Interferon-inducible protein 204 (p204) binds to microbial DNA to elicit inflammatory responses and induce interferon production. The DNA-binding affinity of the p204 HIN1 domain has been characterized and its crystal structure has been determined. Surface-charge distribution together with sequence alignment suggests that the p204 HIN domain uses its L12 and L45 loops for DNA binding.
Keywords: interferon-inducible protein 204, p204, HIN domain, DNA binding
Abstract
Interferon-inducible protein 204 (p204) binds to microbial DNA to elicit inflammatory responses and induce interferon production. p204 also modulates cell proliferation and differentiation by regulating various transcription factors. The C-terminal HIN domains in p204 are believed to be responsible for DNA binding, but the binding mode is not fully understood. The DNA-binding affinity of the p204 HIN1 domain has been characterized and its crystal structure has been determined, providing insight into its interaction with DNA. Surface-charge distribution together with sequence alignment suggests that the p204 HIN domain uses its L12 and L45 loops for DNA binding.
1. Introduction
Microbial nucleic acids are potent inducers of inflammatory responses, interferon production and cell death. Upon the recognition of microbial DNA in the cytoplasm and the nucleus, interferon-inducible (IFI) proteins either assemble into the multi-component inflammasome to activate interleukin maturation and pyroptosis (Fernandes-Alnemri et al., 2009 ▸; Hornung et al., 2009 ▸; Roberts et al., 2009 ▸; Bürckstümmer et al., 2009 ▸; Kerur et al., 2011 ▸) or induce interferon production in coordination with the cGAS–STING axis (Unterholzner et al., 2010 ▸; Almine et al., 2017 ▸).
IFI proteins are also known as PYHIN proteins owing to their bipartite domain organization. They usually contain an N-terminal PYD domain that is required for inflammasome assembly, followed by one or two C-terminal hematopoietic, interferon-inducible and nuclear localization (HIN) domains that recognize DNA, especially double-stranded DNA (dsDNA) (Cridland et al., 2012 ▸; Yin et al., 2015 ▸). HIN is a domain of about 200 amino acids. It consists of two oligonucleotide/oligosaccharide-binding (OB) folds, which are well established nucleic acid-binding motifs (Theobald et al., 2003 ▸). The HIN domains comprise a family of fast-evolving proteins. From a single gene in the common ancestor of marsupials and placental mammals, they have evolved into three distinct clades in mammals: HIN-A, HIN-B and HIN-C (Cridland et al., 2012 ▸; Ludlow et al., 2005 ▸). All three clades share highly similar sequences and three-dimensional structures, but their functions can be very different from each other, ranging from DNA binding to protein oligomerization (Jin et al., 2012 ▸; Yin et al., 2013 ▸).
Interferon-inducible protein 204 (p204) is a murine PYHIN protein. It is the mouse ortholog of human interferon-inducible protein 16 (IFI16). Together, p204 and IFI16 have been shown to activate the inflammasome and induce interferon production upon the recognition of viral DNA both in the cytoplasm and the nucleus (Unterholzner et al., 2010 ▸; Kerur et al., 2011 ▸; Orzalli et al., 2012 ▸). More recently, p204 has been shown to be necessary for canonical lipopolysaccharide (LPS)-induced Toll-like receptor 4 (TLR4) signaling (Yi et al., 2018 ▸). In addition to their role in host defense and inflammatory responses, p204 and IFI16 also modulate cell proliferation, differentiation and signaling, potentially by regulating tissue-specific transcription factors (Zhao et al., 2015 ▸; Luan et al., 2008 ▸).
p204 is composed of an N-terminal PYRIN domain and two HIN domains at its C-terminus (Fig. 1 ▸ a). The HIN1 domain belongs to the HIN-A clade. Among the HIN domains with known structures, the p204 HIN1 domain is most closely related to the mouse p202 HIN1 domain, followed by the IFI16 HIN1 domain (Cridland et al., 2012 ▸), both of which also belong to the HIN-A clade. However, members of the HIN family display divergent behaviors even when they share high sequence similarity. The p202 HIN1 domain has been shown to preferentially bind to dsDNA over ssDNA (Roberts et al., 2009 ▸), while the IFI16 HIN1 domain prefers cruciform and superhelical DNA over linear duplexes (Brázda et al., 2012 ▸). The p202 HIN2 domain, a HIN-B-type domain, does not bind to DNA at all (Yin et al., 2013 ▸), while the IFI16 HIN2 domain, another HIN-B-type domain, binds to dsDNA with micromolar affinity (Jin et al., 2012 ▸). Therefore, it is crucial to experimentally characterize the structure of each HIN domain in order to fully understand its function without any ambiguity.
