Abstract
Gene expression is regulated by numerous elements including enhancers, insulators, transcription factors, and architectural proteins. Regions of DNA distal to the transcriptional start site, called enhancers, play a central role in the temporal and tissue-specific regulation of gene expression through RNA polymerase II. The identification of enhancers and other cis regulatory elements has largely been possible due to advances in next generation sequencing technologies. Enhancers regulate gene expression through chromatin loops mediated by architectural proteins such as YY1, CTCF, the cohesin complex, and LDB1. Additionally, enhancers can be transcribed to produce non-coding RNAs termed enhancer RNAs that likely participate in transcriptional regulation. The central role of enhancers in regulating gene expression implicates them in both normal physiology but also many disease states. The importance of enhancers is evident by the suggested role of SNPs, duplications, and other alterations of enhancer function in many diseases, ranging from cancer to atherosclerosis to chronic kidney disease. Although much progress has been made in recent years, the field of enhancer biology and our knowledge of the cis regulome remains a work in progress. This review will highlight recent seminal studies which demonstrate the role of enhancers in normal physiology and disease pathogenesis.
Keywords: enhancers, transcriptional regulation, cis regulatory elements
Introduction:
The proper regulation of gene expression is required for the billions of cells within a metazoan to properly develop and perform their proscribed function(s). Both cis and trans genetic elements can regulate gene expression. Cis elements are regulatory chromatin regions that act upon other areas of the same chromosome, whereas trans elements act between elements on different chromosomes with the classic example being DNA-binding proteins such as transcription factors. Some important cis or trans elements which we will discuss further include promoters, enhancers, transcription factors, CCCTC-binding factor (CTCF) and the cohesin complex (Figure 1). All of these DNA elements or proteins play a central role in proper gene expression. The three-dimensional nature of chromatin interactions and how these interactions play a role in regulating gene expression is still being uncovered and will also be discussed. Here we aim to describe several aspects of transcriptional regulation, including the early discovery of regulatory elements, how the field studies these elements, how we currently understand the role of nuclear architecture in controlling gene expression, and the role of regulatory elements in promoting development, normal physiology, and disease. While we attempt to share a comprehensive account of what is currently understood in this field only a subset of topics is reviewed to provide a high-level overview of this burgeoning field. We will focus on key recent discoveries that had a significant impact on our understanding of transcriptional regulatory elements.
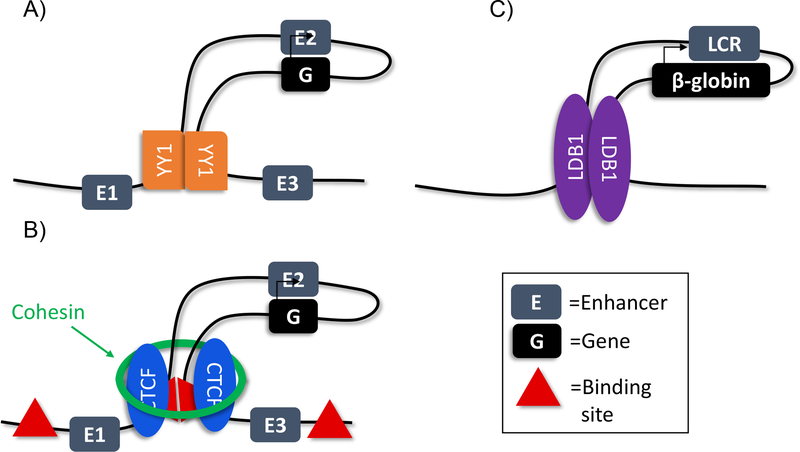
Figure 1.
Trans factors involved in chromatin looping.
A) YY1 dimerization increases chromatin looping and enhancer-promoter interaction
B) Cohesin and CTCF are enriched at chromatin loop anchors
C) LDB1 is enriched at the loop anchors at the β-globin locus
Teaching point: Chromatin looping may be due to multiple different mechanisms
What are enhancers?
Enhancers are cis regulatory elements containing DNA binding motifs for specific transcription factors that cooperate with enhancers to drive gene expression independent of distance or orientation. This is distinct from promoters, which are required to be immediately adjacent to the transcriptional start site (TSS) of a gene (Figure 2). Transcription factor binding at an enhancer element can drive either activation or repression of nearby genes. The precise molecular mechanism of this remains unclear, as we will discuss later in this review. Enhancers can function at a great distance, up to hundreds or thousands of kilobases away from their target gene (114). These regulatory elements are either intragenic or extragenic, and are highly tissue specific, making them versatile molecular “switches” of gene expression. As technological advances have been made in experimental methods (detailed in Table 1), the enhancer field has expanded correspondingly.
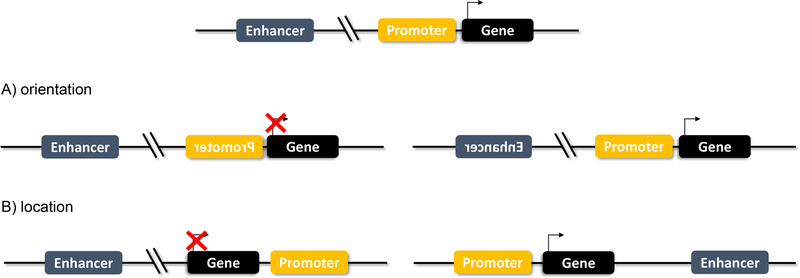
Figure 2.
Enhancers vs Promoters
A) Enhancers function independent of orientation. If an enhancer is flipped, it maintains its gene regulation function. A flipped promoter, however, will block gene transcription.
B) Enhancers function independent of location. An enhancer that is moved downstream or upstream remains functional. A promoter that is no longer directly adjacent to the TSS will not drive transcription.
Teaching point: Enhancers function independent of orientation and location, unlike promoters.
Table 1:
Techniques used to study gene regulatory elements.
| Technique | Purpose | Limitations |
|---|---|---|
RNA-seq (29, 51)
|
Identify changes in transcriptional profile between different conditions |
|
ChIP-seq (Chromatin Immunoprecipitation) (8)(10) (44)
|
Identify elements based on their histone modification or TF binding. See Figure 3 for typical signatures. |
|
ATAC-seq (Assay for Transposase Accessible Chromatin) (15)
|
Identify areas of accessible chromatin: areas likely to be functional regions actively involved in transcription |
|
|
Chromosome Conformation(40) (104) Cross link chromatin, digest, and ligate chromatin
|
Identify variety of interactions between one or many regulatory regions and their associated genes based on specific technology used
|
|
STARR-seq (Self Transcribing Active Regulatory Region)(3)
|
Identify the effect of many different putative regulatory sequences on overall gene expression. |
|
CAGE-seq (Cap Analysis of Gene Expression)(2)
|
Identify 5’ ends of nascent transcripts and determine transcriptional direction. |
|
| CRISPR/Cas9-based | Identify specific sequences necessary for enhancer function. |
|
Teaching point: a wide variety of techniques utilizing next generation sequence have been developed that allow us to study the role enhancers play in gene regulation.
Enhancers were first identified in 1981 using plasmid-based reporter assays (7, 65). A 72bp viral sequence, known to promote efficient transcription, and the β-globin gene were cloned into a plasmid and transfected into HeLa cells. A 200-fold increase in β-globin transcripts was identified in these cells, indicating that the short sequence aided in enhancing transcription, thus dubbing these sequences “enhancers”. The 72bp sequence enhanced transcription irrespective of orientation and position in relation to the β-globin gene while on the same reporter plasmid. Transfection of three copies of the β-globin gene and two copies of the enhancer sequence on separate plasmids did not increase β-globin transcription. This experiment formally demonstrated the enhancer could not increase β-globin transcription when present on a separate plasmid, and therefore enhancers function in cis and not in trans. Further experimentation showed that multiple copies of the enhancer sequence in tandem increased β-globin expression (7, 65). Another study utilizing additional plasmid reporter assays identified enhancer sequences in the mouse immunoglobulin heavy chain gene that were only present in immune cells, thereby defining enhancers as critical to tissue-specific gene regulation (6, 42). These experiments led to the commonly accepted definition of enhancers: DNA elements that augment gene expression in cis and act independent of orientation and position (Figure 2). These are the features that DNA elements must possess to be defined as enhancers. A caveat to these experiments is that while they identified a new class of DNA element, they were limited by the plasmid-based technology, which required prior knowledge of the sequences they were interrogating. In addition, by isolating the sequence from its native chromatin state, plasmid-based approaches are limited to identifying the potential of a DNA sequence to act as an enhancer, while not definitively establishing the sequence acts as an enhancer in vivo.
