Endophytes are particularly fascinating because of their multifaceted lifestyle, i.e., they may exist as either free-living soil microbes or saprobic ones or pathogens. Endophytic communities of halophytes may be different than those in other plants because salinity acts as an environmental filter. At the same time, they may contribute to the host’s adaptation to adverse environmental conditions, which may be of importance in agriculture.
KEYWORDS: 16S rRNA and ITS amplicon sequencing, endophyte, halophyte, microbial community structure, soil salinity
ABSTRACT
We examined Salicornia europaea, a nonmycorrhizal halophyte associated with specific and unique endophytic bacteria and fungi. The microbial community structure was analyzed at two sites differing in salinization history (anthropogenic and naturally saline site), in contrasting seasons (spring and fall) and in two plant organs (shoots and roots) via 16S rRNA and internal transcribed spacer amplicon sequencing. We observed distinct communities at the two sites, and in shoots and roots, while the season was of no importance. The bacterial community was less diverse in shoot libraries than in roots, regardless of the site and season, whereas no significant differences were observed for the fungal community. Proteobacteria and Bacteroidetes dominated bacterial assemblages, and Ascomycetes were the most frequent fungi. A root core microbiome operational taxonomic unit belonging to the genus Marinimicrobium was identified. We detected a significant influence of the Salicornia bacterial community on the fungal one by means of cocorrespondence analysis. In addition, pathways and potential functions of the bacterial community in Salicornia europaea were inferred and discussed. We can conclude that bacterial and fungal microbiomes of S. europaea are determined by the origin of salinity at the sites. Bacterial communities seemed to influence fungal ones, but not the other way around, which takes us closer to understanding of interactions between the two microbial groups. In addition, the plant organs of the halophyte filter the microbial community composition.
IMPORTANCE Endophytes are particularly fascinating because of their multifaceted lifestyle, i.e., they may exist as either free-living soil microbes or saprobic ones or pathogens. Endophytic communities of halophytes may be different than those in other plants because salinity acts as an environmental filter. At the same time, they may contribute to the host’s adaptation to adverse environmental conditions, which may be of importance in agriculture.
INTRODUCTION
Halophytes are salt-tolerant plants that usually grow and survive in environments with salt concentrations as high as 1 M (1). They may play a role in the remediation of salt-affected (2) and heavy-metal-contaminated (3) environments and are widely utilized as an option to tackle the worldwide problem of decreasing area of cultivated lands (4).
The salt accumulating obligate halophyte Salicornia europaea L. (Amaranthaceae) is commonly found in coastal and inland salt marshes (5) and is cultivated in some countries for its culinary values (6). Its geographical distribution spans four continents: North America, Asia, Africa, and Europe (7). Research on halophyte plants is of particular interest today due to their diverse strategies to survive in such harsh environments. Halophytes developed several primary and secondary mechanisms to cope with salinity (8, 9). The primary mechanisms include osmotic adjustment, the exclusion of Na+ from the cell or plant tissue, or the isolation of Na+ in the vacuole (8). The secondary mechanisms consist of associated microorganisms that ameliorate plant growth and fitness, particularly under stress conditions (9). These microorganisms include arbuscular mycorrhizal fungi (AMF) that are known to protect plants under stress conditions by providing nutrients and maintain a better ion balance (10). However, some halophytes belong to typical nonmycorrhizal plants, e.g., Armeria maritima, Limonium vulgare, Juncus gerardii, and Triglochin maritima (11). Only a few publications revealed very low (<1%) colonization of these halophytes by AMF (11–13). However, even these results can be dubious due to the possibility of spores coming from adjacent mycorrhizal plants being nonspecifically attached to otherwise-noncolonized roots. Given the uncertainty of AMF associations with the roots of S. europaea, specific and unique endophytes inhabiting this halophyte could compensate for missing symbiotic protection against, e.g., abiotic stress or nutrient deficiency (14, 15). Endophytes are a group of microorganisms (include bacteria, fungi, and actinomycetes) that colonize the internal tissues of healthy living plant hosts without causing harm or symptoms of disease (16). They coevolve in their plant host to adapt themselves in the plant environment. Some endophytes produce phytochemicals, bioactive secondary compounds to increase plant growth and development, as well as improve plant host fitness during abiotic and biotic stress conditions (reviewed in references 9 and 17).
In this study, we assessed two sites differing in salinization history. One of them is a naturally saline site where brines emerge from Zechstein salt deposits, giving the plants an opportunity to coevolve with halophilic microorganisms for a long time (18). At the other site, salinity originated from soda factory wastewater ponds and has lasted for only 50 years (19). The two sites are only 40 km from one another, and the soil physicochemistry is similar, apart from the salinity origin. This system gives a unique opportunity to observe differences due to the various times of plant host-microbiome coevolution. Therefore, we first hypothesized that the Salicornia endophytic community composition at the two sites would be different, but a set of common microorganisms (a core microbiome) can be delineable.
Plants can provide niches for microbes under unfavorable environmental conditions and assist the microbes in reducing environmental stress. There is clear evidence on the role of endophytes from stressed environments and the benefits their host receives in this association (20). This is why our second hypothesis is that S. europaea, belonging to a small group of nonmycorrhizal plants, would bear a unique assemblage of bacterial and fungal endophytes being halotolerant that perform various ecological roles in protecting the host plant under saline conditions.
Although metagenomic studies on bacterial or fungal endophytes of crop species have been performed (21–23), few reports have discussed the composition of endophytic community in species with the ability to accumulate salts (24, 25), and no studies have simultaneously analyzed the two communities. Thus, our third and last hypothesis evaluated whether there was an influence of the bacterial endophytic community on the fungal one and vice versa.
To address all three hypotheses, we used a culture-independent (Illumina sequencing of 16S rRNA and internal transcribed spacer [ITS] amplicons) approach to simultaneously analyze the bacterial and fungal endophytic community in S. europaea. The results obtained can contribute to agriculture technologies, leading to enhanced production of nonmycorrhizal crops in saline soils.
RESULTS
Soil physicochemical analysis.
Table 1 lists the soil characteristics of the two sampling sites, site 1 (S1) and site 2 (S2), during two seasons: fall and spring. The pH was neutral and nearly the same at both sites during the two seasons. In addition, carbonates (%) and total organic carbon (TOC; %) were similar at both sites. The S1 soil had significantly higher Na+, Cl–, CaCO3, Mg2+, and ECe values (two-way analysis of variance [ANOVA], P < 0.05) and significantly lower levels of Ca2+ (two-way ANOVA, P < 0.05) than S2 soil. There were no significant differences in other physicochemical properties between the S1 and S2 soils.
TABLE 1.
Chemical parameters of soils from the two sampling areas: site 1 and site 2 in fall 2015 and spring 2016
| Parameter | Mean ± SEM (n = 9)a
|
|||
|---|---|---|---|---|
| Site 1 |
Site 2 |
|||
| Fall 2015 | Spring 2016 | Fall 2015 | Spring 2016 | |
| ECe | 100.5 ± 27.6B | 51.1 ± 12.7A | 76.0 ± 19.5C | 59.7 ± 12.2A |
| pHe | 6.8 ± 0.1A | 7.8 ± 0.1B | 6.9 ± 0.1A | 7.3 ± 0.1B |
| Na+ (g ⋅ dm−3) | 21.5 ± 7.9A | 9.2 ± 2.4C | 11.8 ± 7.4B | 7.4 ± 2.1CB |
| Cl− (g ⋅ dm−3) | 65.3 ± 21.6A | 30.8 ± 5.9C | 44.1 ± 13.4B | 34.2 ± 5.6C |
| Ca2+ (g ⋅ dm−3) | 4.2 ± 3.5B | 0.9 ± 0.2C | 8.1 ± 3.3A | 7.6 ± 1.5A |
| K+ (g ⋅ dm−3) | 0.4 ± 0.2A | 0.2 ± 0.0C | 0.2 ± 0.2B | 0.2 ± 0.1C |
| Mg2+ (g ⋅ dm−3) | 0.5 ± 0.2A | 0.2 ± 0.1B | 0.3 ± 0.2B | 0.1 ± 0.0C |
| SO42− (g ⋅ dm−3) | 0.3 ± 0.085A | 0.8 ± 0.2B | 0.1 ± 0.1A | 0.6 ± 0.3C |
| HCO3− (g ⋅ dm−3) | 0.1 ± 0.0A | 0.2 ± 0.1A | 0.1 ± 0.0A | 0.1 ± 0.0A |
| SP (%) | 94.5 ± 14.1A | 83.0 ± 9.3B | 89.4 ± 10.5B | 133.1 ± 48.6C |
| TOC (%) | 5.9 ± 2.5A | 4.8 ± 3.1A | 7.5 ± 5.5B | 3.3 ± 2.4A |
| CaCO3 (%) | 39.4 ± 7.1A | 33.9 ± 9.4A | 28.4 ± 10.5B | 23.1 ± 2.0C |
Values labeled with the same superscript capital letters are not significantly different (P ≤ 0.05). Abbreviations: ECe, electrical conductivity of the saturated extract; TOC, total organic carbon; SP, saturation percentage.