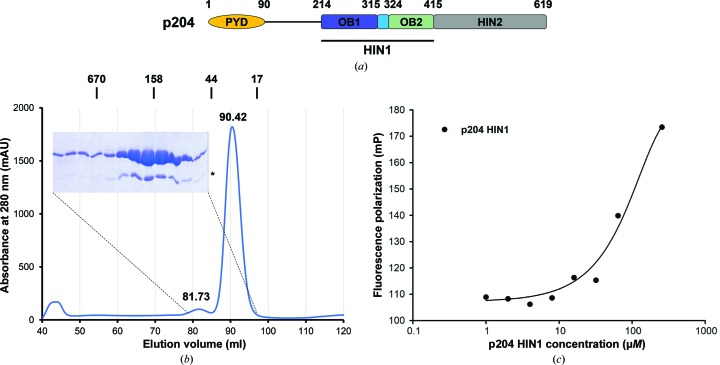
Figure 1.
Characterization of the p204 HIN1 domain. (a) Domain organization of p204. The HIN1 domain used in this study is indicated by a horizontal bar. (b) Size-exclusion chromatography profile of p204 HIN1. The elution positions of standards are marked above the profile and are labeled in kDa. A Coomassie Blue-stained SDS–PAGE gel covering the peak fractions is shown. An asterisk denotes residual SUMO protein. (c) Fluorescence polarization measurement of the DNA-binding affinity between p204 HIN1 and 20-mer dsDNA.
2. Materials and methods
2.1. Macromolecule production and fluorescence polarization
The mouse Ifi204 gene that encodes p204 was purchased from Open Biosystems (now part of Dharmacon). The region that encodes the p204 HIN1 domain (p204 HIN1; Ser211–Arg415) was cloned into a pSMT3 vector between the BamHI and SalI sites using standard PCR protocols (Table 1 ▸). The sequence was validated by Sanger sequencing (Genewiz). The expression plasmid was transformed into Escherichia coli BL21-CodonPlus (DE3) RIPL cells (Stratagene/Agilent). The bacterial culture was induced using 0.4 mM IPTG when the optical density (OD600) reached 0.6–0.7 and the cells were left to grow at 20°C overnight. The cell pellet was resuspended in lysis buffer (50 mM sodium phosphate pH 7.4, 300 mM NaCl, 5 mM β-mercaptoethanol, 20 mM imidazole). The cells were lysed by sonication and the cell debris was removed by centrifugation at 15 000 rev min−1 at 4°C for 45 min. His-SUMO-p204 HIN1 was purified from the supernatant by affinity chromatography using Ni–NTA resin (Qiagen). Fractions that contained His-SUMO-p204 HIN1 were pooled together and incubated with Ulp1 [1:1000(w:w)] for 2 h on ice. The mixture was loaded onto a Heparin HP column (GE Life Sciences) to separate p204 HIN1 from His-SUMO. Heparin column fractions containing p204 HIN1 were pooled and concentrated before loading onto a HiLoad 10/600 column (GE Life Sciences) pre-equilibrated with running buffer consisting of 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM dithiothreitol (DTT). The peak fractions were collected, pooled and concentrated to 17.9 mg ml−1.
Table 1. Macromolecule-production information.
| Source organism | Mus musculus (mouse) |
| DNA source | cDNA clone MGC 18551 |
| Forward primer† | 5′-CGCGGATCCTCACAACCCCAGAATCAGAATATTC-3′ |
| Reverse primer† | 5′-ACGCGTCGAC TCATCTCTTTGATATCTTGATGAAACTGTG-3′ |
| Cloning vector | pSMT3 |
| Expression vector | pSMT3 |
| Expression host | E. coli |
| Complete amino-acid sequence of the construct produced | SSQPQNQNIPRGAVLHSEPLTVMVLTATDPFEYESPEHEVKNMLHATVATVSQYFHVKVFNINLKEKFTKKNFIIISNYFESKGILEINETSSVLEAAPDQMIEVPNSIIRNANASPKICDIQKGTSGAVFYGVFTLHKKTVNRKNTIYEIKDGSGSIEVVGSGKWHNINCKEGDKLHLFCFHLKTIDRQPKLVCGEHSFIKISKR |
The BamHI site in the forward primer and the SalI site in the reverse primer are underlined. The stop codon in the reverse primer is shown in bold. The BamHI site in the forward primer doubles as a Ulp1 cleavage site in the translated fusion protein. Cleavage by Ulp1 leaves an additional serine at the N-terminus.
