Abstract
Mitochondria are descendants of endosymbiotic bacteria and retain essential prokaryotic features such as a compact circular genome. Consequently, in mammals, mitochondrial DNA is subjected to bidirectional transcription that generates overlapping transcripts, which are capable of forming long double-stranded RNA structures1,2. However, to our knowledge, mitochondrial double-stranded RNA has not been previously characterized in vivo. Here we describe the presence of a highly unstable native mitochondrial double-stranded RNA species at single-cell level and identify key roles for the degradosome components mitochondrial RNA helicase SUV3 and polynucleotide phosphorylase PNPase in restricting the levels of mitochondrial double-stranded RNA. Loss of either enzyme results in massive accumulation of mitochondrial double-stranded RNA that escapes into the cytoplasm in a PNPase-dependent manner. This process engages an MDA5-driven antiviral signalling pathway that triggers a type I interferon response. Consistent with these data, patients carrying hypomorphic mutations in the gene PNPT1, which encodes PNPase, display mitochondrial double-stranded RNA accumulation coupled with upregulation of interferon-stimulated genes and other markers of immune activation. The localization of PNPase to the mitochondrial inter-membrane space and matrix suggests that it has a dual role in preventing the formation and release of mitochondrial double-stranded RNA into the cytoplasm. This in turn prevents the activation of potent innate immune defence mechanisms that have evolved to protect vertebrates against microbial and viral attack.
Bidirectional transcription of mitochondrial DNA (mtDNA) is an extreme example of convergent transcription in mammalian cells owing to symmetrical synthesis of both the heavy (H) and the light (L) strand encoded RNAs. Notably, nearly the entire L-strand transcript undergoes rapid RNA decay by the RNA degradosome3. This decay process probably prevents the formation of potentially deleterious mitochondrial double-stranded RNA (mtdsRNA). Indeed, among different cellular compartments, mitochondrial RNA (mtRNA) is known to be especially immunogenic4. Cellular nucleic acid sensors must discriminate viral nucleic acids from the vast excess of often biochemically indistinguishable cellular RNA and DNA as part of the innate immune response5. To achieve this, nucleic acid metabolism is pivotal in suppressing immune responses to self nucleic acids6. Recently, numerous pathways have been shown to suppress mtDNA sensing by preventing its escape into the cytoplasm7,8. We sought to determine whether mitochondria are also a source of dsRNA in vivo, and in so doing uncovered a pathway that suppresses the formation of immunostimulatory mtdsRNA.
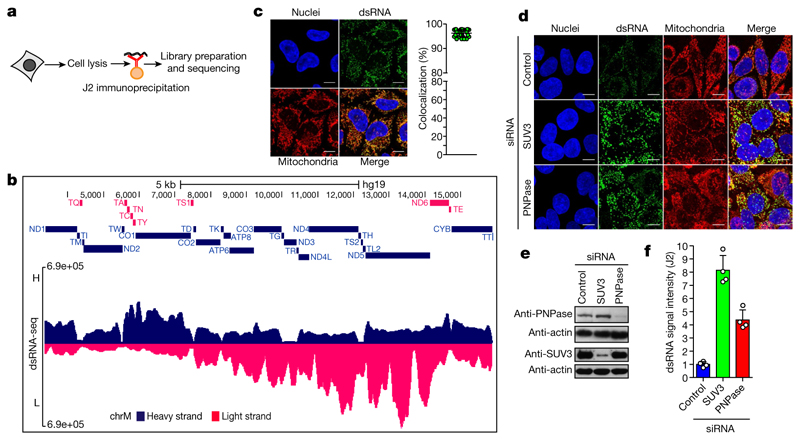
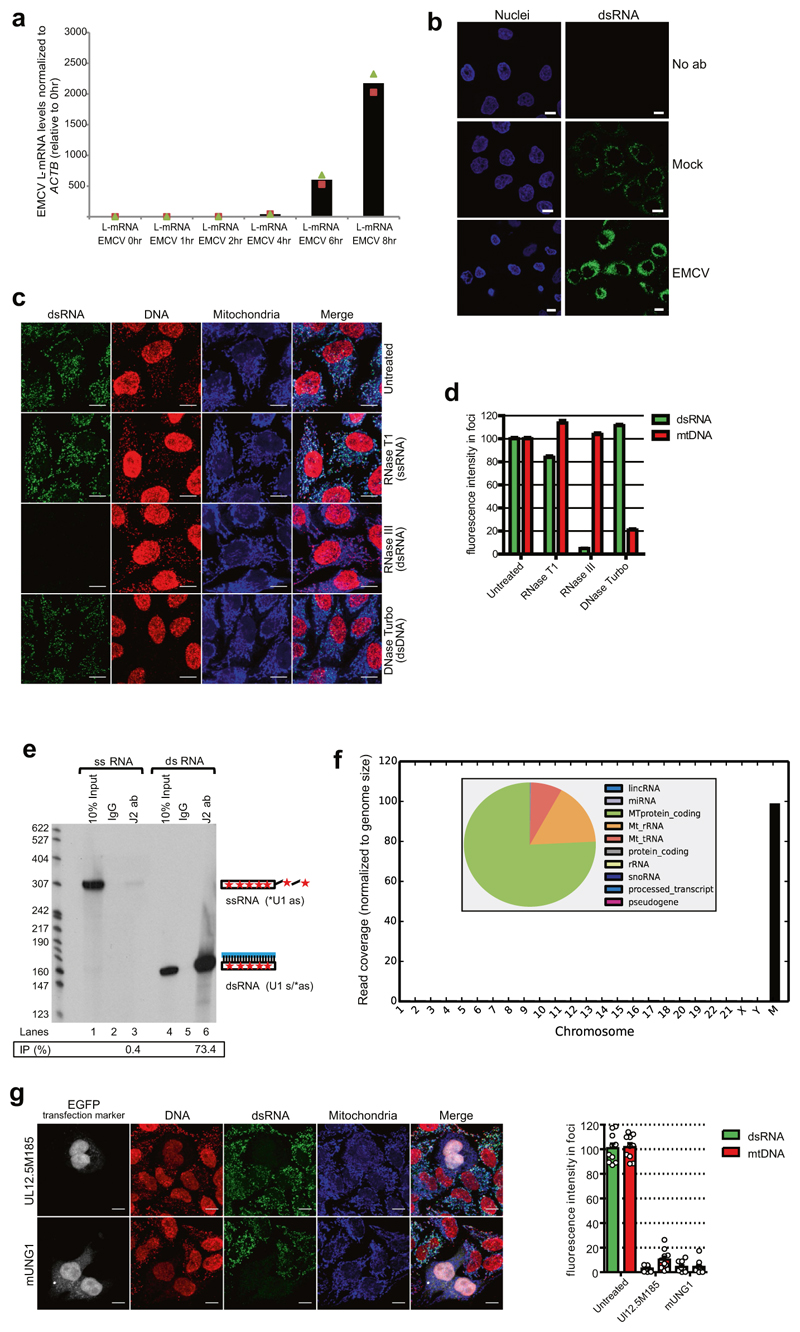
We used a monoclonal antibody (J2) specific for dsRNA that is widely used to detect viral dsRNA in animals and plants9. As shown previously, HeLa cell infection with the positive-strand RNA virus, encephalomyocarditis virus (EMCV) resulted in strong cytoplasmic dsRNA signals9 (Extended Data Fig. 1a, b). Notably, weaker immunofluorescence signals were also observed in uninfected HeLa cells suggesting the existence of cellular dsRNA. To further characterize these cellular immunofluorescence signals, fixed cells were pre-treated with structure-specific RNases. Immunofluorescence signals were sensitive to dsRNA-specific RNase III but not single-stranded RNA (ssRNA)-specific RNase T1 or TURBO DNase confirming the presence of dsRNA at a single-cell level (Extended Data Fig. 1c, d). We then verified the specificity of J2 for dsRNA in vitro using ss- or dsRNA immunoprecipitation experiments (Extended Data Fig. 1e). We next performed J2-immunoprecipitation-based dsRNA sequencing (dsRNA-seq) to identify selected cellular dsRNA (Fig. 1a). Notably, the mitochondrial genome generates nearly all detectable cellular dsRNA with 99% of the reads attributable to the mitochondrial genome (Extended Data Fig. 1f). Furthermore, the RNA sequencing profile showed widespread reads from both the H- and L-strand of mtDNA, implying the presence of intermolecular dsRNA (Fig. 1b). This was confirmed by immunofluorescence as 95% of J2 foci colocalized with mitochondria (Fig. 1c). To rule out potential artefacts caused by the expression of mitochondrial pseudogenes integrated in the nuclear genome, we performed dsRNA staining in mtDNA-depleted HeLa cells obtained by either expressing the herpes simplex virus 1 (HSV-1) protein UL12.5M185 or human uracil–N-glycosylase (mUNG1)10. A lack of J2 signal confirmed that the dsRNA identified in our experiments can be wholly attributed to the mitochondrial genome (Extended Data Fig. 1g).
Fig. 1. Mitochondria form dsRNA that is suppressed by the RNA degradosome.
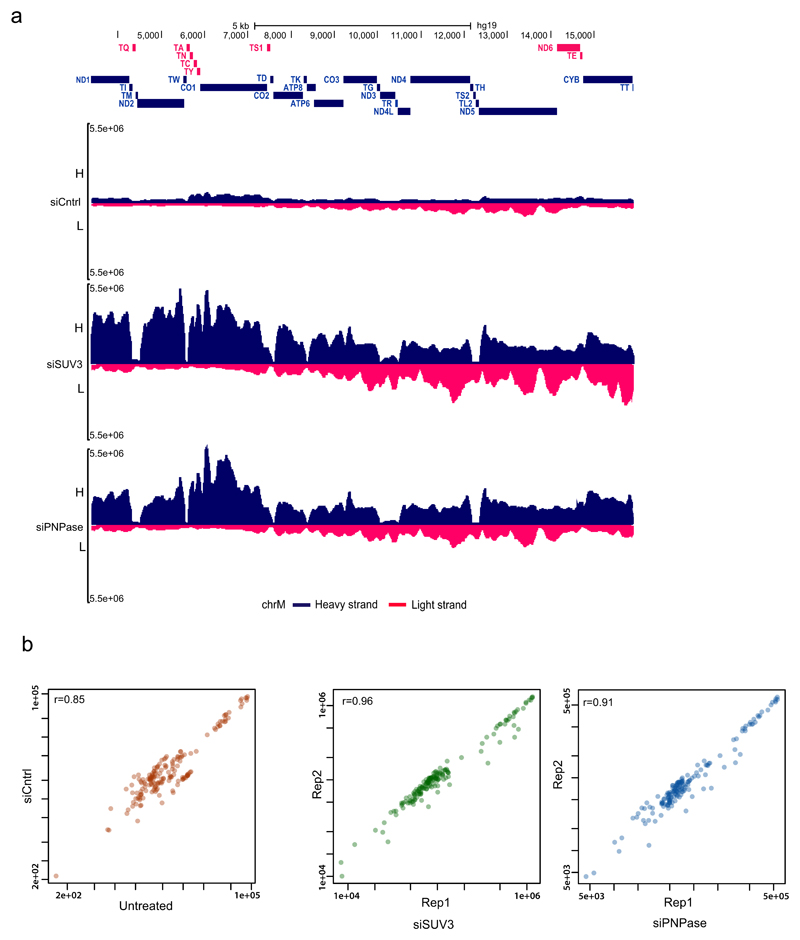
a, dsRNA-seq experimental approach. b, dsRNA-seq reads across the mitochondrial genome spanning protein coding region in untreated HeLa cells. H-strand genes are shown as blue bars and L-strand as red bars. Short tRNA genes are denoted with T as the first letter. Data are representative of two experiments. c, Immunostaining of dsRNA in HeLa cells with anti-dsRNA (J2) antibody. Mitochondria and nuclei are stained with MitoTracker Deep Red and Hoechst, respectively. Scale bars, 10 μm. Graphs quantify co-localization of dsRNA foci with mitochondria. Data are mean ± s.d. from 29 cells. d, Anti-dsRNA (J2) staining in HeLa cells depleted for PNPase or SUV3 by siRNA as in c. Different imaging settings were applied in panel c and d so that the J2 intensity of control cells varies. e, Western blot showing PNPase or SUV3 depletion. Blots are representative of four experiments. f, Quantification of dsRNA levels in PNPase- or SUV3-depleted cells. Data are mean ± s.d. from four experiments. For gel source data, see Supplementary Fig. 1.
