Abstract
Proteins in the cell have to be eliminated once their function is no longer desired or they become damaged. Most regulated protein degradation is achieved by a large enzymatic complex called the proteasome. Many proteasome substrates are targeted for degradation by the covalent attachment of ubiquitin molecules. Ubiquitinated proteins can be bound by the proteasome, but for proteolysis to occur the proteasome needs to find a disordered tail somewhere in the target at which it initiates degradation. The initiation step contributes to the specificity of proteasomal degradation. Here, we review how the proteasome selects initiation sites within its substrates and discuss how the initiation step affects physiological processes.
Keywords: proteasome, ubiquitin, protein degradation, initiation region, protein unfolding
Introduction
To maintain cellular homeostasis, protein abundance is regulated by adjusting the rates of synthesis and degradation. A large fraction of regulated intracellular protein degradation is performed by the ubiquitin proteasome system (UPS), which plays pivotal roles in a variety of cellular processes including cell cycle regulation, membrane trafficking, and DNA repair.1, 2, 3 Proteins are targeted to proteasomal degradation through conjugation of the small globular protein ubiquitin to Lys residues in the substrate.4, 5, 6 The specificity and processivity of the UPS ensure proper cellular function and thus the UPS is indispensable for cells to survive.
The proteasome is a large protein complex with a molecular weight of ~2.5 MDa and consists of a 20S core particle (CP) and 19S regulatory particles (RP, also known as PA700) (Fig. 1) (Unverdorben P et al, PubMed ID: 24706844). In cells, the CP is predominantly capped at either or both ends by a specific RP, forming the holoenzyme known as the 26S proteasome.7 The CP possesses the proteolytic sites at the surface of an internal chamber.8 The RP contains three well‐characterized ubiquitin receptors (Rpn1, Rpn10, and Rpn13) and thus is responsible for substrate recognition.9 Once bound to the proteasome, substrate is unfolded by the action of six ATPase subunits (Rpt1–6) located in the RP and translocated from the RP into the CP, where it is cleaved into peptides of 3–8 amino acids.10 The ubiquitin chain is removed by the deubiquitinating activity of subunit Rpn11 in the RP as the substrate is pulled into the proteasome particle.11, 12, 13, 14 In addition to the stoichiometric proteasome subunits, several accessory proteins, such as the deubiquitinases Ubp6 (USP14 in mammals) and UCH37 (not present in yeast) and the shuttle factors Rad23 (HR23A/B), Dsk2 (UBQLNs) and Ddi1 (DDI1/2), bind transiently to the proteasome to play auxiliary roles in degradation.15 The proteasome also cooperates with some upstream factors including the ubiquitin‐selective chaperone Cdc48 (p97/VCP).16, 17
Figure 1.
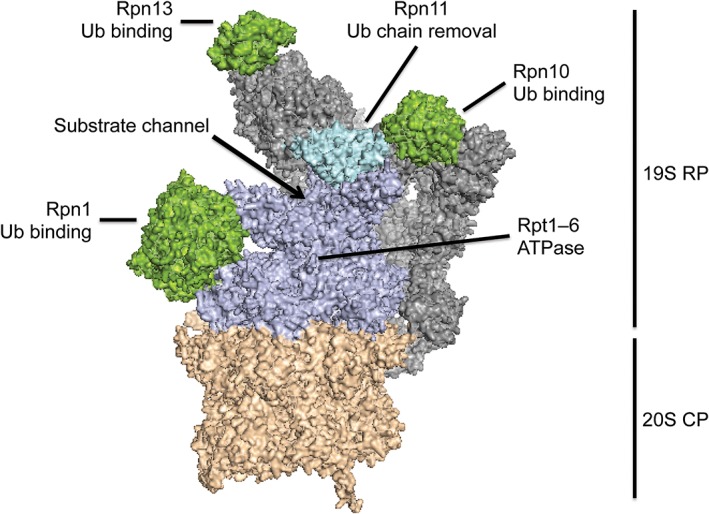
Structure of the 26S proteasome (based on PDB: 4CR2). The 26S proteasome consists of a 20S CP (orange) and a 19S RP (gray or indicated colors). The CP is responsible for proteolysis. The RP contains the ATPase subunits Rpt1–6 (purple), the deubiquitinase Rpn11 (cyan), and the ubiquitin receptors Rpn1, Rpn10, and Rpn13 (green). Substrates enter the proteasome through a channel at the center of a ring formed by the ATPase subunits.
The mechanism of proteasomal degradation can be divided into three steps, substrate recognition, unfolding, and proteolysis. Most proteins are targeted to the proteasome by the covalent attachment of multiple ubiquitin molecules through a cascade of three classes of enzymes acting sequentially: ubiquitin‐activating enzymes (E1), ubiquitin‐conjugating enzymes (E2), and ubiquitin ligases (E3). The E1 activates ubiquitin by catalyzing the formation of a ubiquitin adenylate. The activated ubiquitin is transferred to the E2. Finally, the E3 recognizes a short sequence or degron in the target protein and mediates the transfer of ubiquitin from the E2 to the target. The ubiquitin moieties are attached through isopeptide bonds between their C‐termini and amino groups in the substrate, usually the ε‐amino groups of Lys residues. Ubiquitin itself contains seven lysine residues (Lys 6, Lys 11, Lys 27, Lys 29, Lys 33, Lys 48, and Lys 63) as well as an α‐amino group at the N‐terminus (Met 1), and the ubiquitination process typically forms poly‐ubiquitin chains on proteins, as the first ubiquitin tag becomes itself modified and so on.18 Chains formed by ubiquitin molecules linked through Lys 48 are the most abundant in cells and represent the canonical degradation signal that targets substrates to the proteasome.19 The next most common linkage is through Lys 63, and chains of this type are associated with membrane trafficking, as are tags consisting of a single ubiquitin molecule.6, 20 Chains in which ubiquitin moieties are linked C‐ to N‐terminus through peptide bonds form protein complexes in signaling cascades. Branched chains in which ubiquitin moieties are tagged at two Lys residues are also observed. For example, chains with Lys 11 and Lys 48 linkages are made during cell cycle regulation.21 However, there is not a strict one‐to‐one correspondence between ubiquitin modification and cellular process and, for example, multiple monoubiquitin and Lys 63 chains can also target proteins for proteasomal degradation in some circumstances.6, 22
On the proteasome, ubiquitin chains are recognized by the three stoichiometric proteasome subunits mentioned above, through one (yeast) or two (human) ubiquitin‐interacting motif domains in Rpn10, the pleckstrin‐like receptor for ubiquitin domain of Rpn13, and the T1 site of Rpn1. Cells encode additional proteins that seem to function as supplementary substrate receptors. These shuttle receptors bind to ubiquitin chains through ubiquitin‐associated domains and are recruited to the proteasome via their N‐terminal ubiquitin‐like domains.23, 24, 25, 26, 27, 28, 29, 30, 31 The receptors perform largely overlapping functions though mutation of individual receptors affects degradation of subsets of proteasome substrates.32, 33, 34
Protein binding to the proteasome is not sufficient for degradation. A substrate is degraded only when the proteasome is able to engage it at a disordered region in the protein to initiate degradation. The length, location, and amino acid sequence of the disordered region determine how well it is recognized by the proteasome and thus how rapidly a protein is degraded. In this review, we highlight the mechanism by which the proteasome initiates degradation. We review the advances of the field from early conceptualization to recent progress, including application to disease treatment.
