Abstract
The processes and mechanisms of virus infection fate decisions that are the result of a dynamic virus-immune system interaction with either an efficient effector response and virus elimination or an alleviated immune response and chronic infection are poorly understood. Here, we characterized the host response to acute and chronic lymphocytic choriomeningitis virus (LCMV) infections by gene coexpression network analysis of time-resolved splenic transcriptomes. First, we found an early attenuation of inflammatory monocyte/macrophage prior to the onset of T cell exhaustion, and second, a critical role of the XCL1-XCR1 communication axis during the functional adaptation of the T cell response to the chronic infection state. These findings not only reveal an important feedback mechanism that couples T cell exhaustion with the maintenance of a lower level of effector T cell response but also suggest therapy options to better control virus levels during the chronic infection phase.
Overwhelming infections with noncytopathic viruses can lead to chronic infections. The fate of a virus infection can be fundamentally categorized as acute or chronic according to its temporal relationship with the host organism (Virgin et al. 2009). In humans, acute infections are usually resolved within a few weeks. In contrast, chronic infections are not resolved and, instead, develop when innate and adaptive immune responses are not sufficient to eliminate the invading virus during the primary infection phase. Examples for this latter case are infections with the Human Immunodeficiency Virus (HIV) or the Hepatitis B and C viruses (HBV, HCV) that can establish persistence in their hosts with different probabilities and pathogenic consequences (Rehermann and Nascimbeni 2005; Feinberg and Ahmed 2012).
A hallmark of an overwhelming infection is the down-regulation of immune effector mechanisms to avoid immunopathology. Indeed, the simultaneous presence of a widespread virus infection and strong cytotoxic effector cell responses can induce massive cell and tissue destruction and may directly threaten the life of the infected host (Rouse and Sehrawat 2010). This threat is sensed by incompletely understood mechanisms, and several suppressive immune components are activated that adapt the host response to the viral threat. Among these are T cell exhaustion by deletion (Moskophidis et al. 1993) and functional impairment (Zajac et al. 1998; Wherry et al. 2003; Barber et al. 2006), the generation of monocyte-derived suppressor cells (MDSCs) (Cai et al. 2013; Norris et al. 2013) and regulatory cell subsets (Wilson et al. 2012). They exert their function via distinct processes including interaction with inhibitory ligands, the production of soluble immunosuppressive factors like IL10 and Indoleamine 2,3-dioxygenase, and the delivery of suppressive signals by cell–cell contacts to conventional T cells and to antigen-presenting cells, thus influencing effector functions directly or indirectly (Barber et al. 2006; Day et al. 2006; Doering et al. 2012; Ng et al. 2013).
Despite the various suppressive mechanisms induced during a chronic virus infection, the effector T cell shutdown is only partial and some T cell functionality remains that restrains the expansion of a persisting virus (Schmitz et al. 1999; He et al. 2016; Im et al. 2016; Leong et al. 2016). The processes however that mediate the transition toward a lower level response are not well understood. We therefore sought to analyze on a systems level infection-fate-specific gene signatures and adaptive processes of the host to an overwhelming virological threat. The infection of mice with lymphocytic choriomeningitis virus (LCMV) was used as a model system. It enabled reproducibly establishing acute and chronic infections, and exhibited several fundamental features within the virus-immune system's crosstalk that hold true for human infections as well (Wilson and Brooks 2010).
Results
Spleen gene coexpression networks in acute and chronic viral infections
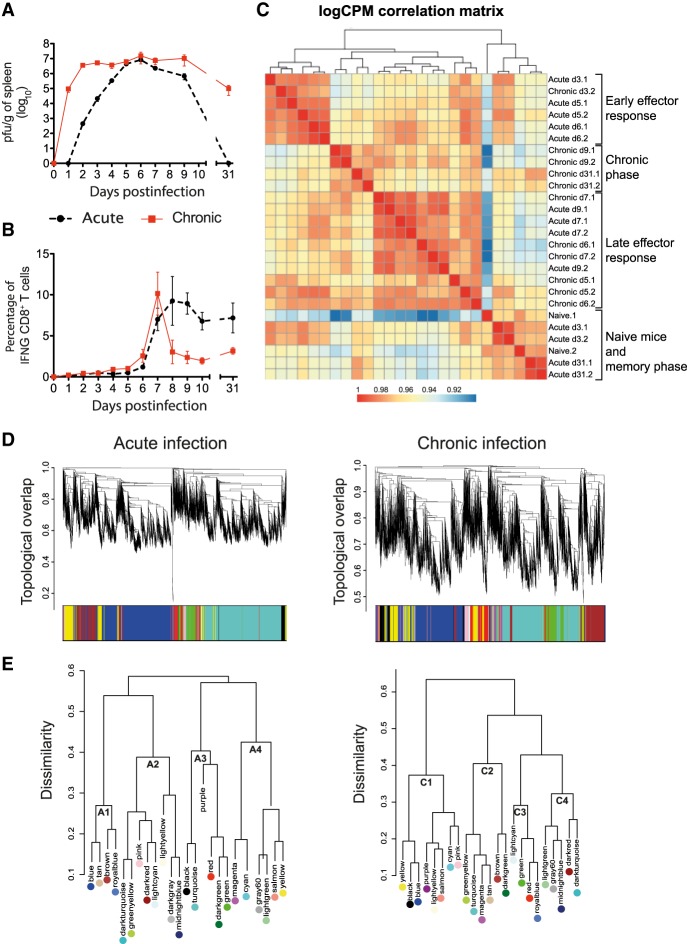
The development of an acute or a chronic virus infection is the result of a coordinated response of the immune system toward the invading microbe. Although several of the individual components that contribute to this fate decision have been described, a systemic global view is still lacking. Thus, we characterized transcriptome changes at the level of the spleen during the course of viral infections with different outcomes. C57BL/6 mice were infected with a low-dose (2×102 plaque-forming units; acute infection) or a high-dose (2×106 plaque-forming units; chronic infection) of LCMV strain Docile (LCMVDoc). Virus titers, virus-specific CD4+ and CD8+ T cell responses, and spleen-derived total messenger RNA (mRNA) transcriptomes were determined longitudinally by titration, intracellular cytokine staining (ICS), and RNA-seq, respectively. Because of the different inoculum size, virus titers in spleen increased later in acute than in chronic infection, however, similar levels were observed at d5 postinfection (PI) (Fig. 1A). As previously described (Moskophidis et al. 1993), low-dose infection resulted in virus clearance, whereas high-dose infection led to virus persistence shown as high viral titers in spleen at d31 PI. The interferon gamma (IFNG)-producing CD8+ and CD4+ T cells expanded at d6-d7, and their percentages remained high at d31 when virus was undetectable after low-dose infection (Fig. 1B; Supplemental Fig. S1C). In high-dose infected mice, IFNG-producing CD8+ T cells also increased at d6-d7 PI, however, their number dropped at d7-d9, which is indicative of CD8+ T cell exhaustion. This latter conclusion was verified by observing a concomitant drop of TNF-production as well as an increase of PDCD1 (also known as PD-1) and HAVCR2 (also known as TIM-3) expression levels that were maintained at day 15 PI (Supplemental Fig. S1A,B; Wherry et al. 2007). Altogether this strengthens the previous suggestion that accumulative virus loads rather than the virus load per se determine the observed differences in the immune responses and infection fates (Bocharov 1998; Côrtes et al. 2018).
Figure 1.
Kinetics of virus expansion, CD8+ T cell response, and transcriptome analysis in acute and chronic LCMV infections. Virus titers (A) and percentages of GP33-specific IFNG-producing CD8+ T cells (B) in spleen. The mean ± SEM is shown. (C) Correlation matrix heatmap of gene expression across acute and chronic infection samples. The color scale indicates the degree of correlation. (D) Hierarchical clustering dendrogram for all differentially expressed genes (lines) obtained by WGCNA. The branches correspond to modules of highly coexpressed groups of genes. Colors below the dendrogram indicate the module to which each gene was assigned. (E) Hierarchical clustering dendrogram of dissimilarity based on module eigengenes.
