SUMMARY
Human erythropoiesis serves as a paradigm of physiologic cellular differentiation. This process is also of considerable interest for better understanding anemias and identifying new therapies. Here, we apply deep transcriptomic and accessible chromatin profiling to characterize a faithful ex vivo human erythroid differentiation system from hematopoietic stem and progenitor cells. We reveal stage-specific transcriptional states and chromatin accessibility during various stages of erythropoiesis, including 14,260 differentially expressed genes and 63,659 variably accessible chromatin peaks. Our analysis suggests differentiation stage-predominant roles for specific master regulators, including GATA1 and KLF1. We integrate chromatin profiles with common and rare genetic variants associated with erythroid cell traits and diseases, finding that variants regulating different erythroid phenotypes likely act at variable points during differentiation. In addition, we identify a regulator of terminal erythropoiesis, TMCC2, more broadly illustrating the value of this comprehensive analysis to improve our understanding of erythropoiesis in health and disease.
In Brief
Ludwig et al. chart the dynamic transcriptional and chromatin landscapes as hematopoietic stem and progenitor cells differentiate into mature red blood cells. This multi-omic profiling reveals dynamic transcription factor activities and human genetic variation that modulate this process.
Graphical Abstract
INTRODUCTION
Erythropoiesis describes the process of proliferation and differentiation of hematopoietic stem and progenitor cells (HSPCs) through distinct functionally and morphologically defined stages to produce enucleate reticulocytes. In the circulation, these cells mature further into red blood cells (RBCs), which are the key transporters of oxygen and carbon dioxide for cellular respiration (Nandakumar et al., 2016; Sankaran and Weiss, 2015). In adult humans, approximately 2 million RBCs are produced every second in the bone marrow through the tightly coordinated process of erythropoiesis, making RBCs the most abundant cell type in the human body (Palis, 2014). Congenital as well as acquired defects may lead to various forms of anemia, the lack of sufficient RBCs, causing significant morbidity and mortality (Sankaran and Weiss, 2015).
Genetic and cell biological approaches have provided important insights into normal RBC differentiation and how this process is perturbed in different blood diseases, such as anemias (Arlet et al., 2014; Giani et al., 2016; Khajuria et al., 2018; Ludwig et al., 2014, 2016). Our knowledge of this process has been further enhanced by comprehensive transcriptomic and proteomic profiling of distinct stages of neonatal and adult erythropoiesis (An et al., 2014; Gautier et al., 2016; Li et al., 2014; Yan et al., 2018). Additional efforts have recorded the accessible chromatin landscape (Buenrostro et al., 2018; Corces et al., 2016) and epigenetic marks such as histone modifications (Huang et al., 2016) at select stages and investigated the role of genetic variation and chromatin accessibility in murine cellular models (Behera et al., 2018). However, we lack a detailed characterization of the accessible chromatin landscape and transcription factor dynamics throughout the entire process of human erythroid differentiation, even though such characterization would facilitate a more comprehensive understanding of gene regulatory dynamics and their relation to human genetic variation and disease.
Here, we leverage the assay for transposase accessible chromatin using sequencing (ATAC-seq) (Corces et al., 2016, 2017) to assess eight populations that chart the dynamic open chromatin landscapes of HSPCs undergoing the process of erythroid differentiation. We integrate our data with matched deep transcriptomic profiles, results from genome-wide association studies (GWAS), and mutations resulting in human disease, to facilitate a systems-level understanding of the molecular circuits governing erythropoiesis in human health and disease. We show that our chromatin accessibility profiles yield a detailed description of the regulatory elements and transcription factor dynamics that control gene expression during human erythropoiesis. The intersection with variants identified in GWAS and Mendelian diseases reveal the differential and putative temporal contributions of genetic variants in accessible chromatin in affecting human erythroid traits and pathologies. Overall, our integrated analysis framework provides a comprehensive resource to advance our understanding of gene regulation and will expedite downstream functional and validation studies of regulators, as exemplified by our use of this dataset to identify a role for TMCC2 in terminal human erythropoiesis.
RESULTS
An Epigenomic and Transcriptional Time Course of Human Erythroid Differentiation
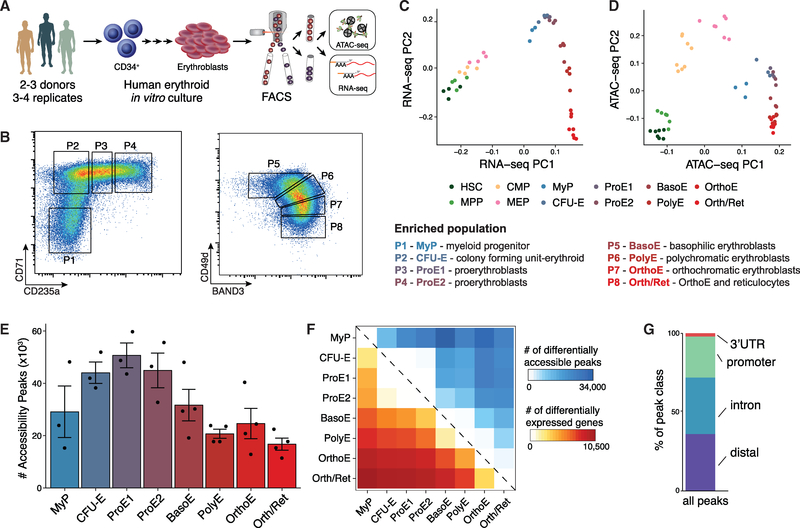
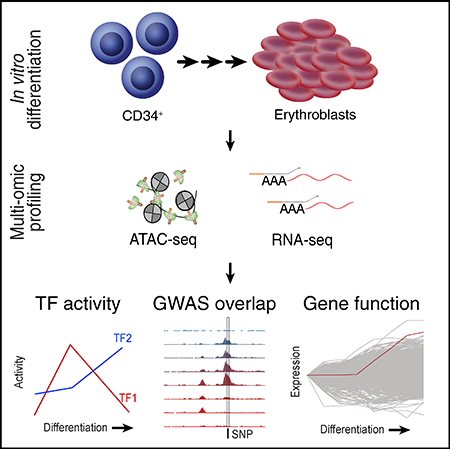
To obtain a comprehensive and integrative picture of the chromatin and transcriptional landscape of human adult erythropoiesis, we differentiated CD34+ HSPCs from healthy donors using an established three-phase erythroid differentiation protocol that allows the efficient production of mature enucleated reticulocytes (Giani et al., 2016; Hu et al., 2013). We then applied flow cytometry-activated cell sorting (FACS) using well-characterized erythroid surface markers CD71, CD235a, CD49d, and Band 3 (encoded by the SLC4A1 gene) across multiple stages of the differentiation process to enrich for cells at different stages of maturation (Hu et al., 2013), validated purity by morphology, and processed each population (P1-P8) using ATAC-seq (FAST-ATAC) and RNA sequencing (RNA-seq) (Smart-seq2) (Figures 1A, 1B, and S1A) (Corces et al., 2016; Picelli et al., 2014). Each population was processed in three or four replicates using cultured cells from two or three healthy adult human donors, resulting in a total of 28 paired RNA-seq and ATAC-seq libraries. Overall, our ATAC-seq libraries were sequenced to an average depth of 21.5 million aligned reads/sample (mean 75.3 million/population), and the RNA-seq libraries were sequenced to an average depth 26.1 million aligned reads/sample (mean 91.1 million/population) (Tables S1 and S2).
Figure 1. An Epigenomic and Transcriptional Time Course of Human Erythroid Differentiation.
(A) Schematic outline of generating matched transcriptome and open chromatin profiles across human erythropoiesis.
(B) FACS gating scheme showing expression of surface markers CD71, CD235a, CD49d, and BAND3 used to sort indicated populations P1-P8 (P1, MyP; P2, CFU-E; P3, ProE1; P4, ProE1; P5, BasoE; P6, PolyE; P7, OrthoE; P8, Ortho/Ret).
(C and D) Principal-components plots of (C) RNA-seq data and (D) ATAC-seq data, colored by FACS-sorted populations. Color code indicated below.
(E) Number of chromatin accessibility peaks across populations. Error bars represent SEM number of peaks per population between replicates.
(F) Heatmap showing relative numbers of differentially accessible peaks (top, blue shades) and differentially expressed genes (bottom, red shade) between populations.
Stage-Specific Chromatin Accessibility and Transcriptional Variability in Human Erythropoiesis
To yield a complete accessible chromatin and transcriptional trajectory during human erythroid differentiation, we included published profiles of paired sequencing data from early hematopoietic stem and progenitor populations, including hematopoietic stem cells (HSCs), multipotential progenitors (MPPs), common myeloid progenitors (CMPs), and megakaryocyte erythroid progenitors (MEP)s (Corces et al., 2016). Indeed, principal-component analysis on our paired datasets shows a continuous trajectory of erythroid differentiation and high concordance between replicates (Figures 1C, 1D, S1B, and S1C). Population P1 was most similar to myeloid progenitor cells (MyP) that did not commit to the erythroid lineage in our in vitro culture but were included in many downstream analyses. Population P2 was enriched for colony-forming unit-erythroid cells (CFU-E), P3 and P4 for proerythroblasts (ProE1 and ProE2), P5 for basophilic erythroblasts (BasoE), P6 for polychromatic erythroblasts (PolyE), and P7 for orthochromatic erythroblasts (OrthoE), which were further enriched with reticulocytes in P8 (Orth/Ret), as confirmed by morphology as well as comparison with published transcriptional profiles (Yan et al., 2018) (Figures S1A and S2A), providing an independent validation for this dataset of human erythroid differentiation. Overall, chromatin accessibility profiles were of high quality (Figure S1D) and across populations, we detected 63,659 variably accessible chromatin peaks and 14,260 differentially expressed genes (Figures 1E and 1F). The majority of identified peaks mapped to distal, intronic, and promoter gene regions, with a comparable distribution with those in previous reports profiling human T lymphocytes (Figures 1G and S1D) (Qu et al., 2015).