Sequencing results.
A total of 1,841,573 high-quality sequences were retrieved after denoising, merging, and chimera checking. The number of reads per sample ranged from 2,031 to 36,430. After separating bacterial and fungal reads, 236,835 bacterial sequences (2 to 13,125 per sample) and 260,575 fungal sequences (6 to 14,969 per sample) were left. We eliminated 3 bacterial and 15 fungal libraries due to not reaching the required number of sequences (500 for bacterial and 300 for fungal libraries).
Alpha- and beta-diversity in S. europaea endophytic communities.
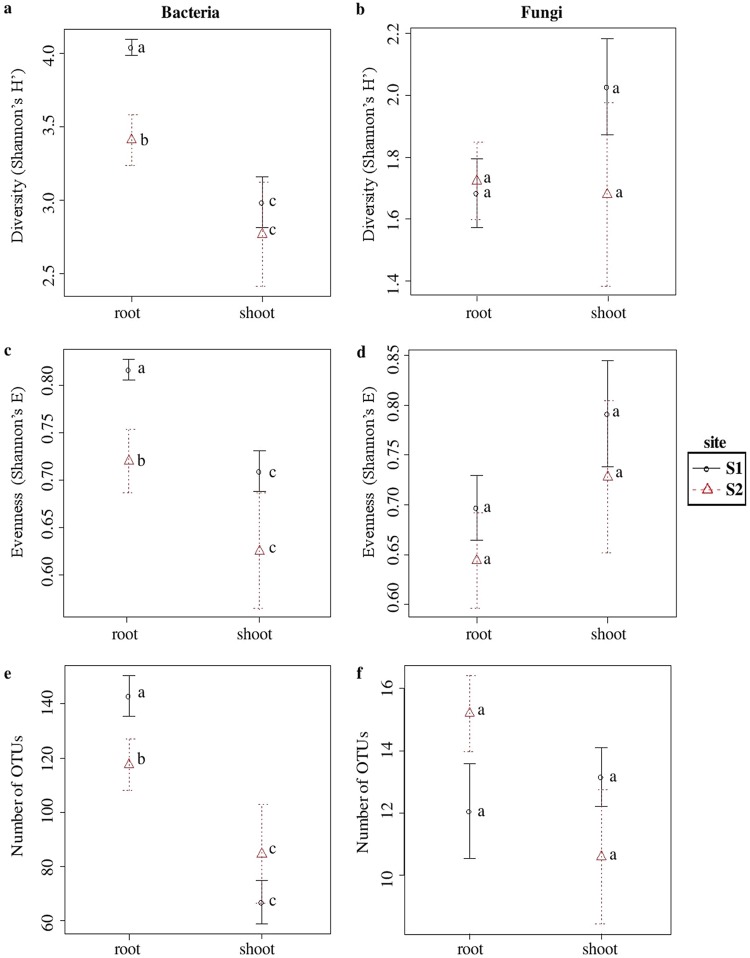
No differences due to the season were observed in any of measured alpha-diversity indices both for bacterial and fungal communities. The bacterial Shannon’s diversity (H′) and evenness (E), as well as species richness, were lower in libraries derived from S. europaea shoots than in those from roots, regardless of site (Fig. 1a, c, and e). Differences were significant (ANOVA, P < 0.05) in the case of the S1 site, while for S2 they were not significant (ANOVA, P > 0.05). Unlike bacterial communities, no significant differences due to organs and sites were found for fungi. However, a tendency of greater diversity in shoots was visible for S2 libraries (Fig. 1b, d, and f).
FIG 1.
Species richness, diversity, and evenness in different test sites and plant organs for OTUs constructed at 0.03 dissimilarity for bacterial and fungal sequences. (a and b) Shannon’s H′; (c and d) Shannon’s E; (e and f) observed number of OTUs. Robust ANOVA test with the Tukey’s post hoc analysis was used to assess the significance of differences between test sites and plant organs. Variants labeled with the same letters are not significantly different (P ≤ 0.05).
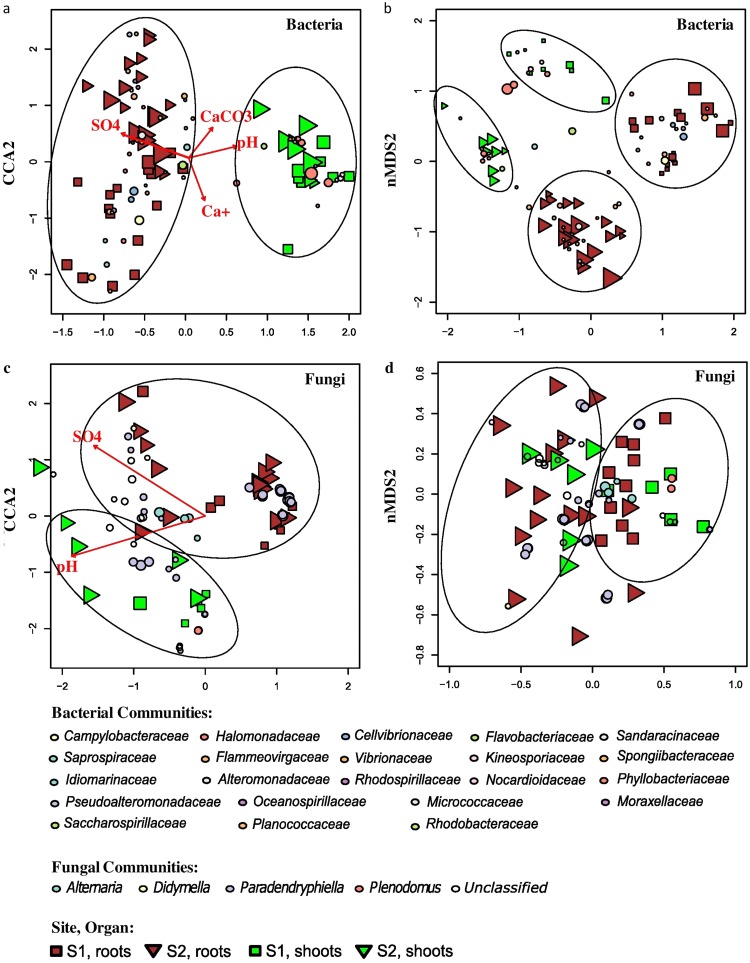
Beta-diversity analysis revealed that bacterial communities clustered according to site and organ in nonmetric multidimensional scaling (NMDS) (Fig. 2b), the grouping was significant (permutational analysis of variance [PERMANOVA], P < 0.01), and the differences in variance were not significant (PERMDISP, P > 0.01). Notably, the samples did not cluster according to the season. The distance between bacterial communities in the ordination plot for the roots was larger compared to the shoots. The canonical correspondence analysis (CCA) showed that Ca+, SO42+, CaCO3, and pH were the significant physicochemical variables influencing the community structure (Fig. 2a; P < 0.01). These variables explained 22.65% of the variance in the communities. The variations observed in the endophytic bacterial communities of the roots in S1 and S2 were distinctly high compared to the community present in the shoots.
FIG 2.
Analysis of log-transformed and Wisconsin double-standardized Bray-Curtis dissimilarity matrix for endophytic bacterial and fungal communities associated with S. europaea, respectively. (a and c) CCA (canonical correspondence analysis); (b and d) NMDS (nonmetric multidimensional scaling analysis). Circles represent OTUs; their fill color denotes consensus taxonomy at the family level (bacteria) and genus level (fungi). The size reflects abundance. Squares, S1 samples; triangles, S2 samples; green, shoots; brown, roots. Arrows in the CCA graphs denote soil parameters that were significantly associated with the community structure.
Unlike bacterial ones, fungal communities clustered according to the site only (Fig. 2d), and this grouping was significant (PERMANOVA, P < 0.01), while variance was homogenous (PERMDISP, P > 0.01). pH and SO42+ were the only physicochemical variables that significantly influenced the communities (Fig. 2c; P < 0.01). These variables explained 7.07% of the variance in the communities. In CCA, the fungal community variations were explained only by plant organs and not by the two different sites, confirming the results of PERMANOVA.
Core microbiome.
A single bacterial OTU12 (a member of the Marinimicrobium genus) was found to exceed the abundance threshold in all root samples and might comprise Salicornia roots core microbiome. However, there were neither bacterial nor fungal operational taxonomic units (OTUs) with abundance greater than 0.5% in all samples.
Bacterial and fungal community composition.