The Initiation Region of Proteasomal Degradation
Identification of the initiation region: Ubiquitination is not sufficient
Ubiquitination serves as the proteasome targeting signal, but it does not always lead to rapid degradation.19, 35 Substrates have to reach the proteolytic sites inside the proteasome through a channel, and the pore at its entrance has a diameter of approximately 13 Å.36, 37 Thus, folded proteins must be unraveled and threaded through the substrate channel to the proteolytic sites to be degraded. Early studies exploring how the proteasome unfolds proteins and initiates proteolysis analyzed the degradation of artificial substrates constructed from well‐defined building blocks. At their center were tightly folded domains derived from the ribonuclease barnase or dihydrofolate reductase (DHFR). The domains can be stabilized against unfolding by tightly binding ligands, barstar for barnase, and methotrexate for DHFR.38, 39, 40, 41 The proteins were then targeted to the proteasome by artificial degradation tags at their N‐termini composed of four ubiquitin domains fused to each other in frame through short linkers. Thus, the resulting hybrid proteins consisted entirely of compact domains. The proteasome was only able to degrade these targets if an unstructured region was also attached to their C‐termini.42 These and other experiments showed that the proteasome degrades proteins by engaging them at an unstructured region and then pulling them from there into the substrate channel and on to the proteolytic sites, unraveling any folded domains in the process.43 Thus, the proteasome degrades proteins sequentially from an initiation site that does not have to coincide with the ubiquitin tag.42, 43
Experiments investigating the regulation of ornithine decarboxylase (ODC) came to similar conclusions. ODC is degraded without ubiquitination but requires the cofactor antizyme.44 The proteasome recognizes ODC at an unstructured region at its C‐terminus and initiates its degradation there.45, 46 This C‐terminal region can induce the degradation of other proteins when attached to their C‐termini.47 Degradation of ODC itself requires binding of antizyme, to induce a conformational change that exposes the C‐terminal tail and to provide an additional interaction surface for proteasome binding.48 These observations led to the identification of initiation regions or initiation sites of proteasomal degradation: a disordered region at which the proteasome engages its substrates and initiates unfolding and degradation [Fig. 2(a,b)]. This initiation step can explain the behavior of physiological proteins. For example, Ubp6, mentioned above as an accessory factor in degradation, binds to the proteasome near the entrance to the substrate channel but escapes degradation of itself because it lacks efficient initiation regions.31
Figure 2.
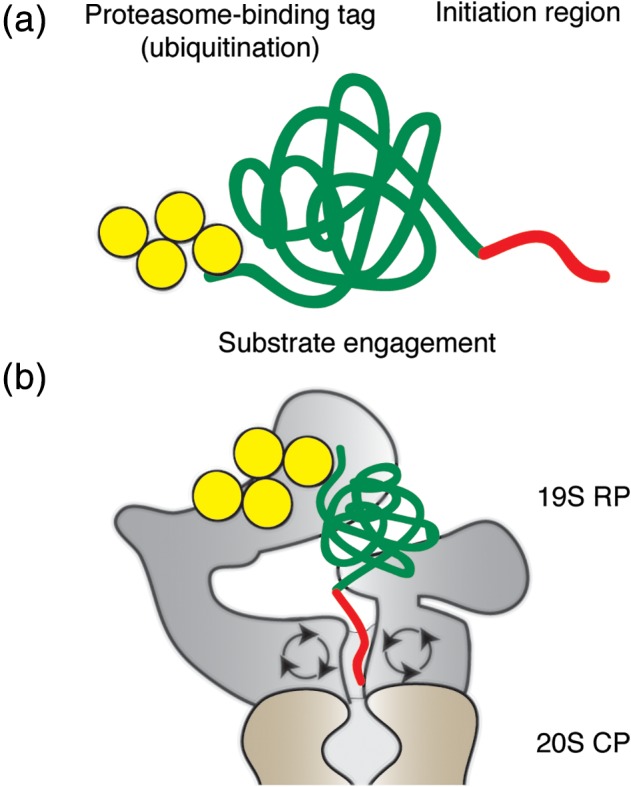
The proteasomal degradation signal has two parts. (a) Schematic representation of a proteasome substrate. A substrate protein (green) has to contain a proteasome‐binding tag (typically poly‐ubiquitin chains, yellow) and an unstructured region (red) for efficient degradation. (b) Substrate engagement by the proteasome. The proteasome recognizes a substrate via the ubiquitin tag and engages it at the disordered region for unfolding and translocation from the 19S RP to 20S CP.
Length: Be long enough to reach the proteasome
The proteasome is able to initiate degradation not only at N‐ or C‐terminal tails but also at internal disordered regions within a protein.49, 50 Interestingly, the disordered regions have to be much longer to allow efficient proteasomal degradation when they are located internally than as tails.50 This observation is compatible with a mechanism in which the disordered region has to engage a receptor in a channel before translocation and unfolding can occur. Recent high‐resolution structures of the proteasome and elegant biochemical experiments show that this is indeed how proteasomal protein degradation takes place.51, 52 The proteasome engages its substrates with loops (pore‐1 loops) in the proteasome's ATPase subunits. The loops contain Tyr residues that face the substrate channel and change their orientation during the ATPase cycle, apparently moving the substrate through channel. These Tyr loops are located some 35 Å from the entrance to the channel.9 This arrangement is compatible with the experimental observation that initiation regions at the C‐terminus of a protein have to be some 30 amino acids long and much longer when flanked by folded domains to allow the proteasome to engage the substrate effectively [Fig. 3(a)].31, 42, 47, 53, 54, 55, 56 After the proteasome grabs the initiation region, the ubiquitin chain is removed from substrate by the deubiquitinase Rpn11, which is located above the central pore of the proteasome and repositions upon substrate engagement.9, 11, 12, 13, 14
Figure 3.