To characterize changes of transcriptional profiles from spleens during acute or chronic infection, we isolated total mRNAs from naive mice (d0) and from mice at d3, d5, d6, d7, d9, and d31 after low-dose or high-dose of LCMVDoc infection, and determined time-resolved transcriptomes by RNA-seq. Hierarchical clustering across all samples revealed four main groups corresponding to different infection phases (Fig. 1C). The first group, naive mice and mice in memory phase, is represented by samples from acute infected mice at days 3 and 31, and uninfected mice (d0), indicating that spleen transcriptome profiles return to that of naive mice once the infection is resolved. Early effector responses (group 2) and late effector responses (group 3) are represented by samples from d5-d6 of acute plus d3 of chronic infection and d7-d9 of acute plus d5-d7 of chronic infection, respectively. This indicates that at a tissue level, central host responses are similar in acute and chronic infections albeit, owing to the different inoculum size, they appear at different time points. Finally, d9 and d31 samples from chronic infection, that is once CD8+ T cell exhaustion is established, cluster in a separate group 4 representing a specific chronic infection phase. Thus, distinct virus infection phases exhibit separated transcriptome signatures in the lymphoid tissue.
Taking noninfected mice as a reference point, the analysis of differentially expressed genes after acute and chronic infection showed a similar number of up-regulated and down-regulated genes from d5 to d9 (Supplemental Fig. S1D). Appreciable differences between both types of infection were found only at early (d3) and late (d31) time points when acute infected mice showed fewer differentially expressed genes. This reflects the different host response kinetics between the two types of infection at early time points and the continuous deviation of the chronic infection state from the basal level of transcription of naive mice. Gene Ontology (GO) analysis of all genes differentially expressed at any time point postinfection showed identical enriched terms in acute and chronic infections representing general biological processes such as “cell cycle,” “immune response,” and “programmed cell death.”
To explore the dynamics of the spleen transcriptomes during both infection courses, we applied weighted gene correlation network analysis (WGCNA) and constructed two signed coexpression networks (Zhang and Horvath 2005). Topological overlap was used to measure the connection strength between genes based on shared network neighbors, identifying genes with similar patterns of connections, and subsequently defining modules of highly coexpressed genes (Fig. 1D). Finally, the eigengene was extracted for each module to represent the gene expression profile. Twenty-three modules were identified in both acute and chronic infections (Supplemental Fig. S2). They were color-coded for identification purposes. Of note is that the same color in modules from acute and chronic networks does not imply any similarity between them. The number of genes per module ranged from 15 to 4952 (Supplemental Fig. S5).
To obtain a global view on the similarity of the module expression patterns, we generated a hierarchical clustering dendrogram based on the module's eigengenes and defined four clusters for each infection outcome (A1 to A4, acute infection; C1 to C4, chronic infection) (Fig. 1E; Supplemental Fig. S2). Clusters A1, A4, and C2 include modules with up-regulated expression profiles at d3-d7 PI reflecting the complexity of the host effector response, whereas the modules from clusters A3 and C1 show an early down-regulation of expression. Clusters A2 and C4 contain modules with bimodal expression profiles having the first peak at an early time point postinfection when the host response is triggered and a second peak at d9 when the infection outcome is already determined. Finally, the expression pattern of the modules from Cluster C3 was characteristic of chronic infection showing an initial down-regulation but subsequent gradual up-regulation until d31.
Cellular and humoral adaptive responses are disassociated in chronic infection
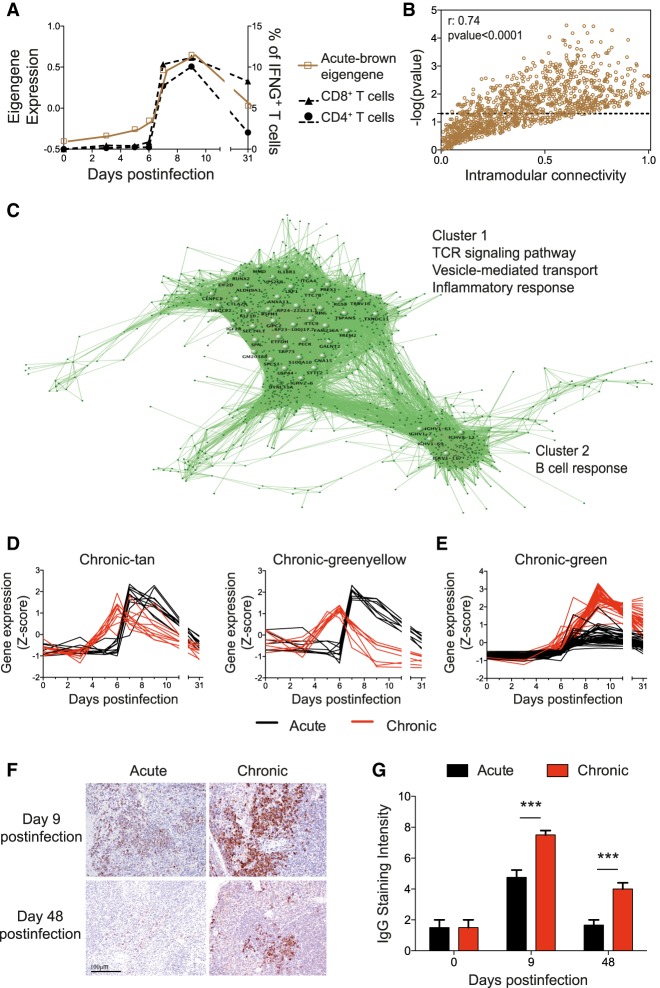
CD8+ T cells play a major role in the control of an LCMV infection as well as in exhaustion during chronic infections (Oldstone 2006). We therefore tested whether the behavior of virus-specific T cells was mirrored by a gene expression pattern of similar kinetics. For this we correlated the module eigengenes with the kinetics of IFNG-producing GP33-specific CD8+ T cells (Supplemental Fig. S3A). The acute-brown module significantly correlated with the virus-specific CD8+ and CD4+ T cell responses of acute infection (Fig. 2A). To visualize this correlation at the level of the individual genes, we compared the intramodular connectivity (KIM) of each gene of this module with its correlation to the T cell response. A correlation was observed indicating a link between the hub genes (high KIM values) to the T cell response (Fig. 2B). In contrast, no significant correlation between chronic-module eigengenes and the virus-specific CD8+ T cell response of chronic infection was observed (Supplemental Fig. S3A). This indicates that IFNG production is a good marker to identify pathways involved in the T cell response in acute infection but not sufficient to reveal the underlying gene signature of exhaustion.
Figure 2.
Genes related to T and B cell responses from the acute-brown module are disassociated in chronic infection. (A) Acute-brown module eigengene and kinetics of GP33-specific CD8+ and GP61-specific CD4+ T cells. (B) Intramodular connectivity versus GP33-specific CD8+ T cell kinetics correlation significance for acute-brown module genes. (C) Visualization of the acute-brown module genes. Large nodes represent hub genes with high connection densities (TOM > 0.5). (D,E) Expression kinetics of acute-brown module genes related to T cell activation (D) and hub genes related to the B cell response (E) in acute and chronic infections. (F) Representative images of IgG immunohistochemical staining of spleens. Magnification bar: 100 µm. (G) Semiquantification of IgG-positive cells in spleens (score 0–10). Data shown are mean ± SEM of four mice per group and time point. (***) P ≤ 0.001 unpaired two-tailed t-test.
