Summary
Somatic mutations arising in human skin cancers are heterogeneously distributed across the genome, meaning that certain genomic regions (e.g., heterochromatin or transcription factor binding sites) have much higher mutation densities than others. Regional variations in mutation rates are typically not a consequence of selection, as the vast majority of somatic mutations in skin cancers are passenger mutations that do not promote cell growth or transformation. Instead, variations in DNA repair activity, due to chromatin organization and transcription factor binding, have been proposed to be a primary driver of mutational heterogeneity in melanoma. However, as we review here, recent studies indicate that chromatin organization and transcription factor binding also significantly modulate the rate at which UV lesions form in DNA. We propose that local variations in lesion susceptibility may be an important driver of mutational hotspots in melanoma and other skin cancers, particularly at binding sites for ETS transcription factors.
Keywords: mutational heterogeneity, nucleotide excision repair, DNA lesions, pyrimidine dimer, mutation hotspots, melanoma, ETS transcription factors, Nucleosome
1. Introduction
Exposure to ultraviolet (UV) light is the principal etiological agent of melanoma and other forms of skin cancer. UV light causes extensive damage to cellular DNA, primarily in the form of cyclobutane pyrimidine dimers (CPDs) and 6–4 pyrimidine-pyrimidone photoproducts (6–4PPs), which both occur at dipyrimidine sequences (i.e., TT, TC, CT, and CC).[1] Genome sequencing of cutaneous melanomas has revealed that most somatic mutations in these cancers are C>T transitions occurring within dipyrimidine sequences, consistent with these mutations arising from UV-induced DNA lesions.[2–4] Moreover, individuals who are unable to repair CPD and 6–4PP lesions, due to defects in the nucleotide excision repair (NER) pathway (i.e., xeroderma pigmentosum (XP) patients), have up to ~10,000-fold higher rates of skin cancer.[5–7] These findings indicate that UV-induced CPDs and 6–4PPs are the likely cause of a high fraction of somatic mutations in skin cancers, such as melanoma, including many ‘driver’ mutations that contribute to carcinogenesis.[8, 9]
DNA sequencing of individual tumors has revealed that the density of somatic mutations in melanoma and other cancers varies significantly across the genome.[10–12] Regional differences in mutation rates have been attributed to differences in chromatin state (e.g., heterochromatin versus euchromatin), transcription activity, and replication timing.[10, 11, 13, 14] Some of these features (i.e., chromatin state and transcription activity) are thought to influence mutation rates in skin cancers by regulating the efficiency of lesion removal by the NER pathway (e.g., [12, 15, 16]). For example, regions of heterochromatin characterized by repressive histone post-translational modifications are thought to significantly reduce NER activity,[15] leading to the persistence of mutagenic CPD lesions in heterochromatin. Even individual nucleosomes, which are the building blocks of chromatin, can inhibit NER activity,[17–22] thereby potentially stimulating mutagenesis.
While regional differences in mutation rates affect large (i.e., mega-base pair) segments of the genome, significant variations in mutation rates have also been detected at very localized genomic features, such as transcription factor binding sites (TFBS). It has been recently reported that mutation rates are generally elevated at TFBS and other promoter elements in a number of cancers.[23–25] Elevated mutation rates at TFBS may in some cases be due to driver mutations under selection, such as occur in the promoter of the human telomerase reverse transcriptase (TERT) gene.[26–29] However, most mutations at TFBS, even those that are highly recurrent in cancers (e.g., [30, 31]), are likely passenger mutations that are not under selection.
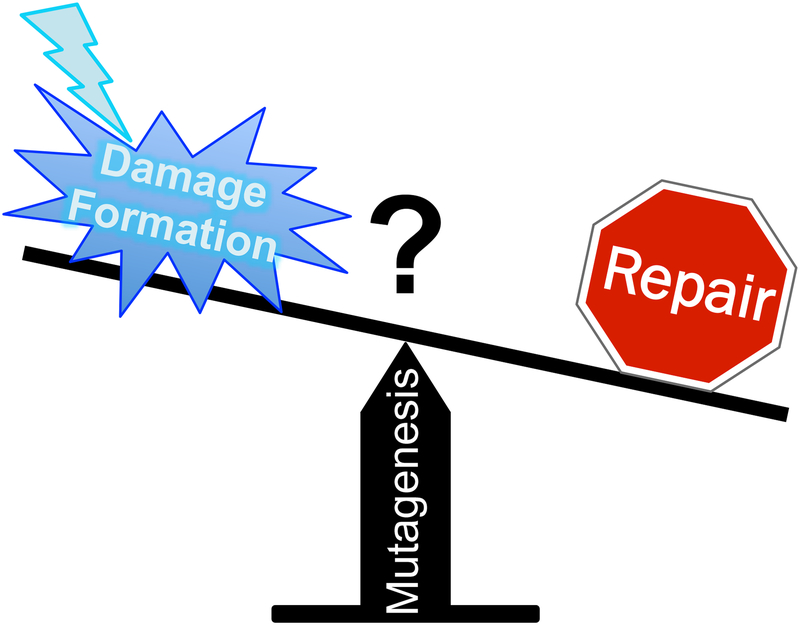
It has been proposed that transcription factor (TF) binding stimulates the frequency of mutations by inhibiting the repair of mutagenic lesions resident in TFBS.[24] Repair inhibition has been proposed as a general mechanism promoting mutagenesis in heterochromatin and TFBS in a variety of cancers (Fig. 1), but this model is primarily based on data measuring the activity of the NER pathway in repairing UV-induced CPDs and 6–4PPs in human skin cells,[15, 32, 33] as well as the distribution of somatic mutations in NER-deficient skin cancers.[25] However, recent studies indicate that chromatin and TF binding can also significantly affect the initial yield of UV damage,[22, 34–36] leading to elevated CPD levels at certain classes of TFBS, which could promote mutagenesis at these sites (Fig. 1). In this article, we evaluate the relative contributions of repair inhibition and UV damage susceptibility to mutational hotspots and heterogeneity in skin cancer.
Figure 1:
Schematic showing the potential importance of UV damage formation (left side of scale) versus repair inhibition (stop sign on right side of scale) in promoting mutagenesis in skin cancers. The prevailing view is that repair inhibition is a primary driver of mutagenesis in skin cancers, but recent studies suggest variations in UV damage formation may also be an important driver of recurrent mutagenesis.
