Abstract
Bacterial endospores can survive harsh environmental conditions and long-term dormancy in the absence of nutrients, but can rapidly germinate under favorable conditions. In the present study, we employed transposon sequencing (Tn-seq) to identify genes with previously uncharacterized roles in spore germination. Identified genes that encoded spore inner membrane proteins were chosen for study of defined mutants, which exhibited delayed germination in several assays in response to varying germinants. Significantly slowed release of DPA indicated that mutants were affected in Stage I of germination. Several mutants exhibited phenotypic traits consistent with failure of a GerA germinant receptor-mediated response, while others appeared to have a more general loss of response to varied germinants. Use of a gerA-lacZ transcriptional fusion and quantitative western blotting of GerAC allowed mutants to be classified based upon normal or decreased gerA transcription and normal or reduced GerA accumulation. Fourteen genes were identified to have newly described roles within Bacillus spore germination. A more complete understanding of this process can contribute to the development of better spore decontamination procedures.
Introduction
Bacterial endospores are capable of extended periods of dormancy while remaining resistant to a variety of chemical and physical decontamination measures [1]. Dormant spores can rapidly germinate when in a suitable environment, returning to a vegetative state [2, 3]. These factors allow endospores produced by certain species of Bacillus and Clostridium to excel as human pathogens, act as potential bioterrorism agents, and contribute to significant food contamination events [4, 5]. Preservation of dehydration of the metabolically inactive spore core is the greatest factor in spore resistance properties and maintenance of spore dormancy [1]. This dormant state is maintained by the inner spore membrane, which exists in a largely non-fluid state [6], and a thick layer of peptidoglycan termed the cortex [7]. Additionally, the accumulation of small molecule solutes within the core, such as calcium dipicolinic acid (DPA), contribute to spore dehydration and resistance properties [1].
When dormant spores sense an environment conducive to vegetative growth, they will rapidly germinate. Environmental sensing is achieved through the action of proteins expressed late in sporulation, termed germinant receptors. Bacillus subtilis encodes three functional Ger receptors: GerA, GerB, and GerK [8]. The GerA receptor responds to amino acids such as L-Alanine and L-Valine, while the GerB and GerK receptors work together to respond to a mixture of L-Asparagine, D-glucose, D-fructose and K+ ions (AGFK) [2]. The mechanism of signal transduction from Ger receptors to other spore components to initiate germination is not well understood, but a major event is the opening of a channel involving SpoVA proteins to release Ca2+-DPA from the spore core [9]. The GerD protein of Bacillus species is required for a rapid response to germinants. Recent work suggests that GerD is essential for the colocalization of Ger receptors in the spore’s inner membrane in a cluster termed the germinosome and probably plays an intermediate role in the signal transduction pathway from germinant-receptor complex to downstream germination effectors [9, 10].
Each Ger receptor is composed of three subunits: A, B, and C. The A subunits are transmembrane proteins featuring sizable domains on each side of the membrane [3, 8]. The B subunit proteins are thought to be integral membrane proteins that may be involved in germinant recognition [11]. The C subunits are lipoproteins attached to the other surface of the membrane [12]. Following triggering of the Ger receptors, water begins to partially rehydrate the spore core, and Ca2+-DPA is released along with other ions contained within the spore core. The spore cortex is then degraded through the action of germination-specific lytic enzymes (GSLEs) which allow the spore core to continue to expand and return to a fully hydrated state [13]. The spore will then resume metabolism and continue through outgrowth, eventually returning to a fully vegetative state.
The goals of the current study were to identify additional genes with potential roles in spore germination. Whereas previous studies have characterized genes whose loss resulted in a near complete block of germination, we sought to find genes with more subtle phenotypes that were potentially missed by previous procedures. Creation of a transposon-insertion mutant library and submission of spores produced by that library to germination conditions, in combination with Transposon Sequencing (Tn-seq)[14], facilitated identification of 61 genes that exerted significant effects on germination efficiency or rate. Among these, 14 genes had not been previously associated with spore germination and had been shown to produce proteins within the spore membrane proteome, and these were selected for further characterization. Defined gene knockout strains demonstrated reduced germination. Further studies implicated certain genes in affecting the overall GerA receptor abundance within the dormant spores.
Materials and methods
Strain constructions
All strains are listed in Table A in S1 File. DNA extracted from a previously constructed Tn-insertion library [15] was transformed into PS832 with selection for spectinomycin resistance (100 μg/mL). Roughly 150,000 independent transformants were pooled from plates to produce a new library.
Mutants lacking single genes were obtained from the Bacillus Genetic Stock Center. Each mutation was a deletion/insertion, with the gene of interest replaced by an erythromycin resistance gene flanked by two loxP sites [16]. The mutations were introduced into B. subtilis strain PS832 by natural transformation with selection for erythromycin (2.5 μg/ml) and lincomycin (12.5 μg/ml) (MLS) resistance.
Chromosomal DNA from B. subtilis strain DPVB724 with a gerB deletion and insertion of a chloramphenicol (3.0 μg/ml) resistance gene was used to transform strains to GerB-. This was done to reduce background detection of GerBC during Western blot quantification of GerAC.
B. subtilis strain DPVB761 with a gerA-lacZ fusion marked with an MLS resistance gene was obtained from the Setlow lab [17, 18]. Since all the putative Ger mutant strains had the same resistance gene marker, it was essential to delete the MLS resistance gene in order to transform the mutant strains with chromosomal DNA carrying the gerA-lacZ fusion. The Cre recombinase was expressed from plasmid pDR244 [16] to stimulate deletion of the resistance gene, leaving an unmarked in-frame deletion mutation. The gerA-lacZ fusion was then introduced by natural transformation into the 14 mutant strains carrying an unmarked deletion, with selection for MLS resistance.
Chromosomal DNA from B. subtilis strain DPVB833, in which gerA expression is under the control of spore-specific promoter PsspD [19], was introduced by natural transformation into strains with an unmarked deletions, with selection for MLS resistance. All mutations were verified by PCR and agarose gel electrophoresis.
Spore preparation
B. subtilis spores, both single strains and the Tn-insertion strain library, were prepared in 2xSG broth [20]. Spores were harvested after 3–4 days incubation at 37°C and washed by shaking in water at 4°C and repeated centrifugation for several days. Purified spores were examined by microscopy and were judged to be >95% phase-bright spores prior to further assay.
To prepare germinated spores from the Tn-insertion library, a 10-ml suspension of dormant spores at an optical density at 600 nm (OD600) of 100 (~3x1011 spores in total) in water was heat-activated at 70°C for 30 min and cooled on ice for 10 min. The spores were then submitted to germination conditions in 50 mM NaPO4 buffer (pH 7.4) with 10 mM L-Valine at 37°C for 45 minutes. OD600 was monitored to ensure progression through germination. Germinated spores were collected by centrifugation at 12,000 g for 5 min at 4°C. After germination, subpopulations of dormant and partially germinated spores (𝛿≥1.25 g/ml) and fully germinated spores (𝛿~1.19 g/ml) were collected following centrifugation through a layer of 43% sodium diatrizoate.
Sequencing of Tn insertion sites
All spore samples were decoated using urea, SDS, and DTT as described previously [21]. DNA was extracted from decoated spores using the Gram-Positive protocol from the Qiagen Blood and Tissue kit. DNA was digested with MmeI and quantified using Qubit (ThermoFisher). Samples were sent to the High-Throughput Sequencing and Genotyping Center at the University of Illinois for library preparation and sequencing. Adaptor ligations were performed including index bar codes as well as flow cell sequences. Following adaptor ligation, samples were PCR amplified for 18 cycles. After amplification, samples were purified and sequenced using an Illumina Hi Seq 2500, yielding reads with 5’ transposon sequence followed by a 16-bp region of genomic DNA. Sequencing read data files were uploaded an analyzed using Geneious (version 10.0) (http://www.geneious.com, [22]). Reads were filtered and trimmed leaving only genomic sequences and mapped to the JH642 B. subtilis genome. Tables were exported from Geneious listing number of reads contained within each annotated gene. Reads within each individual data set were expressed as a function of the total number of reads per million from that sample. Once normalized, both experimental data sets, dormant and germinated, were compared against one another allowing reporting of fold change between samples. DESeq2 was used to determine p-values comparing dormant and germinated samples [23]. Genes with 2-fold higher number of reads in the dormant versus the germinated sample and significant p-values (≤0.05) were selected for further study.
