Abstract
We have cloned cDNAs that encode a complete open reading frame for a calcium channel α1 subunit from Drosophila melanogaster. The deduced 1851 amino acid protein belongs to the superfamily of voltage-gated sodium and calcium channels. Phylogenetic analysis shows that the sequence of this subunit is relatively distant from sodium channel α subunits and most similar to genes encoding the A, B, and E isoforms of calcium channel α1 subunits. To indicate its similarity to this subfamily of vertebrate isoforms, we name this protein Dmca1A, for Drosophila melanogaster calcium channel α1 subunit, type A. Northern blot analysis detected a single 10.5 kb transcript class that is regulated developmentally, with expression peaks in the first larval instar, midpupal, and late pupal stages. In late-stage embryos, Dmca1A is expressed preferentially in the nervous system. Variant transcripts are generated by alternative splicing. In addition, single nucleotide variations between cDNAs and genomic sequence are consistent with RNA editing. Dmca1A maps to a chromosomal region implicated in, and is the likely candidate for, the gene involved in the generation of behavioral, physiological, and lethal phenotypes of the cacophony, nightblind-A, and lethal(1)L13mutants.
Keywords: cDNA sequence, RNA editing, alternative splicing, phenylalkylamine binding site, chromosome aberrations, vital gene
Calcium channels are involved in functions including membrane excitability, synaptic transmission, regulated secretion, and cell differentiation. They conduct currents with heterogeneous conductances, kinetics, and pharmacological sensitivities (Hille, 1992). Calcium channels are hetero-oligomeric assemblies of α1, α2, δ, β, and γ subunits (Campbell et al., 1988;Catterall et al., 1988; Ahlijanian et al., 1990; McEnergy et al., 1991;Witcher et al., 1993; Leveque et al., 1994); the α1 subunit forms the calcium-conducting pore of the channel. Channel diversity is generated by multiple genes, alternative splicing of transcripts from a given gene, and perhaps by combinatorial assembly of variant isoforms of the subunits (reviewed in Hofmann et al., 1994; Stea et al., 1995).
There is evidence for calcium channel diversity in Drosophila melanogaster. Both high- and low-affinity binding of phenylalkylamines were identified in Drosophila head extracts (Pauron et al., 1987; Greenberg et al., 1989). In culturedDrosophila embryonic neurons and myocytes, cell-attached patch-clamp studies have identified currents with variable properties, including inactivating and noninactivating barium currents with differential sensitivity to purified Hololena spider toxin (HoTX) (Leung et al., 1989; Leung and Byerly, 1991). Reconstitution ofDrosophila head membrane extracts into artificial bilayers revealed calcium conductances with eight distinct conductance levels; some classes were sensitive to dihydropyridines and others to phenylalkylamines (Pelzer et al., 1989). Gielow et al. (1995)distinguished whole-cell calcium currents in Drosophilalarval body wall muscles with differential sensitivity to dihydropyridines and amiloride.
Genetic studies of calcium channels are beginning to define the functional significance of various α1 subunits. An α1 subunit (Dmca1D) similar to L-type vertebrate channels has been cloned fromDrosophila melanogaster (Zheng et al., 1995). Mutations in the Dmca1D gene cause embryonic lethality (D. F. Eberl and L. M. Hall, unpublished observations). A partial α1 cDNA sequence from theCaenorhabditis elegans unc-2 locus has similarity to vertebrate non-L-type channels, and mutations in this gene disrupt physiological adaptation to dopamine and serotonin (Schafer and Kenyon, 1995). A single-base deletion mutation leading to a frame shift in a skeletal muscle α1 subunit gene has been found in themuscular dysgenesis mutant mouse (Chaudhari, 1992).
We have been analyzing a genetic locus defined by the courtship song mutant cacophony, the visually defectivenightblind-A mutants, and by lethal(1)L13variants. We report here the identification and molecular analysis of a calcium channel α1 subunit. This is only the fourth such subunit cloned from invertebrates (see above; see also, Grabner et al., 1994). The Dmca1A transcript spans deletion and inversion breakpoints associated with, and is therefore likely a product of the gene responsible for, these genetic variants. In addition to providing further evidence pertaining to invertebrate calcium-channel diversity, this new α1 subunit gene may permit genetically based studies of α1 subunit variation and its connection to these behavioral- and visual system-specific phenotypes.
MATERIALS AND METHODS
cDNA cloning. As part of an analysis of transcripts from the cytogenetic region 11A1–2, known to encode several genes of interest, clone pNB53 was isolated from a 12–24 hr embryonic cDNA library (Brown and Kafatos, 1988). It was subcloned into pBS(+) (Stratagene, La Jolla, CA) after a completeNotI and partial HindIII digestion to generate the clone cSK53. Subclones generated by restriction digestions and by digestion into 200–500 nucleotide fragments with DNAaseI in the presence of manganese (Sambrook et al., 1989) were ligated into pBluescript IISK(+) for sequencing. Additional sequence was obtained from cSK53 with insert-specific primers.
A ClaI fragment of cSK53 corresponding to the region encoding amino acids 603–749 in Dmca1A in Figure 2 was used to probe 1.2 × 106 pfu of a λ gt11Drosophila head cDNA library (Itoh et al., 1986), resulting in the acquisition of 11 clones. One of them, c31, extended the sequence in the 5′ direction but still was missing the 5′ end of the open reading frame. Two probes, corresponding to the regions encoding amino acids 259–637 and 608–1076 in Dmca1A (Fig. 2), were generated by PCR from clones c31 and cSK53, respectively; these were used together to screen 1.2 × 106 pfu from a λ-zapIIDrosophila head cDNA library (DiAntonio et al., 1993) to isolate the 5′ clones cS14a and cS25a, as well as 43 additional partial cDNAs; the latter included cS26a and cS29b. These were excised into pBluescript IIKS(+) and sequenced with vector- and insert-specific primers.
Fig. 2.
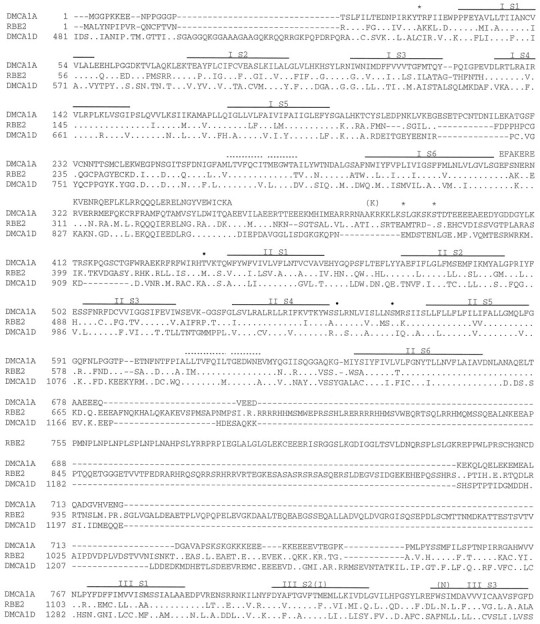
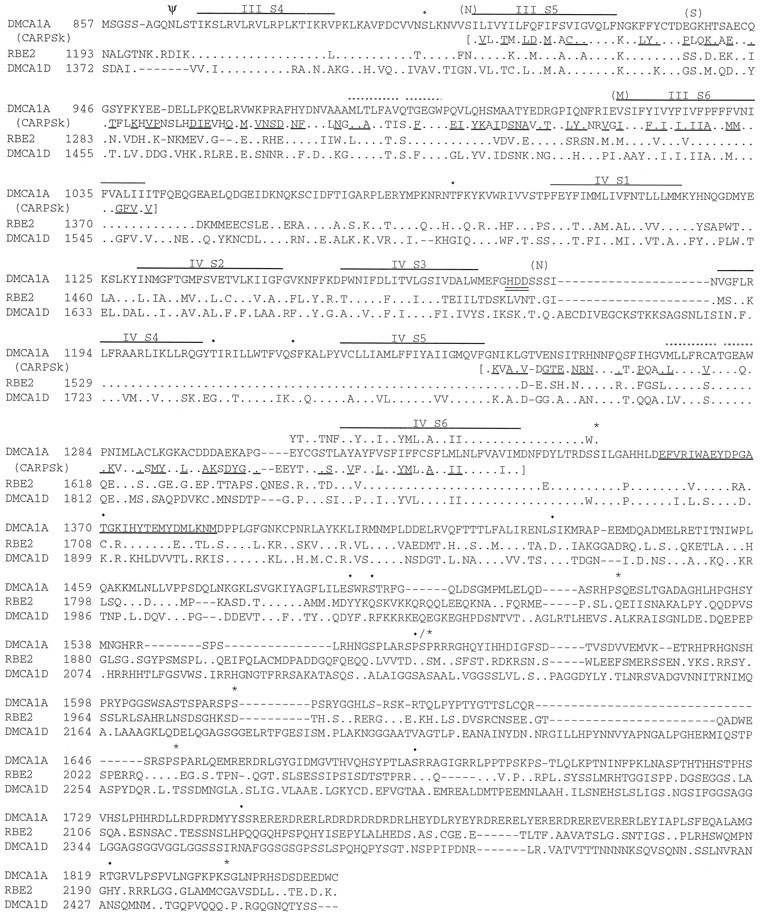
Comparison of the deduced amino acid sequence of Dmca1A with rat brain E (rbE2) and Drosophila Dmca1D sequences. Sequences were aligned with ClustalW software (gap penalty, 20; gap extension, 0.05); the first 480 amino acids of Dmca1D align 5′ to the included sequences and were omitted. Identical amino acids are indicated by dots and gaps, by dashes. Putative transmembrane domains are indicated by a single line overthe Dmca1A sequence. The short segment 1 and 2 regions in each repeat are indicated by dotted lines above the Dmca1A sequence. The sequence of the alternative exon detected in clone c31 is aligned above Dmca1A, just downstream of IS6. The amino acids encoded by the nine bases representing the largest variant at the IVS3–S4 variable region are double-underlined. Amino acid sequence from carp skeletal muscle (CARPSk) identified as containing dihydropyridine binding sites (Grabner et al., 1996) is included withinbrackets and aligned below the IIIS5–S6 and IVS5–S6 domains in Dmca1A. A proposed phenylalkylamine binding sequence (Striessnig et al., 1990) is aligned above the IVS6 region of Dmca1A. A calcium-binding EF-hand structure downstream of IVS6 issingle-underlined in Dmca1A. The potential N-glycosylation site at the N terminus of IIIS4 is marked with Ψ. cAMP kinase sites are marked with *; PKC sites with •. Sites of potential RNA editing are indicated by aligning the unedited codon identity inparentheses above the Dmca1A sequence. The GenBank accession number for Dmca1A is U55776.