Hub genes from coexpression modules are defined by a high KIM value, or alternatively by a high topological overlap, and are predicted to be central for the biological processes they represent (Jeong et al. 2001; Doering et al. 2012). Thus, we visualized the hub genes of the acute-brown module as a network based on their topological overlap values (Fig. 2C). Two clusters were observed that are enriched in the GO terms TCR signaling pathway, vesicle-mediated transport, inflammatory response, and B cell response. Cluster 1 contains key genes in T cell activation, and Cluster 2 contains genes from the B cell response. To analyze how these hub genes in acute infection behave during chronic infection, we compared their intramodular connectivities in both infection outcomes (Supplemental Fig. S3B). Genes from Cluster 1 related to T cell activation were found in two different chronic modules, chronic-tan and chronic-greenyellow (Supplemental Fig. S3B). However, only genes in chronic-tan retained a KIM > 0.6 and therefore represented control points of T cell activation in chronic infection. These genes were up-regulated at day 6 and returned to basal levels at day 31, thus having similar kinetics as in acute infection (Fig. 2D). In contrast, genes in the chronic-greenyellow module showed an up-regulation at day 6 with a subsequent down-regulation at day 9, thus coinciding with T cell exhaustion. To validate these observations and to link the day 9 down-regulation of the chronic-greenyellow genes to T cell exhaustion, we performed qPCR of selected genes from spleens of mice at day 9 after acute or chronic infection (Supplemental Fig. S3C,D). The qPCR results positively correlated with the RNA-seq data. Cd3d and Zap70 from the chronic-greenyellow module that play a critical role in T cell receptor signaling, and that were chosen as examples because of their high KIM values, were significantly down-regulated in chronic infection thus supporting a role during T cell exhaustion.
The B cell response-related genes of Cluster 2 were mainly found in the chronic-green module and retained a high intramodular connectivity value (Supplemental Fig. S3B). Thus, the B cell response during both infection types is controlled by a highly coregulated gene expression. The expression kinetics of these genes during acute and chronic infections were identical, being up-regulated from d7 to d9 and remaining elevated at d31. However, their expression levels were notably higher in chronic infection (Fig. 2E; Supplemental Fig. S3C), suggesting a causal link to the previously described hypergammaglobulinemia of chronic infections (Hunziker et al. 2003). To validate this, spleens from acute and chronic LCMV infections were stained for total IgG and analyzed by immunohistochemistry. IgG production was evident at d9 in both types of infection but was significantly higher in chronic infection. An elevated level of IgG remained visible even at day 48 postinfection when the B cell response of acute infection was comparable to that of naive mice (Fig. 2F,G; Supplemental Fig S4). Taken together, coexpression network analysis from spleen-derived transcriptomes permits the identification of biologically relevant molecular pathways that are characteristic for acute or chronic infections.
Transcriptional network preservation between acute and chronic infections
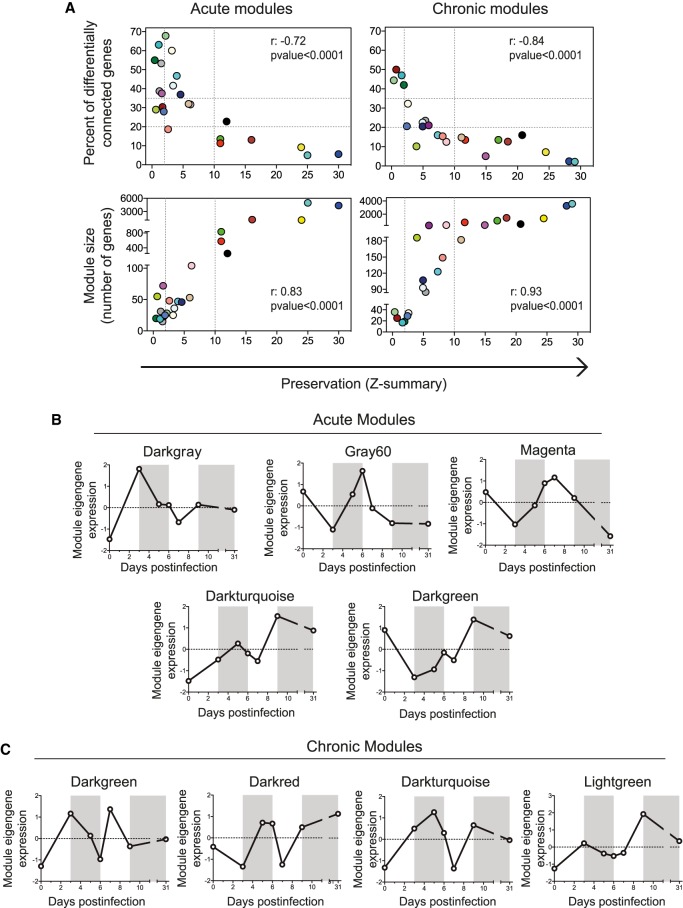
The obtained gene coexpression modules represent the biological processes participating in acute and chronic infections. To identify the common and the specific features for both infection fates, we first determined the gene overlap between the acute and chronic modules by Fisher's exact test (Supplemental Fig. S5A). Eight acute and 12 chronic modules showed a high degree of overlap (P-value < 10−35) with modules from the other group. To then quantify module preservation (the degree to which coexpressed genes in acute infection are also coexpressed in chronic infection), we computed a connectivity-based preservation statistic (Z-summary) that allows scoring the degree of similarity of the connectivity patterns between genes from two networks. Module preservation scores were inversely correlated with the percentage of differentially connected genes (defined as those with a difference in their KIM between acute and chronic networks higher than 0.4) (Fig. 3A). This indicates that the preserved modules, that share similar gene connectivity patterns, also share common hub genes. Furthermore, module preservation scores correlated with the module size (Fig. 3A), indicating that a large number of genes participate in biological pathways that are common in acute and chronic infection. In contrast, the biological pathways that are specific for an infection fate are governed by few genes.
Figure 3.
Module preservation between acute and chronic infections. (A) Module preservation scores were compared to the percentage of differentially connected genes in a module and to module size. Dashed lines indicate the thresholds (Methods). The Spearman's rank correlation coefficients (r) and the P-values are indicated. (B,C) Eigengene expression profiles of acute- (B) and chronic-specific (C) modules.
To further examine the highly preserved modules, we compared their expression patterns (Supplemental Fig. S5B,C). Relevant overlapping hub genes between preserved modules are listed in Supplemental Table S1. With the exception of the acute-yellow and the chronic-brown module that shared genes involved in type I interferon signaling (Irf7, Mx1-2, Oas1a, Ifit1-3), all the other highly preserved acute modules were decomposed into two modules in chronic infection. For instance, the acute-brown module overlapped with the chronic-green and the chronic-tan module, respectively, as described above (Fig. 2). Thirty-six hub genes from the acute-blue module overlapped with chronic-magenta, a module with a decrease in its expression between d7 and d9 when T cell exhaustion appears (Fig 1B). Four of these genes were related to leukocyte chemotaxis (Itgb2, Itgam, Ccr1, and C5ar1). qPCR of spleens from additional animals validated the differential down-regulation of Ccr1 and C5ar1 between acute and chronic infections at days 7 and 9 PI and showed a tendency to decrease for Itgb2 and Itgam (Supplemental Fig. S6A,B). Three of these four genes (Itgb2, Itgam, and C5ar1) are involved in neutrophil chemotaxis. A sustained expansion of neutrophilic MDSCs has been described in LCMV chronic infection (Norris et al. 2013). Therefore, these results may suggest that prior to the appearance of neutrophilic MDSCs, there is an alteration in the migration of these cells concomitant with exhaustion of CD8+ T cells.
The modules with low preservation scores represent biological processes specific of acute or chronic infections, and therefore might provide novel mechanisms influencing infection fates. We identified five acute-specific and four chronic-specific modules (Fig. 3B,C). The respective genes and their connectivity values are listed in Supplemental Table S2. Acute-specific modules showed diverse expression profiles characterized by marked expression peaks at different time points. The included genes represent both innate and adaptive immune pathways highlighting the need of diverse biological responses to properly resolve a viral infection. On the other hand, all chronic infection-specific modules showed a bimodal expression profile with a second peak or drop of expression at days 7–9 PI (Fig. 3C) when T cell exhaustion appears. Thus, genes that participate in early events after infection seem also to play a role in critical events governing chronic infection at later time points. This suggests that the adaptation process to a chronic infection involves not a simple shutdown of genes but is an active process as described at the cellular level for CD8+ T cell exhaustion (Wherry et al. 2007). Group-specific modules are further analyzed in the next sections.