2. Mutations in skin cancer genomes are heterogeneously distributed.
Initial attempts to understand the mechanisms that underlie mutational heterogeneity focused on correlating mutation densities in cancer genomes with larger scale features of the human genome. These efforts revealed replication timing as a key correlate with mutation density in cancer.[13, 14] Similarly, chromosomal regions associated with reduced transcriptional activity or histone modifications indicative of heterochromatin also correlated with higher mutation densities in multiple cancer types including melanoma.[10, 11, 37] This association of melanoma mutagenesis with transcriptional activity suggested that differential NER activity was likely a major determinant governing melanoma mutation distributions. The effects of transcription-coupled NER on melanoma mutagenesis were already evident based upon the bias of melanoma mutations that occur on the non-transcribed strand of genes.[2] Interestingly, the difference in the density of mutations between transcribed and non-transcribed strands of genes is not sufficient to account for the large differences in mutation density seen between highly and lowly transcribed regions.[2, 11] Analysis of mutation densities in cutaneous squamous cell carcinomas similarly indicated that individuals with fully functional NER had higher levels of mutation in poorly transcribed regions of heterochromatin (i.e., high H3K9 tri-methylation levels) than in highly transcribed regions. As in melanomas, the activity of transcription-coupled NER appeared insufficient to explain the regional disparity in mutation. The density of mutation in transcribed regions of these cutaneous squamous cell carcinomas were reduced on both the transcribed and non-transcribed strands, indicating that transcription was by some means facilitating the repair of UV lesions on both DNA strands. Contrastingly, tumors from patients deficient in global genomic NER (due to loss of the XPC protein) lacked the expression-dependent differences in mutation density on the non-transcribed strand, despite maintaining a reduced number of mutations on the transcribed strand of genes, due to the activity of transcription-coupled NER.[16] These results indicate that either transcription or the related more accessible chromatin state in transcribed regions influences the ability of global genomic NER to access and repair lesions on both DNA strands, thereby preferentially protecting these regions from mutagenesis.
3. NER efficiency is influenced by chromatin compaction and correlates with melanoma mutation.
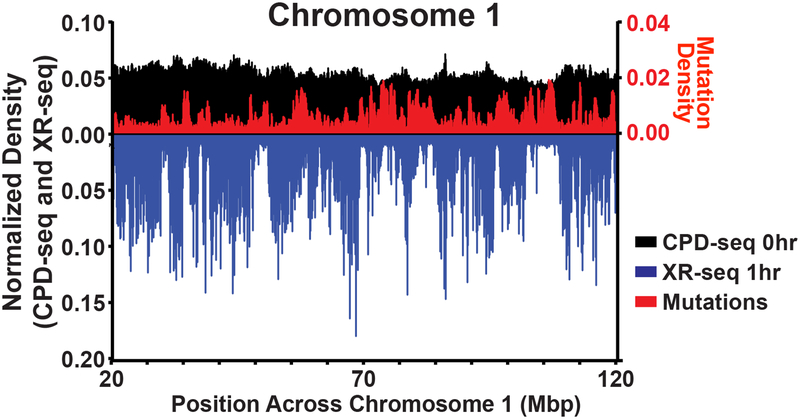
Recent development of high-throughput sequencing methods to map NER repair activity across the human genome has allowed the direct assessment of how large-scale features of chromosomes and chromatin domains influence the efficiency of NER and correlate with mutation density in cancers.[15] The excision repair sequencing (XR-seq) method,[33] which determines the location of ~20–30 nt excision fragments generated during NER-mediated removal of CPDs or 6–4PPs, has confirmed that NER activity is higher on the transcribed strands of genes across the genome due to the action of transcription-coupled repair, but also has revealed how chromatin state influences the kinetics of repair. XR-seq reads, and consequently NER activity, occur more frequently in ChromHMM-defined domains of open chromatin (i.e. active promoters, elongating transcripts, etc.) as well as in chromatin domains containing histone modifications associated with active transcription (i.e. H3K4 mono- and dimethylation, H3K27 acetylation) than in heterochromatic regions.[15] These variations in NER activity inversely correlate with mutation density in skin cancers, which is elevated in heterochromatic regions compared to open chromatin, implying that NER efficiency is likely a primary determinant of mutational heterogeneity in skin cancers. In accordance, lower mutation densities are reported in the genomic regions that are repaired the most efficiently. Moreover, direct comparison of mutation density (i.e. the number of mutations per dipyrimidine sequence) to XR-seq reads (produced 1 hr post-UV exposure and normalized by dipyrimidine sequence context) among 100,000 bp bins across the human genome, reveals a strong negative correlation (Spearman’s rho = −0.6324, p<0.0001) (Fig. 2), indicating that NER efficiency likely governs variation in mutation distribution globally.
Figure 2:
Distribution of CPD formation, repair, and mutation in skin cells. Chromosomes were divided into bins of 100,000bp and CPD lesions, repair, and mutation sites were measured in these regions using CPD-seq, XR-seq, and whole genome sequencing of human melanomas. The number of dipyrimidine sites was also calculated for each region, and was used to normalize the data for sequence context. A portion of Chromosome 1 is shown as an example, although similar trends were observed throughout the genome. Coefficients of variation and two-sided Spearman correlations between XR-seq and melanoma mutations across the entire genome were calculated. In a small number of bins across the genome (not depicted here) very large CPD densities are observed. However, this is likely due to repetitive DNA sequences in the bins resulting in erroneous mapping of CPD-seq reads as opposed to large variation in CPD formation.
3.1. Do other chromatin domains impact CPD formation and mutagenesis?
These initial analyses of genome-wide NER activity and skin cancer mutation distributions assume a uniform distribution of UV lesions across the genome. However, variable lesion formation could also influence the levels of NER activity and mutagenesis in specific genomic regions. Mapping the distribution of CPDs in normal human fibroblasts immediately following exposure to UVC light (by the CPD-seq method[22, 35]) indicates that globally, CPD lesion formation is fairly constant across chromosomes. This is especially true after normalizing CPD-seq from irradiated fibroblasts by the distribution of dipyrimidine sequences in the genome. When the amount of normalized CPD-seq reads is compared between large chromatin regions (i.e. 100,000 bp bins across the human genome), post-irradiation CPD yields display a coefficient of variation of 0.2 (Fig. 2). This relatively small degree of regional variation in CPD levels contrasts with both NER-activity (measured by XR-seq) and melanoma mutation density, which display much greater variation (coefficient of variation = 0.78 and 0.71 respectively). Still, mild regional variation in CPD formation appears to occur across the human genome. This regional variation in CPD formation could be in part due to regional differences in sequence composition and GC content, since these affect the frequency of TT dinucleotides, which are more prone to CPD formation relative to other dipyrimidines (CT, CC, CC).
Similar to its influence on NER efficiency and melanoma mutagenesis, chromatin may also alter regional CPD formation. Morrison and colleagues recently mapped CPD formation in human cells by immunoprecipitating DNA containing CPD lesions and subsequently using high-throughput sequencing to identify damage sites across the genome, and determined that CPD levels significantly vary among large chromatin domains.[38] Significant decreases in CPDs were observed within chromatin containing histone marks associated with open chromatin, while a small positive correlation of CPD formation with H3K9 trimethylation (a marker of heterochromatin) was found. Further analysis led the authors to argue that lamin-associated domains within heterochromatin were particularly susceptible to UV damage, potentially because lamin-associated domains are near the nuclear periphery, and thus may be more exposed to UV than other regions of the genome.[38] These results suggest that where CPDs form may be impacted by more than the sequence content of a region. While additional investigation is needed to fully understand the underlying causes of regional variation in CPD formation, differences in both CPD formation and NER activity likely underlie mutational heterogeneity in human cancer, although the chromatin effects on repair appear to be much more dramatic.