Nineteen of 45 cDNAs isolated from the original screen of the λ-zap library could not be subcloned into pBluescript IIKS(+). Suspecting that some of these might represent 3′ cDNAs, we screened excised filamentous phage supernatants by PCR, using a 5′ insert-specific primer corresponding to the region encoding amino acids 1139–1145 and T3 or T7 vector-specific primers (cf. Chiang et al., 1994) to detect the inclusion and size of potential 3′ clones. Clones cS9a and cS11 were found by this analysis to extend 3′ to the existing cDNAs. We were unable to propagate cDNAs containing the 3′ ends of the open reading frame in plasmid vectors, so they were sequenced directly from PCR products.
Clone c3p1 (which extended 520 bases past the 3′ end of cS9a) was obtained in a screen of an additional 2.5 × 105 pfu from the λ-zap library probed with a PCR product generated with cS9a as template; that probe corresponds to the region encoding amino acids 1556–1802 in Dmca1A. Clone c3p1 was sequenced as described for cS9a and cS11 above.
Preparation of probes and sequencing templates. Probes for cDNA screens were labeled with 32P-dCTP by random priming by either the Random Primer DNA Labeling System (BRL, Grand Island, NY) or Prime-IT II (Stratagene). PCR products were purified for labeling or sequencing either directly with the QIAquickSpin PCR Purification Kit (Qiagen, Chatsworth, CA) or after gel purification with the QIAEXII Gel Purification Kit. All sequencing used double-stranded templates prepared either with the Qiagen Plasmid Kit or by alkaline lysis and LiCl precipitation (Sambrook et al., 1989). Most sequencing was done on an ABI 373A sequencer using vector- or gene-specific primers with the PRISM DyeDeoxy Terminator Cycle Sequencing Kit [Applied Biosystems (ABI), Foster City, CA] and/or using labeled T3 or T7 primers with theTaq Dye Primer Cycle Sequencing kit (ABI). DNA was sequenced at least twice in each direction, except as noted in Results. When sequencing was conducted from PCR products, sequence was derived from at least two independent reactions. Sequence analysis and contig assembly were done by the GCG package of programs (Genetics Computer Group, 1991). Database searches were performed by the BLAST network service at National Center for Biotechnology Information.
RNA preparation and Northern blots. Samples from different developmental stages were grown, collected, and synchronized at 25°C, as described by Ashburner and Thompson (1978). Preparation of poly(A+) mRNA, Northern blots, and hybridization conditions were as described in Zheng et al. (1995). Transcript abundances were quantitated with an UltroScan scanning laser densitometer with GelScan XL software, version 2.1 (Pharmacia, Piscataway, NJ).
In situ hybridization to embryo whole mounts.Whole-mount in situ hybridization to Drosophilaembryos (stage 16) was done as described by Tautz and Pfeifle (1989). A single-stranded 245-base digoxigenin-labeled cDNA probe (corresponding to the region encoding amino acids 970–1052 in Dmca1A in Fig. 2) was prepared and applied as described by Zheng et al. (1995).
Southern blotting. The deletion Df(1)HF368, inversion In(1)N66, and balancer chromosomeIn(1)FM7,B carried in flies that were the DNA source for this experiment are described in Goralski (1985), Kulkarni and Hall (1987), and Lindsley and Zimm (1992). One breakpoint ofIn(1)N66 is in cytogenetic region 11A2; this rearrangement fails to complement the phenotypes of cacophony, nightblind-A, and l(1)L13 mutations (Kulkarni and Hall, 1987; Homyk and Pye, 1989). Df(1)HF368 also is broken in 11A2 and fails to complement these mutations; this deletion removes a portion of the chromosome toward the centromere from 11A2. Preparation of DNA and probe, restriction digestions, blotting, and hybridization were performed as described in Sambrook et al. (1989). Five micrograms of genomic DNA were electrophoresed in each lane. The template for making probe was prepared by digesting the genomic phage clone 320 (cf.Goralski, 1985) with EcoRI, followed by electrophoretic purification of the insert. This genomic clone is homologous to portions of cDNA clones c31, cS14a, cS25a, and cS11 (see Fig. 1). The ultimate autoradiograph was scanned with a ScanJet IIc scanner and DeskScan II software (Hewlett-Packard). The scanned image was filtered to reduce mid-densities (thus reducing background), and the figure was printed from the scanned image by a commercial pictrography service (Pageworks, Cambridge, MA).
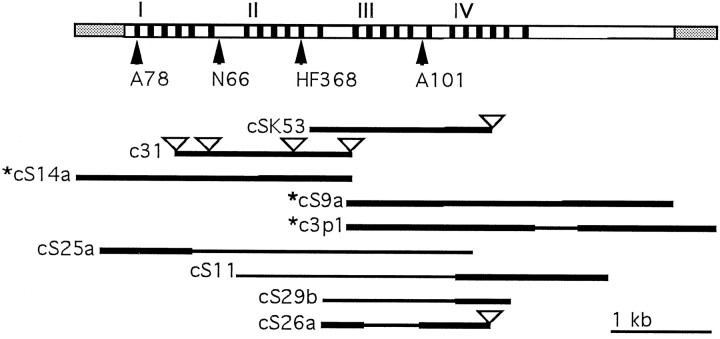
Fig. 1.
Overlapping cDNAs used to deduce the full-length ORF of Dmca1A. The diagram at the top of this figure shows the organization of the Dmca1A calcium channel α1 subunit ORF (inwhite), untranslated regions (UTRs, in gray), and the approximate locations of transmembrane domains (inblack). Roman numerals indicate the beginning of each of the four repeat domains. Below this diagram, bold black lines show cloned cDNAs for which sequence was determined at least twice in each direction. Narrow black lines indicate regions determined by partial sequence analysis. Triangles show the location of unspliced introns in the indicated cDNA, as determined by comparison with genomic sequence and other cDNAs. Sequences used to assemble the full-length Dmca1A sequence shown in Figure 2 are marked with an asterisk. Arrowheads indicate the location of exons that we have mapped to genomic restriction fragments; these, in turn, were shown previously and contemporaneously by RFLP detection (Goralski, 1985; Fig. 7 of the current study) to contain an array of four 11A2 region breakpoints, associated with the following chromosome rearrangements (indicated in abbreviated form in the figure):In(1)A78, In(1)N66, and In(1)A101 are chromosomal inversions that fail to complement l(1)L13, cacophony, andnightblind-A mutants (Kulkarni and Hall, 1987; Homyk and Pye, 1989). Df(1)HF368 is a chromosomal deletion that also fails to complement these mutations (see papers just cited) and removes sequences 5′ to the indicated breakpoint (Goralski, 1985); the sequences removed are centromere-proximal to the HF368breakpoint (i.e., the 5′ end of the coding sequences implied by thetop line is shown to the left, as usual, which makes the centromere end of the X chromosome to the left of this image; the gd gene is located centromere-proximal to, i.e., leftward of, the transcription unit shown). Phage clone 320 (see Materials and Methods) spans a genomic interval that encodes a portion of the channel running from approximately IS5 to IIS5. Clone 320 hybridizes to genomic restriction fragments containing theIn(1)N66 and Df(1)HF368 breakpoints (Fig. 7, below); the total genomic distance between the chromosomal breakpoints flanking the two (more central lesions) just indicated is ∼15 kb (Goralski, 1985); owing to the fact that this gene is especially intron-rich in genomic intervals to the left of the A78 breakpoint and to the right of the A101 one, the coding material and UTR shown (top line) arise from a ∼50 kb genomic interval (Peixoto, Smith, Hall, Hall, unpublished observations). Clone 320 also was determined by Northern blotting to hybridize to an 0.8 kb transcript (Goralski, 1985). Sequence analysis of an 0.8 kb cDNA hybridizing to this clone reveals short regions of identity to the Dmca1A transcript (Smith and Hall, unpublished observations). This implies that the 0.8 kb transcript, which is the only other transcript detected in this region (also see Fig. 4, below), might be an aberrant form of the Dmca1A transcript.
RESULTS
Isolation of cDNAs encoding a new calcium channel α1 subunit
The cDNA encoding the calcium channel α1 subunit reported here was isolated during the analysis of a region of the X chromosome known to contain the gene cacophony (cac) and the interacting genetic variants nightblind-A (nbA) and lethal(1)L13 (also known as l(1)11Aa;Lindsley and Zimm, 1992). The cac locus was mapped cytogenetically using inversions and deletions to the X chromosomal region 11A2 (Kulkarni and Hall, 1987). Genomic phage clones 320 and 0371 (Goralski, 1985) were derived from a chromosome walk through the flanking gastrulation defective (gd) locus (which is ∼10 kb from the left-hand, centromere-proximal end of the putative cac-locus, as depicted in Fig. 1). Clones 320 and 0371 recognized restriction-fragment-length polymorphisms (RFLPs) associated with breakpoints in this region [Goralski (1985); also see Fig. 7, below]. Northern blots of adult RNA probed with fragments of clone 320 recognized two transcripts: 0.8 kb (Goralski, 1985) and >10 kb (our preliminary data, not shown; also see Fig. 1 legend and Fig. 4, below). To clone cDNAs encoding these transcripts, we probed a 12–24 hr embryonic cDNA library (Brown and Kafatos, 1988) with genomic clone 0371 and isolated the cDNA clone cSK53 (Fig. 1). In situ hybridization of cSK53 to salivary gland chromosomes from third-instar larvae mapped this cDNA to the distal portion of region 11A, consistent with the origin of the original genomic probes (data not shown). Northern blotting showed that the cSK53 cDNA corresponds to a subset of the large mRNA transcribed from this X chromosomal region (see below, Fig. 4). The genomic interval that gives rise to the coding (plus untranslated) RNA indicated on the top line of Figure 1 is approximately eight times longer than the amount of sequence so depicted (see Fig. 1 legend).