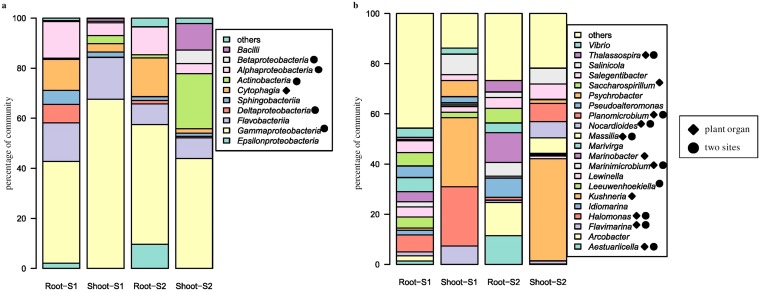
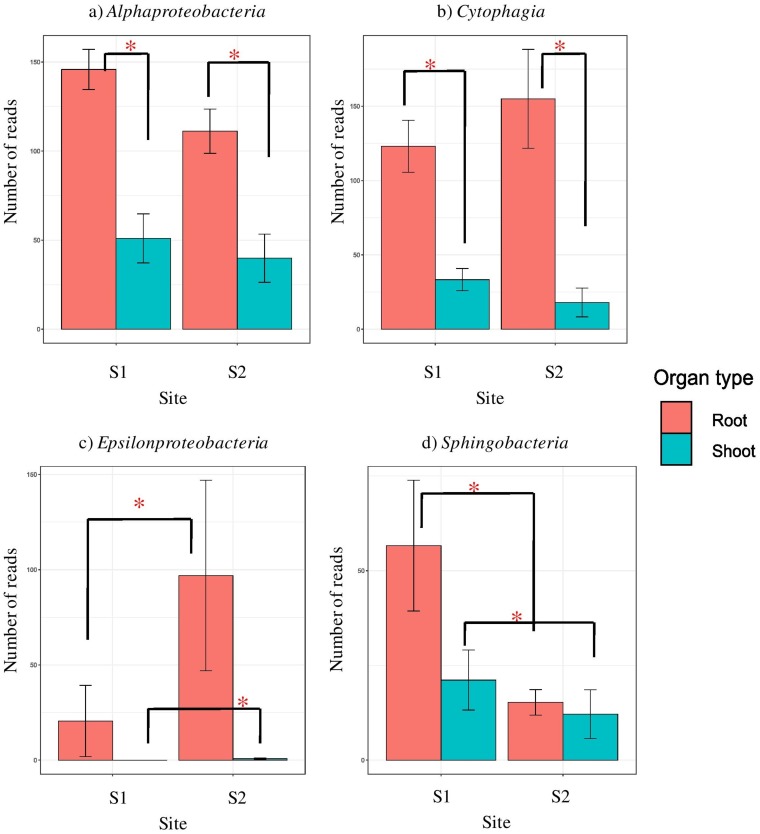
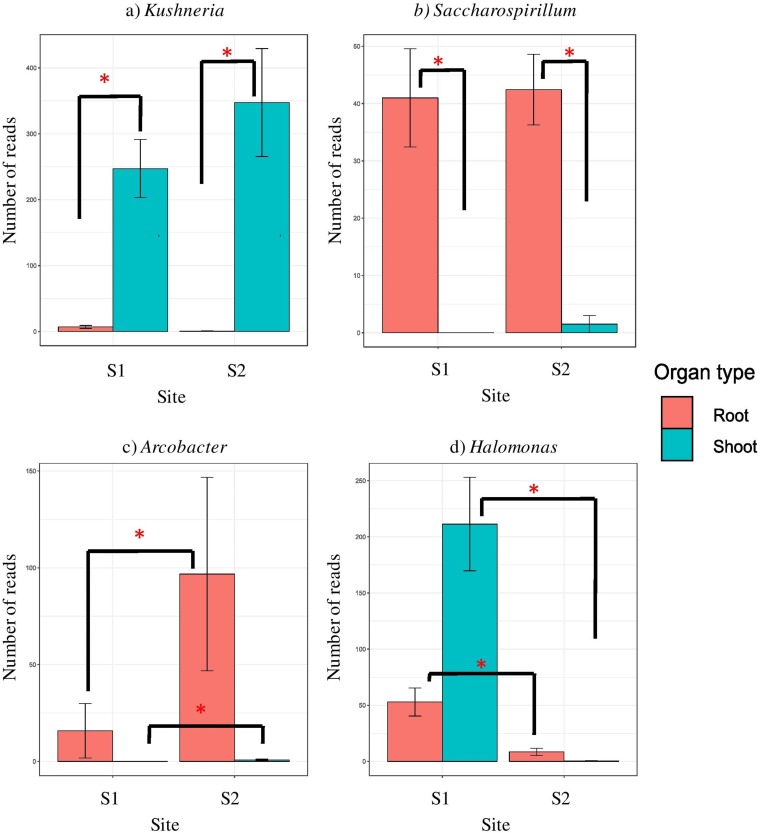
Bacterial libraries were dominated by Proteobacteria- and Bacteroidetes-derived reads (see Fig. S3A in the supplemental material). At the level of class, all the libraries, regardless of site and organ, were composed mainly of Gammaproteobacteria and Flavobacteriia, with minor amounts of Alphaproteobacteria, and Cytophagia (Fig. 3). The latter two classes were more frequent in roots than in shoots (Fig. 4a and b). Certain classes were specific for some sample types. Sphingobacteriia were found only in the S1 libraries (Fig. 4d), while Epsilonproteobacteria were characteristic for S2 (Fig. 4c). Deltaproteobacteria were specific for roots at S1 and Betaproteobacteria as well as Bacilli were characteristic for shoots at S2 (Fig. 3a). At the level of family, Halomonadaceae were much more abundant in shoots, whereas Alteromonadaceae, Cellvibrionaceae, Flammeovirgaceae, Rhodobacteraceae, and Saccharospirillaceae were characteristic for roots. Vibrionaceae and Sandaracinaceae were found exclusively at S1 (Fig. S3B). Thirty-one genera were found in the libraries, and shoot libraries were more similar between sites than root ones. Kushneria was characteristic for and abundant in shoots (Fig. 5a), while Saccharospirillum was a hallmark of roots (Fig. 5b). Halomonas, Levinella, Vibrio, Pseudoalteromonas, and Leuweenhoekiella were found exclusively at S1 (Fig. 5d). Arcobacter was present in minor quantities, but it was more abundant at S2 (Fig. 5c).
FIG 3.
Endophytic bacterial community structure at class (a) and genus (b) levels among the two test sites and plant organs.
FIG 4.
Significantly represented bacterial classes. (a) Alphaproteobacteria; (b) Cytophagia; (c) Epsilonproteobacteria; (d) Sphingobacteriia. Whiskers denote the standard errors of the mean. Significant differences (P < 0.05), assessed using robust ANOVA and Tukey’s HSD test, are indicated by asterisks.
FIG 5.
Significantly represented bacterial genera. (a) Kushneria; (b) Saccharospirillum; (c) Arcobacter; (d) Halomonas. Whiskers denote standard errors of the mean. Significant differences (P < 0.05), assessed using robust ANOVA and Tukey’s HSD test, are indicated by asterisks.
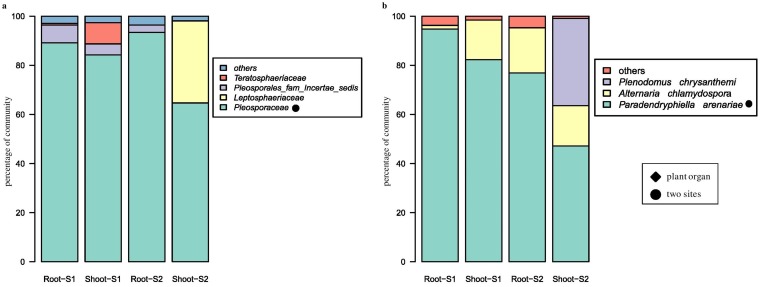
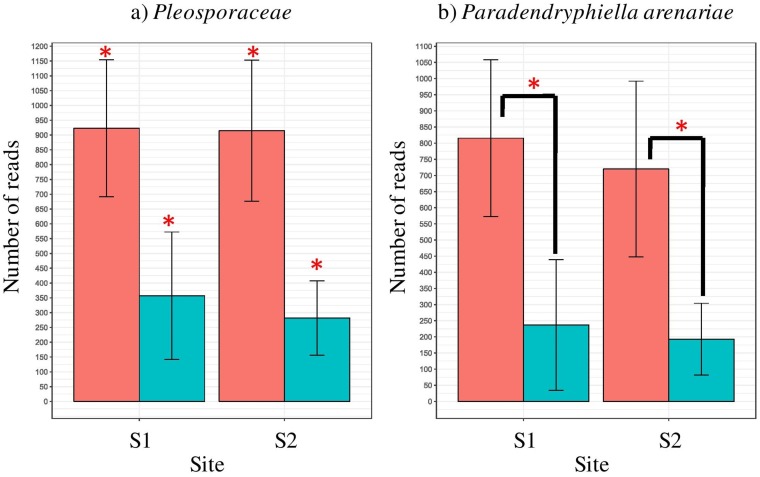
At high phylogenetic levels, no differences between sites and organs were visible in fungal communities: libraries were composed of over 95% Ascomycota reads derived from Dothideomycetes class (Fig. S4A and B). Pleosporaceae was the dominating family in all sample types; however, it was more abundant in roots than in shoots. Pleosporales fam. incertae sedis was found in all sample types but shoots at S2, albeit in small amounts (Fig. 6a). Leptosphaeriaceae, Teratosphaeriaceae, and Didymosphaeriaceae were found exclusively in shoots, the first of them at S2, while the remaining two at S1. Over 80% of fungal reads were classified down to the species level (Fig. 6b). Paradendryphiella arenariae was the only species present in all sample types, but more frequent in roots (Fig. 7b). An unclassified member of the Pleosporaceae made up a significant portion of the community in shoots but was also found in roots at S1 (Fig. 7a), whereas Alternaria chlamydospora was found in shoots and roots at S2.