3.2. Nucleosome structure influences CPD formation and repair.
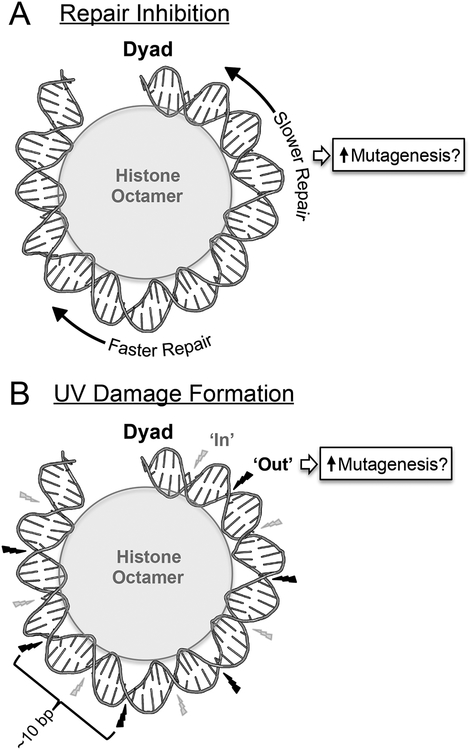
While large compacted chromatin domains appear to more significantly impact the efficiency of NER compared to the amount of CPDs formed, CPD formation can be altered by individual DNA binding proteins bending DNA into a conformation favorable to photoproduct formation.[39–43] Consequently, variable CPD formation may play a greater role in influencing cancer mutagenesis on a local DNA level at specific protein binding sites. Histones are the most abundant DNA binding proteins. The wrapping of the DNA around the histone octamer[44] can significantly affect both the repair and formation of CPD lesions. Repair of CPDs lesions occurs more rapidly near the edges of the nucleosome compared to CPDs located near the central dyad position, indicating that the translational positioning of the lesion within the nucleosome significantly regulates NER activity (Fig. 3A).[17, 18, 22, 45, 46] Likewise, histone-DNA contacts at inward rotational settings in nucleosomes inhibit CPD formation, apparently by constraining the structure and flexibility of the DNA (reviewed in [42]). In contrast, CPD formation is higher at outward facing rotation settings in nucleosomes, since these regions have greater conformational flexibility to facilitate CPD formation (Fig. 3B). As a consequence, the efficiency of CPD formation oscillates across nucleosomes with an ~10 bp periodicity in human chromatin.[41] In contrast, the packaging of DNA into nucleosomes generally inhibits 6–4PP formation, presumably due to the significantly greater structural distortion required to form a 6–4PP.[47–49]
Figure 3:
Schematic showing the effects of nucleosomes on repair inhibition and UV damage formation, and their potential contributions to somatic mutations in skin cancer. (A) NER occurs more slowly near the central dyad axis of the nucleosome, potentially promoting somatic mutation rates in nucleosome centers. (B) UV-induced CPD formation occurs more frequently at outward facing rotational settings (‘Out’ - indicated by black lightning bolt) than inward rotation settings (‘In’ - indicated by smaller gray lightning bolt) in nucleosomal DNA. It is not clear to what extent this damage pattern influences mutagenesis in skin cancers. DNA structure was visualized using pymol from PDB ID: 1ID3. Only half of the nucleosomal DNA is depicted. Adapted from Mao et al.[22]
Genome-wide sequencing based analysis of in vivo CPD formation and repair by our group has confirmed in yeast that CPD repair is inhibited near the nucleosome center and CPD formation oscillates with the expected ~10bp periodicity imposed by the rotational setting of DNA in strongly positioned nucleosomes.[22] Recent analyses of CPD formation in human cell lines indicate that a similar 10bp periodicity of CPD enrichment occurs across human nucleosomes and ultimately results in a striking oscillation of mutations across melanoma genomes.[50, 51] Approximately 150 bp oscillations in mutation density have also been observed in melanoma genomes that appear to coincide with the position of nucleosomes determined by MNase-seq or DNase-seq.[24, 50] This suggests that both nucleosomal influences on CPD formation and inhibition of repair likely influence skin cancer mutation distributions (Fig. 3A). Such effects have the ability to sensitize a large number of nucleotides to mutagenesis; however, more detailed studies are required to determine what influence, if any, variable CPD formation across nucleosomes has on cancer driver mutations and promoting disease progression.
4. Recurrent somatic mutations are frequently associated with transcription factor binding sites (TFBS).
Recurrent somatic mutations in cancer genomes are frequently associated with noncoding sequences such as TFBS (e.g., [4, 28, 52–55]). The best studied examples are mutations in the promoter of the gene encoding telomerase reverse transcriptase (TERT), which occur frequently in melanoma and other cancers.[4, 26, 27, 29] These mutations induce the expression of TERT by creating a new consensus binding site for E26 transformation-specific (ETS) family transcription factors,[56] which subsequently promotes carcinogenesis.[57] Highly recurrent mutations have been identified at other ETS binding sites in sequenced melanoma genomes, some of which affect the expression of the associated genes and are correlated with clinical outcomes.[4, 28, 31, 52, 55, 58] For example, ETS binding site mutations upstream of the succinate dehydrogenase D (SDHD) tumor suppressor gene occur frequently in melanoma tumors and are associated with decreased SDHD expression and worse prognosis in melanoma patients.[52]
However, it is unclear whether other recurrent mutations at TFBS in melanoma are potentially important driver mutations. For example, one of the most frequent noncoding mutations in melanoma occurs at position 49990694 in chromosome 19, which is associated with an ETS binding site in the RPL13A promoter.[28, 31] This single nucleotide is mutated in 47 out of 184 melanomas sequenced by the International Cancer Genome Consortium,[4, 35] and is also frequently mutated in other melanoma data sets.[31] The high frequency of this mutation would normally be diagnostic of a ‘driver’ mutation that promotes the transformation and proliferation of melanoma tumors.[59, 60] However, this TFBS mutation does not appear to affect expression of the RPL13A,[31] which is a gene not known to be associated with cancer. Furthermore, recent experiments by Larsson and colleagues have shown that this RPL13A promoter mutation arises frequently upon repeated UV exposure of human cells, in the absence of any apparent selective advantage.[31] These findings indicate that this recurrent mutation is likely over-represented in tumor genomes because it is extremely susceptible to UV-induced mutagenesis. A number of other highly recurrent mutations associated with TFBS (particularly at ETS binding sites) have been identified in melanoma, even though many of these do not appear to fit the typical profile of a driver mutation.[28, 30, 31, 52, 55, 58, 61, 62] For example, the frequency of recurrent mutations at ETS binding sites in melanoma generally correlates with the overall mutational load of the tumor, unlike other established driver mutations in these cancers (e.g., BRAF or NRAS mutations).[31] A key question then is what molecular mechanism is promoting recurrent mutagenesis at these TFBS?
4.1. Transcription factor binding promotes mutagenesis by inhibiting repair.
A recent bioinformatics study has argued that noncoding somatic mutations are elevated at TFBS because TF binding to a lesion-containing site inhibits repair.[24] This study found elevated mutation frequencies at ‘active’ TFBS (defined as TFBS resident in a DNase I-hypersensitivity region in melanocytes) in a number of cancers, particularly melanoma. To determine whether this might be a consequence of decreased repair activity at TFBS, they analyzed published XR-seq data measuring NER activity in UV irradiated human fibroblasts.[33] While NER activity is high in DNA flanking the TFBS, presumably because the TFBS are located in an accessible DNase I hypersensitivity region, NER activity is considerably lower in the immediate vicinity of the TFBS. Based on these findings, it was proposed that TF binding inhibits NER activity, thereby promoting the persistence of potentially mutagenic UV lesions at TFBS.[24] These findings are consistent with past studies indicating that binding of certain TFs can inhibit repair of UV lesions by the NER machinery in cells and in vitro.[63, 64] Moreover, a recent study by Wong and colleagues indicated that elevated mutation rates at CTCF binding sites in melanoma are also associated with decreased repair activity at CTCF binding sites.[65] They further showed that mutation enrichment at CTCF binding sites, and in core promoter regions in general,[25] is absent in squamous cell carcinomas derived from xeroderma pigmentosum complementation group C (XPC) patients, which are defective in the global genomic NER (GG-NER) pathway, but present in these same tumor types derived from NER-proficient patients,[65] supporting that model that repair inhibition is primary driver of mutation enrichment at CTCF binding sites in melanoma.