Early attenuation of the inflammatory response in chronic infection
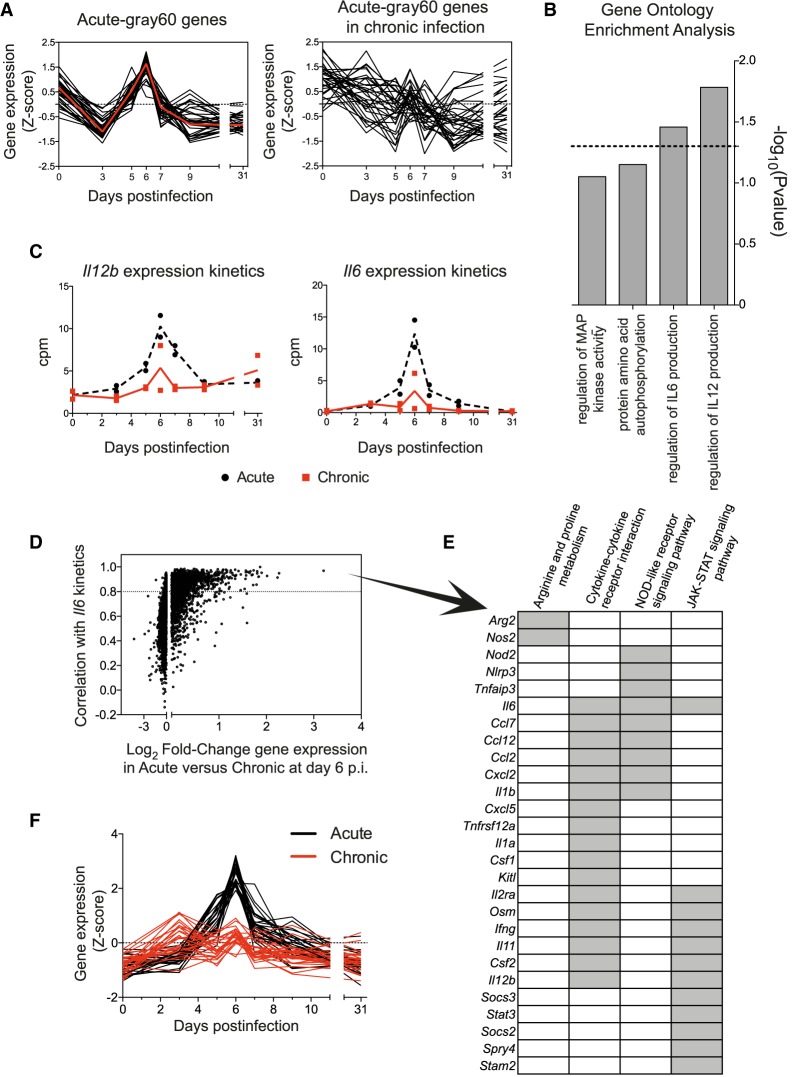
Acute infection-specific modules are expected to participate in the resolution of the virus infection. From the five acute-specific modules (Fig. 3A,B), the acute-gray60 module was most interesting. It has a marked expression peak at day 6 postinfection (Fig. 3B) just before the specific T cell response appeared thus bridging innate immune responses with the adaptive response. All 31 genes of this module are highly coexpressed only during acute infection (Fig. 4A) and enriched for genes involved in the regulation of IL6 and IL12 production (Fig. 4B). Because IL6 and IL12 are pro-inflammatory cytokines with important roles in antiviral host responses, our observations suggested a differential regulation in both infection fates before T cell exhaustion becomes apparent. To verify this hypothesis, we plotted the Il6 and Il12b expression kinetics from our RNA-seq data (Fig. 4C) and validated the Il6 expression results by qPCR including additional infected animals. Both RNA-seq and qPCR gave consistent results showing a peak of Il6 expression at d6 after acute infection that was lacking in chronic infection (Supplemental Fig. S7). Because inflammatory responses are complex responses, we next sought to identify whether Il6 and Il12b were part of a broader cluster of coexpressed genes within their acute-blue module. Indeed, we found 1504 genes in the acute-blue module that correlate to the Il6 expression kinetics with a correlation value greater than 0.8. Moreover, the level of expression of many of these genes was significantly higher in acute than in chronic infection at d6 PI (Fig. 4D). KEGG pathway analysis of the 1504 genes showed enrichment of genes involved in inflammatory pathways such as NOD-like receptor and JAK-STAT signaling pathways, and in cytokine–cytokine receptor interaction (Supplemental Table S3). Furthermore, we identified two genes related to the metabolism of Arginine and Proline (Arg2 and Nos2), a pathway known to play an important role in the regulation of inflammation by monocytes and macrophages (Supplemental Table S3; Burrack and Morrison 2014; Yang and Ming 2014; Liu et al. 2016). A list of 27 representative genes from this submodule with a clear peak of expression at day 6 PI in acute infection, and their expression profiles in acute and chronic infections, are shown in Figure 4, E and F, respectively. Il1b, Arg2, and Nos2 expression kinetics was further confirmed by qPCR including additional infected mice (Supplemental Fig. S7). Together these data show differential spleen monocyte/macrophage characteristics during early time points of acute and chronic virus infections.
Figure 4.
Acute-gray60-specific module unveils a differential regulation of genes related to macrophage inflammatory responses in acute and chronic infections. (A) Normalized expression kinetics of genes from the acute-gray60 module and their corresponding kinetics in chronic infection. Red line represents the kinetics of the eigengene. (B) GO terms enriched in genes from the acute-gray60 module (dashed line mark P = 0.05). (C) RNA-seq-derived Il12b and Il6 expression kinetics in acute and chronic infections. (D) Pearson's correlation between genes of the acute-blue module and Il6 in acute infection is plotted against the log2 fold-change (log2FC) of gene expression between acute and chronic infection at day 6 PI. Dashed line marks a correlation >0.8. (E) List of 27 representative genes with a correlation > 0.8 with Il6 expression kinetics. Gray boxes indicate enriched KEGG pathways to which each gene belongs. (F) Normalized expression kinetics of the 27 genes shown in E.
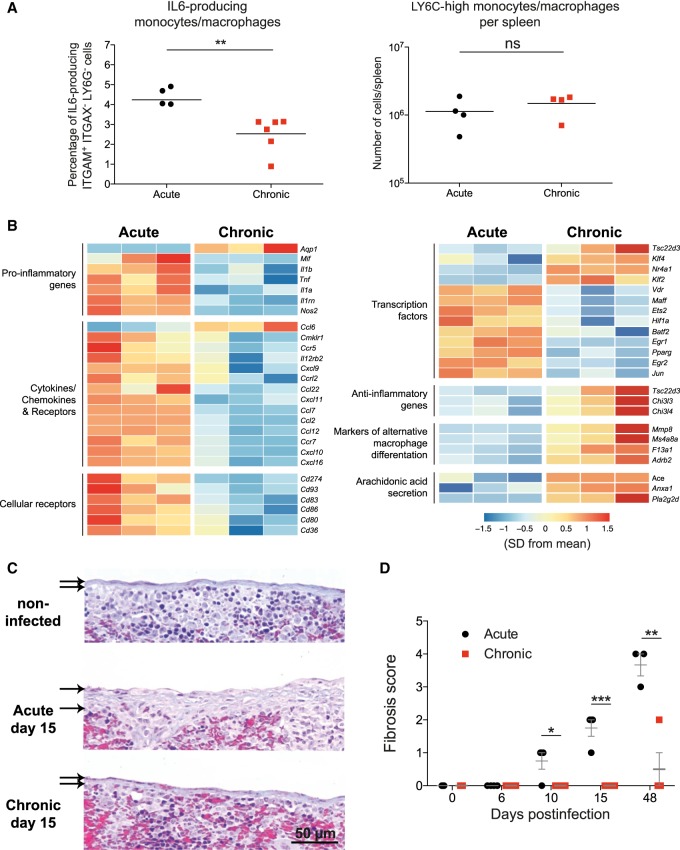
To test the hypothesis of an early differential polarization of monocyte/macrophages during both outcomes of LCMV infection, we characterized spleen monocyte/macrophages at day 6 postinfection by intracellular IL6 staining and RNA-seq. The percentage of IL6-producing monocytes/macrophages from chronic infection were significantly lower than those from acute infection, whereas the number of pro-inflammatory LY6Chi monocytes/macrophages in the spleen were, as described before (Norris et al. 2013), similar in both infections (Fig. 5A). Transcriptional profiling from sorted monocytes/macrophages showed that cells from acute infection expressed higher levels of pro-inflammatory cytokines and chemokines, and cellular receptors and transcription factors associated to M1 macrophages. In contrast, monocytes/macrophages from chronic infection expressed higher levels of transcription factors and genes that exert anti-inflammatory effects and markers of M2 macrophage differentiation (Fig. 5B). These data demonstrate that inflammatory monocytes/macrophages are induced early after an acute LCMV infection and, in contrast, during chronic infection these cells shift to an anti-inflammatory profile before T cell exhaustion becomes evident.