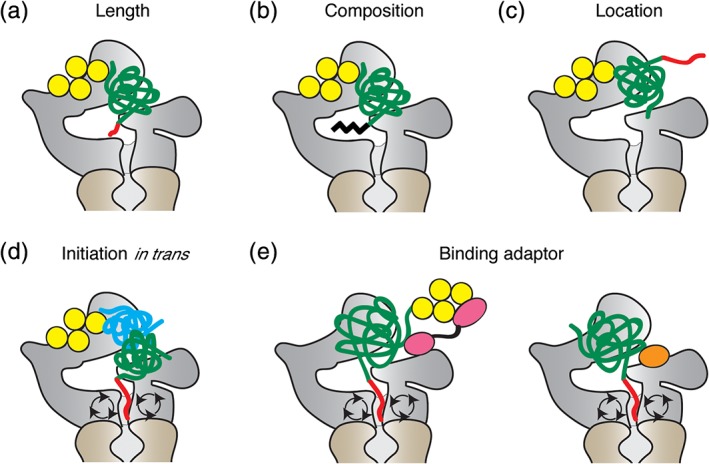
Initiation site selection by the proteasome. (a–c) Requirements for effective initiation regions. The initiation region has to be long enough (a), with a suitable amino acid composition (b), and be located in a position accessible to the proteasome (c). (d) Initiation of proteasomal degradation in trans. The proteasome can recognize a protein complex through ubiquitinated subunits (yellow and blue) but degrades only subunits with accessible initiation regions (green and red). (e) Speculative models of substrate recognition via degradation adaptors. The proteasome may recognize substrates through adaptor proteins (pink and orange) that recognize the ubiquitin chain attached to a substrate or the substrate directly without ubiquitination.
Composition: The proteasome likes sequences with diverse amino acid compositions
The proteasome is able to process almost any protein presented for destruction. At the same time, the surface of the initiation regions to be recognized by the proteasome will vary with the amino acids sequence and the unique chemical features of their side chains. Therefore, it is possible that the proteasome will recognize dissimilar initiation sites with different efficiencies.
An effect of the amino acid sequence of a protein on its degradation by the proteasome was originally proposed based on the observation that the Epstein–Barr virus protein EBNA1 escapes proteolysis and the generation of peptides to be displayed by MHC complexes and thus allows host cells that harbor the provirus in their genome to avoid detection by the immune system.57, 58 It appeared that the stability of EBNA1 was due to the presence of a long stretch of Gly‐Ala repeats within the protein that protected it from proteasomal degradation.59, 60, 61, 62
The proteasome's preference for the amino acid sequence of initiation regions was examined systematically by measuring the degradation of model proteins. An early small‐scale screen revealed that proteins in which the initiation regions have biased amino acid compositions show longer half‐lives than proteins with unbiased sequences in the regions.53 Analysis of ~100 different initiation regions indicated that in addition to compositional complexity, hydrophobicity, charge, and flexibility of the sequence also affect proteasomal recognition; the proteasome prefers hydrophobic and nonpolar amino acid residues and stiffer polypeptide chains, whereas polar, acidic, and structurally flexible sequences are avoided.54
These sequence preferences seem to matter in the cell [Fig. 3(b)]. The ubiquitin‐conjugating enzyme Cdc34 is ubiquitinated on a long disordered region at its C‐terminus, but escapes degradation. The C‐terminal tail is acidic and has biased composition, with 50 out of 130 amino acids being Asp or Glu residues preventing the proteasome form initiating degradation. Similarly, the shuttle receptor Rad23 contains three disordered linker regions of 60–68 amino acids but these regions do not serve as efficient initiation regions because of their biased amino acid composition. Cdc34 and Rad23 are easily degraded when short initiation regions are fused to their C‐termini.50, 53
The sequence preferences are also reflected in the behavior of proteins observed at the system‐level when analyzing protein stability in large‐scale proteomic studies. Proteins containing intrinsically disordered segments have significantly shorter half‐lives than proteins without these features across species.55 Protein stability furthermore correlates with the amino acid composition of the disordered region, as proteins in which the disordered region has strongly biased compositions are as stable as proteins without any disordered regions.53, 55
The proteasome's sequence preferences are most likely due to the effect of these sequences on their recognition in the ATPase subunits during translocation.63, 64 For example, hydrophobic initiation sequences may be preferred because of their interaction with the pore‐1 loops that drive substrate translocation.54, 65 Negatively charged surfaces of the substrate channel that flank the pore‐1 loops may repel acidic sequences.54, 65 Stiffer polypeptide chains may access to the pore‐1 loops more efficiently than more flexible sequences.54 At the same time, the proteasome is able to recognize and degrade a vast number of proteins in cells that do not share any obvious consensus sequence.53 Thus, the proteasome seems to be able to interact with a wide range of features within a stretch of amino acids, and only if pronounced sequence bias dilutes these features does recognition fail.53, 66
Location: Fitting substrate's geometrical arrangement
The proteasome degrades proteins efficiently when it binds to a ubiquitin tag on the substrate and engages it at an initiation region. The arrangement of the proteasome‐binding tag and the initiation region on the substrate presumably has to match the arrangement of their receptors on the proteasome [Fig. 3(c)].56 This relationship can be demonstrated experimentally on model substrates by inserting spacer domains between ubiquitin tag and initiation region to increase the distance between the two. When the initiation site is located too close or too far from the proteasome‐binding tag, the proteasome is unable to initiate degradation of the substrate.56
Do these restrictions on the geometry of degradation signals affect the behavior of physiological proteins? Ubiquitin tag and initiation region have to be close in space, but they can be separated onto different proteins [Fig. 3(d)].67 In protein complexes targeted for destruction, the proteasome degrades the subunits that contain accessible initiation regions while leaving the others intact. An example of this principle may be the remodeling by the proteasome of the protein complexes formed by cyclins, cyclin‐dependent kinases (CDKs), and CDK inhibitors (CKIs). Both cyclins and CKIs are highly reactive proteasome substrates, yet the proteasome is able to extract them one by one from trimeric complex. One explanation for the selective destruction would be that the initiation region on the first subunit to be degraded is placed such that it is recognized more easily than that of the other units, thus competing for degradation more effectively. Once the subunit with the ubiquitin tag is degraded, the remaining subunits are stable until they in turn become ubiquitinated and targeted for destruction.68
At the same time, degradation signal structure may also explain the needs for shuttle receptors. It is possible that one function of these accessory factors is to enhance the degradation of some substrates by presenting them to the proteasome more favorably for initiation than the intrinsic ubiquitin receptors are able to [Fig. 3(e)].