While this is an intriguing mechanism that could potentially explain the elevated levels of somatic mutagenesis at TFBS, a number of important questions related to this mechanism remain to be addressed. First, it is not clear if a TF remains bound to a binding site containing a UV lesion, which presumably is a prerequisite for it to inhibit repair. CPDs and 6–4PPs significantly distort the structure of DNA,[1, 66–68] and these structural distortions could potentially disrupt TF binding. Consistent with this hypothesis, it has been shown that UV-induced CPD lesions severely reduce the affinity of TFs (i.e., AP-1, E2F, NF-κB, NFY, and p53) for a lesion-containing binding site in vitro.[69] 6–4PPs are even more helix distorting than CPDs, and therefore would also be expected to disrupt TF binding. If UV lesions disrupt TF binding in cells, this should render the lesion accessible to the NER machinery and promote repair. However, in some cases, CPD lesions may have no effect or even stimulate TF binding.[70, 71] For example, MeCP2 (methyl CpG binding protein 2) binding to methylated CpG sites in vitro is slightly enhanced by an overlapping CPD lesion.[72] It will be important in future studies to determine what effect UV lesions have on TF occupancy in cells.
Second, it is not clear if TFs significantly affect the rate at which UV damage forms in DNA. Previous studies have shown that many TFs can suppress UV damage formation at their binding sites (reviewed in [42]), presumably by restricting the flexibility and structure of the bound DNA to conformations that are not prone to form CPD lesions. Low UV damage levels at TFBS could provide an alternative explanation for the low NER activity at TFBS, particularly since published analyses of XR-seq data typically do not normalize for differences in initial damage levels (e.g., [24, 25, 65]). However, not all TFs suppress UV damage. A previous study found that both CPDs and 6–4PPs are significantly elevated at CCAAT TFBS in the human PGK1 promoter following UV irradiation in cells.[73] This effect was attributed to TF binding, as normal levels of UV photoproducts were observed when the same DNA sequence was irradiated in vitro, in the absence of TF binding. This finding was recently confirmed and extended by a high-resolution CPD map of UV-irradiated human fibroblasts,[34] which indicated that binding by the Nuclear Transcription Factor Y-B (NFY-B) to CCAAT boxes across the genome is associated with elevated CPD lesions following UV irradiation. Formation of CPD and 6–4PP lesions is also enhanced or repressed at binding sites for other transcription factors,[34, 70, 72, 74–76] indicating that different TFs have distinct effects on the rate of UV photoproduct formation.[32] Higher levels of UV damage induced by binding of NFY or potentially other TFs could presumably promote mutagenesis at these binding sites, although recurrent mutations have not previously been detected at NFY binding sites in melanoma or other cancers.
4.2. ETS transcription factors induce a unique UV damage signature that stimulates recurrent mutations at ETS binding sites in melanoma.
To further investigate the role of TFs in modulating UV damage formation, we recently mapped the distribution of UV-induced CPD lesions across the genome of UV-irradiated human fibroblasts.[35] This CPD map was generated using the CPD-seq method, which identifies the location of CPD lesions at single nucleotide resolution.[22] Analysis of our CPD-seq data revealed significantly higher levels of CPD lesions associated with NFY binding sites, consistent with previous studies.[34, 73] However, elevated CPD lesions at NFY binding sites are not associated with elevated somatic mutations in a cohort of 184 melanoma genomes sequenced by the International Cancer Genome Consortium (ICGC).[4] Our CPD-seq data indicate that NFY binding primarily enhances the formation of TT dimers, which are usually bypassed in an error-free manner by DNA polymerase η,[77, 78] but does not enhance the formation of mutagenic cytosine-containing CPDs (e.g., TC, CC), thus explaining this apparent conundrum.
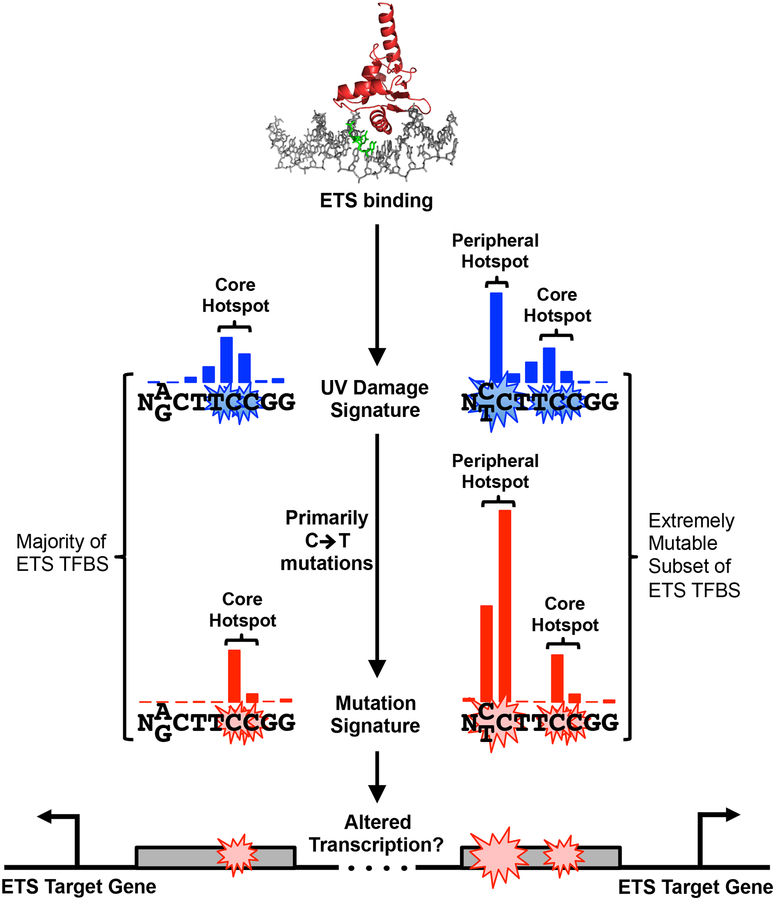
Elevated CPD formation was also detected at binding sites of E26 transformation-specific (ETS) transcription factors, namely ELK4, ETS1, and GABPA.[35] Notably, cytosine-containing mutagenic CPD lesions are also significantly elevated at ETS binding sites. Importantly, CPD lesions were significantly elevated in UV-irradiated cells relative to UV-irradiated naked DNA (both mapped using CPD-seq), indicating that TF binding, not DNA sequence context, is likely the primary driver of elevated CPD formation at these binding sites. Elevated CPD formation primarily occurs at two hotspots in the ETS binding motif (Fig. 4). One of these hotspots occurs at overlapping TC and CC dinucleotides in the core of the ETS motif (‘Core Hotspot’ in Fig. 4); the other hotspot occurs at a specific dinucleotide peripheral to the ETS motif (‘Peripheral Hotspot’ in Fig. 4). This peripheral hotspot only occurs at ~10% of active ETS binding sites that have a dipyrimidine sequence at this location (‘Extremely Mutable’ sites in Fig. 4), but is associated with greatly elevated CPD levels. A recent study by Larsson and colleagues using CPD-seq also found significantly elevated CPD levels at the same hotspots in ETS binding sites in a UV-irradiated melanoma cell line,[36] consistent with the results obtained in fibroblasts.[35] Taken together, these findings indicate that ETS TF binding in cells induces a unique UV damage signature, with highly elevated CPD formation at specific hotspots in the binding motif.