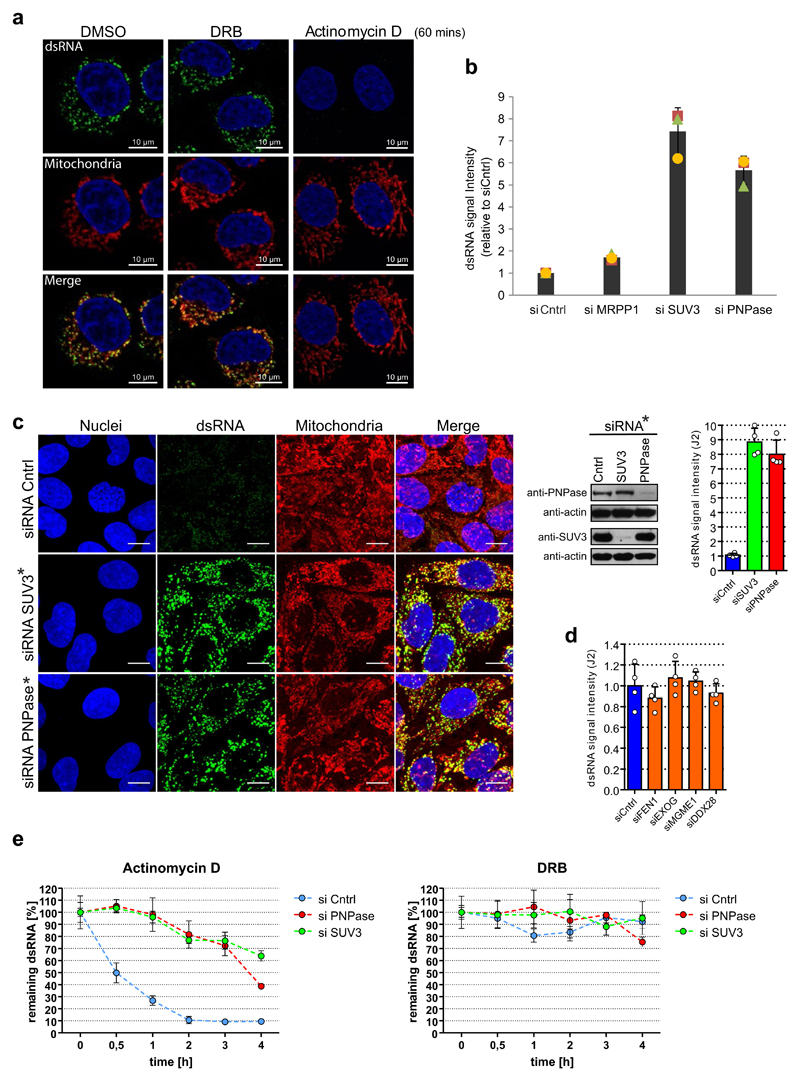
As dsRNA levels are normally suppressed in the cell, presumably to avoid the induction of an interferon response, we investigated mtdsRNA turnover. Actinomycin D (Act-D) treatment, which inhibits mitochondrial transcription, caused a rapid loss of mtdsRNA, unlike the CDK9 inhibitor DRB which inhibits nuclear RNA polymerase II transcription (Extended Data Fig. 2a). To search for factors involved in mtdsRNA suppression, we focused on the SUV3 and PNPase enzymes (encoded by SUPV3L1 and PNPT1 genes, respectively), which are known to be involved in the degradation of L-strand transcripts3. siRNA-mediated depletion of either enzyme resulted in a five- to eightfold increase in dsRNA levels, on the basis of both confocal microscopy (Fig. 1d–f) and flow cytometry (Extended Data Fig. 2b). The same effect was observed with a different set of siRNAs (Extended Data Fig. 2c). Other tested factors involved in the metabolism of mitochondrial nucleic acids had no effect on dsRNA levels (Extended Data Fig. 2d). We next confirmed that this increase in steady-state levels of dsRNA was due to changes in mtdsRNA turnover. Upon Act-D treatment, but not DRB, dsRNA levels in control-siRNA-treated cells were rapidly turned over (half-life of 30 min) whereas dsRNA levels were relatively stable for up to 3 h in either SUV3- or PNPase-depleted cells (Extended Data Fig. 2e).
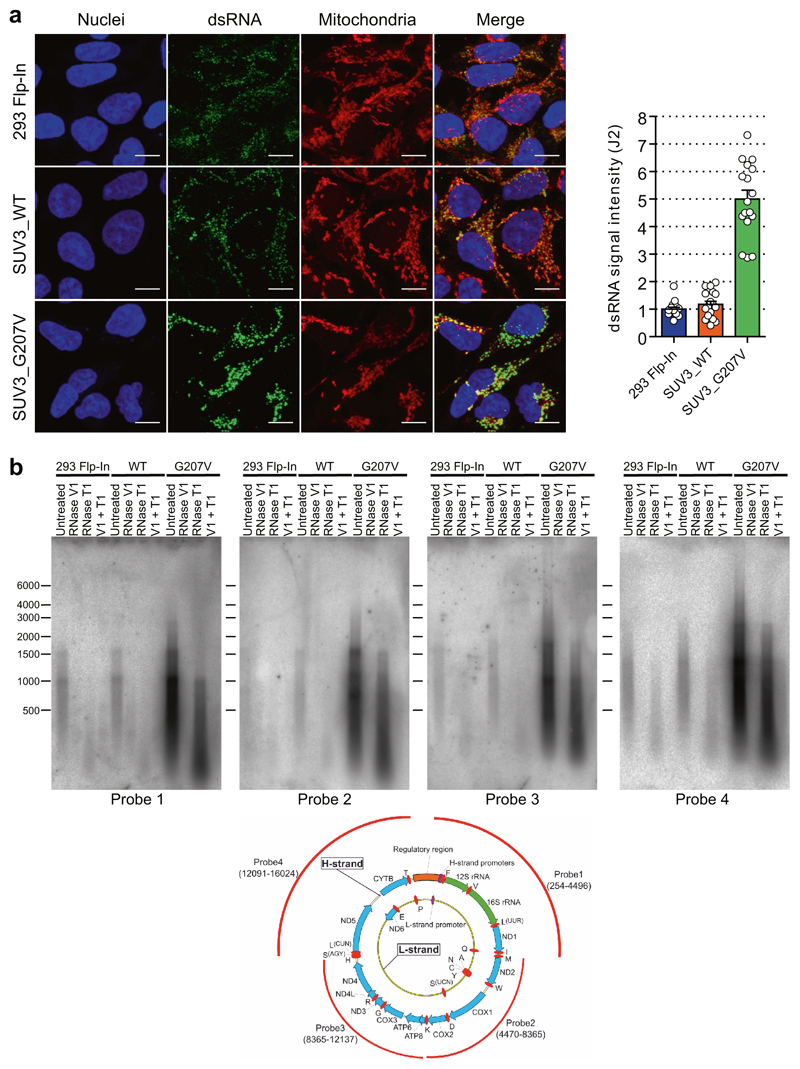
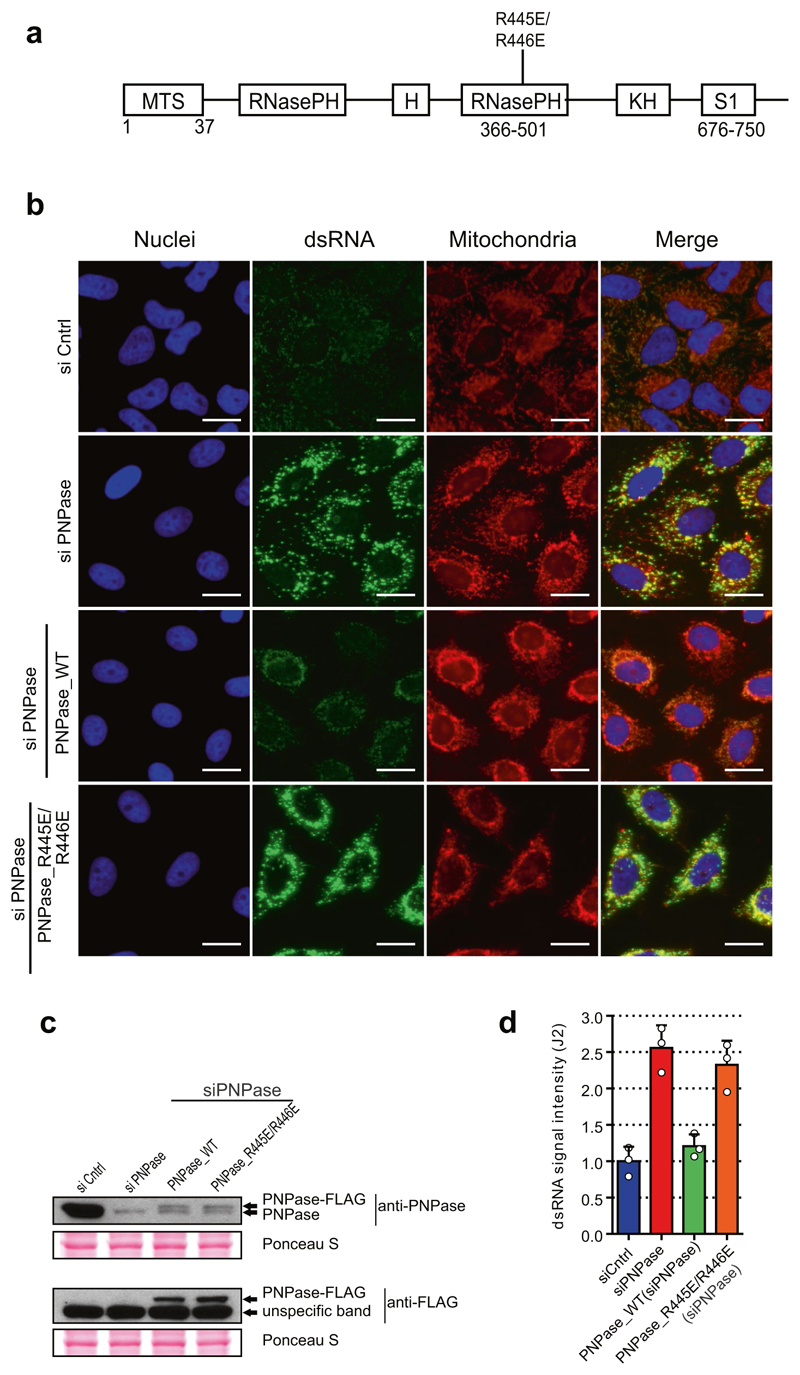
To further understand the mechanism of dsRNA turnover by SUV3 and PNPase, we used their catalytic mutants. Overexpression of a SUV3 transgene carrying an inactivating mutation (G207V) in the Walker A motif of the helicase in HEK 293 cells acted as a dominant-negative protein11 resulting in accumulation of dsRNA (Extended Data Fig. 3a). Furthermore, northern-blot analysis of J2-immunoprecipitated dsRNA isolated from this dominant negative mutant showed the accumulation of long dsRNA species (approximately 1–6 kb) mapping over the entire mitochondrial genome (Extended Data Fig. 3b). Both RNA import and RNA turnover functions have been ascribed to PNPase3,12. Therefore, an R445E/R446E mutant of PNPase, which lacks exonuclease activity without affecting RNA import, was used3,12 (Extended Data Fig. 4a). dsRNA levels accumulating upon PNPase depletion were suppressed by overexpression of siRNA-resistant PNPase but not the R445E/R446E mutant in HeLa cells (Extended Data Fig. 4b–d) and HEK 293 cells (data not shown). Overall, these results implicate the unwinding activity of SUV3 and the exonuclease activity of PNPase in dsRNA turnover. Consistently, J2-immunoprecipitation dsRNA-seq of SUV3- and PNPase-depleted HeLa cells showed substantial accumulation of mtdsRNA as compared to control siRNA, which was highly reproducible (Extended Data Fig. 5a, b).