Germination assays
To quantify change in OD600, purified spores were heat activated at 70°C for 30 minutes, quenched on ice for 5 minutes, and diluted to an OD600 of 0.2 in 50 mM NaPO4 buffer (pH 7.4) containing L-Valine (10 mM) or AGFK (10 mM L-Asparagine, 1 mM D-Glucose, 1 mM D-Fructose, 10 mM KCl). Purified spores were heat activated at 70°C for 30 minutes, quenched on ice for 5 minutes, and stimulated to germinate by dilution to an OD600 of 0.2 in 2xYT (Final concentrations: 8 mg/ml Yeast Extract, 12.8 mg/ml Tryptone, 4 mg/ml NaCl). Changes in OD600 were monitored using a Spectronic Genesys 5 spectrophotometer.
Purified spores of strains with and without overexpressed gerA were heat activated at 70°C for 30 minutes, quenched on ice for 5 minutes, and diluted to OD600 of 0.1 in 25 mM HEPES buffer (pH 7.4). Nutrient germinants were added as described above, and changes in OD600 were monitored using a Tecan M200 plate reader.
To quantify non-nutrient-triggered germination, purified spores at an OD600 of 1.0 were germinated with 1 mM dodecylamine in 20 mM KPO4 buffer (pH 7·4) at 37°C. At indicated times, 1 ml of germinating spore suspensions were centrifuged 15,800 x g for 2 min. The A270 of the supernatant was measured to quantify DPA release, which was expressed as a fraction of the total spore DPA content. Total spore DPA was determined by boiling spores for 30 min and measuring the A270 of the resulting supernatant.
To measure DPA release, spores from each mutant strain were heat activated at 70°C, suspended in 25 mM HEPES buffer (pH 7.4) and submitted to germination conditions with 10 mM L-Valine at 37°C. Aliquots were taken from the germinating spores at indicated times and centrifuged at 10,000 g for 45 seconds. The spore exudate was analyzed to determine the amount of DPA released [24].
To measure release of cortex fragments, mutant spores were heat activated at 70°C, suspended in 25 mM HEPES buffer (pH 7.4) and incubated with 10 mM L-Valine at 37°C. OD600 was recorded and aliquots were taken from the germinating spores at indicated times. Samples were centrifuged, and the spore exudate was subjected to amino acid/amino sugar analysis as previously described [25]. Peaks representing N-acetyl muramic acid (NAM) were identified based on elution times and quantified by integration of peak areas in comparison to known standards.
Dormant spores were observed using phase-contrast microscopy prior to germination, such that there were 70–100 spores per field of view and images were collected for 10 fields per sample. Spores were then heat activated at 70°C for 30 minutes, quenched on ice for 5 minutes, resuspended in 50 mM NaPO4 buffer (pH 7.4) and stimulated to germinate by addition of 10 mM L-Valine for 1 hour, and observed by microscopy again. The image processing toolkit Fiji was combined with segmentation machine learning algorithms [26], in the open-source software project Trainable Weka (Waikato Environment for Knowledge Analysis) Segmentation (TWS)[27]. This prototype segmentation algorithm was used to classify phase-bright and phase-dark spores. To segment the input image data, TWS transforms the segmentation problem into a pixel classification problem in which each pixel can be classified as belonging to a specific segment or class such as phase-bright or phase-dark. A set of input pixels that has been labeled is then used as the training set for a selected classifier. Once the classifier is trained, it can be used to classify either the rest of the input pixels or completely new image data. Data were analyzed for images from 3 fields per sample.
Assay of gerA transcription
B. subtilis strains with a gerA-lacZ transcriptional fusion were grown and sporulated at 37°C in 2xSG medium. Purified spores were chemically decoated, washed, and extracted, and 𝛽-galactosidase activity was assayed using methyl-umbelliferyl-D-galactoside (MUG) as previously described [28–30]. MUG fluorescence was measured in a microplate reader (Tecan) using excitation and emission wavelengths of 365 nm and 450 nm, respectively. Standard solutions of methylumbelliferone were prepared in the same mix of buffers in order to calibrate the fluorescence readings. The average activity of PS832 (wildtype without gerA-lacZ fusion) samples was subtracted from the values for all samples containing the gerA-lacZ fusion, and all readings were normalized to decoated spore OD600 values.
Western blotting
Quantitative western blots were performed on strains carrying a gerB deletion to avoid cross-reactivity with the GerAC antibody. Purified dormant B. subtilis spores (~100 OD600 units) were decoated and proteins were extracted as previously described for western blot analysis [21]. Samples were then serially diluted with 2x SDS-PAGE sample loading buffer and gerA gerB or gerD spore protein extract. TGX Stain-Free Fast Cast premixed acrylamide solution (Bio-Rad) was used, which enabled rapid fluorescent detection of tryptophan residues in proteins directly within gels and blots. The proteins were Trp-modified after separation by a trihalo compound included in the electrophoresis gel, allowing fluorescent visualization and quantitation of proteins on gels and blots immediately after the completion of electrophoresis and transfer. The total protein load and recovery for each lane was measured as the total fluorescence intensity for each lane of the blot. This was followed by probing with anti-GerAC [30] or anti-GerD [30] antibodies via chemiluminescent western blot. Band intensity was normalized to the protein present in each lane. Biorad Image Lab 6.0 was used to perform data analysis of quantitative blots. Dilutions of 1.0, 0.5, 0.25, and 0.125 concentration were blotted. Only the 1.0 and 0.5 concentrations were found to be within the linear range of detection, and these were used for quantification. Quantitative GerAC and GerD western blots were performed in triplicate.
Results
Identification of mutant strains with slowed or reduced germination
Seeking to identify additional genes that contribute to spore germination, Tn-seq was used to reveal genes functioning in the early stages of germination. A library of magellan6x transposon insertions [15] was transformed into a B. subtilis wild type strain, PS832, that is highly efficient at spore formation and germination. An estimated 150,000 independent transformants were pooled into a library from which dormant spores were produced. A sample of this spore library was collected for Tn insertion site sequencing.
Dormant spores were heat-activated and were submitted to germination-inducing conditions with 10 mM L-Valine at 37°C for 45 minutes. A 45% drop of the starting OD600 was observed as an indication of germination. Following germination, a density gradient was used to separate dormant (and possibly partially germinated) spores (≥1.25 g/ml) from fully germinated spores (~1.19 g/ml)[31]. This procedure was performed using two independent dormant spore preparations.
DNA was extracted from the starting spore population and the dormant and germinated spores, and the Tn insertion sites were sequenced as described [15] and mapped to the B. subtilis genome. Sequence data is available at https://www.ncbi.nlm.nih.gov/sra/PRJNA544251. The total library was found to have 5.5 x 104 unique insertions spread over 3,114 genes featuring ≥10 unique insertions per gene. The number of reads within each gene were normalized as a fraction of the total reads obtained for that sample. Normalized data sets from germinated and dormant spores were then compared against one another to determine fold change. Genes with a higher proportion of reads in the dormant population indicate a possible role in germination. DESeq2 was implemented to determine p-values to further differentiate mutant abundance between sample sets [23, 32].
In total, Tn insertions in 61 genes were found to be ≥2-fold underrepresented in the germinated spores compared to those unable to complete germination (Table 1). These included all three genes of the gerA operon, gerE, coat proteins cotH and cotE, and genes from the gerP operon, all of which have known strong effects on germination. Slightly less than half of the genes identified were known previously to have either sporulation or germination defects.