5′-Fluorescein-labeled single-stranded (ss) 20 nt DNA (5′-CCGATGGTTAGTAGCTATCG-3′) was annealed with an equimolar amount of the respective complementary nonlabeled DNA. 25 nM of the dsDNA thus formed was incubated alone or with 1, 2, 4, 8, 16, 32, 64 and 256 µM p204 HIN1. The final volume was 20 µl. Measurements were performed in a 384-well plate using a SpectraMax M5 at 25°C. For each concentration, 11 readings in a 5 min time window were averaged for plotting.
2.2. Crystallization
p204 HIN1 was crystallized using the hanging-drop vapor-diffusion method at 293 K. In brief, 1 µl p204 HIN1 was mixed with 1 µl reservoir solution consisting of 1.33 M ammonium sulfate, 0.05 M sodium acetate pH 4.5, 0.2 M NaCl. Crystals started to appear in two days. A summary of the crystallization conditions is provided in Table 2 ▸.
Table 2. Crystallization.
| Method | Hanging-drop vapor diffusion |
| Plate type | 24-well VDXm plate |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 17.9 |
| Buffer composition of protein solution | 20 mM Tris–HCl pH 8.0, 100 mM NaCl, 5 mM DTT |
| Composition of reservoir solution | 1.33 M ammonium sulfate, 0.05 M sodium acetate pH 4.5, 0.2 M NaCl |
| Volume and ratio of drop | 1 µl:1 µl |
| Volume of reservoir (µl) | 250 |
2.3. Data collection and processing
The crystals were flash-cooled in reservoir solution with 30% glycerol. Diffraction data were collected on beamline X25A at NSLS. Diffraction data were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▸). Diffraction data statistics are summarized in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the highest resolution shell.
| Beamline | X25A, NSLS |
| Wavelength (Å) | 1.10 |
| Temperature (K) | 80 |
| Crystal-to-detector distance (mm) | 350 |
| Oscillation (°) | 1 |
| Oscillation range (°) | 220 |
| Space group | P6522 |
| a, b, c (Å) | 58.38, 58.38, 315.84 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution range (Å) | 49.92–1.94 (2.01–1.94) |
| Total reflections | 536790 (44585) |
| Unique reflections | 24562 (2324) |
| Completeness (%) | 98.52 (95.95) |
| Multiplicity | 21.9 (19.2) |
| 〈I/σ(I)〉 | 33.03 (7.95) |
| R merge | 0.08329 (0.3337) |
| R meas | 0.08349 |
| CC1/2 | 0.985 (0.977) |
| CC* | 0.996 (0.994) |
| Wilson B factor (Å2) | 30.68 |
2.4. Structure solution and refinement
The structure of p204 HIN1 was determined by molecular replacement using the human IFI16 HIN1 domain (PDB entry 2oq0) as a search model using Phaser (McCoy et al., 2007 ▸). The p204 HIN1 and human IFI16 HIN1 domains share 56% sequence identity and 68% similarity. Iterative model building and refinement were carried out using Coot (Emsley et al., 2010 ▸), REFMAC (Murshudov et al., 2011 ▸) and PHENIX (Adams et al., 2010 ▸). Refinement statistics are summarized in Table 4 ▸. The atomic coordinates and structural factors for p204 HIN1 have been deposited in the Protein Data Bank (PDB) with the accession code 6oe9.
Table 4. Structure refinement.