In 1987, a LacZ reporter method was developed in Drosophila to investigate and visualize regulatory genomic elements in situ in the developing fly (77). Reporter studies in Drosophila melanogaster in the early 1990s used the even-skipped (eve) stripe 2 gene to investigate enhancer sequences (4, 99). In these studies, lacZ fused to the eve gene allowed visualization in developing embryos as a readout for enhancer function. eve2 is regulated in trans by the maternal morphogen bicoid and three “gap genes”: hunchback, Kruppel, and giant. These genes, when mutated, cause a loss in Drosophila segments or form “gaps” in development. All four of these bind regions upstream and downstream of the eve2 gene. Mutations in the binding sites for each of these cause variable expansion of the eve stripe 2, indicating that binding of the proteins can either activate or repress eve2 expression. Specifically, it is the combination of the different proteins binding the DNA element at specific levels that either activate and repress expression, suggesting enhancers can be genetic sensors for protein levels. These studies emphasize that enhancers act as genetic “on-off” switches, and this activity can be modulated by protein levels.
While much of early enhancer biology was investigated using Drosophila as a model, additional studies of enhancers were performed in mouse and human genomes in the 1990s. B- and T-lymphocyte experiments using transgenic constructs in mice investigating the immunoglobulin VDJ rearrangement found two cis acting elements that drove recombination (36). These enhancers were also identified as tissue-specific, as one drove DJ recombination in B cells and another VDJ recombination in T cells. Enhancer studies in mouse and human genomes were limited initially however, by the available technology to identify cis regulatory elements until advancements in genome-wide methods allowed identification and characterization of enhancers in mammalian genomes.
Identification and Characterization
Before enhancers were identified as a type of cis regulatory element (CRE), studies using SV40 and Drosophila worked to identify regions of the genome most susceptible to digestion by DNase I (34). These were regions of the genome absent of nucleosomes, and presumably accessible to transcriptional machinery and transcription factors. Studies found that these regions corresponded to both promoters and enhancers. While promoter identification was simpler due to proximity to the TSS, DNase I sensitivity for enhancers was critical to identification of potential distal enhancer sequences. As new technologies were discovered in the late 1990s and early 2000s, in particular high throughput technologies, the enhancer field obtained tools to answer key questions regarding enhancer function. Chromatin immunoprecipitation (ChIP) and real time quantitative PCR (qPCR) led to the discovery of protein enrichment at enhancers, particularly RNA polymerase II and p300 (an important transcriptional coactivator) (110). By overlaying p300-bound sites onto known gene locations, DNase hypersensitive sites, and evolutionarily conserved enhancer sites, Heintzman et al. used ChIP paired with microarray (ChIP-chip) to formulate a histone signature for enhancers (44). Specifically, an enrichment of H3K4me1 at the peak of p300 and a depletion of H3K4me3 helped identify DNA regions with enhancer activity. By contrast, promoters could be distinguished by the presence of high levels of H3K4me3, although they also frequently demonstrate H3K4me1 and p300 occupancy. ChIP-chip in three additional human cell lines further identified H3K27Ac enrichment at putative enhancers, completing a histone signature for enhancers enabling genome-wide analysis for possible enhancer-elements (Figure 3; 43).
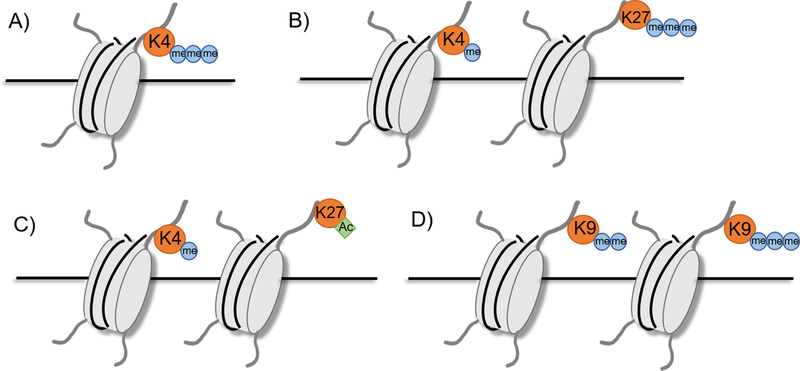
Figure 3.
Characteristic histone marks.
A) Active promoter: H3K4me3
B) Poised enhancer: H3K4me1, H3K27me3
C) Active enhancer: H3K4me1, H3K27ac
D) Silenced region: H3K9me2 or H3K9me3
Teaching point: the activity or lack thereof of enhancers can be inferred by various histone marks.
The establishment of a histone signature of enhancers combined with the advent of next generation sequencing (NGS) revolutionized the field of enhancer biology by permitting their unbiased, genome-wide identification. ChIP coupled with NGS (ChIP-Seq) provides a high-throughput method to study chromatin structure and DNA-protein binding (Table 1). Analysis of p300 enriched regions in human cell lines combined with transgenic mice studies showed that ChIP-Seq can accurately predict enhancers that are driving gene expression at the time point of the experiment (106). ChIP-Seq enabled more in-depth analysis of enhancers and led to the definition of novel enhancer subgroups. Studies in human embryonic stem cells found that p300 and H3K4me1 enriched regions could be divided into two groups, one enriched for H3K27Ac and one enriched for H3K27me3 (84). RNA-seq showed that genes near enhancers enriched for H3K27Ac were highly expressed, while genes near enhancers enriched for H3K27me3 were poorly expressed. As the cells differentiated, however, the enhancers previously enriched for H3K27me3 gained H3K27Ac, establishing that these enhancers are a group of inactive or “poised” enhancers. Enhancers, therefore, can be identified by enrichment of H3K4me1, with active enhancers by H3K27Ac enrichment and poised enhancers by both an absence of H3K27Ac and an enrichment for H3K27me3 (Figure 3). ChIP-Seq analysis of embryonic mouse limbs further fine-tuned our understanding of histone modifications of enhancers by recognizing a lower level of H3K27Ac enrichment signature in inactive developmental enhancers that maintain an open chromatin state (23). This established the paradigm that enhancer presence and activity can be identified using a level of H3K27Ac enrichment, allowing in depth computational study of enhancers in the genome. Studies focusing on DNA methylation have also identified that active enhancers are hypomethylated, providing another chromatin mark for enhancers that can be identified using genome-wide approaches (98, 113). Together, these studies have formed recognizable chromatin signatures that can be used to identify enhancers (Figure 3).
The establishment of a histone signature has allowed more in-depth analysis of enhancers and driven new discoveries. Combination of ChIP-Seq data and the enhancer histone signature led to the discovery of a subset of enhancers bound directly by RNA polymerase II (29, 51). Another group found that 70% of extragenic RNA pol II binding sites overlap with enhancers, based upon histone signature and previously defined long non-coding RNAs (29). Classically, enhancers were believed to be bound by transcription factors to help recruit RNA pol II to promoters to regulate gene expression. The discovery that a subset of enhancers is also occupied by RNA pol II and transcribed to produce a long non-coding RNA, termed enhancer RNAs (eRNAs), has led to new questions about the role of enhancers in transcriptional regulation. More details on eRNAs are discussed later in this review. Nonetheless, based upon a number of studies, eRNA production is now considered an additional marker of active enhancers (2, 51, 117).
Further study of enhancers has led to the identification of regions now referred to as super-enhancers (SE; 112). These are large, highly active enhancer regions that are bound at high levels by key transcription factors and the Mediator complex and represent a small minority of all enhancers active within a given cell type. In embryonic stem cells (ESCs), these regions regulate key pluripotency genes, including Nanog, Oct4 and Sox2. ChIP-Seq data also showed that these regions are separate from other enhancers based on their occupancy by two other key pluripotency factors, Klf4 and Essrb. In B-lymphocytes super-enhancers were identified through their occupancy by the Mediator complex and the lineage-critical transcription factor PU.1. Super-enhancers have since been identified in virtually all cell-types, making them likely a universal feature of mammalian tissues. There is some debate on whether these regions are truly a unique category of enhancer, or simply represent a sub-group of highly active enhancers within close proximity to one another (33, 64, 82). Interestingly, super-enhancers overlap with previously defined locus control regions thought to be the regulatory regions for specific genes, such as β-globin. Super-enhancers have also been associated with a number of diseases, being enriched at oncogenes and trait-associated variants, described later in this review. Together these features indicate that super-enhancers are an important subclass of enhancers that require further study and defining them aids our understanding of enhancer biology (45).
Techniques
As mentioned above, a main reason that we have been able to discover the functions of cis regulatory elements has been the vast improvements in high throughput technologies over the last decade. These approaches permit an unbiased, genome-wide identification of DNA elements with potential enhancer activity. Importantly, none of these approaches can definitively determine which gene(s) a DNA sequence may regulate nor whether they are required for proper gene expression regulation. To address either of these issues requires a genetic approach, which has become increasingly possible due to recent advances in genomic editing, which permits the rapid functional interrogation of chromatin regions with enhancer potential. See Table 1 for a detailed description of the most prominently used NGS-related techniques. For further details or reviews of non-NGS techniques see reviews by Chatterjee (19), Li (60), and Murakawa (68).