Table 1. Genes in which Tn insertions altered germination.
| Gene | p-valuea | Fold-change Sample 1b | Fold-change Sample 2b | Reference for Ger defect |
|---|---|---|---|---|
| cotH | 2.2E-35 | 15.0 | 10.9 | [33] |
| gerAA | 4.4E-31 | 150.1 | 43.7 | [18, 34] |
| gerAC | 2.0E-29 | 279.9 | 73.9 | [18, 34] |
| cotE | 3.8E-24 | 17.0 | 12.0 | [35] |
| ygaC | 2.3E-18 | 4.6 | 4.2 | |
| yqfT | 3.0E-16 | 15.4 | 8.8 | |
| ypzK | 6.3E-16 | 10.1 | 9.7 | |
| yqeF | 2.0E-15 | 3.3 | 5.5 | |
| ymzD-ymcCc | 5.7E-13 | 2.5 | 2.7 | |
| gerPF | 5.8E-13 | 7.4 | 4.1 | [36] |
| safA | 1.1E-11 | 7.3 | 4.1 | [37] |
| pcrB | 2.3E-11 | 3.0 | 2.6 | |
| ylbC | 3.9E-11 | 4.1 | 2.9 | |
| gidA | 1.3E-10 | 4.2 | 2.8 | |
| gerPB | 1.0E-09 | 5.9 | 3.6 | [36] |
| gerPC | 3.1E-09 | 11.7 | 3.9 | [36] |
| nocA | 4.2E-09 | 2.0 | 2.2 | |
| veg | 1.1E-08 | 4.8 | 3.2 | [38] |
| gerE | 1.8E-08 | Infinite | 32.0 | [34] |
| ytoA | 2.2E-08 | 5.1 | 4.1 | |
| ytpA | 1.0E-07 | 5.3 | 4.6 | |
| ytpB | 1.1E-07 | 3.9 | 3.5 | |
| rsbW | 1.2E-07 | 2.2 | 2.5 | |
| yfhD | 2.8E-07 | 4.2 | 4.6 | |
| cotZ | 7.7E-07 | 2.1 | 3.1 | [39] |
| yqhL | 1.1E-06 | 4.3 | 2.5 | |
| kinB | 1.8E-06 | 1.6 | 1.9 | |
| skfC | 1.9E-06 | 7.9 | 3.7 | |
| skfE | 3.8E-06 | 8.4 | 3.7 | |
| gerAB | 5.8E-06 | 181.7 | 73.6 | [18, 34] |
| ymaB | 6.6E-06 | 1.9 | 2.9 | |
| sipT | 7.5E-06 | 2.5 | 2.8 | |
| skfG | 9.7E-06 | 3.5 | 2.3 | |
| phoR | 1.0E-05 | 2.5 | 3.0 | |
| cotN (tasA) | 1.5E-05 | 2.1 | 2.2 | [40] |
| yhbJ | 2.8E-05 | 1.8 | 1.7 | |
| yhaM | 3.3E-05 | 4.6 | 2.5 | |
| ymaF | 3.7E-05 | 8.1 | 2.8 | |
| yabG | 1.7E-04 | 2.5 | 2.3 | [41] |
| spoVID | 1.8E-04 | 2.7 | 2.1 | [42] |
| yqhR | 3.6E-04 | 2.3 | 2.0 | |
| yonF | 4.7E-04 | 1.8 | 3.0 | |
| spoVAF | 4.8E-04 | 2.2 | 1.8 | [43] |
| hfq | 6.7E-04 | 3.2 | 2.5 | |
| yosK | 9.2E-04 | 4.4 | 6.4 | |
| yozE | 9.4E-04 | 3.0 | 4.8 | |
| yopI | 1.1E-03 | 2.3 | 2.8 | |
| flgN | 1.2E-03 | 14.2 | 3.6 | |
| gerD | 1.2E-03 | 2.6 | 1.9 | [34] |
| fliW | 2.0E-03 | 1.9 | 3.4 | |
| yfbJ | 2.0E-03 | 3.1 | 2.3 | |
| ytmO | 2.1E-03 | 4.0 | 3.5 | |
| gerPE | 2.1E-03 | 6.5 | 1.7 | [36] |
| phoP | 2.3E-03 | 3.2 | 3.4 | |
| gerPD | 4.7E-03 | 8.2 | 3.2 | [36] |
| tufA | 5.2E-03 | 5.7 | 3.1 | |
| ispA | 5.7E-03 | 2.9 | 2.8 | |
| yoqL | 1.7E-02 | 6.1 | 3.7 | |
| dnaJ | 4.7E-02 | 2.5 | 2.9 | |
| yaaB (remB) | 5.0E-02 | 7.4 | 2.3 | |
| ytxG | 2.1E-01 | 2.7 | 2.9 | |
| yybT (gdpP) | 2.4E-01 | 7.5 | 2.1 |
a p-value determined using DESeq2 [23] comparing Dormant and Germinated sample read counts.
b Fold change in read counts of Dormant/Germinated samples
c Intergenic region
The identified genes that were not previously implicated in spore germination were cross-referenced against proteins found in the inner spore membrane proteome [44, 45], identifying a group of 14 genes that were studied further (Table 2). The majority of these genes were largely uncharacterized and were annotated with a wide range of putative functions. Many of the genes are not known to be expressed via sporulation-specific regulatory factors but rather are regulated by vegetative cell transcriptional controls.
Table 2. Genes without previously known germination role identified by Tn-seq and in spore membrane proteome.
| Gene | Function | Locus structure | Regulation of expression |
|---|---|---|---|
| dnaJ | Protein quality control | hrcA-grpE-dnaK-dnaJ-yqeTUV | σA, HrcA [46] |
| hfq | RNA chaperone | hfq | Increased protein during transition to stationary phase [47] |
| pcrB | Heptaprenylglyceryl phosphate synthase | pcrB-pcrA-ligA-yerH | LexA regulon [48] |
| phoP | Response regulator, phosphate metabolism | phoPR | σA, σB, σE, CcpA, ScoC [49, 50] |
| phoR | Sensor kinase, phosphate metabolism | phoPR | σA, σB, σE, CcpA, ScoC [49, 50] |
| sipT | Signal peptidase I | sipT | DegU [51] |
| skfE | Export of spore killing factor (SkfA) | skfABCEFGH | Spo0A, AbrB, PhoP [52–54] |
| ygaC | Unknown | ygaCD | |
| ylbC | Unknown | ylbBC | σF [55] |
| yqeF | Unknown | yqeF | |
| yqhL | Unknown | yqhL | mRNA processed by RNase Y [56] |
| ytpA | Phospholipase, Bacilysocin synthesis | ytpAB | σM [57] |
| ytxG | General stress | ytxGHJ | σB, σH [58] |
| yybT (gdpP) | c-di-AMP phosphodiesterase. Functions in DNA damage and acid resistance [59] | yybS-gdpP-rplI | σA, σD-induced antisense RNA [60] |
Characterization of germination of mutant strains
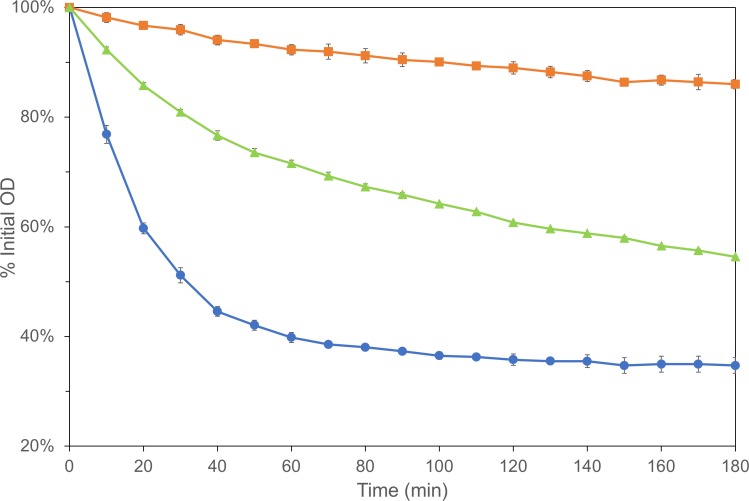
Strains carrying insertion mutations [16] in each of the genes listed in Table 2 were obtained from the Bacillus Genetic Stock Center, and these mutations were transformed into PS832. Mutant strains were characterized with regard to growth rate and sporulation efficiency (Table 3). A number of the mutants exhibited significant growth rate defects, and the ytxG mutant had a severely reduced sporulation frequency. For comparison, gerA mutants grow and sporulate at wild type rates [34]. Purified spores were analyzed using several germination assays to verify the defects indicated by the Tn-seq data in addition to providing insight into potential function of these genes in germination. Spores were germinated with the addition of 10 mM L-Val, and OD600 was monitored; an example assay is shown in Fig 1 (Additional data in Figure A in S1 File). Each mutant strain exhibited a significant germination rate defect in response to L-Val in comparison to the wild type (Table 4). The most severe delays in germination rate were observed for the ylbC, dnaJ, sipT, and hfq mutants. For comparison, a gerA mutant exhibited <1% germination even in the presence of 200 mM L-Ala, which is more strongly stimulatory than 10 mM L-Val [61].