Time-Dependent Modules of Transcriptional Regulation Define Erythroid Differentiation Programs
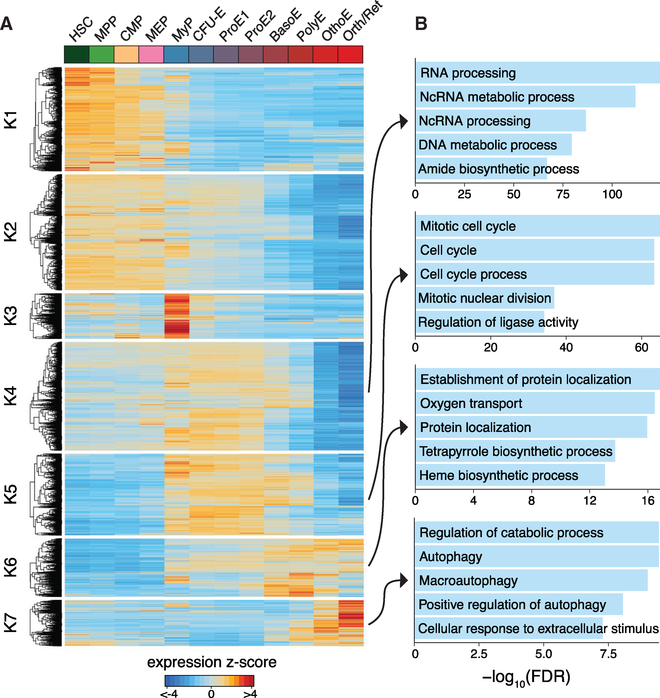
To characterize transcriptional modules throughout human erythroid differentiation, we used k-means clustering to identify co-regulated genes. Using the gap statistic to determine an appropriate number of clusters (Figure S2B), we identified seven clusters of differentially expressed genes showing a wide range of expression patterns and dynamics (Figure 2A; Table S3). From the Z score-normalized gene expression values, we observed population-specific expression primarily in presumed MyP (cluster k3) and at the latest stages of erythroid differentiation (OrthoE/Ret; cluster k7). Most other gene expression programs were broadly expressed in early progenitor populations (clusters k1 and k2) and throughout erythroid differentiation (clusters k4-k6).
Figure 2. Time-Dependent Modules of Transcriptional Regulation Define Erythroid Differentiation Programs.
(A) Heatmap of all differentially expressed genes, clustered by dynamic expression using k-means clustering across hematopoietic progenitor populations (HSC, MPP, CMP, and MEP) and stages of erythropoiesis (MyP, CFU-E, ProE1, ProE2, BasoE, PolyE, OrthoE, and Orth/Ret). Color bar, expression Z score of differentially expressed genes.
(B) Top enriched Gene Ontology biological processes for clusters K4, K5, K6, and K7 of differentially expressed genes. The top five processes are shown for each cluster and their −log10(false discovery rate [FDR]).
To determine the underlying cellular processes governed by these patterns of expression variability, we performed Gene Ontology (GO) analyses to identify biological processes that were enriched within these clusters and that may be involved in regulating erythroid differentiation (Figure 2B; Table S4). RNA, non-coding RNA, and DNA processing-related gene sets were most dominant during the earlier stages of terminal erythroid differentiation (cluster k4). Cell cycle-related processes showed maximum activity around the ProE½ stage (cluster k5), consistent with the high proliferative capacity of these cells. Heme biosynthetic and oxygen transport processes showed highest activity at the BasoE and PolyE stage, reflecting the peak of hemoglobin synthesis (cluster k6). Regulation of catabolic processes dominate during the OrthoE/Ret stage, as cellular organelles are being recycled before final maturation into erythrocytes (cluster k7) (Figure 2B). Overall, our transcriptional analysis revealed highly consistent findings with those described in prior reports (An et al., 2014; Yan et al., 2018).
Dynamics of the Accessible Chromatin Landscape and Enrichment of Transcriptional Regulatory Programs
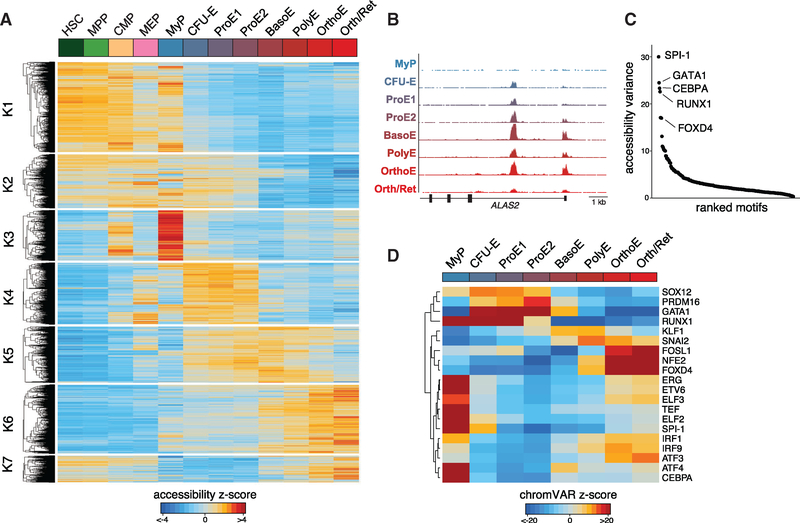
Previous genome-wide analyses of accessible chromatin profiles have revealed a comprehensive repertoire of gene regulatory elements, many of which appear to act in a cell type- and tissue-specific manner (Thurman et al., 2012). However, we have a limited understanding of the accessibility dynamics of these elements during cellular differentiation. Our dense profiling of erythroid populations at different stages of differentiation using ATAC-seq enables the most comprehensive description to date of the dynamic accessible chromatin landscape throughout human erythropoiesis (Figure 3A). Quantification of Tn5 insertion density around CTCF motifs as previously described (Buenrostro et al., 2013) verified high-quality libraries throughout our differentiation system (Figure S3A). Using k-means clustering and gap statistic optimization (Figure S3B), we identify seven clusters of differential accessible peaks, showing similar patterns and dynamics at key erythroid gene loci such as ALAS2 (Figure 3B), similar to what we describe for the transcriptomic profiles described above (Figure 2A).
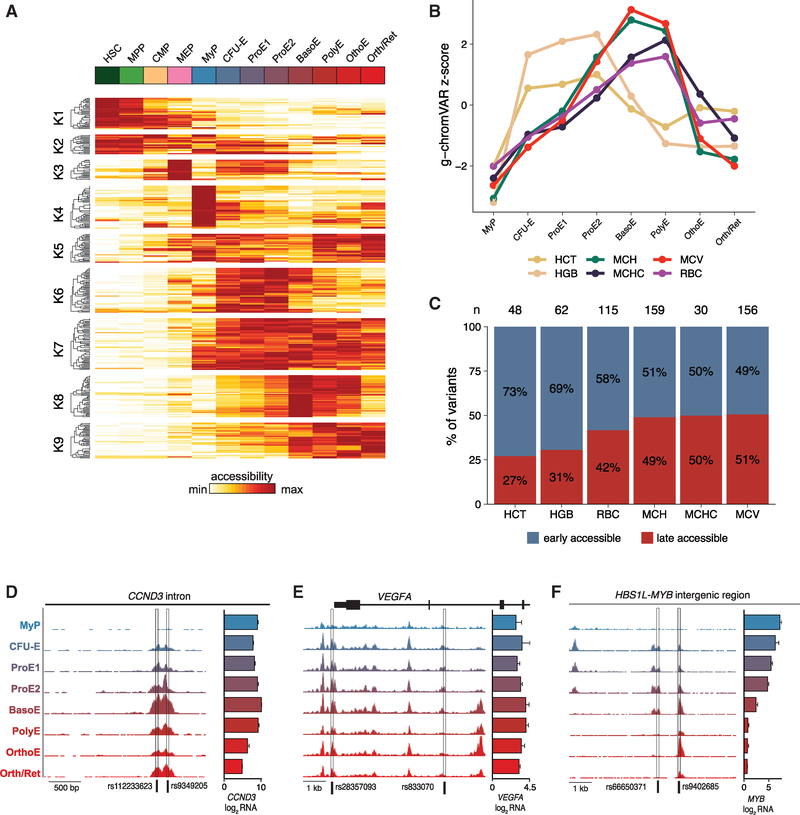
Figure 3. Dynamics of the Open Chromatin Landscape and Enrichment of Transcriptional Regulators.
(A) Heatmap of all differentially accessible peaks, clustered using k-means clustering across hematopoietic progenitor populations (HSC, MPP, CMP, and MEP) and stages of erythropoiesis (MyP, CFU-E, ProE1, ProE2, BasoE, PolyE, OrthoE, and Orth/Ret). Color bar, accessibility Z score of differentially peaks identified by ATAC-seq.
(B) Distribution of accessible peaks in the ALAS2 locus across sampled populations.
(C) Rank order plot of transcription factor binding sites with greatest variability in chromatin accessibility across sampled cell populations.
(D) Heatmap showing temporal changes in chromatin accessibility for the top 20 TFs with greatest accessibility variability between populations profiled. Color bar, chromVAR accessibility deviation Z score.
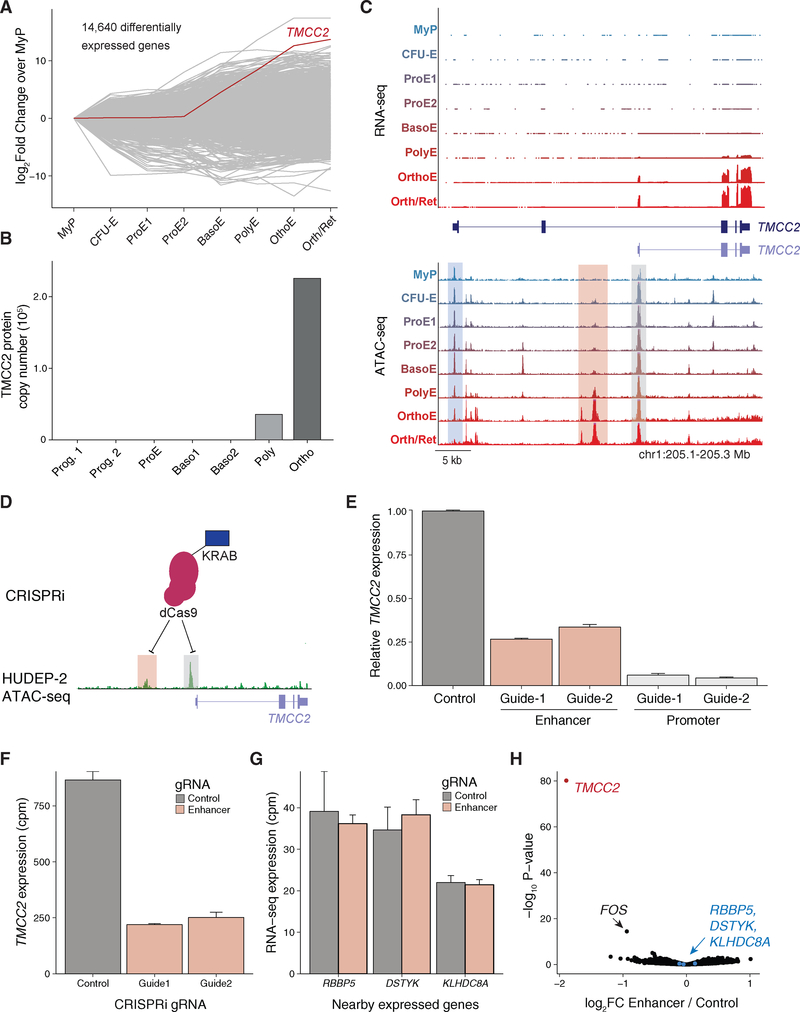
Cluster k1 contained regions of open chromatin showing highest accessibility in HSPCs, which became less accessible during early erythroid differentiation. By contrast, regulatory elements in cluster k2 displayed slower closing dynamics (decreasing accessibility). We also observed profound changes from the ProE½ to the OrthoE stage. Some elements were most accessible around the ProE½ stage but quickly closed (lost accessibility) in BasoE (cluster k4), whereas cluster k5 contained elements remaining accessible throughout the PolyE stage. Interestingly, we also observed elements showing highest accessibility at the OrthoE stage immediately preceding enucleation. This process has been typically associated with increased nuclear condensation and an anticipated loss of accessibility, also supported by an overall decreasing number of accessibility peaks at late stages of differentiation (Figure 1E) (Zhao et al., 2016). Of note, the late stage-specific peaks were enriched in genes displaying the highest levels of induction and expression at the OrthoE stage, as exemplified by the TMCC2 locus, a gene highly induced in terminal erythropoiesis whose role in this process has not been previously studied (Figures 6 and 7).