Enhancers and RNA polymerase II: Potential Mechanism
The mechanism by which enhancers interact with promoters to regulate gene expression remains largely unclear. By the late 1990s, the accepted model of enhancer function was that activator proteins bind the enhancer and recruit an already assembled transcriptional complex to promote gene transcription (47, 88). Biochemical analysis demonstrated that transcriptional complexes are pre-formed in the cell and increased in concentration upon activators binding to enhancers. This supported a model wherein transcription factor binding of an enhancer helped recruit RNA pol II. The rise of ChIP-Seq and other NGS methods, however, has shown that RNA pol II is actually paused at promoters after bidirectionally transcribing short, 20–120 nucleotide, transcripts consistent with the polymerase being “locked” in a promoter-proximal paused state (22, 69). There is evidence that enhancer-promoter interactions are associated with RNA pol II that is paused at the TSS and that it is the release of RNA pol II, potentially mediated by enhancers, that drives transcription (63, 101). Gene expression changes are thus thought to be mediated by the release of the RNA polymerase from the promoter, thereby permitting transcriptional elongation to produce a full-length mRNA.
Enhancers have also been implicated in the transition from transcriptional initiation to elongation by multiple groups (57, 90). At the β-globin locus, allele-specific ChIP assays show that RNA pol II enrichment decreases going from exon 1 to exon 3 when the locus control region is deleted (90). JMJD6 and Brd4 have been shown to regulate RNA pol II proximal pause-release and that the mechanism is regulated by binding to distal enhancers termed “anti-pause enhancers” (57). RNA pol II has been found to be regulated by a number of factors, including Pol II-associated factor 1 (Paf1; 20, 120), which occupies active promoters and enhancers. Paf1 knockdown results in an increase in the release of a paused RNA pol II from highly paused genes indicating that enhancer function to regulate gene expression in a Paf1-mediated release of RNA pol II (20). While the exact mechanism of enhancer function remains uncertain, there is increased evidence that enhancers interact with RNA pol II or with proteins associated with RNA pol II to permit elongation and the production of full-length mRNAs (Figure 4).
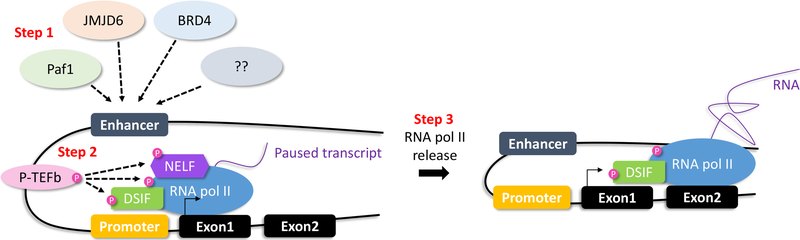
Figure 4.
Enhancers and RNA polymerase II
Proteins, such as Paf1, JMDJ6, BRD4 or others, bind enhancers, and promote PTEFb-mediated phosphorylation of NELF (negative elongation factor), DSIF (DRB sensitive inducing factor) and RNA pol II. RNA pol II is released, and transcriptional elongation occurs.
Teaching point: Enhancers interact with proteins allowing conversion from transcriptional initiation to elongation.
Enhancers in Nuclear Architecture
Nuclear architecture is the three-dimensional organization of chromatin within nuclear space, and its role in transcriptional regulation remains a prominent question. Early understanding of DNA transcription quickly generated different models for transcriptional regulation, one of which was chromatin looping (108). In this model, it was hypothesized that two proteins bind two linear sections of DNA and come together thereby, allowing these two regions and proteins to interact while the intervening chromatin segment was “looped-out” (Figure 1). Support for this model came from studies done in prokaryotes that found that regulation of operons was possibly achieved through a looping mechanism. The rise of chromosome capture technologies has enabled closer study of chromatin interaction, in particular enhancer-promoter interactions, and provided greater evidence for chromatin looping as method of DNA regulation (25, 89, 122). Chromosome Conformation Capture (3C) was the first method invented, through which the interaction between two specific genomic sites can be interrogated. This led to the development of other 3C-based methods, such as Circular Chromosome Conformation Capture (4C) which can study the interactions between one genomic site and any other site, and Hi-C, which uses high throughput sequencing to study the interaction between all genomic regions (methods discussed in Table 1). Hi-C studies have identified large, megabase regions of chromatin more likely to interact within the region than outside the region, termed Topologically Associated Domains (TADs; 30, 75). The ends of these domains are enriched for CCCTC-factor (CTCF) binding sites, stable across multiple cell types, and conserved evolutionarily, suggesting they have a role in chromatin organization in many organisms. ChIA-PET data has further identified another level of chromatin hierarchy called insulated neighborhoods (32, 46). These are chromatin loops formed by CTCF-CTCF homodimers that contain a gene and at least one regulatory element. It is possible that insulated neighborhoods are a sub-division of TADs in the chromatin organization hierarchy. Since these were identified using two different experimental methods, they could be describing the same phenomenon and the differences could be simply an artifact of the resolution of the method. Both of these chromatin structures are shown to constrain enhancer function, preventing or permitting interactions, supporting the theory that a physical interaction between enhancer and promoter is required for transcriptional regulation.
Examination of the β-globin locus drove increased investigation into the connection between chromatin looping and enhancer biology. Experiments at the β-globin locus and its locus control region (LCR) found that promoters for different globin genes compete for the LCR, and that this interaction was based in looping (16). 3C studies in erythroid cells showed that hypersensitive regions in the LCR interact with active promoters, while inactive regions are looped out (101). Loops at the β-globin locus are thus thought to be dynamic. Extensive 4C experiments in Drosophila at various developmental time points have shown that enhancer-promoter interactions in a loop are present across different developmental stages with a paused RNA pol II (41). This finding, in addition to similar findings in mouse and human cells (26, 49), suggests that loops are pre-formed in many cases, ready for transcriptional activation. Thus, at some loci the enhancer-promoter interaction is thought to be pre-formed, regardless of cell type (26, 41, 63), while at other loci the interaction is more dynamic (72, 73, 101), arguing that the mechanisms governing interaction may differ between loci and species. Both sides, however, agree that there is physical interaction between the enhancer and promoter, through a form of looping. While chromosome capture has shown that enhancer-mediated regulation is associated with physical looping interactions between different sections of DNA, the mechanism of this looping is unknown. Interestingly, a number of trans acting factors have been associated with enhancer function and looping, including YY1, CTCF, the cohesin complex and LDB1 (Figure 1; 118).
Yin Yang 1 (YY1) is a DNA-binding zinc finger transcription factor that binds sites present ubiquitously through the genome and can oligomerize. In mice, YY1-null embryos are embryonic lethal while YY1-heterozygous embryos survive with mental and developmental defects, indicating that YY1 plays a critical role during development (31). Interestingly, ChIA-PET data shows YY1 is heavily associated with enhancer-promoter interactions, with YY1 bound to both enhancers and promoters (111). A DNA circularization assay, in which a linear piece of DNA with a YY1 site was incubated with purified YY1 protein and the amount of circular DNA generated was measured, showed that YY1 increased physical interactions, implicating YY1 in looping. Mutations in YY1 binding motifs at the Raf1 and Etv4 locus caused a decrease in enhancer-promoter interactions, supporting the theory that YY1 regulates these interactions. YY1 is also shown to be an interacting partner of Oct4 and is enriched at super-enhancers in ESCs, potentially forming complexes to bind promoters and super-enhancers (Figure 1A; 109). Together, these data support a model in which YY1 mediates the physical enhancer-promoter interaction through a looping based mechanism.
CCCTC-binding factor (CTCF) is a multi-function insulator protein that has binding sites present throughout the genome (5, 78). Although these sites are present in every cell type, examination of super-enhancers active only in thymocytes found CTCF was enriched at these loci in thymocytes, but not in other cell types (48). CTCF, therefore, is enriched at active, cell-type specific super-enhancers. Depletion of CTCF using an auxin-inducible system in ESCs shows a drastic decrease in chromatin loops and enhancer-promoter interactions (74). RNA-seq post auxin-treatment showed that CTCF depletion caused both an upregulation and downregulation of a number of genes. Upregulated genes were more likely to have a CTCF site near an active enhancer, indicating that CTCF may insulate against a particular enhancer-promoter interaction. Deletion of a CTCF site upstream of a super-enhancer and its associated gene, decreases gene expression and, in some cases, changes enhancer-promoter interaction (32). Disruption of CTCF sites has shown an expansion of gene activation at numerous loci, and changing enhancer-promoter interaction, implicating CTCF in the establishment of enhancer-promoter interaction (70, 87, 115). Collectively, this work indicates that CTCF plays a critical role in organizing the genome into regions which restricts which enhancer-promoter interactions can occur (Figure 1B).
The cohesin complex was initially discovered in 1988 because of the role it plays in maintaining sister chromatid cohesion during metaphase of mitosis and meiosis (76). However, recently there have been multiple new identified roles of the cohesin complex, including the regulation of gene expression (86). The cohesin complex has been shown to interact with Mediator and NIPBL to aid in transcriptional regulation by permitting distal CREs, including enhancers, to interact with gene promoters (50). Additionally, the cohesin complex has been found to associate with CTCF to aid in cohesin binding at enhancers (80). Interestingly, depletion of cohesin results in the elimination of chromatin loops, however there is little transcriptional effect (85). One debate that remains in the field is whether cohesin depletion eliminates TAD architecture (85, 92), or is solely limited to inter-TAD loops (93, 96, 124). Disruption of cohesin causes a downregulation of super-enhancer-regulated genes (48). Super-enhancer position, histone modification and transcription remain intact with cohesin disruption, while the enhancer-promoter interactions were weakened. In erythroid cells, however, key enhancer-promoter looping occurs independent of cohesin levels, arguing that cohesin’s role in enhancer-promoter looping is variable (53). This work demonstrates that cohesin plays a central role in permitting enhancer-promoter interactions, and its possible role in higher-order chromatin structures requires further investigation (Figure 1B).