Figure 4:
Model describing the unique UV damage and mutational signature induced by ETS binding. CPD hotspots are associated with the core ETS motif (‘Core Hotspot’) for essentially all ETS binding sites, and a peripheral site (‘Peripheral Hotspot’) at a subset of extremely mutable binding sites, which contain a dipyrimidine at the position of the peripheral CPD hotspot in the ETS motif (positions −3/−4 from the ETS motif midpoint). Mutational hotspots at ETS binding sites in melanomas occur at essentially the same locations in the ETS binding sites, indicating that CPD formation is likely driving mutagenesis at these sites. Blue and red bars indicate the relative frequency of CPD lesions and melanoma mutations at each position in the ETS motif, respectively. Mutations in ETS binding sites may alter the transcription of ETS target genes, hence potentially contributing to carcinogenesis. Structure of the ETS transcription factor GABPA bound to DNA (PDB ID: 1AWC) was created using pymol. Figure adapted from Mao et al.[35]
Notably, UV-irradiation studies conducted in vitro using purified ETS1 protein recapitulated these findings, as UV damage hotspots were induced at similar locations in the ETS motif upon binding by ETS1.[35] ETS1 binding stimulates CPD formation up to ~40-fold higher than unbound DNA in vitro, indicating that ETS1 binding alone, in the absence of other DNA-associated proteins or co-factors, is sufficient to sensitize DNA to UV irradiation.
Analysis of somatic mutations in 184 sequenced melanoma genomes (ICGC) revealed that mutation frequency is significantly elevated at ETS binding sites.[35, 36] Moreover, recurrent mutations are specifically associated with the identified CPD hotspots in ETS binding motifs. The most frequently mutated site is associated with the CPD hotspot at the periphery of the ETS motif (Fig. 4), presumably due to the extremely high induction of UV damage levels at this location upon ETS binding. These findings can explain why highly recurrent mutations are associated with this position in certain ETS binding sites, such as in the RPL13A (see above) or DPH3 promoters.[28, 30, 31, 62] Since these mutations occur at weakly conserved positions at the periphery of the ETS motif, their effect on ETS binding and target gene activity is still unclear. In some cases, these mutations have been reported to alter the expression of a reporter gene (e.g., YAE1D1 promoter mutation),[55] but in other cases have no apparent effect (e.g., KIAA0907, RPL13A, and DPH3).[4, 28, 30, 31, 55, 62] The other site of recurrent mutations is associated with the CPD hotspot at the core ETS motif (‘Core Hotspot’ in Fig. 4). C>T mutations at these locations alter conserved positions in the ETS consensus, and have been reported to disrupt binding by GABPA (an ETS family member) in vitro.[79] It seems likely that recurrent mutations in the core ETS motif would alter gene expression, a concept that has been verified in a few cases (e.g., SDHD and ZNF277 genes).[4, 52, 79]
It will be important in future studies to elucidate the cellular consequences of recurrent ETS binding site mutations, particularly since ETS transcription factors are known oncogenes that regulate the expression of target genes involved in angiogenesis, apoptosis, differentiation, cell migration, and proliferation.[80, 81] The most well known ETS binding site mutations occur in the TERT promoter, and have been linked to carcinogenic transformation in a variety of tumor types, particularly melanoma.[26–29] Since TERT promoter mutations create canonical ETS binding sites from sequences that would be predicted to have only weak (or nonexistent) ETS binding in the absence of these mutations, it is not clear whether ETS-induced UV damage contributes to the etiology of these recurrent mutations.
4.3. ETS binding induces a DNA conformation predisposed to form CPD lesions.
A key question is how the binding of ETS proteins renders DNA susceptible to forming CPD lesions. The formation of CPD lesions involves a 2+2 cycloaddition reaction between the C5-C6 double bounds of adjacent pyrimidines.[82, 83] The typical distance (d ~ 4.5 Å) and torsion angle (η ~ 36°) between the C5-C6 double bonds of adjacent pyrimidines in B-form DNA is unfavorable for CPD formation.[84–86] Analysis of 3D structures of ETS1 or GABPA bound to DNA revealed that the central TC base step in ETS motif (‘Core Hotspot’ in Fig. 4) is shifted toward a more favorable distance and torsion angle upon ETS binding, thereby rendering the DNA more susceptible to forming CPD lesions at this location.[35] Molecular dynamics simulations confirmed that GABPA binding promotes a DNA structure prone to form CPD lesions at the central TC dinucleotide in the ETS motif. While this analysis provides a molecular mechanism for the core CPD hotspot, it is not clear what is the mechanism for the CPD hotspot at the periphery of the ETS motif (Fig. 4).
An implication of these structural studies is that DNA-binding by ETS proteins is inherently mutagenic in cells exposed to UV light. This would seem to be detrimental to terrestrial organisms, due to the ensuing load of UV damage and mutagenesis at ETS binding sites. However, this is likely mitigated by the fact that in metazoan animals, which are the only species in which ETS TFs have been identified,[87] a significant proportion of cells reside in the interior of the body, and thus are shielded from the mutagenic effects of UV exposure.
While this structural analysis focused on ETS1 and GABPA family members, it seems likely that other ETS domain-containing transcription factors (28 family members in humans) will induce similar alterations to the DNA structure to promote CPD formation in UV exposed cells. For example, a previous report revealed that a variant TCF binding site in the FOS promoter, which presumably is bound by the TCF subfamily of ETS transcription factors, showed elevated CPD formation in UV-irradiated cells at positions corresponding to the core CPD hotspot (Fig. 4).[74] One exception to this rule, however, may be the ETS family member ELF1. CPD enrichment at binding sites for ELF1 was significantly diminished compared to other ETS family members (i.e., ELK4, ETS1, and GABPA),[35] indicating that ELF1 binding may not induce CPD formation to the same extent as other ETS family members. This may be in part due to the difference in DNA binding modes of ELF1, which is a class II ETS family member, and ELK4, ETS1, and GABPA, which are all class I ETS family members.[88] Further research is needed to elucidate to what extent UV damage formation and mutagenesis are promoted by different ETS family members.
