Abstract
CD8 T cells can play both a protective and pathogenic role in inflammation and autoimmune development. Recent studies have highlighted the ability of CD8 T cells to function as T follicular helper cells in the germinal center in the context of infection. However, whether this phenomenon occurs in autoimmunity and contributes to autoimmune pathogenesis is largely unexplored. In this study, we show that CD8 T cells acquire a CD4 T follicular helper profile in the absence of functional regulatory T cells in both the IL-2-deficient and scurfy mouse models. Depletion of CD8 T cells mitigates autoimmune pathogenesis in IL-2-deficent mice. CD8 T cells express the germinal center localizing chemokine receptor CXCR5, a principal T follicular helper transcription factor Bcl6, and the T follicular helper effector cytokine IL-21. CD8 T cells localize to the B cell follicle, express B cell co-stimulatory proteins, and promote B cells to differentiate and antibody isotype class-switch. These data reveal a novel contribution of autoreactive CD8 T cells to autoimmune disease, in part, through CD4 follicular-like differentiation and functionality.
Introduction
The development of autoimmunity involves both a breakdown in tolerance control mechanisms and complex interactions between immune cells. Some of these cells promote disease while others act to block this dysregulation. As disease progresses, immune activation is amplified, self-perpetuating the lymphoproliferation, inflammation and self-destruction associated with autoimmunity. The process initiated by inflammatory and antigen signaling promotes T cell proliferation, differentiation, and acquisition of T cell effector functions. Autoimmunity in the IL-2-deficient (KO) mouse model results from reduced T regulatory (Treg) frequency and functionality (1–3), promoting dysregulation of the T effector response. CD4 T helper type 1 (Th1) cells subsequently promote the production of anti-RBC IgG antibodies and bone marrow failure dependent on IFNγ secretion (4–6). While there is a clearly established role for CD4 T helper cells in promoting antibody-mediated disease, the importance of CD8 T cells in these diseases has been less explored.
B cell responses to self-antigen during autoimmune disease are induced and enhanced by germinal center (GC) reactions in the peripheral lymphoid organs (7, 8). Within the GC, activated, antigen-specific B cells undergo clonal expansion, B cell receptor somatic mutation, affinity maturation and antibody class-switching, and differentiate into memory and long-lived plasma cells. GC reactions begin at the border of the B cell follicle and the T cell zone, where CD4 T follicular helper (Tfh) cells interact with B cells (9). CXCR5 upregulation and CCR7 downregulation facilitates migration of CD4 Tfh and activated B cells into the follicle. Engagement of several interactions between the activated CD4 Tfh and B cells (including ICOS-ICOSL and CD40-CD40L) ensures optimal GC reactions and CD4 Tfh cell development by promoting the transcription factor, Bcl6 (10). CD4 Tfh cells are required for the production of high avidity, class-switched antibodies (11).
Abnormal activation of CD4 Tfh cells, or loss of regulation, can promote antibody-mediated autoimmune disease (12–14). Follicular CD4 Tregs are essential inhibitors of GC interactions by mediating T cell help to B cells (7). In the absence of CD4 follicular Tregs, CD4 Tfh expansion results in autoantibody generation and autoimmune disease (13, 15). Similarly, CD8 Tregs also control self-reactive cells and their elimination exacerbates autoimmune disease (16, 17). Like CD4 helper cells, CD8 T cells differentiate into effector subsets based upon their transcription factor expression and cytokine production, and these cytokines may amplify CD4 T helper cell responses or act through mechanisms unique to CD8 T effectors (17). However, regulation and function of distinct CD8 T cell subsets are less clearly delineated as compared to CD4 helper cells.
CD8 T cell effectors located within the GC have recently been described (18–22). In rheumatoid arthritis synovial ectopic follicles, CD8 T cells make up the majority of the infiltrating T cells, and express CD40L, which is important in B cell GC reactions (19). Further, the CD8 T cells are required for the formation and maintenance of ectopic GCs (18). CD8 T cells expressing CXCR5 also develop in chronic viral infection and under inflammatory conditions (20, 21, 23–25). CXCR5+ CD8 T cells are localized in the B cell follicle in human tonsil, and these cells support B cell survival in ex vivo culture (26). CXCR5+ CD8 memory T cells within the GC control viral load in LCMV and SIV infections and express many of the genes associated with CD4 Tfh differentiation and function (20, 23). Together, these data suggest that under specific disease conditions, CD8 T cells may acquire unique functionality within the GC. Whether CD8 T cells function in GC reactions in autoimmune disease is largely unexplored.
Materials and Methods
Mice, Immunizations and ab depletions
BALB/c IL-2-KO mice, WT and IL-2-heterozygous (HET) littermate controls (IL-2-HET and WT; referred to as WT) were used. IL-2-KO autoimmune disease is not gender specific; only age is used to determine data inclusion as specified in the figure legend. BALB/c hemizygous male Foxp3sf/Y (scurfy) mice and heterozygous female FoxPsf/+ (scurfy-HET) mice were purchased from JAX (27). Both gender and age are used to determine data inclusion in scurfy disease; our breeding setup is restricted to hemizygous male mice, age-matched female scurfy-HET littermates were used as controls where indicated. For immunization mice were treated by i.p. injection with 100–200 μg keyhole limpet hemacyanin (KLH) in Complete Freund’s Adjuvant (CFA) at −15 to −26 days, followed by a second i.p. injection at −5 days as previously described (7, 8). CD4 or CD8 depletions were performed by i.p. injection of 20 μg anti-CD4 (GK1.5) or anti-CD8 (2.43) ab per gram weight three times per week, from day 8 to 16. IL-2 depletions were performed by i.p. injection of 20 μg anti-IL-2 (JES6-1A12) ab per gram weight three times per week between days 7 to 15. Antibodies were purchased from the UCSF Monoclonal Antibody Core or Bio X Cell. All mice were bred and maintained in our specific pathogen-free facility in accordance with the guidelines of the Department of Animal Research Services at UC Merced. The UC Merced Institutional Animal Care and Use Committee approved all animal procedures.
Complete blood counts
Cardiac punctures or eye bleeds were performed immediately following cervical dislocation, and blood collected in heparinized tubes (28). Complete blood counts were evaluated within 24 hours on a Hemavet 950 Veterinary Hematology System.
Microscopy and immunofluorescence
Spleens were embedded in Optimal Cutting Temperature (O.C.T.) compound (Fisher Scientific) and snap frozen in the vapor phase of liquid nitrogen. 15 μm sections were generated then fixed with 100% ice cold acetone followed by blocking with PBS/5%BSA. Sections were stained overnight with anti-CD8α-FITC (53-6.7; eBioscience), anti-IgD-PE-Dazzle594 (11-26c.2a, Biolegend), and anti-GL7-eFluor450 (GL-7; eBioscience) and single plane confocal imaged on a Zeiss LSM 880 confocal system with a 10× objective. Confocal images were processed in ImageJ to adjust for contrast and pseudocolored in red, green, and blue. GL-7+ GCs were traced in ImageJ using the freehand selection tool and the area was determined. CD8+ cells within the GC were marked using the multi-point selection tool when a dark center (nucleus) surrounded by CD8 surface staining could be identified. GC CD8 T cells within GC area were quantified manually.