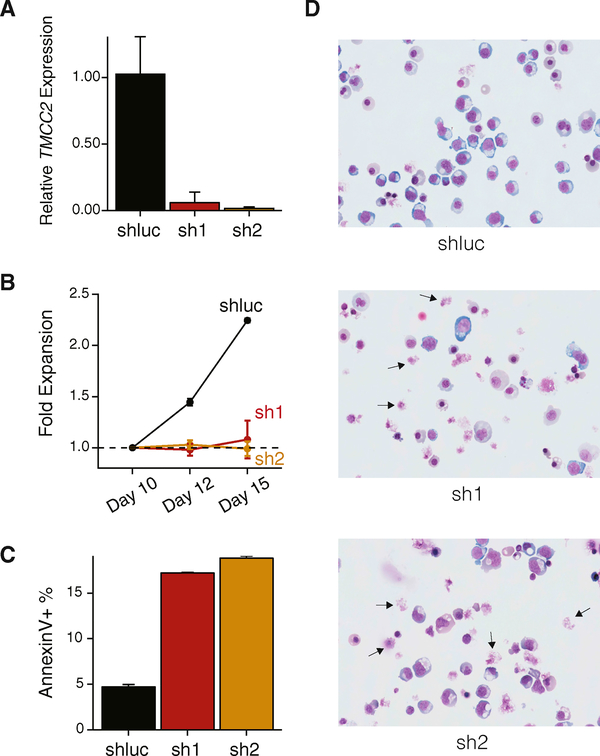
Figure 6. Erythroid-Specific Isoform Expression and Regulation of TMCC2.
(A) Line plot showing 14,640 differentially expressed genes across indicated populations, with TMCC2 highlighted.
(B) TMCC2 protein copy number across human erythroid populations.
(C) TMCC2 locus with corresponding RNA-seq (top) and ATAC-seq data (bottom) across indicated populations. Track plot depicting two isoforms of TMCC2 is shown in the middle. Compared with MyP-PolyE stages, certain regions lose chromatin accessibility (blue shade), whereas others gain accessibility in late erythropoiesis (OrthoE, pink shade). The shorter TMCC2 isoform is the dominant isoform expressed in blood cells.
(D) Schematic showing the KRAB domain fused to dCas9 used to target the enhancer (pink shade) or promoter (gray shade) non-coding DNA regulatory regions in the TMCC2 locus. ATAC-seq tracks of HUDEP-2 cells of the TMCC2 locus are shown in green.
(E) Guide RNAs targeting the enhancer and promoter result in reduction of TMCC2 expression as determined using RT-qPCR in HUDEP-2 cells. Error bars represent ± 1 SD between replicates.
(F and G) Bar plots showing RNA-seq expression counts per million (CPM) for TMCC2 (F) and other expressed genes (G) in the locus when treated with guide RNAs (gRNAs) targeting the enhancer and a non-targeting guide. Error bars represent ± 1 SD between replicates. (G) Pools replicates between gRNAs.
(H) Volcano plot of all genes considered for differential expression between the pooled enhancer gRNAs and a non-targeting control.
Figure 7. TMCC2 Is an Essential Regulator of Terminal Human Erythropoiesis.
(A) Short hairpin RNAs (sh1 and sh2) knockdown TMCC2 expression as determined using RT-qPCR in in vitro cultured primary hematopoietic cells. Error bars represent ± 1 SD between replicates.
(B) Erythroid cells with shRNA knockdown of TMCC2 demonstrate reduced proliferative capacity compared with a non-targeting control (shluc). Day (x axis) represents the day following the start of the culture, representing days 3, 5, and 8 post-infection at the three indicated time points. Error bars represent ± 1 SD between replicates.
(C and D) TMCC2-knockdown cells further show increased frequency of apoptotic annexin V+ cells (C) and impaired erythroid differentiation (D) as shown by altered morphology and an increased frequency of disrupted cells (arrows, 63× magnification, MayGrunwald staining). In (C), error bars represent ± 1 SD between replicates.
Our ATAC-seq dataset enables the use of chromVAR to infer transcription factor (TF) variability and dynamics throughout erythroid development (Schep et al., 2017). In brief, chromVAR aggregates accessible regions sharing the same TF motif, then compares the observed accessibility of all peaks containing that TF motif with a background set of peaks normalizing for known technical confounders. Among the most variably accessible sequence motifs determined, we identified important hematopoietic TF motifs such as from SPI1, GATA1, CEBPA, RUNX1, and FOXD4 (Figures 3C and S3C). Notably, we observed profound differences in the accessibility within our sampled populations, including significant loss of inferred activity of erythroid master regulator GATA1 after the BasoE stage (Figures 3D, S3D, and 4). Similar to regions of open chromatin most accessible at the OrthoE stage (Figure 3A), we identified several TF motifs with highest accessibility at this stage (FOSL1, NFE2, and FOXD4). These findings suggest a possible role for these factors during the most terminal stages of erythropoiesis and warrant further investigation, noting that augmenting motif-based inference with true-positive factor binding via chromatin immunoprecipitation sequencing (ChIP-seq) may enable greater specification of individual factors. Overall, our results provide comprehensive insights into chromatin accessibility dynamics and the distinct TF regulatory programs active during human RBC development.
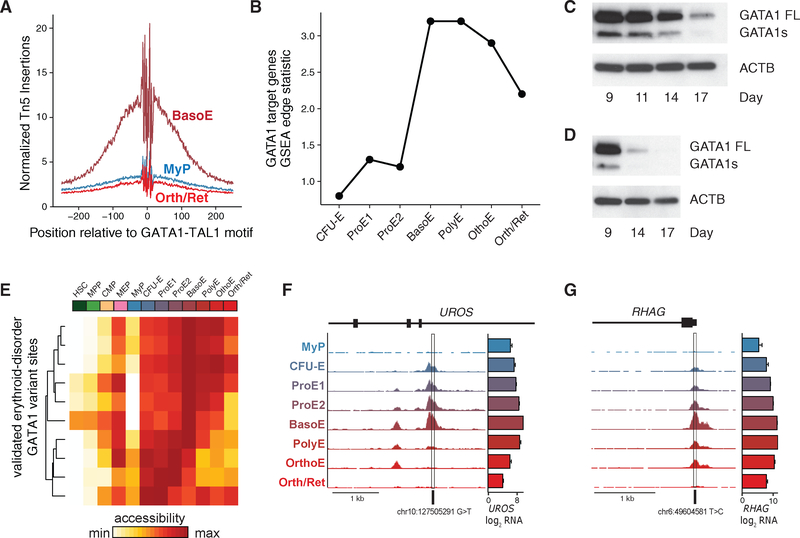
Figure 4. Regulatory Dynamics of Erythroid Master Regulators in Terminal Erythropoiesis.
(A) ATAC-seq footprinting, showing that Tn5 insertion density near GATA1-TAL1 motif is relatively lower in MyP and Orth/Ret compared with BasoE, when GATA1 is most active.
(B) GATA1 transcriptional activity using GSEA edge statistics across indicated populations.
(C and D) Western blot showing decreasing GATA1 protein expression levels in differentiating primary human erythroid cells over time. FL, full-length protein isoform of GATA1; S, short protein isoform of GATA1. Results of culture 1 (C) and culture 2 (D) are shown.
(E) Heatmap of chromatin accessibility intensity per population at loci at which rare variants disrupt GATA1 regulatory elements.
(F and G) Locus-specific examples of rare variants affecting GATA1 motifs are shown in the promoters of (F) uroporphyrinogen III synthase and (G) RH50 antigen, both demonstrating maximal chromatin accessibility in BasoE. Bar graphs indicate mean ± SEM log2 counts per million RNA-seq reads per population.
Regulatory Dynamics of Erythroid Master Regulators during Terminal Erythroid Differentiation
GATA1, TAL1, and KLF1 are master transcriptional regulators of hematopoiesis and are essential for erythroid development (Arnaud et al., 2010; Ludwig et al., 2014; Shivdasani et al., 1995). However, their DNA binding and regulatory dynamics in controlling gene expression in a stage-specific manner has not yet been fully resolved. Here, we investigated the relation between ATAC-seq Tn5 insertion density (i.e., chromatin accessibility) and proximity to the canonical GATA1-TAL1 and KLF1 motifs throughout erythroid differentiation. Chromatin accessibility near canonical GATA1 motifs appeared highest in BasoE, consistent with a peak in expression of canonical GATA1 target genes at this stage as predicted using gene set enrichment analysis (GSEA) (Figures 4A and 4B) (Ludwig et al., 2014; Subramanian et al., 2005). Interestingly, GATA1 activity declined preceding enucleation, suggesting that its expression becomes dispensable at the final stage of erythroid differentiation. Indeed, using western blot analysis, we documented reduced GATA1 protein levels in late-stage erythroid cells in our in vitro culture system (Figures 4C, 4D, S4A, and S4B), consistent with concomitant downregulation of HSP70 protein levels and its protective role in caspase-3-dependent GATA1 cleavage (Gautier et al., 2016; Ribeil et al., 2007) (Figure S4C). In contrast, KLF1 motif accessibility and target gene expression appeared to increase throughout terminal differentiation, consistent with increasing transcript levels and higher chromatin accessibility in the KLF1 locus throughout the late stages compared with the GATA1 locus (Figures S4D–S4G). These findings support the notion that GATA1 and KLF1 play distinct regulatory roles in orchestrating the erythroid maturation program. As such, human KLF1 mutations result in distinct erythroid phenotypes (Arnaud et al., 2010; Borg et al., 2010) compared with GATA1-associated pathologies; the latter are characterized by altered erythroid lineage commitment or impaired differentiation of erythroid progenitors (Campbell et al., 2013; Crispino and Horwitz, 2017; Khajuria et al., 2018; Ludwig et al., 2014; Sankaran et al., 2012a).