Accessory factor Cdc48: The proteasome occasionally needs help
The degradation of some proteins by the proteasome requires the action of the ATPase Cdc48 (p97/VCP) as an accessory factor.69, 70 Cdc48 cooperates with cofactors such as Ufd1 (UFD1L) and Npl4 (NPLOC4), which serve as ubiquitin receptors and regulate Cdc48‐dependent degradation pathways.71, 72, 73, 74 Most of these proteins are associated with membranes or are subunits of protein complexes.69, 70 For example, Cdc48 is a component of endoplasmic‐associated protein degradation where it mediates the transfer of proteins from the retrotranslocation machinery in the endoplasmic reticulum membrane to the proteasome.71, 72 Other examples are the transcription factors Spt23 and Mga2, which are synthesized as membrane‐anchored precursors and become activated when they are liberated by partial degradation by the proteasome in a Cdc48‐dependent manner.75
Biochemically, the role of Cdc48 may be to unfold proteins that do not contain regions that would allow the proteasome to initiate degradation.17, 76, 77 This function could explain the role of Cdc48 in the ubiquitin‐fusion degradation pathway.78, 79, 80, 81 For example, a chimeric protein consisting of ubiquitin domain attached to the N‐terminus of green fluorescent protein is not degraded by the proteasome in vitro unless a disordered region is fused to the protein, but can be unfolded by Cdc48.70, 74, 82 Similarly, Rpb1 (RPB1), a subunit of RNA polymerase II requires Cdc48 for degradation by the proteasome.83 Rpb1 has a long disordered region at its C‐terminus (the C‐terminal domain or CTD), which consists of many copies of a heptad repeat motif and serves as the binding site for cofactors and regulators of transcription. Rpb1 becomes ubiquitinated at the CTD when the polymerase stalls at sites of DNA damage but degradation of Rpb1 requires Cdc48.80, 84, 85, 86, 87 Cdc48 may be needed to unfold ubiquitinated Rpb1 and present it to the proteasome, because initiation at the CTD is prevented by the strong bias of its amino acid sequence.
Ubiquitin‐independent degradation: Another targeting mechanism?
If a protein has a particularly effective initiation region, it might be possible that it is degraded by the proteasome without ubiquitination, just as bacterial ATP‐dependent proteases recognize some substrates at C‐ or N‐terminal targeting signals.88 Indeed, some proteins are degraded in a ubiquitin‐independent manner either because they are recognized directly by the proteasome or because they interact with the proteasome via an adaptor protein [Fig. 3(e)]. The best‐established example is the ubiquitin‐independent degradation of ODC mediated by antizyme as described above.44, 89 Another example may be the degradation of the cytidine deaminase APOBEC3G, which is targeted to the proteasome by ubiquitinated viral infectivity factor (Vif).90 APOBEC3G can be degraded even when all Lys residues in the protein are mutated to prevent ubiquitination, presumably by ubiquitinated Vif acting as a proteasome adaptor.67
Several other proteins have been reported to be degraded in a ubiquitin‐independent manner, including NAD(P)H:quinone‐oxidoreductase‐1, steroid receptor coactivator 3, the CDK inhibitors p21Cip1, p16Ink4a, and p19Arf, p53, IκBα, a regulator of the transcription factor NFκB, and the transcription factor Rpn4, although some of these are also degraded through ubiquitin‐dependent pathways.91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102 A basal level of ubiquitin‐independent degradation of regulatory proteins may be common in signaling networks to allow them to respond rapidly to signals and revert back to steady state after the signal is withdrawn.103, 104 We expect that there are still a large number of substrates of ubiquitin‐independent degradation to be identified.
UPS and neurodegenerative diseases: Lack of initiation regions
Protein aggregation and inclusion body formation underlie the pathology of several neurodegenerative disorders including Huntington's disease (HD), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), Parkinson's disease, and Alzheimer's disease.105, 106, 107, 108 For example, in HD the intensity of the phenotype in mouse models correlates with the accumulation of a protein fragment corresponding to exon 1 of a mutated HTT gene (Htt) in nuclear inclusions, and a gain of toxic function in Htt mutants is implicated in the pathogenesis of HD.109 Despite evidence for the colocalization of ubiquitin and Htt as well as direct ubiquitination of Htt, the protein is not degraded and accumulates.110, 111, 112, 113, 114 Indeed, proteasome subunits are detected in the inclusion bodies formed in HD, suggesting that the proteasome may attempt to clear them.115 Autophagy, which is another bulk degradation process in cells, can also contribute to removal of protein aggregates but recent studies suggest that UPS inhibition has a greater effect on Htt accumulation than autophagy inhibition.116
A possible explanation for the failure of the UPS to eliminate these aggregates is an impairment of the proteasome. Protein aggregates can inhibit the UPS in culture cells and they may do so by clogging up the proteasome.117, 118 However, investigations in vivo have not yet reached a consensus. Several studies in HD animal models did not detect general defects in proteasome activity, while a cryo electron tomography study using an ALS/FTD model found that proteasome particles at the aggregates are in a substrate‐processing conformation, suggesting stalled degradation.119, 120, 121 Another possible reason for the stability of Htt aggregates is that Htt lacks an effective initiation region because the amino acid composition of its sequence is strongly biased. It consists of a short N‐terminal sequence of 18 amino acids, followed by a stretch of at least 23 Gln residues (polyQ) and then a Pro‐rich region of 50 amino acids, and does not allow the proteasome to initiate degradation in in vitro experiments.53, 122 In turn, attaching an effective initiation region to Htt leads to its proteasomal degradation in vitro and in yeast.53, 123 Thus, the pathogenesis of neurodegenerative diseases may in part be linked to the lack of proteasome initiation regions in aggregate‐prone proteins.
Inducible degradation: For a better design of protein knockdown tools
The targeted destruction of proteins in cells is a useful tool to investigate their functions and potentially a powerful therapeutic strategy. Various inducible degradation systems have been developed, including proteolysis‐targeting chimeras (PROTACs), which show promise for clinical use.124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134 Unlike DNA‐ or RNA‐targeting methods, inducible protein degradation systems could be effective for long‐lived proteins and may be able to distinguish between otherwise identical target proteins with different post‐translational modifications. The most common strategy is to induce ubiquitination of target proteins, in the case of PROTACs through small molecules that serve as adaptors that induce the interaction of the target with ubiquitin ligases.125, 127 The design of successful degradation tools is hindered by the fact that we do not fully understand the signals that control ubiquitination or how ubiquitin modifications are interpreted in the cell. In some cases PROTACs fail to induce degradation of a target despite binding efficiently.135, 136 One possible explanation is that once ubiquitinated the target proteins are not presented to the proteasome in a manner that allows effective initiation and thus degradation. Accordingly, taking into account the initiation step of proteasomal degradation may be helpful in designing of inducible degradation systems.