Flow cytometry and cell sorting
Splenocytes and lymphocytes were stained with fluorochrome-conjugated antibodies (eBioscience unless otherwise noted) following incubation with Fc-block (anti-CD16/CD32; 2.4G2). Cell viability was determined by DAPI, Fixable Viability Dye eFluor780 or eFluor506 (eBioscience). For CD8 Tfc identification, cells were stained with anti-CXCR5-biotin (SPRCL5) then stained with Streptavidin-BUV395 (BD Bioscience), anti-CD4 (RM4-5), anti-CD8α (53-6.7), anti-CD278 (ICOS; C398.4A), anti-GL-7 (GL-7), anti-CD279 (PD-1; J43, Biolegend), anti-CD11c (N418), anti-CD11b (M1/70), anti-Ly-6G (Gr-1; RB6-8C5) and anti-CD45R (B220; RA3-6B2) as previously defined (29). For intracellular proteins, cells were stained as above, fixed using the FoxP3/Transcription Factor Fixation/Permeabilization Kit (eBioscience), and stained with anti-Bcl6 (BCL-DWN), and SA-BUV395 (BD Bioscience) or anti-IL-2-PE (JES6-5H4; Biolegend). Flow cytometry was performed on a Becton Dickinson LSR-II and data analyzed using FCS Express with Diva Version 4.07.0005 (DeNovo Software) or FlowJo.Version 10.1 (FlowJo).
Prior to cell sorting, pooled splenocytes and lymphocytes were depleted of non-T cells using EasySep Mouse PE Selection Kit according to the manufacturer’s instructions (Stem Cell Technologies) to remove B220-PE+, CD11c-PE+, CD11b-PE+, and Gr-1-PE+ cells. CD4 Tfh (CXCR5+PD-1hi) and CD8 Tfc (CXCR5+PD-1hi) were sorted from IL-2-KO mice, CD4 Tfh (CXCR5+PD-1hi) were sorted from KLH-immunized mice, and bulk CD4 and CD8 T cells were sorted from WT or IL-2-KO mice as indicated. B cells (CD19+TCRβ−CD11c−CD11b−Gr-1−) were sorted from pooled WT spleens. All sorts were performed with >90% purity on the Aria II cell sorter (BD Biosciences).
RBC ab detection
Serum RBC ab levels were detected as previously described (30). Freshly isolated RBCs were washed three times in PBS, and resuspended to 1% RBCs. 10 μl of 1% RBCs were incubated with anti-mouse IgM-FITC (1:150; on ice) or anti-mouse IgG-FITC (1:50; at 37 °C; Jackson ImmunoResearch). The %RBCs bound by ab was determined by flow cytometry.
T cell stimulations
Harvested cells were stimulated at 37°C with 50 ng/ml PMA and 500 ng/ml ionomycin for 5 h with brefeldin A or monensin added during the final 4 h. IL-2 cytokine production was determined by intracellular flow cytometry. Anti-CD40L (MR1) was added directly to cells during the stimulation as previously described (31). Cells were stained post-stimulation for CD4 Tfh and CD8 Tfc markers (CXCR5+PD-1hi). IL-21 cytokine production was determined in CD4 Tfh and CD8 Tfc as previously described with recombinant mouse IL-21R subunit Fc chimera (R&D Systems) and PE-conjugated F(ab′)2 goat anti-human IgG (Jackson ImmunoResearch) at 4°C (32).
In vitro T cell and B cell culture assays
T and B cell stimulation was performed as previously described (22). Indicated T cell populations were plated at 5×104 cells/well and activated with 5 μg/mL soluble anti-CD3ε (145-2C11, Biolegend) and 1 μg/mL anti-CD28 (37.51, Biolegend) for 72 h. Supernatant from activated T cells were plated with 5×104 sorted WT B cells/well and with 1 μg/mL anti-CD40 (IC10) and 5 μg/mL F(ab′)2 goat anti-mouse IgMμ (Jackson ImmunoResearch) for 6 d. B cell supernatant was analysis for ab production by ELISA.
T cell adoptive transfer assays
IL-2-KO T cells were adoptively transferred as previously described (4). 1×106 CD4 T cells, 2×106 CD8 T cells, or a combination of each were transferred into TCRα-KO mice via eye injection. Two days post cell transfer mice were immunized i.p with 200 μg KLH in CFA. Seven days post cell transfer mice were re-immunized i.p. with 100 μg 4-Hydroxy-3-nitrophenylacetyl (NP) conjugated KLH in IFA.
Ab ELISA
Total IgG was determined by ELISA as described (28). Serum and culture supernatant samples were prepared in PBS/1%BSA. Serum from depletion experiments was prepared at 1:50,000 dilution, a standard curve of purified mouse IgG (Southern Biotech), and stimulated B cell supernatants prepared at 1:50 dilution, and serum from adoptive transfer experiments was diluted to 1:10,000 dilution for IgG2a and IgG2b or 1:100,000 dilution for IgG1. Ab were detected with HRP-conjugated goat anti-mouse IgG, IgG1, IgG2a, IgG2b, or IgG3 (Southern Biotech) then developed with, TMB peroxidase substrate (Vector Laboratories) following manufacturers’ instructions. Plates were stopped with 1 N sulfuric acid and read on a Vector3 1420 Multilabel Counter plate reader (PerkinElmer) at 450 nm. When applicable, a standard curve of purified mouse IgG (Southern Biotech) was used to interpolate IgG concentrations from a sigmoidal standard protein curve.
RNA isolation and analysis of RNA next generation sequencing
CD8 cells were sorted from 12 day old IL-2-KO and WT mice to ≥ 85% purity. Samples were quick frozen and shipped to Expression Analysis Inc. for total RNA isolation using Illumina TrueSeq Stranded Total RNA Sample Preparation Kit. Eight samples were sequenced, four biological replicates each for IL-2-KO and WT mice, producing 2X50 paired-end reads using the Illumina HiSeq 2500 platform. Raw reads were provided by Expression Analysis and were used for further analyses. Adapter removal and quality trimming, at the Q20 level, were performed using Atropos v. 1.1.17 (33) with Python v3.6.2. Read pairs were removed if either read was < 20 bp trimming. Rsubread v1.28.1 (34) was used to perform read alignment reporting up to 10 equally likely mapping locations. Read pairs that could not be aligned together were aligned individually. The genome used for alignment was C57BL/6J of GRCm38/mm10 (GCF_000001635.20). Read summarization was performed on the gene level using featureCounts (35) using annotations from a modified version of the annotations for GRCm38/mm10 containing only protein coding genes. Multi-mapping reads were treated as fractional counts when mapping to several genes, reads mapping across more than one gene and read pairs where ends mapped to different chromosomes were discarded. Genes that had <1 count/million in 3–4 samples were discarded. Remaining gene counts were normalized using TMM (36). Differential expression analysis was performed using limma v3.34.9 (37) with voom transformed read counts (38). Genes were considered differentially expressed if their p-value was < 0.05 after the false discovery rate was controlled (39). Read mapping, summarization, and differential expression analysis were performed using R v3.4.3. Differentially expressed genes were annotated with their biological process Gene Ontology group using Panther 13.1 (40) using the GO Ontology database released on 2018-02-02. Sequence data was uploaded to NCBI GEO accession number: GSE112540; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112540.
Real Time PCR (RT-PCR)
Total RNA was isolated from cells using Qiagen RNeasy (Qiagen) and cDNA synthesis were conducted according to the manufacturer’s instructions with Superscript III First-Strand Synthesis SuperMix for RT-PCR (Invitrogen). RT-PCR analysis was conducted in duplicates using Mx3000P (Stratagene) with Fast SYBR Green RT-PCR master mix (BIO-Rad). Averages of the collected data were normalized to β-actin or HPRT. Relative expressions (ΔΔCt) were calculated to the indicated cell population.
Statistics
GraphPad (Prism) was used to for statistics. Differences between two experimental groups were determined by unpaired Student’s t test unless otherwise noted. Mantel-Cox Logrank test was used to analyze Kaplan-Meier survival curves. Protein concentrations from a standard protein curve were interpolated using a sigmoidal four parameter logistic standard curve with × as log(concentration). Statistically significant differences in adoptive transfer and supernatant culture experiments were determined by one-way ANOVA with Bonferroni correction.