We next hypothesized that rare variants disrupting GATA1 motifs would also be most accessible at stages of maximal GATA1 transcriptional activity. Investigating the chromatin accessibility of 11 variants known to disrupt GATA1 regulatory elements, including several in human Mendelian erythroid disorders (Campagna et al., 2014; Kaneko et al., 2014; Manco et al., 2000; Wakabayashi et al., 2016), we indeed found that BasoE have maximal chromatin accessibility in regulatory regions encompassing many of these variants (Figure 4E; Table S5). For instance, GATA1 element mutations in uroporphyrinogen III synthase (UROS), a heme biosynthetic enzyme implicated in congenital erythropoietic porphyria (Solis et al., 2001), as well as in the core promoter of the erythroid-specific RH50 antigen (Iwamoto et al., 1998), both fell within chromatin peaks which were maximally accessible in BasoE (Figures 4F and 4G). Altogether, these findings emphasize the role of GATA1 in earlier stages of human terminal erythroid differentiation.
Human Genetic Variation in the Context of Chromatin Dynamics
Motivated by our finding of differential GATA1 activity in rare disease-associated erythroid variants, we next investigated whether differential chromatin dynamics could extend more broadly to common variants associated with RBC traits. GWAS have identified thousands of common genetic variants that fall within accessible regions of chromatin in diverse tissues and cell types and have been associated with various human phenotypes, including blood cell traits (Astle et al., 2016). However, it remains an open question as to how and at what stage of differentiation or cell state these variants primarily affect hematopoietic development and traits. Given that the majority of nominated genetic variants fall into gene-regulatory regions and are thought to alter gene expression, we reasoned that our comprehensive open chromatin profiles could provide insights into the precise stages of erythroid development during which such regulation may occur. First, we intersected hematopoietic cell type-specific regions of open chromatin with fine-mapped variants affecting six commonly measured erythroid cell traits nominated by a recent GWAS (Ulirsch et al., 2019). Among the 114,905 individuals comprising this GWAS cohort, 4,327 (3.76%) had hemoglobin concentrations meeting the criteria for clinical anemia as defined by the World Health Organization (2011), suggesting that our analysis may be relevant for better understanding the genetics of both normal variation, as well as disease risk. Overall, 347 of 1,789 (19.4%) fine-mapped GWAS variants with posterior probability (PP) > 0.10 for causal association fell within a chromatin-accessible region in one or more of the characterized populations. Upon k-means clustering of these variants by chromatin dynamics (Figures 5A and S5A; Table S6), we observed that a majority showed maximum accessibility at specific stages of erythroid differentiation rather than being uniformly accessible, suggesting potential windows of active gene regulation.
Figure 5. Dynamic cis-Regulatory Variation across Stages of Erythropoiesis.
(A) All ATAC-seq peaks containing fine-mapped (PP > 0.10) GWAS variants associated with red cell traits, clustered into nine groups. Color bar, chromatin accessibility.
(B) g-chromVAR enrichments of six different red cell traits across stages of erythroid differentiation.
(C) Proportions of fine-mapped regulatory variants, grouped by trait, that are in more accessible chromatin in earlier versus later stages of erythropoiesis. An early-accessible variant was defined as residing in an ATAC-seq peak with higher mean counts per million in the CFU-E, ProE½ populations relative to the BasoE/PolyE populations, and a late-accessible variant was the opposite. (n) indicates the total number of fine-mapped (PP > 0.10) variants per trait.
(D-F) Representative examples of fine-mapped variants located within regions that have maximal chromatin accessibility in (D) late (BasoE-OrthoE), (E) early (CFU-E, ProE½), or (F) both early- and late-stage erythroid populations. HGB, hemoglobin; HCT, hematocrit; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; MCV, mean corpuscular volume; RBC, red blood cell count. Bar graphs indicate mean ± SEM log2 counts per million RNA-seq reads per population.
We next sought to leverage this comprehensive data to more finely assess the stages of erythropoiesis during which RBC trait-associated variants may act using g-chromVAR, a recently developed high-resolution cell type enrichment tool (Ulirsch et al., 2019). Interestingly, cells at the earlier stages of differentiation (CFU-E and ProE½) were significantly enriched for variants regulating hematocrit (HCT) and hemoglobin (HGB) levels (Figure 5B). In contrast, the BasoE and PolyE stages were significantly enriched for variants affecting mean corpuscular volume (MCV), mean corpuscular HGB (MCH), MCH concentration (MCHC), and RBC count (Figure 5B). To dissect this duality, we looked at high-confidence regulatory ATAC-seq peaks containing fine-mapped variants with a combined PP of 0.10 or higher. Although fewer absolute variants were associated with HGB and/or HCT relative to MCV, MCH, and/or RBC count, the density of HGB and/or HCT variants in chromatin-accessible regions was shifted toward earlier stages of erythropoiesis, whereas a more balanced distribution was observed for MCV, MCH, MCHC, and/or RBC count variants (Figures 5C, S5B, and S5C). We note that HCT is calculated as the product of RBC count and MCV, two traits that are negatively correlated, such that when a variant alters the RBC count and MCV in opposing directions, the net effect on HCT is neutralized. Indeed, 41% of fine-mapped variants associated with RBC count were not associated with HGB or HCT (Figure S5D), consistent with previous studies showing higher genetic correlations between HGB and HCT compared with those with a RBC count (Ulirsch et al., 2019). Thus, the enrichment for HGB and/or HCT variants acting in earlier stages of erythropoiesis is likely driven in part by HGB and/or HCT-associated variants affecting the RBC count or MCV alone or in consistent directions, but not in opposite directions.
Our previous work has shown that variants rs112233623 and rs9349205 in CCND3 (Figure 5D), associated with MCV and RBC count, act by modulating the expression levels of cyclin D3, thereby affecting the number of cell divisions during terminal erythropoiesis (Sankaran et al., 2012b; Ulirsch et al., 2019). Although this had pronounced effects on the RBC count and MCV (in opposite directions), HGB and HCT were less affected in Ccnd3-knockout mice, providing an example of how common genetic variants modulate different erythroid traits at distinct stages of erythropoiesis. Here, we show that both rs112233623 and rs9349205 fall within a single chromatin peak that opens up in the BasoE, PolyE, and OrthoE stages (Figure 5D), consistent with the expected differentiation stage of these variants’ phenotypic effects. Similarly, in the KLF1 locus, we identified a region with maximum chromatin accessibility at the later stages of erythropoiesis (BasoE to OrthoE) encompassing rs80326512 (Figure S5E), a fine-mapped (PP = 0.24) variant associated with MCV. When considered together with the expression of KLF1 and its target gene set both peaking during terminal stages (Figures S4D and S4E), these findings suggest that rs80326512 acts primarily during the late stages of erythropoiesis. As a third example, a fine-mapped MCV variant, rs117747069 (PP = 1.00), falls in a region that is both (1) maximally accessible during late stages of erythropoiesis and (2) correlated with alpha-globin expression (Figure S5F) (lotchkova et al., 2016).
Going the other direction, genetic fine-mapping identified two putative causal variants (rs28357093, PP = 0.33; rs833070, PP = 0.28) in the vascular endothelial growth factor A (VEGFA) locus associated with HGB and HCT, both falling in regions with higher chromatin accessibility at earlier stages of erythropoiesis (CFU-E, ProE½) (Figure 5E). VEGFA has been shown to regulate erythropoietin signaling in response to hypoxia (Rehn et al., 2014; Tam et al., 2006), suggesting that VEGFA may preferentially act during early erythropoiesis to modulate expansion of total erythroid cell mass. Other variants in early-accessible regions included HGB-associated rs1175550 (PP = 0.36), a known modifier of SMIM1 that encodes the Vel blood group antigen (Ulirsch et al., 2016), and rs218264 (PP = 0.31) upstream of KIT, an essential receptor tyrosine kinase for hematopoietic progenitor cells (Figures S5G and S5H) (Edling and Hallberg, 2007). Our integrative findings suggest that these variants and associated genes may play a more dominant role in the regulation of HGB and HCT during early rather than late stages of erythroid differentiation.
Finally, certain loci may contain multiple regulatory elements active at distinct stages of erythropoiesis, as exemplified by the HBS1L-MYB intergenic region. One HCT-associated (PP = 0.18) variant from our data, rs66650371, falls within a region with maximal chromatin accessibility at the CFU-E, ProE½ stages (Figure 5F) and has been previously implicated as a functional variant significantly associated with fetal HGB and MYB expression (Stadhouders et al., 2014). In contrast, another fine-mapped HBS1L-MYB intergenic variant associated with several erythroid traits (HCT, PP = 0.27; HGB, PP = 0.15; RBC count, PP = 0.10), rs9402685, falls within a region with increased chromatin accessibility in BasoE to OrthoE populations. Strikingly, both rs66650371 and rs9402685 localize within functional enhancer elements (−84 and −83 kb upstream of the MYB transcription start site) previously shown to block erythroid differentiation and reduce MYB expression upon their deletion (Canver et al., 2017). Although complex linkage disequilibrium (LD) structure prevents us from ruling out the possibility of other causal variants in these two loci, the present analysis expands upon our previous work (Guo et al., 2017) and adds insights into the complex regulation at the HBS1L-MYB region by suggesting that the −84 element likely acts at early stages of erythropoiesis, whereas the −83 element may act at later stages of erythropoiesis. Altogether, our stage-specific matched profiles of ATAC-seq and RNA-seq are beginning to provide a comprehensive resource to delineate specific differentiation stages at which various erythroid phenotypes and genes are most highly regulated, allowing functional follow-up of nominated variants and genes at high temporal and stage-specific resolution.
An Erythroid-Specific Isoform of TMCC2 Is an Essential Regulator of Terminal Human Erythropoiesis
Our dataset provides a rich resource to expedite functional studies of regulators of erythropoiesis. Highlighting one such example, we identified TMCC2 as one of the most strongly induced genes during the terminal stages of erythroid differentiation (Figure 6A). TMCC2 is highly expressed in the human brain and has been implicated in neurodegeneration (Hopkins, 2013), but a role in hematopoietic development has not been previously described (Figure S6A). Of note, TMCC2 showed high expression in whole blood, appeared to be selectively expressed in erythroblasts (Figures S6A and S6B) with maximum protein expression levels at the OrthoE stage (Figure 6B) (Gautier et al., 2016), and was strongly induced in differentiating G1E-ER cells, suggesting a cell type-specific role in erythropoiesis (Figure S6C). Surprisingly, we observed decreasing chromatin accessibility near the promoter of the reference transcript (Figure 6C, bottom, blue shade), but an increase in accessible chromatin at an alternative promoter and nearby putative enhancer in late stages, supporting an isoform (ENST00000329800.7) of TMCC2 that is blood specific (Figure S6D). Indeed, coverage tracks of the RNA-seq data uniquely supported the expression of the shorter isoform, indicating that this transcript may have a specific role in erythropoiesis (Figure 6C, top). To confirm the functional roles of these regulatory elements, we used a CRISPR interference (CRISPRi) strategy, taking advantage of the local heterochromatin formation and functional silencing mediated by a Kruppel-associated box (KRAB) domain fused to a catalytically dead Cas9 (dCas9) (Figures 6D and S6E) (Gilbert et al., 2014). Consistent with a role in TMCC2 gene regulation, we observed significant downregulation of transcript levels as measured by qRT-PCR upon targeting the promoter or enhancer of TMCC2 with two independent guide RNAs in HUDEP-2 cells (Kurita et al., 2013), which was more pronounced when targeting the promoter (Figure 6E). Importantly, the enhancer showed great specificity for TMCC2 regulation as determined using RNA-seq analysis (Figures 6F–6H).