Values in parentheses are for the highest resolution shell.
| Resolution range (Å) | 49.94–1.94 (1.99–1.94) |
| Completeness (%) | 98.1 (95.6) |
| No. of reflections | |
| Working set | 22561 |
| Test set | 2000 |
| Final R cryst | 0.1792 (0.1976) |
| Final R free | 0.2019 (0.2348) |
| No. of non-H atoms | |
| Total | 1843 |
| Protein | 1621 |
| Ligand | 44 |
| Solvent | 178 |
| R.m.s. deviations | |
| Bonds (Å) | 0.012 |
| Angles (°) | 1.27 |
| Average B factors (Å2) | |
| Overall | 43.80 |
| Protein | 42.3 |
| Ligand | 69.9 |
| Water | 51.1 |
| Ramachandran plot | |
| Most favored (%) | 97 |
| Allowed (%) | 3 |
| Outliers (%) | 0 |
3. Results and discussion
3.1. p204 HIN1 binds to dsDNA, but weakly
We expressed and purified the p204 HIN1 domain from a bacterial culture. The elution volume of p204 HIN1 from a size-exclusion chromatography column was 90.42 ml (21.5 kDa), which was consistent with a calculated monomeric molecular weight of 23.1 kDa (Fig. 1 ▸ b). A minor peak that eluted at 81.73 ml also turned out to be p204 HIN1. Its elution volume indicated a molecular weight of 51.3 kDa, which was likely to represent a minor population of p204 HIN1 in a dimeric form.
The p202 HIN1 domain binds to 20-mer dsDNA with a K d value of about 1.7 µM (Yin et al., 2013 ▸). As p204 HIN1 is closely related to the p202 HIN1 domain, we examined the DNA-binding capability of p204 HIN1 using a fluorescence polarization assay. The fluorescence polarization values increased as more HIN1 was added but did not reach saturation even at the highest HIN1 concentration that was tested (256 µM, 5.9 mg ml−1; Fig. 1 ▸ c). Based on the fluorescence polarization results, we estimated that the K d value between p204 HIN1 and 20-mer dsDNA was greater than 150 µM. This is an ∼90-fold decrease in affinity compared with the p202 HIN1 domain, but more closely resembles the affinity of the IFI16 HIN1 domain, which also showed a K d value of greater than 100 µM (Unterholzner et al., 2010 ▸).
3.2. Structure and surface features of the p204 HIN domain
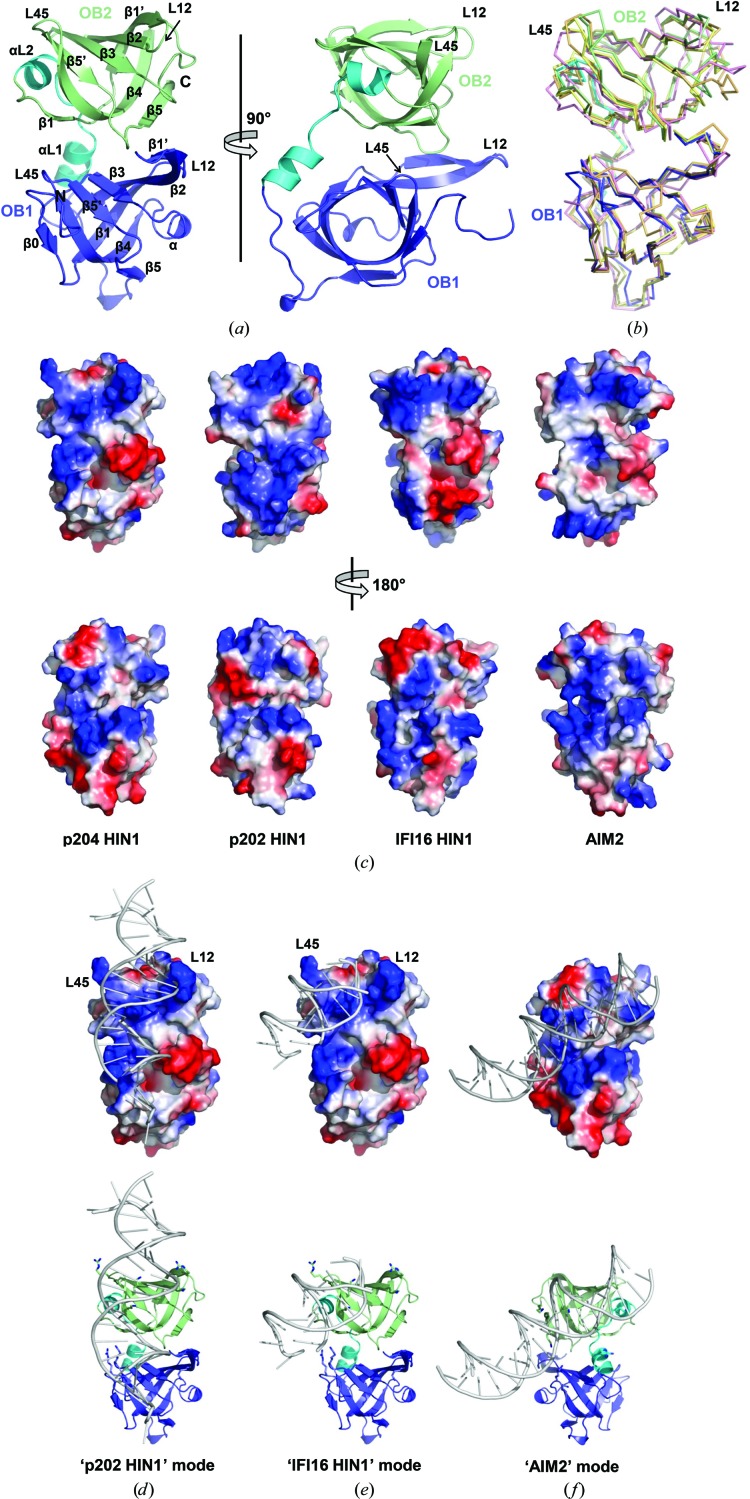