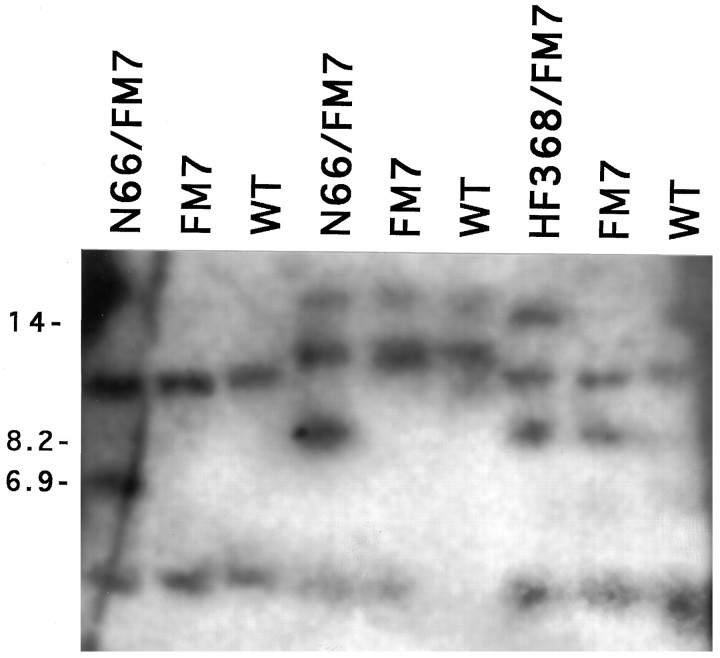
Fig. 7.
Southern blot detection of lethal chromosomal lesions, the breakpoints of which map to the region encoding Dmca1A.Goralski (1985) localized breakpoints of inversion- and deletion-containing chromosomes to a relatively small genomic interval within the cytogenetic region called 11A2 by RFLP detection with multiple restriction enzymes. To confirm elements of these findings, we prepared genomic DNA from In(1)N66 and Df(1)HF368flies heterozygous for the In(1)FM7 balancer chromosome, from homozygous In(1)FM7 flies, or from a wild-type strain (Canton-S). DNA in lanes 1–3 was subjected to restriction digestion with HindIII, in lanes 4–6with BamHI and KpnI, and in lanes 7–9with HindIII and KpnI (following Goralski, 1985). The blot was probed with genomic phage clone 320 (cf. Goralski, 1985), which contains a portion of the Dmca1A ORF extending approximately from transmembrane domains IS5 to IIS5 (see Fig. 1). The novel restriction fragments present in lanes 1, 4, and 7 are as originally detected by Goralski (1985); the size of these fragments (in kb) is indicated in the left margin of the figure.
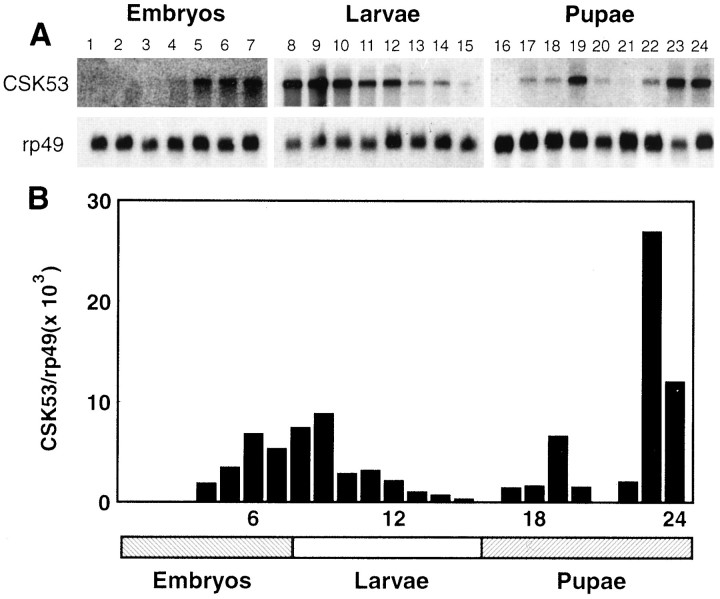
Fig. 4.
Developmental profile of the Dmca1A calcium channel α1 subunit mRNA expression. A, Each lane contains ∼10 μg of poly(A+) RNA. Blots were probed first with a 1 kb EcoRI fragment of Dmca1A cDNA and later reprobed with a ribosomal protein (rp49) cDNA probe (O’Connell and Rosbash, 1984). Exposure time for the Dmca1A autoradiographs was 21 d for the embryonic and pupal blots and 7 d for the larval blot. Exposure times for the rp49-probed blots were 18 hr for embryos and pupae and 6 hr for larvae. The collection times in hours postoviposition for animals in the embryonic (lanes 1–7) and larval (lanes 8–15) stages were lane 1, 0–3 hr;lane 2, 3–6 hr; lane 3, 6–9 hr; lane 4, 9–12 hr; lane 5, 12–15 hr; lane 6, 15–18 hr; lane 7, 18–21 hr; lane 8, 21–36 hr;lane 9, 36–48 hr; lane 10, 48–60 hr; lane 11, 60–72 hr; lane 12, 72–84 hr; lane 13, 84–96 hr; lane 14, 96–108 hr; lane 15, 108–120 hr. The collection times for pupal stages (lanes 16–24) in hours postpuparium formation were lane 16, 0–12 hr; lane 17, 12–24 hr; lane 18, 24–36 hr; lane 19, 36–48 hr; lane 20, 48–60 hr; lane 21, 60–72 hr; lane 22, 72–84 hr;lane 23, 85–96 hr; lane 24, 96–108 hr.B, The autoradiographs were quantitated by scanning densitometry. Dmca1A/rp49 ratios were calculated from the densities of the Dmca1A and rp49 signals for each lane after correction for exposure times.
Sequence analysis showed that cSK53 contains an open reading frame (ORF) encoding a fragment similar to a voltage-sensitive calcium channel α1 subunit (Fig. 2). Several rounds of cDNA isolation from Drosophila head cDNA libraries, starting with a probe derived from cSK53 and continuing in later rounds with probes from newly isolated cDNAs, isolated a total of 57 cDNAs. A subset of these was chosen for further analysis based on length and overlap with other cDNAs (Fig. 1). A 6522 nucleotide cDNA contig—assembled from the overlapping cDNAs cS14a, cS9a, and c3p1—contains a single large ORF of 5553 nucleotides, which encodes a voltage-sensitive calcium channel α1 subunit (Fig. 2). An AUG at nucleotide positions 553–555 is the only in-frame methionine codon between five upstream in-frame stop codons and sequences coding for the first transmembrane domain (IS1). The sequence flanking this methionine codon (UAGA AUG) shows two of four matches to the Drosophilatranslation initiation consensus sequence (C/A AA A/C AUG) (Cavener, 1987). It includes the highly conserved A at the −3 position, and the G at −2 is the second most frequently used nucleotide at this position. Thus, we infer this AUG to be the translation start site. Although we have no evidence to suggest alternate initiation methionines, we cannot rule out the possibility of alternative 5′ exons in transcripts not represented in this analysis.
The first in-frame stop codon is a TAG at nucleotide position 6106. It is followed by two additional in-frame stop codons within the next 50 nucleotides. The 3′ untranslated region of this contig is 416 nucleotides in length. There is no polyadenylation signal or polyadenylated tract in this contig, suggesting that the 3′ untranslated region is incomplete. This suggestion is consistent with the difference between the length of the transcript (10.5 kb) and the assembled contig (6.5 kb).
Structure of the calcium channel α1 subunit protein
The ORF of 5553 nucleotides encodes a protein of 1851 amino acids, with a calculated molecular weight of 212,155. The protein has the canonical structure of voltage-gated calcium channel α1 and sodium channel α subunits, with four internal repeats (I–IV), each containing six presumed membrane-spanning hydrophobic domains (S1–S6). Transmembrane segments S4 of each internal repeat contain positively charged amino acids every third or fourth amino acid, consistent with the postulated role of these segments in sensing and responding to transmembrane voltage changes. In addition, the conserved domains for short segments 1 and 2 (ss1, ss2) in the loop between transmembrane domains S5 and S6 of each repeat are conserved in this protein.
Comparison of both the overall protein and of these conserved domains reveals a strikingly greater similarity to calcium channel α1 subunits than to sodium channel α subunits (see below, Fig. 6). A conserved glutamate present in ss2 in the loop between transmembrane domains S5 and S6 of each repeat is involved in ion selectivity (Kim et al., 1993; Tang et al., 1993a; Yang et al., 1993). Sodium channels contain this glutamate residue only in repeats I and II, whereas calcium channels have this glutamate in all four repeats (Heinemann et al., 1992). Changing the appropriate residue to glutamate in repeats III and IV of a sodium channel converts the ion selectivity of a sodium channel to that of a calcium channel (Heinemann et al., 1992). Conversely, changing the identity of these glutamate residues alters the ion selectivity and conductance of calcium channels (Mikala et al., 1993; Tang et al., 1993a; Yang et al., 1993; Ellinor et al., 1995). The glutamate residues relevant to ion selectivity are conserved in all four ss2 domains of the Dmca1A protein, consistent with identification of this protein as a calcium channel α1 subunit.
Fig. 6.