Table 3. Phenotypic properties of B. subtilis strains.
| Genotype | Doubling timea (min) | Sporulation efficiencyb (%) | DPA releasec (μg/ml/OD) | NAM releasec (nmole/ml/OD) | % phase-dark sporesd |
|---|---|---|---|---|---|
| Wild type | 20 | 66 | 5.3 ± 0.1 | 61.0 ± 14.2 | 95 |
| dnaJ | 31 | 89 | 1.1 ± 0.4* | 25.9 ± 12.7 | 14 |
| hfq | 40 | 63 | 2.2 ± .03* | 30.1 ± 6.2 | 48 |
| pcrB | 31 | 68 | 3.0 ± 0 | 30.3 ± 8.0 | 41 |
| phoP | 44 | 83 | 2.7 ± 0.4* | 46.6 ± 5.8 | 63 |
| phoR | 35 | 54 | 3.9 ± 0.5 | 39.2 ± 9.5 | 86 |
| sipT | 34 | 54 | 2.3 ± 1.0* | 23.5 ± 10.2* | 22 |
| skfE | 27 | 59 | 1.8 ± 0* | 40.6 ± 10.7 | 38 |
| ygaC | 21 | 71 | 3.1 ± 0.4* | 36.6 ± 9.9 | 60 |
| ylbC | 29 | 48 | 1.1 ± 0.3* | 14.3 ± 3.4* | 19 |
| yqeF | 23 | 84 | 1.9 ± 0* | 37.2 ± 9.4 | 50 |
| yqhL | 20 | 53 | 2.6 ± 0.4* | 36.1 ± 8.1 | 69 |
| ytpA | 22 | 95 | 1.63 ± 0* | 17.7 ± 2.7* | 55 |
| ytxG | 31 | 18 | 4.8 ± 0.3 | 50.9 ± 4.8 | 90 |
| yybT | 21 | 69 | 4.7 ± 0.4 | 56.6 ± 14.1 | 92 |
a Growth in 2xSG medium at 37°C
b Heat-resistant count/total viable count after 24 hr incubation on 2xSG medium at 37°C.
c Release of DPA and NAM 30 or 45 min, respectively, after exposure to 10 mM L-Val at 37°C.
* indicates a significant difference from the wild type (T-test, p<0.05). Values are indicative of averages and standard deviations of three biological replicates.
d Spores pixel intensities quantified and classified as described in Materials and Methods after 60 min exposure to 10 mM L-Val at 37°C.
Fig 1. Germination rates of B. subtilis strains.
Purified spores were heat activated, stimulated to germinate by addition of 10 mM L-Val, and shaken at 37°C, during which the OD600 was monitored. Values are averages of three assays and error bars are standard deviations. Each assay was performed on three replicate spore preparations. For the ylbC (■) and phoP (▲) mutants, all points after 10 min are significantly different from those of the wild type(●); P≤0.05.
Table 4. Response of B. subtilis strains to varied germinants.
| Genotype | % OD600 lossa | % DPA released by dodecylamineb | ||
|---|---|---|---|---|
| L-Val (60 mins) |
AGFK (60 mins) |
2xYT (40 mins) |
||
| Wild type | 60 ± 1 | 41 ± 2 | 60 ± 2 | 75 ± 1 |
| dnaJ | 6 ± 0** | 28 ± 10* | 40 ± 3* | 68 ± 4 |
| hfq | 26 ± 5* | 30 ± 1* | 52 ± 2* | 75 ± 1 |
| pcrB | 47 ± 10* | 27 ± 3* | 58 ± 4 | 87 ± 2 |
| phoP | 28 ± 1* | 36 ± 10 | 53 ± 1* | 59 ± 6 |
| phoR | 44 ± 2* | 22 ± 10* | 56 ± 1 | 52 ± 6 |
| sipT | 7 ± 0** | 33 ± 10 | 57 ± 7 | 69 ± 5 |
| skfE | 35 ± 10* | 28 ± 6* | 50 ± 3* | 82 ± 5 |
| ygaC | 22 ± 3* | 37 ± 10 | 55 ± 2 | 67 ± 5 |
| ylbC | 8 ± 1** | 13 ± 2** | 23 ± 8** | 66 ± 3 |
| yqeF | 38 ± 3* | 33 ± 7 | 58 ± 1 | 84 ± 1 |
| yqhL | 33 ± 6* | 37 ± 10 | 54 ± 3 | 71 ± 2 |
| ytpA | 32 ± 3* | 41 ± 3 | 47 ± 3* | 73 ± 4 |
| ytxG | 35 ± 0* | 28 ± 8 | 53 ± 1* | 72 ± 4 |
| yybT | 47 ± 2* | 44 ± 7 | 60 ± 0 | 76 ± 4 |
a Values are averages and standard deviations of assays on three replicate spore preparations. OD600 of purified spore suspension monitored at the indicated time after addition of 10 mM L-Valine, 1X AGFK, or 2xYT while shaking at 37°C.
* indicates a significant difference (T-test, p<0.05)
** indicates a significant difference (T-test, p<0.01) from the wild type.
b Values are averages and standard deviations of assays on three replicate spore preparations. DPA release by purified spore suspension monitored 100 min after addition of 1 mM dodecylamine while shaking at 37°C.
The slowed germination phenotype was further characterized by examining the individual stages of germination using assays for release of dipicolinic acid (DPA) (Stage I) and N-acetylmuramic acid (Stage II)[3]. Spores from each mutant strain were heat-activated, suspended in 25 mM HEPES buffer, and submitted to germination conditions with 10 mM L-Val at 37°C. The amount of DPA released from many of the mutant strains was vastly reduced compared to that of PS832 (Table 3). For comparison, spores of a gerA mutant release ≤1% of their DPA over 120 min in the presence of L-Val [62]. The only strains that were not significantly different from PS832 were the yybT, ytxG, pcrB, and phoR mutants. Mutant strains lacking sipT, ylbC, or ytpA demonstrated NAM release significantly less (p<0.05) than that of PS832 (Table 3, Figure B in S1 File). The rest of the mutants exhibited reduction compared to PS832 but due to high variation among replicates were not found to be significantly different (Table 3, Figure B in S1 File).
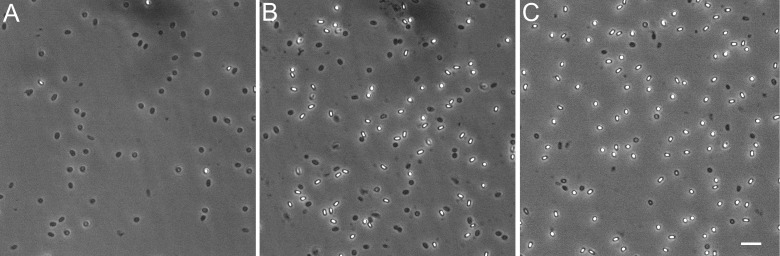
When mutant spore populations exhibit a decreased OD600 change during germination, it could result from a large percentage of the spore population germinating incompletely or from a smaller percentage of the population germinating at a normal rate with the remainder remaining fully dormant. Spores of the various strains were imaged using phase-contrast microscopy, both prior to and one-hour post-germination with 10 mM L-Val (Fig 2), and spores were classified as phase-bright or phase-dark based on pixel intensities (Figure C in S1 File). All spore samples had ≥95% phase-bright spores prior to germination. Post-germination, the wildtype spores had 95% phase-dark spores while most of the mutant strains had significantly decreased percentages of phase-dark spores (Table 3), indicating that much of the mutant strain spore populations did not initiate germination.
Fig 2. Phase-contrast microscopy of germinating B. subtilis spore populations.
Purified spores of B. subtilis wild type and mutant strains were heat-activated and stimulated to germinate by addition of 10 mM L-Val followed by incubation at 37°C for 60 mins. A) PS832 B) ytpA mutant strain C) ylbC mutant strain All panels are the same magnification; the bar in panel C is 5 μm.
Additional assays were performed using the germinants AGFK and 2xYT (Table 4), which began to differentiate the mutants into distinct phenotypic groups. The first group features a reduction in germination rate to all nutrient germinants tested: L-Val, AGFK, and 2xYT, and includes strains with mutations in skfE, ylbC, hfq and dnaJ. The second phenotype includes strains with a significantly delayed L-Val germination response, via a GerA receptor, but otherwise germinate normally in response to rich medium and AGFK, via the GerB and GerK receptors. The following strains featured this phenotype: yybT, ygaC, yqhL, yqeF and sipT. The final group included strains with significantly slower germination rates in response to L-Val but have a delay in response to either AGFK or 2xYT, but not both. Mutants lacking ytpA, phoP, phoR, pcrB and ytxG feature these phenotypes. In addition, like a gerA mutant [63], all mutant strains were capable of normal non-nutrient, non-Ger-receptor-mediated germination, in response to dodecylamine (Table 4).