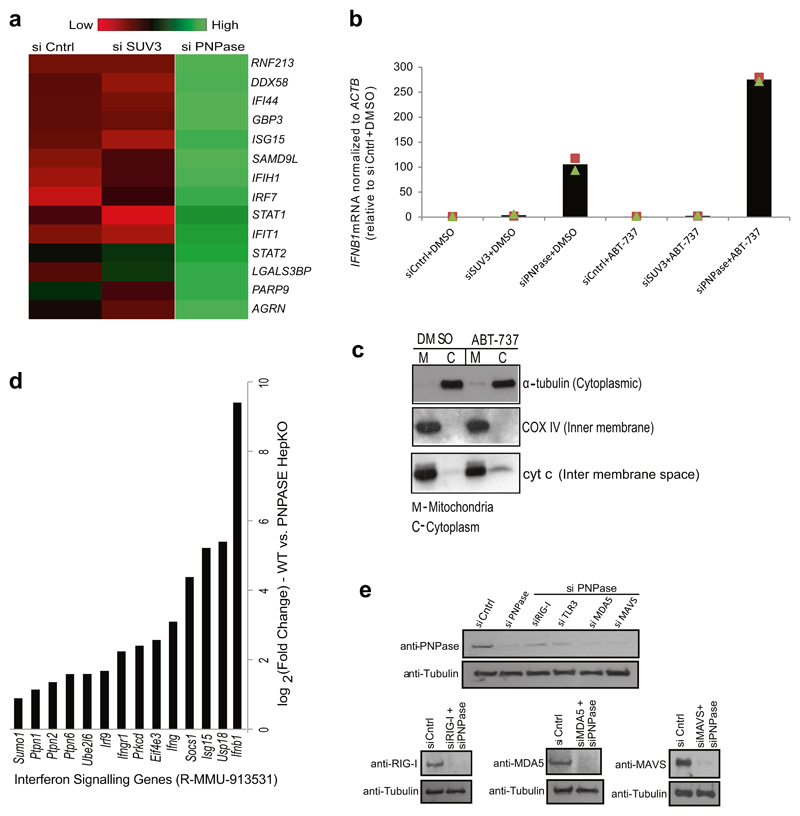
As long dsRNA is a hallmark of viral replication that triggers a type I interferon response, IFNB1 induction was tested in various knockdowns of mitochondrial RNA processing factors. Quantitative PCR with reverse transcription (RT–qPCR) analysis revealed an approximately 90-fold induction of IFNB1 mRNA upon depletion of PNPase but not upon depletion of SUV3 or MRPP1 (Fig. 2a). Consistently, gene-expression profiling revealed activation of interferon-stimulated genes (ISGs) such as genes with direct antiviral activity (for example, IFI44, IFIT1), cytoplasmic RNA sensors DDX58 and IFIH1 (encoding RIG-I and MDA5, respectively) and the transcription factor IRF7 that positively reinforces the antiviral response (Extended Data Fig. 6a). The observation that mtdsRNA activated an interferon response upon depletion of PNPase, but not upon depletion of SUV3, suggested that SUV3-restricted mtdsRNA is either non-immunogenic or somehow concealed from cytosolic dsRNA sensors. We therefore isolated mtRNA from mitochondria depleted of SUV3 or PNPase using a magnetic-activated cell sorting (MACS) approach13 and transfected it into HeLa cells to induce IFNB1 mRNA (Fig. 2b). Notably, mtRNA extracted from either condition triggered a similar IFNB1 induction, which was RNase III sensitive (Fig. 2b). The latter finding confirms that the interferon induction is triggered by mtdsRNA and not by mtDNA. The experiment also excludes the possibility that SUV3-dependent mtds-RNA is non-immunogenic, and led us to explore dsRNA localization. Transmission electron microscopy with immunogold labelling using J2 demonstrated mitochondrial localization of dsRNA in control siRNA samples and substantial accumulation in SUV3-depleted cells (Fig. 2c). By contrast, in PNPase-depleted cells, J2 staining displayed both a mitochondrial and cytoplasmic distribution, indicating the release of mtdsRNA into the cytoplasm (Fig. 2c). Consistently, enhanced mitochondrial outer membrane permeabilization of PNPase-depleted cells using ABT-737 (Bcl-2 inhibitor) resulted in an approximately threefold greater induction of IFNB1 mRNA (Extended Data Fig. 6b). Lack of an interferon response in SUV3-depleted cells with or without ABT-737 treatment suggested that mtdsRNA remains restricted to mitochondria (Extended Data Fig. 6b). We confirmed ABT-737-mediated mitochondrial outer membrane permeabilization through release of intermembrane-space-localized protein cytochrome c into the cytoplasm (Extended Data Fig. 6c).
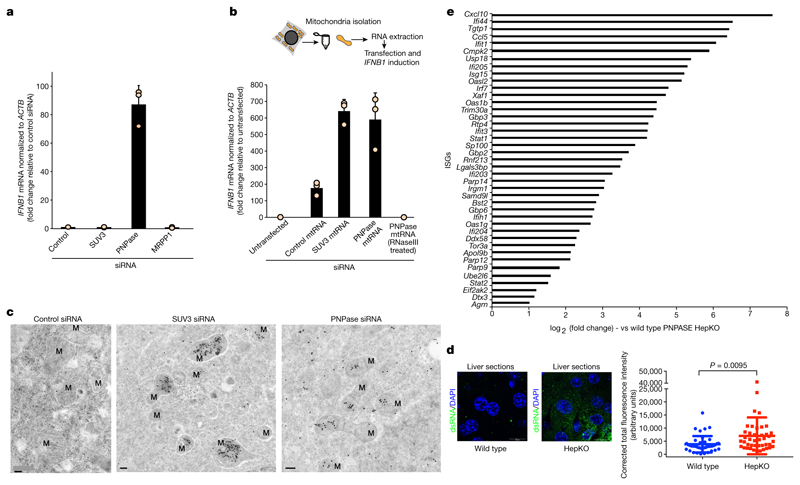
Fig. 2. PNPase suppresses a mtdsRNA-mediated type I interferon response.
a, RT–qPCR analysis of IFNB1 mRNA in HeLa cells treated with the indicated siRNAs. Data are mean ± s.d. from three independent experiments. b, Top, schematic of MACS strategy to purify mtRNA. Bottom, RT–qPCR analysis of IFNB1 mRNA in HeLa cells transfected with mtRNA (using MACS) as indicated. Data are mean ± s.d. from three independent experiments. c, Transmission electron microscopy images of immunogold labelled dsRNA (J2) in cryofixed HeLa cells treated with the indicated siRNAs. Images are representative of two experiments. M, mitochondria. Scale bars, 0.2 μm. d, Left, fluorescence immunohistochemistry staining of dsRNA (J2) in liver sections from wild-type and HepKO mice. Nuclei are stained with DAPI. Right, dsRNA quantification. Data are mean ± s.e.m. from 41 (wild-type) and 42 (HepKO) randomly sampled regions in two liver sections measured. The P value is from a two-sided unpaired t-test with Welch’s correction. e, log2 fold expression change of ISGs in wild-type versus HepKO mice. The ISG list is based on previously published work8.
We wished to extend our results on PNPase-restricted mtdsRNA in HeLa cells to an animal gene knockout model. We therefore used the hepatocyte-specific Pnpt1HepKO (hereafter HepKO) mouse that has a liver-specific knockout of PNPase as previously described12. We consistently observed an accumulation of dsRNA in HepKO liver sections versus controls (Fig. 2d). Notably, HepKO cells showed a gradual loss of mtDNA over time, suggesting an adaptive response to interferon activation (M.T., unpublished results), and probably accounting for the heterogeneous increase in dsRNA levels (Fig. 2d, right). However, differential gene-expression analysis showed upregulation of Ifnb1 and numerous ISGs such as Ifi44, Ifit1, and Cxcl10 in HepKO mice (Fig. 2e, Extended Data Fig. 6d). These results are consistent with activation of a type I interferon response upon loss of PNPase, and support our HeLa cell siRNA-depletion data in murine primary cells.
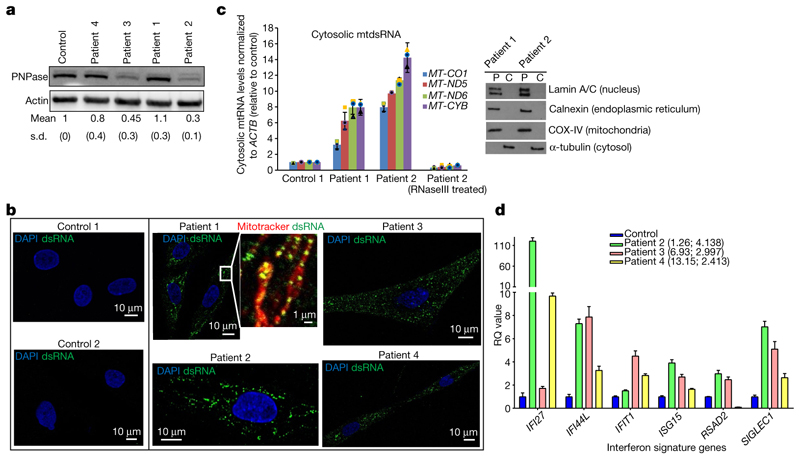
The importance of PNPase in the restriction of mtdsRNA led us to examine primary fibroblast cells from four different patients carrying biallelic hypomorphic mutations in the PNPT1 gene identified by exome sequencing and clinical manifestation (Table 1, Extended Data Table 1). These PNPT1 mutations led to decreased PNPase protein levels in fibroblasts of patients 2, 3 and to some extent 414. However, the homozygous active site mutation (R136H) recorded in patient 1 did not15 (Fig. 3a). Fibroblasts from all four patients demonstrated an accumulation of dsRNA (J2 signal) that was not observed in control cells (Fig. 3b). Moreover, this dsRNA colocalized with mitochondria (Fig. 3b, inset).
Table 1. Summarized data from patients with PNPT1 mutations.
| Patient | PNPT1 mutation | Amino acid change | Effect on PNPase | Outcome | IFNα levels in CSF (fg l−1)a | Neopterin levels in CSFb |
|---|---|---|---|---|---|---|
| 1 | G407A Homozygous | R136H | Abolishes active site | Died aged 2 years | NA | NA |
| 2 | T208C G2137T Heterozygous | S70P D713Y | Reduced protein level | Alive at age 1 year | 603 | 101 |
| 3 | G1495C G1519T Heterozygous | G499R A507S | Reduced protein level | Alive at age 7 years | ND | ND |
| 4 | A1160G Homozygous14 | Q387R | Trimerization defective; reduced protein level | Alive at age13 years | ND | ND |
NA, not available; ND, not determined. See also Extended Data Table 1.
IFNα normal range is <1 fg l−1.
Neopterin normal range is 8−43 nmol l−1.
Fig. 3. Pathological PNPT1 mutations result in mtdsRNA accumulation and activation of ISGs.
a, PNPase western blot in fibroblasts from four patients with mutations in PNPT1 and a control. Quantification is shown as mean and s.d. from four experiments. b, Immunostaining of dsRNA (J2) in fibroblast cell lines from patients with PNPT1 mutations and controls. Mitochondria are stained with MitoTracker Red CMXRos and nuclei with DAPI (blue). Scale bars, 10 μm (main images) and 1 μm (expanded view). Images are representative of two experiments. c, Left, RT–qPCR analysis of cytosolic mtRNA (four loci) in cells with PNPT1 mutations versus control cells. Data are mean ± s.d. from three independent experiments. Right, fraction purity as shown by western blots. Blots are representative of two experiments. P, pellet; C, cytosolic fractions. d, RT–qPCR analysis of six ISGs in whole blood from patients 2, 3 and 4. Ages when tested in decimalized years and interferon score are shown in brackets. The data plotted is relative quantification (RQ) values for each patient, with the error bars representing RQmin and RQmax. Data are from combined 29 control samples (blue bar) and 3 individual patient samples measured in triplicate. For gel source data, see Supplementary Fig. 1.
The identity of this mtdsRNA signal was further established by RT-qPCR analysis showing the presence of mtdsRNA (RNase III-sensitive) in pure cytosolic fractions from cells of patients 1 and 2 (Fig. 3c). Also, for three of the patients (no sample was available from patient 1), we recorded an upregulation of ISGs in peripheral blood (Fig. 3d). Furthermore, in patient 2, IFNα protein measured using a digital enzyme-linked immunosorbent assay (ELISA) was increased (603 fg l−1) in cerebrospinal fluid (CSF), which is equivalent to levels observed in certain cases of viral meningitis16 (Table 1). Patient 2 also showed abnormally high levels of neopterin in the CSF (101 nmol l−1), consistent with a hyperactivated immune response17 (Table 1). Overall, our analysis of patients harbouring hypomorphic mutations in PNPT1 clearly underlines the importance of preventing cytosolic sensing of mtdsRNA.
