Abstract
RNA-binding proteins (RBPs) and miRNAs are critical gene expression regulators that interact with one another in cooperative and antagonistic fashions. We identified Musashi1 (Msi1) and miR-137 as regulators of a molecular switch between self-renewal and differentiation. Msi1 and miR-137 have opposite expression patterns and functions, and Msi1 is repressed by miR-137. Msi1 is a stem–cell protein implicated in self-renewal while miR-137 functions as a proneuronal differentiation miRNA. In gliomas, miR-137 functions as a tumor suppressor while Msi1 is a prooncogenic factor. We suggest that the balance between Msi1 and miR-137 is a key determinant in cell fate decisions and disruption of this balance could contribute to neurodegenerative diseases and glioma development. Genomic analyses revealed that Msi1 and miR-137 share 141 target genes associated with differentiation, development, and morphogenesis. Initial results pointed out that these two regulators have an opposite impact on the expression of their target genes. Therefore, we propose an antagonistic model in which this network of shared targets could be either repressed by miR-137 or activated by Msi1, leading to different outcomes (self-renewal, proliferation, tumorigenesis).
Keywords: Musashi1, miR-137, glioblastoma, neurogenesis, RNA-binding protein, miRNA
INTRODUCTION
RNA-binding proteins (RBPs) and miRNAs function in the same regulatory space and interact in multiple ways. RBPs participate in the processing of specific pre-miRNAs (Treiber et al. 2017; Nussbacher and Yeo 2018). RBPs and miRNAs can bind overlapping or nearby regulatory elements and modulate the expression of shared target transcripts in an antagonistic fashion (Gardiner et al. 2015; Neelamraju et al. 2015; Yano et al. 2015; Banelli et al. 2017; Lennox et al. 2018; Rajgor 2018). On the other hand, they can act as partners to repress the expression of a common target (Ciafrè and Galardi 2013; Zhao et al. 2018). Although most cases described so far involve a single transcript, there are examples suggesting that the relationship between RBPs and miRNAs can be more complex. For instance, seven mammalian microRNAs preferentially colocalize with Pumilio (PUM) binding sites and have binding motifs that reverse complements the PUM recognition site; PUM binding to transcripts enhances miRNA-binding efficiency and subsequently increases decay of shared targets (Jiang and Lou 2013). Many more of these types of complex interactions are predicted to occur (Preusse et al. 2015). RBP-miRNA interactions are particularly important in the context of the nervous system; both regulators play critical roles in neurogenesis and brain development, and alterations affecting their function or expression levels contribute to neurological disorders and brain tumors (Gardiner et al. 2015; Neelamraju et al. 2015; Yano et al. 2015; Banelli et al. 2017; Lennox et al. 2018; Rajgor 2018).
Musashi1 (Msi1) is an evolutionarily conserved RBP and stem cell marker that plays a critical role in neurogenesis by regulating the balance between self-renewal and differentiation (Glazer et al. 2012). Msi1 modulates the expression of its target genes by binding to (G/A)UnAGU (n = 1–3) motifs located mainly in the 3′-UTR of target genes (Toda et al. 2001; Uren et al. 2015; Iwaoka et al. 2017). Msi1 is required for proper development of the brain as Msi1−/− mice develop obstructive hydrocephalus and ependymal abnormalities (Sakakibara et al. 2002). In experimental models of hypoxia and ischemic injury, it has been shown that Msi1-enriched progenitor cells are recruited to the site of injury to reconstitute neural networks and possibly prevent neuronal cell death (Yagita et al. 2001, 2002; Takasawa et al. 2002; Tonchev et al. 2003, 2005; Zhang et al. 2004; Shimmyo et al. 2007). Similarly, in adult epileptic tissue, there is an increased expansion of neural progenitor cells that display high expression of Msi1 (Crespel et al. 2005).
Musashi1 is highly expressed in a variety of tumor types including brain tumors (medulloblastoma and gliomas) (Kudinov et al. 2017). Msi1 expression is higher in medulloblastoma than in adult cerebellum and is significantly elevated in high-risk groups 3 and 4. Furthermore, survival analysis revealed a significant association of Msi1 immuno-reactivity with poor overall survival (OS) and progression-free survival (PFS) (Vo et al. 2012b). A similar scenario is encountered in gliomas, with Msi1 expression being particularly high and prevalent in the most aggressive type, glioblastoma multiforme (GBM) (Toda et al. 2001; Ma et al. 2008; Dahlrot et al. 2013). Supporting its prooncogenic roles, several groups have shown that Msi1 high expression is necessary to maintain cancer phenotypes and influences tumor growth (Kudinov et al. 2017). Msi1 is also implicated in radio- and chemo-resistance (de Araujo et al. 2016). Moreover, Msi1 target analysis by cross-linking immunoprecipitation (CLIP) and RNP immunoprecipitation (RIP) identified more than 1000 transcripts whose biological functions support Msi1 involvement in apoptosis, cell cycle, proliferation, adhesion, invasion, migration, and DNA-repair (de Sousa Abreu et al. 2009; Vo et al. 2012b; Uren et al. 2015).
We have previously shown that Msi1 is regulated by several tumor suppressor miRNAs, including miR-137 (Vo et al. 2011). Contrary to Msi1, miR-137 promotes differentiation and prevents glioblastoma development. We show here a novel aspect of this antagonistic relationship; our results indicate that these two regulators share a large number of interconnected target genes implicated in neurogenesis and glioblastoma growth. We propose a model in which Msi1 and miR-137 regulate this network of targets in different directions (activation vs repression); by turning up or down the expression levels of their components, Msi1 and miR-137 contribute to cell fate decisions (self-renewal, differentiation, or tumorigenesis).
RESULTS AND DISCUSSION
Gene regulatory networks are well-documented in the case of transcription factors and miRNAs. The complex interactions between these two regulators and their target genes take place in different organisms, affect many biological processes and contribute to diseases and cancer (Martinez and Walhout 2009; Arora et al. 2013). In the case of miRNA-RBPs, the network aspect has not been fully explored yet. Although they have been shown to interact in multiple ways, the large majority of examples described so far involve a single shared target transcript.
Musashi1 is regulated by a set of “tumor suppressor” miRNAs that encompass miR-34a, miR-101, miR-128, miR-137, and miR-138 (Vo et al. 2011). Like Musashi1, these miRNAs function in the interesting intersection of neurogenesis-brain tumor development. They promote neuronal differentiation and display dramatic changes in expression during this process (Chang et al. 2011; Santos et al. 2016; Rajman and Schratt 2017). In the cancer scenario, these miRNAs often show reduced expression in GBM in comparison to normal brain and mimics transfection affects cancer relevant phenotypes (Qiu et al. 2013; Rathod et al. 2014; Tamim et al. 2014; Sun et al. 2015; Liu et al. 2016; Shan et al. 2016; Shea et al. 2016; Tian et al. 2016; Rooj et al. 2017). We inquired if these miRNAs and Msi1 have a more complex relationship, meaning if their antagonistic functions in neurogenesis and glioma development come as a result, at least in part, via their impact on shared target transcripts. To test that, we downloaded from Targetscan (Agarwal et al. 2015) the list of predicted targets for all five miRNAs, built a Msi1 target list from previous CLIP and RIP analyses (de Sousa Abreu et al. 2009; Vo et al. 2012b; Uren et al. 2015) and determined the overlap. Corroborating our expectations, we observed that these miRNAs potentially share a large number of targets with Msi1 and also among themselves; in all cases with the exception of miR-138, the observed overlaps are statistically significant (Supplemental Fig. S1; Supplemental Tables S1, S2).
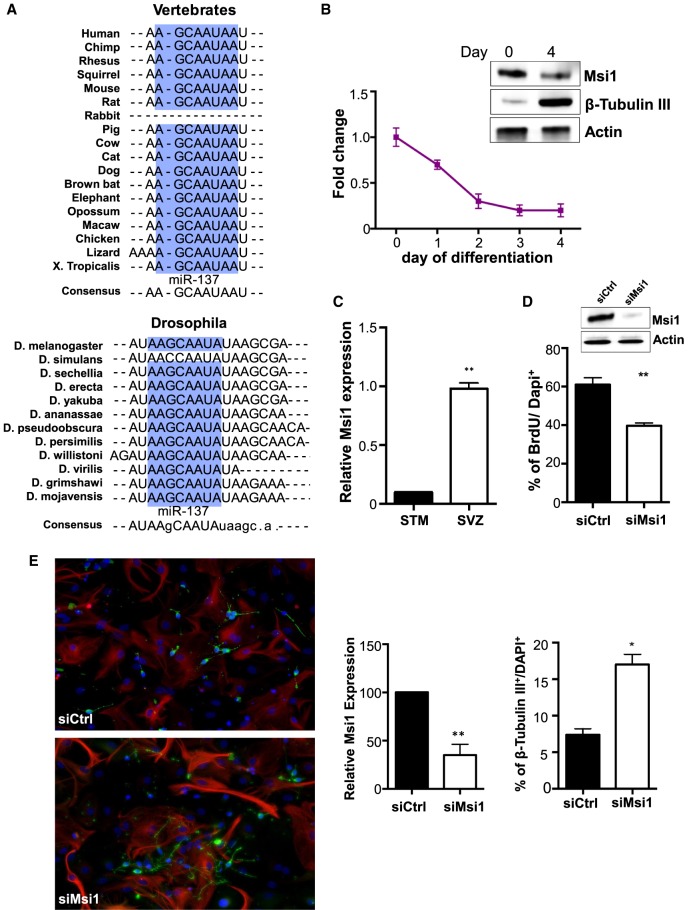
We decided to focus on miR-137–Msi1 association due to the fact that Musashi regulation by miR-137 is the only one among the miRNAs we studied to be highly conserved. In Figure 1A, we show that the miR-137 binding site in Msi1's 3′-UTR is not only conserved among all vertebrates, from zebrafish till humans, but also across the genus Drosophila. This grade of conservation suggests that Musashi regulation by miR-137 is a critical component in the balance between self-renewal and differentiation.