To determine if spores from mutant strains were blocked in germination or if they were simply severely delayed, spores were plated and colonies that appeared over a 48-hour period were counted. After 24 hours, all strains produced cfu/OD600 values similar to that of the wild type strain, and none of the strains produced a >4% increase in colonies after the first 24 hours (Table B in S1 File), indicating that the defects were a significantly slowed germination process and not death of the spores.
Expression of the GerA receptor in mutant strains
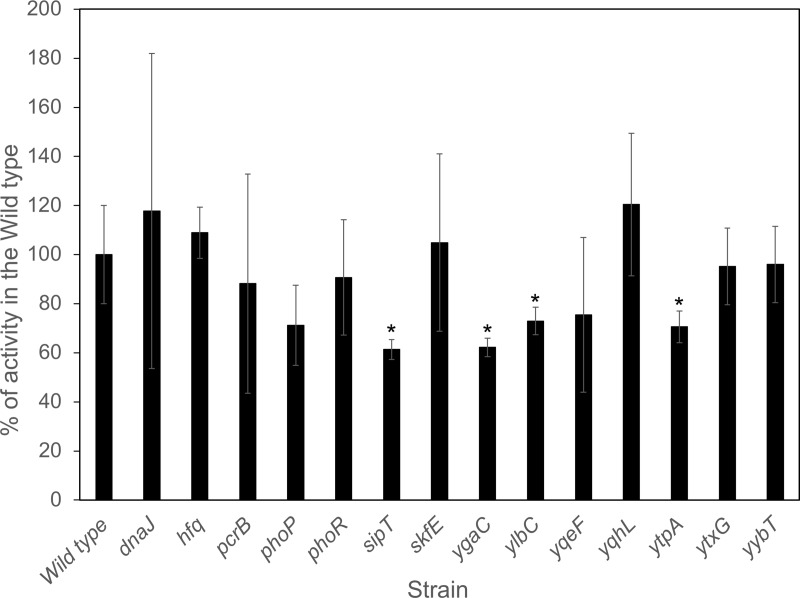
Decreased germination in response to L-Val can result from a low abundance of the GerA receptor [29, 30]. A gerA-lacZ transcriptional fusion was used to determine if germination defects were correlated to reduced gerA transcription. Mutant strains lacking sipT, ytpA, ylbC, or ygaC showed a significant decrease in gerA transcription in comparison to the wildtype (Fig 3). To determine if this was a general effect on 𝜎G-dependent transcription, the effects of these mutation on the expression of pbpF and sspB were examined using lacZ fusions. The expression of these two genes was unaffected by these mutations (Figure D in S1 File).
Fig 3. Expression of a gerA-lacZ transcriptional fusion.
Purified spores carrying a gerA-lacZ transcriptional fusion were decoated and lysed, and extracts were assayed for β-galactosidase. Values are expressed as a percentage of that detected in DPVB761, the wild type strain containing the gerA-lacZ fusion. Values are averages of assays on three replicate spore preparations and error bars are standard deviations. * indicates a significant difference from the wild type (p ≤ 0.05).
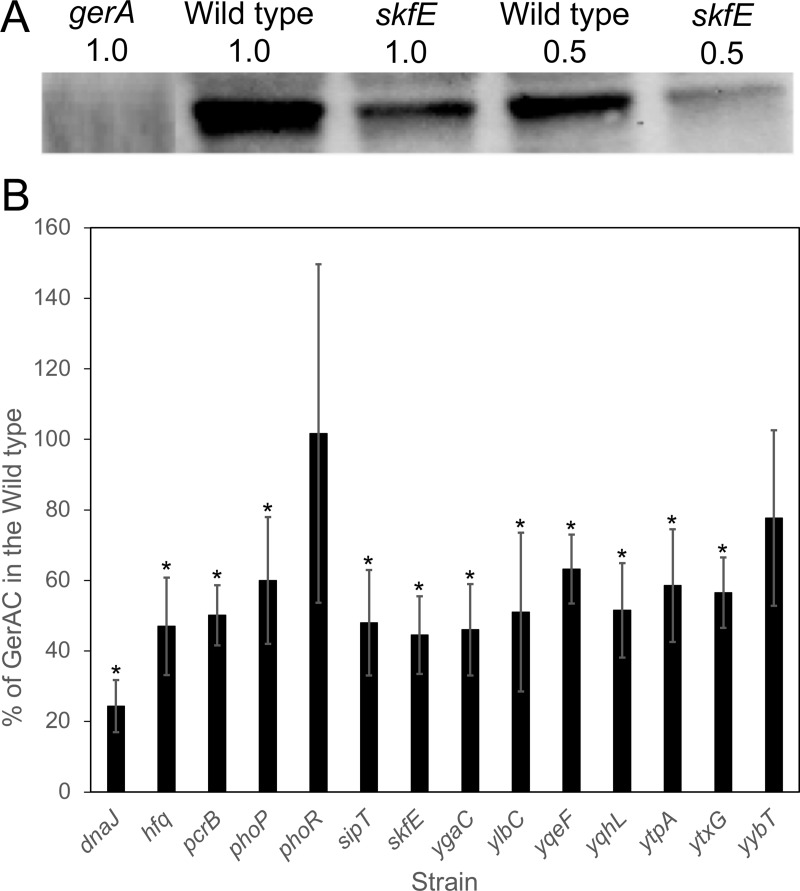
Quantitative GerAC western blots were performed to determine the amount of GerA receptor in spores of all strains. An example Western blot is in Fig 4A, and additional blots are in Figure E in S1 File. Many of the mutant strains exhibited significant decreases in GerAC abundance; the most significant being a 75% decrease in a dnaJ mutant (Fig 4B). The abundance of GerD was also determined using quantitative Western blots for spores of all mutant strains, as a GerD deficiency could result in reduced germination efficiency. All the strains contained amounts of GerD similar to that of the wild type, suggesting that GerD remains unaffected in these mutant strains (Figure F in S1 File).
Fig 4. GerAC is reduced in the spores of several B. subtilis mutant strains.
Equal quantities of spore suspensions were decoated and broken, and proteins were extracted, serially diluted, separated on SDS-PAGE, and transferred to PVDF membrane as described previously [21]. The membrane was probed with anti-GerAC antibodies [30] (Panel A and Figure E in S1 File). Strain genotype (All strains were also 𝛥gerB.) and sample dilution is indicated above each lane. Protein load and transfer to membrane in each lane was normalized as described in Materials and Methods, and the amount of GerAC detected in each strain was compared to that found in the wild type (Panel B). Error bars indicate standard deviations. * indicates a significant difference from the wild type (p ≤ 0.05).
Overexpression of the GerA receptor in spores has previously been shown to increase the response to GerA-specific germinants [19]. A fusion of the gerA operon to the forespore-specific sspD promoter [19] was introduced into strains in order to determine if GerA overexpression could reverse the germination defects associated with the mutations under study. In almost all cases, GerA overexpression reversed the germination deficiency with L-Val (Table 5), suggesting that decreased GerA abundance made a significant contribution to the reduced germination efficiency in these mutant strains. Strikingly, in the ytxG mutant, overexpression of GerA increased the germination deficiency. As previously observed [19], overexpression of GerA resulted in significant decreases in the germination response to AGFK in all strains (Table C in S1 File). In mutant strains that exhibited reduced germination in response to 2xYT, GerA overexpression reversed this deficiency (Table C in S1 File).
Table 5. Overexpression of gerA suppresses germination defect of multiple mutants.
| Genotype | % OD Lossa | |
|---|---|---|
| without sspDp-gerA | with sspDp-gerA | |
| Wild type | 35 ± 4 | 38 ± 2 |
| 𝛥skfE | 23 ± 5* | 38 ± 3 |
| 𝛥pcrB | 34 ± 7 | 37 ± 1 |
| 𝛥ygaC | 26 ± 1* | 36 ± 1 |
| 𝛥sipT | 9 ± 5* | 41 ± 1 |
| 𝛥ylbC | 7 ± 0* | 37 ± 0 |
| 𝛥hfq | 27 ± 3* | 38 ± 3 |
| 𝛥yqhL | 29 ± 3* | 37 ± 0 |
| 𝛥dnaJ | 12 ± 2* | 36 ± 2 |
| 𝛥yqeF | 28 ± 2* | 31 ± 2 |
| 𝛥phoR | 37 ± 1 | 42 ± 9 |
| 𝛥phoP | 32 ± 1 | 36 ± 0 |
| 𝛥ytxG | 25 ± 6* | 15 ± 4* |
| 𝛥ytpA | 25 ± 0* | 38 ± 2 |
| 𝛥yybT | 37 ± 2 | 37 ± 2 |
a Values are averages and standard deviations of assays on three replicate spore preparations. OD600 of purified spore suspension monitored 45 min after addition of 10 mM L-Valine while shaking at 37°C.