Results
Depletion of either CD4 or CD8 T cells reduces anemia and anti-RBC antibodies, and prolongs survival
We have previously demonstrated that early death in IL-2-KO mice is driven in part by autoimmune hemolytic anemia that requires IFNγ and is CD4 Th1-mediated (5, 6). We found that CD8 T cell numbers were increased on average by 3-fold in the LNs and spleen of IL-2-KO mice (Fig. S1A). Both splenic and LN CD8 T cells expressed decreased CD62L and elevated CD44 and CD69, indicating an activated state (Fig. S1B and S1C). CD8 T cells have been shown to promote and inhibit autoimmunity in multiple models due to both regulatory and effector responses (4, 8, 19, 41). As the population of IL-2-KO CD8 T cells were activated and expanded, we next determined how and to what extent CD8 T cells contributed to autoimmunity in IL-2-KO mice.
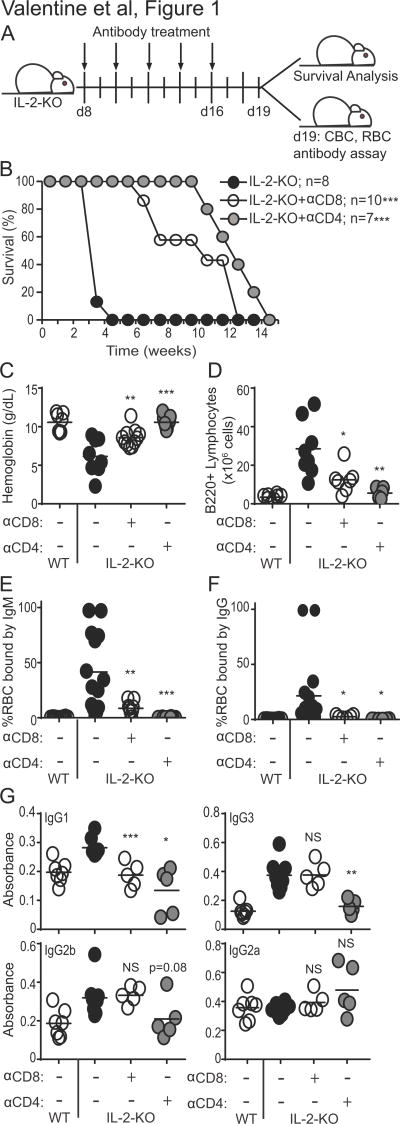
To evaluate the contribution of CD8 T cells to autoimmune disease, we eliminated CD8 T cells or CD4 T cells as a control prior to disease onset by treating with anti-CD4 or anti-CD8 depleting antibodies (Fig. 1A). As expected, depletion of CD4 T cells significantly delayed disease onset with a median survival of 14 weeks, as compared to 19–25 day survival for PBS-treated IL-2-KO mice (Fig. 1B). Surprisingly, depletion of CD8 T cells also significantly prolonged survival, with a median survival of 12 weeks. In concordance with augmented survival, IL-2-KO mice with depleted CD4 or CD8 T cells had increased hemoglobin levels relative to PBS-treated IL-2-KO mice (Fig. 1C). Both T cell depletions also significantly reduced B cell numbers (Fig. 1D) and frequency of IgM and IgG autoantibodies bound to RBCs (Fig. 1E–F) relative to PBS-treated IL-2-KO mice. In the absence of CD8 T cells serum IgG1 was significantly reduced in comparison to PBS-treated IL-2-KO mice (Fig. 1G). While these data confirm that CD4 T cells play a critical role in autoimmune progression, they also reveal the contribution of CD8 T cells to the rapid autoimmunity that occurs in IL-2-KO mice. Furthermore, these data demonstrate that CD8 T cells facilitate enhanced B cell expansion and antibody production in IL-2-KO mice, as these outcomes are significantly reduced when CD8 T cells are eliminated.
Fig. 1. Depletion of IL-2-KO CD4 or CD8 T cells prolongs survival and delays antibody production.
(A) Schematic describing the antibody depletion. Peripheral blood and LN cells were isolated at 18–21 days of age. (B) Kaplan-Meier survival plots. Statistics were performed to test differences relative to untreated IL-2-KO mice. (C) Hemoglobin levels were measured from peripheral blood by CBC at day 19 of age. (D) Total LN B220+ B cell numbers were determined by flow cytometry and cell counting. (E, F) RBCs were stained with anti-mouse IgM-FITC (E) or IgG-FITC (F) and analyzed by flow cytometry to detect the percentage of RBC bound by antibodies. (G) Serum IgG1, IgG3, IgG2b and IgG2a levels were determined by ELISA. (C–G) Each symbol indicates an individual animal. Statistics tests were performed relative to IL-2-KO and unpaired Student’s t-test with a Welch correction; * p<0.05; ** p<0.01; *** p<0.001.
Transcriptional profiling of CD8 T cells during early autoimmunity
To determine mechanistically how CD8 T cells contribute to antibody-mediated autoimmune development in the absence of functional Tregs, we examined early gene dysregulation in IL-2-KO CD8 T cells. We performed RNA sequencing of CD8 T cells sorted from 12 day old mice, as this is the earliest time point at which IL-2-KO CD8 T cells have been shown to be activated (42). Bulk CD8 T cells were sorted from the peripheral LNs of four sets of pooled IL-2-KO and littermate control WT mice. Differential expression analysis identified 2226 genes (1290 upregulated; 936 downregulated) that showed significant (p<0.05) differences in expression in IL-2-KO CD8 T cells relative to WT CD8 T cells (Table S1). Differentially expressed genes were grouped into several biological processes Gene Ontology categories including metabolic processes, cellular processes and biological regulation (Fig. 2A). Several genes upregulated in IL-2-KO CD8 T cells were genes involved in cytolytic function (granzymes, tbx21, fasl) but we also identified differential expression of costimulatory molecules and follicular helper-associated genes such as icos, cd28, il21 and bcl6 (Fig. 2B). Further evaluation of RNA sequencing data confirmed the profiles of cytolytic gene expression in individual IL-2-KO mice (Fig. 2C). Based on the observed reduction in B cell numbers and antibody production in the absence of CD8 T cells (Fig. 1), we focused our analysis on the expression of costimulatory molecules and genes involved in B cells. This analysis revealed a profile of gene expression in CD8 T cells comparable to that described in CD4 Tfh cells (Fig. 2D). IL-2-KO CD8 T cells expressed elevated cxcr5, sh2d1a (SAP), icos, bcl6, il21 and several other genes that define CD4 Tfh cells, as further confirmed by RT-PCR (Fig. 2E). Thus, during early autoimmune initiation, CD8 T cells acquire a gene expression profile associated with varying functional roles, including a CD4 Tfh helper-like role.
Fig. 2. Cytolytic and follicular helper profile of IL-2-KO CD8 T cells.