Phylogenetic analysis of Dmca1A and α1 subunits representative of other calcium channel subfamilies. The channels indicated are rbA1, rat brain class A; M64373 (Starr et al., 1991); rbB1, rat brain class B; M92905 (Dubel et al., 1992); rbE2, rat brain class E; M94172 (Soong et al., 1993); rbC1, rat brain class C; M67516 (Snutch et al., 1991); a1D, human class D; M76558 (Williams et al., 1992); RSkm, rat skeletal muscle; X05921 (Tanabe et al., 1987);Mdla1, Musca larvae; Z31723 (Grabner et al., 1994); Dmca1D, Drosophila; U00690 (Zheng et al., 1995). Transmembrane sequences of each of these channels were concatenated into a single sequence, and the resulting “core” sequence files were aligned with the ClustalW program (default BLOSUM scoring matrix series; gap penalty, 20; gap extension, 0.5) (Thompson et al., 1994). Full-length sequences also were aligned. Small discrepancies between endpoints of reported transmembrane domains were resolved by reference to the multiple alignment in Stea et al. (1995). A sodium channel α subunit sequence (Noda et al., 1986) was included to indicate relatively greater similarity of Dmca1A to calcium channel α1 subunit sequences. A, Distance matrix for calcium channel α1 subunits. The distance between channel sequences, representing the minimum number of nucleotide changes necessary to the observed amino acid differences, was calculated for both the core sequences and for the full-length sequences, using a neighbor-joining algorithm as implemented in ClustalW (Thompson et al., 1994) (ignore gaps = on; multiple substitutions = off). In the bottom left of the matrix, distances are calculated from alignment of transmembrane core sequences. In the top right of the matrix, distances are calculated from alignment of full-length sequences. B, Phylogenetic tree for core sequence alignment. The Retree and Drawtree programs of the Phylip software package (Felsenstein, 1989) were used to display a phylogenetic tree using data from the core sequence alignment shown in the bottom left corner of the distance matrix. Branch lengths between subunits are proportional to divergence. A tree generated from full-length sequence alignments had identical topology to the one shown.
Possible phenylalkylamine binding site in Dmca1A
On the basis of immunoprecipitation of phenylalkylamine-labeled proteolytic peptide fragments, Striessnig et al. (1990) proposed that a fragment of the rabbit skeletal muscle α1 subunit, including transmembrane domain IVS6 and the adjacent intra- and extracellular sequences, functions as a binding site for the phenylalkylamine calcium channel blockers. Combined with previous work suggesting that phenylalkylamines block calcium channels intracellularly, this evidence identified the intracellular portion of the IVS6 transmembrane domain and the adjacent intracellular amino acids as a binding site for phenylalkylamines. Figure 2 shows the proposed phenylalkylamine binding fragment sequence defined by Striessnig et al. (1990) aligned above the Dmca1A sequence. A 17 amino acid sequence bracketing the intracellular junction of the IVS6 transmembrane domain is conserved completely between these two proteins. This conserved region is flanked on the N-terminal side by two conservative amino acid changes (isoleucine to leucine and isoleucine to methionine), preceded by two more identical amino acids (FL). The region extends to within two amino acids of the C-terminal end of the rabbit proteolytic fragment, where there is a nonconservative tryptophan-to-serine change in the Dmca1A sequence, followed by a conserved serine.
Dihydropyridine binding sites are poorly conserved in Dmca1A
Proteolytic fragments containing the IIIS6 and IVS6 transmembrane domains and regions immediately adjacent to them have been shown to bind dihydropyridines (Nakayama et al., 1991; Striessnig et al., 1991). Dihydropyridine sensitivity of an L-type channel is abolished when a portion of the polypeptide overlapping the extracellular end of IVS6 is replaced with non-L-type sequence (Tang et al., 1993b). Conversely, dihydropyridine sensitivity can be conferred upon a non-L-type channel by replacing the IIIS5–S6 and IVS5–S6 regions with sequences from L-type (carp or rabbit) skeletal muscle subunits (Grabner et al., 1996). Because dihydropyridines bind to the channel from the outside (Bangalore et al., 1994), the portions of these fragments that begin in the extracellular domain and enter into the transmembrane segments from the outside are, most likely, involved in dihydropyridine binding.
The sequences from a dihydropyridine-sensitive carp skeletal muscle α1 subunit that confer dihydropyridine sensitivity to chimeric channels (see above) are shown in Figure 2, aligned below Dmca1A in the IIIS5–S6 and IVS5–S6 regions. Certain amino acids in the dihydropyridine-sensitive carp skeletal muscle subunit (underlined in the figure) were identified by Grabner et al. (1996) as potentially relevant to dihydropyridine sensitivity. In the region of IIIS5–S6 and IVS5–S6, there are 102 such amino acids. Of these, Dmca1A is identical to dihydropyridine-sensitive channels at only 18 sites. By comparing all known dihydropyridine-sensitive and -resistant channels, Grabner et al. (1996) also identified 23 positions within these regions where the amino acid was different between dihydropyridine-resistant and -sensitive vertebrate α1 subunit, but 100% identical within the resistant and sensitive subgroups. Of these 23 amino acids, Dmca1A was identical to the dihydropyridine-resistant channels at 20 sites and showed identity to the sensitive channels at only three sites. The lack of correspondence between Dmca1A and dihydropyridine-sensitive channels at these positions suggests that the Drosophila Dmca1A α1 subunit may be insensitive to dihydropyridines.
Calcium binding EF-hand motif in Dmca1A
Calcium and sodium channels often contain in their C-terminal intracellular regions an EF-hand motif, which forms a structure of two α helices flanking a calcium-binding loop (Babitch, 1990). This motif has been correlated functionally with Ca2+-sensitive inactivation of calcium channels (deLeon et al., 1995). A potential EF-hand in Dmca1A, immediately C terminal to transmembrane domain IVS6, is underlined in Figure 2. In the Tufty–Kretsinger test (Tufty and Kretsinger, 1975) the Dmca1A sequence has 12 matches of 16 for residues important for calcium binding. Allowing conservative substitutions increases this match to 14 of 16 positions.
Potential sites of post-translational modification
There are several sites of possible post-translational modification of the Dmca1A protein. A single extracellular site matching the consensus sequence [N]-[∼P]-[S/T]-[∼P] for N-linked glycosylation (cf. Hubbard and Ivatt, 1981) was found at N865 near the N terminus of the IIIS4 transmembrane domain. Nine intracellular consensus sites for cAMP-dependent protein kinase phosphorylation [R/K]-[X]-[X]-[S/T] were found (cf. Krebs and Beavo, 1979): in the N terminus at T31; in the I/II loop at S386 and S392; and in the C terminus at S1348, S1519, S1559, S1616, S1650, and S1836. Fifteen intracellular sites matching the consensus PKC phosphorylation site [S/T]-[X]-[R/K] were found (cf. Woodgett et al., 1986): in the I/II loop at T437; in the IIS4–S5 loop at S552 and S563; in the IIIS4–S5 loop at S900; in the III/IV loop at T1083; in the IVS4–S5 loop at T1209 and S1220; and in the C terminus at T1370, S1432, S1493, S1496, S1559, S1683, S1748, and T1820. The clustering of 13 potential phosphorylation sites in the C terminus suggests that this region may be involved in phosphorylation-dependent modification of calcium channel function.
Of particular interest because of their possible functional significance are five conserved PKC sites that are found in the S4–S5 loops of all non-L-type channels. These sites are not found in any L-type channels and thus may mediate a property that distinguishes these channels functionally. In Dmca1A these sites are S552, S563, S900, T1209, and S1220. Their proximity to the voltage-sensing S4 transmembrane domain is intriguing. There is, in addition, a cAMP-dependent protein kinase phosphorylation site conserved in the segment between IVS6 and the EF-hand in all calcium channels sequenced to date. This site is likely to modulate a function common to all calcium channels.
Alternative exons are different in the region of the β subunit binding site
Calcium channel β subunits interact with α1 subunits to stimulate peak current amplitude, to increase the rate of activation, and to modify the voltage dependence of activation and inactivation inDrosophila (D. Ren, M. Chopra, L. M. Hall, unpublished observations) as well as in other species (Lacerda et al., 1991; Varadi et al., 1991; Neely et al., 1993). Pragnell et al. (1994) identified a conserved 18 amino acid sequence (QQ-E–L-GY–WI—E) in the I–II cytoplasmic linker that binds β subunits. Mutations in this conserved domain inhibit β subunit binding. Analysis of the cDNA clones summarized in Figure 1 shows that alternative splicing in the region encoding this I/II linker in Dmca1A generates α1 subunits with major differences in the β subunit binding domain.
The ORF of clone c31 is interrupted by four unspliced introns (Fig. 1). The introns are bounded by consensus splice-site sequences (cf. Mount et al., 1992) and contain no regions of similarity to overlapping cDNAs. Identity as introns was confirmed by comparison with genomic sequences (A. A. Peixoto, L. A. Smith, L. M. Hall, J. C. Hall, unpublished observations). In addition, in the region immediately downstream of the IS6 transmembrane domain, the c31 ORF diverges from that of cS14a for 116 nucleotides, encoding a 38 amino acid sequence beginning at amino acid 315. Each of these divergent sequences is of the same length and is in frame with the Dmca1A ORF. The two divergent sequences (alternative cassettes) are encoded in separate (albeit nearby) genomic regions (Peixoto, Smith, Hall, Hall, unpublished observations), where they each are flanked by consensus splice-site sequences (cf. Mount et al., 1992). In Figure 2, the c31-encoded amino acid sequence is shown aligned above Dmca1A, which contains the sequence encoded by cS14a. Comparison of these divergent transcripts with representative vertebrate sequences (Table 1) shows that the pattern of similarity to these sequences differs between the exons. The c31-encoded exon is more similar to vertebrate α1 subunits in this region (58–84% identity) than is the cS14a sequence (37–47% identity). The c31 form is most similar to the non-L-type isoforms A, B, and E in this region.
Table 1.
