et al., [12] and Lauron et al., [15].
It should also be mentioned that while we examined FPKM as a confirmation of active gene expression, we were not able to compare expression levels between the two species. Differential gene expression is typically investigated within one species, across a changing variable. While differential gene expression can be investigated across different species, it is usually necessary to have the genome of both species available, which are not available in the case of P. homocircumflexum and P. delichoni. Furthermore, the inability to synchronize infection stages as well as differences in RNA quality and resulting libraries could have confounded our results, making differential expression analysis infeasible. However, since all blood stages were present in the blood samples, we believe that we have acquired a transcriptome that reflects the diversity of developmental stages.
Plasmodium homocircumflexum and P. delichoni had 827 and 683 contigs respectively map to KEGG database pathways, and most notably, 59.4% and 66.2% of mapped contigs in each of the transcriptomes mapped to the purine metabolism pathway. This finding is notable as Plasmodium parasites are purine auxotrophs, requiring the salvage of purines from their host to build nucleic acids during intraerythrocytic stages [15,48,49]. This highlights the importance and evolutionarily conserved nature of the enzymes in this pathway.
The genetic mechanisms of Plasmodium parasite pathogenicity have not been studied significantly in an avian model. With the work presented here, we now have a framework for identifying genes and pathways that are common to all malaria parasites, which will aid in reaching our ultimate goal of elucidating the mechanisms underlying P. homocircumflexum’s pathogenicity. While the RNA quality for P. delichoni was not optimal, we have a completed transcriptome that can be improved upon with higher quality samples, and additional sequencing. Furthermore, this work acts to increase available data on avian Plasmodium parasites, something that is crucial before further in depth analyses can be completed. Further research into avian malaria genes could ultimately aid in understanding human Plasmodium parasite pathogenicity, as well as the ecological impacts of these parasites on bird populations and health.
Supplementary Material
Acknowledgments
We would like to thank Elvin Lauron and Samantha Taffner for allowing us to use their BLAST Parse Script, Cameron Everson and Dr. Scott Roy for their coding expertise/guidance and allowing access to the Roy laboratory server, and Elin Videvall as well as Greg Fedewa for their helpful advice.
Funding
This work used the Vincent J. Coates Genomic Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. This work was also supported by the grant to RNMS, NIH 1SC3GM118210-01A1.
This study was funded by the Research Council of Lithuania (nr. MIP-045/2015) and also supported by the Open Access to research infrastructure of the Nature Research Centre under Lithuanian open access network initiative.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygeno.2018.12.004.
References
- [1].Garnham PCC, Malaria Parasites and Other Haemosporidia, Blackwell Sci Publ Ltd, Oxford, UK, 1966. [Google Scholar]
- [2].World Health Organization, World Malaria Report, World Health Organization, Geneva, 2016. [Google Scholar]
- [3].Atkinson CT, Utzurrum RB, Lapointe DA, Camp RJ, Crampton LH, Foster JT, Giambelluca TW, Glob. Chang. Biol 20 (8) (2014. August) 2426–2436, 10.1111/gcb.12535. [DOI] [PubMed] [Google Scholar]
- [4].Palinauskas V, Žiegytė R, Ilgūnas M, Iezhova TA, Bernotiėne R, Bolshakov C, et al. , Description of the first cryptic avian malaria parasite, Plasmodium homocircumflexum n. sp., with experimental data on its virulence and development in avian hosts and mosquitoes, Int. J. Parasitol. [Internet] 45 (1) (2015) 51–62 January [cited 2017 Mar 2] Available from: http://linkinghub.elsevier.com/retrieve/pii/S0020751914002616. [DOI] [PubMed] [Google Scholar]