FIG 6.
Endophytic fungal community structure at family (a) and species (b) levels among the two test sites and plant organs.
FIG 7.
Significantly represented fungal taxa. (a) Pleosporaceae; (b) Paradendryphiella arenariae. Whiskers denote standard errors of the mean. Significant differences (P < 0.05), assessed using robust ANOVA and Tukey’s HSD test, are indicated by asterisks.
Bacterial metabolic pathways differentially represented in roots and shoots.
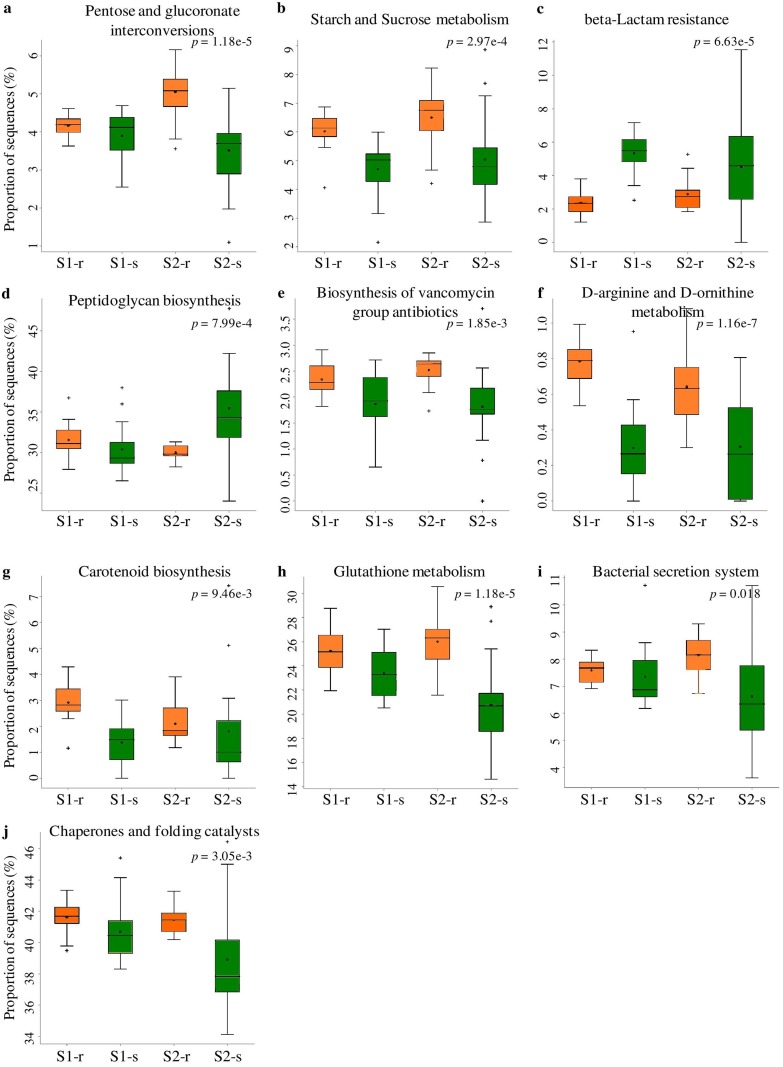
Possible bacterial metagenomes were imputed using PICRUSt with GreenGenes as the underlying database. The nearest sequenced taxon index (NSTI) values were around 0.05 in the shoot and 0.09 in root samples, regardless of site and season. This difference was statistically significant (ANOVA, P < 0.01, F = 23.071). Ten level-3 pathways were differently represented in simulated metagenomes (Fig. 8): two of them belonged to carbohydrate metabolism (pentose-glucuronate interconversions, starch and sucrose metabolism) and three to antibiotic resistance and biosynthesis (beta-lactam resistance, peptidoglycan biosynthesis, and vancomycin biosynthesis), and the remaining ones came from host interactions (bacterial secretion system), amino acid metabolism (arginine and ornithine metabolism), protein biosynthesis (chaperones and folding catalysts), and regulation of redox potential (glutathione metabolism), as well as secondary metabolite biosynthesis (carotenoid biosynthesis). The majority of the pathways were overrepresented in root sample metagenomes; only beta-lactam resistance and peptidoglycan biosynthesis were overrepresented in shoots.
FIG 8.
PICRUSt analysis of bacterial sequences. Box-and-whisker plots for most significantly different KEGG Orthology (KO) categories. Error bars represent standard errors of the mean of each given category abundance in different sample types. For sites S1 and S2, “-r” indicates “root” and “-s” indicates “shoot.”
Interactions between endophytic bacterial and fungal communities.
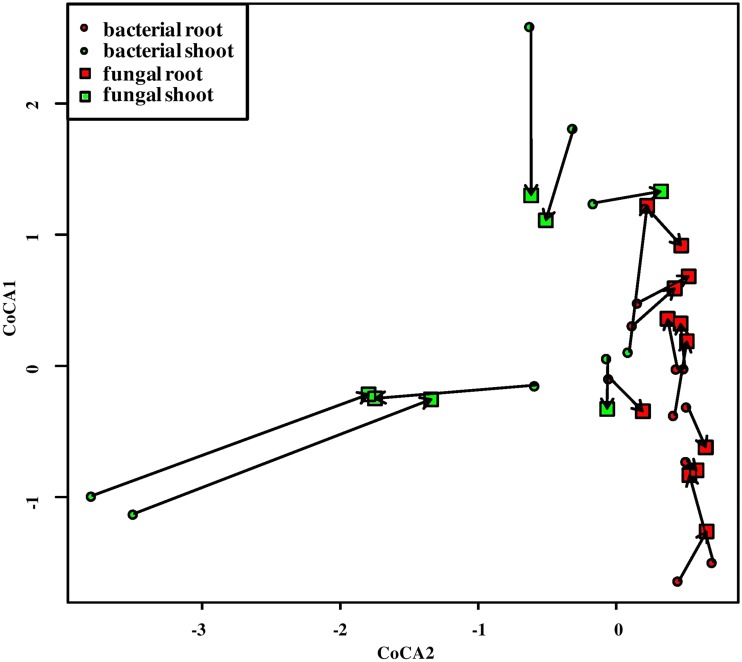
Cocorrespondence analysis (CoCA) indicated that there was a significant influence of the bacterial community on the fungal one and vice versa. In other words, the fungal community might be predicted based on the bacterial one. Leave-one-out cross-validation showed that two first axes were sufficient, and permutational testing demonstrated that only the first one was significant (permutest, P < 0.01). Distances between respective bacterial and fungal communities were significantly greater for shoot samples (1.21) than for roots (0.40) (ANOVA, P < 0.01, F = 13.73; Fig. 9), indicating that the influence was greater in the latter ones. Bacterial community explained 9.91% of the variance in the fungal one, while, in spite of insignificant influence, fungi explained 24.93% of the variance in bacterial assemblages.
FIG 9.
CoCA of bacterial versus fungal communities’ plot of sites. Bacterial sites are displayed as circles, fungal ones as squares. The circle color denotes organ: green, shoots; and red, roots. Respective bacterial and fungal communities are connected with arrows. Arrow length conveys the strength of community association: the longer the arrow, the weaker the association.
DISCUSSION
Salinization history is a critical factor for S. europaea endophytic community structure.
Endophytes can play a major role in plants response to abiotic stresses (e.g., salinity [20]); however, unfavorable environmental factors can affect their diversity and colonization density. In our work, we have observed significant differences in the level of soil salinity between fall and spring at both test sites, but seasons did not affect the distribution of bacterial and fungal endophytes in plant organs. This is contrary to the reports on endophytes of nonhalophytic plants, e.g., Quercus ilex, Tinospora cordifolia, Salix alba, S. caprea, and Betula pendula, where endophytes were investigated (26–28). The reason can be that the interior of the halophyte tissues provides a relatively stable and protective environment for diverse communities of endophytic microbes compared to the saline soil, which is subject to wide fluctuations in the osmotic potential (29).