* indicates a significant difference from the wild type (T-test, p<0.05)
Discussion
The germination and return to growth of bacterial spores is an essential step in the initiation of several diseases and of some causes of food spoilage. This Tn-seq analysis identified 42 B. subtilis genes that had not previously been associated with germination but are required for a highly efficient germination response to L-Val. As the majority of proteins previously found to play major roles in germination are membrane-associated, fourteen of these genes, whose products had also been identified in studies of the spore membrane proteome [44, 45], were further characterized. Well-defined mutations in these genes caused significantly reduced responses to L-Val, and in some cases a decreased response to other nutrient germinants. Several of these strains also exhibited slowed vegetative growth; such a growth defect could certainly alter gene expression and progression through the sporulation process, potentially affecting the germination apparatus. Future work on the specific mechanism by which these mutations alter germination may reveal such effects. For all these mutants, the germination defect appears to largely be a slow initiation of germination rather than a specific slowing of a subsequent step in the germination process. The reduced percentage of spores within the population that do initiate germination appear to progress through Stages I and II of germination at a near normal pace; rates of OD loss and phase darkening are largely mirrored by rates of DPA and NAM release. This suggests that the genes under study play a role in the earliest steps of germination initiation.
Consistent with this idea, many of the mutant strains had reduced abundance of the GerA receptor, indicating effects on receptor expression and stability or membrane incorporation. A GerA deficiency is not surprising, given that the primary screen for identification of these genes was for a reduced response to L-Val, which is recognized via GerA [2]. Based on responses to additional germinant classes, the mutant strains could be separated into distinct phenotypic groups. The first group features a reduction in germination rate with all nutrient germinants tested (L-Val, AGFK, and 2xYT), demonstrating reduced germination efficiency mediated through all receptors: GerA, GerB, and GerK. Mutant strains lacking skfE, ylbC, hfq, or dnaJ fall in this group, which also includes the strains with the greatest decreases in GerA abundance. These genes may play roles in expression or assembly of all Ger receptors or in facilitating signal transduction from germinant receptors to other parts of the germination apparatus. The well-studied function of DnaJ as a protein chaperone [64] might explain its effect on GerAC abundance in spores, and this effect suggests that DnaJ is active in the forespore late into sporulation. While the role of Hfq in Gram-positive species is not as well studied as in some Gram-negative species, its known role as an RNA chaperone [65] might exert post-transcriptional effects on the production of proteins important in the germination process. Interestingly, several other genes found in this study to affect germination (dnaJ, ylbBC, yqeF, yqhL, yybT) have sizable 5’ untranslated regions in their mRNAs, which might be sites for post-transcriptional regulation, or for which antisense RNAs have been identified (See http://subtiwiki.uni-goettingen.de/ for genes yqeF, yqhL, and yybT). A mechanism by which SkfE, which is involved in export of a sibling-killing antimicrobial [66], might affect germination is harder to imagine, but the fact that Tn insertions in two other genes in the skf operon also reduced germination supports the importance of this effect. Interestingly, expression of the skf operon is regulated by PhoPR [52], genes also implicated by this study in altering germination response.
One of the more interesting genes identified for future study may be ylbC, which is likely expressed as the downstream gene in the σF-regulated ylbB-ylbC operon [55]. YlbC contains two conserved domains: an N-terminal cysteine rich secretory “CAP” domain, and a YkwD domain of unknown function that is found only in proteins of spore-forming bacteria [67]. YlbB contains two conserved CBS domains [67], which in at least one case has a role in ATP-binding [68]. Tn insertions in ylbB were significantly underrepresented in the germination screen (p = 0.014) but did not achieve the cutoff of a 2-fold change. Thus, ylbB may function with ylbC, but might be partially redundant with the paralogous yhcV, which is 𝜎G-dependent [55, 69] and encodes one of the most abundant transcripts in the dormant spore [70]. The mechanism by which YlbC affects gerA transcription and GerA abundance is a topic for ongoing study.
A second phenotypic group is composed of mutant strains with significantly delayed germination via a GerA-mediated response but germinate normally through GerB and GerK sensing. Strains lacking yybT, ygaC, yqhL, yqeF, or sipT exhibit this phenotype, and all except the yybT mutant have significantly reduced spore GerA content. The roles of these genes in germination are unclear, as most are relatively uncharacterized. YybT (GdpP) acts as a c-di-AMP phosphodiesterase and exerts pleotropic effects on physiology and gene expression [71–74]. SipT, acting as a signal peptidase [75], could certainly exert effects on assembly of membrane proteins important for germination, including GerA.
The third phenotypic group includes strains with significantly slower germination rates in response to L-Valine but either have a decreased response to AGFK or 2xYT but not both. Mutants lacking ytpA, phoP, phoR, pcrB or ytxG feature these phenotypes. It is not clear how or why a mutant would be deficient in GerA mediated response, have a normal GerB and GerK response, but still be deficient for germination in rich media. The PhoPR mutants have poor vegetative growth and pleotropic effects on gene expression [76, 77], which might exert quite variable effects into the sporulation process. These mutants seemed to exhibit significant variability between multiple spore preparations. Three mutants in this group may exert effects on membrane structure. YtpA is a phospholipase [57, 78], PcrB is a heptaprenylglyceryl phosphate synthase [79], and a ytxG mutant exhibits defects in membrane morphology [80]. Alterations in the spore inner membrane might affect assembly or function of the germination initiation apparatus. None of these three genes are specifically expressed in sporulating cells, and thus their activity levels and effects on germination might be more varied among spore preparations and possibly with regard to different germinants. Interestingly, the ytxG mutant was the only strain in which overexpression of gerA did not correct the L-Val germination defect. This overexpression in the ytxG mutant did decrease AGFK germination, as in other strains, suggesting that the gerA overexpression was successful. Perhaps a membrane defect in this mutant renders Ger protein complexes nonfunctional regardless of their expression level.
Four of the mutants identified here exhibit decreased gerA transcription. The predicted functions of these gene’s products provide no simple explanation for how such an effect on transcription could come about, and the mechanisms may therefore be indirect. The expression of two other 𝜎G-dependent genes, pbpF and sspB, was not decreased in the mutant strains, indicating that this was not an effect on the entire regulon. Altered activity of a transcription factor involved in gerA transcription, SpoVT or YlyA [17, 55, 69, 81, 82], could be an expected pathway for such an effect. Future work should examine the effects of these mutations on other genes within forespore-specific regulons to resolve this.
Among the germination mutants identified in our Tn-seq screen, strains that could complete Stage I of germination but were blocked in Stage II were not present. This may be due to the mutant screening process utilized. Mutants with Tn insertions in cwlD, which should exhibit this phenotype [83, 84], were slightly enriched in our non-germinating spore population, but not above the significance cutoff value used. Spores blocked at stage II were expected to pellet with dormant spores in the density gradient utilized [31]. One possibility is that spores blocked at Stage II were unstable through the time of incubation with germinant, density gradient separation, and subsequent washing, and thus were not efficiently recovered. Utilization of an alternative isolation method might allow identification of mutants with this phenotype.
Supporting information
Contains Table A. B. subtilis strains used in this study; Table B. Long-term germination efficiency of B. subtilis mutant strains; Table C. Spore germination in response to diverse germinants following overexpression of gerA; Figure A. Germination rates of B. subtilis strains; Figure B. Release of DPA and NAM by B. subtilis strains; Figure C. Phase contrast microscopy image pixel intensities during spore germination; Figure D. Expression of 𝛔G-dependent genes in B. subtilis mutant strains; Figure E. GerAC is reduced in the spores of several B. subtilis mutant strains; Figure F. GerD is not reduced in the spores of B. subtilis germination mutants.
(PDF)
Acknowledgments
We thank Alan Grossman for providing the Tn insertion library, Peter Setlow and George Korza for strains and antibodies, and Jennifer Meador-Parton and Isabelle Wal for technical assistance.