- [5].Ilgūnas M, Bukauskaitė D, Palinauskas V, Iezhova TA, Dinhopl N, Nedorost N, et al. , Mortality and pathology in birds due to Plasmodium (Giovannolaia) homocircumflexum infection, with emphasis on the exoerythrocytic development of avian malaria parasites, Malar. J 15 (1) (2016) 256, 10.1186/s12936-016-1310-x (Epub 2016/05/06. PubMed PMID: ; PubMed Central PMCID: PMCPMC4857288). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. , Genome sequence of the human malaria parasite Plasmodium falciparum, [cited 2017 Nov 8] Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3836256/pdf/emss-54165.pdf. [DOI] [PMC free article] [PubMed]
- [7].Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, DeRisi JL, The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. Ward Gary, editor, PLoS Biol. [Internet] 1 (1) (2003) e5 August 18 [cited 2017 Mar 6] Available from: http://dx.plos.org/10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Bohme U, et al. , New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq, Mol. Microbiol 76 (1) (2010) 12–24, 10.1111/j.1365-2958.2009.07026.x (Epub 2010/02/10. PubMed PMID: ; PubMed Central PMCID: PMCPMC2859250). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Martinsen ES, Perkins SL, The diversity of Plasmodium and other haemosporidians: the intersection of taxonomy, phylogenetics and genomics, in: Carlton JM, Perkins SL, Deitsch KW (Eds.), Malaria Parasites: Comparative Genomics, Evolution and Molecular Biology, Caister Academic Press, Norfolk, Virginia, 2013, pp. 1–15. [Google Scholar]
- [10].Videvall E, Cornwallis CK, Palinauskas V, Valkiūnas G, Hellgren O, The avian transcriptome response to malaria infection, Mol. Biol. Evol 32 (5) (2015) 1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bensch S, Rn B, Ck C, Debarry JD, Johansson T, Hellgren O, et al. , The Genome of Haemoproteus tartakovskyi and Its Relationship to Human Malaria Parasites, [cited 2017 Mar 8]; Available from: http://mbio-serv2.mbioekol.lu.se/Malavi/Downloads. [DOI] [PMC free article] [PubMed]
- [12].Videvall E, Cornwallis CK, Ahrén D, Palinauskas V, Valkiūnas G, The Transcriptome of the Avian Malaria Parasite Plasmodium ashfordi Displays Host-Specific Gene Expression, [cited 2017 Feb 9]; Available from: 10.1101/072454. [DOI] [PubMed]
- [13].Hellgren O, Kutzer M, Bensch S, Valkiūnas G, Palinauskas V, Identification and characterization of the merozoite surface protein 1 (msp1) gene in a host-generalist avian malaria parasite, Plasmodium relictum (lineages SGS1 and GRW4) with the use of blood transcriptome, Malar. J [Internet] 12 (2013) [cited 2017 Apr 12];Available from: http://www.malariajournal.com/content/12/1/381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lauron EJ, Oakgrove KS, Tell LA, Biskar K, Roy SW, Sehgal RN, Transcriptome sequencing and analysis of Plasmodium gallinaceum reveals polymorphisms and selection on the apical membrane antigen-1, Malar. J 13 (2014) 382, 10.1186/1475-2875-13-382 (Epub 2014/09/28. PubMed PMID: ; PubMed Central PMCID: PMCPMC4182871). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lauron EJ, Aw Yeang HX, Taffner SM, Sehgal RN, De novo assembly and transcriptome analysis of Plasmodium gallinaceum identifies the Rh5 interacting protein (ripr), and reveals a lack of EBL and RH gene family diversification, Malar. J 14 (2015) 296, 10.1186/s12936-015-0814-0 (Epub 2015/08/06. PubMed PMID: ; PubMed Central PMCID: PMCPMC4524024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bensch S, Pérez-Tris J, Waldenström J, Hellgren O, Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: multiple cases of cryptic speciation? Evolution 58 (7) (2004) 1617–1621 (Epub 2004/09/03. PubMed PMID: ). [DOI] [PubMed] [Google Scholar]
- [17].Palinauskas V, Valkiūnas G, Bolshakov CV, Bensch S, Plasmodium relictum (lineage P-SGS1): effects on experimentally infected passerine birds, Exp. Parasitol 120 (4) (2008) 372–380 Epub 2008/09/24 10.1016/j.exppara.2008.09.001 (PubMed PMID: ). [DOI] [PubMed] [Google Scholar]
- [18].Mantilla JS, Gonzalez AD, Valkiūnas G, Moncada LI, Matta NE, Description and molecular characterization of Plasmodium (Novyella) unalis sp. nov. from the Great Thrush (Turdus fuscater) in highland of Colombia, Parasitol. Res 112 (12) (2013) 4193–4204, 10.1007/s00436-013-3611-0 (Epub 2013/09/ 21. PubMed PMID: ). [DOI] [PubMed] [Google Scholar]