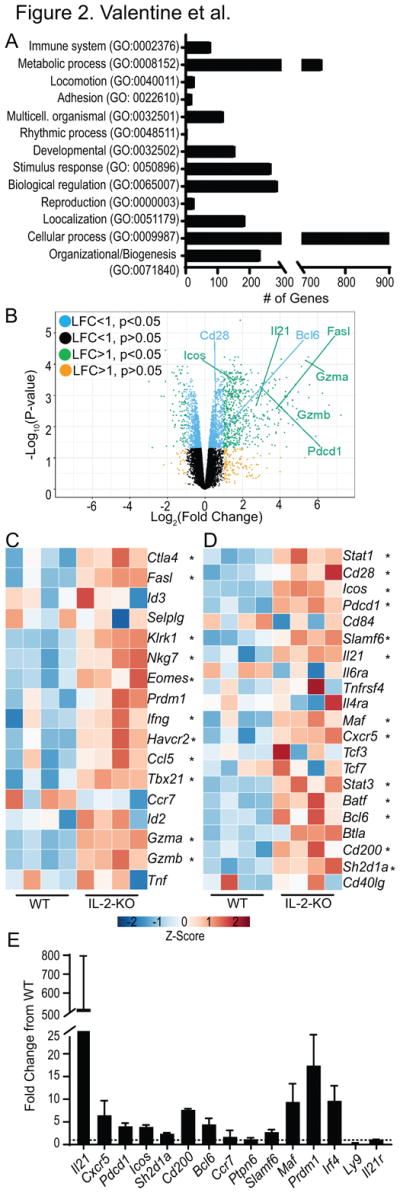
RNA sequencing of four independent WT and IL-2-KO CD8 T cell samples pooled from LN of 12 day old mice. (A) Gene ontology analysis of all 2226 differentially expressed genes in IL-2-KO CD8 T cells relative to WT. (B) Volcano plots displaying log2 fold change gene expression of IL-2-KO relative to WT CD8 T cells versus log10 p-value. Select differentially expressed genes are labeled in the plot. Data is organized by color to indicate both log fold change (LFC) and p-value. (C, D) Heat maps showing CD8 T cell expression data. Color indicates gene expression by Z-score, * indicates IL-2-KO gene expression with statistical significance relative to WT. Differential expression of select cytolytic-associated genes (C), and CD4 T follicular helper-associated genes (D). (E) mRNA expression of select genes in two independent experiments from 12 day old IL-2-KO CD8 T cells relative to WT CD8 T cells. Dashed line indicates a fold change of 1, error bars indicate standard deviation.
CD8 T follicular cells (Tfc) develop during systemic autoimmune disease
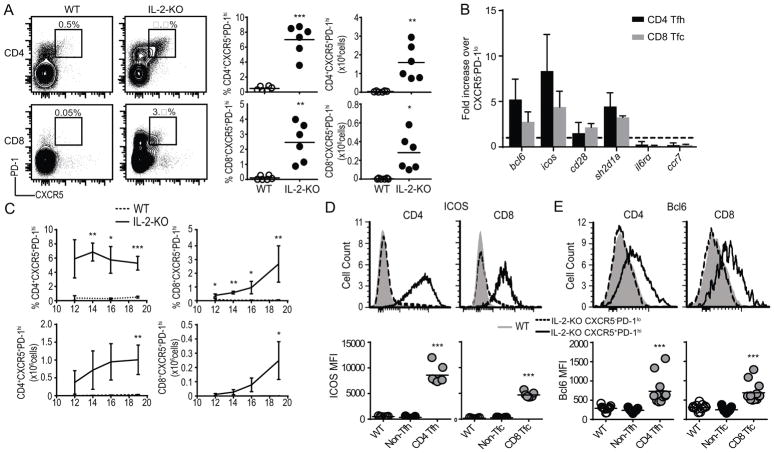
We next evaluated protein expression of CD4 Tfh cell associated genes in IL-2-KO CD8 T cells. In naïve mice, a small population (0.1–0.5%) of CXCR5+PD-1hi CD4 Tfh cells has been described (43, 44). We used this population and percentage range, in addition to fluorescence-minus-one controls, to confirm our gating strategy for CD8 T cells. Using these stringent conditions, a very small population of ≤0.2% CD8 T cells expressed CXCR5 and PD-1 in naïve WT mice. In contrast, the same markers were significantly expanded among both CD4 and CD8 T cells in IL-2-KO LN (7.0±1.8% of CD4 T cells, and 2.5±1.2% of CD8 T cells; Fig. 3A). Similarly, CXCR5+PD-1hi CD4 and CXCR5+PD-1hi CD8 T cells were significantly increased in frequency and total number in the spleens of IL-2-KO mice relative to naïve WT (Fig. S2A). To confirm select gene expression patterns identified in the bulk CD8 T cell RNA sequencing, we performed RT-PCR analysis of IL-2-KO CD4 Tfh and CD8 Tfc cells relative to IL-2-KO CD4 non-Tfh cells and CD8 non-Tfc cells respectively. CD8 Tfc cell mRNA levels of bcl6, icos, cd28, sh2d1a, il6ra, and ccr7 were comparable to CD4 Tfh cells (Fig. 3B). Thus, based on their surface phenotype and gene expression profile, we defined the CXCR5+PD-1hi CD8 T cells as CD8 T follicular (Tfc) cells.
Fig. 3. CD8 T cells express markers of follicular helpers.
(A) Flow cytometric analysis of CXCR5 and PD-1 expression on CD4 or CD8 T cells from 18–21 day old IL-2-KO or WT LN gated on B220−CD11c−CD11b−GR-1−. Representative flow plots, frequency, and total number of CXCR5+PD-1hi CD4 and CD8 T cells are shown. (B) RT-PCR comparing relative gene expression in IL-2-KO CD4 Tfh or CD8 Tfc cells (CXCR5+PD-1hi) to IL-2-KO CD4 CXCR5−PD-1lo (non-Tfh) or CD8 CXCR5−PD-1lo (non-Tfc) cells respectively. Dashed line indicated fold change of 1, error bars indicate standard deviation. (C) Percent and total number of CD4 Tfh and CD8 Tfc cells in IL-2-KO and WT mice from 12–20 days of age. (D, E) Representative flow plots and mean fluorescence intensity (MFI) quantification of surface expression of ICOS (D) and Bcl6 (E) in WT naïve bulk CD4 and CD8 T cells, IL-2-KO CD4 non-Tfh or CD8 non-Tfc, and IL-2-KO CD4 Tfh and CD8 Tfc cells. (A, D–E) Each symbol indicates an individual animal. (A, C–E) Data is representative of 3–6 independent experiments. (B) Data is representative of 2 independent experiments. Statistics: unpaired Student’s t-test relative to WT with a Welch correction; * p<0.05; ** p<0.01, *** p≤0.001.
A detectable population of CD8 Tfc cells was identified in IL-2-KO mice at day 12 by flow cytometry, and continued to expand in both frequency and total number over time (Fig. 3C). Although, CXCR5+PD-1hi T cells comprise only a small fraction of the expanded CD4 and CD8 T cell population observed in IL-2-KO mice (Fig. S2B). We next evaluated CD8 Tfc cells for the expression of other proteins known to be involved in B cell interactions within the follicle. ICOS is highly expressed during CD4 Tfh cell differentiation and promotes the expression of Bcl6, a master regulator of CD4 Tfh fate that promotes CD4 Tfh:B cell interactions (45, 46). IL-2-KO CD8 Tfc cells expressed increased ICOS and Bcl6, similar to IL-2-KO CD4 Tfh cells, in comparison to both naïve WT CD8 T cells and IL-2-KO CD8 non-Tfc cells (Fig. 3D–E, Fig. S2C and S2D). Together, mRNA and surface expression of effector proteins that typically describe CD4 Tfh cells confirms the identity of CD8 Tfc cells in systemic autoimmunity.