FIGURE 1.
Musashi1's role in neurogenesis. (A) TargetScan predictions show that miR-137 binding site in Msi1's 3′-UTR is conserved among vertebrates and in the Drosophila genus. Highly conserved nucleotides are shown in blue boxes. (B) Msi1 mRNA expression decreases during NSC differentiation in vitro (left panel). Western blot analysis shows that Msi1 protein levels are higher in NSCs in comparison to differentiated cells. β-Tubulin III was used as a neuronal marker (right panel). (C) Msi1 expression is higher in cells located in subventricular zone (SVZ) in comparison to the cells in the striatum (STM). (D) Western analysis shows effective siRNA knockdown of Msi1 in NSCs, siMsi1 versus siCtrl (top panel). Quantification of the percentage of NSCs that incorporated BrdU following transfection with siMsi1 and siRNA control (bottom panel). (E) NSCs were transfected with siMsi1 or siControl and then transferred to differentiation media. Cells were immunostained with β-Tubulin III (red) 4 d later (left panel). Nuclei were stained with DAPI1 (blue). qRT-PCR analysis showing Msi1 knockdown levels obtained in NSCs (panel in the middle). Quantification of neurons 4 d after transfer to differentiation media (right panel). Experiment was performed using biological triplicates. The data are showed as mean ± standard deviation. (*) P-value > 0.05, (**) P < 0.001, (***) P < 0.0001.
The importance of miR-137 goes beyond neurogenesis. miR-137 has been implicated in multiple neurological disorders; for instance, single nucleotide polymorphism (SNP) analysis established miR-137 as a marker for schizophrenia susceptibility (for review, see Yin et al. 2014). Reduced miR-137 expression has been observed in brain tissue of patients with depression and suicidal behavior (Zhao et al. 2013). Moreover, miR-137 has been shown to be deregulated in a cellular model of Huntington Disease (Kozlowska et al. 2013) and in Alzheimer's disease (Saraiva et al. 2017). In the context of tumorigenesis, miR-137 has been defined as a tumor suppressor miRNA in numerous cancer types, including gliomas (Gao et al. 2015; Chang et al. 2016; Dong et al. 2016; Han et al. 2016; Neault et al. 2016; Sakaguchi et al. 2016; Wu et al. 2016). In the particular case of gliomas, miR-137 expression levels predict survival and transfection of miR-137 mimics into glioma cells decreased proliferation, invasion and growth (Chen et al. 2012; Li et al. 2013; Tamim et al. 2014).
We have previously shown that miR-137 expression increases as murine neural stem cells (NSCs) differentiate (Santos et al. 2016). Analysis of Msi1 expression during neurogenesis showed a profile opposite to the one described for miR-137 (Fig. 1B). Msi1 expression levels in the SVZ, a neural stem cell niche, and in the STM, a differentiated tissue of murine brains, are the inverse of what we reported for miR-137 (Santos et al. 2016). Msi1 expression is higher in the SVZ where self-renewing and proliferating stem cells are located (Fig. 1C). Similarly, analysis of The Cancer Genome Atlas (TCGA) glioma samples showed that Msi1 and miR-137 display opposite patterns of expression and these patterns are strongly correlated (R = −0.45; P-value = 2.2 × 10−26) (Supplemental Fig. S1D). These results support the hypothesis of miR-137 being a critical regulator of Msi1 expression.
miR-137 is required for lineage progression and neural differentiation. We have shown that blockage of miR-137 by antagomiRs disrupts the production of neuronal cells while miR-137 mimics transfection has the opposite effect (Santos et al. 2016). Moreover, transfection of miR-137 mimics in NSCs decreased self-renewal and proliferation (Santos et al. 2016). A similar analysis was conducted with Msi1, and we observed that in respect to control, Msi1 knockdown reduced NSCs proliferation (Fig. 1D) and increased the number of neurons when NSCs were induced to differentiation (Fig. 1E).
Msi1 and miR-137 shared targets are implicated in development, differentiation, and cell migration
To expand on the potential antagonism between Msi1 and miR-137, we generated a list of identified miR-137 targets by compiling the results of our previous studies (Tamim et al. 2014; Santos et al. 2016). Additionally, we incorporated the validated set of targets described on miRTarBase (Hsu et al. 2011). A comparison to Msi1 targets list identified 141 potential shared targets (Supplemental Tables S3, S4; Supplemental Fig. S1C). To evaluate the significance of this overlap, we compared the target sets of three RBPs (Msi1, hnRNPH1, and IGF2BP3) (Supplemental Table S1) and miRNAs (conserved families) (Supplemental Table S5). First, we looked at miRNAs that share a large number of targets with Msi1, hnRNPH1 and IGF2BP3 (minimum 10% of total identified targets for each RBP). Next, we evaluated the nature of these overlaps. More specifically, we checked the number of miRNA binding sites per transcript and determined for each RBP-miRNA overlap the number of transcripts targeted only by a small number of miRNAs (less than three, and less than five). miR-137 is not only among the miRNAs sharing the largest number of targets with Msi1 but also has the largest number of shared targets with a small number of miRNA sites. This trend was not observed in the analysis with the other two RBPs we analyzed (Supplemental Table S6).
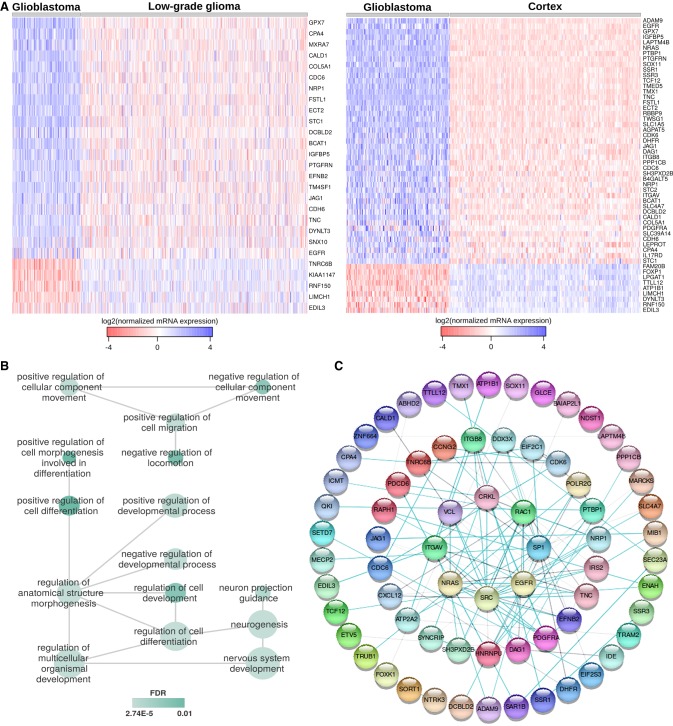
We analyzed the expression levels of Msi1/miR-137 shared targets in GBM, low-grade glioma (LGG) and normal brain using samples from TCGA and Genotype-Tissue Expression (GTEx) project. For the ones we observed consistent expression values across samples sets, we determined that the large majority of them show increased expression in GBM in comparison to both normal brain and LGG (Fig. 2A; Supplemental Table S7).
FIGURE 2.
Msi1 and miR-137 shared targets are associated with neurogenesis and gliomagenesis. (A) Heatmap showing the expression of miR-137/Msi1 shared targets in GBM versus LGG samples from the TCGA data repository and GBM versus healthy (frontal) cortex samples from GTEx. We show only the genes with consistent expression values across sample sets. (B) Gene Ontology (GO) analysis (biological process) of Msi1/miR-137 shared targets. Associations between most relevant GO terms according to REVIGO (Supek et al. 2011). (C) Predicted protein network according to STRING (Szklarczyk et al. 2015) showing the associations between Msi1/miR-137 common targets.
Gene ontology and pathway analyses showed that Msi1/miR-137 shared targets are preferentially associated with differentiation, morphogenesis, development and migration (Fig. 2B; Supplemental Table S4). Moreover, network analysis shows that a large number of shared targets are highly interconnected. Major nodes include SRC, EGFR, NRAS, PDGFRα, and SP1 (Fig. 2C). These five genes are known key players in neurogenesis and gliomagenesis. EGFR and PDGFRα are members of the epidermal growth factor and receptor tyrosine kinase families, respectively, and are established drivers of gliomagenesis (Aldape et al. 2015). SRC is a member of the Src family of kinases (SFKs), which are nonreceptor tyrosine kinases. SFKs interact with various cell surface receptors including EGFR and PDGFRα (Han et al. 2014). SRC inhibitors have been explored to treat glioblastoma (Eom et al. 2016). NRAS, a member of the RAS family of proto-oncogenes, is a GTPase that is frequently mutated in tumors and is known to promote cell proliferation and inhibit apoptosis (Serna-Blasco et al. 2018). SP1 is a transcription factor that regulates the expression of a large number of target genes and has been implicated in cell growth, differentiation, apoptosis, and tumor development. Its increased expression is observed in multiple tumor types and SP1 targeting has been pursued as a therapeutic strategy (Vizcaíno et al. 2015).