- [19].Ilgūnas M, Palinauskas V, Iezhova TA, Valkiūnas G, Molecular and morphological characterization of two avian malaria parasites (Haemosporida: Plasmodiidae), with description of Plasmodium homonucleophilum n. sp, Zootaxa 3666 (2013) 49–61 (Epub 2013/01/01. PubMed PMID: ). [DOI] [PubMed] [Google Scholar]
- [20].Walther EL, Valkiūnas G, Gonzalez AD, Matta NE, Ricklefs RE, Cornel A, et al. Description, molecular characterization, and patterns of distribution of a wide-spread New World avian malaria parasite (Haemosporida: Plasmodiidae), Plasmodium (Novyella) homopolare sp. nov. Parasitol. Res 2014;113(9):3319–32. doi: 10.1007/s00436-014-3995-5. (Epub 2014/07/01. PubMed PMID: ). [DOI] [PubMed] [Google Scholar]
- [21].Perkins G, Frey T, Recent structural insight into mitochondria gained by microscopy, Micron [Internet] 31 (1) (2000) 97–111 [cited 2017 Apr 10]; Available from: http://www.sciencedirect.com/science/article/pii/S0968432899000657. [DOI] [PubMed] [Google Scholar]
- [22].Martinsen ES, Paperna I, Schall JJ, Morphological versus molecular identification of avian Haemosporidia: an exploration of three species concepts, Parasitol. Int 133 (3) (2006) 279 September 2 [cited 2017 Mar 2];Available from http://www.ncbi.nlm.nih.gov/pubmed/16740182. [DOI] [PubMed] [Google Scholar]
- [23].Pérez-Tris J, Hellgren O, Križanauskienė A, Waldenström J, Secondi J, Bonneaud C, et al. , Within-host speciation of malaria parasites. Buckling A, editor, PLoS One [Internet] 2 (2) (2007) e235 February 21 [cited 2017 Apr 10] Available from: http://dx.plos.org/10.1371/journal.pone.0000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Durrant KL, Beadell JS, Ishtiaq F, Graves GR, Olson SL, Gering E, et al. , Avian hematozoa in South America: a comparison of temperate and tropical zones, Ornithol. Monogr [Internet] 60 (2006) 98–111 January [cited 2017 Apr 10] Available from: http://www.bioone.org/doi/abs/10.2307/40166831. [Google Scholar]
- [25].Valkiūnas G, Ilgūnas M, Bukauskaitė D, Žiegytė R, Bernotienė R, Jusys V, et al. , Plasmodium delichoni n. sp.: description, molecular characterisation and remarks on the exoerythrocytic merogony, persistence, vectors and transmission, Parasitol. Res 115 (7) (2016) 2625–2636. [DOI] [PubMed] [Google Scholar]
- [26].Snow D, Perrins CM (Eds.), The birds of the Western Palearctic, Oxford University Press, Oxford, 1998. [Google Scholar]
- [27].Perez-Tris J, Hellgren O, Krizanauskiene A, Waldenstrom J, Secondi J, Bonneaud C, et al. Within-host speciation of malaria parasites . PLoS ONE. 2007;2(2):e235. doi: 10.1371/journal.pone.0000235. (Epub 2007/02/22. PubMed PMID: ; PubMed Central PMCID: PMCPMC1794596). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Szöllősi E, Rosivall B, Hasselquist D, Török J, The effect of parental quality and malaria infection on nestling performance in the Collared Flycatcher (Ficedula albicollis), J. Ornithol Springer Verlag 150 (2009) 519–527, 10.1007/s10336-008-0370-2. [DOI] [Google Scholar]
- [29].Dimitrov D, Palinauskas V, Iezhova TA, Bernotienė R, Ilgūnas M, Bukauskaitė D, Zehtindjiev P, Ilieva M, Shapoval AP, Bolshakov CV, Markovets MY, Bensch S, Valkiūnas G, Exp. Parasitol 148 (2015. January) 1–16, 10.1016/j.exppara.2014.11.005. [DOI] [PubMed] [Google Scholar]
- [30].Iezhova TA, Valkiūnas G, Bairlin F, Vertebrate host specificity of two avian malaria parasites of the subgenus Novyella: Plasmodium nucleophilum and Plasmodium vaughani, J. Parasitol 91 (2005) 458–461, 10.1645/GE-3377RN. [DOI] [PubMed] [Google Scholar]
- [31].Martinez C, Marzec T, Smith CD, Tell LA, Sehgal RN, Identification and expression of maebl, an erythrocyte-binding gene, in Plasmodium gallinaceum, Parasitol. Res 112 (3) (2013) 945–954, 10.1007/s00436-012-3211-4 (Epub 2012/12/12. PubMed PMID: ; PubMed Central PMCID: PMCPMC3581715). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mueller O, Lightfoot S, Schroeder A, RNA Integrity Number (RIN) – Standardization of RNA Quality Control Application, [cited 2017 Nov 8]; Available from: https://www.agilent.com/cs/library/applications/5989-1165EN.pdf.