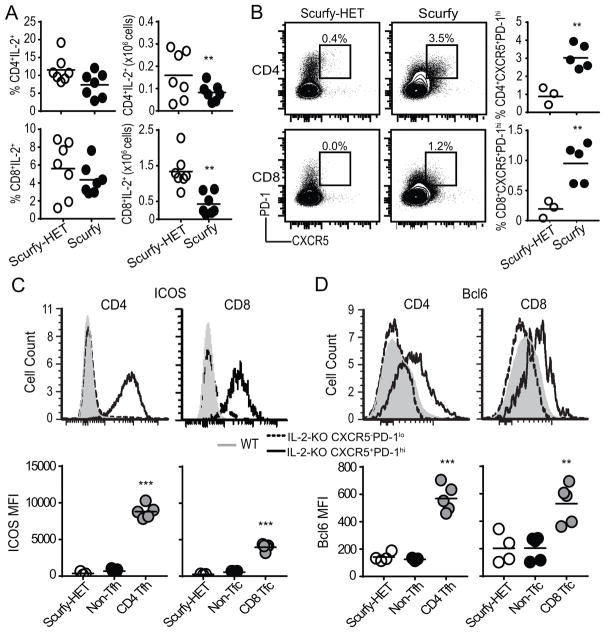
Next, we validated that CD8 Tfc cells develop under IL-2-sufficient autoimmune conditions (47, 48). Scurfy mice lack functional Treg development resulting in early systemic autoimmune disease (48). We first confirmed that despite a reduction in the total number of IL-2 producing CD4 and CD8 T cells in the scurfy mice, the frequency of IL-2 producing cells was comparable to WT mice (Fig. 4A) (47). We identified a significantly expanded population of CXCR5+PD-1hi CD4 Tfh cells (3.0±0.6%) and CXCR5+PD-1hi CD8 Tfc cells (1.0±0.3%) in scurfy LN compared to scurfy-HET LNs (Fig. 4B). Depleting IL-2 in scurfy mice using a neutralizing antibody or by genetic cross to the IL-2-KO background resulted in a higher percentage of CD4 Tfh and CD8 Tfc cells as compared to scurfy mutants but not IL-2-KO mice (data not shown). Similar to IL-2-KO mice, CD8 Tfc cells in scurfy mice expressed ICOS and Bcl6 (Fig. 4C–D). Together, these data demonstrate even under IL-2 sufficient conditions, CD8 Tfc cells can be identified during autoimmunity induced by the absence of functional Tregs. However, differences in CD8 Tfc cell frequency between the IL-2-KO and scurfy mice suggest that IL-2 may play a role in regulating the expansion of this distinct CD8 T cell population.
Fig. 4. Autoimmune Scurfy mice develop CD8 Tfc cells.
Flow cytometric analysis of Scurfy and Scurfy-HET littermate control LNs, (A) Quantification of IL-2 producing CD4 and CD8 T cell frequency and total cell number after PMA and ionomycin stimulation. (B) CXCR5 and PD-1 expression on CD4 or CD8 T cells from 18–19 day old Scurfy or scurfy-HET gated on live B220−CD11c−CD11b−GR-1−. Representative flow plots, frequency, and total number of CXCR5+PD-1hi CD4 and CD8 T cells are shown. (C, D) Representative flow plots and quantification of MFI of expression of ICOS (C) and Bcl6 (D) in Scurfy-HET naïve bulk CD4 and CD8 T cells, Scurfy CD4 non-Tfh or CD8 non-Tfc, and Scurfy CD4 Tfh and CD8 Tfc (CXCR5+PD-1hi) following stimulation with PMA and ionomycin. Each symbol indicates an individual animal. Data is representative of 3–6 independent experiments. Statistics: unpaired Student’s t-test relative to scurfy-HET (A–B, D) with a Welch correction in (C): * p<0.05; *** p≤0.001.
GC localization of CD8 T cells
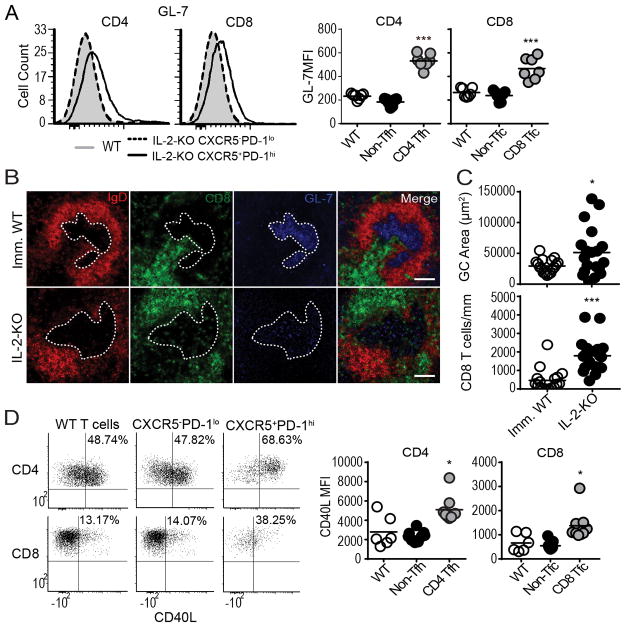
CXCR5 expression by B cells and CD4 Tfh cells allows co-localization of these cells into the follicle, providing a site for productive T: B cell interactions. During chronic viral infection, CXCR5+CD8 T cells have been reported both within the B cell follicle, and primarily excluded from the follicle (20, 21, 23). CXCR5+CD8 T cells localize to the follicle in ectopic GCs, but not in the spleen and LN during arthritis and influenza infection (18, 22). We investigated whether CD8 T cells enter the B cell follicle during systemic autoimmunity. As IL-2-KO mice develop abnormal GC structure during late stage disease (42, 49), we selected early disease-stage IL-2-KO mice with normal, albeit large, splenic gross morphology. To determine if CD8 Tfc cells were capable of GC localization we evaluated GL-7 expression, a known GC specific marker (50). Both IL-2-KO CD4 Tfh and CD8 Tfc cells express elevated levels of GL-7 (Fig. 5A). To examine CD8 T cell localization within IL-2-KO GCs, we compared immunized WT spleens with IL-2-KO spleens. GCs were defined as GL-7+ GC B cells within IgD+ B cell follicles (Fig 5B). IL-2-KO spleens had significantly larger GC areas compared to immunized WT spleens. Nonetheless, 3.9-fold more CD8 T cells were present within a comparable area of IL-2-KO GC relative to those observed in immunized WT GCs (Fig. 5C). To determine if CD8 Tfc cells are capable of providing B cell help we tested for the expression of CD154 (CD40L), as CD40:CD40L interactions are known to be crucial for B cell activation and antibody class-switching in normal and autoimmune settings (51, 52). IL-2-KO CD8 Tfc cells were found to express CD40L upon stimulation (Fig. 5D). These results indicate that IL-2-KO CD8 T cells are capable of co-expressing B cell zone specific markers (CXCR5 and GL-7), localizing to the GC, and expressing helper proteins which may promote CD8 Tfc and B cell interactions that influence autoimmune disease.
Fig. 5. CD8 T cells localize to the GC during autoimmune disease.
(A) Representative flow plots and MFI quantification of GL-7 expression IL-2-KO and WT splenocytes in the indicated populations. (B) Immunoflourescence staining of B cells (red, IgD), GCs (blue, GL-7), and CD8 (green, CD8α) of spleens from 18–21 day old IL-2-KO and KLH-immunized WT (Imm. WT) mice. White dotted lines indicate the GC outline and white measurement bar indicated 100 μm. (C) Quantification of GC area (μm2) and CD8 T cell (per mm2). (D) Representative flow plots and MFI quantification of CD40L expression in IL-2-KO and WT CD4 and CD8 T cells stimulated with PMA and ionomycin. (A, C–D) Each symbol indicates an individual animal. (A) Data from 4 independent experiments. (B–C) Data are representative of 5 spleens and 19 GCs from IL-2-KO mice or 4 spleens and 17 GCs in KLH imm. WT mice. (D) Data are representative of three independent experiments. Statistics: unpaired Student’s t-test relative to WT (A) with a Welch correction in (C–D); * p<0.05; *** p≤0.001.