4.4. Do ETS transcription factors inhibit repair?
While ETS binding appears to significantly enhance CPD lesion formation, whether these proteins also promote mutagenesis by inhibiting subsequent repair of UV-induced CPD or 6–4PP lesions is unclear. The original study by Lopez-Bigas and colleagues suggested that there is decreased NER activity at active ETS binding sites in UV-irradiated cells, based on their analysis of published XR-seq data of CPD and 6–4PP lesions at a single repair time point (1 hour repair) in UV-irradiated human fibroblasts.[24, 33] However, our reanalysis of CPD XR-seq data revealed that repair activity is actually elevated at ETS binding sites.[35] This trend was consistent across multiple repair time points (i.e., 1, 4, and 8 hours), indicating that repair activity is higher at ETS binding sites, presumably due in part to higher initial damage levels. This discrepancy is likely due to the fact that the original study analyzed repair at both known and ‘discovered’ ETS binding sites,[24] while our study focused on known binding sites that fit the canonical ETS binding motif.[35] Further analysis indicated that decreased repair activity was specifically associated with these ‘discovered’ binding sites, which primarily consisted of GC-rich sequences that lacked any known ETS binding motif, and thus are unlikely to represent real ETS binding sites (see [35] for more details).
However, these analyses do not rule out the possibility that ETS binding may stimulate mutagenesis not just by enhancing UV damage formation, but also by inhibiting the subsequent repair of these UV lesions, as it is difficult to accurately measure repair at ETS binding sites using XR-seq data alone, due to higher initial damage levels at these sites. Testing this hypothesis will likely require additional high-resolution genomics studies to measure the cumulative removal of CPD lesions across ETS binding sites at different repair time points. It will also be important to test whether ETS binding inhibits repair of UV damage in vitro. However, it is important to note that Larsson and colleagues have shown that mutation frequency is still elevated at ETS motifs in squamous cell carcinomas derived from NER-defective XPC patients, albeit to a lesser extent than in repair-proficient tumors.[31] Moreover, they have recently shown that UV irradiation of repair-deficient fibroblasts in vitro still induces a mutation hotspot at the RPL13A ETS binding site.[36] These observations indicate that differences in repair efficiency are not required for ETS binding to stimulate UV mutagenesis; however, repair inhibition due to ETS binding may amplify mutagenesis at these binding sites.
4.5. Are UV damage levels associated with mutagenesis at other TF binding sites?
While the greatest enrichment of somatic mutations in melanoma was detected at ETS binding sites, there is also enrichment of somatic mutations at binding sites for other transcription factors, particularly the CCCTC-binding factor CTCF.[35] Previous studies reported elevated mutations at CTCF binding sites in a variety of cancers, including melanoma.[24, 53, 65, 89] Mutational enrichment of CTCF binding sites is likely associated with decreased NER activity at these sites,[24, 65] particularly since we and others have shown that CPD formation is not generally elevated at CTCF binding sites,[34, 35]. However, 6–4PP formation is elevated at specific locations within and adjacent to CTCF binding sites,[34] and could, hence, contribute to mutagenesis. In general, NER activity is lower at non-ETS TFBS, a phenomenon that correlates with broadly elevated levels of somatic mutations around these binding sites.[35] However, somatic mutation frequency in melanoma is decreased at certain TFBS, particularly binding sites of the Fos/Jun family of transcription factors. We have shown that lower mutation frequency at these Fos/Jun binding sites is correlated with decreased CPD (and mCPD) levels.[35] These findings suggest that DNA-binding by Fos and Jun transcription factors may protect their binding sites from UV-induced mutagenesis by suppressing the formation of UV damage.
In general, the frequency of mutagenic CPD formation across different classes of TFBS is significantly correlated with somatic mutation frequency at these binding sites in melanoma.[35] This indicates that the effect of TF binding on UV damage formation is a critical factor regulating UV-induced mutagenesis in skin cancers. It will be important in future studies to further explore the mechanism by which the binding of different TFs regulates UV damage formation and repair.
5. Conclusions and outlook
Much attention has been focused on the role of DNA repair in shaping the frequency and genomic distribution of somatic mutations in human cancers; however, recent studies highlight the potential significance of variable lesion formation in promoting mutagenesis in cancer. This is most evident in skin cancers such as melanoma, where there is accumulating evidence that TFs and chromatin can modulate the formation of mutagenic UV lesions. However, it will be important to determine whether TFs and chromatin affect the formation and repair of DNA lesions induced by other sources, such as alkylating agents and reactive oxygen species. Recent advances in technologies to map different classes of DNA lesions across the genome[34, 90–92] should provide new insights.
While significant progress has been made in understanding how DNA binding proteins modulate UV damage formation, it is not yet clear to what extent the resulting variations in mutation density affect cancer progression or chemotherapy resistance. The overall differences in mutation levels across nucleosomes are relatively modest; in contrast, ETS TFs dramatically elevate mutation frequencies at their binding sites, resulting in highly recurrent mutations. Determining experimentally whether these binding site mutations provide a proliferative advantage for melanoma or other skin cancers will be of significant importance. Characterizing whether specific ETS TFBS are more prone to mutation in skin cancers may help address this question. Since ETS transcription factors are downstream targets of the BRAF and NRAS signaling pathways,[93, 94] which are frequently dysregulated in melanoma,[95, 96] it will be important to determine whether activated BRAF or NRAS signaling in tumors modulates UV damage formation and mutagenesis at ETS binding sites. In parallel, it will be important to investigate whether ETS binding site mutations affect BRAF and/or NRAS signaling in melanoma, as well as the efficacy of BRAF inhibitors.
Acknowledgments
We thank Dr. Peng Mao and Dr. Michael Smerdon for helpful comments. Research in the Roberts and Wyrick laboratories are supported by grants from the NIEHS (R01ES028698 and R21ES029302 to J.J.W. and R21ES027937 to J.J.W. and S.A.R.) and NCI (R01CA218112 to S.A.R.). The authors declare that they have no conflicts of interest.