To interrogate a potential functional role and the relevance of TMCC2 in RBC differentiation, we introduced two independent short hairpin RNAs (shRNAs sh1 and sh2, both targeting the 3′ end of the gene) by lentiviral infection to knockdown its expression using our primary erythroid culture system. Both shRNAs resulted in pronounced reduction of TMCC2 mRNA levels compared with a non-targeting control (shluc) as measured by qRT-PCR (Figure 7A). Knockdown of TMCC2 resulted in altered cell proliferation (Figure 7B) and reduced levels of cellular viability as indicated by flow cytometry with altered forward and side scatter properties (Figure S7A). We also noted increased cell apoptosis as measured by annexin V staining and use of DNA binding dyes (Figures 7C, S7A, and S7B). We did not observe significant alterations in the expression of erythroid markers CD71 and CD235a in knockdown compared with control cells (shluc) (Figure S7B). However, terminal erythroid maturation appeared significantly impaired as demonstrated by altered cellular morphology and an increased frequency of “disrupted” cells on Cytospin images of TMCC2-knockdown cells (Figure 7D, arrows). As such, the observed decrease in proliferation (Figure 7B) may also be in part a consequence of increased cell lysis in these late stages of erythroid differentiation. Finally, from population-based sequencing studies of healthy individuals (ExAC) (Lek et al., 2016), we observed fewer than expected loss of function (observed = 3, expected = 12) and missense (observed = 216, expected = 330) variants in TMCC2, suggesting that mutations in this gene are poorly tolerated and selected against (constraint metric z = 3.07). Taken together, these results identify an erythroid-specific isoform of TMCC2 as an essential regulator of human terminal erythroid differentiation.
DISCUSSION
Here, we paired RNA-seq and ATAC-seq to provide deep transcriptomic profiles and describe the dynamic accessible chromatin landscape across distinct stages of adult human erythroid differentiation. Our work provides a rich resource for functional studies and complements previously published transcriptomic and proteomic datasets (An et al., 2014; Gautier et al., 2016; Li et al., 2014; Yan et al., 2018). Although prior studies have performed epigenomic analyses at specific stages (Buenrostro et al., 2018; Corces et al., 2016; Huang et al., 2016), our comprehensive analysis across the entire process of human erythroid differentiation provides substantial insights. We comprehensively characterized differential regions of accessible chromatin across erythroid development and identified regulatory elements with maximum accessibility specific to PolyE and OrthoE, stages associated with nuclear condensation and overall decreasing accessibility (Figures 1E and 6). Furthermore, we inferred the TF dynamics and their expression programs that govern the molecular circuits of RBC differentiation (Figures 3 and 4), inferred their maximum activity, and identified TF motifs most accessible in polychromatic and OrthoEs, although their exact roles and transcriptional programs remain to be functionally validated. Although the chromatin accessibility profiles identified in this work enable robust inference of accessibility peaks and global TF dynamics, these data have limited capacity to identify site-specific TF binding (via TF-foot-printing) or nucleosome positioning. Thus, additional assays applied in our in vitro system, such as ChIP-seq of specific factors, may further reveal insights into dynamic chromatin states during erythropoiesis.
Integration of ATAC-seq profiles with summary statistics from GWAS and Mendelian erythroid disorders further enable the nomination of genetic variants acting in gene regulatory elements and imply a potential window of activity (Figures 4 and 5). Our analyses suggest that regulatory elements and the variants harbored within may show maximum activity at different stages of erythroid differentiation, thereby affecting distinct erythroid traits or causing different blood disease phenotypes. Variants in regulatory regions associated with HGB and HCT appeared to be primarily accessible and regulate gene expression at the early stages of RBC development. In contrast, RBC count and MCV were primarily affected by regulatory elements containing variants most accessible in late stage erythroblasts, as showcased by our previous work characterizing the role of cyclin D3 in terminal mouse and human erythropoiesis (Sankaran et al., 2012b). We nominate multiple additional gene regulatory elements containing common variants associated with distinct erythroid traits and suggest stage-specific activities, suggesting that our data may aid in the identification of non-coding variants underlying GWAS results.
Because ~90% of variants identified in GWAS (including hematopoietic traits) are in the non-coding genome (Ulirsch et al., 2019), cell type-specific epigenomic profiling provides a powerful means for more precise annotations of functionally relevant variants underlying human phenotypes. As such, we anticipate that our resource will inform functional validation studies, which may further include the use of chromatin conformation capture, massively parallel reporter assays, and genome editing approaches via CRISPR and related methods (Canver et al., 2017; Ulirsch et al., 2016; Wakabayashi et al., 2016). As an example, our data identified a blood-specific isoform of TMCC2 that is highly induced at the PolyE/OrthoE stage (Figure 6). Knockdown of TMCC2 resulted in pronounced proliferation and differentiation defects, enabling us to identify this gene as an essential regulator of terminal human erythropoiesis (Figure 7). In summary, our data provide a comprehensive resource on the expression dynamics, TF elements and their transcriptional programs, as well as their relation to genetic variation to inform functional studies of human erythroid differentiation.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for further information or reagents should be directed to and will be fulfilled by the Lead Contact, Vijay G. Sankaran (sankaran@broadinstitute.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human erythroid cell culture
Human adult CD34+ hematopoietic stem and progenitor cells were obtained from the Fred Hutchinson Hematopoietic Cell Processing and Repository (Seattle, USA). No human subjects were involved in this study. The CD34+ samples were deidentified and approval for use of these samples was provided by the Institutional Review Board and Biosafety Committees at Boston Children’s Hospital. Donor characteristics: ID R003221 (Female, 65 years old), ID R003498 (Female, 46 years old), ID R003172 (Male, 54 years old). These cells were differentiated into mature erythroid cells utilizing a three-phase culture protocol (Giani et al., 2016; Hu et al., 2013). In phase 1 (day 0 – 7), cells were cultured at a density of 105 - 106 cells per milliliter (ml) in IMDM supplemented with 2% human AB plasma, 3% human AB serum, 1% penicillin/streptomycin, 3 IU/ml heparin, 10 μg/ml insulin, 200 μg/ml holo-transferrin, 1 IU erythropoietin (Epo), 10 ng/ml stem cell factor (SCF) and 1 ng/ml IL-3. In phase 2 (day 7 – 12), IL-3 was omitted from the medium. In phase 3 (day 12 – 18), cells were cultured at a density of 106 cells per milliliter, with both IL-3 and SCF omitted from the medium and the holo-transferrin concentration was increased to 1 mg/ml. Cells were cultured at 37°C and 5% CO2.
HUDEP-2 cell culture
HUDEP-2 cells were maintained in StemSpan SFEM II medium (Stem Cell Technologies) supplemented with hSCF (50 ng/ml), erythropoietin (3 IU/ml), dexamethasone (10−6 M) and doxycycline (1 μg/ml). For erythroid differentiation cells were cultured in differentiation media (IMDM, 2% human AB plasma, 3% human AB serum, 1 mg/mL Holo-human transferrin, 3 IU/mL heparin, 10 μg/mL insulin and 3 IU/mL erythropoietin. Cells were cultured at 37°C and 5% CO2.
293T cell culture
293T cells (ATCC) were maintained in DMEM (GIBCO) supplemented with 10% FBS, 1% penicillin/streptomycin and 2 mM L-glutamine. Cells were cultured at 37°C and 5% CO2.
METHOD DETAILS
Flow cytometry analysis and apoptosis
For flow cytometry analysis, in vitro cultured erythroid cells were washed in FACS buffer (1% FBS in PBS) before antibody staining. The following antibodies at indicated dilutions were used in this study: 1:50 APC-conjugated CD235a (Glycophorin A, clone HIR2, 50–153-69, eBioscience), 1:50 FITC-conjugated CD71 (OKT9, 14–0719-82, eBioscience), 1:50 APC-conjugated CD49d (9F10, Biolegend, 304308) and 1:200 FITC-conjugated BAND3 (An et al., 2014; Hu et al., 2013). For live/ dead cell discrimination Sytox Blue was used according to the manufacturer’s instructions (Thermo Fisher, S34857). For apoptosis analysis, BV421-Annexin V (BD Bioscience, 563973) was used in combination with Sytox Orange according to the manufacturer’s instructions (Thermo Fisher, S34861). FACS analysis was conducted on a BD Bioscience Fortessa flow cytometer at the Whitehead Institute Flow Cytometry core. Data were analyzed using FlowJo software v10.4.2.
Flow cytometry activated cell sorting
In vitro cultured erythroid cells were washed in FACS buffer and stained as described above using antibodies against CD235a, CD71, CD49d and BAND3. Sytox Blue (Thermo Fisher, S34857) was used for live/ dead cell discrimination. Cells were sorted using a Sony SH800 sorter with a 100 μm chip at the Broad Institute Flow Cytometry facility.
May-Grünwald-Giemsa staining
50,000 – 100,000 cells were harvested, washed once at 300 × g for 5 min, resuspended in 200 μLof FACS Buffer, and spun onto poly-L-lysine coated microscope slides with a Shandon 4 (Thermo Scientific) cytocentrifuge at 300 rpm for 4 min. When visibly dry slides were transferred into May-Grünwald solution (Sigma-Aldrich) for 5 min, rinsed 4 times for 30 s in water, and transferred to Giemsa solution (Sigma-Aldrich) for 15 min. Slides were washed as described above, dry mounted with coverslips, and examined. All images shown were taken using a Metafer slide scanning platform and software (Metasystems) at 63X magnification.
FAST-ATAC-seq
For ATAC-seq library preparations 10,000–15,000 cells were washed in PBS, pelleted by centrifugation and lysed and tagmented in 1x TD buffer, 2.5 μl Tn5 (Illumina), 0.01% Digitonin (Promega, G9441), 0.3x PBS in a 50 μl reaction volume as described (Corces et al., 2016). Samples were incubated at 37°C for 30 min at 300 rpm. Tagmented DNA was purified using the MinElute PCR kit (QIAGEN). The complete eluate underwent PCR, as follows. After initial extension, 5 cycles of pre-amplification using indexed primers (Buenrostro et al., 2015) and NEBNext High-Fidelity 2X PCR Master Mix (NEB) were conducted, before the number of additional cycles was assessed by quantitative PCR using SYBR Green. Typically, 7–9 additional cycles were run. The final library was purified using a MinElute PCR kit (QIAGEN) and quantified using a Qubit dsDNA HS Assay kit (Invitrogen) and a High Sensitivity DNA chip run on a Bioanalyzer 2100 system (Agilent).