To expand on the analysis of shared Msi1/miR-137 targets, we selected a set of genes with little or no information regarding their participation in neurogenesis and glioblastoma development (FSTL1, PTGRFN, STC1, CDH6, DCBLD2) and evaluated via siRNA knockdown their impact on the proliferation of U251 glioblastoma cells and neuronal differentiation using the neuroblastoma BE-(2)-C as a model. DCBLD2 and STC1 knockdown decreased cell proliferation while knockdown of FSTL1 induced neuronal differentiation (Supplemental Fig. S4A,B). Discoidin, CUB, and LCCL domain containing 2 (DCBLD2) is a receptor tyrosine kinase that is aberrantly expressed in a variety of tumors and linked to invasiveness and poor prognostic outcome (Hofsli et al. 2008; Kikuta et al. 2017; Martinez-Romero et al. 2018; Raman et al. 2018). Stanniocalcin-1 (STC1) is secretory glycoprotein that has been linked to chemo-resistance, metastasis and increased cell proliferation in many tumor types including gastric cancer, hepatocellular carcinoma, colorectal cancer, and acute myeloid leukemia (Abaza et al. 2016; Rezapour et al. 2016; Chan et al. 2017; Wang et al. 2019); in glioblastoma, STC1 functions as a stimulator for stem-like traits of GBM cells (Li et al. 2018). Follistatin-like 1 (FSTL1) is a secreted glycoprotein implicated in development cardiovascular disease, cancer, and arthritis (Mattiotti et al. 2018). Analysis of the TCGA glioma data indicated that these three genes show higher levels of expression in glioblastoma (grade IV) in comparison to grades II and III. Moreover, glioblastoma patients displaying high expression of these genes have a worse prognosis (Supplemental Fig. S4C,D). Overall, these results are in line with the hypothesis that Msi1/miR-137 shared targets are “activated” in cancer cells and contribute to tumorigenesis.
Msi1 and miR-137 regulatory impact on shared targets
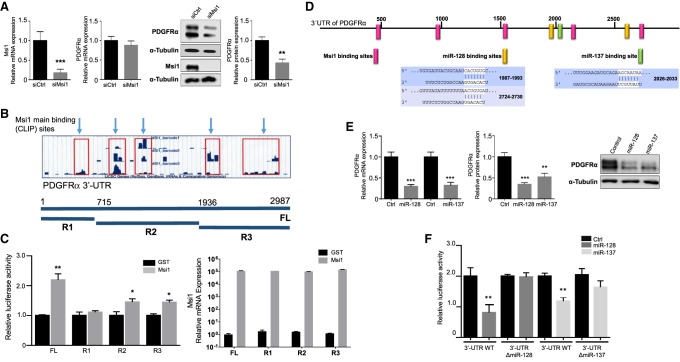
Considering Msi1 and miR-137 opposite roles in neurogenesis and tumorigenesis, we expect most of the shared targets to be regulated in opposite directions. To test that, we selected initially PDGFRα, one of the major nodes of the network described above. Msi1 was initially described to be a repressor of translation (Imai et al. 2001), but several studies later demonstrated that Msi1 also functions as an activator of translation in addition to roles in mRNA decay, splicing and polyadenylation (Kudinov et al. 2017). We have previously shown that Msi1 knockdown affects the profile of PDGFRα mRNA in polysomal gradients, suggesting that Msi1 increases its translation (Uren et al. 2015). In support, we show that Msi1 knockdown produces minimal changes at the mRNA level but drastically affects PDGFRα protein levels (Fig. 3A). Changes in PDGFRα protein levels are corroborated in U251 Msi1 KO cells (Supplemental Fig. S2A). Our CLIP analysis (Uren et al. 2015) indicates that Msi1 has multiple binding sites along PDGFRα 3′-UTR (Fig. 3B). We prepared four luciferase reporters, the first one containing the full-length 3′-UTR of PDGFRα and the other three containing adjacent fragments named R1, R2, and R3. An increase in luciferase activity was observed when the full-length 3′-UTR reporter was cotransfected with a vector expressing Msi1; in the cases of clones R2 and R3, Msi1 transgenic expression produced only a partial increase in reporter activity (Fig. 3B,C). The results suggest that binding of Msi1 to different sites along the 3′-UTR is necessary to properly regulate PDGFRα expression.
FIGURE 3.
PDGFRα is regulated by Msi1, miR-128, and miR-137. (A) U251 cells were transfected with siCtrl or siMsi1 for 72 h. Msi1 (left panel) and PDGFRα (middle panel) mRNA expression levels were determined by qRT-PCR. Protein levels of Msi1 and PDGFRα were evaluated by western blot, using α-Tubulin as loading control (right panel). Experiment was performed with biological and technical triplicates. (B) UCSC Genome Browser plots of three experimental replicas of Msi1 iCLIP analysis. Light blue arrows show Msi1 binding sites (iCLIP sites) in the 3′-UTR of PDGFRα. In the middle section, we show a diagram representing the inserts cloned into a luciferase reporter; full-length (FL) contains the entire 3′-UTR of PDGFRα, R1 (from nucleotides 1 to 715), R2 (from nucleotides 716 to 1936), and R3 (from nucleotides 1937 to 2987). (C) Results of luciferase assays. Cells were transfected with combinations of the luciferase reporter constructs shown in B and Msi1 or GST expressing vectors. Figure on the right shows Msi1 expression levels measured by qRT-PCR in each transfection. Experiment was performed with biological and technical triplicates. (D) Diagram shows predicted miR-128 and miR-137 binding sites in the 3′-UTR of PDGFRα according to TargetScan and main Msi1 binding sites identified by CLIP. Base pair interactions between the mRNA and the miRNA seed region are displayed as vertical lines. (E) PDGFRα mRNA levels measured by qRT-PCR of U251 cells transfected with miRNA mimics (control, miR-128, or miR-137) (left panel). Western blot analysis of PDGFRα levels in U251 cells transfected with miRNA mimics (control, miR-128, or miR-137). α-Tubulin was using like a loading control. The two PDGFRα bands detected in all western blots are likely due to differences in glycosylation (Ip et al. 2018). (F) Regulation of PDGFRα by miR-128 and miR-137 was validated using luciferase assays. Three luciferase constructs were used. The first one contains the wild-type sequence of PDGFRα 3′-UTR. In the other two, the predicted binding motifs for miR-137 or miR-128 were deleted. Experiment was performed with biological and technical triplicates. Data was analyzed with Student's t-test and are presented as the mean ± deviation standard. (*) P < 0.05, (**) P < 0.001, (***) P < 0.0001.
PDGFRα 3′-UTR also contains predicted binding sites for a large number of miRNAs. miR-34a inhibits colon cancer proliferation and metastasis by repressing PDGFRα (Li et al. 2015). Similarly, overexpression of miR-140-5p prevented ovarian cancer cell proliferation and induced apoptosis via PDGFRα regulation (Lan et al. 2015). Relevant to our study, miR-137 and miR-128, another Msi1 regulator, have conserved binding sites in PDGFRα 3′-UTR (Fig. 3D). Transfection of GBM cells with miR-128 or miR-137 showed a reduction in PDGFRα mRNA and protein levels (Fig. 3E; Supplemental Fig. S2B). The regulation of PDGFRα by each miRNA was validated via luciferase assays. We used the luciferase reporter described above and prepared additional constructs by deleting miR-137 and miR-128 predicted binding sites. Reduction in luciferase activity was observed when the PDGFRα 3′-UTR luciferase reporter was cotransfected with either miR-128 or miR-137 mimics. The cotransfections were repeated, this time with luciferase reporters with deleted miRNA binding sites. miR-128 was no longer able to inhibit the expression of the reporter while the effect was diminished in the case of miR-137 (Fig. 3F).
It could be argued that Msi1 positive impact on PDGFRα expression could take place via repression of miR-128 and -137 binding to their target sequences. This possibility is very unlikely since typically miR-128 and -137 display very low levels of expression in GBM. In any case, we conducted luciferase assays using constructs lacking the binding sites for miR-128 and -137. Deletion of either miR-128 or miR-137 binding sites did not affect Msi1 positive affect on the expression of the luciferase reporter vector; values for luciferase activity were very similar to the ones obtained with the wild-type 3′-UTR construct (Supplemental Fig. S2D).