CD8 Tfc cells promote B cell antibody class-switch
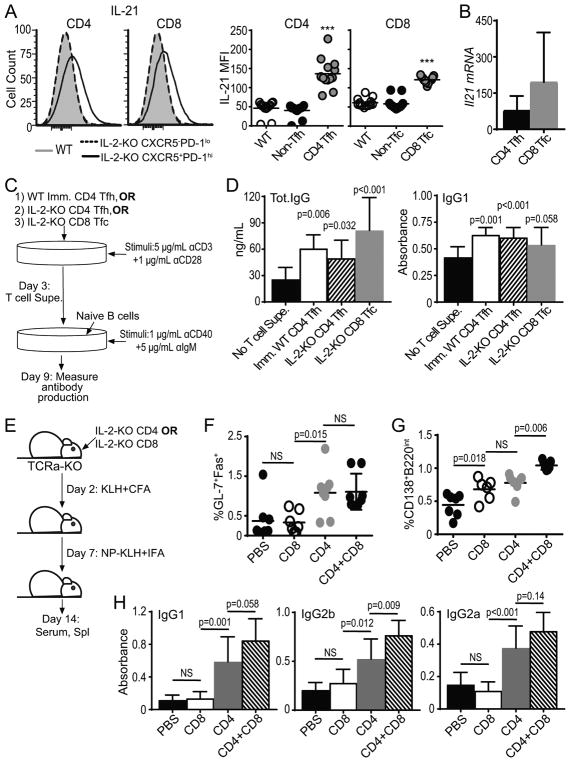
To determine the influence of IL-2-KO CD8 Tfc cells on B cell activities, we examined CD8 Tfc cell production of the cytokine IL-21, an effector cytokine produced by CD4 Tfh cells (10). Stimulation of CD8 Tfc cells yielded similar levels of IL-21 (average MFI of 120), as compared to IL-2-KO CD4 Tfh cells (average MFI of 136) and significantly more than naïve WT cells or IL-2-KO non-Tfc cells (Fig. 6A). Consistent with higher IL-21 production, both CD4 Tfh cells and CD8 Tfc cells expressed higher levels of IL-21 mRNA as compared to their CXCR5−PD-1lo counterparts (Fig. 6B). We next evaluated whether CD8 Tfc cells secreted proteins capable of influencing B cell antibody class-switching using in vitro culture assays. WT B cells were cultured with and without αIgM and αCD40 plus supernatant from activated T cells (Fig. 6C). Supernatant from IL-2-KO CD8 Tfc cells induced significant amounts of total IgG and IgG1 production by B cells, comparable to levels produced by B cells with immunized WT CD4 Tfh and IL-2-KO CD4 Tfh supernatants (Fig. 6D). These data confirm that IL-2-KO CD8 Tfc cells are capable of inducing antibody class-switch recombination, independent of CD4 Tfh cells.
Fig. 6. IL-2-KO CD8 Tfc cells promote antibody class-switch by B cells.
(A) IL-2-KO and WT lymphocytes were stimulated with PMA and ionomycin. CD8 and CD4 T cells were gated on the indicated populations and analyzed for IL-21 expression by flow cytometry. (B) IL-21 mRNA expression in sorted IL-2-KO CD4 Tfh or CD8 Tfc cells relative to IL-2-KO CD4 non-Tfh or CD8 non-Tfc respectively. (C) Schematic describing the assay for T cell stimulation and B cell antibody induction. B cell supernatant was analyzed for total IgG and IgG1 by ELISA. (D) Total IgG concentration or IgG1 levels of stimulated B cells with and without stimulated IL-2-KO CD4 Tfh, IL-2-KO CD8 Tfc, or Imm. WT CD4 Tfh supernatants were determined by ELISA. (E) Schematic describing IL-2-KO T cell transfer and B cell induction. Sorted IL-2-KO CD4 or CD8 T cells were adoptively transferred to TCRα-KO mic, immunized with KLH in CFA and reimmunized with NP-KLH in IFA. (F) B220+GL-7+Fas+ GC B cells frequency and (G) B220intCD138+ plasma cell frequency from TCRα-KO recipient spleens. (H) IgG1, IgG2b and IgG2a levels determined by ELISA from TCRα-KO recipient serum. (A, F, G) Each symbol indicates an individual animal. Data representative of in 4–6 independent experiments. Statistics: unpaired Student’s t-test relative to WT with a Welsh correction (A) and ordinary one-way ANOVA with select comparisons and a Bonferroni correction (D, F–H); NS= not significant, * p<0.05; ** p<0.01, *** p≤0.001.
To next evaluate the impact of CD8 T cells on B cell responses in vivo, we adoptively transferred IL-2-KO CD4 T cells, CD8 T cells or a combination of both into TCRα-KO mice that where then immunized with KLH and reimmunized with NP-KLH (Fig. 6E). Fourteen days post transfer Fas+GL-7+ GC B cell expansion was measured relative to PBS-treated TCRα-KO mice. Transferred IL-2-KO CD4 T cells alone produced significant GC B cell expansion, while transfer of IL-2-KO CD8 T cells alone did not. A combination of IL-2-KO CD4 and CD8 T cells yielded similar GC B cell frequency as compared to IL-2-KO CD4 T cells alone (Fig 6F). IL-2-KO CD4 and CD8 T cells, when transferred independently induced a similar plasma cell expansion. Transfer of IL-2-KO CD4 and CD8 T cells together induced significantly more plasma cells than either population alone (Fig. 6G). Interestingly, when transferred alone, CD8 T cells did not induce antibody class-switching. However, when transferred with CD4 T cells, CD8 T cells promoted an increased in IgG1 and significantly increased IgG2b as compared to the transfer of CD4 T cells alone (Fig. 6H). Together these data demonstrate that CD8 T cells act synergistically with CD4 T cells to enhance B cells differentiation and specific class-switching. CD8 Tfc cells therefore have the potential to provide a helper-like interaction within the GC to facilitate B cell antibody production during systemic autoimmune disease.
Discussion
In this study, we provide the first evidence that a new class of CD8 T follicular cells develop during autoimmune disease, in the absence of functional Tregs. In the setting of inflammation and autoimmunity, CD8 Tfc cells acquire CD4 Tfh-like functionality by producing Tfh effector cytokines and co-receptors and promoting B cell antibody class-switching. CD8 Tfc cells promote antibody class-switch recombination at a level comparable to CD4 Tfh cells in vitro, and synergize with CD4 Tfh cell responses in vivo. Our findings concur with recent reports describing the generation of similar CD8 Tfc cells during chronic infection (20, 21, 23, 53).
CD8 Tfc cells affected class-switch recombination in B cells through a cell-secreted factor in the supernatant. One likely factor is the cytokine IL-21, as it is produced by IL-2-KO CD8 Tfc cells. IL-21 is known to induce plasma cell differentiation, immunoglobulin production and class-switching. In vitro-derived CXCR5+CD8 cells also produce IL-21 that promotes influenza-specific IgG production that is reduced in IL-21R deficient B cells (22). In addition to the role of IL-21 in developing CD4 Tfh cell populations, other CD4 Tfh secreted cytokines promote specialized antibody class-switch; for example, IFNγ supports IgG2a, and IL-4 supports IgG1 switching (54, 55). While IL-2-KO disease has been described as a Th1 mediated disease (5), IFNγ-mediated class-switching to IgG2a was only detected when IL-2-KO CD4 T cells were transferred alone and only moderately affected by co-transfer with CD8 T cells (Fig. 6H). In contrast, both CD8 depletion and co-transfer of CD4 and CD8 T cells most significantly impacted class-switching to IgG1. Autoimmune interactions in the IL-2-KO mouse, and specifically in the GC, may be governed by a combination of cytokines including IL-21 and IL-4, in contrast to the IFNγ production found systemically. Additionally, elevated CD40L and ICOS expression by IL-2-KO CD8 Tfc cells was observed and may promote B cell function via direct cell:cell interactions. In IL-2-KO mice, CD8 T cells localize to the B cell follicle and can be identified within the GC. CD8 T cells migrate into proximity with B cells, providing the localization necessary for influencing GC B cell reactions.