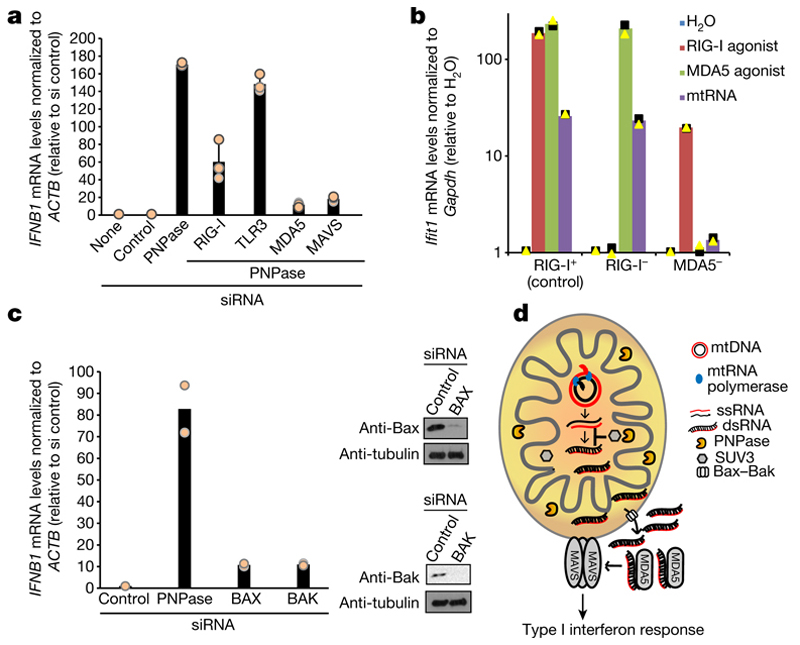
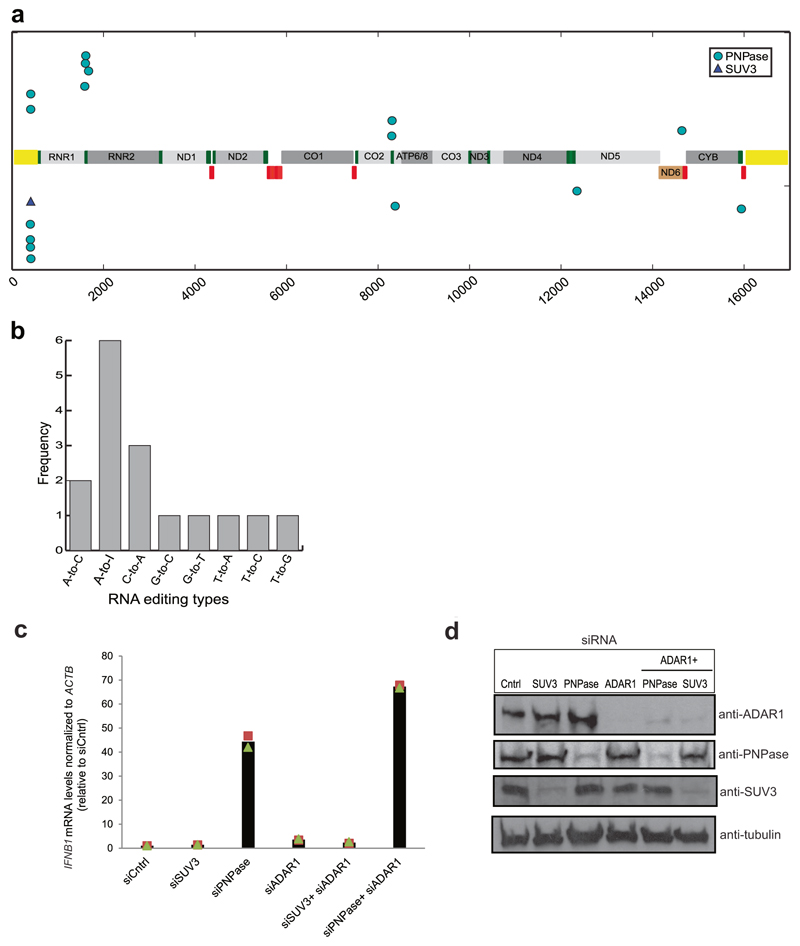
We sought to determine the mechanism of interferon activation by mtdsRNA in the context of PNPase deficiency. We tested the involvement of the RNA sensors RIG-I, MDA5 and TLR3 in this process. In PNPase-depleted HeLa cells, siRNA knockdown of MDA5 or (to a lesser extent) RIG-I abrogated the interferon response, but knockdown of TLR3 did not (Fig. 4a). These data implicate MDA5 as the primary sensor of mtdsRNA. MDA5 signals via the mitochondrial antiviral signalling protein (MAVS) to induce type I interferons so MAVS knockdown also abrogated IFNB1 induction (Fig. 4a, Extended Data Fig. 6e). We further confirmed these results by transfecting mtRNA isolated from PNPase-depleted HeLa cells (as in Fig. 2b) into RIG-I-deficient (Ddx58−/−) or MDA5-deficient (Ifih1−/−) murine embryonic fibroblasts (MEFs) (Fig. 4b). Ifih1−/− cells, but not Ddx58−/− cells, failed to upregulate mRNA levels of the ISG Ifit1 in response to mtds-RNA. This strongly suggests that the mtdsRNA-induced interferon response is mediated through the MDA5–MAVS axis. The possibility that mtdsRNA release into cytoplasm involves Bcl2-associated X protein (Bax)–Bcl-2 homologous antagonist/killer (Bak), as in the case of mtDNA7,18 was also investigated. Notably, depletion of Bax–Bak prevented IFNB1 mRNA induction after PNPase depletion, suggesting that mtdsRNA release depends on Bax–Bak pores (Fig. 4c). As a final test for the escape of mtdsRNA into the cytoplasm after PNPase depletion, we tested for dsRNA editing by the adenosine deaminase ADAR119. Notably, 16 mitochondrial RNA editing sites (including six adenosines to inosines) were observed within the RNA sequencing data of PNPase depleted cells, whereas only one was observed in the SUV3-depleted sample (Extended Data Fig. 7a, b). Concurrent depletion of ADAR1 and PNPase enhanced the observed interferon response by 1.5-fold, suggesting that ADAR1 acts as a feedback suppressor of the innate immune response, which is activated by mtdsRNA (Extended Data Fig. 7c, d). Overall, these mechanistic data on mtdsRNA formation, export and engagement with dsRNA sensors can be summarized in a model in which the escape of mtdsRNA into the cytoplasm triggers an ‘inappropriate’ type I interferon response (Fig. 4d).
Fig. 4. MDA5 is the primary sensor of cytosolic mtdsRNA released in a Bax–Bak dependent fashion.
a, RT–qPCR analysis of IFNB1 expression in HeLa cells transfected with indicated siRNAs. Data are mean ± s.d. from three independent experiments. b, RT–qPCR analysis of Ifit1 expression in Ddx58+/− (control, RIG-I+), Ddx58−/− (RIG-I−) and Ifih1−/− (MDA5−) immortalized MEFs transfected with mtRNA, RIG-I (ppp-IVT-RNA99nt) or MDA5 (CIP-EMCV RNA) specific agonists. Data are the mean from two independent experiments. Values are plotted on a logarithmic scale. c, Left, RT–qPCR analysis of IFNB1 mRNA in HeLa cells treated with the indicated siRNAs. Data are mean from two independent experiments. Right, western blot of siRNA depletion efficiency. Blots are representative of two experiments. For gel source data, see Supplementary Fig. 1. d, Model of mtdsRNA suppression by the RNA degradosome. Loss of PNPase causes accumulation of mtdsRNA and release of mtdsRNA into the cytoplasm in a Bax–Bak dependent manner. PNPase restricts mtdsRNA in matrix (together with SUV3) and IMS. MDA5 acts as the primary mtdsRNA sensor transducing an interferon response through the MAVS signalling pathway.
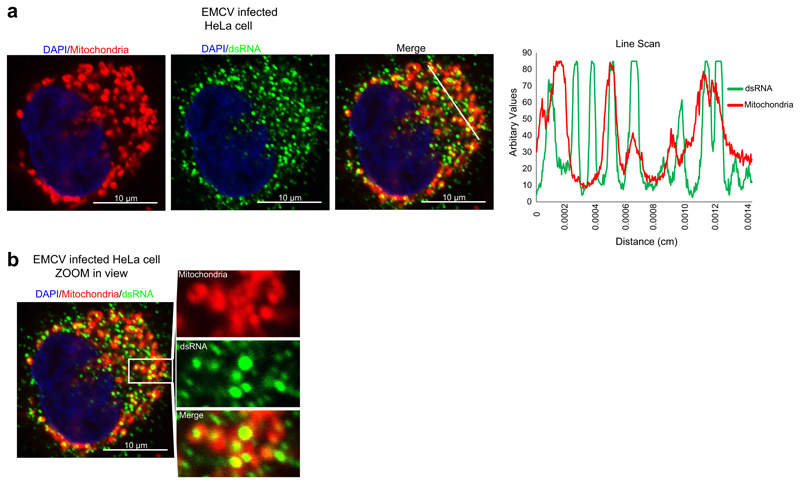
Our findings highlight an important function of PNPase which is underscored by its embryonic lethality in knockout mice and its identification as an essential fitness gene in CRISPR screens of human cell lines12,20. We considered it plausible that dysregulation of such an important pathway might induce an innate immune response, consistent with a disease class referred to as the type I interferonopathies16. Indeed, we show that biallelic hypomorphic mutations in PNPT1 cause mtdsRNA accumulation and immune activation. We suggest that mtdsRNA is a key mitochondrial-derived agonist of the innate immune system, a role until now mainly attributed to mtDNA21. Of note, genetic variants in MDA5 have been implicated in a number of human pathologies, both monogenic, and complex22–27. It is plausible that mtdsRNA mislocalization into the cytosol triggers an innate immune response upon viral infection, as dsRNA accumulation is detectable upon viral infection of mammalian cells9. Notably, dsRNA accumulation upon EMCV infection in HeLa cells partially colocalizes with mitochondria (Extended Data Fig. 8). It is possible that cellular mtdsRNA accumulation and the escape of mtdsRNA into the cytoplasm upon viral infection prime an antiviral response, as shown for mtDNA8. Overall, our results demonstrate a fundamental role of mitochondrial RNA processing in preventing the accumulation of deleterious self nucleic acid such as dsRNA that would otherwise trigger innate immunity.
Methods
In vitro experiments were not randomized, in vivo experiments were randomized. The investigators were blinded to allocation during in vivo experiments (immunohistochemistry) and outcome assessment. No statistical methods were used to predetermine sample size.
Antibodies and reagents
The following antibodies were obtained commercially: mouse anti-ADAR1 mAb (sc-73408, Santa Cruz), rabbit anti-PNPT1 (ab96176, Abcam; sc-49315, Santa Cruz), mouse anti-dsRNA mAb J2 (10010500, Scions), anti-DNA (61014, Progen), rabbit anti-SUV3 (A303-055A, Bethyl Laboratories)11, mouse anti-RIG-I mAb (Alme-1) (AG-20B-0009, AdipoGen), rabbit anti-COX IV (3E11, Cell Signaling), rabbit anti-cytochrome c (NB100-91732, Novus Biologicals), rabbit anti-calnexin (2433, Cell Signaling), mouse anti-lamin A/C (4C11, Cell Signaling), donkey anti-mouse IgG (H+L) conjugated with Alexa Fluor 488 (A-21202, Lifetech), goat anti-mouse IgM conjugated with Alexa Fluor 555 (A-21426, Thermo Fisher Scientific), normal mouse IgG2a (sc-3878, Santa Cruz), mouse anti-MDA5 (in house from J.R.), rabbit anti-MAVS (ALX-210-929-C100, Enzo Life Sciences), rabbit anti-Bax (2772T, Cell Signaling), rabbit anti-Bak (6947T, Cell Signaling), rabbit anti-OXA1L (HPA003531, Sigma), mouse anti-α-Tubulin (T5168, Sigma), mouse anti-actin (ab8226, Abcam; A5441, Sigma), rabbit anti-Flag (PA1-984B, Thermo Fisher Scientific), HRP secondary anti-mouse (ab6728, Abcam; A9044, Sigma)11, HRP secondary anti-rabbit (A0545, Sigma; ab6721, Abcam)11, goat anti-mouse IgG (20 nm gold) preadsorbed (ab27242, Abcam). ABT-737 (sc-207242, Santa Cruz), 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (D1916, Sigma) or Act D (A1410, Sigma), digitonin (D141, sigma), MitoTracker Deep Red (M22426, Thermo Fisher Scientific), MitoTracker Red CMXRos (9082, Cell Signaling). ppp-IVT-RNA99nt and CIP-EMCV-RNA were provided by J.R.28.
Cell culture, siRNA transfection and western blotting
HeLa (ATCC), hSUV3_WT/hSUV3_G207V HEK 293 cells11, PNPase_WT/PNPase_R445E-R446E HeLa cells, or HEK 293 Flp-In T-Rex cells (Thermo Fisher Scientific), MEFs (Ddx58+/−, Ddx58−/−, Ifih1−/−)29 and skin fibroblasts were grown as a monolayer at 37 °C, under 5% CO2 in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific). Skin fibroblasts were isolated from skin biopsies of controls and PNPT1 patients. Fibroblast medium was supplemented with 2 mM l-glutamine, 2.5 mM pyruvate, 100 μg ml−1 streptomycin, 100 U ml−1 penicillin at 37 °C. Silencing of genes of interest was performed using stealthRNA or other siRNAs (Extended Data Table 2) with Lipofectamine RNAiMAX (Thermo Fisher Scientific) in HeLa cells according to the manufacturer’s instructions. The stealthRNA oligonucleotides and siRNAs were used at a final concentration of 20 nM. For double siRNA treatments, each siRNA was used at final concentration of 20 nM. Cells were harvested three days after transfection unless stated otherwise. For Flp-In cells, expression of exogenous genes was induced by addition of tetracycline to the culture medium at a concentration of 25 ng ml−1. For western blots, total protein cell extracts were prepared in lysis solution (10 mM Tris, 140 mM NaCl, 5 mM EDTA, 1% (v/v) Triton X-100, 1% (w/v) deoxycholate, 0.1% (w/v) SDS) except for fibroblasts where lysis solution (50 mM Tris, 300 mM NaCl, 10 mM MgCl2, 0.5% NP-40, 2 mM DTT, Protease Inhibitor Cocktail 1X (Roche)) was used. Protein concentration was determined by the Bradford method. Protein extracts (30 μg per lane) were separated by SDS–PAGE and transferred to a nitrocellulose membrane (Protran, Whatman GmbH). Western blotting was performed according to standard protocols.