LIM domain-binding protein 1 (LDB1) is a nuclear protein that is part of DNA-binding complexes in many cell types and necessary for hematopoietic stem cell maintenance (56). In erythroid cells, LDB1 associates with other hematopoietic factors to form a transcriptional transactivating complex (107). ChIP analysis shows that LDB1 and the associated factors together bind at the β-globin locus, while 3C identifies LDB1 as the anchor of a chromatin loop that brings the LCR and β-globin regions together (13, 97). When LDB1 is tethered to the β-globin promoter, in the absence of GATA1 (a transcription factor critical for β-globin and LCR looping) β-globin transcription is activated (27) and in adult erythroblasts previously silenced fetal globin genes can be transcribed (28). Thus, forced looping through LDB1 activates transcription even in sub-optimal conditions, indicating looping is a critical step of gene expression regulation. Interestingly, although cohesin and Mediator are thought to be critical for chromatin looping, ChIP analysis shows that LDB1 is essential for looping in erythroid cells and can function independent of cohesin and Mediator (53). LDB1’s function as an erythroid specific looping factor makes it a highly attractive locus for further studying additional questions about how enhancers mediate gene expression (Figure 1C).
Despite the support these studies provide for the model of chromatin looping, no clear mechanism has yet been established. Instead, the multitude of experimental examples describing different mechanisms of looping suggest that there may be multiple mechanisms of chromatin looping that aid enhancer-promoter interaction.
eRNAs
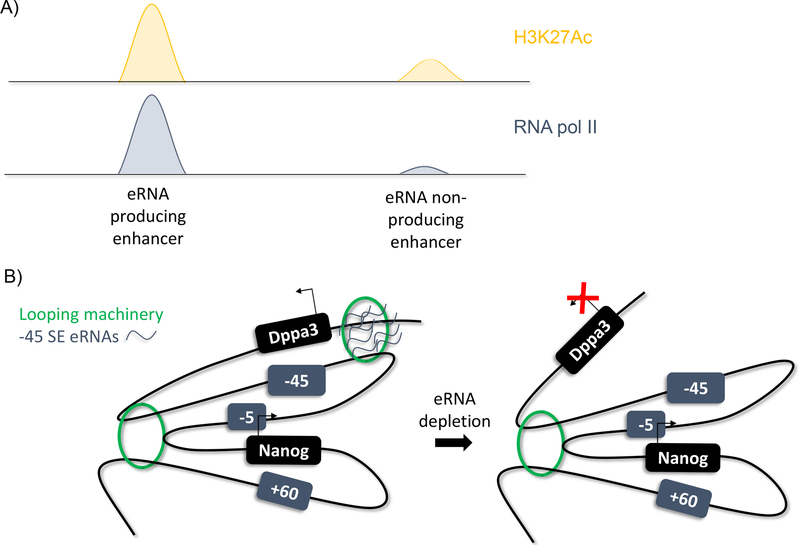
Enhancer RNAs, or eRNAs, are a subtype of long non-coding RNA. They were first postulated in a 1992 study in erythroid cells that found that DNase hypersensitive regions of the genome, identified as enhancers, were also transcribed to produce a ncRNA (102). Later, ChIP-Seq analysis of the mouse and human genomes identified enhancer regions through histone signatures and found that these sites are also bound by RNA pol II (Figure 5A; 29, 51, 79). These RNA sequences were identified at extragenic and intragenic enhancers in both the sense and anti-sense directions. Further, they were not classically polyadenylated, differentiating them from mRNA and promoter RNAs. Studies identifying super-enhancers found that these enhancers are also bidirectionally transcribed (45, 83). In addition, eRNA producing enhancers are found to be highly enriched for H3K27Ac and the DNA hydroxylase Tet1 and decreased DNA methylation, allowing computational identification of eRNA producing enhancers (83). eRNA synthesis is also positively correlated with enhancer activity, indicating they are a hallmark of highly-active enhancers.
Figure 5.
eRNAs
A) ChIP-Seq analysis identifies eRNA producing enhancers by enrichment for H3K27Ac and RNA pol II
B) Model of eRNA function at Nanog function. eRNAs appear to facilitate or stabilize chromatin loops between specific enhancer:gene promoter combinations.
Teaching Point: eRNAs can be identified through ChIP-Seq analysis and may play a role in regulation gene expression changes
The biological functions of eRNAs are still debated. One suggested role is transcription factor trapping. When short RNA sequences were tethered at six enhancer sites known to be bound by YY1, YY1 binding at these enhancers was increased, as measured by ChIP-qPCR (95). This change was specifically due to the RNA sequences as no change in YY1 occupancy was observed at enhancers not tethered to an RNA sequence. Thus, short RNA transcripts, such as eRNAs, help maintain transcription factor binding to DNA and potentially ensure transcription through this mechanism. In human breast cancer cells, 17β-estradiol (E2) binding of oestrogen receptor-α (ER-α) induces eRNA transcription at the E2/ER-α bound enhancer sequences of numerous E2 upregulated genes (59). siRNA knockdown of these eRNAs decreases transcription of the genes they regulate. Gene expression was rescued when the eRNAs of the specific enhancer were provided, but not with any other eRNA, suggesting that the function of the eRNA is sequence-specific. Another study found that eRNA depletion in neuronal cells prevented the release of the RNA pol II associated protein Negative Elongation Factor (NELF; 91). NELF establishes RNA pol II pausing by binding nascent transcripts and RNA pol II. A subunit of NELF, NELF-E, contains an RNA recognition motif (RRM) that is shown to bind both nascent transcripts and eRNAs through experiments using biotinylated eRNAs. Perturbation of the RRM reduced NELF occupancy at the promoter and mRNA production, indicating that the RRM is an important motif for both RNA pol II pausing and elongation. shRNA-mediated depletion of eRNAs reduced the levels of elongating RNA polymerase II, and thus, eRNAs potentially play a role as decoy nascent transcripts for NELF release via interaction with the RRM. Lastly, investigation of the MyoD locus identified differential roles for eRNAs of proximal versus distal enhancers (66). Proximal enhancer eRNAs were critical for MyoD expression, while distal enhancer eRNAs were important for downstream myogenic gene activation. Depletion of these eRNAs also reduced chromatin accessibility, suggesting a role of eRNAs in chromatin remodeling. Despite these findings, some still suggest that eRNAs have little to no function, based on their low-levels of transcription and poor evolutionary conservation, and may just be a side-effect of promiscuous transcription by RNA pol II at the promoter (60). By contrast, our studies at the extended Nanog locus demonstrate otherwise (Figure 5B). The extended Nanog locus contains three associated super-enhancers (SE), the −5 SE, the −45 SE and the +60 SE. The −45 SE regulates not only Nanog expression but Dppa3 expression as well, which lies upstream of Nanog and the −45 SE. Depletion of the −45 SE eRNAs reduced interaction between the −45 SE and Dppa3, implying that the eRNAs are responsible for maintaining enhancer-promoter interaction (12).
The biological function of eRNAs remains unclear and, as with much of enhancer biology, could be highly variable between enhancer regions. Allele-specific studies and directional studies would provide greater insight into the value of eRNAs, and the potential differential roles of sense versus anti-sense transcripts. Overall, there is ample evidence that at some loci eRNAs play important roles in gene regulation.
Development
Early development is marked by numerous differentiations of pluripotent cells down different lineages. Pluripotent cells, capable of differentiation into any of the three germ layers (endoderm, mesoderm, ectoderm) are maintained by a group of pluripotency genes, including Nanog, Oct4 and Sox2. Pluripotency genes are regulated by a number of identified super-enhancers. Nanog is a blastocyst stage master transcription factor important for pluripotency. As mentioned above, it is regulated by three super-enhancers (12). CRISPR-mediated deletion of each super-enhancer caused a variable change in Nanog, implicating a unique role for each enhancer in its regulation of Nanog expression. The −5 super-enhancer (SE), in particular, is a key Nanog regulator, as homozygous deletion of the enhancer causes a loss of pluripotency. In addition, a key finding of this study was that the −45 SE is able to regulate two different genes, Nanog and Dppa3.