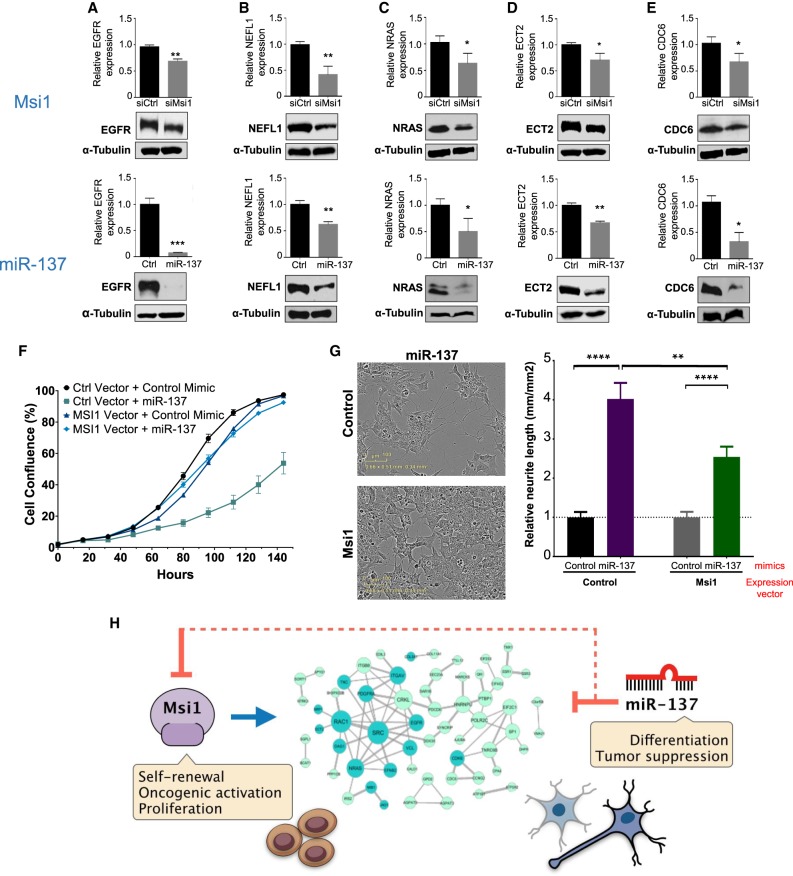
To further test our antagonistic model, we evaluated a set of shared targets of Msi1/miR-137 by western blot. Control and U251 GBM cells were transfected with Msi1 siRNA or miR-137 mimics and changes in protein levels were measured 48 h later. Treatment with Msi1 siRNA or miR-137 mimics decreased protein expression, suggesting that these target transcripts are repressed by miR-137 and activated by Msi1 (Fig. 4A–E).
FIGURE 4.
Msi1 and miR-137 have opposite regulatory effects on the expression of shared target genes. (A–E) Western blot analysis showing EGFR, NEFL1, NRAS, ECT2, and CDC6 expression in U251 cells transfected with Msi1 siRNA (siMsi1) or control siRNA (upper panel) and miR-137 or control mimics (Ctrl) (bottom panel). α-Tubulin was included as a loading control. (F) Graph shows proliferation curves obtained with the IncuCyte live imaging system of U251 cells transfected/infected with control mimics and control vector, miR-137 mimics and control vector, miR-137 mimics and Msi1 expressing vector. Experiment was performed using biological and technical triplicates. (G) BE(2)C cells were transfected with combinations of miRNA mimics and expression vectors, and impact on differentiation (neurite outgrowth) was measured 5 d later with Incucyte. Pictures on the left show aspect of transfected cells. Graphs on the right show relative neurite length in each condition. Experiment was performed using biological duplicates and technical quadruplicates. Data was analyzed using Student's t-test and are presented as the mean ± standard deviation t-test. (*) P < 0.05, (**) P < 0.001, (***) P < 0.0001. (H) Msi1 and miR-137 and their opposite impact on cell fate decisions. In our antagonistic model, miR-137 drives differentiation (tumor suppression) using a double negative switch: first, by direct inhibition of Msi1 and second, by repressing the expression of their shared targets. On the other hand, Msi1 positive impact on the expression of shared targets is central in its function in self-renewal, proliferation, and tumorigenesis.
Target repression by miR-137 could depend on Msi1 inhibition. To rule out this possibility and show direct repression of shared targets by miR-137, we conducted experiments in a U251 Msi1 KO line (de Araujo et al. 2016). U251 wild-type cells and U251 Msi1 KO were transfected with either control or miR-137 mimics and later, we measured by qRT-PCR the impact on mRNA levels of a list of Msi1/miR-137 shared targets. In all cases analyzed, we observed a reduction in mRNA levels upon miR-137 mimics transfection and the levels of repression in each case were similar between U251 Msi1 KO and U251 wild-type cells (Supplemental Fig. S3). We also measured the distances between the closest Msi1 and miR-137 binding sites in shared transcripts to determine if they tend to overlap or be in close proximity, what could suggest a direct competition between these regulators to bind to their targets. The median value is 220 nt. Although mRNAs form secondary structure that potentially could approximate binding sites, based on our current data, we do not favor the idea that Msi1 and miR-137 often compete for binding.
A model of RBP-miRNA antagonism and its potential implication in neurogenesis and gliomagenesis
Ectopic expression of miR-137 in GBM cells disrupts several cancer related phenotypes (Tamim et al. 2014; Sun et al. 2015). Similar results were observed upon Msi1 knockdown (Uren et al. 2015). To support the model of antagonism between Msi1 and miR-137, we determined if Msi1 is able to counteract or decrease miR-137 effect on cell proliferation and differentiation. Transfection of U251 cells with miR-137 mimics strongly reduced cell proliferation. However, when cells were also infected with a lentiviral construct containing Msi1 ORF, the negative effect of miR-137 mimics on cell proliferation was almost entirely neutralized by Msi1 (Fig. 4F). Similarly, we used the neuroblastoma BE-(2)-C line as a model to evaluate the impact of miR-137 versus Msi1 on neuronal differentiation. Cells were transfected/infected with miR-137 or control mimics in combination with Msi1 expressing lentivirus or control. After 5 d, we used the Incucyte system to measure neurite outgrowth as an indicator of differentiation. miR-137 effectively induced neurite formation but the effect was diminished in the presence of Msi1 (Fig. 4G).
Down-regulation of miR-137 is observed in multiple cancer types and in many cases, including glioblastoma, often as a result of hypermethylation of its promoter (Chen et al. 2011; Bier et al. 2013; Kang et al. 2015; Zhang et al. 2018). We suggest that down-regulation of miR-137 could function as an important event that will enhance the expression of Msi1 and their shared targets in glioma cells, promoting “stemness” and oncogenic activation (Fig. 4H).
Dramatic changes in gene expression take place during neurogenesis and gliomagenesis. The regulatory network modulated by Msi1 and miR-137 is likely part of a larger one involving other miRNAs and RBPs. In fact, we have already shown that miR-137 share a large number of targets with miR-124 and -128 and these three miRNAs work synergistically to regulate neurogenesis (Santos et al. 2016). miR-128 is also a regulator of Msi1 (Vo et al. 2011) and they potentially share several targets (Supplemental Table S1). Based on expression and distribution of binding sites, HafezQorani et al. (2016) predicted potential interactions involving miRNAs and RBPs. In particular, they show that HuR and Msi1 binding sites co-occur in a large number of transcripts. Cooperative interactions between these RBPs were suggested based on the fact that transcripts containing Msi1 sites within 200 nucleotides (nt) of the HuR sites are more destabilized upon HuR depletion (HafezQorani et al. 2016). Interestingly, we have shown previously that HuR positively regulates the expression of Msi1 and they show strong expression correlation in GBM samples (Vo et al. 2012a).
Conclusion
In summary, we propose that Msi1 and miR-137 modulate cell fate decisions by acting primarily on a shared network of target genes. In our antagonistic model, miR-137 drives differentiation (tumor suppression) using a double negative switch: first, by direct inhibition of Msi1 and second, by repressing the expression of their shared targets. On the other hand, Msi1 positive impact on the expression of shared targets is central for its function in self-renewal and proliferation (oncogenic activation) (Fig. 4H). Functional studies are required to identify Msi1/miR-137 main mediators of their roles in neurogenesis, self-renewal and brain tumor development.
MATERIALS AND METHODS
Cell culture
U251 and T98G glioblastoma cells, neuroblastoma BE-(2)-C and HT1080 fibrosarcoma cells were maintained in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (Thermo Fisher), 100 units/mL penicillin, 100 µg/mL streptomycin and 1 mM sodium pyruvate at 37°C in a 5% CO2 incubator. U251 Musashi1 Knockout line was described in de Araujo et al. (2016).
To prepare a U251 line stably expressing Musashi1, we transduced cells with a lentiviral vector containing Msi1 ORF (pReceiver-Lv242) (GeneCopoeia). A control line containing the empty vector was also prepared. A total of 2000 cells were seeded in 96-well cell plates and later transduced with lentivirus; a multiplicity of infection (MOI) of 10 per cell was used. After 24 h, cells were selected with 0.5 µg/mL puromycin. The transduction efficiency was determined daily with an inverted fluorescent microscope and was quantified by measuring the GFP-expressing cells as a percentage of the total number of visible cells.
NSC culture
All animal care and experimental procedures were approved by the UTHSCSA Institutional Animal Care and Use Committee (Protocol #13091X). SVZs were dissected under a stereoscope from 2-mo-old Swiss-Webster mice (Charles River Laboratories) and dissociated into single-cell suspension with 0.25% papain (Worthington Biochemical Corporation) and 12 mg/mL DNase (Sigma) in DMEM at 37°C for 45 min. Cells were washed 2× in DMEM by centrifugation and plated in a six-well plate (Corning) in the presence of N5 medium (DMEM/F-12/N-2), 5% fetal bovine serum (FBS, Thermo Fisher), 20 ng/mL epidermal growth factor (EGF, Thermo Fisher), 20 ng/mL basic fibroblast growth factor (bFGF, Peprotech), and 35 mg/mL bovine pituitary extract (Thermo Fisher). Media were changed every 2 d. After cells were confluent, they were transfected as described below. Differentiation of NSCs was induced by removing EGF, FGF, and FBS from the media.
siRNA transfection
Cells were transiently transfected with 25 nM of nontargeting control or Msi1 siRNA, using Lipofectamine RNAiMAX (Invitrogen). Cells were harvested after 72 h and subsequently used in assays described below. The following siRNAs were obtained from the Invitrogen Stealth RNAi collection: MSI1HSS106732, MSI1HSS106733, and MSI1HSS106734. Invitrogen siRNA negative control was # Cat. 12935-300. The following siRNAs are a part from the collection ThermoFisher Silencer Select: FSTL1 (s22034) cat# 4427037, PTGRFN (s11462) cat# 4427037, STC1 (s13549) cat# 4427037, CDH6 (s2784) cat# 4427037, DCBLD2 (s43617) cat# 4427037.