We crystallized the p204 HIN1 domain in space group P6522. There is one copy of p204 HIN1 in each asymmetric unit. We determined the crystal structure of p204 HIN1 using the structure of the IFI16 HIN1 domain (Liao et al., 2011 ▸) as a template and refined it to a resolution of 1.94 Å with R work and R free values of 17.9% and 20.2%, respectively (Table 4 ▸). In p204 HIN1, the two OB folds are connected by two short α-helices to form one entity (Fig. 2 ▸ a). The OB1–OB2 interface buries an area of 542 Å2, with contributions from multiple residues. Both OB folds adopted the canonical OB fold, although the α-helix is not well formed in OB2 (Fig. 2 ▸ a and Supplementary Fig. S1). The two OB folds are orientated in the same direction. The loops between β-strands 1 and 2 (L12) and between β-strands 4 and 5 (L45) in both OB folds point towards the same side of the HIN1 molecule (Fig. 2 ▸ a). The overall structure of p204 HIN1 is very similar to those of other HIN domains (Fig. 2 ▸ b), with pairwise Cα r.m.s.d. values of around 1 Å. The L12 and L45 loops and their adjacent regions display the greatest structural divergence (Fig. 2 ▸ b).
Figure 2.
Structural analysis of p204 HIN1. (a) Cartoon representations of the p204 HIN1 crystal structure in two orthogonal orientations. HIN1 is colored as in Fig. 1 ▸(a): OB1 in blue, helical linker in cyan and OB2 in pale green. (b) Superposition of several HIN domains. p204 HIN1 is colored as in Fig. 2 ▸(a). The human AIM2 HIN domain (PDB entry 3rn2) is in pink, the mouse p202 HIN1 domain (PDB entry 4l5r) is in orange and the human IFI16 HIN1 domain (PDB entry 4qgu) is in yellow. (c) Surface-charge distributions of the p204 HIN1, p202 HIN1, IFI16 HIN1 and AIM2 HIN domains in two orientations. The top and lower panels are rotated vertically by 180°. (d, e, f) Potential DNA-binding modes of p204 HIN1 obtained using structural superposition and sequence-conservation information. In (d) and (e) p202 HIN1 is in the same orientation, while in (f) it is rotated vertically by 180°. Upper panels, electrostatic potential surfaces; lower panels, cartoon representations with conserved Lys and Arg residues shown in stick representation.