Studies of enhancers regulating the gene Sox2 further increased our understanding of the role of different enhancers on temporally specific gene expression (123). Sox2 is associated with two proximal regulatory regions and three distal enhancers making up a distal regulatory region. Deletion of one of the proximal regions, SRR1, has no effect on pluripotency, thus questioning its role in regulating Sox2 expression. Distal enhancers, however, are found to loop in and interact with the proximal enhancers Sox2 in ESCs but not mouse embryonic fibroblasts (MEFs), and deletion of this region dramatically reduces Sox2 expression in ESCs. These cells are also impaired in their ability to form ectoderm. Interestingly, although deletion of SRR1 is largely dispensable in ESCs, it is required in neural cells. Thus, a gene can be regulated by multiple enhancers simultaneously and these can potentially be cell-type specific, providing a mechanism for genes with multiple functions in different cell types.
Another important developmental process shown to be regulated by enhancers is limb development. Limb development is regulated by two Hox clusters, HoxA and HoxD, which are modulated by upstream enhancer regions (9, 63). Using chromosomal capture technology, the gene desert region surrounding the HoxD locus was identified as a complex regulatory region (63). Multiple and simultaneous interactions were found, and only deletion of the entire regulatory region caused major limb defects. Microdeletions within this regulatory landscape mimicked human developmental defects, indicating that the regulatory region is critical for proper limb development (62). The HoxA cluster is similarly regulated by an upstream enhancer region (9). This region was found to interact solely with the 5’ end of the HoxA cluster that is expressed during limb development. Transgenic analysis of candidate enhancers showed specific expression of each enhancer mimicked the segmental expression of the HoxA genes, providing a mechanism by which these genes are turned on in specific regions. In both clusters, multiple interactions are observed between the enhancer region and the cluster, indicating a complex and potentially redundant regulatory mechanism.
Although these are just a few examples of enhancers in development, they show that enhancers are key to the temporal-spatial and tissue-specific regulation of gene expression during development. Given the multiple roles some genes play, enhancer modulation of expression is likely key to when and where genes are active, contributing heavily to the maintenance of normal development.
Normal Physiology and Disease
In addition to studying the role enhancers play in development, additional focus has been aimed on determining how enhancers contribute to normal physiology. An interesting study investigated the regulation of the angiotensin II (AngII) response by enhancers in vascular smooth muscle cells (VSMCs; 24). They performed ChIP-Seq to identify changes in enhancer activity and found thousands of changes in H3K27Ac occupancy in cells treated with AngII. Additionally, they observed several key transcription factor motifs present in upregulated enhancers that have been implicated in the angiotensin response (AP1, ETS, and STAT). RNA-sequencing of VSMCs treated with AngII revealed that 35.7% of genes upregulated in response to AngII treatment were associated with AngII upregulated enhancers. Using an in-silico model, they were able to assess the function of genes near 584 identified AngII upregulated super-enhancers. From that, they discovered that genes proximal to these enhancers had promoters with increased H3K27Ac. Additionally, RNA-seq data showed these genes were upregulated significantly more compared to upregulated genes not associated with an enhancer (24). This study provides a model in which epigenetic changes allow enhancers to help facilitate the large scale, downstream response to Angiotensin II in vascular smooth muscle cells.
Enhancers have also been shown to play a role in lactation (116). Both mammary gland-specific and universal cytokine-responsive enhancers are occupied by STAT5 (the downstream transcription factor of prolactin) during lactation. However, upon cessation of lactation, only the mammary-specific enhancers become unoccupied. This response is due to the ability of these enhancers to sense STAT5 concentrations. Genes responsible for differentiation of mammary tissue (which need to be downregulated upon cessation of lactation to stop gland formation and milk production) are associated with enhancers that are highly sensitive to STAT5. These enhancers are rapidly decommissioned upon loss of prolactin signaling. Other genes responsible for cellular maintenance are associated with enhancers that are less sensitive to STAT5 changes, setting up a system in which shifts in prolactin signaling preferentially affect milk production and epithelial differentiation (116). This represents an important example where a physiological process is dictated by enhancers acting to sense the levels of a specific transcription factor, STAT5.
While the normal functions of enhancers and other transcriptional regulatory elements in development are still an active area of investigation, several studies have indicated that alterations of enhancer function can lead to disease. The first studies on this topic came in 1983, when Kioussis et al showed that translocation of a DNA region that is normally hypermethylated and insensitive to DNase near the β-globin gene leads to thalassemia (52). They postulated that the translocation of a normally inactive locus in close proximity to the β-globin gene leads to the inactivation of β-globin, either by the actions of the inactive locus suppressing β-globin or by the positional effect of moving important activating sequences further away from the β-globin locus.
Fast forward 35 years, we now know much more about the role of enhancers in disease. In 2004, Andersson et al. used CAGE to identify enhancers via their bidirectional transcription. They found that 64% of putative enhancers were associated within 500kb of a TSS. Additionally, they found that on average, a TSS was associated with 4.9 enhancers, and one enhancer was associated with 2.4 TSSs (2). It is important to note, however, that these associations do not explicitly prove that enhancers regulate the transcription of their respective genes. A more recent study further described the role of super-enhancers in the control of cell identity and disease by combining data from 1,675 genome-wide studies across 86 human samples (45). In summary, they inspected 5,303 disease-causing SNPs, and found that 64% are located in enhancer regions, a surprisingly large number given that most genome-wide association studies have focused solely on coding region mutations. Furthermore, they showed that super-enhancers are more likely to be enriched for disease-associated SNPs than regular enhancers. This enrichment ranges from 0.4× more enrichment for disease-associated SNPs in super-enhancers genome-wide, to 2.0× more enrichment for Multiple Sclerosis-related SNPs. Additionally, these SNPs tend to occur only in cell types relevant to the disease. Their studies focused on a few key diseases to get a better understanding of how enhancers may have an effect on disease. They identified 5 SNPs in super-enhancers of brain tissue from patients with Alzheimer’s disease, 13 SNPs genes associated with Th-cell biology in patients with Type 1 Diabetes Mellitus, and 22 SNPs in super-enhancers associated with B-cell function of patients with Systemic Lupus Erythematosus (45). The presence of SNPs in such a wide variety of diseases reveals an area in need of continued research. Future studies may identify and characterize truly pathogenic mutations in enhancers.
Chronic Kidney Disease (CKD) is another disease that has shown some interesting connections to enhancer dysfunction. Numerous genome-wide association studies of CKD, a vastly heterogeneous disease, revealed SNPs in both coding and non-coding regions. Brandt et al sought to determine if some of the SNPs in the non-coding regions might be located in enhancer regions (14). By performing 4C on healthy patient samples, they looked for interactions between genes and areas where CKD associated SNPs exist. They identified 304 target genes that co-localized with CKD SNP regions. Subsequently, by cross referencing both human and mouse databases, they were able to find 23 genes that, when silenced, play a role in CKD. In summation of their work, they proposed that dysregulation in the expression of a gene profile that is part of the regulation of kidney homeostasis in healthy individuals is likely a contributing factor in CKD etiology (14).
The increasing number of disease-associated SNPs that have been found to be in enhancer regions makes the continued study of enhancer biology critical. Furthermore, several studies have indicated that expression quantitative trait loci (eQTLs) containing enhancer regions are common in humans (35, 100). eQTLs are regions of DNA with high genetic variation that is specifically associated with a variation in gene expression. The high prevalence of genetic variation in cis regulatory regions makes the further study of both the physiological and pathological functions of these elements necessary as we aim to have a complete understanding of transcriptional regulation.
Cancer
While many studies suggest that enhancers may be involved in a vast number of diseases, the literature reveals that enhancers certainly contribute to cancer development through a variety of mechanisms (Figure 6). Chen et al performed a genome-wide analysis of 8,928 tumor samples across 33 cancer types and identified 4,691 enhancers present in at least 10% of all cancer types (21). While they found a vast amount of genetic variation from patient to patient, overall, they observed enhancer activation amongst all cancer types. Additionally, they found this activation signature to be associated with increased aneuploidy, and not mutational load. They proposed a model in which increased open chromatin allows enhancers to be transcribed but may also allow long-range DNA interactions that could make the genome more susceptible to rearrangement (Figure 6A). Furthermore, they use patient outcome to determine if enhancer involvement is correlated with patient survival. They conclude that indeed, enhancer involvement in cancer appears to play a comparable role in outcome to mutations in protein coding sequences (21). One of the key findings of this study is that alterations in enhancers may promote cancer independently of their ability to regulate gene expression.
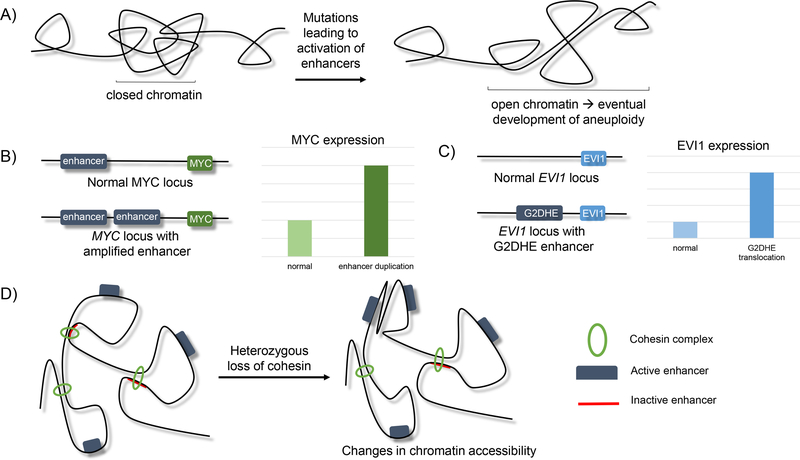
Figure 6.