Comparison of alternative exons encoding the β subunit binding domain
| rbA1 | rbB1 | rbE2 | rbC1 | a1D | RSk | |
|---|---|---|---|---|---|---|
| cS14a | 15 | 16 | 15 | 18 | 18 | 14 |
| 0.39 | 0.42 | 0.39 | 0.47 | 0.47 | 0.37 | |
| c31 | 32 | 32 | 30 | 22 | 23 | 22 |
| 0.84 | 0.84 | 0.79 | 0.58 | 0.61 | 0.58 |
Alternative exons encoded by cDNAs cS14a and c31 were aligned with the mammalian calcium channel subunit sequences named in the top row. The top number in each cell represents the number of amino acid identities at 38 positions; the bottom number is the proportion of the 38 residues that are identical between a given Drosophilasubsequence (second and third rows) and the mammalian ones. Abbreviations for the mammalian α1 channel subtypes are as indicated in Figure 6.
The c31 exon contains the first 17 amino acids of the conserved β subunit binding domain. The final conserved glutamate (E) is encoded by the first codon of the downstream exon. Interestingly, the c31-encoded exon has 100% conservation of the nine amino acids required for β subunit binding, whereas the cS14a-encoded exon has only a 4/9 match, with the tyrosine (Y), tryptophan (W), isoleucine (I), and terminal glutamate (E) being conserved. If the cS14a exon is incorporated into a functional α1 subunit, this subunit might not bind β subunits or may be involved in differential interactions with β isoforms.
Transcript diversity in the IVS3–S4 extracellular region
As summarized in Figure 3, sequencing of six cDNAs from different libraries revealed substantial heterogeneity in the IVS3–S4 loop. Some of this sequence diversity may arise from incomplete splicing, because the sequence downstream of the common region in cS26a and cSK53 contains no large ORF and begins with 5/6 or 6/6 matches to Drosophila 5′ consensus splice-site sequences (Mount et al., 1992). Relative to cS26a, cSK53 contains six additional in-frame nucleotides before the start of the presumed unspliced exon.
Fig. 3.
Alignment of cDNA sequences at the variable region encoding the IVS3–S4 loop. Sequences matching 5′ splice-site consensus sequences are double-underlined, and the sequence from inferred introns is in lower case. These intron junctions begin large unspliced introns (see Fig. 1) 3′ to the variable region in these cDNAs. Underlined A and G nucleotides in the completely spliced cDNAs correspond to an A in genomic sequence and likely reflect RNA editing at this position. The conceptually translated protein sequence, determined by the predominant G nucleotide at the edited position, is aligned underneath; use of the A nucleotide at this position changes this codon identity from Ser (S) to Asn (N). Terminal residues of the IVS3 and IVS4 transmembrane domains are solid-underlined, and the variable HDD amino acid sequence is marked with a dotted underline.
Additional heterogeneity in the length of the cDNAs changes the number of amino acids in the IVS3–S4 loop from 9 to 10 or 12. Clone cS9a is the shortest (encoding the 9 amino acid IVS3–S4 loop). Clone c3p1 is slightly longer, containing an in-frame insertion of three nucleotides that are not present in cS9a but are found in both cS11 and cS26a. The latter two clones contain identical in-frame insertions of nine nucleotides; these have identical sequence to the six nucleotides in cSK53 plus the three in c3p1. The nine nucleotides found within cS11 and cS26a encode the amino acids HDD. This variable HDD segment is included as amino acids 1181–1183 in Dmca1A (Fig. 2).
Possible post-transcriptional modifications of the Dmca1A transcript
Additional single nucleotide differences were detected at seven positions in the cDNAs (Table 2). In each case, the differences are between guanosine and adenosine nucleotides. Each difference, except the one at nucleotide 1691, causes an amino acid change. The positions and amino acid differences involving these A- versus G-containing codons are presented in Table 2 and are shown in context in Figure 2. We examined the corresponding genomic sequence at these seven positions (Peixoto, Smith, Hall, Hall, unpublished observations); in all cases, the relevant nucleotide in the genomic sequence was an adenosine.
Table 2.
Positions and codon identity of possible RNA editing sites
| 1691 | 2997 | 3069 | 3269 | 3361 | 3597 | 4106 | |
|---|---|---|---|---|---|---|---|
| cSK53 | ATA | AAT | AAC | AGT | ATG | ||
| c31 | AAG | ||||||
| cS14a | AAA | ATA | |||||
| cS9a | ATG | AGT | AGC | GGT | GTG | AGC | |
| c3p1 | AAT | AGC | AGT | ATG | AAC | ||
| genome | AAA | ATA | AAT | AAC | AGT | ATG | AAC |
| 380 | 815 | 839 | 906 | 937 | 1016 | 1185 | |
| K>K | I>M | N>S | N>S | S>G | M>V | N>S |
The position of the variable nucleotide in the assembled contig is indicated at the top of the table. Codon sequences of the cDNAs and genomic sequence are shown with the relevant adenosine or guanosine indicated in bold font. Aligned at the bottom of a given column are the amino acid position number from Figure 2 and the amino acids encoded by unedited and edited codons, respectively.
Alteration of adenosines in genomic DNA to guanosines in cDNA is thought to reflect RNA editing by deamination of adenosine to inosine (reviewed by Bass, 1993). A double-stranded RNA-specific deaminase activity has been reported in Drosophila (reported in Bass, 1993). Although a few of these changes may be attributable to genetic polymorphism, the observed changes are uniformly consistent with this mechanism of RNA editing. It seems likely that a majority of these adenosine-to-guanosine differences are caused by a deamination mechanism similar to that observed in vertebrates.
Temporal pattern of expression of Dmca1A
We used quantitative Northern blotting to determine the developmental profile of expression of the Dmca1A calcium channel α1 subunit. As shown in Figure 4A, the probe used in these studies recognized a single major mRNA species of 10.5 kb at each stage tested. To correct for apparent differences in expression because of variations in RNA recovery, we reprobed the blot in Figure 4A with a cDNA encoding a widely expressed ribosomal protein (rp49) (O’Connell and Rosbash, 1984), and we determined the relative expression of Dmca1A and rp49 by scanning densitometry, as summarized in Figure 4B.
There are three peaks of expression during development. The first peak begins to rise in mid-to-late embryo stages (Fig. 4B, lanes 5–7) and reaches a peak during the first larval instar (Fig. 4B, lanes 8–9). Expression then declines over the remaining larval instars but begins to rise again after pupariation. There is a second peak in midpupal stages and a final peak in late pupae just before adult eclosion.
Spatial pattern of expression of Dmca1A
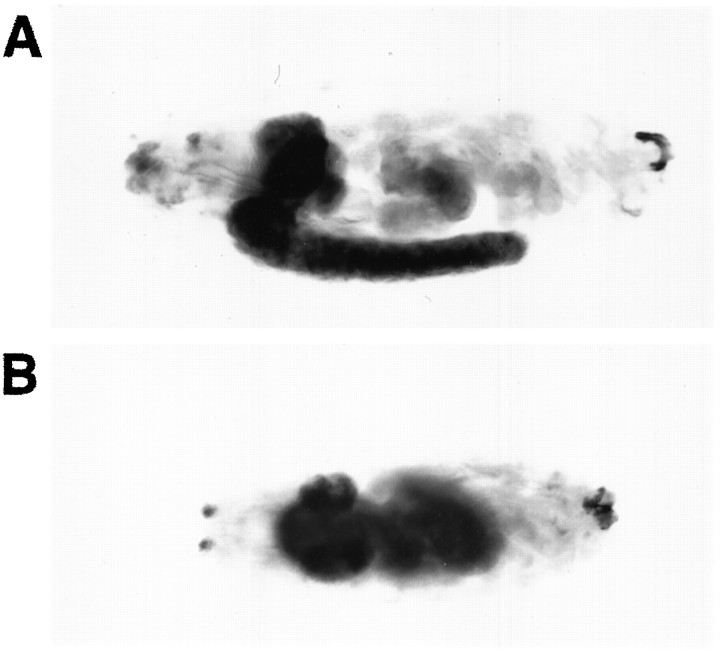
To determine where the message for the Dmca1A calcium channel α1 subunit is expressed, we used a digoxigenin-labeled antisense DNA probe on relatively late-stage embryos (equivalent to lanes 5 or6 in Fig. 4). As shown in Figure 5, this α1 subunit RNA is expressed widely in the embryonic nervous system. Intense, dark staining is seen in the dorsal cerebral hemispheres as well as throughout the ventral nerve cord. In addition, as shown in Figure 5B, bilaterally symmetric, lightly stained nerves can be seen extending anteriorly from the CNS toward the region of the antennomaxillary complex at the extreme anterior end of the animal.
Fig. 5.
Expression of Dmca1A mRNA in the embryonic nervous system. A single-stranded antisense DNA probe labeled with digoxigenin was hybridized to whole-mounted stage 16 embryos. The darkly stained areas represent regions of RNA expression. A, Side view in which anterior is to the left and dorsal isup. B, View of the dorsal surface in which anterior is to the left.
Evolutionary relationship of Dmca1A to other calcium channel α1 subunits
To examine the relationship between Dmca1A and other α1 subunits, we generated a phylogenetic tree containing the invertebrate channels and representative members of each of the six classes of vertebrate channels (Fig. 6). The structure of this tree is consistent with those reported previously for the relationship of the α1 subunits (Grabner et al., 1994; Stea et al., 1995). The Dmca1A sequence branches at the most ancestral node of the non-L-type channels, indicating that it is less similar to any of the vertebrate class A, B, or E subunit sequences than they are to each other and implying that the diversification of the vertebrate non-L-type lineage occurred after the evolutionary divergence of vertebrate and invertebrate lineages.
A partial α1 subunit sequence from the unc-2 gene ofC. elegans was reported recently (Schafer and Kenyon, 1995). Inclusion of the extant sequence in a similar analysis indicates that the C. elegans protein occupies the same branch of the tree as Dmca1A (data not shown). Dmca1A and Unc-2 could represent orthologous channels in these two species or could be representative of separate invertebrate α1 subunits that diverged after the evolutionary separation of vertebrate and invertebrate lineages.