Although the frameworks are highly similar, the surface-charge distribution varies among the different HIN domains, giving rise to multiple modes of engaging dsDNA (Jin et al., 2012 ▸; Yin et al., 2013 ▸; Ni et al., 2016 ▸; Fig. 2 ▸ c). Phylogenetic analysis places p204 HIN1 closer to the p202 HIN1 domain (Cridland et al., 2012 ▸), but p204 HIN1 is less positively charged, especially in its OB1 region (Fig. 2 ▸ c). Its surface-charge distribution more closely resembles that of the IFI16 HIN1 domain (Fig. 2 ▸ c). Lys220 in the loop preceding OB1, β0, Lys250 in OB1 L12, Lys292 in OB1 L45, Arg353 and Lys354 in OB2 L12, and Lys394, Arg398 and Lys401 in OB2 L45 form two distinct positively charged patches that may potentially interact with dsDNA in the way that the p202 HIN1 and IFI16 HIN1 domains bind to dsDNA (Figs. 2 ▸ d and 2 ▸ e). OB2 L12 and L45 are optimally positioned to clamp dsDNA between them (Figs. 2 ▸ d and 2 ▸ e). OB1 L45 may further stabilize the HIN1–DNA interaction. Indeed, most of the proposed residues are conserved in p202 or IFI16 and have been experimentally shown to directly contact dsDNA (Supplementary Fig. S1). Alternatively, p204 HIN1 may use the opposite surface to bind dsDNA in the manner employed by the AIM2 and IFI16 HIN2 domains (Jin et al., 2012 ▸). In this case, Lys279 and Lys280 in OB1 β4, Arg320 and Lys327 in the linker region and Lys385, Lys411 and Lys414 in OB4 β4 and β5 are positioned in the correct location for interaction with the DNA backbone (Fig. 2 ▸ f and Supplementary Fig. S1).
HIN-domain proteins have rapidly evolved to undertake multiple roles in the innate immune response, host defense, and cell differentiation and proliferation (Schattgen & Fitzgerald, 2011 ▸; Zhao et al., 2015 ▸). Although they share high sequence similarity and overall structural organization, the functions of HIN domains span from binding DNA in at least two different ways to mediating self-oligomerization (Jin et al., 2012 ▸; Yin et al., 2013 ▸). Biochemical and structural studies are necessary to delineate the function of each HIN domain. Based on the results of this study, we conclude that the HIN1 domain of interferon-inducible protein p204 binds to dsDNA, albeit with low affinity. p204 HIN1 adopts the canonical HIN structure of two OB folds. Our analysis of the surface-charge distribution of p204 HIN1 suggests that p204 HIN1 is closely related to the IFI16 HIN1 and p202 HIN1 domains.
During the preparation of this manuscript, we noticed that another p204 HIN1 domain structure had been released in the PDB with accession code 5yzp. Except for OB1 L12, which moved by about 5 Å, the two structures superpose well with each other, with a Cα r.m.s.d. value of 0.48 Å (Supplementary Fig. S2).
Supplementary Material
PDB reference: HIN1 domain of interferon-inducible protein 204, 6oe9
Supplementary Figures. DOI: 10.1107/S2053230X19007167/ek5016sup1.pdf
Acknowledgments
We thank Madhurima Bhattacharya and Erin Kassebaum for help with the fluorescence polarization assay and the Yin laboratory for criticism of the manuscript.
Funding Statement
This work was funded by National Institutes of Health, National Institute of Allergy and Infectious Diseases grant R00AI108793 to Qian Yin. Florida State University grant .
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Almine, J. F., O’Hare, C. A. J., Dunphy, G., Haga, I. R., Naik, R. J., Atrih, A., Connolly, D. J., Taylor, J., Kelsall, I. R., Bowie, A. G., Beard, P. M. & Unterholzner, L. (2017). Nature Commun. 8, 14392. [DOI] [PMC free article] [PubMed]
- Brázda, V., Coufal, J., Liao, J. C. C. & Arrowsmith, C. H. (2012). Biochem. Biophys. Res. Commun. 422, 716–720. [DOI] [PubMed]