We report that autoimmune disease is delayed in IL-2-KO mice in the absence of CD8 T cells. Expansion of CD8 Tfc cells is also delayed in comparison to the observed expansion of CD4 Tfh cell during disease progression. The kinetics suggest that CD4 T cell dysfunction proceeding CD8 Tfc cell expansion may promote a transition to more rapid, lethal autoimmunity. Our reported differences in frequency of CD4 Tfh and CD8 Tfc cells in the IL-2-KO and scurfy mouse models also suggest a role for IL-2 in CD8 Tfc cell development. The absence of IL-2 contributes to a lymphoproliferative disorder in IL-2-KO mice that is not seen in scurfy mice, which may partially account for observed differences (48). However, IL-2 is known to suppress CD4 Tfh cell differentiation via Bcl6 expression in vivo (56). Thus, the difference in CD8 Tfc cell frequency between IL-2-KO mice and scurfy mice may be explained in part by the reduced numbers of IL-2 expressing cells in addition to differences in lymphoproliferation. As scurfy mice produce fairly normal levels of IL-2 (Fig. 4A; (47)), other factors in addition to IL-2 loss likely contribute to CD8 Tfc cell development.
As a reduction or impairment of Tregs is the driving defect underlying autoimmunity in both IL-2-KO (2), and scurfy mice (47), the expansion of CD8 Tfc cells in both models is likely due, in part, to the breakdown in immune tolerance mechanisms that precedes autoimmune disease. During chronic inflammation and situations of high localized antigen, especially when immune regulation is compromised, CD4 Tfh cells expand (13, 16). Cellular expansion and inflammation occurs in the IL-2-KO mouse because of reduced function and frequency of FoxP3+ Treg cells (2). Both CD4 and CD8 Treg cells have been identified as essential regulators of GC tolerance (7, 8). The absence of CD4 T follicular regulatory cells have been shown to promote autoimmune disease and aging via increased CD4 Tfh cells, GC B cells and antibody class-switching (7, 57). Impaired Treg function may similarly allow for the development of CD8 Tfc cells during chronic inflammatory conditions.
Together, this study adds to a growing body of research supporting a helper role for CD8 T cells during chronic antigen exposure and inflammation, including chronic viral infections, cancer and autoimmune disease. Our data provides a unique perspective on the role of CD8 T cells in GC interactions that may promote, amplify, or shift the autoimmune disease process. Future studies are needed to reveal the overlapping and distinct immune stages, roles and influences of CD4 Tfh and CD8 Tfc cells during autoimmune disease progression. The identification of CD8 Tfc cell interactions within the GC provides many avenues for continuing research, including defining the contribution of CD8 Tfc cells to immunity and disease. An understanding of the types of antibodies generated and the contribution to affinity maturation throughout the kinetics of CD8 Tfc cell development may unveil a new paradigm for GC reactions and a deeper insight into strategies for manipulating these processes.
Supplementary Material
Acknowledgments
The authors thank Dr. Jennifer Manilay for scientific advice and critical evaluation of our manuscript, Roy Hoglund and the staff members of the UC Merced Department of Animal Research Services for animal husbandry care, the UC Merced Stem Cell Instrumentation Foundry for their assistance in cell sorting, Anh Diep, Haword Cha and Karina Arroyo for experimental assistance and technical support.
Funding: This work was supported by the National Institutes of Health Grant R00HL090706 to K.K.H. and NHLBI Mentored Career Development Award K01HL130753 to A.E.B., an Aplastic Anemia and MDS International Foundation Grant to K.M.V., and by the National Science Foundation Grant No. ACI-1429783 to T.J.L.
Footnotes
Conflict-of-interest disclosure: The authors have declared that no conflict of interest exists.
References
- 1.Hoyer KK, Dooms H, Barron L, Abbas AK. Interleukin-2 in the development and control of inflammatory disease. Immunol Rev. 2008;226:19–28. doi: 10.1111/j.1600-065X.2008.00697.x. [DOI] [PubMed] [Google Scholar]
- 2.Barron L, Dooms H, Hoyer KK, Kuswanto W, Hofmann J, O’Gorman WE, Abbas AK. Cutting edge: mechanisms of IL-2-dependent maintenance of functional regulatory T cells. J Immunol. 2010;185:6426–6430. doi: 10.4049/jimmunol.0903940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002;17:167–178. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- 4.Gravano DM, Al-Kuhlani M, Davini D, Sanders PD, Manilay JO, Hoyer KK. CD8+ T cells drive autoimmune hematopoietic stem cell dysfunction and bone marrow failure. J Autoimmun. 2016;75:58–67. doi: 10.1016/j.jaut.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoyer KK, Kuswanto WF, Gallo E, Abbas AK. Distinct roles of helper T-cell subsets in a systemic autoimmune disease. Blood. 2009;113:389–395. doi: 10.1182/blood-2008-04-153346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoyer KK, Wolslegel K, Dooms H, Abbas AK. Targeting T cell-specific costimulators and growth factors in a model of autoimmune hemolytic anemia. J Immunol. 2007;179:2844–2850. doi: 10.4049/jimmunol.179.5.2844. [DOI] [PubMed] [Google Scholar]
- 7.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, Dong C. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HJ, Verbinnen B, Tang X, Lu L, Cantor H. Inhibition of follicular T-helper cells by CD8(+) regulatory T cells is essential for self tolerance. Nature. 2010;467:328–332. doi: 10.1038/nature09370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vinuesa CG, Cyster JG. How T cells earn the follicular rite of passage. Immunity. 2011;35:671–680. doi: 10.1016/j.immuni.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Förster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alvarez Arias DA, Kim HJ, Zhou P, Holderried TA, Wang X, Dranoff G, Cantor H. Disruption of CD8+ Treg activity results in expansion of T follicular helper cells and enhanced antitumor immunity. Cancer Immunol Res. 2014;2:207–216. doi: 10.1158/2326-6066.CIR-13-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastian M, Lopez-Ocasio M, Metidji A, Rieder SA, Shevach EM, Thornton AM. Helios Controls a Limited Subset of Regulatory T Cell Functions. J Immunol. 2016;196:144–155. doi: 10.4049/jimmunol.1501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, Roberts IS, Copley RR, Bell JI, Cornall RJ, Goodnow CC. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 15.La Cava A, Ebling FM, Hahn BH. Ig-reactive CD4+CD25+ T cells from tolerized (New Zealand Black x New Zealand White)F1 mice suppress in vitro production of antibodies to DNA. J Immunol. 2004;173:3542–3548. doi: 10.4049/jimmunol.173.5.3542. [DOI] [PubMed] [Google Scholar]
- 16.Kim HJ, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, Kaygusuz Y, Meissner T, Holderried TA, Chan S, Kastner P, Haining WN, Cantor H. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350:334–339. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gravano DM, Hoyer KK. Promotion and prevention of autoimmune disease by CD8+ T cells. J Autoimmun. 2013;45:68–79. doi: 10.1016/j.jaut.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Kang YM, Zhang X, Wagner UG, Yang H, Beckenbaugh RD, Kurtin PJ, Goronzy JJ, Weyand CM. CD8 T cells are required for the formation of ectopic germinal centers in rheumatoid synovitis. J Exp Med. 2002;195:1325–1336. doi: 10.1084/jem.20011565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner UG, Kurtin PJ, Wahner A, Brackertz M, Berry DJ, Goronzy JJ, Weyand CM. The role of CD8+ CD40L+ T cells in the formation of germinal centers in rheumatoid synovitis. J Immunol. 1998;161:6390–6397. [PubMed] [Google Scholar]