NSCs transfections were performed as described previously (Santos et al. 2016). Briefly, 105 cells were plated into a 24-well plate coated with poly-l-Ornithine (Sigma) and Laminin (Thermo Fisher). Cells were transiently transfected with control siRNA or Msi1 siRNA (MSI1HSS106732, MSI1HSS106733, MSI1HSS106734; Life Technologies, Thermo Fisher) using X-Treme siRNA transfection reagent (Roche). Experiments were performed using biological and technical replicates.
microRNAs mimics transfection
Cells were transfected with 5–25 nM of miR-128 (cat. no. MSY0000424, QIAGEN), miR-137 (cat. no. MSY0000429, QIAGEN), or miR-control mimics (cat. no. 1027280, QIAGEN), using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's instructions. Transfected cells were used in different assays described below.
RNA extraction, cDNA synthesis, and qRT-PCR
Total RNA from cells was isolated using TRIzol Reagent (Invitrogen). In the case of NSCs, total RNA was extracted from cells using the mirVana RNA Isolation System (Ambion), according to the manufacturer's instructions. cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol. Quantitative RT-PCR (qRT-PCR) was performed using a 7500 Real-Time PCR System (Applied Biosystems). TaqMan Universal PCR Master Mix (Applied Biosystems) and the following probes: Msi1 Hs01045894; PDGFRα Hs00998018; EGFR Hs01076078; ECT2, 00978168; NEFL1 Hs0019624; CDC6 Hs00154374; NRAS Hs00180035; GAPDH Hs99999905, FSTL1 Hs00907496_m1, PTGRFN Hs01385989_m1, STC1 Hs00174970_m1, CDH6 Hs00191832_m1, DCBLD2 Hs00294635_m1 were used. Data were acquired using the SDS 2.0.1 software package (Applied Biosystems) and analyzed with the 2−ΔΔCT method with GAPDH as a normalization control. For all comparisons, biological and technical triplicates were used.
Reverse transcription of miRNAs was performed with the High Capacity cDNA Reverse Transcription Kit (Thermo Fischer Scientific) and the miRNA-specific reverse primer. qRT-PCR was done using the TaqMan probes Hsa-miR-128 TR002216; hsa-miR-137, RT001129 and RNU48, TM 001006, from the TaqMan MicroRNA Assay (Applied Biosystems).
Luciferase reporter constructs
Luciferase reporter constructs containing the full-length 3′-UTR of PDGFRα or regions (nucleotides 1 to 715, R1), (nucleotides 716 to 1936, R2), and (nucleotides 1937 to 2987, R3) were cloned into the pmirGLO vector (Promega) upstream of the Firefly luciferase ORF.
Deletion of miR-128 and miR-137 sites in PDGFRα 3′-UTR was performed using the Quick Change Mutagenesis Kit (Agilent Technologies). The following primers were used: miR-128 poor conserved site, forward (GAAACATGGGCTGTGGCAGATGATGCTTTGG), reverse (CCAAAGCATCATCTGCCACAGCCCATGTTTC); miR-128 conserved site forward (GTATCACTGCCTTCGCCACAGGCACATTAAC), reverse (GTTAATGTGCCT GTGGCGAAGGCAGTGATAC); miR-137 forward (GCAGATGATGCTTTGGAAGACTACCTACTGGTGTAATC), reverse (GTAGGTAGTCAAGTACTTTAGCATCATCTGCCGATAGCAC).
Luciferase assay
HT1080 cells were transiently cotransfected in 96 well-plates with 45 ng of luciferase reporter plasmids described above and 15 ng of pcDNA3 (Invitrogen) constructs expressing either Msi1 or GST (control) using GeneJammer (Agilent Technologies). Forty-eight hours after transfection, cells were harvested and assayed for luciferases activity using the Dual Luciferase Assay System (Promega). Firefly luminescence activity was normalized using Renilla luminescence activity as an internal loading control. Biological and technical triplicates were used.
A similar experiment was performed to measure the impact of microRNAs on PDGFRα expression. HT1080 cells were cotransfected with 50 ng/µL of the PDGFRα-3′-UTR luciferase reporter vector (wild-type or miRNA site deletions) and 10 nmol of the pertinent miRNA or control mimic, using Lipofectamine 3000 (Invitrogen). Forty-eight hours after transfection, cells were harvested and assayed for luciferases activity as above. Firefly luminescence activity was normalized using Renilla luminescence activity as an internal loading control. Biological and technical triplicates were used.
Immunoblot analysis
Analysis of GBM cells
Cells were harvested in RIPA buffer (1% Nonidet P-40, 0.5% deoxycholate, and 0.1% sodium dodecylsulfate [SDS] in phosphate-buffered saline [PBS]) containing a protease inhibitor cocktail Complete Mini, EDTA Free (Roche) and subsequently sonicated (Branson model W-350). Extracts were centrifuged at 12,000 rpm for 10 min at 4°C and supernatant was recovered. Lysates (30 µg) were denatured in 2% SDS, 10 mM dithiothreitol, 60 mMTris-hydrochloric acid (Tris-HCl, pH 6.8), and 0.1% bromophenol blue and loaded onto a 15% polyacrylamide/SDS gel. Gel was transferred to a PVDF membrane that was latter blocked for 1 h at room temperature in PBS containing 5% nonfat dry milk. Next, the membrane was incubated overnight at 4°C in PBS-T containing the primary antibody. The membrane was washed in PBS-T, incubated with the secondary antibody conjugated to horseradish peroxidase for 1 h at room temperature, and then washed in PBS-T.
Antibodies
Anti-Msi1 (EP1302) (Abcam, ab52865; 1:2000 in 5% milk-TBST); anti-PDGF Receptor α (D13C6) XP (Cell Signaling, #5241; 1:1000 in 5% BSA-TBST); anti-EGF Receptor (C74B9) (Cell signaling #2646; 1:2000 in 5%BSA-TBST); anti-ECT2 (G-4) (Santa Cruz, sc-514750; 1:1000 5% milk-TBST); anti-NEFL1 (Cell signaling, #2835; 1:1000 in 5% milk-TBST), Anti-CDC6 (C42F7) (Cell signaling, #3387; 1:500 in 5% BSA-TBST); Anti-NRAS (27H5) (Cell signaling, #3339; 1:1000 in 5% BSA-TBST); anti-α Tubulin (GeneTex, GTX102078); Anti-GST (B-14) HRP (Santa Cruz, sc-138; 1:5000 in 5% milk-TBST).
Analysis of mouse samples
Cells were lysed in RIPA buffer and proteins were resolved on SDS-PAGE, and transferred to poly(vinylidene) difluoride membranes (GE Healthcare), which were blocked and incubated with anti-Msi1 antibody (1:1000; EMD Millipore) or anti-ß-Tubulin III (Tuj1, 1:500, Covance Inc.) followed by incubation with an anti-rabbit horseradish peroxide (HRP)-conjugated secondary antibody. As endogenous control, membranes were incubated with anti-β-Actin (Abcam).
The immunoblots were developed using the Immobilon Western Chemiluminescent HRP Substrate (Millipore).
Cell proliferation assay
U251 cells were transduced with either a lentiviral vector expressing Msi1 or control vector. Subsequently, cells were transfected with 10 nM of miRNA-137 or control mimics and transferred to an Essen ImageLock cell culture plate (Essen BioScience). Percentage confluence was monitored for 5 d using the high definition automated imaging system from IncuCyte (Essen BioScience) following the manufacturer's direction. Experiments were performed using biological and technical triplicates. Statistical analyses were carried out using GraphPad software and data are presented as mean ± standard deviation.
Immunocytochemistry
NSC cultures were fixed with 4% Paraformaldehyde (PFA) for 20 min, washed and blocked with 10% Normal Donkey Serum (NDS) in PBS-T for an hour prior to overnight incubation with primary antibody anti-β-Tubulin III (Tuj1, 1:500, Covance, Inc.). Cells were then stained with Alexa 488- or Alexa 546-conjugated secondary antibodies (Invitrogen, Inc., Thermo Fisher) and the nuclei were stained with 40,6-diamidino-2-phenylindole (DAPI) (Sigma).
Quantification of neurite outgrowth
Ninety-six hours after transfection, cells were imaged by an IncuCyte at 10× or 20× magnification. Neurite outgrowth was quantified on an IncuCyte ZOOM Imaging System (Essen BioScience). A neurite definition specific to BE-(2)-C cells was created using the NeuroTrack software module (Essen BioScience). Experiments were performed using biological duplicates and technical quadruplicates.
Cell counting
The experiments were done with three biological replicates. DAPI1 nuclei were identified, and then each nucleus was checked for Tuj1. For each triplicate, a minimum of five fields were assessed for DAPI-positive nuclei and anti-Tuj1. The proportion of Tuj1+ was determined for all 15 fields (five fields for each triplicate). Some samples were counted several times to assess the reliability of counts.