Proposed models of alteration of enhancer function leads to cancer development.
A) Activation of enhancers results in the opening of chromatin, leading to the greater likelihood of chromosomal rearrangements that ultimately lead to aneuploidy (21).
B) Duplication of enhancer sequences that regulate MYC leads to increased MYC expression that drives epithelial cancers (121).
C) Translocation of G2DHE (a GATA2 enhancer) near the EVI1 locus leads to overexpression of the EVI1 oncogene, promoting myeloid and B-cell leukemic development (119).
D) Heterozygous loss of cohesin leads to changes in nuclear architecture that can alter enhancer-promoter interactions and lead to gene expression changes that promote myeloid transformation.
Teaching point: there are many routes that enhancer perturbation can lead to cancer development.
A study focusing on human epithelial cancers provides a more direct model in which enhancers may contribute to disease. Zhang et al focus specifically on samples with MYC upregulation (121). They found that samples containing amplification of MYC-regulating enhancers had significantly higher MYC expression than samples without amplification of MYC-related enhancers (Figure 6B). In fact, the expression levels of MYC in the enhancer-amplified samples was comparable to samples that had amplification of the MYC coding region itself. These apparent copy number responses were verified in vitro by comparing lung adenocarcinoma cells with single and duplicate expression of a specific MYC enhancer. Additionally, they showed that repression of the enhancer region not only results in a decreased expression of MYC target genes but impedes the growth of lung adenocarcinoma (121). These experiments provide evidence that focal amplification of enhancer containing regions can drive oncogene expression.
Another mechanism by which enhancers may promote cancer development was identified by Yamazaki et al in 2014 (119). Patients with inv(3)(q21;q26) and t(3,3)(q21;q26) driven leukemias have overexpression of a common proto-oncogene, EVI1. EVI1 is a transcription factor with many roles including directly regulating the transcription of target genes (GATA2, PBX1, PTEN), binding and inhibiting the action of other transcription factors (RUNX1, PU.1, GATA1), interacting with epigenetic modifying enzymes, and regulating centromere duplication. The chromosomal rearrangements identified in patients with leukemia result in an enhancer for GATA2 (G2DHE) being in closer proximity to the EVI1 locus (Figure 6C). Because the mislocalization of the G2DHE element leads to increased expression of EVI1, they hypothesized that this enhancer is responsible for the upregulation of EVI1 seen in patients with these rearrangements. They developed a BAC mouse model for this rearrangement and observed that myeloid and B-cell leukemias developed with the expression of 3q21q26. The leukemias they observed were similar to leukemias identified using a retrovirus to drive overexpression of EVI1 in mice. Upon deletion of G2DHE within the BAC model, leukemia development was eliminated but EVI1 expression was not completely abolished, indicating that there is likely a threshold of EVI1 needed to drive leukemogenesis (119). The results of this study suggest that to promote leukemia in this chromosomal rearrangement model, the G2DHE element acts to increase the expression of EVI1 instead of GATA2. This study revealed a novel way for chromosomal translocations to promote cancer.
While enhancers have been implicated in numerous cancer types, other cis regulatory elements may be involved in cancer development as well. Mutations in the cohesin complex seem to play an important role in myeloid malignancies. While most of these studies focus on acute myeloid leukemia (AML), cohesin mutations have been identified in myelodysplastic syndromes, myeloproliferative neoplasms, and chronic myelogenous leukemia as well (55). In a 2013 study of AML, The Cancer Genome Atlas Research Network revealed a previously underappreciated amount of genetic variation in the cohesin complex (17). In their study of 200 adult patients with de novo AML, they found that 9.1% had mutations in members of the cohesin complex. Interestingly, all of the mutations were heterozygous and nearly 50% of the samples were euploid, which suggests that cohesin’s function in mitosis remains intact and its role in nuclear architecture is more likely being affected by these mutations (17). Cohesin depletion leading to disrupted nuclear architecture has been identified by increased transcription factor motif accessibility (67, 105) and decreased H3K27me3 (37). These studies indicate that mutations in the cohesin complex seem to promote malignant transformation, at least in a myeloid cell specific manner, likely via alteration of enhancer-promoter interactions (Figure 6D).
From the studies discussed above, it is evident that cis regulatory elements, or the factors which regulate their ability to interact with gene promoters, can be important factors in driving cancer development. Although cancer development can occur through a variety of mechanisms (gene amplification, chromosomal rearrangement, gene mutation/haploinsufficiency) the downstream consequence of alteration of the function of these regulatory elements is the same. Changes in transcriptional profile are an imperative driving component of cancer development. While each context certainly varies, we can use this overall knowledge to advance our continued study of both cancer development and therapies.
Therapeutics
The knowledge that we now have about enhancers and their role in promoting disease brings us to a new question. Since we know the alteration of enhancer elements can lead to disease, is there a way we can target them with a therapeutic strategy? A few studies have begun answering that exact question. One group focused on MYC-upregulated cancers by targeting BRD4. BRD4 is an important BET transcription factor that binds acetylated chromatin and promotes transcription by recruiting other factors like Mediator. Lovén et al showed that upon treatment of multiple myeloma cells with JQ1, BRD4 is globally depleted, as are its recruited elements, including Mediator and RNA polymerase II (61). However, upon further inspection, they identified that the depletion appears to be most selective for super-enhancers. Genes that are regulated by these targeted super-enhancers are expressed higher and more specifically in multiple myeloma cells, compared to the same genes in non-malignant cells and other genes regulated by typical enhancers. They found the same preferential depletion of super-enhancers in glioblastoma multiforme and small cell lung cancer. Even though super-enhancers are larger than typical enhancers, when adjusted for size they find that super-enhancers are occupied by Mediator, BRD4, and H3K27Ac an order of magnitude more than typical enhancers. They suggest this as the cause for the selective depletion of BRD4 and other factors from super-enhancers compared to typical enhancers and the rest of the genome (61). Since super-enhancers have been implicated in a number of cancers, this study provides a possible treatment strategy of enhancer-targeting therapies that would normalize the transcriptional misregulation apparent in enhancer-driven cancers.
The idea of using a BET inhibitor that might preferentially target super-enhancer dependent transcriptional changes was expanded upon by Peeters et al in their study of Juvenile Idiopathic Arthritis (JIA) (81). After the identification of inflammation-associated SNPs in the super-enhancers of T-cells of synovial fluid from patients with JIA, they treated these cells with JQ1. They found that treatment significantly reduced the expression of enhancer associated genes, an effect that they did not observe in healthy controls. Additionally, functional aspects of the disease were altered as they observed decreased cytokine production from these T cells (81).
Some early phase I and II studies on the effects of BET inhibitors on a variety of disease (multiple myeloma, lymphoma(1), acute leukemia(11), prediabetes(94), and atherosclerotic cardiovascular disease (71)) have begun to uncover the toxicity and efficacy of BET inhibitors. While these early studies seem promising, we must proceed with caution, as there have been reports of resistance to BET inhibitors in cases of acute myeloid leukemia (38) and pancreatic cancer (54).
Other efforts have begun to investigate whether therapeutic editing of an enhancer may help patients with sickle cell disease and beta thalassemia. BCL11A plays a role in the development switch from Ɣ (fetal) to β (adult) hemoglobin. Patients with sickle cell or beta thalassemia do better when they have higher levels of Ɣ hemoglobin. Canver et al employed a CRISPR mutagenesis screen to identify key nucleotides of BCL11A that regulate the Ɣ to β switch. By then using sgRNAs to target these sequences, they were able to drive fetal hemoglobin induction in primary erythroid precursors (18). While these data is exciting for the field of therapeutic editing, further work to understand the possible off-target effects of targeting BCL11A is needed before studies may proceed to a higher level.
These experiments are only the beginning of what could be a world of new treatment options that target disease-driving enhancers. Some unanswered questions to consider include: Will enhancer targeting therapies continue to preferentially target disease-associated enhancers in human patients? How might enhancer targeting therapy be combined with other therapeutic strategies for the best outcome? What side effects (short vs. long term) might occur as a result of targeting transcriptional regulation in vivo?
Conclusion
Transcriptional regulation is a complex process with numerous interacting elements. Here we have attempted to describe the roles of these players and, specifically, enhancers. The field has learned a great amount in the last 40 years about the function of regulatory elements. Our knowledge of the role of enhancers in the regulation of transcription has exploded through the advent of new technologies to permit their unbiased identification genome-wide. The advances of high throughput NGS techniques allow us to identify enhancers genome-wide and further interrogate their functions. We have seen that enhancers play a role in the activation of transcription of certain genes, can have an effect both proximally and distally, and know that these actions vary from cell to cell. We have discovered some of the key proteins that promote these interactions (YY1, CTCF, and the cohesin complex). This knowledge has been used to study the depletion of each of these factors, identifying the downstream changes in both transcription and nuclear architecture. Finally, we have learned that enhancers are essential for promoting development and maintaining normal physiology. The multitude of disease found to be associated with changes in enhancer sequences further emphasize the importance of these elements in maintaining the imperative balance of transcription across all cells of the human body. This highlights that enhancers likely represent a new and exciting area of research for disease causing variants.