Plasmid transfection and establishing of stable cell lines
Plasmid transfections were performed with TranIT2020 (Mirus) according to the manufacturer’s instructions. HeLa cells were plated on glass coverslips 24 h after transfection with plasmids encoding UL12.5M185 and mUNG1, and after one day of culturing were subjected to an immunofluorescence procedure as described in the ‘Immunofluorescence labelling’ section. The stable inducible cell lines were established using plasmids pRS946 (PNPase_WT), pRS950 (PNPase_R445E/R446E) and HeLa Flp-In T-Rex cells (gift from M. Hentze) as described previously11. The identity of HeLa Flp-In T-Rex cells was confirmed using STR profiling by DSMZ (Germany).
Mice
Hepatocyte-specific Pnpt1HepKO (HepKO) mice were generated by breeding AlbCRE/WT/Pnpt1neo-flox/neo-flox with AlbWT/WT/Pnpt1neo-flox/neo-flox as described12. Mice are housed, bred and studied in accordance with an approved protocol consistent with the UCLA Chancellor’s Animal Research Committee (ARC) policies and procedures, as stated in Laboratory Animals in Teaching and Research (rev. 1998), the provisions of the NIH Guide for the Care and Use of Laboratory Animals and all applicable state and federal regulations.
Identification of PNPT1 mutations
Exome sequencing was performed on genomic DNA (1 μg) isolated from blood leukocytes. Exons were captured by the in-solution enrichment methodology (SureSelect Human All Exon Kits Version 3, Agilent) using biotinylated oligonucleotide probe library (Human All Exon v3 50 Mb, Agilent). Each genomic DNA was then sequenced as paired-end 75 bases (Illumina HISEQ2000, Illumina). After demultiplexing, sequences were aligned to the reference human genome hg19 using the Burrows–Wheeler Aligner (v.0.7.12). Downstream processing was carried out with the Genome Analysis Toolkit (GATK 3.7), SAMtools (v.1.4), and Picard (v.2.9.0-1), following documented best practices (https://www.broadinstitute.org/gatk/guide/topic?name=best-practices). Variant calls were made with the GATK Unified Genotyper. The annotation process was based on the latest release of the Ensembl database (version 75), dbsnp (version 140), 1,000 genome project (version 2013/05/02), Gnomad (version 2.0.2) and EVS (version ESP6500SI-V2). Variants were annotated and analysed using the Polyweb software interface designed by the Bioinformatics platform of University Paris Descartes. Sequences were filtered against SNPs (>0.1% frequency) reported in public (dbSNP, 1,000 genomes and Exome Variant Server) and in-house databases including intergenic and non-coding region variants. Only homozygous variations were considered for patient 1, born to consanguineous parents, resulting in a list of 14 genes with only PNPT1, encoding a mitochondrial protein. Targeted exome sequencing using a panel of known genes for mitochondrial disorders was performed for patient 2 and two heterozygous PNPT1 mutations were identified. Exome sequencing was performed for patient 3 and her non-consanguineous parents. The same filtering was used, identifying only one gene with two compound heterozygous mutations in PNPT1. DNA sequencing confirmed these mutations as well as their segregation with the disease in the families. All these variations were predicted to be deleterious by several software packages (Extended Data Table 1). Informed consent for diagnostic and research studies was obtained for all subjects in accordance with the Declaration of Helsinki protocols and all studies were approved by local Institutional Review Boards in Paris such as the human research participants ethics committee, Comité de Protection des Personnes, Ile de France II.
Targeted ISG RNA expression in total blood
Whole blood was collected into PAXgene tubes, total RNA extracted using a PreAnalytix RNA isolation kit and RNA concentration assessed using a spectrophotometer (FLUOstar Omega, Labtech). RT–qPCR analysis was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems), and cDNA derived from 40 ng total RNA. To generate a standard six probe interferon score, TaqMan probes for IFI27 (Hs01086370_m1), IFI44L (Hs00199115_m1), IFIT1 (Hs00356631_g1), ISG15 (Hs00192713_m1), RSAD2 (Hs01057264_m1) and SIGLEC1 (Hs00988063_m1) were used. The relative abundance of each target transcript was normalized to the expression level of HPRT1 (Hs03929096_g1) and 18S (Hs999999001_s1), and assessed with the Applied Biosystems StepOne Software v2.1 and DataAssist Software v.3.01. For all six probes, individual data were expressed relative to a single calibrator. RQ (relative quantification) is equal to 2−ΔΔCt, that is, the normalized fold change relative to the control data. The median fold change of the six genes compared to the median of 29 previously collected healthy controls was used to create an interferon score for each individual, with an abnormal interferon score being defined as greater than 2 s.d. above the mean of the control group, that is, 2.466. The experiment was performed in triplicate from one blood sample obtained from each individual.
Quantification of IFNα in CSF by Simoa assay
Simoa IFNα assay was developed using a Quanterix Homebrew Simoa assay and two autoantibodies specific for IFNα isolated and cloned from two patients mutated in APS1 (causing autoimmune polyendocrinopathy with candidiasis and ectodermal dysplasia, APECED) patients as recently described16,30. The 8H1 antibody clone was used as a capture antibody after coating on paramagnetic beads (0.3 mg ml−1), and the 12H5 was biotinylated (biotin/antibody ratio of 30:1) and used as the detector. Recombinant IFNα17/αI (PBL Assay Science) was used as a standard curve after cross-reactivity testing. The limits of detection were calculated by the mean value of all blank runs + 3 s.d. and was 0.23 fg ml−1.
Reverse transcription and real-time qPCR analysis
Total RNA was treated with TurboDNase (Ambion) and reverse-transcribed using SuperScript Reverse Transcriptase III (Invitrogen) with oligo (dT)20 for INFB1 mRNA, Ifit1 mRNA, L-mRNA (EMCV). qPCR was performed with 2× Sensimix SYBR mastermix (Bioline) and analysed on a Corbett Research Rotor-Gene GG-3000 machine.
Immunofluorescence of HeLa cells infected with EMCV or transcription inhibitors and skin fibroblasts
HeLa cells were grown on a coverslip in a 6-well plate 24 h before treatment. HeLa cells were infected with EMCV at multiplicity of infection (MOI) of 1 for the indicated time point. For transcription inhibitor treatment, cells were treated with dimethylsulfoxide (DMSO) or Act-D or DRB at the indicated concentrations for 60 min. For J2 immunofluorescence on HeLa or skin fibroblasts, cells were incubated with MitoTracker Red CMXRos (100 nM) for 30 min at 37 °C before fixing in 4% PFA in PBS. Cells were washed three times with PBS and permeabilized with 0.25% Triton X-100 in PBS. Cells were then washed with 0.05% Tween20–PBS and incubated with 3% BSA in PBS for 30 min at room temperature. Primary antibodies anti-dsRNA (J2) were used at 1:200 in 3% BSA in PBS for 1 h at room temperature. Cells were washed three times with 0.05% Tween20–PBS and then incubated with secondary donkey anti-mouse IgG (H+L) conjugated with Alexa Fluor 488 at (1:300) concentration. Cells were then washed three times with 0.05% Tween20-PBS and twice with PBS, and mounted with Vectashield mounting media with DAPI (Vector Laboratories). Z-stack images were collected with a FluoView1000 confocal microscope (Olympus) using a UPLSAPO 60.0× / 1.35 oil objective. Images were analysed using ImageJ and prepared using OMERO software.
Immunofluorescence of nuclease treated samples
HeLa cells were plated on glass coverslips one day before fixation. Mitochondria-specific dye MitoTracker Deep Red (200 nM) was added to culture 1 h before fixation. Cells were washed twice with PBS and fixed in 5% (v/v) formaldehyde, 0.25% (w/v) Triton X-100 and Hoechst 33342 (2 μg ml−1) in PBS for 30 min in room temperature. Cells were washed three times with PBS. Following enzymes were used: RNase T1 (EN0541, Thermo Fisher Scientific, concentration 100 U ml−1), RNase III (M0245S, NEB, concentration 40 U ml−1), TURBO DNase (AM2238, Thermo Fisher Scientific, concentration 40 U ml−1). Enzymes were added in PBS containing 5 mM MgCl2. Samples were incubated in 37 °C for 30 min and washed three times with PBS. Cells were incubated with 3% (w/v) BSA in PBS for 30 min. Primary antibodies anti-dsRNA (2.5 μg ml−1) and anti-DNA (0.5 μg ml−1) were used in 3% (w/v) BSA 16 h at 4 °C. Cells were washed three times with PBS and secondary goat IgG anti-mouse IgG2a conjugated with Alexa Fluor 488 and goat anti-mouse IgM conjugated with Alexa Fluor 555 (Thermo Fisher Scientific) were used at 2 μg ml−1 concentration in 3% (w/v) BSA. Cells were incubated for 1 h at room temperature and washed three times with PBS and mounted. Slides were imaged with a FluoView1000 confocal microscope (Olympus) and with ScanR fluorescence microscopy system (Olympus) (using UPlanSApo 20.0× objective) adapted for high throughput image acquisition. The latter was used for quantitative fluorescent signal analysis. Quantification was performed for at least 1,755 cells per condition. Images were analysed using ScanR_2.7.2 analysis software (Olympus). The same microscope instrument settings were used for all samples.
RNA polymerase inhibition
For Extended Data Fig. 2e, HeLa cells were treated with siRNA for 3 days in 384-well format. Prior to fixation cells were treated for a given time with inhibitors of transcription: actinomycin D (0.5 μg ml−1), DRB (100 μM). Detailed procedure is described in ‘siRNA transfection in 384-well format’ and ‘Immunofluorescence labelling’ sections. Quantitative analysis of dsRNA fluorescent signal was performed with ScanR fluorescence microscopy system. This analysis was performed for at least 500 cells per replica per condition. Images were analysed using ScanR_2.7.2 analysis software (Olympus).
siRNA transfection in 384-well format
Cells were reverse transfected in 384-well microplates (781946, Greiner Bio-One) using siRNA (final concentration 20 nM) and Lipofectamine RNAiMAX according to the manufacturer’s instructions (Thermo Fisher Scientific). Cells were plated with the Multidrop Combi Reagent Dispenser (Thermo Fisher Scientific). After 72 h, cells were subjected to an immunofluorescence procedure described in the ‘Immunofluorescence labelling’ section. Cells were left in PBS for imaging. All PBS washes were performed with 405 LS Microplate Washer (BioTek) and all other solutions were added with the Multidrop Combi Reagent Dispenser.
Immunofluorescence labelling
One hour before fixation, mitochondria-specific dye MitoTracker Deep Red (200 nM) was added to the culture. Cells were washed twice with PBS and fixed for 30 min with PBS solution containing 5% (v/v) formaldehyde, 0.25% (w/v) Triton X-100 and Hoechst 33342 (2 μg/mL). Cells were washed three times with PBS and incubated with 3% (w/v) BSA in PBS for 30 min. Primary antibodies against anti-dsRNA (2.5 μg ml−1) were used in 3% (w/v) BSA overnight at 4 °C. Cells were washed three times with PBS and secondary goat IgG anti-mouse IgG2a conjugated with Alexa Fluor 488 and goat anti-mouse IgM conjugated with Alexa Fluor 555 (Thermo Fisher Scientific) were used at 2 μg ml−1 concentration in 3% (w/v) BSA. Cells were incubated 1 h at room temperature and washed three times with PBS. Cover slips were mounted with ProLong Gold Antifade Mountant (P36930, Thermo Fisher Scientific) or left in PBS if imaged with a ScanR fluorescence microscopy system. If the samples were subjected to quantitative analysis, the same microscope instrument settings were applied.
Co-localization of dsRNA with mitochondria
Cells were subjected to staining as described in the ‘Immunofluorescence labelling’ section. Z-stack images of microscopic slides were collected with a FluoView1000 confocal microscope (Olympus) using a PLANAPO 60.0× 1.40 oil objective. XY optical resolution of images was 215 nm. Images were analysed using Imaris v.7.2.3 software (Bitplane). Object-based colocalization of spots was performed. Colocalization of J2 spots with mitochondria was based on fluorescence intensity from MitoTracker. Quantification was performed for 29 randomly selected cells.
High-throughput fluorescence imaging
Data presented on Fig. 1f, Extended Data Figs. 2c, d, 4d were obtained using a ScanR fluorescence microscopy system (Olympus, UPlanSApo 20.0× objective). Images were analysed using ScanR 2.7.2 analysis software (Olympus). Quantification of fluorescent signal was performed for at least 400 cells per replica per condition.