Abbreviations:
- 6–4PPs
6–4 pyrimidine-pyrimidone photoproducts
- CPD
cyclobutane pyrimidine dimer
- CPD-seq
cyclobutane pyrimidine dimer sequencing
- ETS
E26 transformation-specific
- NER
nucleotide excision repair
- TERT
telomerase reverse transcriptase
- TF
transcription factor
- TFBS
transcription factor binding site
- XR-seq
excision repair sequencing
- UV
ultraviolet light
References
- [1].Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T, DNA repair and Mutagenesis, ASM Press, Washington, D.C. 2006. [Google Scholar]
- [2].Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, Stratton MR, Nature 2010, 463, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, Zhang H, Zeid R, Ren X, Cibulskis K, Sivachenko AY, Wagle N, Sucker A, Sougnez C, Onofrio R, Ambrogio L, Auclair D, Fennell T, Carter SL, Drier Y, Stojanov P, Singer MA, Voet D, Jing R, Saksena G, Barretina J, Ramos AH, Pugh TJ, Stransky N, Parkin M, Winckler W, Mahan S, Ardlie K, Baldwin J, Wargo J, Schadendorf D, Meyerson M, Gabriel SB, Golub TR, Wagner SN, Lander ES, Getz G, Chin L, Garraway LA, Nature 2012, 485, 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch AM, Kakavand H, Alexandrov LB, Burke H, Jakrot V, Kazakoff S, Holmes O, Leonard C, Sabarinathan R, Mularoni L, Wood S, Xu Q, Waddell N, Tembe V, Pupo GM, De Paoli-Iseppi R, Vilain RE, Shang P, Lau LMS, Dagg RA, Schramm SJ, Pritchard A, Dutton-Regester K, Newell F, Fitzgerald A, Shang CA, Grimmond SM, Pickett HA, Yang JY, Stretch JR, Behren A, Kefford RF, Hersey P, Long GV, Cebon J, Shackleton M, Spillane AJ, Saw RPM, Lopez-Bigas N, Pearson JV, Thompson JF, Scolyer RA, Mann GJ, Nature 2017, 545, 175. [DOI] [PubMed] [Google Scholar]
- [5].DiGiovanna JJ, Kraemer KH, J Invest Dermatol 2012, 132, 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bradford PT, Goldstein AM, Tamura D, Khan SG, Ueda T, Boyle J, Oh KS, Imoto K, Inui H, Moriwaki S, Emmert S, Pike KM, Raziuddin A, Plona TM, DiGiovanna JJ, Tucker MA, Kraemer KH, J Med Genet 2011, 48, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lehmann AR, McGibbon D, Stefanini M, Orphanet J Rare Dis 2011, 6, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L, Cell 2012, 150, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cancer Genome Atlas N., Cell 2015, 161, 1681.26091043 [Google Scholar]
- [10].Schuster-Bockler B, Lehner B, Nature 2012, 488, 504. [DOI] [PubMed] [Google Scholar]
- [11].Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortes ML, Auclair D, Saksena G, Voet D, Noble M, DiCara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau DA, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll SA, Mora J, Lee RS, Crompton B, Onofrio R, Parkin M, Winckler W, Ardlie K, Gabriel SB, Roberts CW, Biegel JA, Stegmaier K, Bass AJ, Garraway LA, Meyerson M, Golub TR, Gordenin DA, Sunyaev S, Lander ES, Getz G, Nature 2013, 499, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Polak P, Lawrence MS, Haugen E, Stoletzki N, Stojanov P, Thurman RE, Garraway LA, Mirkin S, Getz G, Stamatoyannopoulos JA, Sunyaev SR, Nature biotechnology 2014, 32, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hodgkinson A, Chen Y, Eyre-Walker A, Human mutation 2012, 33, 136. [DOI] [PubMed] [Google Scholar]
- [14].Woo YH, Li WH, Nature communications 2012, 3, 1004. [DOI] [PubMed] [Google Scholar]
- [15].Adar S, Hu J, Lieb JD, Sancar A, Proceedings of the National Academy of Sciences of the United States of America 2016, 113, E2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zheng CL, Wang NJ, Chung J, Moslehi H, Sanborn JZ, Hur JS, Collisson EA, Vemula SS, Naujokas A, Chiotti KE, Cheng JB, Fassihi H, Blumberg AJ, Bailey CV, Fudem GM, Mihm FG, Cunningham BB, Neuhaus IM, Liao W, Oh DH, Cleaver JE, LeBoit PE, Costello JF, Lehmann AR, Gray JW, Spellman PT, Arron ST, Huh N, Purdom E, Cho RJ, Cell reports 2014, 9, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wellinger RE, Thoma F, The EMBO journal 1997, 16, 5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu X, Smerdon MJ, The Journal of biological chemistry 2000, 275, 23729. [DOI] [PubMed] [Google Scholar]
- [19].Hara R, Mo J, Sancar A, Molecular and cellular biology 2000, 20, 9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kosmoski JV, Ackerman EJ, Smerdon MJ, Proceedings of the National Academy of Sciences of the United States of America 2001, 98, 10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nag R, Smerdon MJ, Mutation research 2009, 682, 13. [DOI] [PubMed] [Google Scholar]
- [22].Mao P, Smerdon MJ, Roberts SA, Wyrick JJ, Proceedings of the National Academy of Sciences of the United States of America 2016, 113, 9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kaiser VB, Taylor MS, Semple CA, PLoS genetics 2016, 12, e1006207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, Lopez-Bigas N, Nature 2016, 532, 264. [DOI] [PubMed] [Google Scholar]
- [25].Perera D, Poulos RC, Shah A, Beck D, Pimanda JE, Wong JW, Nature 2016, 532, 259. [DOI] [PubMed] [Google Scholar]
- [26].Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA, Science 2013, 339, 957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R, Science 2013, 339, 959. [DOI] [PubMed] [Google Scholar]
- [28].Fredriksson NJ, Ny L, Nilsson JA, Larsson E, Nature genetics 2014, 46, 1258. [DOI] [PubMed] [Google Scholar]
- [29].Heidenreich B, Kumar R, Mutation research 2017, 771, 15. [DOI] [PubMed] [Google Scholar]
- [30].Colebatch AJ, Di Stefano L, Wong SQ, Hannan RD, Waring PM, Dobrovic A, McArthur GA, Papenfuss AT, Oncotarget 2016, 7, 66569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fredriksson NJ, Elliott K, Filges S, Van den Eynden J, Stahlberg A, Larsson E, PLoS genetics 2017, 13, e1006773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hu J, Selby CP, Adar S, Adebali O, Sancar A, The Journal of biological chemistry 2017, 292, 15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hu J, Adar S, Selby CP, Lieb JD, Sancar A, Genes & development 2015, 29, 948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hu J, Adebali O, Adar S, Sancar A, Proceedings of the National Academy of Sciences of the United States of America 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mao P, Brown AJ, Esaki S, Lockwood S, Poon GMK, Smerdon MJ, Roberts SA, Wyrick JJ, Nature communications 2018, 9, 2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Elliott K, Bostrom M, Filges S, Lindberg M, Van den Eynden J, Stahlberg A, Clausen AR, Larsson E, PLoS genetics 2018, (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Polak P, Karlic R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahovicek K, Stamatoyannopoulos JA, Sunyaev SR, Nature 2015, 518, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Garcia-Nieto PE, Schwartz EK, King DA, Paulsen J, Collas P, Herrera RE, Morrison AJ, The EMBO journal 2017, 36, 2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Becker MM, Wang JC, Nature 1984, 309, 682. [DOI] [PubMed] [Google Scholar]
- [40].Brown DW, Libertini LJ, Suquet C, Small EW, Smerdon MJ, Biochemistry 1993, 32, 10527. [DOI] [PubMed] [Google Scholar]
- [41].Gale JM, Nissen KA, Smerdon MJ, Proceedings of the National Academy of Sciences of the United States of America 1987, 84, 6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mao P, Wyrick JJ, Roberts SA, Smerdon MJ, Photochemistry and photobiology 2017, 93, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pehrson JR, Cohen LH, Nucleic acids research 1992, 20, 1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ, Nature 1997, 389, 251. [DOI] [PubMed] [Google Scholar]