Figure 5.
Early shift from inflammatory to pro-resolving monocytes/macrophages in chronic infection to prevent fibrosis. (A) Percentage of IL6-producing ITGAM+ ITGAX− LY6G− (also known as CD11b+ CD11c− LY6G−) cells and number of LY6C high monocytes/macrophages per spleen at day 6 postinfection. (B) Heatmap illustrating the relative expression of genes in monocyte/macrophages from acute and chronic infected mice at day 6 PI. (C) Representative images of Masson's trichrome staining of spleens. Thickness of the splenic capsules are marked by arrows. Magnification bar: 50 µm. (D) Semiquantification of fibrosis in spleens. (A–D) Data shown are the mean ± SEM of n = 4 to 6 mice. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001 unpaired two-tailed t-test.
The monocytic cell population of chronic infection had an up-regulated expression of genes involved in arachidonic acid secretion such as Pla2g2d (phospholipase A2, group IID), an anti-inflammatory lipid mediator (Miki et al. 2013). Because these molecules play an important role in the resolution of inflammation (Sugimoto et al. 2016), one might expect a tissue protective role of these cells in chronic infection. To test this idea, we quantified the level of fibrosis in spleens from acute and chronic LCMV infections by Masson's trichrome staining. Fibrosis, a feature of insufficient resolution of chronic inflammation, was evident as early as d10 in acute infection, and remained elevated even at days 15 and 48, when the virus infection is resolved (Fig. 5C,D). In contrast, fibrosis was only observed in chronic infection in one of four animals at d48. Together this suggests that besides the known role of suppressive monocytic cells to dampen virus T cell responses in chronic infection, they also might participate in inflammation resolution via expression of pro-resolving lipid mediators that finally lead to fibrosis prevention.
The XCL1-XCR1 communication axis links T cell exhaustion with effector cell maintenance
Cytokines and chemokines are major components of the signaling network that regulates the coordinated immune response against a viral infection (Cameron and Kelvin 2003). Thus, we next investigated which cytokines/chemokines-encoding hub genes were present in acute- and chronic-specific modules, and therefore might participate in processes involved in infection fate regulation. From all the identified hub genes (Supplemental Table S2), of particular interest was the chemokine-encoding gene Xcl1 from the chronic-darkturquoise module. XCL1 is mainly produced by NK and activated CD8+ T cells, and promotes the recruitment of dendritic cells expressing the receptor XCR1 (XCR1+ DCs), which are implicated in the priming and boosting of cytotoxic responses to cross-presented antigens (Kroczek and Henn 2012; Brewitz et al. 2017).
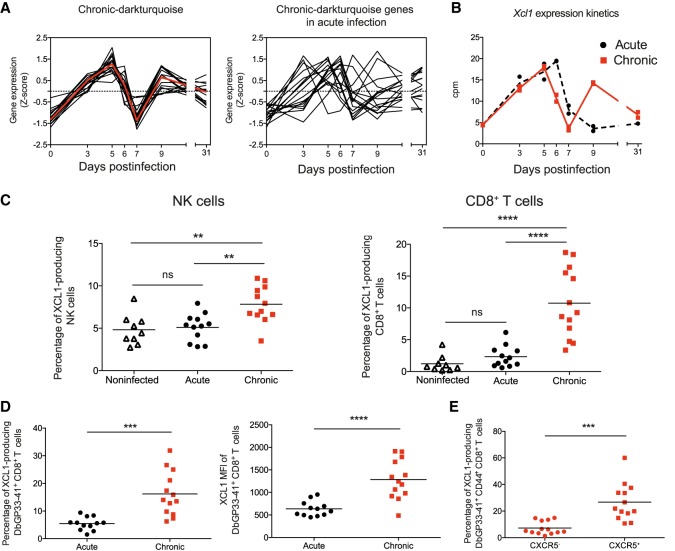
All 17 genes of the chronic-darkturquoise module are highly coexpressed in chronic infection and deregulated in acute infection (Fig. 6A). They show a “two-peak” behavior with an expression peak at d5 and a second up-regulation from d7 to d9 PI. The second peak of expression of Xcl1 was verified by qPCR using additional infected mice (Supplemental Fig. S8A). In contrast to chronic infection, Xcl1 showed only a single early peak of expression at d6 in acute infection, returning to basal levels from d7 PI (Fig. 6B). To investigate the role of XCL1 during the establishment of the chronic infection phase, we first analyzed the chemokine expression by NK and CD8+ T cells at d9 by intracellular staining. Both NK and CD8+ T cells showed a higher expression of XCL1 in chronic infection compared to acute infection or uninfected mice (Fig. 6C). XCL1 was highly produced by LCMV-specific CD8+ T cells once exhaustion was established (Fig. 6D; Supplemental Fig. S8B), and particularly abundant in the subpopulation of CXCR5+ cells (Fig. 6D). Because virus-specific CXCR5+ CD8+ T cells have recently been shown to play a major role in the control of virus replication during chronic infections (He et al. 2016; Im et al. 2016), our data now show that they also play an important role in the immune adaptation process during exhaustion.
Figure 6.
Chronic-darkturquoise-specific module reveals an important role of XCL1 in chronic infection. (A) Normalized expression kinetics of genes from the chronic-darkturquoise module and their corresponding kinetics in acute infection. (B) Xcl1 expression kinetics obtained from RNA-seq analysis. (C) Percentages of XCL1-producing NK and CD8+ T cells at day 9 PI. Data shown are the mean ± SEM from n = 11 to 13 mice representative of three independent experiments (significance determined using one-way ANOVA). (D,E) MFI of XCL1 in DbGP33-41+ CD8+ T cells (D, right) and percentages of XCL1-producing DbGP33-41+ CD8+ T cells (D, left) and CXCR5- or CXCR5+ DbGP33-41+ CD44+ CD8+ T cells (E) at day 9 PI. Data shown are the mean ± SEM from n = 11 to 13 mice (significance determined using unpaired two-tailed t-test). (ns) not significant; (**) P ≤ 0.01; (***) P ≤ 0.001; (****) P ≤ 0.0001.