Musashi1, miR-137 expression correlation
To evaluate if Musashi1 and miR-137 expression profiles in gliomas are anticorrelated, mRNA and miRNA expression data was downloaded from The Cancer Genome Atlas (TCGA). mRNA expression data was normalized using Upper Quartile normalized FPKM, as provided in the Genomics Data Commons (GDC) data portal. miRNA expression data was normalized using reads per million miRNA mapped, as provided in the GDC data portal. In total, 508 subjects had both mRNA and miRNA expression data. A square root transformation was applied to both Musashi1 and miR-137 expression for variance stabilization. The Pearson correlation coefficient and P-value were computed between the transformed expression of Musashi1 and miR-137.
miRNA and RBP target identification and comparison
miRNA targets
Target predictions for broadly conserved miRNA families were obtained from Targetscan (Agarwal et al. 2015). An expanded list of miR-137 targets was generated using data sets published in (Tamim et al. 2014; Santos et al. 2016) and validated targets from miRTarBase (Hsu et al. 2011). A list of miRNA target sites per transcript was generated and the number of miRNAs regulating each transcript was calculated and used in further analyses.
RBP targets
A list of Musashi1 targets identified by RIP or CLIP were generated using data sets described in de Sousa Abreu et al. (2009), Vo et al. (2012b), and Uren et al. (2015). Target lists for hnRNPH1 and IGF2BP3 were obtained from Ennajdaoui et al. (2016) and Uren et al. (2016).
miRNA-RBP target overlap
We compared miRNAs and RBPs target sets to identify miRNAs sharing a large number of targets with each RBP (10% or more of the total number of identified targets). Next, we evaluate each RBP-miRNA overlap to determine the number of transcripts containing just a small number of miRNA binding sites (less than 3 or less than 5 miRNA binding sites for conserved miRNA families). Significance of each overlap was calculated using Fisher's exact test (P-value <0.01).
miR-137 and Msi1 shared target analysis
Gene Ontology (GO) Enrichment Analysis was carried out using PANTHER (Thomas et al. 2003). REVIGO (Supek et al. 2011) was used to compile and link similar GO terms. Network analysis was performed using STRING (version 10.5) (Szklarczyk et al. 2015).
To compare the expression profile of Msi1 and miR-137 shared target genes in cortex, LGG and GBM, we used the following procedure. Gene read counts from 156 glioblastoma (GBM) and 511 low-grade glioma (LGG) primary tumors were directly obtained from the TCGA data repository (https://portal.gdc.cancer.gov/), while gene read counts from 287 healthy (frontal) cortex samples were obtained from GTEx release V7 (https://www.gtexportal.org/home/). To process expression data, read counts were first normalized using the RLE method of DESeq2 (Love et al. 2014). Then, we separately performed differential expression analyses to compare GBM with (frontal) cortex and LGG samples. All comparisons were performed using DESeq2 and only genes presenting |log2fold change| ≥ 1 and FDR < 0.05 were considered to be differentially expressed. Next, we analyzed a list of 141 genes targeted by both miR-137 and Msi1. Finally, after log-transforming normalized data, heatmaps of differentially expressed targets were created using R scripts.
Survival and expression analyses of Msi1/miR-137 shared targets using TCGA glioma data were performed using resources in Gliovis (Bowman et al. 2017).
To analyze the distances between miR-137 and Msi1 target sites, we obtained the coordinates of miR-137 sites from TargetScan. Msi1 sites were selected by filtering only iCLIP sites at 3′-UTR regions and corresponding to targets shared between MsI1 and miR-137. Distances were calculated between each miR-137 site and the closest Msi1 site.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The Owens Foundation (L.O.F.P. and E.K.) and National Institutes of Health (NIH) 2R01 HG006015 (L.O.F.P.). G.H. was supported by an internal funding program of the National Institute of Cancer (Mexico). B.R.C. was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo—FAPESP (2013/25483-4 and 2013/07159-5) and G.D.A.G. (2017/19541-2). M.X.V. is a student in the Biomedical Sciences Program at UNAM and was supported by the National Council of Science and Technology (CONACyT ID 270268), Financial Aid for Postgraduate Study Program (PAEP), Mexican College for Cancer Research (C-MIC; CONACyT ID 295466), and Programa de Doctorado en Ciencias Biomédicas, Facultad de Medicina, Universidad Nacional Autónoma de México (CONACyT ID 270268). A.K. was supported by an NIH Supplement to 2R01 HG006015 and the Greehey Foundation.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.069211.118.
REFERENCES
- Abaza HMH, Elmougy MI, El Maraghy HMA, Mahmoud HM. 2016. Stanniocalcin1 gene expression in patients with acute leukemia: impact on response to therapy and disease outcome. Int J Lab Hematol 38: 81–89. 10.1111/ijlh.12445 [DOI] [PubMed] [Google Scholar]
- Agarwal V, Bell GW, Nam J-W, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4: e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. 2015. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol 129: 829–848. 10.1007/s00401-015-1432-1 [DOI] [PubMed] [Google Scholar]
- Arora S, Rana R, Chhabra A, Jaiswal A, Rani V. 2013. MiRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics 288: 77–87. 10.1007/s00438-013-0734-z [DOI] [PubMed] [Google Scholar]
- Banelli B, Forlani A, Allemanni G, Morabito A, Pistillo MP, Romani M. 2017. MicroRNA in glioblastoma: an overview. Int J Genomics 2017: 7639084 10.1155/2017/7639084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier A, Giladi N, Kronfeld N, Lee HK, Cazacu S, Xiang C, Poisson L, Ana C, Slavin S, Yalon M, et al. 2013. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget 4: 665–676. 10.18632/oncotarget.928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M. 2017. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol 19: 139–141. 10.1093/neuonc/now247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KK, Leung CO, Wong CC, Ho DW, Chok KS, Lai CL, Ng IO, Lo RC. 2017. Secretory Stanniocalcin 1 promotes metastasis of hepatocellular carcinoma through activation of JNK signaling pathway. Cancer Lett 403: 330–338. 10.1016/j.canlet.2017.06.034 [DOI] [PubMed] [Google Scholar]
- Chang SJ, Weng SL, Hsieh JY, Wang TY, Chang MD, Wang HW. 2011. MicroRNA-34a modulates genes involved in cellular motility and oxidative phosphorylation in neural precursors derived from human umbilical cord mesenchymal stem cells. BMC Med Genomics 4: 65 10.1186/1755-8794-4-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X, Zhang H, Lian S, Zhu W. 2016. miR-137 suppresses tumor growth of malignant melanoma by targeting aurora kinase A. Biochem Biophys Res Commun 475: 251–256. 10.1016/j.bbrc.2016.05.090 [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen X, Zhang M, Fan Q, Luo S, Cao X. 2011. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig Dis Sci 56: 2009–2016. 10.1007/s10620-010-1536-3 [DOI] [PubMed] [Google Scholar]
- Chen L, Wang X, Wang H, Li Y, Yan W, Han L, Zhang K, Zhang J, Wang Y, Feng Y, et al. 2012. miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur J Cancer 48: 3104–3111. 10.1016/j.ejca.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Ciafrè SA, Galardi S. 2013. microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol 10: 934–942. 10.4161/rna.24641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespel A, Rigau V, Coubes P, Rousset MC, de Bock F, Okano H, Baldy-Moulinier M, Bockaert J, Lerner-Natoli M. 2005. Increased number of neural progenitors in human temporal lobe epilepsy. Neurobiol Dis 19: 436–450. 10.1016/j.nbd.2005.01.020 [DOI] [PubMed] [Google Scholar]
- Dahlrot RH, Hansen S, Hjelmborg J, Kristensen BW. 2013. Prognostic value of Musashi-1 in gliomas. J Neuroncology 115: 453–461. 10.1007/s11060-013-1246-8 [DOI] [PubMed] [Google Scholar]
- de Araujo PR, Grothi A, da SA E, Tonapi SS, Burns SC, Qiao M, Uren PJ, Yuan Z-M, Bishop AJR, Penalva LOF. 2016. Musashi1 impacts radio-resistance in glioblastoma by controlling DNA-protein kinase catalytic subunit. Am J Pathol 186: 2271–2278. 10.1016/j.ajpath.2016.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, Burns SC, Ko D, Burton TL, Vo DT, Chennasamudaram S, Le S-Y, Shapiro BA, et al. 2009. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J Biol Chem 284: 12125–12135. 10.1074/jbc.M809605200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Jin M, Li Y, Ren P, Liu J. 2016. MiR-137 acts as a tumor suppressor in papillary thyroid carcinoma by targeting CXCL12. Oncol Rep 35: 2151–2158. 10.3892/or.2016.4604 [DOI] [PubMed] [Google Scholar]