Conclusions
Ubiquitin tags target proteins to the proteasome but proteolysis requires that the proteasome engage its substrates at a disordered region to initiate degradation. The initiation step contributes to the specificity of proteasomal degradation. However, there are gaps in our understanding of how initiation regions function in cells. It is still hard to map initiation regions on a proteome‐wide scale, and the interplay between ubiquitination and initiation region remains elusive. The concept of proteasomal initiation has been developed mostly using model substrates, and it is necessary to translate existing knowledge to the behavior of natural proteins. Future studies will reveal the contribution of initiation regions in physiological processes and lead to a better understanding of protein degradation mechanisms in the UPS.
Acknowledgments
This work was supported by the National Institutes of Health through Grants R01GM124501 and R21CA196456 as well as the Welch Foundation through Grant F‐1817. A.M. is a paid consultant for Kymera Therapeutics.
Significance statement: The ubiquitin proteasome system surveils the proteome. Proteasome substrates are tagged with the small protein ubiquitin, but ubiquitination also signals other cellular fates. Proteolysis requires a disordered region in the substrate at which the proteasome initiates degradation, and this step contributes to target selection. Proteasome failure is associated with neurodegenerative diseases, and methods to induce protein degradation artificially could be a powerful therapeutic strategy.
Abbreviations: CDK, cyclin‐dependent kinases; CKI, CDK inhibitor; CP, core particle; CTD, C‐terminal domain; DHFR, dihydrofolate reductase; ODC, ornithine decarboxylase; PROTAC, proteolysis targeting chimera; RP, regulatory particles; UPS, the ubiquitin proteasome system; Vif, viral infectivity factor.
References
- 1. Schwartz AL, Ciechanover A (2009) Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol 49:73–96. [DOI] [PubMed] [Google Scholar]
- 2. Ravid T, Hochstrasser M (2008) Diversity of degradation signals in the ubiquitin‐proteasome system. Nat Rev Mol Cell Biol 9:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murata S, Yashiroda H, Tanaka K (2009) Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol 10:104–115. [DOI] [PubMed] [Google Scholar]
- 4. Finley D (2009) Recognition and processing of ubiquitin‐protein conjugates by the proteasome. Annu Rev Biochem 78:477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collins GA, Goldberg AL (2017) The logic of the 26S proteasome. Cell 169:792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229. [DOI] [PubMed] [Google Scholar]
- 7. Rousseau A, Bertolotti A (2018) Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol 19:697–712. [DOI] [PubMed] [Google Scholar]
- 8. Tanaka K (2009) The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci 85:12–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A (2018) Structure and function of the 26S proteasome. Annu Rev Biochem 87:697–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nussbaum AK, Dick TP, Keilholz W, Schirle M, Stevanović S, Dietz K, Heinemeyer W, Groll M, Wolf DH, Huber R, Rammensee H‐G, Schild H (1998) Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc Natl Acad Sci U S A 95:12504–112509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao T, Cohen RE (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419:403–407. [DOI] [PubMed] [Google Scholar]
- 12. Verma R, Aravind L, Oania R, McDonald WH, Yates JR, Koonin EV, Deshaies RJ (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298:611–615. [DOI] [PubMed] [Google Scholar]
- 13. Pathare GR, Nagy I, Śledź P, Anderson DJ, Zhou H‐J, Pardon E, Steyaert J, Förster F, Bracher A, Baumeister W (2014) Crystal structure of the proteasomal deubiquitylation module Rpn8‐Rpn11. Proc Natl Acad Sci U S A 111:2984–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Worden EJ, Padovani C, Martin A (2014) Structure of the Rpn11‐Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol 21:220–227. [DOI] [PubMed] [Google Scholar]
- 15. Yu H, Matouschek A (2017) Recognition of client proteins by the proteasome. Annu Rev Biophys 46:149–173. [DOI] [PubMed] [Google Scholar]
- 16. Bodnar N, Rapoport T (2017) Toward an understanding of the Cdc48/p97 ATPase. F1000Research 6:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saeki Y (2017) Ubiquitin recognition by the proteasome. J Biochem 161:mvw091. [DOI] [PubMed] [Google Scholar]
- 18. Deshaies RJ, Joazeiro CAP (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78:399–434. [DOI] [PubMed] [Google Scholar]
- 19. Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hicke L (2001) Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2:195–201. [DOI] [PubMed] [Google Scholar]
- 21. Meyer H‐J, Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157:910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh‐e A, Tanaka K (2009) Lysine 63‐linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J 28:359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391:715–718. [DOI] [PubMed] [Google Scholar]
- 24. Watkins JF, Sung P, Prakash L, Prakash S (1993) The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin‐like domain required for biological function. Mol Cell Biol 13:7757–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, Feng MT, Tübing F, Dittmar GAG, Finley D (2002) Proteasome subunit Rpn1 binds ubiquitin‐like protein domains. Nat Cell Biol 4:725–730. [DOI] [PubMed] [Google Scholar]
- 26. Chen L, Madura K (2002) Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol 22:4902–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saeki Y, Sone T, Toh‐e A, Yokosawa H (2002) Identification of ubiquitin‐like protein‐binding subunits of the 26S proteasome. Biochem Biophys Res Commun 296:813–819. [DOI] [PubMed] [Google Scholar]
- 28. Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin‐proteasome system. Cell 118:99–110. [DOI] [PubMed] [Google Scholar]
- 29. Kim I, Mi K, Rao H (2004) Multiple interactions of Rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell 15:3357–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Elsasser S, Chandler‐Militello D, Müller B, Hanna J, Finley D (2004) Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem 279:26817–26822. [DOI] [PubMed] [Google Scholar]
- 31. Yu H, Kago G, Yellman CM, Matouschek A (2016) Ubiquitin‐like domains can target to the proteasome but proteolysis requires a disordered region. EMBO J 35:1522–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee B‐H, Shi Y, Zhang N, de Poot SAH, Tuebing F, Sun S, Vannoy J, Tarasov SG, Engen JR, Finley D, Walters KJ (2016) Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351:aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peth A, Uchiki T, Goldberg AL (2010) ATP‐dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell 40:671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hamazaki J, Hirayama S, Murata S (2015) Redundant roles of Rpn10 and Rpn13 in recognition of ubiquitinated proteins and cellular homeostasis. PLoS Genet 11:e1005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Petroski MD, Deshaies RJ (2003) Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol Cell 11:1435–1444. [DOI] [PubMed] [Google Scholar]