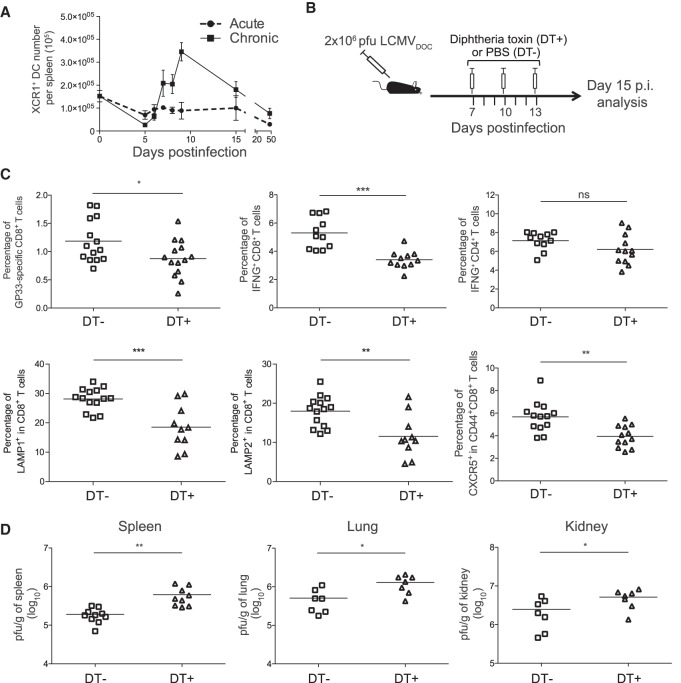
Coinciding with the second peak of Xcl1 expression, the quantification of XCR1+ DCs showed a clear increase of these cells in the spleens at d9 in chronic infection (Fig. 7A; Supplemental Fig. S8C). To further analyze the impact of this XCL1-XCR1 axis, we depleted XCR1+ DC in chronically infected XCR1-DTRvenus mice (Yamazaki et al. 2013) with diphtheria toxin (DT) (Fig. 7B; Supplemental Fig. S8D). Although the virus-specific CD4+ T cell response was not affected, XCR1+ DC depletion led to a significant reduction in the percentage of GP33-specific CD8+ T cells at day 15 PI and lower percentages of LAMP1 (also known as CD107a), LAMP2 (also known as CD107b), and IFNG-producing cells than in untreated animals (Fig. 7C). Moreover, the percentage of CXCR5+ CD8+ T cells was also significantly decreased, suggesting an interdependence between this CD8+ T cell subset and cross-presenting XCR1+ DCs. Consistently, the viral titers in spleen, lung, and kidney were higher in mice lacking XCR1+ DCs (Fig. 7D). Together these results demonstrate that the XCL1-XCR1 communication axis is (1) an important component of immune system adaptation to an overwhelming virus threat, and (2) an important component of virus control in the chronic infection phase.
Figure 7.
XCR1+ dendritic cells maintain antiviral T cell responses and control virus titers. (A) Number of XCR1+ dendritic cells in spleen. (B) Schematic representation of DT treatment regimen for depletion of XCR1+ dendritic cells. (C) Percentages of DbGP33-41+ CD8+ T cells, IFNG-producing CD8+ and CD4+ T cells, LAMP1+ and LAMP2+ CD8+ T cells, and CXCR5+ CD44+ CD8+ T cells at day 15 PI from chronic infected mice nontreated (DT−) or treated (DT+) with DT. (D) Virus titers in spleen, lung, and kidney at day 15 PI from chronic infected mice nontreated (DT−) or treated (DT+) with DT. (C,D) The mean ± SEM is shown. (ns) nonsignificant; (*) P ≤ 0.05; (**) P ≤ 0.001; (***) P ≤ 0.0001 unpaired two-tailed t-test. Data are representative of two or three independent experiments.
Discussion
Nonlethal pathogenic viral infections are either acutely resolved or become chronic. Although this fate decision is made during the primary infection phase, it is incompletely understood, and a systems view is lacking. Here, we used time-resolved spleen transcriptomes from acutely or chronically LCMV-infected mice in combination with weighted gene coexpression network analysis to decipher differentiating host responses and biological pathways. We demonstrate an early attenuation of inflammatory monocyte/macrophage responses prior to T cell exhaustion and the involvement of the XCL1-XCR1 communication axis in virus containment during chronic infection. Thus, the response of a host to an overwhelming infection threat is complex and subtle, involving both immune-stimulatory and immune-suppressive pathways.
Acute and chronic infection fates are readily distinguished by their sequential splenic transcriptomes. The whole transcriptomes reveal differentiating traits at day 9 postinfection when acute infection starts to be resolved. However, close inspection of the coexpression modules demonstrates an attenuation of inflammatory monocytic cells at day 6 before T cell exhaustion becomes apparent. Thus, the decision point to down-regulate immune responses toward an overwhelming virus threat is already taken before T cells shut off their effector functions. Whether this decision point lies even earlier remains to be determined. So far, early innate immune responses represented, that is, by modules containing type I IFN genes are very similar in acute and chronic infections, and differences come up only from day 6 onward. In addition to these global characteristics, we were also able to link specific genes and pathways to critical, already established phenotypes associated with both infection fates including the effective LCMV-specific T cell response as well as T cell exhaustion and hypergammaglobulinemia. Together this validates our systems approach for analyzing virus infection fates and leaves a rich comprehensive data set for further, more specific analyses.
Infection-fate-specific module information provides a firm basis to reveal specific, physiologically relevant changes in the infected host organism. An intriguing observation was that the infection-fate-specific modules contained rather few genes. This notion may not only help to define diagnostic biomarker for predicting infection outcomes but also hint to regulators of pathways that may direct phenotype evolution over time. Indeed, the acute-gray60 module with its 31 genes that have an expression peak at day 6, is a good example. Starting from a GO enrichment analysis of its genes, we subsequently identified important differences in early monocytic cell polarization with an M1-type profile in acute infection and cells with an attenuated phenotype during chronic infection. Previous studies have described a skewing of monocytic cells toward MDSCs in chronic LCMV infections of mice (Norris et al. 2013) and in HIV, HBV, and HCV infections in humans (Cai et al. 2013; Qin et al. 2013; Pallett et al. 2015). MDSCs appear during the primary infection phase and have been characterized, for example, by transcriptome analysis at day 14 post-chronic LCMV clone 13 infection, a few days after T cell exhaustion becomes apparent (Norris et al. 2013). Our own RNA-seq results from isolated monocytic cells at day 6 postinfection now point to an even earlier phenotypic shift with an expression of genes linked to alternative macrophage differentiation and anti-inflammation. This early monocytic cell skewing was associated with histological changes in the spleen. Acutely infected mice develop spleen fibrosis that is maintained even when the virus is already well controlled, but in chronic infection, fibrosis was almost absent (Fig. 5C,D). Although the direct evidence for a causal link between the splenic monocyte/macrophage phenotypes and fibrosis development is still missing, the known features of tissue macrophages in tissue injury, fibrosis, and repair are well in line with our observations (Sugimoto et al. 2016; Wynn and Vannella 2016).
Concomitant with CD8+ T cell exhaustion during chronic infection, XCR1+ DC number increase in spleen. Depletion of XCR1+ DCs resulted in reduction of virus-specific CD8+ T cells and a concurrent increase in viral loads (Fig. 7C,D). However, because down-regulation of T cell effector function by exhaustion is a mechanism to avoid immunopathology (Cornberg et al. 2013), as is attenuation of inflammatory macrophages and generation of MDSCs (Medzhitov et al. 2012), why should a host organism at the same time expand a subtype of antigen-presenting cells that are highly potent in priming and boosting novel T cell responses? This fundamental issue may simply highlight two important aspects within the in vivo virus-immune system crosstalk. First, the immune system has to shut down when confronted with an overwhelming viral threat. A high-dose LCMVDoc infection requires an adaptation of the immune response and viral load dynamics such that it levels below a life-threatening situation. This is achieved by a variety of immune-suppressive mechanisms of which T cell exhaustion and the generation of MDSCs are important components. Second, the immune system does not surrender completely to an overwhelming threat but maintains partial control and restricts virus overloads. One possible way for this seems to be a feedback regulation involving cells from the population of virus-specific CD8+ T cells that become exhausted. Indeed, the population of LCMV-specific CD8+ T cells that becomes exhausted contains cells that produce XCL1 (Fig. 6E) and thus can set up XCR1+ DC recruitment and cytotoxic T cell stimulation and effector cell maintenance. Overall, this argues for the operation of virus-threat-sensing host mechanisms that adapt the immune system effectors to minimize damage and the XCL1-XCR1 axis of being part of a functional adaptation to the chronic infection phase.
How may these observations be exploited for host benefit? Recent publications have shown that virus-specific CXC5+ CD8+ T cells expand most efficiently after immune checkpoint inhibition (He et al. 2016; Im et al. 2016). They are, besides NK cells, the main producer of XCL1 in the chronic infection phase (Fig. 6C). Furthermore, XCL1 is elevated in HIV-infected elite controllers (Jacobs et al. 2017) and its receptor XCR1 is solely expressed on cross-presenting DCs (Dorner et al. 2009) that are specifically up-regulated during the chronic infection phase (Fig. 7A; Supplemental Fig. S8C). Therefore, enforcing the XCL1-XCR1 axis in an immunotherapeutic intervention seems a promising strategy to better control chronic virus infections. One option in this direction would be the already established targeting of immunogens to XCR1+ DCs via linkage to XCL1 itself or to a XCR1-targeting antibody (Fossum et al. 2015; Hartung et al. 2015). Combining such immunotherapeutic vaccination with checkpoint inhibition may then allow effector cell expansion enhanced by a positive feedback loop and thus a greater increase in the effector cell to virus ratio.