The structure of the L-type branch of the tree is also consistent with that reported for the relationship of the L-type vertebrate channel classes and the Mdla1 subunit cloned from housefly larva (Grabner et al., 1994); the Mdla1 subunit is on a branch arising at the most ancestral node of this clade. It has been reported previously that, when compared with known vertebrate sequences, the Dmca1D α1 subunit is most similar to class D subunits (Zheng et al., 1995). In this analysis, we find additionally that Mdla1 and Dmca1D are more similar to each other than to any of the L-type vertebrate α1 subunits. Also, Mdla1 and Dmca1D are less similar to the vertebrate C, D, and Sk subunits than the latter three are to each other and occupy a branch at the most ancestral node on the L-type side of the tree.
Dmca1A maps to chromosomal breakpoints that define a locus containing the interacting genetic variants cacophony, nightblind-A, and l(1)L13
Goralski (1985) molecularly mapped chromosomal breakpoints that were later found to damage or remove functions associated with thecac, nbA, and l(1)L13 variants (Kulkarni and Hall, 1987; Homyk and Pye, 1989). We have confirmed the original RFLP data (Goralski, 1985) for a subset of the relevant chromosome aberrations by Southern blotting that compared the banding patterns from inversion- and deletion-bearing flies with those of control flies devoid of chromosomal lesions near thecac/nbA/l(1)L13 locus (Fig. 7). We also have mapped portions of the Dmca1A cDNA to this genomic region (Peixoto, Smith, Hall, Hall, unpublished observations; also see legend to Fig.1). Combining the findings from the current Fig. 7 (Goralski, 1985; Peixoto, Smith, Hall, Hall, unpublished data), we infer that thel(1)L13-minus (hence cac- andnbA-minus) lesions are almost certainly within the Dmca1A locus. In particular (and as is summarized in Fig. 1), the deletionDf(1)HF368 has a breakpoint within 2 kb of transmembrane domain IIS5 and removes sequences 5′ to this; the inversionIn(1)A78 has a breakpoint within 4 kb of the putative transcription initiation site and the first transmembrane domain IS1;In(1)N66 has a breakpoint within 2 kb of the alternatively spliced exons in the I–II loop; and In(1)A101 has a breakpoint within 2 kb of the IIIss1–ss2 domain.
DISCUSSION
Dmca1A participates in generation of calcium channel diversity
The sequence and deduced structure of Dmca1A places it in the superfamily of voltage-sensitive calcium and sodium channels. The protein sequence is more similar to calcium channel α1 subunits than to sodium channel α subunits, both overall and within conserved transmembrane and ss1–ss2 motifs. Key glutamate residues in the ss2 motifs that have been implicated in ion selectivity are present in a pattern that is conserved perfectly in calcium channels and is required for ion selectivity. Near-perfect conservation of a motif implicated in phenylalkylamine binding implies that Dmca1A is sensitive to this class of calcium channel-specific pharmacological agents. In combination, this evidence clearly establishes that Dmca1A belongs to the family of calcium channel α1 subunits. Dmca1A maps to a different chromosome from the Dmca1D gene (Zheng et al., 1995) and encodes a structurally distinct α1 subunit. Thus, in Drosophila, as in vertebrates, one source of calcium channel diversity involves separate genes encoding distinct α1 subunits.
Zheng et al. (1995) presented evidence for variant transcripts from the Dmca1D locus. The diversity of cDNA sequences reported here demonstrates further that alternative splicing plays a role in generation of calcium channel diversity in invertebrates. The 116 nucleotide alternative exons in the I–II loop encode different amino acid sequences. These differences in a motif important for interaction with β subunits (cf. Pragnell et al., 1994) imply that isoforms encoded by these alternative exons might exhibit different affinities for β subunit interactions.
The pattern of variable nucleotide insertions at the extracellular IVS3–S4 loop is consistent with nonexclusive differential inclusion of three- and six-base exons, generating variants differing by zero, three, six, or nine bases. Differential inclusions of small exons (15 or 3 bases in length) have been reported for transcripts from the mammalian NCAM gene (Santoni et al., 1989; Reyes et al., 1991). In Dmca1A, the variable splice site is only five amino acids N terminal to the IVS4 transmembrane domain, which forms part of the voltage sensor in these channels. In addition to the splice variants in this region, a possible RNA editing site (see below) has been found between the variable splice region and the S4 voltage sensor. Thus, in this small region there is the potential for significant transcript variability. This raises the intriguing possibility that these differences might play a role in modulating the voltage dependence of calcium channels containing Dmca1A.
Direct sequence analysis of PCR products derived from RNA from whole flies indicates that each of the alternatively spliced transcripts described here is expressed at detectable levels (L. A. Smith and J. C. hall, unpublished observations). Of the two forms at the I–II loop, the c31-encoded exon, containing a perfectly conserved β subunit binding motif, is predominant. At the IVS3–S4 variable region, the shortest form is predominant. In Drosophila, optional or differential splicing of exons of the parasodium channel α subunit occurs at six known sites (Loughney et al., 1989; Thackeray and Ganetzky, 1994; O’Dowd et al., 1995), whereas theShaker potassium channel gene generates multiple developmentally regulated classes of transcripts (Kamb et al., 1988;Pongs et al., 1988; Schwartz et al., 1988; Mottes and Iverson, 1995). Although the known splicing-generated transcript diversity of Dmca1A is not as extensive as for these well characterized Drosophilaion channels, further analysis of the new calcium channel gene and its products may reveal additional instances of alternative splicing.
The Dmca1A mRNA seems to be post-transcriptionally modified (Table 2). This is the first evidence for RNA editing of a neurobiologically relevant gene in Drosophila (and in any invertebrate, to our knowledge). In vertebrates, adenosine deamination is often dependent on an editing site-complementary sequence in an adjacent intron (Higuchi et al., 1993; Lomeli et al., 1994). For five of the seven potential editing sites inferred for Dmca1A, preliminary analysis of genomic sequence has detected flanking intronic complementary sequences, ranging in length from seven to nine nucleotides and, in each case, perfectly centered on the relevant adenosine residue (Peixoto, Smith, Hall, and Hall, unpublished observations). Six of the seven adenosine-to-guanosine differences result in changes of codon identity, suggesting that RNA editing may contribute to functional diversity of the Dmca1A protein. The apparent preferential localization near conserved transmembrane domains implies that these amino acid changes might be relevant to regulated functions of the Dmca1A calcium channels. The apparent lack of editing in the embryonic cDNA cSK53 suggests that editing may be stage- or tissue-specific.
Northern blot analysis of the first cloned Drosophilacalcium channel α1 subunit detected three size classes of mRNA in heads: a major band at 9.5 kb and two minor bands at 10.2 and 12.5 kb (Zheng et al., 1995). Because the RNA encoding this subunit undergoes extensive alternative splicing, it was not possible to determine whether the minor bands were minor splice forms or represented distinct members of a calcium channel α1 subunit gene family. The results reported here suggest that the 10.2 kb band previously detected with a Dmca1D probe (Zheng et al., 1995) might encode the Dmca1A subunit. Dmca1A is expressed throughout the embryonic nervous system; the relatively head-enriched expression of the 10.2 kb message (seen with the Dmca1D probe) implies predominantly neural expression in the adult. The slight discrepancy in size (10.2 vs 10.5 kb measured in this study) could be attributable to the difficulty in accurately estimating the size of high-molecular-weight RNAs.
Comparison of pharmacological motifs of Dmca1A and Dmca1D
Conservation of a proposed phenylalkylamine binding site near the 3′ end of the IVS6 transmembrane domain suggests that Dmca1A may bind phenylalkylamines. Relatively poor conservation of amino acids in the proposed dihydropyridine binding sites suggests that this α1 subunit does not bind dihydropyridines, consistent with the phylogenetic similarity to the non-L-type channels. Because both Dmca1A and Dmca1D α1 subunits similarly are conserved in the proposed phenylalkylamine binding region, both may contribute to the phenylalkylamine binding activity found in Drosophilaextracts (Pauron et al., 1987; Greenberg et al., 1989). On the basis of sequence analysis, it was suggested initially that Dmca1D might encode the predominant dihydropyridine-insensitive calcium channel inDrosophila heads (Zheng et al., 1995). However, recent electrophysiological studies show that Dmca1D encodes a dihydropyridine-sensitive current in larval muscle (D. Ren, H. Xu, G. Feng, M. Chopra, L. M. Hall, unpublished observations) consistent with its structural similarity to L-type channels. If Dmca1A is expressed in muscles, it is a candidate for encoding the amiloride-sensitive current (Gielow et al., 1995).
Dmca1A may be encoded by a gene defined by behavioral, physiological, and lethal mutations
The cacophony, nightblind-A, andlethal(1)L13 mutations all map by deletion analysis to the same genetic interval (Kulkarni and Hall, 1987). Breakpoints of certain physically lesioned inversion chromosomes that fail to complementcacophony, nightblind-A, and lethal(1)L13mutations not only map genetically to the sites of these mutations (see introductory remarks) but now also have been mapped molecularly to the Dmca1A-encoding locus (Figs. 1, 7). Although chromosomal breakpoints can induce spreading effects that cause perturbation of neighboring genes not directly disrupted by the genetic lesion, the fact that all of these breakpoints disrupt the Dmca1A locus (Fig. 1) strongly suggests involvement of this calcium channel α1 subunit gene in the generation of the physiological, behavioral, and lethal phenotypes associated with the cac, nbA, and l(1)L13mutants. It follows that cac, nbA, and l(1)L13are likely to be Dmca1A mutants and that these genetic variants likely define a single gene.