- 20.Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, Zotos D, Fenix KA, Atnerkar A, Preston S, Chipman JG, Beilman GJ, Allison CC, Sun L, Wang P, Xu J, Toe JG, Lu HK, Tao Y, Palendira U, Dent AL, Landay AL, Pellegrini M, Comerford I, McColl SR, Schacker TW, Long HM, Estes JD, Busslinger M, Belz GT, Lewin SR, Kallies A, Yu D. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol. 2016;17:1187–1196. doi: 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- 21.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, Xu L, Chen X, Hao Y, Wang P, Zhu C, Ou J, Liang H, Ni T, Zhang X, Zhou X, Deng K, Chen Y, Luo Y, Xu J, Qi H, Wu Y, Ye L. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature. 2016;537:412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 22.Yang R, Masters AR, Fortner KA, Champagne DP, Yanguas-Casas N, Silberger DJ, Weaver CT, Haynes L, Rincon M. IL-6 promotes the differentiation of a subset of naive CD8+ T cells into IL-21-producing B helper CD8+ T cells. J Exp Med. 2016;213:2281–2291. doi: 10.1084/jem.20160417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Nasti TH, Sharpe AH, Freeman GJ, Germain RN, Nakaya HI, Xue HH, Ahmed R. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537:417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao L, Jia L, Bai L, He L, Yang B, Wu C, Li H. Phenotypic and functional characteristics of IL-21-expressing CD8(+) T cells in human nasal polyps. Sci Rep. 2016;6:30362. doi: 10.1038/srep30362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang J, Champion CI, Wei B, Liu G, Kelly KA. CD8(+)CXCR5(+) T cells regulate pathology in the genital tract. Infect Dis Obstet Gynecol. 2013;2013:813238. doi: 10.1155/2013/813238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quigley MF, Gonzalez VD, Granath A, Andersson J, Sandberg JK. CXCR5+ CCR7− CD8 T cells are early effector memory cells that infiltrate tonsil B cell follicles. Eur J Immunol. 2007;37:3352–3362. doi: 10.1002/eji.200636746. [DOI] [PubMed] [Google Scholar]
- 27.Godfrey VL, Wilkinson JE, Russell LB. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am J Pathol. 1991;138:1379–1387. [PMC free article] [PubMed] [Google Scholar]
- 28.Isakson SH, Katzman SD, Hoyer KK. Spontaneous autoimmunity in the absence of IL-2 is driven by uncontrolled dendritic cells. J Immunol. 2012;189:1585–1593. doi: 10.4049/jimmunol.1200342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meli AP, King IL. Identification of mouse T follicular helper cells by flow cytometry. Methods Mol Biol. 2015;1291:3–11. doi: 10.1007/978-1-4939-2498-1_1. [DOI] [PubMed] [Google Scholar]
- 30.Lee NJ, Rigby RJ, Gill H, Boyle JJ, Fossati-Jimack L, Morley BJ, Vyse TJ. Multiple loci are linked with anti-red blood cell antibody production in NZB mice -- comparison with other phenotypes implies complex modes of action. Clin Exp Immunol. 2004;138:39–46. doi: 10.1111/j.1365-2249.2004.02560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchhoff D, Frentsch M, Leclerk P, Bumann D, Rausch S, Hartmann S, Thiel A, Scheffold A. Identification and isolation of murine antigen-reactive T cells according to CD154 expression. Eur J Immunol. 2007;37:2370–2377. doi: 10.1002/eji.200737322. [DOI] [PubMed] [Google Scholar]
- 32.Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS One. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Didion JP, Martin M, Collins FS. Atropos: specific, sensitive, and speedy trimming of sequencing reads. PeerJ. 2017;5:e3720. doi: 10.7717/peerj.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 36.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 40.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huseby ES, Liggitt D, Brabb T, Schnabel B, Ohlén C, Goverman J. A pathogenic role for myelin-specific CD8(+) T cells in a model for multiple sclerosis. J Exp Med. 2001;194:669–676. doi: 10.1084/jem.194.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadlack B, Lohler J, Schorle H, Klebb G, Haber H, Sickel E, Noelle RJ, Horak I. Generalized autoimmune disease in interleukin-2-deficient mice is triggered by an uncontrolled activation and proliferation of CD4+ T cells. Eur J Immunol. 1995;25:3053–3059. doi: 10.1002/eji.1830251111. [DOI] [PubMed] [Google Scholar]
- 43.Meli AP, Fontés G, Avery DT, Leddon SA, Tam M, Elliot M, Ballesteros-Tato A, Miller J, Stevenson MM, Fowell DJ, Tangye SG, King IL. The Integrin LFA-1 Controls T Follicular Helper Cell Generation and Maintenance. Immunity. 2016;45:831–846. doi: 10.1016/j.immuni.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, Crotty S, Craft J. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 2010;185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma R, Sharma PR, Kim YC, Leitinger N, Lee JK, Fu SM, Ju ST. IL-2-controlled expression of multiple T cell trafficking genes and Th2 cytokines in the regulatory T cell-deficient scurfy mice: implication to multiorgan inflammation and control of skin and lung inflammation. J Immunol. 2011;186:1268–1278. doi: 10.4049/jimmunol.1002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng L, Sharma R, Gaskin F, Fu SM, Ju ST. A novel role of IL-2 in organ-specific autoimmune inflammation beyond regulatory T cell checkpoint: both IL-2 knockout and Fas mutation prolong lifespan of Scurfy mice but by different mechanisms. J Immunol. 2007;179:8035–8041. doi: 10.4049/jimmunol.179.12.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wrenshall LE, Smith DR, Stevens ET, Miller JD. Influence of interleukin-2 deficiency on the generation of autoimmune B cells. J Autoimmun. 2007;29:125–133. doi: 10.1016/j.jaut.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinstein JS, Herman EI, Lainez B, Licona-Limón P, Esplugues E, Flavell R, Craft J. TFH cells progressively differentiate to regulate the germinal center response. Nat Immunol. 2016;17:1197–1205. doi: 10.1038/ni.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H, Liu J, Cui X, Zuo Y, Zhang Z, Li Y, Tao R, Pang J. Increased frequency of circulating follicular helper T cells in lupus patients is associated with autoantibody production in a CD40L-dependent manner. Cell Immunol. 2015;295:46–51. doi: 10.1016/j.cellimm.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 53.Xing J, Zhang C, Yang X, Wang S, Wang Z, Li X, Yu E. CXCR5(+)CD8(+) T cells infiltrate the colorectal tumors and nearby lymph nodes, and are associated with enhanced IgG response in B cells. Exp Cell Res. 2017 doi: 10.1016/j.yexcr.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 54.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohr E, Cunningham AF, Toellner K-M, Bobat S, Coughlan RE, Bird RA, MacLennan ICM, Serre K. IFN-γ produced by CD8 T cells induces T-bet–dependent and –independent class switching in B cells in responses to alum-precipitated protein vaccine. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17292–17297. doi: 10.1073/pnas.1004879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sage PT, Ron-Harel N, Juneja VR, Sen DR, Maleri S, Sungnak W, Kuchroo VK, Haining WN, Chevrier N, Haigis M, Sharpe AH. Suppression by TFR cells leads to durable and selective inhibition of B cell effector function. Nat Immunol. 2016;17:1436–1446. doi: 10.1038/ni.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.