Data Availability
Sequence data has been deposited to the NCBI GenBank and can be accessed via https://www.ncbi.nlm.nih.gov/sra/PRJNA544251. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101(3):514–25. 10.1111/j.1365-2672.2005.02736.x [DOI] [PubMed] [Google Scholar]
- 2.Moir A. How do spores germinate? J Appl Microbiol. 2006;101(3):526–30. 10.1111/j.1365-2672.2006.02885.x [DOI] [PubMed] [Google Scholar]
- 3.Setlow P. Spore germination. Curr Opin Microbiol. 2003;6(6):550–6. [DOI] [PubMed] [Google Scholar]
- 4.Mallozzi M, Viswanathan VK, Vedantam G. Spore-forming Bacilli and Clostridia in human disease. Future Microbiology. 2010;5(7):1109–23. 10.2217/fmb.10.60 [DOI] [PubMed] [Google Scholar]
- 5.Setlow P, Johnson EA. Spores and Their Significance. Food Microbiology: Fundamentals and Frontiers, Third Edition. 2007:35–67. [Google Scholar]
- 6.Cowan AE, Olivastro EM, Koppel DE, Loshon CA, Setlow B, Setlow P. Lipids in the inner membrane of dormant spores of Bacillus species are largely immobile. Proc Natl Acad Sci USA. 2004;101(20):7733–8. 10.1073/pnas.0306859101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koshikawa T, Beaman TC, Pankratz HS, Nakashio S, Corner TR, Gerhardt P. Resistance, germination, and permeability correlates of Bacillus megaterium spores successively divested of integument layers. J Bacteriol. 1984;159(2):624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross C, Abel-Santos E. The Ger receptor family from sporulating bacteria. Curr Issues Mol Biol. 2010;12(3):147–58. [PMC free article] [PubMed] [Google Scholar]
- 9.Vepachedu VR, Setlow P. Analysis of interactions between nutrient germinant receptors and SpoVA proteins of Bacillus subtilis spores. FEMS Microbiol Lett. 2007;274(1):42–7. 10.1111/j.1574-6968.2007.00807.x [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Jin K, Ghosh S, Devarakonda P, Carlson K, Davis A, et al. Structural and functional analysis of the GerD spore germination protein of Bacillus species. J Mol Biol. 2014;426(9):1995–2008. 10.1016/j.jmb.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie G, Lowe CR. Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J Bacteriol. 2007;189(12):4375–83. 10.1128/JB.00110-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igarashi T, Setlow P. Interaction between individual protein components of the GerA and GerB nutrient receptors that trigger germination of Bacillus subtilis spores. J Bacteriol. 2005;187(7):2513–8. 10.1128/JB.187.7.2513-2518.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popham DL, Bernhards CB. Spore Peptidoglycan In: Driks A, Eichenberger P, editors. The Bacterial Spore: From Molecules to Systems. Washington, D.C.: ASM Press; 2015. p. In Press. [Google Scholar]
- 14.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6(10):767–72. 10.1038/nmeth.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson CM, Grossman AD. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol. 2014;93(6):1284–301. 10.1111/mmi.12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koo BM, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, et al. Construction and Analysis of Two Genome-Scale Deletion Libraries for Bacillus subtilis. Cell Syst. 2017;4(3):291–305 e7. 10.1016/j.cels.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feavers IM, Foulkes J, Setlow B, Sun D, Nicholson W, Setlow P, et al. The regulation of transcription of the gerA spore germination operon of Bacillus subtilis. Molec Microbiol. 1990;4:275–82. [DOI] [PubMed] [Google Scholar]
- 18.Zuberi AR, Moir A, Feavers IM. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene. 1987;51(1):1–11. [DOI] [PubMed] [Google Scholar]
- 19.Cabrera-Martinez RM, Tovar-Rojo F, Vepachedu VR, Setlow P. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J Bacteriol. 2003;185(8):2457–64. 10.1128/JB.185.8.2457-2464.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leighton TJ, Doi RH. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971;246(10):3189–95. [PubMed] [Google Scholar]
- 21.Stewart KA, Setlow P. Numbers of individual nutrient germinant receptors and other germination proteins in spores of Bacillus subtilis. J Bacteriol. 2013;195(16):3575–82. 10.1128/JB.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson WL, Setlow P. Sporulation, germination, and outgrowth In: Harwood CR, Cutting SM, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd; 1990. p. 391–450. [Google Scholar]
- 25.Dowd MM, Orsburn B, Popham DL. Cortex peptidoglycan lytic activity in germinating Bacillus anthracis spores. J Bacteriol. 2008;190(13):4541–8. 10.1128/JB.00249-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. ACM SIGKDD Explorations Newsletter. 2009;11(1):10–8. 10.1145/1656274.1656278 [DOI] [Google Scholar]
- 28.Ghosh S, Scotland M, Setlow P. Levels of germination proteins in dormant and superdormant spores of Bacillus subtilis. J Bacteriol. 2012;194(9):2221–7. 10.1128/JB.00151-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez-Peralta A, Stewart KA, Thomas SK, Setlow B, Chen Z, Li YQ, et al. Effects of the SpoVT regulatory protein on the germination and germination protein levels of spores of Bacillus subtilis. J Bacteriol. 2012;194(13):3417–25. 10.1128/JB.00504-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez-Peralta A, Zhang P, Li YQ, Setlow P. Effects of sporulation conditions on the germination and germination protein levels of Bacillus subtilis spores. Appl Environ Microbiol. 2012;78(8):2689–97. 10.1128/AEM.07908-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Setlow B, Melly E, Setlow P. Properties of spores of Bacillus subtilis blocked at an intermediate stage in spore germination. J Bacteriol. 2001;183(16):4894–9. 10.1128/JB.183.16.4894-4899.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson DG, Chen W, Storey JD, Gresham D. Design and analysis of Bar-seq experiments. G3 (Bethesda). 2014;4(1):11–8. 10.1534/g3.113.008565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naclerio G, Baccigalupi L, Zilhao R, De Felice M, Ricca E. Bacillus subtilis spore coat assembly requires cotH gene expression. J Bacteriol. 1996;178(15):4375–80. 10.1128/jb.178.15.4375-4380.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moir A, Lafferty E, Smith DA. Genetics analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype with map location. J Gen Microbiol. 1979;111(1):165–80. 10.1099/00221287-111-1-165 [DOI] [PubMed] [Google Scholar]
- 35.Zheng L, Donovan WP, Fitz-James PC, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes & Dev. 1988;2:1047–54. [DOI] [PubMed] [Google Scholar]
- 36.Behravan J, Chirakkal H, Masson A, Moir A. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J Bacteriol. 2000;182(7):1987–94. 10.1128/jb.182.7.1987-1994.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takamatsu H, Kodama T, Nakayama T, Watabe K. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J Bacteriol. 1999;181(16):4986–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukushima T, Ishikawa S, Yamamoto H, Ogasawara N, Sekiguchi J. Transcriptional, functional and cytochemical analyses of the veg gene in Bacillus subtilis. J Biochem. 2003;133(4):475–83. 10.1093/jb/mvg062 [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Fitz-James PC, Aronson AI. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J Bacteriol. 1993;175(12):3757–66. 10.1128/jb.175.12.3757-3766.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serrano M, Zilhao R, Ricca E, Ozin AJ, Moran CP Jr., Henriques AO. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J Bacteriol. 1999;181(12):3632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuwana R, Okuda N, Takamatsu H, Watabe K. Modification of GerQ reveals a functional relationship between Tgl and YabG in the coat of Bacillus subtilis spores. J Biochem. 2006;139(5):887–901. 10.1093/jb/mvj096 [DOI] [PubMed] [Google Scholar]
- 42.Beall B, Driks A, Losick R, Moran CP Jr. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J Bacteriol. 1993;175(6):1705–16. 10.1128/jb.175.6.1705-1716.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Valdespino A, Li Y, Setlow B, Ghosh S, Pan D, Korza G, et al. Function of the SpoVAEa and SpoVAF proteins of Bacillus subtilis spores. J Bacteriol. 2014;196(11):2077–88. 10.1128/JB.01546-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Barat B, Ray WK, Helm RF, Melville SB, Popham DL. Membrane Proteomes and Ion Transporters in Bacillus anthracis and Bacillus subtilis Dormant and Germinating Spores. J Bacteriol. 2019;201(6). 10.1128/JB.00662-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng L, Abhyankar W, Ouwerling N, Dekker HL, van Veen H, van der Wel NN, et al. Bacillus subtilis Spore Inner Membrane Proteome. J Proteome Res. 2016;15(2):585–94. 10.1021/acs.jproteome.5b00976 [DOI] [PubMed] [Google Scholar]