Fluorescent immunohistochemistry
Fluorescent immunohistochemistry staining of 4 μm-thick formalin-fixed, paraffin-embedded (FFPE) tissue sections was performed on livers from sex-matched (female) six-week-old wild-type C57BL/6 and Pnpt1HepKO (HepKO) littermate mice on a pure C57BL/6 background. FFPE slides were deparaffinized by immersion in 100% xylene, two times for 5 min and rehydrated twice in fresh 100% ethanol, 95% ethanol, 70% ethanol, and 50% ethanol for 3 min each. Sections were washed in double distilled H2O and permeabilized with 0.1% Triton X-100 in 1× PBS. Heat-induced antigen retrieval (HIER) was performed by heating sections in 1 mM EDTA at 95 °C in a pressure cooker for 20 min and then 20 min of cooling at room temperature. Sections were incubated for 12 h at 4 °C in blocking buffer (5% goat serum + 0.3% Triton X-100 + 3% BSA diluted in 1× PBS) and subsequently incubated with mouse anti-dsRNA J2 antibody, diluted 1:200 in blocking buffer, for 2 h at room temperature. Sections were washed in 0.1% Triton X-100 in 1× PBS three times for 10 min and incubated with secondary antibody goat anti-mouse Alexa Fluor 488 at 1:200 for 1 h at room temperature and washed again with 1× PBS–Tween. All processed slides were mounted in Prolong gold antifade mount with DAPI (Invitrogen cat # P36931). Some slides were processed in the absence of primary antibody to verify specificity of labelling.
Imaging of immunohistochemistry samples
All images were obtained with a 100× objective on a Leica TCS-SP8 inverted spectral confocal microscope (Leica Microsystems) equipped with a 405 nm blue diode laser, argon laser (5 lines), and white light laser for excitation. Further image processing of maximal z-projection images of 4 μM thick liver sections showing 488 and DAPI overlay was performed on LAS X v.3.30 software. Identical settings were used to obtain fluorescent images within datasets. Brightness and contrast for final images were adjusted equally across datasets using Photoshop CC 2017. Confocal laser-scanning microscopy was performed at the CNSI Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA.
Quantification of immunohistochemistry samples
To quantify J2 dsRNA immunofluorescence in liver sections from wild-type C57BL/6 and Pnpt1HepKO (HepKO) mice, a single in-focus plane was acquired at 100× at 20–21 locations across the tissue selected using a random coordinate generator. Quantifications were performed using ImageJ software, by drawing an outline around tissue and measuring area, integrated density and mean fluorescence. Additionally, background readings were measured on secondary-only tissue samples. To calculate the corrected total fluorescence intensity (CTFI) we used the following formula: CTFI = integrated density − (area of selected tissue × mean fluorescence of background readings). Scatter plot and statistical analysis (two-sided unpaired t-tests with Welch’s correction) were performed using GraphPad Prism 7.
Flow cytometry analysis
Cells were trypsinized, washed with PBS and fixed with 4% formaldehyde diluted in PBS for 20 min at room temperature. Cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min followed by incubation in 1% BSA (Sigma, A7030) in PBS for 1 h. Primary antibodies (anti-dsRNA (J2) or normal mouse IgG2a (Iso)) were used at 2.5 μg ml−1 in 1% BSA for 1 h at room temperature. Secondary donkey anti mouse IgG (H+L) conjugated with Alexa Fluor 488 were used at 2.2 μg ml−1 concentration for 1 h at room temperature. Cells were rinsed three times with FACS buffer (0.5% BSA in PBS with 2mM EDTA). Data were acquired with a FACSCalibur (BD Biosciences) flow cytometer. Data were analysed in FlowJo (TreeStar).
Immunoprecipitation of dsRNA
Protein G Dynabeads were washed and resuspended in NET-2 buffer. 5 μg of anti-dsRNA mAb (J2) was bound to 100 μl of beads for 1 h at room temperature on a Thermoshaker. Conjugated beads were washed three times with NET-2 Buffer. 80–90% confluent HeLa cells from 10 cm2 plate (×2) were washed with 10 ml of cold PBS. Cells were scraped and transferred to a falcon and spun at 500g at 4 °C, 5 min. Cell pellet from one 10-cm2 plate was lysed in 1 ml of NP-40 lysis buffer and transferred to an eppendorf and incubated on ice for 5 min. Following a spin at 17,000g at 4 °C for 5 min, supernatant was carefully transferred to a new eppendorf. Total RNA was harvested from 10% input lysate using Trizol reagent. For immunoprecipitation, lysate was diluted 1:4 in NET-2 buffer and supplemented with 10 units of RNase free TurboDNase (Ambion) at 10 mM MgCl2 per 1 ml of mix. 100 μl of J2-Dynabeads was added to 1 ml of above lysate and left for 1–2 h at 4 °C. Following magnetic separation, beads were washed twice with 1 ml of high salt washing buffer (HSWB). Beads were transferred to a new tube with NET-2 buffer and washed twice with the same buffer. J2-bound dsRNA was extracted with Trizol reagent. The RNA samples were sent for sequencing (described in the ‘RNA-sequencing’ section). NET-2 buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.5% NP-40), NP-40 lysis buffer (50 mM Tris-Cl pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40), high salt wash buffer (50 mM Tris-Cl pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.5% DOC, 0.1% SDS).
dsRNA isolation for northern blot
HEK 293 cells from 150-cm2 plate (×3) were used. Cell pellet was lysed in 4.5 ml of NP-40 lysis buffer and kept on ice for 5 min. Lysate was transferred to 1.5 ml eppendorf and centrifuged at 20,000g at 4 °C for 5 min. The supernatant was then transferred to 15 ml tube. Lysate was diluted 1:4 in NET-2 buffer and supplemented with 12 units of RNase free TurboDNase (Ambion) and 10 mM MgCl2 per 1ml of final mix. RNases were added (RNase T1-1U from 1 U μl−1, RNase V1-1U from 0.1 U μl−1 (Life Technologies)) and incubated at 37 °C for 10 min. 100 μl of J2-Dynabeads were added to 15 ml of above lysate and left on a rotor at 4 °C for 1–2 h. Beads were spun at 3,000g at 4 °C, 3 min. Supernatant was discarded and beads transferred to 1.5 ml tube, washed twice with 1 ml of HSWB and washed twice with NET-2 buffer. J2-bound dsRNA was extracted with Trizol reagent. RNA samples were used for northern blot.
Northern blot analysis
dsRNA after immunoprecipitation was purified by TRI Reagent (Sigma) using the manufacturer’s protocol. 20% of dsRNA eluate was dissolved in denaturing solution and run on a 1% agarose gel as described previously11. Subsequently, RNA was transferred to Amersham Hybond-N+ membrane (GE Healthcare Life Sciences) and UV cross-linked. For detection of mitochondrial transcripts probes were labelled with [α-32P] dATP (Hartmann Analytic) using a DECAprime II Kit (Ambion). PCR products corresponding to the following fragments of human mtDNA were used as templates: 254–4469 (Probe 1), 4470–8365 (Probe 2), 8365–12137 (Probe 3), 12091–16024 (Probe 4). Hybridizations were performed in PerfectHyb Plus buffer (Sigma) at 65 °C. Membranes were exposed to PhosphorImager screens (FujiFilm), which were scanned following exposure by a Typhoon FLA 9000 scanner (GE Healthcare). Data were analysed by Multi Gauge v.3.0 software (FujiFilm).
Probes for RNA protection assay (RPA)
U1 snRNA antisense fragment was amplified from pGEM4-tU1 (S. Murphy, University of Oxford) by PCR using the following primers: tU1 forward, AGCTCGGATCCATACTTACCTGGCAGGGGAGATA; tU1 reverse, ATTCATTAATGCAGCTGGCTT. According to the manual of StrataClone Blunt PCR cloning kit (Agilent Genomics), the PCR product was cloned as pSC-B-tU1_RPA. T7 transcription was performed using [α-32P]UTP and XhoI-digested pSC-B-tU1_RPA to label the antisense tU1 RNA. The radio-labelled RNA was purified from 6% denaturing gel.
In vitro J2 immunoprecipitation assay
In brief, 2,000 cps [α-32P]UTP-labelled antisense tU1 RNA was incubated with 10 μg of purified HeLa nuclear RNA followed by RNase protection analysis (RPA)31. After RPA, dsRNA was immuno-precipitated with 5 μg of J2 antibody conjugated protein G beads (Thermo Fisher Scientific) in NET-2 buffer. [α-32P]-labelled antisense tU1 RNA was used as ssRNA substrate. The antibody beads were washed with NET-2 buffer several times and then incubated with Trizol (Thermo Fisher Scientific) to purify immunoprecipitated RNAs. The RNAs were analysed on 8% denaturing gel.
mtRNA isolation and treatment of cells
Mitochondria were isolated from HeLa cells using magnetic cell separation procedure as described by the manufacturer (Mitochondria Isolation Kit; MACS, Miltenyl Biotec). RNA was purified from mitochondria using Trizol reagent (Sigma) and was treated with RNasefree TurboDNase (Ambion) according to manufacturer’s instructions. 1 μg of mtRNA was transfected into HeLa cells and 300 ng was transfected into MEFs in a 1:3 ratio with Lipofectamine 2000 in 12-well plates with cells at 80% confluency. For enzymatic treatment, 1 μg of mtRNA was incubated with RNase III as per the manufacturer’s instructions. 100 ng of ppp-IVT-RNA99nt and CIP-EMCV-RNA were transfected in MEFs in 12-well plates using Lipofectamine 2000 in a 1:3 ratio. Total RNA from HeLa cells or MEFs was extracted 20 h after transfection for IFNB1 or Ifit1 mRNA quantification, respectively. In Extended Data Fig. 6b, HeLa cells were treated with ABT-737 (10 μM) or DMSO 65 h after siRNA transfection and incubated for a further 8 h. Total RNA was isolated using Trizol for IFNB1 mRNA quantification.
Separation of cytoplasm and mitochondria fractions
Mitochondrial and cytoplasmic fractions in Extended Data Fig. 6c were prepared as previously described32. Purity of fractions was tested by western blot according to standard protocols.
Detection of mtRNA in cytosolic extracts
Cytosolic extracts (Fig. 3c) were prepared using digitonin extraction as described previously8. Digitonin (Sigma) at 25 μg ml−1 was used for control and patient fibroblast cells. Purity of cytosolic fractions were tested by western blot, Lamin A/C was probed as a nuclear loading control, Calnexin was probed for endoplasmic reticulum, COX IV was probed for mitochondria and Tubulin was probed as a cytoplasmic control. RT–qPCR was performed on RNA isolated from cytosolic fractions using random hexamers for cDNA synthesis followed by qPCR using mtDNA-specific primers (Extended Data Table 2) normalized to ACTB mRNA levels.
RNA sequencing
RNA sequencing was performed by the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics, University of Oxford. Input RNA samples were ribo-depleted with Ribo-Zero rRNA-removal kit (Human/Mouse/Rat, EpiCentre RZH110424). Immunoprecipitated RNA samples were not ribo-depleted. Libraries were prepared with the NEBNext Ultra Directional RNA Library Prep Kit for Illumina, v1.0 (cat. no. E7420) according to the manufacturer’s guidelines. Libraries were sequenced on an Illumina HiSeq-2000 with 100-bp paired-end reads, v3 chemistry.
Microarray method
Hepatocytes were isolated from perfused livers of two PNPASE (Pnpt1) liver-specific knockout C57BL/6 mice (HepKO; one male aged 12.9 weeks, one female aged 4.29 weeks, two independent experiments) and two sex-matched wild-type littermate mice. Total RNA was extracted from hepatocytes using TriZol Reagent (Invitrogen cat. #15596026) and the Qiagen RNeasy Mini Kit (Qiagen, cat. #74104). Labelled complementary RNA was generated using 200 ng of total RNA from each sample and the Agilent Low RNA Input Linear Amplification labelling kit. Each labelled sample was hybridized against its gender-matched sample in fluor-reversed pairs of arrays to an Agilent 4×44k Mouse Whole Genome Microarray. The arrays were scanned using the Agilent DNA Microarray Scanner, and data were extracted using the Agilent Feature Extraction (v.9.5.1.1) software using the standard Agilent protocol except without Lowess normalization. The fluor-reversed pairs were combined into Experiments in Rosetta Resolver 7.1 to produce the male and female signature gene ratios. We performed age and gender-matched differential expression analysis and generated a list of signature genes with significant differential expression in both the male and female cohorts. Both male and female cohorts showed very similar results. For simplicity, only fold expression changes in female mice were shown.
Interferon reactome methods
The gene set for interferon signalling (encompassing IFNα/β/γ signalling and interferon-stimulated antiviral response) was extracted from the Reactome database under pathway ID R-MMU-91353133. An additional antiviral innate immune response gene set was curated from recent studies investigating the role of mitochondrial DNA in innate immunity8. Corresponding genes from the set were compared to the HepKO signature genes (adjusted P < 0.05, significant in both female and male matched pairs). Overlapping genes were plotted using the male and female matched log2(fold change). Both male and female data showed very similar results. For simplicity, only fold expression change data from female mice were shown.