- [45].Smerdon MJ, Thoma F, Cell 1990, 61, 675. [DOI] [PubMed] [Google Scholar]
- [46].Tijsterman M, de Pril R, Tasseron-de Jong JG, Brouwer J, Molecular and cellular biology 1999, 19, 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gale JM, Smerdon MJ, Photochemistry and photobiology 1990, 51, 411. [DOI] [PubMed] [Google Scholar]
- [48].Mitchell DL, Nguyen TD, Cleaver JE, The Journal of biological chemistry 1990, 265, 5353. [PubMed] [Google Scholar]
- [49].Suquet C, Mitchell DL, Smerdon MJ, The Journal of biological chemistry 1995, 270, 16507. [DOI] [PubMed] [Google Scholar]
- [50].Brown AJ, Mao P, Smerdon MJ, Wyrick JJ, Roberts SA, PLoS genetics 2018, 14, e1007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pich O, Muinos F, Sabarinathan R, Reyes-Salazar I, Gonzalez-Perez A, Lopez-Bigas N, Cell 2018, 175, 1074. [DOI] [PubMed] [Google Scholar]
- [52].Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W, Nature genetics 2014, 46, 1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Katainen R, Dave K, Pitkanen E, Palin K, Kivioja T, Valimaki N, Gylfe AE, Ristolainen H, Hanninen UA, Cajuso T, Kondelin J, Tanskanen T, Mecklin JP, Jarvinen H, Renkonen-Sinisalo L, Lepisto A, Kaasinen E, Kilpivaara O, Tuupanen S, Enge M, Taipale J, Aaltonen LA, Nature genetics 2015, 47, 818. [DOI] [PubMed] [Google Scholar]
- [54].Melton C, Reuter JA, Spacek DV, Snyder M, Nature genetics 2015, 47, 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Araya CL, Cenik C, Reuter JA, Kiss G, Pande VS, Snyder MP, Greenleaf WJ, Nature genetics 2016, 48, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bell RJ, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, Choi S, Hong C, He D, Pekmezci M, Wiencke JK, Wrensch MR, Chang SM, Walsh KM, Myong S, Song JS, Costello JF, Science 2015, 348, 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chiba K, Lorbeer FK, Shain AH, McSwiggen DT, Schruf E, Oh A, Ryu J, Darzacq X, Bastian BC, Hockemeyer D, Science 2017, 357, 1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhang W, Bojorquez-Gomez A, Velez DO, Xu G, Sanchez KS, Shen JP, Chen K, Licon K, Melton C, Olson KM, Yu MK, Huang JK, Carter H, Farley EK, Snyder M, Fraley SI, Kreisberg JF, Ideker T, Nature genetics 2018, 50, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pon JR, Marra MA, Annu Rev Pathol 2015, 10, 25. [DOI] [PubMed] [Google Scholar]
- [60].Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin MA, Gerstein M, Nature reviews. Genetics 2016, 17, 93. [DOI] [PubMed] [Google Scholar]
- [61].Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, Chung J, Wang NJ, Kakavand H, Mann GJ, Thompson JF, Wiesner T, Roy R, Olshen AB, Gagnon A, Gray JW, Huh N, Hur JS, Busam KJ, Scolyer RA, Cho RJ, Murali R, Bastian BC, Nature genetics 2015, 47, 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Denisova E, Heidenreich B, Nagore E, Rachakonda PS, Hosen I, Akrap I, Traves V, Garcia-Casado Z, Lopez-Guerrero JA, Requena C, Sanmartin O, Serra-Guillen C, Llombart B, Guillen C, Ferrando J, Gimeno E, Nordheim A, Hemminki K, Kumar R, Oncotarget 2015, 6, 35922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Conconi A, Liu X, Koriazova L, Ackerman EJ, Smerdon MJ, The EMBO journal 1999, 18, 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gao S, Drouin R, Holmquist GP, Science 1994, 263, 1438. [DOI] [PubMed] [Google Scholar]
- [65].Poulos RC, Thoms JA, Guan YF, Unnikrishnan A, Pimanda JE, Wong JW, Cell reports 2016, 17, 2865. [DOI] [PubMed] [Google Scholar]
- [66].Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor JS, Kang C, Proceedings of the National Academy of Sciences of the United States of America 2002, 99, 15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Taylor JS, Garrett DS, Cohrs MP, Biochemistry 1988, 27, 7206. [DOI] [PubMed] [Google Scholar]
- [68].Yokoyama H, Mizutani R, International journal of molecular sciences 2014, 15, 20321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tommasi S, Swiderski PM, Tu Y, Kaplan BE, Pfeifer GP, Biochemistry 1996, 35, 15693. [DOI] [PubMed] [Google Scholar]
- [70].Kwon Y, Smerdon MJ, Mutation research 2005, 577, 118. [DOI] [PubMed] [Google Scholar]
- [71].Kwon Y, Smerdon MJ, The Journal of biological chemistry 2003, 278, 45451. [DOI] [PubMed] [Google Scholar]
- [72].Cannistraro VJ, Taylor JS, Nucleic acids research 2010, 38, 6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pfeifer GP, Drouin R, Riggs AD, Holmquist GP, Molecular and cellular biology 1992, 12, 1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tornaletti S, Pfeifer GP, Journal of molecular biology 1995, 249, 714. [DOI] [PubMed] [Google Scholar]
- [75].Wang Y, Gross ML, Taylor JS, Biochemistry 2001, 40, 11785. [DOI] [PubMed] [Google Scholar]
- [76].Aboussekhra A, Thoma F, The EMBO journal 1999, 18, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F, The EMBO journal 1999, 18, 3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Johnson RE, Prakash S, Prakash L, Science 1999, 283, 1001. [DOI] [PubMed] [Google Scholar]
- [79].Zhang T, Xu M, Makowski MM, Lee C, Kovacs M, Fang J, Willems E, Trent JM, Hayward NK, Vermeulen M, Brown KM, Cancer research 2017, 77, 1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Findlay VJ, LaRue AC, Turner DP, Watson PM, Watson DK, Adv Cancer Res 2013, 119, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC, Nat Rev Cancer 2017, 17, 337. [DOI] [PubMed] [Google Scholar]
- [82].Cadet J, Anselmino C, Douki T, Voituriez L, J Photochem Photobiol B 1992, 15, 277. [DOI] [PubMed] [Google Scholar]
- [83].Schreier WJ, Gilch P, Zinth W, Annu Rev Phys Chem 2015, 66, 497. [DOI] [PubMed] [Google Scholar]
- [84].Schreier WJ, Schrader TE, Koller FO, Gilch P, Crespo-Hernandez CE, Swaminathan VN, Carell T, Zinth W, Kohler B, Science 2007, 315, 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Law YK, Azadi J, Crespo-Hernandez CE, Olmon E, Kohler B, Biophys J 2008, 94, 3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Johnson AT, Wiest O, J Phys Chem B 2007, 111, 14398. [DOI] [PubMed] [Google Scholar]
- [87].Degnan BM, Degnan SM, Naganuma T, Morse DE, Nucleic acids research 1993, 21, 3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hollenhorst PC, McIntosh LP, Graves BJ, Annual review of biochemistry 2011, 80, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Guo YA, Chang MM, Huang W, Ooi WF, Xing M, Tan P, Skanderup AJ, Nature communications 2018, 9, 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Mao P, Brown AJ, Malc EP, Mieczkowski PA, Smerdon MJ, Roberts SA, Wyrick JJ, Genome research 2017, 27, 1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Hu J, Lieb JD, Sancar A, Adar S, Proceedings of the National Academy of Sciences of the United States of America 2016, 113, 11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Wu J, McKeague M, Sturla SJ, J Am Chem Soc 2018. [DOI] [PubMed] [Google Scholar]
- [93].Foulds CE, Nelson ML, Blaszczak AG, Graves BJ, Molecular and cellular biology 2004, 24, 10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Tetsu O, McCormick F, Clin Transl Med 2017, 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Platz A, Egyhazi S, Ringborg U, Hansson J, Mol Oncol 2008, 1, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA, Nature 2002, 417, 949. [DOI] [PubMed] [Google Scholar]