- 46.Wetzstein M, Volker U, Dedio J, Lobau S, Zuber U, Schiesswohl M, et al. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J Bacteriol. 1992;174(10):3300–10. 10.1128/jb.174.10.3300-3310.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dambach M, Irnov I, Winkler WC. Association of RNAs with Bacillus subtilis Hfq. PLoS One. 2013;8(2):e55156 10.1371/journal.pone.0055156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, et al. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol. 2005;187(22):7655–66. 10.1128/JB.187.22.7655-7666.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puri-Taneja A, Paul S, Chen Y, Hulett FM. CcpA causes repression of the phoPR promoter through a novel transcription start site, P(A6). J Bacteriol. 2006;188(4):1266–78. 10.1128/JB.188.4.1266-1278.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaushal B, Paul S, Hulett FM. Direct regulation of Bacillus subtilis phoPR transcription by transition state regulator ScoC. J Bacteriol. 2010;192(12):3103–13. 10.1128/JB.00089-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tjalsma H, Bolhuis A, van Roosmalen ML, Wiegert T, Schumann W, Broekhuizen CP, et al. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12(15):2318–31. 10.1101/gad.12.15.2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allenby NE, Watts CA, Homuth G, Pragai Z, Wipat A, Ward AC, et al. Phosphate starvation induces the sporulation killing factor of Bacillus subtilis. J Bacteriol. 2006;188(14):5299–303. 10.1128/JB.00084-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, et al. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50(5):1683–701. [DOI] [PubMed] [Google Scholar]
- 54.Strauch MA, Bobay BG, Cavanagh J, Yao F, Wilson A, Le Breton Y. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J Bacteriol. 2007;189(21):7720–32. 10.1128/JB.01081-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, et al. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358(1):16–37. 10.1016/j.jmb.2006.01.059 [DOI] [PubMed] [Google Scholar]
- 56.DeLoughery A, Lalanne JB, Losick R, Li GW. Maturation of polycistronic mRNAs by the endoribonuclease RNase Y and its associated Y-complex in Bacillus subtilis. Proc Natl Acad Sci U S A. 2018;115(24):E5585–E94. 10.1073/pnas.1803283115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eiamphungporn W, Helmann JD. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol. 2008;67(4):830–48. 10.1111/j.1365-2958.2007.06090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersohn A, Brigulla M, Haas S, Hoheisel JD, Volker U, Hecker M. Global analysis of the general stress response of Bacillus subtilis. J Bacteriol. 2001;183(19):5617–31. 10.1128/JB.183.19.5617-5631.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285(1):473–82. 10.1074/jbc.M109.040238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo Y, Helmann JD. A sigmaD-dependent antisense transcript modulates expression of the cyclic-di-AMP hydrolase GdpP in Bacillus subtilis. Microbiology. 2012;158(Pt 11):2732–41. 10.1099/mic.0.062174-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Atluri S, Ragkousi K, Cortezzo DE, Setlow P. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J Bacteriol. 2006;188(1):28–36. 10.1128/JB.188.1.28-36.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yi X, Liu J, Faeder JR, Setlow P. Synergism between different germinant receptors in the germination of Bacillus subtilis spores. J Bacteriol. 2011;193(18):4664–71. 10.1128/JB.05343-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Setlow B, Cowan AE, Setlow P. Germination of spores of Bacillus subtilis with dodecylamine. J Appl Microbiol. 2003;95(3):637–48. [DOI] [PubMed] [Google Scholar]
- 64.Straus D, Walter W, Gross CA. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of s32. Genes & Dev. 1990;4:2202–9. [DOI] [PubMed] [Google Scholar]
- 65.Kavita K, de Mets F, Gottesman S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr Opin Microbiol. 2018;42:53–61. 10.1016/j.mib.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301(5632):510–3. 10.1126/science.1086462 [DOI] [PubMed] [Google Scholar]
- 67.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D3. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou R, Cusumano C, Sui D, Garavito RM, Kroos L. Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP. Proc Natl Acad Sci U S A. 2009;106(38):16174–9. 10.1073/pnas.0901455106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steil L, Serrano M, Henriques AO, Volker U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiol. 2005;151(Pt 2):399–420. 10.1099/mic.0.27493-0 [DOI] [PubMed] [Google Scholar]
- 70.Korza G, Camilleri E, Green J, Robinson J, Nagler K, Moeller R, et al. Analysis of the Messenger RNAs in Spores of Bacillus subtilis. J Bacteriol. 2019. 10.1128/JB.00007-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gundlach J, Mehne FM, Herzberg C, Kampf J, Valerius O, Kaever V, et al. An Essential Poison: Synthesis and Degradation of Cyclic Di-AMP in Bacillus subtilis. J Bacteriol. 2015;197(20):3265–74. 10.1128/JB.00564-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gundlach J, Rath H, Herzberg C, Mader U, Stulke J. Second Messenger Signaling in Bacillus subtilis: Accumulation of Cyclic di-AMP Inhibits Biofilm Formation. Front Microbiol. 2016;7:804 10.3389/fmicb.2016.00804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis sigma(M) in beta-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 2012;83(3):623–39. 10.1111/j.1365-2958.2011.07953.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gandara C, Alonso JC. DisA and c-di-AMP act at the intersection between DNA-damage response and stress homeostasis in exponentially growing Bacillus subtilis cells. DNA Repair (Amst). 2015;27:1–8. 10.1016/j.dnarep.2014.12.007 [DOI] [PubMed] [Google Scholar]
- 75.Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev. 2000;64(3):515–47. 10.1128/mmbr.64.3.515-547.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Allenby NE, O'Connor N, Pragai Z, Ward AC, Wipat A, Harwood CR. Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis. J Bacteriol. 2005;187(23):8063–80. 10.1128/JB.187.23.8063-8080.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antelmann H, Scharf C, Hecker M. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J Bacteriol. 2000;182(16):4478–90. 10.1128/jb.182.16.4478-4490.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tamehiro N, Okamoto-Hosoya Y, Okamoto S, Ubukata M, Hamada M, Naganawa H, et al. Bacilysocin, a novel phospholipid antibiotic produced by Bacillus subtilis 168. Antimicrob Agents Chemother. 2002;46(2):315–20. 10.1128/AAC.46.2.315-320.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guldan H, Matysik FM, Bocola M, Sterner R, Babinger P. Functional assignment of an enzyme that catalyzes the synthesis of an archaea-type ether lipid in bacteria. Angew Chem Int Ed Engl. 2011;50(35):8188–91. 10.1002/anie.201101832 [DOI] [PubMed] [Google Scholar]
- 80.Meeske AJ, Rodrigues CD, Brady J, Lim HC, Bernhardt TG, Rudner DZ. High-Throughput Genetic Screens Identify a Large and Diverse Collection of New Sporulation Genes in Bacillus subtilis. PLoS Biol. 2016;14(1):e1002341 10.1371/journal.pbio.1002341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bagyan I, Hobot J, Cutting S. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J Bacteriol. 1996;178(15):4500–7. 10.1128/jb.178.15.4500-4507.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Traag BA, Ramirez-Peralta A, Wang Erickson AF, Setlow P, Losick R. A novel RNA polymerase-binding protein controlling genes involved in spore germination in Bacillus subtilis. Mol Microbiol. 2013;89(1):113–22. 10.1111/mmi.12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Popham DL, Meador-Parton J, Costello CE, Setlow P. Spore peptidoglycan structure in a cwlD dacB double mutant of Bacillus subtilis. J Bacteriol. 1999;181(19):6205–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sekiguchi J, Akeo K, Yamamoto H, Khasanov FK, Alonso JC, Kuroda A. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which effects germination in Bacillus subtilis. J Bacteriol. 1995;177(19):5582–9. 10.1128/jb.177.19.5582-5589.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains Table A. B. subtilis strains used in this study; Table B. Long-term germination efficiency of B. subtilis mutant strains; Table C. Spore germination in response to diverse germinants following overexpression of gerA; Figure A. Germination rates of B. subtilis strains; Figure B. Release of DPA and NAM by B. subtilis strains; Figure C. Phase contrast microscopy image pixel intensities during spore germination; Figure D. Expression of 𝛔G-dependent genes in B. subtilis mutant strains; Figure E. GerAC is reduced in the spores of several B. subtilis mutant strains; Figure F. GerD is not reduced in the spores of B. subtilis germination mutants.
(PDF)
Data Availability Statement
Sequence data has been deposited to the NCBI GenBank and can be accessed via https://www.ncbi.nlm.nih.gov/sra/PRJNA544251. All other relevant data are within the paper and its Supporting Information files.