- 36. Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 268:533–539. [DOI] [PubMed] [Google Scholar]
- 37. Groll M, Glickman MH, Finley D, Bajorek M, Köhler A, Moroder L, Rubin DM, Huber R (2000) A gated channel into the proteasome core particle. Nat Struct Biol 7:1062–1067. [DOI] [PubMed] [Google Scholar]
- 38. Martínez JC, Filimonov VV, Mateo PL, Schreiber G, Fersht AR (1995) A calorimetric study of the thermal stability of barstar and its interaction with barnase. Biochemistry 34:5224–5233. [DOI] [PubMed] [Google Scholar]
- 39. Huang S, Murphy S, Matouschek A (2000) Effect of the protein import machinery at the mitochondrial surface on precursor stability. Proc Natl Acad Sci U S A 97:12991–12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eilers M, Schatz G (1986) Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature 322:228–232. [DOI] [PubMed] [Google Scholar]
- 41. Huang S, Ratliff KS, Schwartz MP, Spenner JM, Matouschek A (1999) Mitochondria unfold precursor proteins by unraveling them from their N‐termini. Nat Struct Biol 6:1132–1138. [DOI] [PubMed] [Google Scholar]
- 42. Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A (2004) An unstructured initiation site is required for efficient proteasome‐mediated degradation. Nat Struct Mol Biol 11:830–837. [DOI] [PubMed] [Google Scholar]
- 43. Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A (2001) ATP‐dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell 7:627–637. [DOI] [PubMed] [Google Scholar]
- 44. Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A (1992) Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360:597–599. [DOI] [PubMed] [Google Scholar]
- 45. Zhang M, Pickart CM, Coffino P (2003) Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin‐independent substrate. EMBO J 22:1488–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang M, MacDonald AI, Hoyt MA, Coffino P (2004) Proteasomes begin ornithine decarboxylase digestion at the C terminus. J Biol Chem 279:20959–20965. [DOI] [PubMed] [Google Scholar]
- 47. Takeuchi J, Chen H, Coffino P (2007) Proteasome substrate degradation requires association plus extended peptide. EMBO J 26:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu H‐Y, Chen S‐F, Hsieh J‐Y, Chou F, Wang Y‐H, Lin W‐T, Lee P‐Y, Yu Y‐J, Lin L‐Y, Lin T‐S, Lin C‐L, Liu G‐Y, Tzeng S‐R, Hung H‐C, Chan N‐L (2015) Structural basis of antizyme‐mediated regulation of polyamine homeostasis. Proc Natl Acad Sci U S A 112:11229–11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nasmyth K, Peters JM, Uhlmann F (2000) Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288:1379–1385. [DOI] [PubMed] [Google Scholar]
- 50. Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A (2011) Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun 2:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de la Peña AH, Goodall EA, Gates SN, Lander GC, Martin A (2018) Substrate‐engaged 26S proteasome structures reveal mechanisms for ATP‐hydrolysis‐driven translocation. Science 362:eaav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dong Y, Zhang S, Wu Z, Li X, Wang WL, Zhu Y, Stoilova‐McPhie S, Lu Y, Finley D, Mao Y (2019) Cryo‐EM structures and dynamics of substrate‐engaged human 26S proteasome. Nature 565:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fishbain S, Inobe T, Israeli E, Chavali S, Yu H, Kago G, Babu MM, Matouschek A (2015) Sequence composition of disordered regions fine‐tunes protein half‐life. Nat Struct Mol Biol 22:214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu H, Singh Gautam AK, Wilmington SR, Wylie D, Martinez‐Fonts K, Kago G, Warburton M, Chavali S, Inobe T, Finkelstein IJ, Babu MM, Matouschek A (2016) Conserved sequence preferences contribute to substrate recognition by the proteasome. J Biol Chem 291:14526–14539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van der Lee R, Lang B, Kruse K, Gsponer J, Sánchez de Groot N, Huynen MA, Matouschek A, Fuxreiter M, Babu MM (2014) Intrinsically disordered segments affect protein half‐life in the cell and during evolution. Cell Rep 8:1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Inobe T, Fishbain S, Prakash S, Matouschek A (2011) Defining the geometry of the two‐component proteasome degron. Nat Chem Biol 7:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald‐Mullen PM, Klein G, Kurilla MG, Masucci MG (1995) Inhibition of antigen processing by the internal repeat region of the Epstein–Barr virus nuclear antigen‐1. Nature 375:685–688. [DOI] [PubMed] [Google Scholar]
- 58. Young LS, Rickinson AB (2004) Epstein‐Barr virus: 40 years on. Nat Rev Cancer 4:757–768. [DOI] [PubMed] [Google Scholar]
- 59. Sharipo A, Imreh M, Leonchiks A, Imreh S, Masucci MG (1998) A minimal glycine‐alanine repeat prevents the interaction of ubiquitinated I kappaB alpha with the proteasome: a new mechanism for selective inhibition of proteolysis. Nat Med 4:939–944. [DOI] [PubMed] [Google Scholar]
- 60. Zhang M, Coffino P (2004) Repeat sequence of Epstein‐Barr virus‐encoded nuclear antigen 1 protein interrupts proteasome substrate processing. J Biol Chem 279:8635–8641. [DOI] [PubMed] [Google Scholar]
- 61. Hoyt MA, Zich J, Takeuchi J, Zhang M, Govaerts C, Coffino P (2006) Glycine‐alanine repeats impair proper substrate unfolding by the proteasome. EMBO J 25:1720–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Daskalogianni C, Apcher S, Candeias MM, Naski N, Calvo F, Fåhraeus R (2008) Gly‐Ala repeats induce position‐ and substrate‐specific regulation of 26 S proteasome‐dependent partial processing. J Biol Chem 283:30090–30100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Beckwith R, Estrin E, Worden EJ, Martin A (2013) Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat Struct Mol Biol 20:1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, Förster F, Schulten K, Baumeister W, Sakata E (2017) Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc Natl Acad Sci U S A 114:1305–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen S, Wu J, Lu Y, Ma Y‐B, Lee B‐H, Yu Z, Ouyang Q, Finley DJ, Kirschner MW, Mao Y (2016) Structural basis for dynamic regulation of the human 26S proteasome. Proc Natl Acad Sci U S A 113:12991–12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK (2001) Sequence complexity of disordered protein. Proteins 42:38–48. [DOI] [PubMed] [Google Scholar]
- 67. Prakash S, Inobe T, Hatch AJ, Matouschek A (2009) Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol 5:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Verma R, McDonald H, Yates JR, Deshaies RJ (2001) Selective degradation of ubiquitinated Sic1 by purified 26S proteasome yields active S phase cyclin‐Cdk. Mol Cell 8:439–448. [DOI] [PubMed] [Google Scholar]