Mutant alleles of l(1)L13 cause late embryonic lethality. The first expression peak of the Dmca1A transcript begins in late embryogenesis, consistent with a requirement for Dmca1A function at this developmental stage. The diversity of the Dmca1A transcript suggests that the complicated complementation interations of cac, nbA, and l(1)L13 mutations (Kulkarni and Hall, 1987) could be attributable to isoform-specific lesions. In adults, thecac mutation causes defects in the male courtship song (Kulkarni and Hall, 1987), and nbA mutants exhibit increased light thresholds for optomotor and phototactic behaviors (Heisenberg and Götz, 1975) as well as defects in the shape and amplitude of the electroretinogram (Homyk and Pye, 1989). The particular song defect exhibited by cac males—larger than normal numbers of cycles within a given “burst” of tone—could be rationalized in terms of modified calcium channel function [cf. Hille (1992), Chapter 5]. The cellular etiology of the abnormal singing behavior is difficult to speculate on, because it could involve defects in neural or muscular physiology (or even anatomy), yet it is difficult to imagine a non-neural etiology for the abnormal ERG in nbA mutants. Taken together, this analysis implies involvement of the Dmca1A voltage-dependent calcium channel in visual transduction and may suggest involvement of calcium-dependent beating or bursting cells in the generation of the rhythmic wingbeat behavior underlying the generation of courtship song. Further experiments on the molecular etiologies of these three types of mutants may reveal how variation within the Dmca1A gene can cause either severe and rather global neurobiological problems or more subtle ones involving these discrete elements of behavior and physiology.
Footnotes
This work was supported by National Institutes of Health (NIH) Grant GM-21473 to J.C.H. and by NIH Merit Award HL-39369 and NIH Javits Award NS-16204 to L.M.H. We thank Thomas J. Goralski for supplying the “starter” genomic clones and Barry Ganetzky, Stephen F. Goodwin, and Christopher Miller for comments on this manuscript.
Correspondence should be addressed to Dr. Jeffrey C. Hall, Department of Biology, 235 Bassine Building, Brandels University, 415 South Street, Waltham, MA 02254-9110.
Dr. Neumann’s present address: Bolt, Beranek, and Newman, 70 Fawcett Street, Cambridge, MA 02138.
REFERENCES
- 1.Ahlijanian MK, Westenbroek RE, Catterall WA. Subunit structure and localization of dihydropyridine-sensitive calcium channels in mammalian brain, spinal cord, and retina. Neuron. 1990;4:819–832. doi: 10.1016/0896-6273(90)90135-3. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner M, Thompson JN., Jr . The laboratory culture of Drosophila. In: Ashburner M, Wright TFR, editors. The genetics and biology of Drosophila, Vol 2A. Academic; New York: 1978. pp. 1–109. [Google Scholar]
- 3.Babitch J. Channel hands. Nature. 1990;346:321–322. doi: 10.1038/346321b0. [DOI] [PubMed] [Google Scholar]
- 4.Bangalore R, Baindur N, Rutledge A, Triggle DJ, Kass RS. L-type calcium channels: asymetrical intramembrane binding domain revealed by variable length, permanently charged 1,4-dihydropyridines. Mol Pharmacol. 1994;46:660–666. [PubMed] [Google Scholar]
- 5.Bass BL. RNA editing: new uses for old players in the RNA world. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor Laboratory; Plainview, NY: 1993. pp. 383–418. [Google Scholar]
- 6.Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- 7.Campbell KP, Leung AT, Sharp AH. The biochemistry and molecular biology of the dihydropyridine-sensitive calcium channel. Trends Neurosci. 1988;11:425–430. doi: 10.1016/0166-2236(88)90193-2. [DOI] [PubMed] [Google Scholar]
- 8.Catterall WA, Seagar MJ, Takahashi M. Molecular properties of dihydropyridine-sensitive calcium channels in skeletal muscle. J Biol Chem. 1988;263:3535–3538. [PubMed] [Google Scholar]
- 9.Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhari N. A single nucleotide deletion in the skeletal muscle-specific calcium channel transcript of muscular dysgenesis (mdg) mice. J Biol Chem. 1992;267:25636–25639. [PubMed] [Google Scholar]
- 11.Chiang PW, Martin T, Osemlak-Hanzlik M, Karnit DM. Rapid PCR-based method to directionally pull out longer cDNA fragments from cDNA libraries. Biotechniques. 1994;18:37–40. [PubMed] [Google Scholar]
- 12.deLeon M, Wang Y, Jones L, Perez-Reyes E, Wei X, Soong TW, Snutch TP, Yue DT. Essential Ca2+-binding motif for Ca2+-sensitive inactivation of L-type Ca2+ channels. Science. 1995;270:1502–1506. doi: 10.1126/science.270.5241.1502. [DOI] [PubMed] [Google Scholar]
- 13.DiAntonio A, Burgess RW, Chin AC, Deitcher DL, Scheller RH, Schwarz TL. Identification and characterization of Drosophila genes for synaptic vesicle proteins. J Neurosci. 1993;13:4924–4935. doi: 10.1523/JNEUROSCI.13-11-04924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubel SJ, Starr TV, Hell J, Ahlijanian MK, Enyeart JJ, Catterall WA, Snutch TP. Molecular cloning of the α1 subunit of an omega-conotoxin-sensitive calcium channel. Proc Natl Acad Sci USA. 1992;89:5058–5062. doi: 10.1073/pnas.89.11.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellinor PT, Yang J, Sather WA, Zhang JF, Tsien RW. Ca2+ channel selectivity at a single locus for high-affinity Ca2+ interactions. Neuron. 1995;15:1121–1132. doi: 10.1016/0896-6273(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. PHYLIP-phylogeny inference package (version 3.2). Cladistics. 1989;5:164–166. [Google Scholar]
- 17.Genetics Computer Group (1991) Program manual for the GCG package, Version 7.
- 18.Gielow ML, Gu GG, Singh S. Resolution and pharmacological analysis of the voltage-dependent calcium channels of Drosophila larval muscles. J Neurosci. 1995;15:6085–6093. doi: 10.1523/JNEUROSCI.15-09-06085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goralski TJ (1985) A molecular analysis of the female sterile locus gastrulation defective (gd) ofDrosophila melanogaster. PhD thesis, Indiana University, Bloomington, IN.
- 20.Grabner M, Bachmann A, Rosenthal F, Striessnig J, Schultz C, Tautz D, Glossman H. Insect calcium channels. Molecular cloning of an α1-subunit from housefly (Musca domestica) muscle. FEBS Lett. 1994;339:189–194. doi: 10.1016/0014-5793(94)80413-3. [DOI] [PubMed] [Google Scholar]
- 21.Grabner M, Wang Z, Hering S, Striessnig J, Glossman H. Transfer of 1,4-dihydropyridine sensitivity from L-type to class A (BI) calcium channels. Neuron. 1996;16:207–218. doi: 10.1016/s0896-6273(00)80037-9. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg RM, Streissnig J, Koza A, Devay P, Glossman H, Hall LM. Native and detergent-solubilized membrane extracts from Drosophila heads contain binding sites for the phenylalkylamine calcium channel blockers. Insect Biochem. 1989;19:309–322. [Google Scholar]
- 23.Heinemann SH, Terlau H, Stuhmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–443. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 24.Heisenberg M, Götz KG. The use of mutations for the partial degradation of vision in Drosophila melanogaster. J Comp Physiol [A] 1975;98:217–241. [Google Scholar]
- 25.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 26.Hille B. Ionic channels of excitable membranes, 2nd Ed. Sinauer; Sunderland, MA: 1992. [Google Scholar]
- 27.Hofmann F, Biel M, Flockerzi V. Molecular basis for Ca2+ channel diversity. Annu Rev Neurosci. 1994;17:399–418. doi: 10.1146/annurev.ne.17.030194.002151. [DOI] [PubMed] [Google Scholar]
- 28.Homyk T, Jr, Pye Q. Some mutations affecting neural or muscular tissues alter the physiological components of the electroretinogram in Drosophila. J Neurogenet. 1989;5:37–48. doi: 10.3109/01677068909167263. [DOI] [PubMed] [Google Scholar]
- 29.Hubbard SC, Ivatt RP. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- 30.Itoh N, Slemmon JR, Kawke DH, Williamson R, Morita E, Itakura K, Roberts E, Shively JE, Crawford GD, Salvaterra PM. Cloning of Drosophila choline acetyltransferase cDNA. Proc Natl Acad Sci USA. 1986;83:4081–4085. doi: 10.1073/pnas.83.11.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamb A, Tseng-Crank J, Tanouye MA. Multiple products of the Drosophila Shaker gene may contribute to potassium channel diversity. Neuron. 1988;1:421–430. doi: 10.1016/0896-6273(88)90192-4. [DOI] [PubMed] [Google Scholar]
- 32.Kim MS, Morii T, Sun LX, Imoto K, Mori Y. Structural determinants of ion selectivity in brain calcium channels. FEBS Lett. 1993;318:145–148. doi: 10.1016/0014-5793(93)80009-j. [DOI] [PubMed] [Google Scholar]
- 33.Krebs EG, Beavo JA. Phosphorylation-dephosphorylation of enzymes. Annu Rev Biochem. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- 34.Kulkarni SJ, Hall JC. Behavioral and cytogenetic analysis of the cacophony courtship song mutant and interacting genetic variants in Drosophila melanogaster. Genetics. 1987;115:461–475. doi: 10.1093/genetics/115.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacerda AE, Kim HS, Ruth P, Perez-Reyes E, Flockerzi V, Hofmann F, Birnbaumer L, Brown AM. Normalization of current kinetics by interaction between the alpha 1 and beta subunits of the skeletal muscle dihydropyridine Ca2+ channel. Nature. 1991;352:527–530. doi: 10.1038/352527a0. [DOI] [PubMed] [Google Scholar]
- 36.Leung HT, Byerly L. Characterization of single calcium channels in Drosophila nerve and muscle cells. J Neurosci. 1991;11:3047–3059. doi: 10.1523/JNEUROSCI.11-10-03047.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung HT, Branton WD, Phillips HS, Jan L, Byerly L. Spider toxins selectively block calcium currents in Drosophila. Neuron. 1989;3:767–772. doi: 10.1016/0896-6273(89)90245-6. [DOI] [PubMed] [Google Scholar]