While we have certainly increased our knowledge of cis regulatory elements in recent years, there are still lingering questions. How exactly do enhancers interact with RNA pol II to regulate transcription? Additionally, how do enhancers and other cis regulatory elements control or participate in chromatin looping? Are super-enhancers truly a separate entity or are they more correctly classified as a sub-type of enhancers? Are eRNAs truly a separate regulatory mechanism or are they simply a byproduct of nearby gene transcription? Lastly, how can we use what we know about the role that enhancers and other cis regulatory elements play in disease to develop targeted therapies to improve patient outcomes? Hopefully, the answers to these outstanding questions will be elucidated by continued emphasis on studying cis regulatory elements in coming years.
Cross References
Genomic DNA expression determined by epigenetic factors (DNA methylation, histone modification, lncRNA)
Genome-wide association studies
Genome sequencing and exome sequencing
QTL and eQTL linkage studies to identify genes associated with complex functions (congenic mapping and positional cloning)
Didactic Synopsis.
Major Teaching Points:
Transcriptional regulation is a complex process controlled by numerous elements which act either in cis or trans.
Enhancers are DNA elements containing transcription factor binding motifs that play an important role in transcriptional regulation in a distance and orientation independent manner.
Technological advances have been essential in identifying enhancers and their role in regulating gene expression.
Enhancers regulate gene expression through interactions with RNA polymerase II by regulating transcriptional elongation.
Enhancers operate by being in close physical proximity to the genes they regulate within the nucleus. This is mediated by chromatin looping mediated by architectural proteins such as YY1, CTCF, LDB1, and the cohesin complex.
SNPs, duplications, and other alterations in enhancer function have been implicated in a wide variety of diseases.
Bibliography
- 1.Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, et al. 2016. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 3(4):e196–204 [DOI] [PubMed] [Google Scholar]
- 2.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature. 507(7493):455–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. 2013. Genome-Wide Quantitative Enhancer Activity Maps Identified by STARR-seq. Science. 339(6123):1074–77 [DOI] [PubMed] [Google Scholar]
- 4.Arnosti DN, Barolo S, Levine M, Small S. 1996. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 122(1):205–14 [DOI] [PubMed] [Google Scholar]
- 5.Arzate-Mejía RG, Recillas-Targa F, Corces VG. 2018. Developing in 3D: the role of CTCF in cell differentiation. Development. 145(6): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerji J, Olson L, Schaffner W. 1983. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 33(3):729–40 [DOI] [PubMed] [Google Scholar]
- 7.Banerji J, Rusconi S, Schaffner W. 1981. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 27(2):299–308 [DOI] [PubMed] [Google Scholar]
- 8.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, et al. 2007. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell. 129(4):823–37 [DOI] [PubMed] [Google Scholar]
- 9.Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M. 2013. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 9(12):e1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 120(2):169–81 [DOI] [PubMed] [Google Scholar]
- 11.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, et al. 2016. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 3(4):e186–95 [DOI] [PubMed] [Google Scholar]
- 12.Blinka S, Reimer MH Jr, Pulakanti K, Rao S. 2016. Super-Enhancers at the Nanog Locus Differentially Regulate Neighboring Pluripotency-Associated Genes. Cell Rep. 17(1):19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, et al. 2004. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struct. Mol. Biol 11(1):73–80 [DOI] [PubMed] [Google Scholar]
- 14.Brandt MM, Meddens CA, Louzao-Martinez L, van den Dungen NAM, Lansu NR, et al. 2018. Chromatin Conformation Links Distal Target Genes to CKD Loci. J. Am. Soc. Nephrol 29(2):462–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10(12):1213–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulger M, Groudine M. 1999. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 13(19):2465–77 [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, et al. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med 368(22):2059–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, et al. 2015. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 527(7577):192–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee S, Ahituv N. 2017. Gene Regulatory Elements, Major Drivers of Human Disease. Annu. Rev. Genomics Hum. Genet 18:45–63 [DOI] [PubMed] [Google Scholar]
- 20.Chen FX, Woodfin AR, Gardini A, Rickels RA, Marshall SA, et al. 2015. PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell. 162(5):1003–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Li C, Peng X, Zhou Z, Weinstein JN, et al. 2018. A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell. 173(2):386–99.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 322(5909):1845–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotney J, Leng J, Oh S, DeMare LE, Reilly SK, et al. 2012. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 22(6):1069–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S, Senapati P, Chen Z, Reddy MA, Ganguly R, et al. 2017. Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells. Nat. Commun 8(1):1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekker J 2006. The three “C” s of chromosome conformation capture: controls, controls, controls. Nat. Methods 3(1):17–21 [DOI] [PubMed] [Google Scholar]
- 26.de Laat W, Duboule D. 2013. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 502(7472):499–506 [DOI] [PubMed] [Google Scholar]
- 27.Deng W, Lee J, Wang H, Miller J, Reik A, et al. 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 149(6):1233–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng W, Rupon JW, Krivega I, Breda L, Motta I, et al. 2014. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 158(4):849–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, et al. 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8(5):e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 485(7398):376–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol 19(10):7237–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, et al. 2014. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 159(2):374–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dukler N, Gulko B, Huang Y-F, Siepel A. 2016. Is a super-enhancer greater than the sum of its parts? Nat. Genet 49(1):2–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elgin SC. 1988. The formation and function of DNase I hypersensitive sites in the process of gene activation. J. Biol. Chem 263(36):19259–62 [PubMed] [Google Scholar]
- 35.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, et al. 2008. Genetics of gene expression and its effect on disease. Nature. 452(7186):423–28 [DOI] [PubMed] [Google Scholar]
- 36.Ferrier P, Krippl B, Blackwell TK, Furley AJ, Suh H, et al. 1990. Separate elements control DJ and VDJ rearrangement in a transgenic recombination substrate. EMBO J. 9(1):117–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher JB, Peterson J, Reimer M, Stelloh C, Pulakanti K, et al. 2017. The cohesin subunit Rad21 is a negative regulator of hematopoietic self-renewal through epigenetic repression of Hoxa7 and Hoxa9. Leukemia. 31(3):712–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fong CY, Gilan O, Lam EYN, Rubin AF, Ftouni S, et al. 2015. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 525(7570):538–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, et al. 2016. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 354(6313):769–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gavrilov A, Eivazova E, Priozhkova I, Lipinski M, Razin S, Vassetzky Y. 2009. Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods Mol. Biol 567:171–88 [DOI] [PubMed] [Google Scholar]
- 41.Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, et al. 2014. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 512(7512):96–100 [DOI] [PubMed] [Google Scholar]
- 42.Gillies SD, Morrison SL, Oi VT, Tonegawa S. 1983. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 33(3):717–28 [DOI] [PubMed] [Google Scholar]
- 43.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 459(7243):108–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet 39(3):311–18 [DOI] [PubMed] [Google Scholar]
- 45.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, et al. 2013. Super-enhancers in the control of cell identity and disease. Cell. 155(4):934–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hnisz D, Day DS, Young RA. 2016. Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell. 167(5):1188–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hori R, Carey M. 1994. The role of activators in assembly of RNA polymerase II transcription complexes. Curr. Opin. Genet. Dev 4(2):236–44 [DOI] [PubMed] [Google Scholar]
- 48.Ing-Simmons E, Seitan VC, Faure AJ, Flicek P, Carroll T, et al. 2015. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 25(4):504–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, et al. 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 503(7475):290–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 467(7314):430–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature. 465(7295):182–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kioussis D, Vanin E, deLange T, Flavell RA, Grosveld FG. 1983. Beta-globin gene inactivation by DNA translocation in gamma beta-thalassaemia. Nature. 306(5944):662–66 [DOI] [PubMed] [Google Scholar]
- 53.Krivega I, Dean A. 2017. LDB1-mediated enhancer looping can be established independent of mediator and cohesin. Nucleic Acids Res. 45(14):8255–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar K, Raza SS, Knab LM, Chow CR, Kwok B, et al. 2015. GLI2-dependent c-MYC upregulation mediates resistance of pancreatic cancer cells to the BET bromodomain inhibitor JQ1. Sci. Rep 5:9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leeke B, Marsman J, O’Sullivan JM, Horsfield JA. 2014. Cohesin mutations in myeloid malignancies: underlying mechanisms. Exp. Hematol. Oncol 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L, Jothi R, Cui K, Lee JY, Cohen T, et al. 2011. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat. Immunol 12(2):129–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu W, Ma Q, Wong K, Li W, Ohgi K, et al. 2013. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 155(7):1581–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X, Zhang Y, Chen Y, Li M, Zhou F, et al. 2017. In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell. 170(5):1028–43.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, Notani D, Ma Q, Tanasa B, Nunez E, et al. 2013. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 498(7455):516–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W, Notani D, Rosenfeld MG. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet 17(4):207–23 [DOI] [PubMed] [Google Scholar]