Immuno-electron microscopy
HeLa cells were grown in a 6-well plate and siRNA treated for 65 h, trypsinized and pelleted for 1 min at 1,000g in cell culture media containing 5% FBS and 20% BSA. Cells were then immediately cryofixed using a Leica EM PACT2 high pressure freezer and then further processed as described34, except that tannic acid was omitted from the freeze substitution medium. Blocks were sectioned on a Leica UC7 ultramicrotome using a diamond knife (Diamtome). 90-nm sections were transferred to 200-mesh Nickel grids and then immunolabelled as follows: grids were floated section side down on a 20 μl droplet of blocking solution (10% goat serum in TBS) for 15 min, then blotted and incubated on a droplet of primary (J2) antibody (diluted 1:25 in buffer A (1% BSA, 1% goat serum, 0.01% Tween-20 in TBS)) for 2 h at room temperature. Grids were washed by passing them over five droplets of buffer A, 5 min each, then incubated with secondary antibody (ab27242, goat anti-mouse conjugated to 20 nm gold) diluted 1:10 in buffer A for 90 min at room temperature, then washed by passing over three droplets of water. Sections were then post-stained for 10 min with uranyl acetate and Reynold’s lead citrate and imaged on a Tecnai 12 transmission electron microscope (FEI) equipped with a Gatan OneView CMOS camera.
Statistical analysis
Unless otherwise stated, the figures present the mean values of at least three independent experiments with s.d. or s.e.m. For analyses with n > 10, individual data points are shown. The mean is reported when n = 2, and no other statistics were calculated for these experiments.
Bioinformatics: mapping of sequencing reads and data visualization
Paired-end reads for each sample were mapped to the human genome reference assembly GRh37/hg19 (build 37.2, Feb 2009) using the Bowtie2 alignment software35. Prior to alignment, adaptor sequences were trimmed using Cutadapt 1.8.3, discarding reads with less than ten bases. An in-house Perl script was used to remove the reads left unpaired (code available upon request). SAMtools 1.236 was used to process aligned reads to only include uniquely mapped reads with no more than two mismatches. Data were scaled to library size (genomeCoverageBed) using Bedtools37. Bigwig track files were generated from the Bowtie2 output files using UCSC bedGraphToBigWig tool38. Correlations among different samples for chrM were calculated with R. Data from replicates (n = 2 for each condition) except untreated and control-siRNA-treated samples (r = 0.85) were then merged and viewed on the UCSC genome browser (Extended Data Fig. 5a, b). For the chromosome-wise read coverage plot of dsRNA-seq, the number of filtered reads mapping for each chromosome were counted. These numbers were then normalized to the size of the respective chromosome. For plotting the distribution of reads belonging to different RNA species, reads mapped to each Ensembl biotype annotations were counted for using bedtools and then normalized to the size of the genome. The heat map was generated with the R package gplots using a subset of significantly altered ISGs identified by gene expression analysis on total RNA sequencing (input RNA) from control siRNA (siRNA targeting the luciferase gene) and two knockdown conditions (siSUV3 and siPNPase).
To screen for RNA editing candidates in chrM, the REDItoolDenovo.py script from the REDItools package39 was used. To avoid the risk of using unreliable editing sites, the output of REDItools was passed through two stringent filters. First, all editing sites were removed that had less than ten edited counts and a total read coverage <50 for all the knockdown libraries. Second, only those sites in which the frequency of editing by siRNA targeting SUV3 and PNPase was at least 1.5-fold that of control siRNA were considered. The resulting editing sites were then filtered for known SNPs in the mitochondrial transcriptome40 to obtain reliable RNA editing candidates.
Quantification of Neopterin levels in CSF
CSF pterins were analysed by reverse-phase high-performance liquid chromatography with fluorescence detection according to41.
Extended Data
Extended Data Fig. 1. Characterization of anti-dsRNA J2 antibody and mtDNA depletion results in loss of mtdsRNA formation.
a, RT–qPCR analysis of L-mRNA expression in encephalomyocarditis virus (EMCV) infected HeLa cells at MOI 1 at the indicated time points after infection. Data are from two independent experiments. b, Confocal microscopy images of uninfected or EMCV-infected HeLa cells at multiplicity of infection (MOI) of 1, 8 h after infection stained with anti-dsRNA (J2) antibody (green) and DAPI (nuclei stained blue). Images are representative of two experiments. Scale bars, 10 μm. c, Immunostaining of dsRNA (green) and DNA (red) in HeLa cells treated with indicated nucleases before staining. Signal from J2 antibody is specific for RNA but not for DNA and is sensitive only to RNase III treatment. Images are representative of three experiments. Scale bars, 10 μm. d, Quantification of fluorescence signal from HeLa cells treated as in c. Data are mean ± s.e.m. from 4,095, 1,755, 4,766 and 5,585 cells for the untreated, RNase T1, RNase III and DNase Turbo groups, respectively. e, Autoradiogram showing substrate specificity of J2 on the basis of immunoprecipitation efficiency for uniformly 32P-radiolabelled ssRNA and dsRNA substrates. Signals were visualized and quantified by PhosphorImager. The level of immunoprecipitation signal is shown and expressed as the percentage of input. Images and data are representative of two experiments. For gel source data, see Supplementary Fig. 1. f, Chromosome-wise coverage plot of dsRNA-seq reads. Inset, read distribution of dsRNA-seq on the basis of RNA class biotypes. g, Left, dsRNA and DNA staining of HeLa cells transfected with constructs encoding the indicated proteins, the expression of which results in mtDNA depletion. Plasmids encoding mtDNA-depletion factors co-express EGFP from an independent promoter, which enables identification of transfected cells. Mitochondria were stained using anti-OXA1L antibody. Scale bars, 10 μm. Right, quantitative analysis of fluorescence signal from HeLa cells. Data are mean ± s.e.m. from ten cells.
Extended Data Fig. 2. RNA degradosome components SUV3 and PNPase are involved in mtdsRNA turnover.
a, HeLa cells treated with DMSO, DRB (100 μM) and actinomycin-D (0.5 μg ml−1) for 60 min and stained with anti-dsRNA (J2) antibody (green). Mitochondria were stained with MitoTracker Red CMXRos and nuclei with DAPI (blue) (representative of two experiments). b, Flow cytometric analysis of dsRNA levels in HeLa cells treated with the indicated siRNAs. Cells were labelled with J2 antibody or an isotype control. Data are mean ± s.d. from three independent experiments. c, Left, detection of dsRNA with J2 antibody in HeLa cells after depletion of PNPase or SUV3 by On-TARGETplus siRNAs (indicated with an asterisk and listed in Extended Data Table 2). Mitochondria were stained with MitoTracker Deep Red. Nuclei are stained with Hoechst (blue). Scale bars, 10 μm. Right top, western blot showing PNPase or SUV3 depletion. Blots are representative of four experiments. For gel source images, see Supplementary Fig. 1. Far right top, Quantification of dsRNA levels in HeLa cells depleted of PNPase or SUV3. Data are mean ± s.d. from four independent experiments. d, Quantitative analysis of fluorescent signals from dsRNA in HeLa cells with depleted enzymes involved in mitochondrial nucleic acids metabolism. Data are mean ± s.d. from four independent experiments. e, HeLa cells were transfected with siRNA specific for PNPase, SUV3, or non-targeting control. Prior to fixation, cells were treated for indicated times with inhibitors of transcription: actinomycin-D (0.5 μg ml−1) or DRB (100 μM). Immunostaining of dsRNA was performed and cells were imaged using a fluorescent microscope screening station. Data are mean ± s.d. from four independent experiments.
Extended Data Fig. 3. Unwinding activity of SUV3 is required to suppress mtdsRNA accumulation.
a, Left, confocal images of HEK 293 cells expressing stably integrated wild-type SUV3 (hSUV3_WT) and the catalytically inactive (G207V) dominant negative form (hSUV3_G207V) stained with J2 ab (green). Mitochondria stained with MitoTracker Deep Red (red). Nuclei stained with Hoechst (blue). Right, quantitative analysis of fluorescence signal from HEK 293 cells in the above experiment. Data are mean ± s.e.m. from 16 cells. b, Top, northern blots of J2 immunoprecipitated dsRNA from hSUV3_WT and hSUV3_G207V overexpressing HEK 293 cell lines with four different probes spanning the entire mitochondrial genome. Bottom, diagram depicts positions of probes on mitochondrial genome. Blots are representative of two experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 4. Exonuclease activity of PNPase is required to suppress mtdsRNA formation.
a, Diagram of PNPase domain structure showing the position of the R445E/R446E mutation in the RNasePH domain. b, Immunostaining of dsRNA (green) in HeLa stable cell lines transfected with siRNA specific for PNPase or non-targeting control siRNA. Depletion of endogenous PNPase was rescued by expression of siRNA-resistant PNPase-FLAG protein (wild-type or mutated (RNA-degradation deficient version of PNPase (R445E/R446E) was expressed)). Mitochondria are stained with MitoTracker Deep Red. Nuclei are stained with Hoechst (blue). Scale bars, 20 μm. c, Western-blot analysis of PNPase in HeLa cells treated as in b. Exogenous, siRNA-resistant PNPase is expressed as a FLAG fusion. Blots are representative of three experiments. d, Quantitative analysis of fluorescent signals from dsRNA in HeLa treated as in b. Data are mean ± s.d. from three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 5. dsRNA-seq of HeLa cells following siRNA depletion of SUV3 and PNPase.
a, dsRNA-seq reads across the mitochondrial genome spanning entire protein coding region (~3.5–16 kb) following siRNA treatment. Data are from two independent experiments. H-strand genes are shown as blue bars and L-strand as red bars. Short tRNA genes are denoted with T as the first letter. b, Correlation plots of J2 immunoprecipitation dsRNA-seq replicates. Pearson correlation coefficients are calculated and shown on each plot.
Extended Data Fig. 6. Upregulation of ISGs in HeLa and murine cells following loss of PNPase accentuated by mitochondrial outer membrane permeabilization.
a, Heat map of ISGs generated from a subset extracted from a list of significantly expressed genes in siRNA treated HeLa cells. Gene expression is depicted by colour intensity. Green denotes upregulation and red downregulation. b, RT–qPCR analysis of IFNB1 mRNA in HeLa cells treated with indicated siRNAs and then 8 h of treatment with vehicle or ABT-737. Data are the mean of two independent experiments. c, Western-blot analysis of the cytochrome c (cyt c) release into the cytoplasm of HeLa cells treated with vehicle or ABT-737 for 8 h. Subcellular fractionation purity confirmed by relevant markers. Blots are representative of two experiments. d, log2(fold change) expression levels of ISGs and genes involved in interferon signalling in HepKO versus wild-type female mice. ISG list is based on the Reactome database33. e, Western blot of whole-cell extracts from cells treated with the indicated siRNAs. Blots are representative of two experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 7. RNA editing of cytoplasmic mtRNA.
a, RNA editing sites mapped on the RNA transcriptome of SUV3 and PNPase depleted cells are shown. Each dot represents an editing event. Dots on the upper panel denote editing events on the H-strand and dots on the lower panel denote editing on the L-strand. Triangle denotes single SUV3 editing site. Yellow bars denote the D-loop region. Short red bars denote tRNA genes on the L-strand and green bars denote tRNA genes on the H-strand. b, Frequency of dinucleotide RNA editing sites mapped in the PNPase depleted samples. c, RT–qPCR analysis of IFNB1 mRNA levels in indicated siRNA-treated cells. Data are the mean from two independent experiments. d, Western blot of ADAR1, SUV3 and PNPase in cell treated with the indicated siRNAs. Blots are representative of two experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 8. EMCV infection results in dsRNA accumulation that partially overlaps with mitochondria.
a, Left, confocal images of EMCV-infected HeLa cell at MOI 1, 6 h after infection, stained with anti-dsRNA (J2) antibody. Mitochondria are stained with MitoTracker Red CMXRos and nucleus with DAPI. Right, line scan RGB profile for the region of interest (ROI) selected with a white line is shown on the right. Data are representative of two experiments. b, Expanded view of the ROI of an EMCV-infected HeLa cell showing colocalization of dsRNA with mitochondria. Image is representative of two experiments. Scale bars, 10 μm.