The diversity of endophytic fungi classified to the genus level in our study was low compared to the diversity seen in endophytic bacterial communities. Previous data revealed that bacterial communities in the rhizosphere exhibit greater richness than endophytes in the organs of halophytes (26, 27). The composition of fungal communities can be affected, as in the case of bacteria, by the soil characteristics. The type of soil acts as a filter for microbial species and, since most endophytes derive from soils, the soil characteristics play an important role (30). While the pH and soil texture are often correlated with the soil bacterial community composition, the fungal community is rather associated with the changes in soil nutrient status (31). Analysis of root endophytic fungal colonization in the halophytes Salicornia patula and Arthrocnemum macrostachyum revealed that fungal colonization was affected by the differences in soil salinity and host age (32). The relatively low microbial diversity in the shoots of S. europaea may be due to the selection imposed by a large salt concentration in this organ.
The differences in bacterial community composition in roots and rhizosphere of S. europaea at the test sites (S1 and S2) investigated in this study were also observed by Szymańska et al. using the phospholipid fatty acid analysis method (33). These researchers observed a higher microbial biomass at S1, where the salinity is of natural origin and occurred earlier than at S2.
Because endophytes originate predominantly from the soil, we show that the differences in origin of salinity and ecological characteristics of the test sites may influence endophytic community structure and that soil properties may be an important selection factor shaping their pools (31, 32). Supporting this view, a clear influence of soil properties on the existence of unique endophytes of S. europaea, specific to the geographical regions in different countries, such as Japan (34), Slovenia (12), Canada (35), China (25, 36, 37), and Poland (38), was demonstrated. On the other hand, seasons did not influence S. europaea endophytic community structure in spite of differences in salinity. This might mean that it was not the salinity itself, but the salinization history, which was causing this variation. Alternatively, the salinity effect might have been so strong that it overrode the influence of seasons.
Differences in the origin of salinity (natural versus anthropogenic) and in consequence the time of microbial community development might be a cause of only Marinimicrobium OTUs being detected as a core microbiome in roots of S. europaea. Bacteria of the genus Marinimicrobium (Gammaproteobacteria) are halophilic and/or halotolerant, are strictly aerobic (39), and were never found to live in planta. The representatives of this genus were previously isolated from hypersaline surface sediments of the southern arm of Great Salt Lake (Utah) (40), tidal flat sediment of the South Sea in South Korea (the Korea Strait) (39), and the marine solar saltern of the Yellow Sea, South Korea (41). Further work is required to determine the role of Marinimicrobium in halophytes.
Salinity acts as an environmental filter for S. europaea bacterial endophytic community both in shoots and roots.
The overall patterns of endophyte abundance, richness, and composition were not only influenced by test sites but also differed between organs. A similar situation was observed in many other plants, e.g., Tinospora cardifolia (27). In the present study, we analyzed both the roots and shoots and found higher endophytic community diversity in the former. These results are consistent with those reported by Mora-Ruiz et al. in the halophyte Arthrocnemum macrostachyum (42). The greater species richness in roots might be due to their contact with soil, given that most endophytes are derived from soil (43) and migrate from the roots (primary site of interaction between plants and soil) to other tissues (44). Although Salicornia is a nonmycorrhizal plant, the abundance and specificity of fungal root endophytes can substitute for the presence of typical mycorrhizae by providing specific functions, e.g., mediating in nutrient transfer from soil into the plant.
Bacterial root endophytes of S. europaea were dominated by Proteobacteria, specifically class Gammaproteobacteria. The greater abundance of Proteobacteria identified among halotolerant endophytes compared to rhizospheric soil was also observed in studies of Szymanska et al. involving S. europaea (45) and another halophytic plant, Aster tripolium L. (33), analyzed using culture-dependent methods. The most abundant phyla of endophytic bacteria observed in our work were also commonly found in other halophytes, e.g., Halimione portulacoides or Salsola imbricate (46, 47). Thus, we suggest that they may play an important role in the ecology of halophytes. The genus Kushneria (family Halomonadaceae) was found to be characteristic for the shoots of S. europaea. This genus is halophilic and mainly found in saline and hypersaline habitats (48). The bacterial microbiome composition in the rhizosphere of Salicornia plants and bulk soils collected by Mapelli et al. (49) from hypersaline ecosystems in Tunisia unveiled the occurrence of the high diversity of Halomonadaceae members. That study suggests that Kushneria acts as a plant growth promoter that is able to fix atmospheric nitrogen, produce ammonia, display protease activity, and play a role as a biocontrol agent. We found Saccharospirillum to be characteristic for the roots of S. europaea. This genus belongs to the family Saccharospirillaceae and is moderately halophilic, mesophilic, and facultatively alkaliphilic, growing at salinities of 0.5 to 5 % (wt/vol) NaCl, with an optimum at 2 % (50). Moderate halophilic nature may explain its higher abundance in roots than in the salt-accumulating shoots.
The PICRUSt tool was used to evaluate potential functions of the bacterial community, which may enable us to understand how S. europaea associated microbiome is able to adapt to high-salt environments and also predict potential ecological roles of the observed organisms. However, the results of such an analysis need to be interpreted with care, since this tool passes on all the biases inherent to amplicon sequencing, as well as depends on a database for unknown organism genome reconstruction. Ten metabolic pathways were differently represented between plant organs. Namely, pathways related to carbohydrate metabolism, e.g., sucrose metabolism, were more common in roots. This might be caused by the greater diversity of complex carbohydrates in roots (51) that would need various enzymes to be utilized by bacteria (52). The complex carbohydrates might be digested to simpler sugars serving as osmolytes and participating in osmotic adjustment (53). Likewise, exposure to frequent salt stress may explain why the bacterial endophytes in this study have an abundance of genes associated with dormancy/sporulation or stress proteins (54). Endophytes escape plant’s defense mechanisms by activating genes involved in glutathione metabolism. Glutathione maintains the proper oxidation state of protein thiols and protects cells from the action of low-pH, chlorine compounds and oxidative as well as osmotic stresses (55). Carotenoid pigments are essential for photosynthetic growth in higher plants and protection against photooxidation (56). The endophytic bacteria may probably contribute by increasing the carotenoid content in the host (57). The simulated bacterial metagenome contained a high number of genes encoding enzymes potentially involved in the detoxification of reactive oxygen species (ROS), particularly in the regulation of redox potential. Plants produce a range of ROS in response to abiotic stress (e.g., salt stress) or colonizing microorganisms that elicit an oxidative burst. Since we cultured many potentially pathogenic fungi from the plant material used in this study (B. U. Furtado, S. Szymańska, and K. Hrynkiewicz, unpublished work), we suggest that mitigation of excess oxidative stress by endophytic bacteria may be beneficial for the host. The greater frequency of oxidative stress response genes in root bacterial communities suggests that the plant response to bacteria is the more important source of stress than salinity, which is higher in shoots (58). However, why the genes involved in the rest of pathways are apparently more common in roots than in shoots still remains to be elucidated.
The root endophytic fungi were dominated by phylum Ascomycota, and among them, the family Pleosporaceae turned out to be the most abundant in this study. This is consistent with other studies where pleosporalean fungi were found to be the dominant colonizers in halophytes (34) and in plants grown under arid conditions (59). These fungi are common endophytes in both coastal and inland arid soils (60). The references mentioned above suggest an important role of endophytes from the family Pleosporaceae in halophytic plants. However, mutualistic interaction between these fungi and plants are not confirmed so far. Among the fungi, Paradendryphiella arenariae was the only species present in all sample types in this study but found more frequently in roots. This finding is consistent with earlier reports on this species that was isolated from S. europaea roots in Japan (34). Alternaria chlamydospora was found in the shoots and roots only at the less-saline site, S2. Muhsin and Booth studied the composition of Alternaria assemblages in six different halophytes—Atriplex patula L., Glaux maritima L., Hordeum jubatum L., Puccinellia nuttalliana, S. europaea, and Suaeda depressa—throughout the growing season. Alternaria alternata and A. chlamydospora turned out to be specific for S. europaea (35).
Notably, some endophytic microorganisms found in our study were only reported from marine environments to date. These are the predominant Salicornia endophytic bacteria belonging to Gammaproteobacteria, such as Aestuaricella, Marinimicrobium, Pseudoalteromonas, and Salegentibacter. We also describe fungal endophytes, such as Paradendryphiella, Neodevriesia, and Neocamarosporium, that were reported in many other halophytic plants (see Table S1A and B in the supplemental material).
The fungal community may be predicted based on the bacterial one.