RNA-seq
Cells were lysed in RLT lysis buffer (QIAGEN) supplemented with beta-mercaptoethanol and RNA was isolated using a RNeasy Micro kit (QIAGEN) according to the manufacturer’s instructions. An on-column DNase digestion was performed before RNA was quantified using a Qubit RNA HS Assay kit (Invitrogen). 1–10 ng of RNA were used as input to a modified SMART-seq2 (Picelli et al., 2014) protocol and after reverse transcription, 8–9 cycles of PCR were used to amplify transcriptome library. Quality of whole transcriptome libraries was validated using a High Sensitivity DNA Chip run on a Bioanalyzer 2100 system (Agilent), followed by library preparation using the Nextera XT kit (Illumina) and custom index primers according to the manufacturer’s instructions. Final libraries were quantified using a Qubit dsDNA HS Assay kit (Invitrogen) and a High Sensitivity DNA chip run on a Bioanalyzer 2100 system (Agilent).
Sequencing
All libraries were sequenced using Nextseq High Output Cartridge kits and a Nextseq 500 sequencer (Illumina). Libraries were consistently sequenced paired end (2×38bp).
Western blotting
Cells were washed twice in PBS, resuspended in RIPA lysis buffer (Santa Cruz Biotechnology) supplemented with 1x Complete Protease Inhibitor Cocktail (Roche) and incubated for 30 min on ice. After centrifugation at 14,000 rpm for 10 min at 4°C to remove cellular debris, the remaining supernatant was transferred to a new tube, supplemented with 4x Laemmli sample buffer (Bio-Rad) and beta-mercaptoethanol and incubated for 10 min at 95°C. Equal amounts of proteins were separated by gel electrophoresis using the Mini-PROTEAN gel system and Tris/Glycine/SDS running buffer (Bio-Rad). Subsequently, proteins were transferred onto a PVDF membrane using Tris/Glycine transfer buffer supplemented with methanol (Bio-Rad). Membranes were blocked with 3% BSA-TBST for 1 h and probed with GATA1 goat polyclonal antibody (M-20, sc-1234, Santa Cruz Biotechnology) at a 1:500 dilution or b-actin mouse monoclonal (AC-15, Sigma) at a 1:5,000 dilution in 3% BSA-TBST for 1 h at room temperature or overnight at 4°C. Membranes were washed four times with TBST, incubated with donkey anti-mouse or anti-goat peroxidase-coupled secondary antibodies (715–035-150 and 705–035-147, Jackson ImmunoResearch) at a 1:10,000 dilution in 3% BSA-TBST for 1 h at room temperature, washed three times with TBST and incubated for 1 min with Western Lightning Plus-ECL substrate (PerkinElmer). Proteins were visualized by exposure to scientific imaging film (Kodak).
Lentiviral shRNA vectors and infection
The shRNA constructs targeting human TMCC2 (sh1 and sh2, SHCLNG-NM_014858) were obtained from the Mission shRNA collection (Sigma-Aldrich). The sequences of the shRNAs used in this study are
sh1: CCGGGCAAGTGTTCGAGAAGAAGAACTCGAGTTCTTCTTCTCGAACACTTGCTTTTTTG
sh2: CCGGCCTGACTGAGCTTCATCAGAACTCGAGTTCTGATGAAGCTCAGTCAGGTTTTTTG
As controls, the lentiviral vectors pLKO-GFP and shluc were used. For production of lentiviruses, 293T cells were transfected with the appropriate viral packaging and genomic vectors (pVSV-G and pDelta8.9) using FuGene 6 reagent (Promega) according to the manufacturer’s protocol. The medium was changed the day after transfection to phase I medium (described above). After 24–30 h, this medium was collected and filtered using an 0.22 μm filter immediately before infection of primary hematopoietic cells. The cells were mixed with viral supernatant supplemented with cytokines in the presence of 8 μg/ml polybrene (Millipore) in a 6-well plate at a density of 250,000–500,000 cells per well. The cells were spun at 2,000 rpm. for 90 min at 22°C and left in viral supernatant overnight. The medium was replaced the morning after infection. Selection of infected cells was started 24 h after infection with 1 μg/ml puromycin for up to 48 h. Infection efficiency of pLKO-GFP-infected cells was assessed by measuring the frequency of GFP+ cells by flow cytometry 48–72 h post infection. Typically, the frequency of GFP+ cells was between 40%−70%.
Construction of the lentiviral CRISPRi constructs
We utilized the LentiCRISPRv2 backbone for constructing the CRISPRi constructs (Sanjana et al., 2014). The sgRNA scaffold in this construct was replaced by sgRNA-(F+E)-combined optimized scaffold from the sgOpti plasmid (Addgene # 85681) (Fulco et al., 2016). The puromycin resistance gene was replaced with the GFP gene using BamHI and SacII sites. To achieve better expression in hematopoietic cells, the EF1α promoter was replaced by the MSCV promoter using Gibson assembly. The SpCas9 was replaced with the KRAB-dCas9 from the pHR-SFFV-KRAB-dCas9-P2A-mCherry construct using BamHI sites (Addgene: #60954) (Gilbert et al., 2014). A scheme of the vector construct is shown in Figure S6E. The sequences of the guide RNAs used in this study are:
Control / non-targeting guide: ATCGCGAGGACCCGTTCCGCC
TMCC2 promoter gRNA1: CCTGGCAAAGCATATTACAT
TMCC2 promoter gRNA2: GTATAGTTTCCATGAGCCCA
TMCC2 enhancer gRNA1: GTCGTGCTGCAGGTGAAGTG
TMCC2 enhancer gRNA2: CCATCCTTCAGAGTAAACAG
CRISPRi experiments in HUDEP-2 cells
The cells were infected with all-in-one CRISPRi lentviral constructs containing KRABdcas9 driven by the MSCV promoter linked to GFP (by a self-cleaving 2A peptide sequence from porcine teschovirus) and an U6 promoter driving respective guide RNAs targeting the TMCC2 gene locus. HUDEP-2 cells were infected as described above and GFP+ cells were FACS sorted 48 h post-infection and expanded for an additional 6 days. HUDEP-2 cells were then differentiated in erythroid differentiation media as described above for 72 h and RNA was extracted using a RNeasy Mini kit (QIAGEN) for gene expression analysis. RNA-seq libraries were constructed for three replicates of the non-targeting gRNA and two replicates for each of the on-target enhancer gRNAs. Differential gene expression analysis was performed as stated below for the FACS-sorted primary cells.
Quantitative RT-PCR
Isolation of RNA was performed using the RNeasy Mini Kit (QIAGEN). An on-column DNase (QIAGEN) digestion was performed according to the manufacturer’s instructions. RNA was quantified by a NanoDrop spectrophotometer (Thermo Scientific). Reverse transcription was carried out using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using the CFX384 Real-Time PCR system and iQ SYBR Green Supermix (Bio-Rad). Quantification was performed using the ΔΔCT method. Normalization was performed using ACTB mRNA as a standard. The primers used for quantitative RT-PCR are:
ACTB forward: 5′-AGAAAATCTGGCACCACACC-3′
ACTB reverse: 5′-GGGGTGTTGAAGGTCTCAAA-3′
TMCC2 forward: 5′-GCAGCGATGATGAGTGCTC-3′
TMCC2 reverse: 5′-GTGCATTGGACTTAGGGCTCC-3′
TMCC2 forward 2: 5′-CTACATGACCCAGTGCCTGC-3′
TMCC2 reverse 2: 5′-CTCCTGCTTCAGGTTCGTCA-3′
QUANTIFICATION AND STATISTICAL ANALYSIS
Data processing and read alignment
For each sequencing library generated in this study, libraries were sequenced on an Illumina NextSeq 500 and demultiplexed using the bcl2fastq program. For each library, raw .fastq reads were aligned using either Bowtie2 version 2.3.3 (ATAC-seq) (Langmead and Salzberg, 2012) or STAR version 2.5.1b (RNA-seq) (Dobin et al., 2013) to the hg19 reference genome. Chromatin accessibility peaks were called using MACS2 using custom parameters for ATAC-seq (-nomodel-nolambda-keep-dup all-call-summits) (Zhang et al., 2008). Gene expression counts were summarized using the-quantMode functionality in STAR. Downstream processing of sequencing data was performed using Samtools (Li et al., 2009). Per population ATAC and RNA-seq values were visualized as heat-maps were generated using Complexheatmap (Gu et al., 2016).
Principal component analyses
To establish a uniform feature set for the ATAC-seq data, we used all 1 bp summits from the MACS2 peak calls per population and expanded them a uniform 250 bp, similar to a strategy implemented previously (Corces et al., 2016). Summits overlapping the same window were iteratively centered at the summit with the strongest signal (measured by −log10 FDR) until all summits were accounted. Gene × sample and Peak × sample matrices were log2 counts-per-million normalized and then transformed to Z scores before the first two principle components were computed via the implicitly restarted Lanczos bidiagonalization algorithm (irlba).
Differential gene expression and chromatin occupancy
To test for differential gene expression from our RNA-seq data and differential chromatin accessibility in individual loci, we used the DEseq2 method (Love et al., 2014). Statistically significant genes varying between two populations were identified at an independent hypothesis weighting (IHW) value of 0.01 (Ignatiadis et al., 2016). A total of 56 comparisons between populations were performed (28 for ATAC-seq; 28 for RNA-seq).
Gene set enrichments
Gene set enrichments of 7 clusters of differentially expressed genes were performed using the Functional Mapping and Annotation of Genome-Wide Associations (FUMA) platform, using all protein-coding genes as background model and requiring a minimum overlap of 2 genes and an FDR-adjusted p < 0.01 for each gene-set. Only Gene Ontology (GO) biological processes were considered.
Downstream ATAC-seq analyses
All motif analyses were performed using the motifmatchr package as part of the chromVAR suite of tools (Schep et al., 2017). For insertion footprinting analyses, reads were offset by +4/−5 base pairs as originally described (Buenrostro et al., 2013). Transcription factor activity scoring was performed using combined replicates for each population for both the data generated in this study (Figures 3C and 3D) and augmented with populations previously described (Corces et al., 2016) (Figures S3C and S3D). We note that as chromVAR builds a background peak set based on the population intensities, deviation z-scores are not absolute but relative to the populations considered.
Integration with Genome-Wide Association Studies
We obtained GWAS summary statistics and fine-mapping results of 6 erythroid traits (HGB, HCT, MCV, RBC count, MCH, and MCHC), which were performed on ~115,000 individuals of European ancestry from the UK Biobank, as described extensively elsewhere (Ulirsch et al., 2019). For g-chromVAR enrichment analysis, the set of all fine-mapped variants with PP > 0.001 for each trait were used as input.