- Bürckstümmer, T., Baumann, C., Blüml, S., Dixit, E., Dürnberger, G., Jahn, H., Planyavsky, M., Bilban, M., Colinge, J., Bennett, K. L. & Superti-Furga, G. (2009). Nature Immunol. 10, 266–272. [DOI] [PubMed]
- Cridland, J. A., Curley, E. Z., Wykes, M. N., Schroder, K., Sweet, M. J., Roberts, T. L., Ragan, M. A., Kassahn, K. S. & Stacey, K. J. (2012). BMC Evol. Biol. 12, 140. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fernandes-Alnemri, T., Yu, J.-W., Datta, P., Wu, J. & Alnemri, E. S. (2009). Nature (London), 458, 509–513. [DOI] [PMC free article] [PubMed]
- Hornung, V., Ablasser, A., Charrel-Dennis, M., Bauernfeind, F., Horvath, G., Caffrey, D. R., Latz, E. & Fitzgerald, K. A. (2009). Nature (London), 458, 514–518. [DOI] [PMC free article] [PubMed]
- Jin, T., Perry, A., Jiang, J., Smith, P., Curry, J. A., Unterholzner, L., Jiang, Z., Horvath, G., Rathinam, V. A., Johnstone, R. W., Hornung, V., Latz, E., Bowie, A. G., Fitzgerald, K. A. & Xiao, T. S. (2012). Immunity, 36, 561–571. [DOI] [PMC free article] [PubMed]
- Kerur, N., Veettil, M. V., Sharma-Walia, N., Bottero, V., Sadagopan, S., Otageri, P. & Chandran, B. (2011). Cell Host Microbe, 9, 363–375. [DOI] [PMC free article] [PubMed]
- Liao, J. C. C., Lam, R., Brazda, V., Duan, S., Ravichandran, M., Ma, J., Xiao, T., Tempel, W., Zuo, X., Wang, Y.-X., Chirgadze, N. Y. & Arrowsmith, C. H. (2011). Structure, 19, 418–429. [DOI] [PMC free article] [PubMed]
- Luan, Y., Lengyel, P. & Liu, C.-J. (2008). Cytokine Growth Factor Rev. 19, 357–369. [DOI] [PMC free article] [PubMed]
- Ludlow, L. E., Johnstone, R. W. & Clarke, C. J. (2005). Exp. Cell Res. 308, 1–17. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ni, X., Ru, H., Ma, F., Zhao, L., Shaw, N., Feng, Y., Ding, W., Gong, W., Wang, Q., Ouyang, S., Cheng, G. & Liu, Z.-J. (2016). J. Mol. Cell Biol. 8, 51–61. [DOI] [PubMed]
- Orzalli, M. H., DeLuca, N. A. & Knipe, D. M. (2012). Proc. Natl Acad. Sci. USA, 109, E3008–E3017. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Roberts, T. L., Idris, A., Dunn, J. A., Kelly, G. M., Burnton, C. M., Hodgson, S., Hardy, L. L., Garceau, V., Sweet, M. J., Ross, I. L., Hume, D. A. & Stacey, K. J. (2009). Science, 323, 1057–1060. [DOI] [PubMed]
- Schattgen, S. A. & Fitzgerald, K. A. (2011). Immunol. Rev. 243, 109–118. [DOI] [PubMed]
- Theobald, D. L., Mitton-Fry, R. M. & Wuttke, D. S. (2003). Annu. Rev. Biophys. Biomol. Struct. 32, 115–133. [DOI] [PMC free article] [PubMed]
- Unterholzner, L., Keating, S. E., Baran, M., Horan, K. A., Jensen, S. B., Sharma, S., Sirois, C. M., Jin, T., Latz, E., Xiao, T. S., Fitzgerald, K. A., Paludan, S. R. & Bowie, A. G. (2010). Nature Immunol. 11, 997–1004. [DOI] [PMC free article] [PubMed]
- Yi, Y.-S., Jian, J., Gonzalez-Gugel, E., Shi, Y.-X., Tian, Q., Fu, W., Hettinghouse, A., Song, W., Liu, R., He, M., Qi, H., Yang, J., Du, X., Xiao, G., Chen, L. & Liu, C.-J. (2018). EBioMedicine, 29, 78–91. [DOI] [PMC free article] [PubMed]
- Yin, Q., Fu, T.-M., Li, J. & Wu, H. (2015). Annu. Rev. Immunol. 33, 393–416. [DOI] [PMC free article] [PubMed]
- Yin, Q., Sester, D. P., Tian, Y., Hsiao, Y. S., Lu, A., Cridland, J. A., Sagulenko, V., Thygesen, S. J., Choubey, D., Hornung, V., Walz, T., Stacey, K. J. & Wu, H. (2013). Cell Rep. 4, 327–339. [DOI] [PMC free article] [PubMed]
- Zhao, H., Gonzalezgugel, E., Cheng, L., Richbourgh, B., Nie, L. & Liu, C. (2015). Genes Dis. 2, 46–56. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: HIN1 domain of interferon-inducible protein 204, 6oe9
Supplementary Figures. DOI: 10.1107/S2053230X19007167/ek5016sup1.pdf