Prior studies on the endophytic community in plants focused either on bacteria or fungi. However, as no organism thrives in isolation, the interaction of bacterial and fungal communities in different plant species can be of great relevance. The significant influence of the Salicornia bacterial community on the fungal one was found by means of cocorrespondence analysis (CoCA). Bacteria may positively affect fungal activity by producing cellulases and pectinases that increase accessibility of substrates to the fungi (61). In addition, bacteria may also decompose solutes that are toxic to particular fungi (61) and may increase the nitrogen levels available for them (62). Many researchers hypothesized that a mutualistic relationship exists, in which the bacteria utilizing fungal exudates supply nitrogen to fungi (63). Several cases of stimulation of spore germination by spore-associated bacteria have been reported (64, 65). Finally, a multitude of antifungal strategies has been revealed in bacteria, including the production of inhibitory factors such as HCN, antibiotics, lytic enzymes, and volatiles, as well as nutrient-sequestering factors such as iron-chelating siderophores (66–68). Fungi also influence bacteria by facilitating the penetration of bacteria into leaf tissue, where both groups of microorganisms can degrade specific polymers into smaller molecules that are subsequently assimilated (61). Furthermore, fungal hyphae can act as a carrier for bacterial transport, enabling bacteria to colonize new niches faster (69). The exact mechanism and direction of the two communities' influence on one another still remains to be elucidated and would require further studies, such as metatranscriptomic ones.
Conclusion.
Our results highlight the fact that the history of salinization, and not the salinity itself, is the factor causing differences in community structure between investigated sites. Saccharospirillum was characteristic for the roots, and Kushneria was found mainly in the shoots, while Paradendryphiella arenariae was the most common among fungal endophytes of S. europaea. Moreover, the Salicornia response to microbes seemed to be a more important source of oxidative stress for bacteria than salinity. Finally, our results showed that the bacterial and fungal communities interact with each other, which led us to speculate that they are codependent, which initiates the need for further research.
MATERIALS AND METHODS
Site description and plant sample collection.
Two saline sites were sampled during two seasons (fall 2015 and spring 2016) (see Fig. S1 in the supplemental material). The sites are located in Kujawy region in Central Poland. Site 1 (S1; N52°53′, E18°47′) is located in the vicinity of three brine concentration towers in the Spa Park in Ciechocinek. This natural area favors the salt-marsh vegetation since it is associated with the occurrence of salt springs and saline groundwater in connection with Zechstein rock-salt deposits that are uplifted in the form of salt domes (18). Site 2 (S2; N52°48′, E18°15′) is a meadow located next to waste ponds of the soda factory in Inowroclaw. The long-term deposition of semifluid waste from the factory into sedimentary ponds situated directly on permeable grounds has led to groundwater pollution (19). Three plots (10 × 10 m, biological replicates) were chosen at random at each site. Three blocks of soil (20 × 20 × 20 cm) were randomly sampled from each plot in each of the two time points: fall (F; 21 September 2015) and spring (S; 9 June 2016). The blocks were immediately transferred to the laboratory and processed. The plants were gently uprooted and washed with sterile distilled water and surface sterilized according to Szymańska et al. (45) (Fig. S2). Approximately 500 mg of shoots and roots per each variant of the experiment was weighed and stored in 2-ml vials at –80°C for further processing.
Physicochemical analysis of soils.
The soil samples were air dried and passed through a 2-mm mesh sieve. The soil was analyzed according to the following methods. The TOC content was determined using a CNS Variomax analyzer, the CaCO3 content was determined by the Scheibler volumetric method (70), the electrical conductivity (ECe) was determined by conductometry, the pHe was determined potentiometrically, and the saturation percentage (SP) was determined gravimetrically as described by van Reeuwijk (71). The concentrations of ions in the extract were determined as follows: Na+, K+, Ca2+, and Mg2+ by the AAS method, Cl– argentometrically, SO42– turbidimetrically, and HCO3– acidimetrically (71).
DNA extraction, library construction, and sequencing.
Portions (500 mg) of frozen plant tissue samples were homogenized with a bead beater (FastPrep-24 MP Biomedicals), and total DNA extraction was performed using a DNeasy plant minikit (Qiagen) according to the producer’s protocol. The DNA was quantified fluorometrically (Qubit 2.0), and the quality was assessed spectrophotometrically (NanoDrop 2000).
A two-step PCR (72) was carried out in order to generate bacterial 16S rRNA and fungal ITS amplicon libraries. Amplification, purification, quantification, and sequencing were performed as described earlier by Thiem et al. (73). Briefly, in the first-PCR-round V3-V4 fragments of bacterial 16S rRNA genes and ITS regions of eukaryotic ribosomal DNA (rDNA) were amplified with the 357f/786r primer pair (72), and ITS1 and ITS2 (74) were amplified with M13/M13R overhangs using metagenomic DNA as the template. In the second round, the amplicons were converted to the Illumina sequencing library by PCR with M13/M13R primers bearing barcodes and P5/P7 adapters. Libraries were purified twice with SPRI beads (GE Healthcare), quantified using the KAPA library quantification kit for Illumina (Roche), and sequenced using MiSeq (Illumina) with custom sequencing primers being high-pressure liquid chromatography-purified versions of the first-round primers for R1 and R2 and reverse complement of the reverse first-round primer for the index read.
Bioinformatics and statistical analysis.
Raw reads were denoised and merged using the DADA2 R package to yield amplicon sequence variants (ASVs) and information on their abundance (75). Then, the denoised sequences were separated into bacterial and nonbacterial (ITS) sequences via classification with the naive Bayesian classifier implemented in mothur (76) using the SILVA v.123 database (77, 78). Sequences that were unclassified were regarded as fungal and then processed separately.
Bacterial sequences were processed as described earlier by Gołębiewski et al. (72), while fungal sequences were processed with ITSx (79), and all fungal ITS1 sequences were used in the downstream analysis. The 16S rRNA ASVs processing pipeline was implemented in mothur v.1.39 and consisted of alignment against SILVA v.123, screening for sequences covering the desired region of the alignment (from positions 6428 to 22440), filtering all-gap- and terminal gap-containing columns from the alignment, and the removal of residual noise via preclustering and chimera identification using UCHIME. The final set of sequences was then clustered to 0.03 dissimilarity OTUs with the Opti-MCC algorithm (80), and an OTU table was constructed. The denoised fungal sequences (ASVs) were dereplicated, and OTUs were constructed using vsearch (81) at a 0.03 dissimilarity level. Singletons, as well as doubletons (OTUs consisting of one or two sequences only), were removed from both data sets. The sequences were classified with a naive Bayesian classifier (82) using SILVA v.123 (bacteria) and ITS1 extracted from UNITE v.7 (fungi), and the nonbacterial and nonfungal sequences were removed from the respective sets. The final data were subsampled to 500 (bacteria) and 300 (fungi) sequences per sample 20 times; OTUs were constructed as described earlier, and the resulting OTU tables were averaged over the 20 subsamples.
The core microbiome was delineated by using the get.coremicrobiome function of mothur with an abundance threshold of 0.5%. The lower threshold was used because no core OTUs were identified at the standard 1% threshold (83).
Statistical analyses were performed using the vegan package v.2.4-5 (39) in R (v3.4.4). The observed OTUs, Shannon’s diversity (H′), and Chao1 indices were calculated using the diversity and estimateR functions of the vegan package, while Shannon’s evenness was computed by dividing the H′ by the logarithm of the number of OTUs. NMDS analysis was performed with meta-MDS using Bray-Curtis and Morisita-Horn distances calculated from Wisconsin-normalized data. Permutational analysis of variance (PERMANOVA; adonis function) was performed to test whether endophytic bacterial and fungal communities were significantly different among tissues or sample sites, and PERMDISP (betadisper) was used to check for homogeneity of variance. Canonical correlation analysis (CCA) was used for community-constrained ordination, and soil parameters were selected as constraints in CCA. Influence of communities on one another was assessed using CoCA (84) implemented in the cocorresp R package (85). A total of 999 permutations were used in permutational tests.
Prediction of putative function sets was performed with PICRUSt as described earlier in Thiem et al. (73).
Data availability.
The bacterial and fungal sequence data generated in this study using MiSeq have been deposited and are available in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA412808.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Sonia Szymańska, Michał Złoch, and Dominika Thiem for conducting the initial plant sample collection. We are also grateful to the BestPass consortium for their valuable input.
B.U.F. participated in all analyses and wrote the first version of the manuscript. M.G. performed the bioinformatic and statistical analyses and participated in preparation of manuscript. M.S. prepared the sequencing libraries and performed sequencing. P.H. performed the soil chemical analysis. K.H. designed and managed all experiments, as well as participated in preparation of the manuscript. All authors revised the manuscript and approved the publication.
This project has received funding from the European Union’s Horizon 2020 Research and Innovation Program under Marie Skłodowska-Curie grant agreement 676480.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00305-19.