In conclusion, coexpression network analysis of time-resolved splenic transcriptome data revealed fundamental features of infection fate regulation. Several links between gene transcripts and immunological phenotypes were established that include an early attenuation of inflammatory monocytic cells and an involvement of the XCL1-XCR1 axis in the functional adaptation of the immune response in a chronic virus infection. This not only provides a unique data set to generate and test novel hypotheses about immune system functioning but also suggests directions for immunotherapeutic strategies to better control or cure persistent infections. An inspection of the results in human infections and cancer is clearly indicated.
Methods
Animals, infections, and in vivo cell depletion
Mice were purchased from Charles River Laboratories and maintained under specific pathogen-free conditions at in-house facilities. All animal work was conducted according to the guidelines from Generalitat de Catalunya and approved by the ethical committee for animal experimentation at Parc de Recerca Biomèdica de Barcelona (CEEA-PRBB, Spain). LCMV strain Docile was used for mouse infections. For details on mouse strains, virus quantification, and in vivo cell depletion, see Supplemental Information.
Flow cytometry, sorting, and immunohistochemistry
For flow cytometry analysis and sorting of cells, spleens were harvested, and single-cell suspensions were generated. For immunohistochemistry, spleens were formalin fixed and paraffin embedded. For details, see Supplemental Methods and Supplemental Table S4.
RNA-seq library preparation and sequencing
Total RNA from samples was isolated and submitted for sequencing to the Genomics Unit of the Centre for Genomic Regulation (CGR, PRBB). For details, see Supplemental Information.
RNA-seq bioinformatic analysis
Reads mapping against the Mus musculus reference genome (GRCm38) was done using the GEMtools RNA-seq pipeline (http://gemtools.github.io/docs/rna_pipeline.html) and quantified with Flux Capacitor (http://confluence.sammeth.net/display/FLUX/Home) with the Mus musculus GENCODE annotation M2 version (https://www.gencodegenes.org/). Normalization was performed with the edgeR TMM method (Robinson and Oshlack 2010). Pairwise Pearson correlation coefficients (PCC) were calculated for comparison among transcriptomes of spleens from uninfected (n = 2, day 0), acute (n = 2 per time point), or chronic (n = 2 per time point) infected mice. Hierarchical clustering across all samples was based on pairwise Pearson correlation coefficients among RNA-seq libraries. Differential expression analysis was performed with the “robust” version of the edgeR R package (Zhou et al. 2014). Genes with a false discovery rate (FDR) < 5% were considered significant. Differentially expressed genes in acute and chronic infection time series (n = 13,971) were used to construct a coexpression network with the WGCNA R package for each data set (Langfelder and Horvath 2008). First, a signed weighted adjacency matrix was calculated with the “blockwiseModules” function using the following parameters: power = 30, TOMtype = “signed”, minModuleSize = 15, mergeCutHeight = 0.25, reassignThreshold = 0, networkType = “signed”, numericLabels = TRUE, pamRespectsDendro = FALSE, nThreads = 7, maxBlockSize = 17000. The power law of 30 was selected to meet the scale-free topology assumption with the pickSoftThreshold function. Then, genes were clustered into network modules using average linkage hierarchical clustering and the topological overlap measure (TOM) as proximity. Each of the identified modules was summarized by its module eigengene (the first principal component), which represents the weighted average expression profile of all module genes (Langfelder and Horvath 2008). To identify intramodular hub genes inside a given module, the intramodular connectivity (KIM) was calculated for each gene and VisANT (http://visant.bu.edu/) was used for network visualizations (TOM > 0.3). Module preservation and module overlap were calculated with functions “modulePreservation” and “userListEnrichment”, respectively. Viral loads, CD4+, and CD8+ T cell levels were correlated with the module eigengenes with Pearson correlation coefficients. Gene Ontology (GO) enrichment analysis was performed with DAVID (http://david.ncifcrf.gov/) (Huang et al. 2009).
Module preservation analysis
Z-summary, implemented within WGCNA, with 100 random permutations of the data, was used as a connectivity-based preservation statistic able to determine whether the connectivity pattern between genes in a reference network is similar to that in a test network. A Z-summary score < 2 indicates no evidence of preservation, 2 < Z-summary < 10 implies weak preservation, and Z-summary > 10 suggests strong preservation. Highly preserved modules were defined as those with a preservation Z-summary value greater than 10 and a percentage of differentially connected genes less than 20%. Group-specific modules were defined as those with a preservation Z-summary value less than 2 and a percentage of differentially connected genes greater than 35%.
Quantitative real-time PCR
Total RNA from spleens was used to perform quantitative real-time PCR using SYBR select master mix (Thermo Fisher Scientific). Primers for all genes were designed with Primer Express 3.0 (Applied Biosystems). For details, see Supplemental Information.
Statistical analysis
Two-tailed t-test and ANOVA analyses were performed using GraphPad Prism 6.0. P-values below 0.05 were considered significant and were indicated by asterisks: (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001; (****) P ≤ 0.0001. Nonsignificant differences were indicated as “ns.” Fisher's exact test was used to quantify overlap between modules. Correlation coefficients (r) were calculated using the Spearman rank correlation test.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE107880.
Supplementary Material
Acknowledgments
We thank Mónica Pérez (CReSA-IRTA), Rita De Giuli-Buehrer (Kantonsspital St. Gallen), and the Flow Cytometry unit (Universitat Pompeu Fabra) for their excellent technical support. This work was supported by a grant from the Spanish Ministry of Economy, Industry and Competitiveness and FEDER Grant No. SAF2016-75505-R (AEI/MINEICO/FEDER, UE), the Russian Science Foundation Grant 18-11-00171, the Uehara Memorial Foundation (to T.K.), Grant-in-Aid for Scientific Research B 17H04088 from the Japan Society for the Promotion of Science (JSPS), (to T.K.), and the grant of the National Bioinformatics Institute (INB), PRB2-ISCIII (PT13/0001/0044 to A.E.-C.).
Author contributions: A.M., G.B., and J.A. conceived the project; J.A., M.P., G.R., E.V., V.C., and C.P.-C. performed experiments; A.E.-C. and S.H. performed bioinformatics’ analysis; D.A., B.L., and T.K. provided tools; A.M. and J.A. designed and supervised the study; J.A., A.E.-C., and M.P. analyzed the data; A.M., J.A., and M.P. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.241372.118.