- Ennajdaoui H, Howard JM, Sterne-Weiler T, Blencowe BJ, Penalva LOF, Sanford JR. 2016. IGF2BP3 modulates the interaction of invasion-associated transcripts with RISC. Cell Rep 15: 1876–1883. 10.1016/j.celrep.2016.04.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom K-Y, Cho BJ, Choi EJ, Kim J-H, Chie EK, Wu H-G, Kim IH, Paek SH, Kim J-S, Kim IA. 2016. The effect of chemoradiotherapy with SRC tyrosine kinase inhibitor, PP2 and temozolomide on malignant glioma cells in vitro and in vivo. Cancer Res Treat 48: 687–697. 10.4143/crt.2014.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Liu L, Li S, Zhang X, Chang Z, Zhang M. 2015. Inhibition of cell proliferation and metastasis of human hepatocellular carcinoma by miR-137 is regulated by CDC42. Oncol Rep 34: 2523–2532. 10.3892/or.2015.4261 [DOI] [PubMed] [Google Scholar]
- Gardiner AS, Twiss JL, Perrone-bizzozero NI. 2015. Competing interactions of RNA-binding proteins, microRNAs, and their targets control neuronal development and function. Biomolecules 5: 2903–2918. 10.3390/biom5042903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer RI, Vo DT, Penalva LOF. 2012. Musashi1: an RBP with versatile functions in normal and cancer stem cells. Front Biosci 17: 54–64. 10.2741/3915 [DOI] [PubMed] [Google Scholar]
- HafezQorani S, Lafzi A, De Bruin RG, Van Zonneveld AJ, Van Der Veer EP, Son A, Kazan H. 2016. Modeling the combined effect of RNA-binding proteins and microRNAs in post-transcriptional regulation. Nucl Acids Res 44: e83 10.1093/nar/gkw048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Zhang W, Yang X, Wheeler CG, Langford CP, Wu L, Filippova N, Friedman GK, Ding Q, Fathallah-Shaykh HM, et al. 2014. The role of Src family kinases in growth and migration of glioma stem cells. Int J Oncol 45: 302–310. 10.3892/ijo.2014.2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Bi Y, Bi H, Diao C, Zhang G, Cheng K, Yang Z. 2016. miR-137 suppresses the invasion and procedure of EMT of human breast cancer cell line MCF-7 through targeting CtBP1. Hum Cell 29: 30–36. 10.1007/s13577-015-0124-4 [DOI] [PubMed] [Google Scholar]
- Hofsli E, Wheeler TE, Langaas M, Lægreid A, Thommesen L. 2008. Identification of novel neuroendocrine-specific tumour genes. Br J Cancer 99: 1330–1339. 10.1038/sj.bjc.6604565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. 2011. MiRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 39: D163–D169. 10.1093/nar/gkq1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. 2001. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol 21: 3888–3900. 10.1128/MCB.21.12.3888-3900.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip CKM, Ng PKS, Jeong KJ, Shao SH, Ju Z, Leonard PG, Hua X, Vellano CP, Woessner R, Sahni N, et al. 2018. Neomorphic PDGFRA extracellular domain driver mutations are resistant to PDGFRA targeted therapies. Nat Commun 9: 4583 10.1038/s41467-018-06949-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaoka R, Nagata T, Tsuda K, Imai T, Okano H, Kobayashi N, Katahira M. 2017. Structural insight into the recognition of r(UAG) by Musashi-1 RBD2, and construction of a model of Musashi-1 RBD1-2 bound to the minimum target RNA. Molecules 22: E1207 10.3390/molecules22071207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lou X. 2013. Alternative polyadenylation in glioblastoma multiforme and changes in predicted RNA binding protein profiles. Omi A J Interact Biol 17: 136–149. 10.1089/omi.2012.0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang N, Choi SY, Kim YK, Yoo IR, Han DH, Lee DS, Kim YS, Hong SH, Kang JH, Lee KY, et al. 2015. Silencing of miR-137 by aberrant promoter hypermethylation in surgically resected lung cancer. Lung Cancer 89: 99–103. 10.1016/j.lungcan.2015.05.011 [DOI] [PubMed] [Google Scholar]
- Kikuta K, Kubota D, Yoshida A, Qiao Z, Morioka H, Nakamura M, Matsumoto M, Chuman H, Kawai A, Kondo T. 2017. Discoidin, CUB and LCCL domain-containing protein 2 (DCBLD2) is a novel biomarker of myxo fibrosarcoma invasion identified by global protein expression profiling. Biochim Biophys Acta Proteins Proteom 1865: 1160–1166. 10.1016/j.bbapap.2017.06.023 [DOI] [PubMed] [Google Scholar]
- Kozlowska E, Krzyzosiak WJ, Koscianska E. 2013. Regulation of Huntingtin gene expression by miRNA-137, -214, -148a, and their respective isomiRs. Int J Mol Sci 14: 16999–17016. 10.3390/ijms140816999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudinov AE, Karanicolas J, Golemis EA, Boumber Y. 2017. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin Cancer Res 23: 2143–2154. 10.1158/1078-0432.CCR-16-2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H, Chen W, He G, Yang S. 2015. miR-140-5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed Pharmacother 75: 117–122. 10.1016/j.biopha.2015.07.035 [DOI] [PubMed] [Google Scholar]
- Lennox AL, Mao H, Silver DL. 2018. RNA on the brain: emerging layers of post-transcriptional regulation in cerebral cortex development. Wiley Interdiscip Rev Dev Biol 7: 1–18. 10.1002/wdev.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KK-W, Yang L, Pang JC, Chan AK, Zhou L, Mao Y, Wang Y, Lau K, Poon WS, Shi Z, et al. 2013. MIR-137 suppresses growth and invasion, is downregulated in oligodendroglial tumors and targets CSE1L. Brain Pathol 23: 426–439. 10.1111/bpa.12015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wnag Y, Lu S, Zhang Z, Meng H, Liang L, Zhang Y, Song B. 2015. miR-34a inhibits colon cancer proliferation and metastasis by inhibiting platelet-derived growth factor receptor α. Mol Med Rep 12: 7072–7078. 10.3892/mmr.2015.4263 [DOI] [PubMed] [Google Scholar]
- Li Y, He Z-C, Zhang X-N, Liu Q, Chen C, Zhu Z, Chen Q, Shi Y, Yao X-H, Cui Y-H, et al. 2018. Stanniocalcin-1 augments stem-like traits of glioblastoma cells through binding and activating NOTCH1. Cancer Lett 416: 66–74. 10.1016/j.canlet.2017.11.033 [DOI] [PubMed] [Google Scholar]
- Liu N, Zhang L, Wang Z, Cheng Y, Zhang P, Wang X, Wen W, Yang H, Liu H, Jin W, et al. 2016. MicroRNA-101 inhibits proliferation, migration and invasion of human glioblastoma by targeting SOX9. Oncotarget 8: 19244–19254. 10.18632/oncotarget.13706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y-H, Mentlein R, Knerlich F, Kruse ML, Mehdorn HM, Held-Feindt J. 2008. Expression of stem cell markers in human astrocytomas of different WHO grades. J Neurooncol 86: 31–45. 10.1007/s11060-007-9439-7 [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Walhout AJM. 2009. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. BioEssays 31: 435–445. 10.1002/bies.200800212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Romero J, Bueno-Fortes S, Martín-Merino M, De Molina AR, De Las Rivas J. 2018. Survival marker genes of colorectal cancer derived from consistent transcriptomic profiling. BMC Genomics 19: 857 10.1186/s12864-018-5193-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiotti A, Prakash S, Barnett P, Van Den Hoff MJB. 2018. Follistatin-like 1 in development and human diseases. Cell Mol Life Sci 75: 2339–2354. 10.1007/s00018-018-2805-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neault M, Mallette FA, Richard S. 2016. MiR-137 modulates a tumor suppressor network-inducing senescence in pancreatic cancer cells. Cell Rep 14: 1966–1978. 10.1016/j.celrep.2016.01.068 [DOI] [PubMed] [Google Scholar]
- Neelamraju Y, Hashemikhabir S, Janga SC. 2015. The human RBPome: from genes and proteins to human disease. J Proteomics 127: 61–70. 10.1016/j.jprot.2015.04.031 [DOI] [PubMed] [Google Scholar]
- Nussbacher JK, Yeo GW. 2018. Systematic discovery of RNA binding proteins that regulate microRNA levels. Mol Cell 69: 1005–1016. 10.1016/j.molcel.2018.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preusse M, Marr C, Saunders S, Maticzka D, Lickert H, Backofen R, Theis F. 2015. SimiRa: a tool to identify coregulation between microRNAs and RNA-binding proteins. RNA Biol 12: 998–1009. 10.1080/15476286.2015.1068496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Huang D, Yin D, Li F, Li X, Kung HF, Peng Y. 2013. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochim Biophys Acta 1832: 1697–1707. 10.1016/j.bbadis.2013.05.015 [DOI] [PubMed] [Google Scholar]
- Rajgor D. 2018. Macro roles for microRNAs in neurodegenerative diseases. Noncoding RNA Res 3: 154–159. 10.1016/j.ncrna.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajman M, Schratt G. 2017. MicroRNAs in neural development: from master regulators to fine-tuners. Development 144: 2310–2322. 10.1242/dev.144337 [DOI] [PubMed] [Google Scholar]
- Raman P, Maddipati R, Lim KH, Tozeren A. 2018. Pancreatic cancer survival analysis defines a signature that predicts outcome. PLoS One 13: e0201751 10.1371/journal.pone.0201751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod SS, Rani SB, Khan M, Muzumdar D, Shiras A. 2014. Tumor suppressive miRNA-34a suppresses cell proliferation and tumor growth of glioma stem cells by targeting Akt and Wnt signaling pathways. FEBS Open Bio 4: 485–495. 10.1016/j.fob.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezapour S, Bahrami T, Hashemzadeh S, Estiar MA, Nemati M, Ravanbakhs R, Feizi MA, Kafil HS, Pouladi N, Ghojazadeh M, et al. 2016. STC1 and NF-κB p65 (Rel A) is constitutively activated in colorectal cancer. Clin Lab 62: 463–469. 10.7754/Clin.Lab.2015.150827 [DOI] [PubMed] [Google Scholar]