- [33].Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. , Full-length transcriptome assembly from RNA-Seq data without a reference genome, Nat. Biotechnol [Internet] 29 (7) (2011) 644–652 May 15 [cited 2017 Mar 2]Available from: http://www.nature.com/doifinder/10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2, Nat. Methods [Internet] 9 (4) (2012) 357–359 March 4 [cited 2017 Mar 9]Available from: http://www.nature.com/doifinder/10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Böhme U, Otto TD, Cotton JA, Steinbiss S, Sanders M, Oyola SO, et al. , Complete avian malaria parasite genomes reveal features associated with lineage-specific evolution in birds and mammals, Genome Res, 28 Cold Spring Harbor Laboratory Press, 2018, pp. 547–560,, 10.1101/gr.218123.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Huang Y, Niu B, Gao Y, Fu L, Li W, CD-HIT Suite: a web server for clustering and comparing biological sequences, Bioinforma. Appl. Note [Internet] 26 (510) (2010) 680–682 [cited 2017 Mar 2]Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2828112/pdf/btq003.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M, Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research, Bioinformatics [Internet] 21 (18) (2005) 3674–3676 September 15 [cited 2017 Apr 24];Available from: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- [38].Das S, Hertrich N, Perrin AJ, Withers-Martinez C, Collins CR, Jones ML, et al. , Processing of Plasmodium falciparum merozoite surface protein MSP1 activates a spectrin-binding function enabling parasite egress from RBCs, Cell Host Microbe [Internet] 18 (4) (2015) 433–444 October 14 [Cited 2017 Oct 10]; Available from http://www.ncbi.nlm.nih.gov/pubmed/26468747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Beeson JG, Drew DR, Boyle MJ, Feng G, Fowkes FJI, Richards JS. Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria. FEMS Microbiol. Rev [Internet]. 2016;40(3):343–72. May [cited 2017 Feb 9] Available from: http://www.ncbi.nlm.nih.gov/pubmed/26833236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lamarque M, Bastien Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, Morlon-Guyot J, et al. , The RON2-AMA1 Interaction is a Critical Step in Moving Junction-Dependent Invasion by Apicomplexan Parasites, [cited 2017 Oct 11]; Available from: http://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1001276&type=printable. [DOI] [PMC free article] [PubMed]
- [41].Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, Moch JK, et al. , Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion, Proc. Natl. Acad. Sci. U S A [Internet] 108 (32) (2011) 13275–13280. August 9 [cited 2017 Oct 11]Available from http://www.ncbi.nlm.nih.gov/pubmed/21788485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Videvall E, Plasmodium parasites of birds have the most AT-rich genes of eukaryotes, Microb. Genomics Microbiol. Soc 4 (2018), 10.1099/mgen.0.000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Josling GA, Petter M, Oehring SC, Gupta AP, Dietz O, Wilson DW, et al. , A Plasmodium falciparum bromodomain protein regulates invasion gene expression, Cell Host Microbe, 17 Elsevier, 2015, pp. 741–751,, 10.1016/j.chom.2015.05.009. [DOI] [PubMed] [Google Scholar]
- [44].Opitz C, Soldati D, “The glideosome”: a dynamic complex powering gliding motion and host cell invasion by Toxoplasma gondii, Mol Microbiol, 45 Blackwell Science Ltd, 2002, pp. 597–604,, 10.1046/j.1365-2958.2002.03056.x. [DOI] [PubMed] [Google Scholar]
- [45].Baum J, Richard D, Healer J, Rug M, Krnajski Z, Gilberger T-W, et al. , A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites, J. Biol. Chem. Am. Soc. Biochem. Mol. Biol 281 (2006) 5197–5208, 10.1074/jbc.M509807200. [DOI] [PubMed] [Google Scholar]
- [46].Baker RP, Wijetilaka R, Urban S, Simpson K, Cowman A, Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria, PLoS Pathog. Public Libr. Sci 2 (2006) e113,, 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Santos J, Graindorge A, Soldati-Favre D, New insights into parasite rhomboid proteases, Mol. Biochem. Parasitol 182 (2012) 27–36, 10.1016/j.molbiopara.2011.11.010. [DOI] [PubMed] [Google Scholar]
- [48].de Koning HP, Bridges DJ, Burchmore RJS, Purine and pyrimidine transport in pathogenic protozoa: From biology to therapy, FEMS Microbiol. Rev [Internet] 29 (5) (2005) 987–1020 (November 1 [cited 2017 Oct 11] Available from: https://academic.oup.com/femsre/article-lookup/doi/10.1016/j.femsre.2005.03.004). [DOI] [PubMed] [Google Scholar]
- [49].Hyde JE. Targeting purine and pyrimidine metabolism in human apicomplexan parasites. Curr. Drug Targets [Internet]. 2007;8(1):31–47. January [cited 2017 Oct 11]Available from: http://www.ncbi.nlm.nih.gov/pubmed/17266529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.