- 38.Leveque C, ElFar O, Martin-Moutot N, Sato K, Kato R, Takahashi M, Seagar M. Purification of the N-type calcium channel associated with syntaxin and synaptotagmin. A complex implicated in synaptic vesicle exocytosis. J Biol Chem. 1994;269:6306–6312. [PubMed] [Google Scholar]
- 39.Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. Academic; San Diego: 1992. [Google Scholar]
- 40.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 41.Loughney K, Kreber R, Ganetzky B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Genetics. 1989;134:847–858. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- 42.McEnery MW, Snowman AM, Sharp AH, Adams ME, Snyder SH. Purified omega-conotoxin GVIA receptor of rat brain resembles a dihydropyridine-sensitive L-type calcium channel. Proc Natl Acad Sci USA. 1991;88:11095–11099. doi: 10.1073/pnas.88.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikala G, Bahinski A, Yatani A, Tang S, Schwartz A. Differential contribution by conserved glutamate residues to an ion-selectivity site in the L-type Ca2+ channel pore. FEBS Lett. 1993;335:265–269. doi: 10.1016/0014-5793(93)80743-e. [DOI] [PubMed] [Google Scholar]
- 44.Mottes JR, Iverson LE. Tissue-specific alternative splicing of hybrid Shaker/lacZ genes correlates with kinetic differences in Shaker k+ currents in vivo. Neuron. 1995;14:613–623. doi: 10.1016/0896-6273(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 45.Mount SM, Burks C, Hertz G, Stormo GD, White O, Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakayama H, Taki M, Striessnig J, Glossman H, Catterall WA, Kanaoka Y. Identification of 1,4-dihydropyridine binding regions within the α1 subunit of skeletal muscle Ca2+ channels by photoaffinity labeling with diazipine. Proc Natl Acad Sci USA. 1991;88:9203–9207. doi: 10.1073/pnas.88.20.9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neely A, Wei X, Olcese R, Birmbaumer L, Stefani E. Potentiation by the beta subunit of the ratio of the ionic current to the charge movement in the cardiac calcium channel. Science. 1993;262:575–578. doi: 10.1126/science.8211185. [DOI] [PubMed] [Google Scholar]
- 48.Noda M, Ikeda T, Kayano T, Suzuki H, Takeshima H, Kurasaki M, Takahashi H, Numa S. Existence of distinct sodium channel messenger RNAs in rat brain. Nature. 1986;320:188–192. doi: 10.1038/320188a0. [DOI] [PubMed] [Google Scholar]
- 49.O’Connell PO, Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Dowd DK, Gee JR, Smith MA. Sodium current density correlates with expression of specific alternatively spliced sodium channel mRNAs in single neurons. J Neurosci. 1995;15:4005–4012. doi: 10.1523/JNEUROSCI.15-05-04005.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pauron D, Qar J, Barhanin J, Fournier D, Cuany A, Pralavorio M, Berge JB, Lazdunski M. Identification and affinity labeling of very high affinity binding sites for the phenylalkylamine series of Ca2+ channel blockers in the Drosophila nervous system. Biochemistry. 1987;26:6311–6315. doi: 10.1021/bi00394a003. [DOI] [PubMed] [Google Scholar]
- 52.Pelzer S, Barhanin J, Pauron D, Trautwein W, Lazdunski M, Pelzer D. Diversity and novel pharmacological properties of Ca2+ channels in Drosophila head membranes. EMBO J. 1989;8:2365–2371. doi: 10.1002/j.1460-2075.1989.tb08365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pongs O, Kecskemethy N, Muller R, Krah-Jengens I, Baumann A, Kiltz HH, Canal I, Llamazares S, Ferrus A. Shaker encodes a family of putative potassium channel proteins in the nervous system of Drosophila. EMBO J. 1988;7:1087–1096. doi: 10.1002/j.1460-2075.1988.tb02917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pragnell M, De Waard M, Mori Y, Tanabe T, Snutch TP, Campbell KP. Calcium channel β-subunit binds to a conserved motif in the I–II cytoplasmic linker of the α1-subunit. Nature. 1994;368:67–70. doi: 10.1038/368067a0. [DOI] [PubMed] [Google Scholar]
- 55.Reyes AA, Small SJ, Akeson R. At least 27 alternatively spliced forms of the neural cell adhesion molecule mRNA are expressed during rat heart development. Mol Cell Biol. 1991;11:1654–1661. doi: 10.1128/mcb.11.3.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd Ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 57.Santoni MJ, Barthels D, Vopper G, Boned A, Goridis C, Wille W. Differential exon usage involving an unusual splicing mechanism generates at least eight types of NCAM cDNA in mouse brain. EMBO J. 1989;8:385–392. doi: 10.1002/j.1460-2075.1989.tb03389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schafer WR, Kenyon CJ. A calcium channel homologue required for adaptation to dopamine and serotonin in Caenorhabditis elegans. Nature. 1995;375:73–78. doi: 10.1038/375073a0. [DOI] [PubMed] [Google Scholar]
- 59.Schwartz T, Tempel B, Papazian D, Jan YN, Jan LY. Multiple potassium channel components are produced by alternative splicing at the Shaker locus in Drosophila. Nature. 1988;331:137–142. doi: 10.1038/331137a0. [DOI] [PubMed] [Google Scholar]
- 60.Snutch TP, Tomlinson WJ, Leonard JP, Gilbert MM. Distinct calcium channels are generated by alternative splicing and are differentially expressed in the mammalian CNS. Neuron. 1991;7:45–57. doi: 10.1016/0896-6273(91)90073-9. [DOI] [PubMed] [Google Scholar]
- 61.Soong TW, Stea A, Hodson CD, Dubel SJ, Vincent SR, Snutch TP. Structure and functional expression of a member of the low voltage-activated calcium channel family. Science. 1993;260:1133–1136. doi: 10.1126/science.8388125. [DOI] [PubMed] [Google Scholar]
- 62.Starr TV, Prystay W, Snutch TP. Primary structure of a calcium channel that is highly expressed in the rat cerebellum. Proc Natl Acad Sci USA. 1991;88:5621–5625. doi: 10.1073/pnas.88.13.5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stea A, Soong TW, Snutch TP. Voltage-gated calcium channels. In: North RA, editor. Ligand- and voltage-gated ion channels. CRC; Boca Raton, FL: 1995. pp. 114–151. [Google Scholar]
- 64.Striessnig J, Glossman H, Catterall WA. Identification of a phenylalkylamine binding region within the α1 subunit of skeletal muscle Ca2+ channels. Proc Natl Acad Sci USA. 1990;87:9108–9112. doi: 10.1073/pnas.87.23.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Striessnig J, Murphy BJ, Catterall WA. Dihydropyridine receptor of L-type Ca2+ channels: identification of binding domains for [3H](+)-PN200–110 and [3H]-azidopine within the α1 subunit. Proc Natl Acad Sci USA. 1991;88:10769–10773. doi: 10.1073/pnas.88.23.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanabe T, Takeshima H, Mikami A, Flockerzi V, Takahashi H, Kangawa K, Kojima M, Matsuo H, Hirose T, Numa S. Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature. 1987;328:313–318. doi: 10.1038/328313a0. [DOI] [PubMed] [Google Scholar]
- 67.Tang S, Mikala G, Babinski A, Yatani A, Varadi G, Schwartz A. Molecular localization of ion selectivity sites within the pore of a human L-type cardiac calcium channel. J Biol Chem. 1993a;268:13026–13029. [PubMed] [Google Scholar]
- 68.Tang S, Yatani A, Bahinski A, Mori Y, Schwartz A. Molecular localization of regions in the L-type calcium channel critical for dihydropyridine action. Neuron. 1993b;11:1013–1021. doi: 10.1016/0896-6273(93)90215-d. [DOI] [PubMed] [Google Scholar]
- 69.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 70.Thackeray JR, Ganetzky B. Developmentally regulated alternative splicing generates a complex array of Drosophila para sodium channel isoforms. J Neurosci. 1994;14:2569–2578. doi: 10.1523/JNEUROSCI.14-05-02569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tufty RM, Kretsinger RH. Troponin and parvalbumin calcium binding regions predicted in myosin light chain and T4 lysozyme. Science. 1975;187:167–169. doi: 10.1126/science.1111094. [DOI] [PubMed] [Google Scholar]
- 73.Varadi G, Lory P, Schultz D, Varadi M, Schwartz A. Acceleration of activation and inactivation by the beta subunit of the skeletal muscle calcium channel. Nature. 1991;352:159–162. doi: 10.1038/352159a0. [DOI] [PubMed] [Google Scholar]
- 74.Williams ME, Feldman DH, McCue AF, Brenner R, Velicelebi G, Ellis SB, Harpold MM. Structure and functional expression of α1, α2, and β subunits of a novel human neuronal calcium channel subtype. Neuron. 1992;8:71–84. doi: 10.1016/0896-6273(92)90109-q. [DOI] [PubMed] [Google Scholar]
- 75.Witcher DR, De Waard M, Sakamoto J, Franzini-Armstrong C, Pragnell M, Kahl SD, Campbell KP. Subunit identification and reconstitution of the N-type Ca2+ channel complex purified from brain. Science. 1993;261:486–489. doi: 10.1126/science.8392754. [DOI] [PubMed] [Google Scholar]
- 76.Woodgett JR, Gould KC, Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986;161:177–84. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- 77.Yang J, Ellinor PT, Sather WA, Zhang JF, Tsien RW. Molecular determinants of Ca2+ selectivity and ion permeation in L-type Ca2+ channels. Nature. 1993;366:158–161. doi: 10.1038/366158a0. [DOI] [PubMed] [Google Scholar]
- 78.Zheng W, Feng G, Ren D, Eberl DF, Hannan F, Dubald M, Hall LM. Cloning and characterization of a calcium channel α1 subunit from Drosophila melanogaster with similarity to the rat brain type D isoform. J Neurosci. 1995;15:1132–1143. doi: 10.1523/JNEUROSCI.15-02-01132.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]