REFERENCES
- 1.Ushakova SA, Kovaleva NP, Gribovskaya IV, Dolgushev VA, Tikhomirova NA. 2005. Effect of NaCl concentration on productivity and mineral composition of Salicornia europaea as a potential crop for utilization NaCl in LSS. Adv Sp Res 36:1349–1353. doi: 10.1016/j.asr.2004.09.017. [DOI] [Google Scholar]
- 2.Oliveira V, Gomes NCM, Cleary DFR, Almeida A, Silva AMS, Simões MMQ, Silva H, Cunha Â. 2014. Halophyte plant colonization as a driver of the composition of bacterial communities in salt marshes chronically exposed to oil hydrocarbons. FEMS Microbiol Ecol 90:647–662. doi: 10.1111/1574-6941.12425. [DOI] [PubMed] [Google Scholar]
- 3.Smillie C. 2015. Salicornia spp. as a biomonitor of Cu and Zn in salt marsh sediments. Ecol Indic 56:70–78. doi: 10.1016/j.ecolind.2015.03.010. [DOI] [Google Scholar]
- 4.Abrol YP, Wild A. 2004. Soils, land, and food: managing the land during the twenty-first century. Ann Bot 93:785–786. doi: 10.1093/aob/mch104. [DOI] [Google Scholar]
- 5.Flowers TJ, Colmer TD. 2008. Salinity tolerance in halophytes. New Phytol 179:945–63. doi: 10.1111/j.1469-8137.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- 6.Ventura Y, Sagi M. 2013. Halophyte crop cultivation: the case for salicornia and sarcocornia. Environ Exp Bot 92:144–153. doi: 10.1016/j.envexpbot.2012.07.010. [DOI] [Google Scholar]
- 7.Kadereit G, Ball P, Beer S, Mucina L, Sokoloff D, Teege P, Yaprak AE, Freitag H. 2007. A taxonomic nightmare comes true: phylogeny and biogeography of glassworts (Salicornia L., Chenopodiaceae). Taxon 56:1143–1170. doi: 10.2307/25065909. [DOI] [Google Scholar]
- 8.Blumwald E. 2000. Sodium transport and salt tolerance in plants. Curr Opin Cell Biol 12:431–434. doi: 10.1016/S0955-0674(00)00112-5. [DOI] [PubMed] [Google Scholar]
- 9.Hardoim PR, van Overbeek LS, Berg G, Pirttilä M, Compant S, Campisano A, Döring M, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A. 2015. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santander C, Aroca R, Ruiz-Lozano JM, Olave J, Cartes P, Borie F, Cornejo P. 2017. Arbuscular mycorrhiza effects on plant performance under osmotic stress. Mycorrhiza 27:639–657. doi: 10.1007/s00572-017-0784-x. [DOI] [PubMed] [Google Scholar]
- 11.Hildebrandt U, Janetta K, Ouziad F, Renne B, Nawrath K, Bothe H. 2001. Arbuscular mycorrhizal colonization of halophytes in Central European salt marshes. Mycorrhiza 10:175–183. doi: 10.1007/s005720000074. [DOI] [Google Scholar]
- 12.Sonjak S, Udovič M, Wraber T, Likar M, Regvar M. 2009. Diversity of halophytes and identification of arbuscular mycorrhizal fungi colonising their roots in an abandoned and sustained part of Sečovlje salterns. Soil Biol Biochem 41:1847–1856. doi: 10.1016/j.soilbio.2009.06.006. [DOI] [Google Scholar]
- 13.Maciá-Vicente JG, Ferraro V, Burruano S, Lopez-Llorca LV. 2012. Fungal assemblages associated with roots of halophytic and non-halophytic plant species vary differentially along a salinity gradient. Microb Ecol 64:668–679. doi: 10.1007/s00248-012-0066-2. [DOI] [PubMed] [Google Scholar]
- 14.Hardoim PR, van Overbeek LS, van Elsas JD. 2008. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Jha Y, Subramanian RB, Patel S. 2011. Combination of endophytic and rhizospheric plant growth promoting rhizobacteria in Oryza sativa shows higher accumulation of osmoprotectant against saline stress. Acta Physiol Plant 33:797–802. doi: 10.1007/s11738-010-0604-9. [DOI] [Google Scholar]
- 16.Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F, Kim Y-O, Redman RS. 2008. Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2:404–416. doi: 10.1038/ismej.2007.106. [DOI] [PubMed] [Google Scholar]
- 17.Zhang HW, Song YC, Tan RX. 2006. Biology and chemistry of endophytes. Nat Prod Rep 23:753–771. doi: 10.1039/b609472b. [DOI] [PubMed] [Google Scholar]
- 18.Piernik A, Hulisz P. 2011. Soil-plant relations in inland and anthropogenic saline habitats. Eur J Plant Sci Biotechnol 5:37–43. [Google Scholar]
- 19.Hulisz P, Piernik A. 2013. Soils affected by soda industry in Inowrocław. Polish Society of Soil Science, Toruń, Poland. [Google Scholar]
- 20.Lata R, Chowdhury S, Gond S, White J Jr. 2018. Induction of abiotic stress tolerance in plants by endophytic microbes. Lett Appl Microbiol 66:268–276. doi: 10.1111/lam.12855. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Yang J, Wang E, Li B, Yuan H. 2015. Effects of growth stage and fulvic acid on the diversity and dynamics of endophytic bacterial community in Stevia rebaudiana Bertoni leaves. Front Microbiol 6:867. doi: 10.3389/fmicb.2015.00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sessitsch A, Hardoim P, Döring J, Weilharter A, Krause A, Woyke T, Mitter B, Hauberg-Lotte L, Friedrich F, Rahalkar M, Hurek T, Sarkar A, Bodrossy L, van Overbeek L, Brar D, van Elsas JD, Reinhold-Hurek B. 2012. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant Microbe Interact 25:28–36. doi: 10.1094/MPMI-08-11-0204. [DOI] [PubMed] [Google Scholar]
- 23.Mendoza JR, Kok CR, Stratton J, Bianchini A, Hallen-Adams HE. 2017. Understanding the mycobiota of maize from the highlands of Guatemala, and implications for maize quality and safety. Crop Prot 101:5–11. doi: 10.1016/j.cropro.2017.07.009. [DOI] [Google Scholar]
- 24.Ma X, Song X, Li X, Fu S, Li M, Liu Y. 2018. Characterization of microbial communities in pilot-scale constructed wetlands with Salicornia for treatment of marine aquaculture effluents. Archaea 2018:7819840. doi: 10.1155/2018/7819840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi YW, Lou K, Li C, Wang L, Zhao ZY, Zhao S, Tian CY. 2015. Illumina-based analysis of bacterial diversity related to halophytes Salicornia europaea and Sueada aralocaspica. J Microbiol 53:678–685. doi: 10.1007/s12275-015-5080-x. [DOI] [PubMed] [Google Scholar]
- 26.Collado J, Platas G, González I, Peláez F. 1999. Geographical and seasonal influences on the distribution of fungal endophytes in Quercus ilex. New Phytol 144:525–532. doi: 10.1046/j.1469-8137.1999.00533.x. [DOI] [PubMed] [Google Scholar]
- 27.Mishra A, Gond SK, Kumar A, Sharma VK, Verma SK, Kharwar RN, Sieber TN. 2012. Season and tissue type affect fungal endophyte communities of the Indian medicinal plant Tinospora cordifolia more strongly than geographic location. Microb Ecol 64:388–398. doi: 10.1007/s00248-012-0029-7. [DOI] [PubMed] [Google Scholar]
- 28.Hrynkiewicz K, Szymańska S, Piernik A, Thiem D. 2015. Ectomycorrhizal community structure of Salix and Betula spp. at a saline site in central Poland in relation to the seasons and soil parameters. Water Air Soil Pollut 226:99. doi: 10.1007/s11270-015-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brachmann A, Parniske M. 2006. The most widespread symbiosis on earth. PLoS Biol 4:e239. doi: 10.1371/journal.pbio.0040239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garbeva P, van Veen JA, van Elsas JD. 2004. Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42:243–270. doi: 10.1146/annurev.phyto.42.012604.135455. [DOI] [PubMed] [Google Scholar]
- 31.Lauber CL, Strickland MS, Bradford MA, Fierer N. 2008. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415. doi: 10.1016/j.soilbio.2008.05.021. [DOI] [Google Scholar]
- 32.Maciá-Vicente JG, Nau T, Piepenbring M. 2016. Low diversity and abundance of root endophytes prevail throughout the life cycle of an annual halophyte. Mycol Progress 15:1303–1311. doi: 10.1007/s11557-016-1241-5. [DOI] [Google Scholar]