- 69. Meyer H, Bug M, Bremer S (2012) Emerging functions of the VCP/p97 AAA‐ATPase in the ubiquitin system. Nat Cell Biol 14:117–123. [DOI] [PubMed] [Google Scholar]
- 70. Xia D, Tang WK, Ye Y (2016) Structure and function of the AAA+ ATPase p97/Cdc48p. Gene 583:64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ye Y, Meyer HH, Rapoport TA (2003) Function of the p97–Ufd1–Npl4 complex in retrotranslocation from the ER to the cytosol. J Cell Biol 162:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA (2004) A membrane protein complex mediates retro‐translocation from the ER lumen into the cytosol. Nature 429:841–847. [DOI] [PubMed] [Google Scholar]
- 73. Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, Saeki Y (2017) In vivo ubiquitin linkage‐type analysis reveals that the Cdc48‐Rad23/Dsk2 axis contributes to K48‐linked chain specificity of the proteasome. Mol Cell 66:488–502. [DOI] [PubMed] [Google Scholar]
- 74. Bodnar NO, Rapoport TA (2017) Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169:722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S (2001) Mobilization of processed, membrane‐tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin‐selective chaperone. Cell 107:667–677. [DOI] [PubMed] [Google Scholar]
- 76. Olszewski MM, Williams C, Dong KC, Martin A (2019) The Cdc48 unfoldase prepares well‐folded protein substrates for degradation by the 26S proteasome. Commun Biol 2:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Beskow A, Grimberg KB, Bott LC, Salomons FA, Dantuma NP, Young P (2009) A conserved unfoldase activity for the p97 AAA‐ATPase in proteasomal degradation. J Mol Biol 394:732–746. [DOI] [PubMed] [Google Scholar]
- 78. Wójcik C, Rowicka M, Kudlicki A, Nowis D, McConnell E, Kujawa M, DeMartino GN (2006) Valosin‐containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N‐end rule and ubiquitin‐fusion degradation pathway substrates in mammalian cells. Mol Biol Cell 17:4606–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chou T‐F, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, Patricelli MP, Hodder P, Rosen H, Deshaies RJ (2011) Reversible inhibitor of p97, DBeQ, impairs both ubiquitin‐dependent and autophagic protein clearance pathways. Proc Natl Acad Sci U S A 108:4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ghislain M, Dohmen RJ, Levy F, Varshavsky A (1996) Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin‐mediated proteolysis in Saccharomyces cerevisiae . EMBO J 15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 81. Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270:17442–17456. [DOI] [PubMed] [Google Scholar]
- 82. Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG (2000) Short‐lived green fluorescent proteins for quantifying ubiquitin/proteasome‐dependent proteolysis in living cells. Nat Biotechnol 18:538–543. [DOI] [PubMed] [Google Scholar]
- 83. Verma R, Oania R, Fang R, Smith GT, Deshaies RJ (2011) Cdc48/p97 mediates UV‐dependent turnover of RNA Pol II. Mol Cell 41:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wilson MD, Harreman M, Svejstrup JQ (2013) Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim Biophys Acta Gene Regul Mech 1829:151–157. [DOI] [PubMed] [Google Scholar]
- 85. Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP (2008) Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc Natl Acad Sci U S A 105:19649–19654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Huibregtse JM, Yang JC, Beaudenon SL (1997) The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin‐protein ligase. Proc Natl Acad Sci U S A 94:3656–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ribar B, Prakash L, Prakash S (2007) ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA‐damaged yeast cells. Mol Cell Biol 27:3211–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Olivares AO, Baker TA, Sauer RT (2015) Mechanistic insights into bacterial AAA+ proteases and protein‐remodelling machines. Nat Rev Microbiol 14:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Erales J, Coffino P (2014) Ubiquitin‐independent proteasomal degradation. Biochim Biophys Acta Mol Cell Res 1843:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dang Y, Siew LM, Zheng Y‐H (2008) APOBEC3G is degraded by the proteasomal pathway in a Vif‐dependent manner without being polyubiquitylated. J Biol Chem 283:13124–13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Moscovitz O, Tsvetkov P, Hazan N, Michaelevski I, Keisar H, Ben‐Nissan G, Shaul Y, Sharon M (2012) A mutually inhibitory feedback loop between the 20S proteasome and its regulator, NQO1. Mol Cell 47:76–86. [DOI] [PubMed] [Google Scholar]
- 92. Li X, Lonard DM, Jung SY, Malovannaya A, Feng Q, Qin J, Tsai SY, Tsai M‐J, O'Malley BW (2006) The SRC‐3/AIB1 coactivator is degraded in a ubiquitin‐ and ATP‐independent manner by the REGγ proteasome. Cell 124:381–392. [DOI] [PubMed] [Google Scholar]
- 93. Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387:296–299. [DOI] [PubMed] [Google Scholar]
- 94. Lu Z, Hunter T (2010) Ubiquitylation and proteasomal degradation of the p21 Cip1 , p27 Kip1 and p57 Kip2 CDK inhibitors. Cell Cycle 9:2342–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM (2007) Ubiquitin‐independent degradation of cell‐cycle inhibitors by the REGγ proteasome. Mol Cell 26:843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tsvetkov P, Reuven N, Shaul Y (2010) Ubiquitin‐independent p53 proteasomal degradation. Cell Death Differ 17:103–108. [DOI] [PubMed] [Google Scholar]
- 97. Krappmann D, Wulczyn FG, Scheidereit C (1996) Different mechanisms control signal‐induced degradation and basal turnover of the NF‐kappaB inhibitor IkappaB alpha in vivo. EMBO J 15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- 98. Krappmann D, Scheidereit C (2005) A pervasive role of ubiquitin conjugation in activation and termination of IκB kinase pathways. EMBO Rep 6:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mathes E, Wang L, Komives E, Ghosh G (2010) Flexible regions within IκBα create the ubiquitin‐independent degradation signal. J Biol Chem 285:32927–32936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Xie Y, Varshavsky A (2001) RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A 98:3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ju D, Xie Y (2006) Identification of the preferential ubiquitination site and ubiquitin‐dependent degradation signal of Rpn4. J Biol Chem 281:10657–10662. [DOI] [PubMed] [Google Scholar]
- 102. Ju D, Xie Y (2004) Proteasomal degradation of RPN4 via two distinct mechanisms, ubiquitin‐dependent and ‐independent. J Biol Chem 279:23851–23854. [DOI] [PubMed] [Google Scholar]
- 103. O'Dea EL, Barken D, Peralta RQ, Tran KT, Werner SL, Kearns JD, Levchenko A, Hoffmann A (2007) A homeostatic model of IκB metabolism to control constitutive NF‐κB activity. Mol Syst Biol 3:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Loriaux PM, Hoffmann A (2013) A protein turnover signaling motif controls the stimulus‐sensitivity of stress response pathways. PLoS Comput Biol 9:e1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ross CA, Poirier MA (2004) Protein aggregation and neurodegenerative disease. Nat Med 10:S10–S17. [DOI] [PubMed] [Google Scholar]