Next, to nominate high-confidence fine-mapped variants located in strong chromatin peaks, we took the consensus peak set for all stages of erythropoiesis, performed row and column quantile normalization on the counts matrix, and kept only peaks that had a maximum count in the top 80% for at least one of the eight sampled cell populations. We then overlapped fine-mapped variants with posterior probability (PP) > 0.10 with this subset of strong peaks.
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw sequencing data reported in this paper is GEO: GSE115684 and is available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115684. AUCSC Track Hub for the ATAC-seq data generated for these eight populations is available here: https://s3.amazonaws.com/atacerythropoiesis/hub.txt. A git repository containing code for analyses described here and processed data files is available at www.github.com/sankranlab/erythroid-profiling.
Supplementary Material
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| FITC anti-human CD71, clone OKT9 | eBioscience | Cat#: 14–0719-82; RRID: AB_467338 |
| APC anti-human CD235a, clone HIR2 | eBioscience | Cat#: 17–9987-42; RRID: AB_2043823 |
| APC anti-human CD49d, clone 9F10 | Biolegend | Cat#: 304308; RRID: AB_2130041 |
| FITC anti-human BAND3 | Laboratory of Narla Mohandas; Hu et al., 2013 | N/A |
| Goat polyclonal anti-GATA1 (M-20) | Santa Cruz Biotechnology | Cat#: sc-1234; RRID: AB_2263157 |
| Mouse monoclonal anti-ACTB (AC-15) | Sigma-Aldrich | Cat#: A1978; RRID: AB_476692 |
| Anti-goat peroxidase-coupled antibody | Jackson ImmunoResearch | Cat#: 715–035-147; RRID: AB_2313587 |
| Anti-mouse peroxidase-coupled antibody | Jackson ImmunoResearch | Cat#: 715–035-150; RRID: AB_2340770 |
| Biological Samples | ||
| Human CD34+ hematopoietic stem and progenitor cells, adult | Fred Hutchinson Cancer Research Center | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| PBS | GIBCO | Cat#: 10010–023 |
| Dulbecco’s Modified Eagle Medium-High Glucose (DMEM) | GIBCO | Cat#: 11965–118 |
| Iscove’s Modified Dulbecco’s Medium (IMDM) | GIBCO | Cat#: 12440–053 |
| Opti-MEM | GIBCO | Cat#: 31985–062 |
| StemSpan SFEM II medium | STEMCELL Technologies | Cat#: 09655 |
| Human AB serum | Atlanta Biologicals | Cat#: S40110 |
| Human AB plasma | SeraCare | Cat#: 1810–0001 |
| Recombinant Erythropoetin (EPO) | Amgen | Cat#: NDC 55513–267-10 |
| Interleukin 3 (IL-3) | Peprotech | Cat#: 200–03 |
| Stem cell factor (SCF) | Peprotech | Cat#: 300–07 |
| Human Holo-Transferrin | Sigma-Aldrich | Cat#: T0665 |
| Dexamethasone Sodium Phosphate | Mylan | Cat#: NDC67457–420-10 |
| Heparin | Hospira | Cat#: NDC 00409–2720-01 |
| Doxycycline | Sigma-Aldrich | Cat#: D3072 |
| Fetal Bovine Serum (FBS) | Atlanta Biologicals | Cat#: S11150 |
| Penicillin-Streptomycin | GIBCO | Cat#: 15140–122 |
| FuGENE 6 Transfection Reagent | Promega | Cat#: E2691 |
| Polybrene Infection/Transfection reagent | Millipore | Cat#: TR-1003-G |
| SYTOX Blue Dead Cell Stain | Thermo Fisher | Cat#: S34857 |
| SYTOX Orange Dead Cell Stain | Thermo Fisher | Cat#: S34861 |
| Buffer RLT | QIAGEN | Cat#: 79216 |
| 2-mercaptoethanol | Sigma | Cat#: M6250 |
| Recombinant Ribonuclease Inhibitor (40U/ul) | Clontech | Cat#: 2313B |
| Digitonin | Promega | Cat#: G9441 |
| Trehalose Solution, 1M, Sterile | Life Sciences Advanced Technologies | Cat#: TS1M-100 |
| dNTP mix (10mM) | Thermo Fisher | Cat#: R0193 |
| Magnesium Chloride | Sigma-Aldrich | Cat#: M1028–10X1ML |
| Buffer EB | QIAGEN | Cat#: 19086 |
| TE Buffer | Thermo Fisher | Cat#: 12090015 |
| UltraPure DNase/RNase-Free Distilled Water | Thermo Fisher | Cat#: 10977015 |
| Ethanol absolute, anhydrous, KOPTEC USP, Multi-compendial (200 Proof) | VWR | Cat#: 89125–186 |
| SYBR Green I Nucleic Acid Gel Stain | Thermo Fisher | Cat#: S7563 |
| May-Grünwald solution | Sigma-Aldrich | Cat#: 63590–500ML |
| Giemsa stain | Sigma-Aldrich | Cat#: GS500–500ML |
| RIPA lysis buffer | Santa Cruz Biotechnology | Cat#: sc-24948A |
| cOmplete Mini Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#: 11836153001 |
| 4x Laemmli sample buffer | Bio-Rad | Cat#: 1610747 |
| Mini-PROTEAN TGX gels | Bio-Rad | Cat#: 4561083 |
| Tris/Glycine/SDS | Bio-Rad | Cat#: 1610732 |
| Tris/Glycine transfer buffer | Bio-Rad | Cat#: 1610734 |
| Methanol | Sigma-Aldrich | Cat#: 322415–2L |
| PVDF membrane | Bio-Rad | Cat#: 1620255 |
| Bovine serum albumin | Sigma-Aldrich | Cat#: A2153–100G |
| Clarity Western ECL substrate | Bio-Rad | Cat#: 1705060 |
| Amersham Hyperfilm ECL | GE Healthcare | Cat#: 28906838 |
| Critical Commercial Assays | ||
| BV421-Annexin V | BD Bioscience | Cat#: 563973 |
| RNeasy Micro Kit | QIAGEN | Cat#: 74004 |
| RNase-Free DNase Set | QIAGEN | Cat#: 79254 |
| iScript cDNA synthesis kit | Bio-Rad | Cat#: 1708891 |
| iQ SYBR Green Supermix | Bio-Rad | Cat#: 1708880 |
| MinElute PCR Purification Kit | QIAGEN | Cat#: 28004 |
| NEBNext® High-Fidelity 2X PCR Master Mix | New England Biolabs | Cat#: M0541L |
| Maxima H-minus RT (200 u/uL) | Thermo Fisher | Cat#: EP0752 |
| KAPA HiFi HotStart PCR ReadyMix | Kapa Biosystems | Cat#: KK2602 |
| Agencourt AMPure XP | Beckman-Coulter | Cat#: A63881 |
| Agencourt RNA Clean XP | Beckman-Coulter | Cat#: A63987 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher | Cat#: Q32854 |
| Qubit RNA HS Assay Kit | Thermo Fisher | Cat#: Q32852 |
| Bioanalyzer High Sensitivity DNA Analysis | Agilent | Cat#: 5067–4626 |
| E-Gel EX Gel, 2% | Thermo Fisher | Cat#: G402002 |
| Tn5 enzyme from Nextera DNA Library Preparation Kit | Illumina | Cat#: FC-121–1031 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#: FC-131–1096 |
| NextSeq 500/550 High Output Kit v2.5 (75 Cycles) | Illumina | Cat#: 20024906 |
| Deposited Data | ||
| Raw and processed data | This paper | GEO: GSE115684 |
| Code and processed data | This paper | https://github.com/sankaranlab/erythroid-profiling |
| Human reference genome UCSC build 19, hg19 | University of California Santa Cruz | https://genome.ucsc.edu/cgi-bin/hgGateway |
| Human erythroblast RNA-seq expression profiles | Yan et al., 2018 | GEO: GSE107218 |
| Human hematopoietic progenitor ATAC-seq datasets | Corces et al., 2016 | GEO: GSE74310 |
| GTEx RNA-seq expression profiles | dbGAP | https://gtexportal.org/home/ |
| Experimental Models: Cell Lines | ||
| 293T cells | ATCC | Cat#: CRL-3216 |
| HUDEP-2 | Kurita et al., 2013 | N/A |
| Sequencing Indexing primer info for NexteraXT and ATAC library preparation | Buenrostro et al., 2015 | N/A |
| 3′ SMART RT primer (Smart-seq2) 5 ′- AAGCAGTGG TATCAACGCAGAGTACT(30)VN - 3’ | IDT | N/A |
| Template switching oligo (Smart-seq2) 5′ - AAGCAG TGGTATCAACGCAGAGTACrGrG+G - 3′ | Exiqon | N/A |
| IS PCR Primer (Smart-seq2) 5′ - AAGCAGTGGTATC AACGCAGAGT - 3′ | IDT | N/A |
| See Table S7 for additional oligonucleotide sequences used in this study | This paper | N/A |
| Recombinant DNA | ||
| TMCC2 shRNA1 (sh1) Bacterial Glycerol Stock 5′ - CCGGGCAAGTGTTCGAGAAGAAGAACTC GAGTTCTTCTTCTCGAACACTTGCTTTTTTG - 3′ | Sigma-Aldrich | Cat#: SHCLNG-NM_014858; TRCN0000130364 |
| TMCC2 shRNA2 (sh2) Bacterial Glycerol Stock 5′ - CCGGCCTGACTGAGCTTCATCAGAACTCG AGTTCTGATGAAGCTCAGTCAGGTTTTTTG - 3′ | Sigma-Aldrich | Cat#: SHCLNG-NM_014858; TRCN0000130626 |
| Luciferase shRNA Control Plasmid DNA | Sigma-Aldrich | Cat#: SHC007 |
| Software and Algorithms | ||
| Python version 3.6 | Python Software Foundation | https://www.python.org/downloads/ |
| R version 3.4 | The R Foundation | https://www.r-project.org |
| bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| ComplexHeatmap | Gu et al., 2016 | https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| g-chromVAR | Ulirsch et al., 2019 | https://caleblareau.github.io/gchromVAR/ |
| chromVAR | Schep et al., 2017 | https://bioconductor.org/packages/release/bioc/html/chromVAR.html |
| DEseq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| MACS2 | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| FlowJo V10.4.2 | FlowJo | https://www.flowjo.com/ |
Highlights.
Integrated accessible chromatin and transcriptome analysis of human erythropoiesis
Inference of differentiation stage-specific transcription factor activities
Mapping of genetic variants underlying diseases and traits to regulatory regions
Identification of TMCC2 as a regulator in terminal erythropoiesis
ACKNOWLEDGMENTS
We thank members of the Sankaran, Regev, and Aryee laboratories for valuable comments and the Whitehead Institute and Broad Institute Flow Cytometry facilities for assistance with cell sorting and flow cytometric analysis. We thank Ryo Kurita and Yukio Nakamura for providing HUDEP-2 cells. E.L.B. received support from the Howard Hughes Medical Institute Medical Research Fellows program. K.C. received support from NIH training grant T32GM007753. J.D.B. received support from the Broad Institute Fellows program and the Allen Institute Distinguished Investigator Award. This research was supported by NIH grant P01 DK32094 (N.M.), the Howard Hughes Medical Institute (A.R.), the Klarman Cell Observatory (A.R.), NIH grants R01 DK103794 and R33 HL120791 (V.G.S.), and the New York Stem Cell Foundation (NYSCF; V.G.S.). V.G.S. is a NYSCF-Robertson Investigator.