References
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439: 682–687. 10.1038/nature04444 [DOI] [PubMed] [Google Scholar]
- Bocharov GA. 1998. Modelling the dynamics of LCMV infection in mice: conventional and exhaustive CTL responses. J Theor Biol 192: 283–308. 10.1006/jtbi.1997.0612 [DOI] [PubMed] [Google Scholar]
- Brewitz A, Eickhoff S, Dähling S, Quast T, Bedoui S, Kroczek RA, Kurts C, Garbi N, Barchet W, Iannacone M, et al. 2017. CD8+ T cells orchestrate pDC-XCR1+ dendritic cell spatial and functional cooperativity to optimize priming. Immunity 46: 205–219. 10.1016/j.immuni.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack KS, Morrison TE. 2014. The role of myeloid cell activation and arginine metabolism in the pathogenesis of virus-induced diseases. Front Immunol 5: 428 10.3389/fimmu.2014.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Qin A, Guo P, Yan D, Hu F, Yang Q, Xu M, Fu Y, Zhou J, Tang X. 2013. Clinical significance and functional studies of myeloid-derived suppressor cells in chronic hepatitis C patients. J Clin Immunol 33: 798–808. 10.1007/s10875-012-9861-2 [DOI] [PubMed] [Google Scholar]
- Cameron MJ, Kelvin DJ. 2003. Cytokines and chemokines—their receptors and their genes: an overview. Adv Exp Med Biol 520: 8–32. 10.1007/978-1-4615-0171-8_2 [DOI] [PubMed] [Google Scholar]
- Cornberg M, Kenney LL, Chen AT, Waggoner SN, Kim SK, Dienes HP, Welsh RM, Selin LK. 2013. Clonal exhaustion as a mechanism to protect against severe immunopathology and death from an overwhelming CD8 T cell response. Front Immunol 4: 475 10.3389/fimmu.2013.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côrtes FH, de Paula HHS, Bello G, Ribeiro-Alves M, de Azevedo SSD, Caetano DG, Teixeira SLM, Hoagland B, Grinsztejn B, Veloso VG, et al. 2018. Plasmatic levels of IL-18, IP-10, and activated CD8+ T cells are potential biomarkers to identify HIV-1 elite controllers with a true functional cure profile. Front Immunol 9: 1576 10.3389/fimmu.2018.01576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443: 350–354. 10.1038/nature05115 [DOI] [PubMed] [Google Scholar]
- Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. 2012. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37: 1130–1144. 10.1016/j.immuni.2012.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Güttler S, Hutloff A, Mages HW, Ranke K, Schaefer M, et al. 2009. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 31: 823–833. 10.1016/j.immuni.2009.08.027 [DOI] [PubMed] [Google Scholar]
- Feinberg MB, Ahmed R. 2012. Born this way? Understanding the immunological basis of effective HIV control. Nat Immunol 13: 632–634. 10.1038/ni.2351 [DOI] [PubMed] [Google Scholar]
- Fossum E, Grødeland G, Terhorst D, Tveita AA, Vikse E, Mjaaland S, Henri S, Malissen B, Bogen B. 2015. Vaccine molecules targeting Xcr1 on cross-presenting DCs induce protective CD8+ T-cell responses against influenza virus. Eur J Immunol 45: 624–635. 10.1002/eji.201445080 [DOI] [PubMed] [Google Scholar]
- Hartung E, Becker M, Bachem A, Reeg N, Jäkel A, Hutloff A, Weber H, Weise C, Giesecke C, Henn V, et al. 2015. Induction of potent CD8 T cell cytotoxicity by specific targeting of antigen to cross-presenting dendritic cells in vivo via murine or human XCR1. J Immunol 194: 1069–1079. 10.4049/jimmunol.1401903 [DOI] [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. 2016. Follicular CXCR5- expressing CD8+ T cells curtail chronic viral infection. Nature 537: 412–428. 10.1038/nature19317 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hunziker L, Recher M, Macpherson AJ, Ciurea A, Freigang S, Hengartner H, Zinkernagel RM. 2003. Hypergammaglobulinemia and autoantibody induction mechanisms in viral infections. Nat Immunol 4: 343–349. 10.1038/ni911 [DOI] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537: 417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs ES, Keating SM, Abdel-Mohsen M, Gibb SL, Heitman JW, Inglis HC, Martin JN, Zhang J, Kaidarova Z, Deng X, et al. 2017. Cytokines elevated in HIV elite controllers reduce HIV replication in vitro and modulate HIV restriction factor expression. J Virol 91: e02051-16 10.1128/JVI.02051-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási AL, Oltvai ZN. 2001. Lethality and centrality in protein networks. Nature 411: 41–42. 10.1038/35075138 [DOI] [PubMed] [Google Scholar]
- Kroczek RA, Henn V. 2012. The role of XCR1 and its ligand XCL1 in antigen cross-presentation by murine and human dendritic cells. Front Immunol 3: 14 10.3389/fimmu.2012.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, et al. 2016. CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol 17: 1187–1196. 10.1038/ni.3543 [DOI] [PubMed] [Google Scholar]
- Liu C, Rajapakse AG, Riedo E, Fellay B, Bernhard M-C, Montani J-P, Yang Z, Ming X-F. 2016. Targeting arginase-II protects mice from high-fat-diet-induced hepatic steatosis through suppression of macrophage inflammation. Sci Rep 6: 20405 10.1038/srep20405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP. 2012. Disease tolerance as a defense strategy. Science 335: 936–941. 10.1126/science.1214935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Yamamoto K, Taketomi Y, Sato H, Shimo K, Kobayashi T, Ishikawa Y, Ishii T, Nakanishi H, Ikeda K, et al. 2013. Lymphoid tissue phospholipase A2 group IID resolves contact hypersensitivity by driving antiinflammatory lipid mediators. J Exp Med 210: 1217–1234. 10.1084/jem.20121887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskophidis D, Lechner F, Pircher H, Zinkernagel RM. 1993. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature 362: 758–761. 10.1038/362758a0 [DOI] [PubMed] [Google Scholar]
- Ng CT, Snell LM, Brooks DG, Oldstone MBA. 2013. Networking at the level of host immunity: immune cell interactions during persistent viral infections. Cell Host Microbe 13: 652–664. 10.1016/j.chom.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris BA, Uebelhoer LS, Nakaya HI, Price AA, Grakoui A, Pulendran B. 2013. Chronic but not acute virus infection induces sustained expansion of myeloid suppressor cell numbers that inhibit viral-specific T cell immunity. Immunity 38: 309–321. 10.1016/j.immuni.2012.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MBA. 2006. Viral persistence: parameters, mechanisms and future predictions. Virology 344: 111–118. 10.1016/j.virol.2005.09.028 [DOI] [PubMed] [Google Scholar]
- Pallett LJ, Gill US, Quaglia A, Sinclair LV, Jover-Cobos M, Schurich A, Singh KP, Thomas N, Das A, Chen A, et al. 2015. Metabolic regulation of hepatitis B immunopathology by myeloid-derived suppressor cells. Nat Med 21: 591–600. 10.1038/nm.3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Cai W, Pan T, Wu K, Yang Q, Wang N, Liu Y, Yan D, Hu F, Guo P, et al. 2013. Expansion of monocytic myeloid-derived suppressor cells dampens T cell function in HIV-1-seropositive individuals. J Virol 87: 1477–1490. 10.1128/JVI.01759-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann B, Nascimbeni M. 2005. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol 5: 215–229. 10.1038/nri1573 [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse BT, Sehrawat S. 2010. Immunity and immunopathology to viruses: What decides the outcome? Nat Rev Immunol 10: 514–526. 10.1038/nri2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283: 857–860. 10.1126/science.283.5403.857 [DOI] [PubMed] [Google Scholar]
- Sugimoto MA, Sousa LP, Pinho V, Perretti M, Teixeira MM. 2016. Resolution of inflammation: What controls its onset? Front Immunol 7: 160 10.3389/fimmu.2016.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Wherry EJ, Ahmed R. 2009. Redefining chronic viral infection. Cell 138: 30–50. 10.1016/j.cell.2009.06.036 [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 77: 4911–4927. 10.1128/JVI.77.8.4911-4927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27: 670–684. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Wilson EB, Brooks DG. 2010. Translating insights from persistent LCMV infection into anti-HIV immunity. Immunol Res 48: 3–13. 10.1007/s12026-010-8162-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB, Kidani Y, Elsaesser H, Barnard J, Raff L, Karp CL, Bensinger S, Brooks DG. 2012. Emergence of distinct multiarmed immunoregulatory antigen-presenting cells during persistent viral infection. Cell Host Microbe 11: 481–491. 10.1016/j.chom.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Vannella KM. 2016. Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44: 450–462. 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki C, Sugiyama M, Ohta T, Hemmi H, Hamada E, Sasaki I, Fukuda Y, Yano T, Nobuoka M, Hirashima T, et al. 2013. Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1. J Immunol 190: 6071–6082. 10.4049/jimmunol.1202798 [DOI] [PubMed] [Google Scholar]
- Yang Z, Ming XF. 2014. Functions of arginase isoforms in macrophage inflammatory responses: impact on cardiovascular diseases and metabolic disorders. Front Immunol 5: 533 10.3389/fimmu.2014.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188: 2205–2213. 10.1084/jem.188.12.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: Article17 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- Zhou X, Lindsay H, Robinson MD. 2014. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res 42: e91 10.1093/nar/gku310 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.