- 106. Dantuma NP, Bott LC (2014) The ubiquitin‐proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front Mol Neurosci 7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hipp MS, Park S‐H, Hartl FU (2014) Proteostasis impairment in protein‐misfolding and ‐aggregation diseases. Trends Cell Biol 24:506–514. [DOI] [PubMed] [Google Scholar]
- 108. Schmidt M, Finley D (2014) Regulation of proteasome activity in health and disease. Biochim Biophys Acta Mol Cell Res 1843:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zuccato C, Valenza M, Cattaneo E (2010) Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev 90:905–981. [DOI] [PubMed] [Google Scholar]
- 110. DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277:1990–1993. [DOI] [PubMed] [Google Scholar]
- 111. Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG, Graham KC, Goldberg YP, Gietz RD, Pickart CM, Hayden MR (1996) Huntingtin is ubiquitinated and interacts with a specific ubiquitin‐conjugating enzyme. J Biol Chem 271:19385–19394. [DOI] [PubMed] [Google Scholar]
- 112. Douglas PM, Dillin A (2010) Protein homeostasis and aging in neurodegeneration. J Cell Biol 190:719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Jana NR, Zemskov EA, Wang G, Nukina N (2001) Altered proteasomal function due to the expression of polyglutamine‐expanded truncated N‐terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet 10:1049–1059. [DOI] [PubMed] [Google Scholar]
- 114. Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE (2001) Accumulation of mutant Huntingtin fragments in aggresome‐like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell 12:1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wyttenbach A, Carmichael J, Swartz J, Furlong RA, Narain Y, Rankin J, Rubinsztein DC (2000) Effects of heat shock, heat shock protein 40 (HDJ‐2), and proteasome inhibition on protein aggregation in cellular models of Huntington's disease. Proc Natl Acad Sci U S A 97:2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Li X‐J, Li H, Li S (2010) Clearance of mutant huntingtin. Autophagy 6:663–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin‐proteasome system by protein aggregation. Science 292:1552–1555. [DOI] [PubMed] [Google Scholar]
- 118. Bennett EJ, Bence NF, Jayakumar R, Kopito RR (2005) Global impairment of the ubiquitin‐proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell 17:351–365. [DOI] [PubMed] [Google Scholar]
- 119. Bett JS, Goellner GM, Woodman B, Pratt G, Rechsteiner M, Bates GP (2006) Proteasome impairment does not contribute to pathogenesis in R6/2 Huntington's disease mice: exclusion of proteasome activator REGγ as a therapeutic target. Hum Mol Genet 15:33–44. [DOI] [PubMed] [Google Scholar]
- 120. Maynard CJ, Bottcher C, Ortega Z, Smith R, Florea BI, Diaz‐Hernandez M, Brundin P, Overkleeft HS, Li J‐Y, Lucas JJ, Dantuma NP (2009) Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc Natl Acad Sci U S A 106:13986–13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Guo Q, Lehmer C, Martínez‐Sánchez A, Rudack T, Beck F, Hartmann H, Pérez‐Berlanga M, Frottin F, Hipp MS, Hartl FU, Edbauer D, Baumeister W, Fernández‐Busnadiego R (2018) In situ structure of neuronal C9orf72 poly‐GA aggregates reveals proteasome recruitment. Cell 172:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Fiumara F, Fioriti L, Kandel ER, Hendrickson WA (2010) Essential role of coiled coils for aggregation and activity of Q/N‐rich prions and PolyQ proteins. Cell 143:1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rousseau E, Kojima R, Hoffner G, Djian P, Bertolotti A (2009) Misfolding of proteins with a polyglutamine expansion is facilitated by proteasomal chaperones. J Biol Chem 284:1917–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Neklesa TK, Tae HS, Schneekloth AR, Stulberg MJ, Corson TW, Sundberg TB, Raina K, Holley SA, Crews CM (2011) Small‐molecule hydrophobic tagging–induced degradation of HaloTag fusion proteins. Nat Chem Biol 7:538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ (2001) Protacs: chimeric molecules that target proteins to the Skp1‐Cullin‐F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A 98:8554–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Fulcher LJ, Hutchinson LD, Macartney TJ, Turnbull C, Sapkota GP (2017) Targeting endogenous proteins for degradation through the affinity‐directed protein missile system. Open Biol 7:170066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Bondeson DP, Mares A, Smith IED, Ko E, Campos S, Miah AH, Mulholland KE, Routly N, Buckley DL, Gustafson JL, Zinn N, Grandi P, Shimamura S, Bergamini G, Faelth‐Savitski M, Bantscheff M, Cox C, Gordon DA, Willard RR, Flanagan JJ, Casillas LN, Votta BJ, den Besten W, Famm K, Kruidenier L, Carter PS, Harling JD, Churcher I, Crews CM (2015) Catalytic in vivo protein knockdown by small‐molecule PROTACs. Nat Chem Biol 11:611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Banaszynski LA, Chen L, Maynard‐Smith LA, Ooi AGL, Wandless TJ (2006) A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hatakeyama S, Watanabe M, Fujii Y, Nakayama KI (2005) Targeted destruction of c‐Myc by an engineered ubiquitin ligase suppresses cell transformation and tumor formation. Cancer Res 65:7874–7879. [DOI] [PubMed] [Google Scholar]
- 130. Robinson MS, Sahlender DA, Foster SD (2010) Rapid inactivation of proteins by rapamycin‐induced rerouting to mitochondria. Dev Cell 18:324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Caussinus E, Kanca O, Affolter M (2012) Fluorescent fusion protein knockout mediated by anti‐GFP nanobody. Nat Struct Mol Biol 19:117–121. [DOI] [PubMed] [Google Scholar]
- 132. Ma Y, Gu Y, Zhang Q, Han Y, Yu S, Lu Z, Chen J (2013) Targeted degradation of KRAS by an engineered ubiquitin ligase suppresses pancreatic cancer cell growth in vitro and in vivo. Mol Cancer Ther 12:286–294. [DOI] [PubMed] [Google Scholar]
- 133. Portnoff AD, Stephens EA, Varner JD, DeLisa MP (2014) Ubiquibodies, synthetic E3 ubiquitin ligases endowed with unnatural substrate specificity for targeted protein silencing. J Biol Chem 289:7844–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wilmington SR, Matouschek A (2016) An inducible system for rapid degradation of specific cellular proteins using proteasome adaptors. PLoS One 11:e0152679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime‐Figueroa S, Wang J, Hamman BD, Ishchenko A, Crews CM (2018) Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol 25:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Huang HT, Dobrovolsky D, Paulk J, Yang G, Weisberg EL, Doctor ZM, Buckley DL, Cho JH, Ko E, Jang J, Shi K, Choi HG, Griffin JD, Li Y, Treon SP, Fischer ES, Bradner JE, Tan L, Gray NS (2018) A chemoproteomic approach to query the degradable kinome using a multi‐kinase degrader. Cell Chem Biol 25:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]