- Rooj AK, Ricklefs F, Mineo M, Nakano I, Chiocca EA, Bronisz A, Godlewski J. 2017. MicroRNA-mediated dynamic bidirectional shift between the subclasses of glioblastoma stem-like cells. Cell Rep 19: 2026–2032. 10.1016/j.celrep.2017.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M, Hisamori S, Oshima N, Sato F, Shimono Y, Sakai Y. 2016. miR-137 regulates the tumorigenicity of colon cancer stem cells through the inhibition of DCLK1. Mol Cancer Res 137: 354–362. 10.1158/1541-7786.MCR-15-0380 [DOI] [PubMed] [Google Scholar]
- Sakakibara SI, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. 2002. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci 99: 15194–15199. 10.1073/pnas.232087499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MCT, Tegge AN, Correa BR, Mahesula S, Kohnke LQ, Qiao M, Ferreira MAR, Kokovay E, Penalva LOF. 2016. miR-124, -128, and -137 orchestrate neural differentiation by acting on overlapping gene sets containing a highly connected transcription factor network. Stem Cells 34: 220–232. 10.1002/stem.2204 [DOI] [PubMed] [Google Scholar]
- Saraiva C, Esteves M, Bernardino L. 2017. MicroRNA: basic concepts and implications for regeneration and repair of neurodegenerative diseases. Biochem Pharmacol 141: 118–131. 10.1016/j.bcp.2017.07.008 [DOI] [PubMed] [Google Scholar]
- Serna-Blasco R, Sanz-Álvarez M, Aguilera Ó, García-Foncillas J. 2018. Targeting the RAS-dependent chemoresistance: the Warburg connection. Semin Cancer Biol 54: 80–90. 10.1016/j.semcancer.2018.01.016 [DOI] [PubMed] [Google Scholar]
- Shan Z, Tian R, Zhang M, Gui Z, Wu J, Ding M, Zhou X-F, He J. 2016. miR128-1 inhibits the growth of glioblastoma multiforme and glioma stem-like cells via targeting BMI1 and E2F3. Oncotarget 7: 78813–78826. 10.18632/oncotarget.12385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea A, Harish V, Afzal Z, Chijioke J, Kedir H, Dusmatova S, Roy A, Ramalinga M, Harris B, Blancato J, et al. 2016. MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics. Cancer Med 5: 1917–1946. 10.1002/cam4.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmyo Y, Kihara T, Akaike A, Niidome T, Sugimoto H. 2007. Trophism of neural progenitor cells to embryonic stem cells: neural induction and transplantation in a mouse ischemic stroke model. J Neurosci Res 85: 1851–1862. 10.1002/jnr.21319 [DOI] [PubMed] [Google Scholar]
- Sun J, Zheng G, Gu Z, Guo Z. 2015. MiR-137 inhibits proliferation and angiogenesis of human glioblastoma cells by targeting EZH2. J Neurooncol 122: 481–489. 10.1007/s11060-015-1753-x [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Nieves Š, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasawa K, Kitagawa K, Yagita Y, Sasaki T, Tanaka S, Matsushita K, Ohstuki T, Miyata T, Okano H, Hori M, et al. 2002. Increased proliferation of neural progenitor cells but reduced survival of newborn cells in the contralateral hippocampus after focal cerebral ischemia in rats. J Cereb Blood Flow Metab 22: 299–307. 10.1097/00004647-200203000-00007 [DOI] [PubMed] [Google Scholar]
- Tamim S, Vo DT, Uren PJ, Qiao M, Bindewald E, Kasprzak WK, Shapiro BA, Nakaya HI, Burns SC, Araujo PR, et al. 2014. Genomic analyses reveal broad impact of miR-137 on genes associated with malignant transformation and neuronal differentiation in glioblastoma cells. PLoS One 9: e85591 10.1371/journal.pone.0085591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, et al. 2003. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 31: 334–341. 10.1093/nar/gkg115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Mingyi M, Qiu X, Qiu Y, Tian T, Mingyi M, Qiu X, Qiu Y. 2016. MicroRNA-101 reverses temozolomide resistance by inhibition of GSK3β in glioblastoma. Oncotarget 7: 79584–79595. 10.18632/oncotarget.12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, Kawase T, Kawakami Y, Okano H, Uyemura K. 2001. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia 34: 1–7. 10.1002/glia.1034 [DOI] [PubMed] [Google Scholar]
- Tonchev AB, Yamashima T, Zhao L, Okano HJ, Okano H. 2003. Proliferation of neural and neuronal progenitors after global brain ischemia in young adult macaque monkeys. Mol Cell Neurosci 23: 292–301. 10.1016/S1044-7431(03)00058-7 [DOI] [PubMed] [Google Scholar]
- Tonchev AB, Yamashima T, Sawamoto K, Okano H. 2005. Enhanced proliferation of progenitor cells in the subventricular zone and limited neuronal production in the striatum and neocortex of adult macaque monkeys after global cerebral ischemia. J Neurosci Res 81: 776–788. 10.1002/jnr.20604 [DOI] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Plessmann U, Harlander S, Daiß JL, Eichner N, Lehmann G, Schall K, Urlaub H, Meister G. 2017. A compendium of RNA-binding proteins that regulate microRNA biogenesis. Mol Cell 66: 270–284. 10.1016/j.molcel.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Uren PJ, Vo DT, De Araujo R, Pötschke R, Burns SC, Bahrami-Samani E, Qiao M, Abreu RDS, Nakaya HI, Correa BR, et al. 2015. RNA-binding protein Musashi1 is a central regulator of adhesion pathways in glioblastoma. Mol Cell Biol 35: 2965–2978. 10.1128/MCB.00410-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren PJ, Bahrami-Samani E, de Araujo PR, Qiao M, Burns SC, Smith AD, Penalva LOF. 2016. High-throughput analyses of hnRNP H1 dissects its multi-functional aspect. RNA Biol 13: 400–411. 10.1080/15476286.2015.1138030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno C, Mansilla S, Portugal J. 2015. Sp1 transcription factor: a long-standing target in cancer chemotherapy. Pharmacol Ther 152: 111–124. 10.1016/j.pharmthera.2015.05.008 [DOI] [PubMed] [Google Scholar]
- Vo DT, Qiao M, Smith AD, Burns SC, Brenner AJ, Penalva LOF. 2011. The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA Biol 8: 817–828. 10.4161/rna.8.5.16041 [DOI] [PubMed] [Google Scholar]
- Vo D, Abdelmohsen K, Martindale J, Qiao M, Tominaga K, Burton T, Gelfond J, Brenner A, Patel V, Trageser D, et al. 2012a. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol Cancer Res 10: 143–155. 10.1158/1541-7786.MCR-11-0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo DT, Subramaniam D, Remke M, Burton TL, Uren PJ, Gelfond JA, de Sousa Abreu R, Burns SC, Qiao M, Suresh U, et al. 2012b. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am J Pathol 181: 1762–1772. 10.1016/j.ajpath.2012.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qi Z, Zhou M, Yang W, Hu R, Li G, Ma X, Zhang Z. 2019. Stanniocalcin-1 promotes cell proliferation, chemoresistance and metastasis in hypoxic gastric cancer cells via Bcl-2. Oncol Rep 41: 1998–2008. 10.3892/or.2019.6980 [DOI] [PubMed] [Google Scholar]
- Wu D-C, Zhang M-F, Su S-G, Fang H-Y, Wang X-H, He D, Xie Y-Y, Liu X-H. 2016. HEY2, a target of miR-137, indicates poor outcomes and promotes cell proliferation and migration in hepatocellular carcinoma. Oncotarget 7: 38052–38063. 10.18632/oncotarget.9343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita Y, Kitagawa K, Ohtsuki T, Takasawa K, Miyata T, Okano H, Hori M, Matsumoto M. 2001. Neurogenesis by progenitor cells in the ischemic adult rat hippocampus. Stroke 32: 1890–1896. 10.1161/01.STR.32.8.1890 [DOI] [PubMed] [Google Scholar]
- Yagita Y, Kitagawa K, Sasaki T, Miyata T, Okano H, Hori M, Matsumoto M. 2002. Differential expression of Musashi1 and nestin in the adult rat hippocampus after ischemia. J Neurosci Res 69: 750–756. 10.1002/jnr.10342 [DOI] [PubMed] [Google Scholar]
- Yano M, Ohtsuka T, Okano H. 2015. RNA-binding protein research with transcriptome-wide technologies in neural development. Cell Tissue Res 359: 135–144. 10.1007/s00441-014-1923-8 [DOI] [PubMed] [Google Scholar]
- Yin J, Lin J, Luo X, Chen Y, Li Z, Ma G, Li K. 2014. MiR-137: a new player in schizophrenia. Int J Mol Sci 15: 3262–3271. 10.3390/ijms15023262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang RZ, Yao Y, Liu ZH, Lian ZG, Zou YJ, Wei YK. 2004. Proliferation and differentiation of neural stem cells in adult rats after cerebral infarction. Chin Med Sci J 26: 8–11. [PubMed] [Google Scholar]
- Zhang W, Chen JH, Shan T, Aguilera-Barrantes I, Wang LS, Huang THM, Rader JS, Sheng X, Huang YW. 2018. miR-137 is a tumor suppressor in endometrial cancer and is repressed by DNA hypermethylation. Lab Invest 98: 1397–1407. 10.1038/s41374-018-0092-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LX, Li HZ, Guo RY, Ma T, Hou RY, Ma XW, Du YF. 2013. miR-137, a new target for post-stroke depression? Neural Regen Res 8: 2441–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chen D, Cai Y, Zhang F, Xu J, Zhao X, Chen D, Cai Y, Zhang F, Xu J. 2018. RBPvsMIR: a computational pipeline to identify competing miRNAs and RNA-binding protein pairs regulating the shared transcripts. Genes 9: E426 10.3390/genes9090426 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.