Extended Data Table 1. Clinical table of patients carrying PNPT1 mutations.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |||
|---|---|---|---|---|---|---|
| Nucleotide substitutions | c.407G>A hom | c.208T>C het | c.2137G>T het | c.1495G>C het | c.1519G>T het | c.1160A>G hom ref14 |
| Amino acid | p.Arg136His | p.Ser70Pro | p.Asp713Tyr | p.Gly499Arg | p.Ala507Ser | p.Gln387Arg |
| SIFT | Deleterious (score: 0, median: 3.48) | Tolerated (score: 0.1, median: 3.48) | Deleterious (score: 0.01, median: 3.47 | Deleterious (score: 0.0, median: 3.49) | Deleterious (score: 0.0, median: 3.49) | ref14 |
| Mutation Taster | Disease causing (p-value:1) | Disease causing (p-value:0.996) | Disease causing (p-value:1) | Disease causing (p-value:1) | Disease causing (p-value:1) | ref14 |
| Polyphen | Probably damaging with score of 1.0 (sensitivity: 0.0; specificity: 1.0) | Probably damaging with score of 0.588 (sensitivity: 0.81; specificity: 0.83) | Probably damaging with score of 0.998 (sensitivity: 0.18; specificity: 0.98) | Probably damaging with score of 0.994 (sensitivity: 0.46; specificity: 0.96) | Probably damaging with score of 0.913 (sensitivity: 0.69; specificity: 0.90) | ref14 |
| Splicing prediction | Predicted change at 5’ss (-1G>C): -54.0% MaxEnt: -100.0%; NNSPLICE:49.1%: HSF:-12.9% | |||||
| ClinVar | RCV000197606.4 | RCV000033022.3 | ||||
| dbSNP | rs746356243 | - | - | - | - | - |
| gnomAD | 0.0000244 (6 hets /245,972 alleles) | Not recorded (>230,000 alleles) | Not recorded (>230,000 alleles) | Not recorded (>230,000 alleles) | 0.0002241 (62 hets/276,714 alleles) | Not recorded (>230,000 alleles) |
| Clinical features | Intrauterine growth retardation | Truncal hypotonia | Hypotonia | ref14 | ||
| Congenital cataract | Nystagmus | Abnormal eye movements | ||||
| Feeding difficulties | Failure to thrive | Sensory neuropathy | ||||
| Deafness | Sensorineural deafness | Deafness | ||||
| Truncal hypotonia | Hypertonia of the lower limbs | Leucodystrophy | ||||
| Nystagmus | ||||||
| Death at age 2 years | Alive at age 1 years | Alive at age 7 years | Alive at age 13 years | |||
| Brain Imaging | Anomalies of putamen and basal ganglia | Delayed myelination | Delayed myelination; abnormal corpus callosum | ref14 | ||
| Metabolic workup | NMR spectroscopy: lactate peak | NMR spectroscopy: lactate peak | NMR spectroscopy: No lactate elevation | ref14 | ||
| Hyper-lactatemia | Hyper-lactatemia | |||||
| Hyper-lactatorachia | ||||||
| NA | High neopterin in CSF (101 nmol/l) | ND | ND | |||
| Interferon score* (blood analysis) | NA | 4.138 | 2.997 | 2.413 | ||
| RC analysis | Multiple RC deficiency in muscle and fibroblasts | Multiple RC deficiency in fibroblasts | Normal RC activities in fibroblasts | ref14 | ||
het = heterozygous; hets = heterozygotes; hom = homozygous; RC = respiratory chain; NA = not available; ND = not determined; *Normal interferon score value is <2.466. SIFT, Sorting intolerant from tolerant (http://sift.bii.a-star.edu.sg/www/SIFT_aligned_seqs_submit.html)
Extended Data Table 2. Oligonucleotide primers and siRNAs used in the study.
| Primer (qRT-PCR) | Gene | Sequence (5’-3’) |
|---|---|---|
| IFN-β forward | IFNB1 | ATGACCAACAAGTGTCTCCTCC |
| IFN-β reverse | IFNB1 | GCTCATGGAAAGAGCTGTAGTG |
| L-mRNA EMCV fwd | L-mRNA EMCV | GCGCACTCTCTCACTTTTGA |
| L-mRNA EMCV rev | L-mRNA EMCV | TCGAAAACGACTTCCATGTCT |
| β-actin mRNA forward | ACTB | CTGTGGCATCCACGAAACTA |
| β-actin mRNA reverse | ACTB | AGTACTTGCGCTCAGGAGGA |
| COX1 forward | MT-CO1 | ACGTTGTAGCCCACTTCCAC |
| COX1 reverse | MT-CO1 | TGGCGTAGGTTTGGTCTAGG |
| ND5 forward | MT-ND5 | TCGAAACCGCAAACATATCA |
| ND5 reverse | MT-ND5 | CAGGCGTTTAATGGGGTTTA |
| ND6 forward | MT-ND6 | CCAATAGGATCCTCCCGAAT |
| ND6 reverse | MT-ND6 | AGGTAGGATTGGTGCTGTGG |
| CYB forward | MT-CYB | AGACAGTCCCACCCTCACAC |
| CYB reverse | MT-CYB | GGTGATTCCTAGGGGGTTGT |
| ms Ifit1 forward | Ifit1 | CAAGGCAGGTTTCTGAGGAG |
| ms Ifit1 reverse | Ifit1 | GACCTGGTCACCATCAGCAT |
| ms Gapdh forward | Gapdh | GACTTCAACAGCAACTCCCAC |
| ms Gapdh reverse | Gapdh | TCCACCACCCTGTTGCTGTA |
| siRNA | Gene (alternative name) | List of the StealthRNA oligos |
| siPNPase | PNPT1 (PNPase) | HSS131758 |
| siSUV3 | SUPV3L1 (SUV3) | HSS110378 |
| siMRPP1 | TRMT10C (MRPP1) | HSS123550 |
| siMDA5 | IFIH1 (MDA5) | HSS127414 |
| siTLR3 | TLR3 | HSS110815 |
| siDDX28 | DDX28 | HSS125053 |
| siEXOG | EXOG | HSS115058 |
| siMGME1 | MGME1 | HSS132389 |
| si FEN1 | FEN1 | HSS103629 |
| siRNA | Gene | siRNA Forward Strand |
| siCntrl | Luciferase | GAUUAUGUCCGGUUAUGUAUU |
| siRIG-I | DDX58 (RIG-I) | ACGGAUUAGCGACAAAUUUAA |
| siADAR1 | ADAR (ADAR1) | sc-37657 |
| ON-TARGETplus Human BAK | BAK | L-003305-00-0005 |
| ON-TARGETplus Human BAX | BAX | L -003308-01-0005 |
| ON-TARGETplus Human MAVS | MAVS | L-024237-00-0005 |
| ON-TARGETplus Human SUV3 * | SUPV3L1 | L-017841-01-0020 |
| ON-TARGETplus Human PNPase * | PNPT1 | L-019454-01-0020 |
Supplementary Material
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-018-0363-0.
Acknowledgements
We acknowledge V. Bondet for CSF IFNα data. We also thank E. Johnson and A. Pielach from the Dunn School Bioimaging facility for electron microscopy work and M. Alexeyev for sharing plasmids encoding UL12.5M185 and mUNG1 (Addgene #70109 and #70110, respectively). This work was supported by funding to N.J.P. (Wellcome Trust Investigator Award (107928|Z|15|Z), ERC Advanced Grant (339270)) and to M.T. (National Institutes of Health (NIH) (GM073981)). Y.J.C. acknowledges funding from the European Research Council (GA 309449: fellowship), a state subsidy managed by the National Research Agency (ANR, France) under the Investments for the Future (ANR-10-IAHU-01), and an ANR grant CE17001002 to Y.J.C. and D.D. Y.J.C. and D.D. thank ImmunoQure AG for sharing of the antibodies used to assess IFNα protein levels in the Simoa assay. Studies were supported by grant 2014/13/D/NZ2/01114 (to R.J.S.) from the National Science Centre, Poland. Experiments were carried out with the use of CePT infrastructure financed by the European Union through the European Regional Development Fund (Innovative economy 2007–13, Agreement POIG.02.02.00-14-024/08-00).
Footnotes
Reporting summary. Further information on experimental design is available in the Nature Research Reporting Summary linked to this paper.
Data availability. Mouse microarray data have been deposited in the Gene Expression Omnibus (GEO) under accession numbers GSE94957 and GSE109210. Source Data for graphical representations obtained from the PNPase HepKO mouse microarray (Fig. 2d, e, Extended Data Fig. 6d) are available with the online version of the paper. Gel source images are presented in Supplementary Fig. 1.
Reviewer information Nature thanks C. Reis e Sousa, S. Riis Paludan, G. Shadel and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author contributions A.Dh. conceived the study, designed and performed most of the experiments and drafted the paper. S.D. performed the bioinformatics analysis. L.S.B., R.J.S. and A.Dz. performed the experiments in Fig. 1c–f and Extended Data Figs. 1c, d, g, 2c–e, 3, 4. L.J. and M.T. generated the immunofluorescence and gene expression data on PNPase HepKO. A.R., A.M. and M.S. provided patient fibroblasts and the clinical data. C.T. provided the PNPase western blot (Fig. 3a). Y.J.C. and G.I.R. generated the blood ISG expression data. D.D. provided the IFNα CSF data. T.N. provided Extended Data Fig. 1e. C.R.A. performed FACS analysis. J.R. provided the MEF KO cells and agonists. A.Dh. discussed and interpreted results with inputs from N.J.P., R.J.S. and the other authors. N.J.P. and A.Dh. wrote the paper with input from the other authors.
Competing interests The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41586-018-0363-0.
Reprints and permissions information is available at http://www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aloni Y, Attardi G. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc Natl Acad Sci USA. 1971;68:1757–1761. doi: 10.1073/pnas.68.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young PG, Attardi G. Characterization of double-stranded RNA from HeLa cell mitochondria. Biochem Biophys Res Commun. 1975;65:1201–1207. doi: 10.1016/s0006-291x(75)80357-3. [DOI] [PubMed] [Google Scholar]
- 3.Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Human mitochondrial RNA decay mediated by PNPase–hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013;41:1223–1240. doi: 10.1093/nar/gks1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGlasson S, Jury A, Jackson A, Hunt D. Type I interferon dysregulation and neurological disease. Nat Rev Neurol. 2015;11:515–523. doi: 10.1038/nrneurol.2015.143. [DOI] [PubMed] [Google Scholar]
- 7.Rongvaux A, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West AP, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spadafora D, Kozhukhar N, Chouljenko VN, Kousoulas KG, Alexeyev MF. Methods for efficient elimination of mitochondrial DNA from cultured cells. PLoS ONE. 2016;11:e0154684. doi: 10.1371/journal.pone.0154684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szczesny RJ, et al. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornig-Do HT, et al. Isolation of functional pure mitochondria by superparamagnetic microbeads. Anal Biochem. 2009;389:1–5. doi: 10.1016/j.ab.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 14.Vedrenne V, et al. Mutation in PNPT1, which encodes a polyribonucleotide nucleotidyltransferase, impairs RNA import into mitochondria and causes respiratory-chain deficiency. Am J Hum Genet. 2012;91:912–918. doi: 10.1016/j.ajhg.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matilainen S, et al. Defective mitochondrial RNA processing due to PNPT1 variants causes Leigh syndrome. Hum Mol Genet. 2017;26:3352–3361. doi: 10.1093/hmg/ddx221. [DOI] [PubMed] [Google Scholar]
- 16.Rodero MP, et al. Detection of interferon alpha protein reveals differential levels and cellular sources in disease. J Exp Med. 2017;214:1547–1555. doi: 10.1084/jem.20161451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murr C, Widner B, Wirleitner B, Fuchs D. Neopterin as a marker for immune system activation. Curr Drug Metab. 2002;3:175–187. doi: 10.2174/1389200024605082. [DOI] [PubMed] [Google Scholar]
- 18.White MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mannion NM, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hart T, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 21.West AP, Shadel GS. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol. 2017;17:363–375. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oda H, et al. Aicardi-Goutières syndrome is caused by IFIH1 mutations. Am J Hum Genet. 2014;95:121–125. doi: 10.1016/j.ajhg.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice GI, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 25.Rutsch F, et al. A specific IFIH1 gain-of-function mutation causes Singleton- Merten syndrome. Am J Hum Genet. 2015;96:275–282. doi: 10.1016/j.ajhg.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lincez PJ, Shanina I, Horwitz MS. Reduced expression of the MDA5 gene IFIH1 prevents autoimmune diabetes. Diabetes. 2015;64:2184–2193. doi: 10.2337/db14-1223. [DOI] [PubMed] [Google Scholar]
- 27.Gorman JA, et al. The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat Immunol. 2017;18:744–752. doi: 10.1038/ni.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hertzog J, et al. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur J Immunol. 2018 doi: 10.1002/eji.201847483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deddouche S, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. eLife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer S, et al. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell. 2016;166:582–595. doi: 10.1016/j.cell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nojima T, Dienstbier M, Murphy S, Proudfoot NJ, Dye MJ. Definition of RNA polymerase II CoTC terminator elements in the human genome. Cell Reports. 2013;3:1080–1092. doi: 10.1016/j.celrep.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimura N, Yano M. Separation of the inner and outer mitochondrial membrane in HeLa cells. Bio Protoc. 2014;4:e1299. [Google Scholar]
- 33.Croft D, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson E, et al. Correlative in-resin super-resolution and electron microscopy using standard fluorescent proteins. Sci Rep. 2015;5 doi: 10.1038/srep09583. 9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Picardi E, Pesole G. REDItools: high-throughput RNA editing detection made easy. Bioinformatics. 2013;29:1813–1814. doi: 10.1093/bioinformatics/btt287. [DOI] [PubMed] [Google Scholar]
- 40.Hodgkinson A, et al. High-resolution genomic analysis of human mitochondrial RNA sequence variation. Science. 2014;344:413–415. doi: 10.1126/science.1251110. [DOI] [PubMed] [Google Scholar]
- 41.Blau N, et al. Cerebrospinal fluid pterins and folates in Aicardi–Goutieres syndrome: a new phenotype. Neurology. 2003;61:642–647. doi: 10.1212/01.wnl.0000082726.08631.e7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.