Footnotes
DECLARATION OF INTERESTS
A.R. is a founder and equity holder in Celsius Therapeutics and an SAB member of the ThermoFisher Scientific and Syros Pharmaceuticals.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.05.046.
REFERENCES
- An X, Schulz VP, Li J, Wu K, Liu J, Xue F, Hu J, Mohandas N, and Gallagher PG (2014). Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 123, 3466–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlet JB, Ribeil JA, Guillem F, Negre O, Hazoume A, Marcion G, Beuzard Y, Dussiot M, Moura IC, Demarest S, et al. (2014). HSP70 sequestration by free α-globin promotes ineffective erythropoiesis in β-thalassaemia. Nature 514, 242–246. [DOI] [PubMed] [Google Scholar]
- Arnaud L, Saison C, Helias V, Lucien N, Steschenko D, Giarratana MC, Prehu C, Foliguet B, Montout L, de Brevern AG, et al. (2010). A dominant mutation in the gene encoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am. J. Hum. Genet 87, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima MA, et al. (2016). The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167, 1415–1429.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera V, Evans P, Face CJ, Hamagami N, Sankaranarayanan L, Keller CA, Giardine B, Tan K, Hardison RC, Shi J, and Blobel GA (2018). Exploiting genetic variation to uncover rules of transcription factor binding and chromatin accessibility. Nat. Commun 9, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg J, Papadopoulos P, Georgitsi M, Gutiérrez L, Grech G, Fanis P, Phylactides M, Verkerk AJ, van der Spek PJ, Scerri CA, et al. (2010). Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat. Genet 42, 801–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, and Greenleaf WJ (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, Majeti R, Chang HY, and Greenleaf WJ (2018). Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173, 1535–1548.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna DR, de Bie CI, Schmitz-Abe K, Sweeney M, Sendamarai AK, Schmidt PJ, Heeney MM, Yntema HG, Kannengiesser C, Grand-champ B, et al. (2014). X-linked sideroblastic anemia due to ALAS2 intron 1 enhancer element GATA-binding site mutations. Am. J. Hematol 89, 315–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AE, Wilkinson-White L, Mackay JP, Matthews JM, and Blobel GA (2013). Analysis of disease-causing GATA1 mutations in murine gene complementation systems. Blood 121, 5218–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Lessard S, Pinello L, Wu Y, Ilboudo Y, Stern EN, Needleman AJ, Galactéros F, Brugnara C, Kutlar A, et al. (2017). Variant-aware saturating mutagenesis using multiple Cas9 nucleases identifies regulatory elements at trait-associated loci. Nat. Genet 49, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet 48, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD, and Horwitz MS (2017). GATA factor mutations in hematologic disease. Blood 129, 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edling CE, and Hallberg B (2007). c-Kit—a hematopoietic cell essential receptor tyrosine kinase. Int. J. Biochem. Cell Biol 39, 1995–1998. [DOI] [PubMed] [Google Scholar]
- Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, and Engreitz JM (2016). Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EF, Ducamp S, Leduc M, Salnot V, Guillonneau F, Dussiot M, Hale J, Giarratana MC, Raimbault A, Douay L, et al. (2016). Comprehensive proteomic analysis of human erythropoiesis. Cell Rep. 16, 1470–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani FC, Fiorini C, Wakabayashi A, Ludwig LS, Salem RM, Jobaliya CD, Regan SN, Ulirsch JC, Liang G, Steinberg-Shemer O, et al. (2016). Targeted application of human genetic variation can improve red blood cell production from stem cells. Cell Stem Cell 18, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Guo MH, Nandakumar SK, Ulirsch JC, Zekavat SM, Buenrostro JD, Natarajan P, Salem RM, Chiarle R, Mitt M, Kals M, et al. (2017). Comprehensive population-based genome sequencing provides insight into hematopoietic regulatory mechanisms. Proc. Natl. Acad. Sci. U S A 114, E327–E336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins PC (2013). Neurodegeneration in a Drosophila model for the function of TMCC2, an amyloid protein precursor-interacting and apolipoprotein E-binding protein. PLoS ONE 8, e55810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu J, Xue F, Halverson G, Reid M, Guo A, Chen L, Raza A, Galili N, Jaffray J, et al. (2013). Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood 121, 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan GC, et al. (2016). Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell 36, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis N, Klaus B, Zaugg JB, and Huber W (2016). Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13, 577–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- lotchkova V, Huang J, Morris JA, Jain D, Barbieri C, Walter K, Min JL, Chen L, Astle W, Cocca M, et al. ; UK10K Consortium (2016). Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat. Genet 48, 1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto S, Omi T, Yamasaki M, Okuda H, Kawano M, and Kajii E (1998). Identification of 5′ flanking sequence of RH50 gene and the core region for erythroid-specific expression. Biochem. Biophys. Res. Commun 243, 233–240. [DOI] [PubMed] [Google Scholar]
- Kaneko K, Furuyama K, Fujiwara T, Kobayashi R, Ishida H, Harigae H, and Shibahara S (2014). Identification of a novel erythroid-specific enhancer for the ALAS2 gene and its loss-of-function mutation which is associated with congenital sideroblastic anemia. Haematologica 99, 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, et al. (2018). Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173, 90–103.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita R, Suda N, Sudo K, Miharada K, Hiroyama T, Miyoshi H, Tani K, and Nakamura Y (2013). Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS ONE 8, e59890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. ; Exome Aggregation Consortium (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hale J, Bhagia P, Xue F, Chen L, Jaffray J, Yan H, Lane J, Gallagher PG, Mohandas N, et al. (2014). Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E. Blood 124, 3636–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA, et al. (2014). Altered translation of GATA1 in Diamond-Blackfan anemia. Nat. Med 20, 748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig LS, Khajuria RK, and Sankaran VG (2016). Emerging cellular and gene therapies for congenital anemias. Am. J. Med. Genet. C. Semin. Med. Genet 172, 332–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manco L, Ribeiro ML, Máximo V, Almeida H, Costa A, Freitas O, Barbot J, Abade A, and Tamagnini G (2000). A new PKLR gene mutation in the R-type promoter region affects the gene transcription causing pyruvate kinase deficiency. Br. J. Haematol 110, 993–997. [DOI] [PubMed] [Google Scholar]
- Nandakumar SK, Ulirsch JC, and Sankaran VG (2016). Advances in understanding erythropoiesis: evolving perspectives. Br. J. Haematol 173, 206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J (2014). Primitive and definitive erythropoiesis in mammals. Front. Physiol 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Qu K, Zaba LC, Giresi PG, Li R, Longmire M, Kim YH, Greenleaf WJ, and Chang HY (2015). Individuality and variation of personal regulomes in primary human T cells. Cell Syst. 1, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehn M, Kertész Z, and Cammenga J (2014). Hypoxic induction of vascular endothelial growth factor regulates erythropoiesis but not hematopoietic stem cell function in the fetal liver. Exp. Hematol 42, 941–944.e1. [DOI] [PubMed] [Google Scholar]
- Ribeil JA, Zermati Y, Vandekerckhove J, Cathelin S, Kersual J, Dussiot M, Coulon S, Moura IC, Zeuner A, Kirkegaard-Sørensen T, et al. (2007). Hsp70 regulates erythropoiesis by preventing caspase-3-mediated cleavage of GATA-1. Nature 445, 102–105. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, and Weiss MJ (2015). Anemia: progress in molecular mechanisms and therapies. Nat. Med 21, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Ghazvinian R, Do R, Thiru P, Vergilio J-A, Beggs AH, Sieff CA, Orkin SH, Nathan DG, Lander ES, and Gazda HT (2012a). Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J. Clin. Invest 122, 2439–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Ludwig LS, Sicinska E, Xu J, Bauer DE, Eng JC, Patterson HC, Metcalf RA, Natkunam Y, Orkin SH, et al. (2012b). Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev. 26, 2075–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, and Greenleaf WJ (2017). chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Mayer EL, and Orkin SH (1995). Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373, 432–434. [DOI] [PubMed] [Google Scholar]
- Solis C, Aizencang GI, Astrin KH, Bishop DF, and Desnick RJ (2001). Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J. Clin. Invest 107, 753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Aktuna S, Thongjuea S, Aghajanirefah A, Pourfarzad F, van Ijcken W, Lenhard B, Rooks H, Best S, Menzel S, et al. (2014). HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Invest 124, 1699–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BYY, Wei K, Rudge JS, Hoffman J, Holash J, Park SK, Yuan J, Hefner C, Chartier C, Lee J-S, et al. (2006). VEGF modulates erythropoiesis through regulation of adult hepatic erythropoietin synthesis. Nat. Med 12, 793–800. [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. (2012). The accessible chromatin landscape of the human genome. Nature 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulirsch JC, Nandakumar SK, Wang L, Giani FC, Zhang X, Rogov P, Melnikov A, McDonel P, Do R, Mikkelsen TS, and Sankaran VG (2016). Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell 165, 1530–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulirsch JC, Lareau CA, Bao EL, Ludwig LS, Guo MH, Benner C, Satpathy AT, Kartha VK, Salem RM, Hirschhorn JN, et al. (2019). Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nat. Genet 51, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi A, Ulirsch JC, Ludwig LS, Fiorini C, Yasuda M, Choudhuri A, McDonel P, Zon LI, and Sankaran VG (2016). Insight into GATA1 transcriptional activity through interrogation of cis elements disrupted in human erythroid disorders. Proc. Natl. Acad. Sci. USA 113, 4434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2011). Haemoglobin concentrations for the diagnosis of anaemia and assessment of severity.https://apps.who.int/iris/bitstream/handle/10665/85839/WHO_NMH_NHD_MNM_11.1_eng.pdf?ua=1.
- Yan H, Hale J, Jaffray J, Li J, Wang Y, Huang Y, An X, Hillyer C, Wang N, Kinet S, et al. (2018). Developmental differences between neonatal and adult human erythropoiesis. Am. J. Hematol 93, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Mei Y, Schipma MJ, Roth EW, Bleher R, Rappoport JZ, Wickrema A, Yang J, and Ji P (2016). Nuclear condensation during mouse erythropoiesis requires caspase-3-mediated nuclear opening. Dev. Cell 36, 498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw sequencing data reported in this paper is GEO: GSE115684 and is available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115684. AUCSC Track Hub for the ATAC-seq data generated for these eight populations is available here: https://s3.amazonaws.com/atacerythropoiesis/hub.txt. A git repository containing code for analyses described here and processed data files is available at www.github.com/sankranlab/erythroid-profiling.