- 61.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, et al. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 153(2):320–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitter D, Chiaie BD, Lüdecke H-J, Gillessen-Kaesbach G, Bohring A, et al. 2010. Genotype-phenotype correlation in eight new patients with a deletion encompassing 2q31.1. Am. J. Med. Genet. A 152A(5):1213–24 [DOI] [PubMed] [Google Scholar]
- 63.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, et al. 2011. A Regulatory Archipelago Controls Hox Genes Transcription in Digits. Cell. 147(5):1132–45 [DOI] [PubMed] [Google Scholar]
- 64.Moorthy SD, Davidson S, Shchuka VM, Singh G, Malek-Gilani N, et al. 2017. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 27(2):246–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P. 1981. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 9(22):6047–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, et al. 2013. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 51(5):606–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, et al. 2015. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med 212(11):1833–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murakawa Y, Yoshihara M, Kawaji H, Nishikawa M, Zayed H, et al. 2016. Enhanced Identification of Transcriptional Enhancers Provides Mechanistic Insights into Diseases. Trends Genet. 32(2):76–88 [DOI] [PubMed] [Google Scholar]
- 69.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, et al. 2007. RNA polymerase is poised for activation across the genome. Nat. Genet 39(12):1507–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narendra V, Rocha PP, An D, Raviram R, Skok JA, et al. 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 347(6225):1017–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nicholls SJ, Puri R, Wolski K, Ballantyne CM, Barter PJ, et al. 2015. Effect of the BET Protein Inhibitor, RVX-208, on Progression of Coronary Atherosclerosis: Results of the Phase 2b, Randomized, Double-Blind, Multicenter, ASSURE Trial. Am. J. Cardiovasc. Drugs 16(1):55–65 [DOI] [PubMed] [Google Scholar]
- 72.Noordermeer D, Leleu M, Schorderet P, Joye E, Chabaud F, Duboule D. 2014. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. Elife. 3:e02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. 2011. The dynamic architecture of Hox gene clusters. Science. 334(6053):222–25 [DOI] [PubMed] [Google Scholar]
- 74.Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, et al. 2017. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 169(5):930–44.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 485(7398):381–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohkura H, Adachi Y, Kinoshita N, Niwa O, Toda T, Yanagida M. 1988. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 7(5):1465–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Kane CJ, Gehring WJ. 1987. Detection in situ of genomic regulatory elements in Drosophila. Proceedings of the National Academy of Sciences. 84(24):9123–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ong C-T, Corces VG. 2014. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet 15(4):234–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell. 143(1):46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 132(3):422–33 [DOI] [PubMed] [Google Scholar]
- 81.Peeters JGC, Vervoort SJ, Tan SC, Mijnheer G, de Roock S, et al. 2015. Inhibition of Super-Enhancer Activity in Autoinflammatory Site-Derived T Cells Reduces Disease-Associated Gene Expression. Cell Rep. 12(12):1986–96 [DOI] [PubMed] [Google Scholar]
- 82.Pott S, Lieb JD. 2015. What are super-enhancers? Nat. Genet 47(1):8–12 [DOI] [PubMed] [Google Scholar]
- 83.Pulakanti K, Pinello L, Stelloh C, Blinka S, Allred J, et al. 2013. Enhancer transcribed RNAs arise from hypomethylated, Tet-occupied genomic regions. Epigenetics. 8(12):1303–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 470(7333):279–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rao S, Huang S-C, St. Hilaire BG, Engreitz JM, Perez EM, et al. 2017. Cohesin Loss Eliminates All Loop Domains, Leading To Links Among Superenhancers And Downregulation Of Nearby Genes
- 86.Remeseiro S, Losada A. 2013. Cohesin, a chromatin engagement ring. Curr. Opin. Cell Biol 25(1):63–71 [DOI] [PubMed] [Google Scholar]
- 87.Ren G, Jin W, Cui K, Rodrigez J, Hu G, et al. 2017. CTCF-Mediated Enhancer-Promoter Interaction Is a Critical Regulator of Cell-to-Cell Variation of Gene Expression. Mol. Cell 67(6):1049–58.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rippe K, von Hippel PH, Langowski J. 1995. Action at a distance: DNA-looping and initiation of transcription. Trends Biochem. Sci 20(12):500–506 [DOI] [PubMed] [Google Scholar]
- 89.Sati S, Cavalli G. 2017. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma. 126(1):33–44 [DOI] [PubMed] [Google Scholar]
- 90.Sawado T, Halow J, Bender MA, Groudine M. 2003. The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 17(8):1009–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schaukowitch K, Joo J-Y, Liu X, Watts JK, Martinez C, Kim T-K. 2014. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 56(1):29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 551(7678):51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, et al. 2013. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 23(12):2066–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Siebel AL, Trinh SK, Formosa MF, Mundra PA, Natoli AK, et al. 2016. Effects of the BET-inhibitor, RVX-208 on the HDL lipidome and glucose metabolism in individuals with prediabetes: A randomized controlled trial. Metabolism. 65(6):904–14 [DOI] [PubMed] [Google Scholar]
- 95.Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, et al. 2015. Transcription factor trapping by RNA in gene regulatory elements. Science. 350(6263):978–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sofueva S, Yaffe E, Chan W-C, Georgopoulou D, Vietri Rudan M, et al. 2013. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 32(24):3119–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Song S-H, Hou C, Dean A. 2007. A Positive Role for NLI/Ldb1 in Long-Range β-Globin Locus Control Region Function. Mol. Cell 28(5):810–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 480(7378):490–95 [DOI] [PubMed] [Google Scholar]
- 99.Stanojevic D, Small S, Levine M. 1991. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 254(5036):1385–87 [DOI] [PubMed] [Google Scholar]
- 100.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, et al. 2007. Population genomics of human gene expression. Nat. Genet 39(10):1217–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10(6):1453–65 [DOI] [PubMed] [Google Scholar]
- 102.Tuan D, Kong S, Hu K. 1992. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proceedings of the National Academy of Sciences. 89(23):11219–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vanhille L, Griffon A, Maqbool MA, Zacarias-Cabeza J, Dao LTM, et al. 2015. High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq. Nat. Commun 6:6905. [DOI] [PubMed] [Google Scholar]
- 104.van Steensel B, Dekker J. 2010. Genomics tools for unraveling chromosome architecture. Nat. Biotechnol 28(10):1089–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, et al. 2015. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J. Exp. Med 212(11):1819–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 457(7231):854–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wadman IA, Osada H, Grütz GG, Agulnick AD, Westphal H, et al. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16(11):3145–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang J, Giaever G. 1988. Action at a distance along a DNA. Science. 240(4850):300–304 [DOI] [PubMed] [Google Scholar]
- 109.Wang J, Wu X, Wei C, Huang X, Ma Q, et al. 2018. YY1 Positively Regulates Transcription by Targeting Promoters and Super-Enhancers through the BAF Complex in Embryonic Stem Cells. Stem Cell Reports. 10(4):1324–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Q, Carroll JS, Brown M. 2005. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19(5):631–42 [DOI] [PubMed] [Google Scholar]
- 111.Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, et al. 2017. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell. 171(7):1573–88.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, et al. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153(2):307–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wiench M, John S, Baek S, Johnson TA, Sung M-H, et al. 2011. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 30(15):3028–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Williamson I, Hill RE, Bickmore WA. 2011. Enhancers: from developmental genetics to the genetics of common human disease. Dev. Cell 21(1):17–19 [DOI] [PubMed] [Google Scholar]
- 115.Willi M, Yoo KH, Reinisch F, Kuhns TM, Lee HK, et al. 2017. Facultative CTCF sites moderate mammary super-enhancer activity and regulate juxtaposed gene in non-mammary cells. Nat. Commun 8:16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Willi M, Yoo KH, Wang C, Trajanoski Z, Hennighausen L. 2016. Differential cytokine sensitivities of STAT5-dependent enhancers rely on Stat5 autoregulation. Nucleic Acids Res. 44(21):10277–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu H, Nord AS, Akiyama JA, Shoukry M, Afzal V, et al. 2014. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS Genet. 10(9):e1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, et al. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36(24):3573–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yamazaki H, Suzuki M, Otsuki A, Shimizu R, Bresnick EH, et al. 2014. A Remote GATA2 Hematopoietic Enhancer Drives Leukemogenesis in inv(3)(q21;q26) by Activating EVI1 Expression. Cancer Cell. 25(4):415–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu M, Yang W, Ni T, Tang Z, Nakadai T, et al. 2015. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 350(6266):1383–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, et al. 2016. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet 48(2):176–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet 38(11):1341–47 [DOI] [PubMed] [Google Scholar]
- 123.Zhou HY, Katsman Y, Dhaliwal NK, Davidson S, Macpherson NN, et al. 2014. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 28(24):2699–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, et al. 2014. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U. S. A 111(3):996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]