- 33.Szymańska S, Płociniczak T, Piotrowska-Seget Z, Hrynkiewicz K. 2016. Endophytic and rhizosphere bacteria associated with the roots of the halophyte Salicornia europaea L.: community structure and metabolic potential. Microbiol Res 192:37–51. doi: 10.1016/j.micres.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Okane I, Nakagiri A. 2015. Assemblages of endophytic fungi on Salicornia europaea disjunctively distributed in Japan: towards clarification of the ubiquity of fungal endophytes on halophytes and their ecological roles. Curr Sci 109:62–71. [Google Scholar]
- 35.Muhsin TM, Booth T. 1987. Fungi associated with halophytes of an inland salt marsh, Manitoba, Canada. Can J Bot 65:1137–1151. doi: 10.1139/b87-159. [DOI] [Google Scholar]
- 36.Zhao S, Zhou N, Zhao ZY, Zhang K, Wu GH, Tian CY. 2016. Isolation of endophytic plant growth-promoting bacteria associated with the halophyte Salicornia europaea and evaluation of their promoting activity under salt stress. Curr Microbiol 73:574–581. doi: 10.1007/s00284-016-1096-7. [DOI] [PubMed] [Google Scholar]
- 37.Zhao S, Zhou N, Zhao ZY, Zhang K, Tian CY. 2016. High-throughput sequencing analysis of the endophytic bacterial diversity and dynamics in roots of the halophyte Salicornia europaea. Curr Microbiol 72:557–562. doi: 10.1007/s00284-016-0990-3. [DOI] [PubMed] [Google Scholar]
- 38.Szymańska S, Piernik A, Baum C, Złoch M, Hrynkiewicz K. 2014. Metabolic profiles of microorganisms associated with the halophyte Salicornia europaea in soils with different levels of salinity. Ecoscience 21:114–122. doi: 10.2980/21-2-3705. [DOI] [Google Scholar]
- 39.Lim JM, Jeon CO, Lee JC, Song SM, Kim KY, Kim CJ. 2006. Marinimicrobium koreense gen. nov., sp. nov. and Marinimicrobium agarilyticum sp. nov., novel moderately halotolerant bacteria isolated from tidal flat sediment in Korea. Int J Syst Evol Microbiol 56:653–657. doi: 10.1099/ijs.0.64075-0. [DOI] [PubMed] [Google Scholar]
- 40.Møller MF, Kjeldsen KU, Ingvorsen K. 2010. Marinimicrobium haloxylanilyticum sp. nov., a new moderately halophilic, polysaccharide-degrading bacterium isolated from Great Salt Lake, Utah. Antonie Van Leeuwenhoek 98:553–565. doi: 10.1007/s10482-010-9472-y. [DOI] [PubMed] [Google Scholar]
- 41.Yoon JH, Kang SJ, Jung YT, Oh TK. 2009. Marinimicrobium locisalis sp. nov., isolated from a marine solar saltern, and emended description of the genus Marinimicrobium. Int J Syst Evol Microbiol 59:2260–2263. doi: 10.1099/ijs.0.008458-0. [DOI] [PubMed] [Google Scholar]
- 42.Mora-Ruiz MDR, Font-Verdera F, Orfila A, Rita J, Rosselló-Móra R. 2016. Endophytic microbial diversity of the halophyte Arthrocnemum macrostachyum across plant compartments. FEMS Microbiol Ecol 92:fiw145. doi: 10.1093/femsec/fiw145. [DOI] [PubMed] [Google Scholar]
- 43.Hallmann J, Rodríguez-Kábana R, Kloepper JW. 1999. Chitin-mediated changes in bacterial communities of the soil, rhizosphere and within roots of cotton in relation to nematode control. Soil Biol Biochem 31:551–560. doi: 10.1016/S0038-0717(98)00146-1. [DOI] [Google Scholar]
- 44.Chi F, Shen S-H, Cheng H-P, Jing Y-X, Yanni YG, Dazzo FB. 2005. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol 71:7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szymańska S, Borruso L, Brusetti L, Hulisz P, Furtado B, Hrynkiewicz K. 2018. Bacterial microbiome of root-associated endophytes of Salicornia europaea in correspondence to different levels of salinity. Environ Sci Pollut Res Int 25:25420–25431. doi: 10.1007/s11356-018-2530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fidalgo C, Henriques I, Rocha J, Tacão M, Alves A. 2016. Culturable endophytic bacteria from the salt marsh plant Halimione portulacoides: phylogenetic diversity, functional characterization, and influence of metal(loid) contamination. Environ Sci Pollut Res Int 23:10200–10214. doi: 10.1007/s11356-016-6208-1. [DOI] [PubMed] [Google Scholar]
- 47.Bibi F, Strobel GA, Naseer MI, Yasir M, Al-Ghamdi AAK, Azhar EI. 2018. Microbial flora associated with the halophyte-Salsola imbricate and its biotechnical potential. Front Microbiol 9:65. doi: 10.3389/fmicb.2018.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sánchez-Porro C, de la Haba RR, Soto-Ramírez N, Márquez MC, Montalvo-Rodríguez R, Ventosa A. 2009. Description of Kushneria aurantia gen. nov., sp. nov., a novel member of the family Halomonadaceae, and a proposal for reclassification of Halomonas marisflavi as Kushneria marisflavi comb. nov., of Halomonas indalinina as Kushneria indalinina comb. nov. and of Halomonas avicenniae as Kushneria avicenniae comb. nov. Int J Syst Evol Microbiol 59:397–405. doi: 10.1099/ijs.0.001461-0. [DOI] [PubMed] [Google Scholar]
- 49.Mapelli F, Marasco R, Rolli E, Barbato M, Cherif H, Guesmi A, Ouzari I, Daffonchio D, Borin S. 2013. Potential for plant growth promotion of rhizobacteria associated with Salicornia growing in Tunisian hypersaline soils. Biomed Res Int 2013:248078. doi: 10.1155/2013/248078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Labrenz M, Lawson PA, Tindall BJ, Collins MD, Hirsch P. 2003. Saccharospirillum impatiens gen. nov., sp. nov., a novel gamma-proteobacterium isolated from hypersaline Ekho Lake (East Antarctica). Int J Syst Evol Microbiol 53:653–660. doi: 10.1099/ijs.0.02406-0. [DOI] [PubMed] [Google Scholar]
- 51.Bingham IJ, Stevenson EA. 1993. Control of root growth: effects of carbohydrates on the extension, branching and rate of respiration of different fractions of wheat roots. Physiol Plant 88:149–158. doi: 10.1111/j.1399-3054.1993.tb01772.x. [DOI] [Google Scholar]
- 52.Valerie SJR, Abratt R. 2005. Minireview: sucrose utilization in bacteria: genetic organization and regulation. Appl Microbiol Biotechnol 67:312–321. doi: 10.1007/s00253-004-1885-y. [DOI] [PubMed] [Google Scholar]
- 53.Gil R, Lull C, Boscaiu M, Bautista I, Lidon A, Vicente O. 2011. Soluble carbohydrates as osmolytes in several halophytes from a Mediterranean salt marsh. Not Bot Hort Agrobot Cluj 39:09. doi: 10.15835/nbha3927176. [DOI] [Google Scholar]
- 54.Acosta-Martínez V, Cotton J, Gardner T, Moore-Kucera J, Zak J, Wester D, Cox S. 2014. Predominant bacterial and fungal assemblages in agricultural soils during a record drought/heat wave and linkages to enzyme activities of biogeochemical cycling. Appl Soil Ecol 84:69–82. doi: 10.1016/j.apsoil.2014.06.005. [DOI] [Google Scholar]
- 55.Masip L, Veeravalli K, Georgiou G. 2006. The many faces of glutathione in bacteria. Antioxid Redox Signal 8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 56.Cunningham FX Jr, Pogson B, Sun Z, McDonald KA, DellaPenna D, Gantt E. 1996. Functional analysis of the β and ε lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell Online 8:1613–1626. doi: 10.2307/3870254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ravanello MP, Ke D, Alvarez J, Huang B, Shewmaker CK. 2003. Coordinate expression of multiple bacterial carotenoid genes in canola leading to altered carotenoid production. Metab Eng 5:255–263. doi: 10.1016/j.ymben.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 58.De Oliveira MVV, Intorne AC, Vespoli LDS, Madureira HC, Leandro MR, Pereira TNS, Olivares FL, Berbert-Molina MA, De Souza Filho GA. 2016. Differential effects of salinity and osmotic stress on the plant growth-promoting bacterium Gluconacetobacter diazotrophicus PAL5. Arch Microbiol 198:287–294. doi: 10.1007/s00203-015-1176-2. [DOI] [PubMed] [Google Scholar]
- 59.Khidir HH, Eudy DM, Porras-Alfaro A, Herrera J, Natvig DO, Sinsabaugh RL. 2010. A general suite of fungal endophytes dominate the roots of two dominant grasses in a semiarid grassland. J Arid Environ 74:35–42. doi: 10.1016/j.jaridenv.2009.07.014. [DOI] [Google Scholar]
- 60.Macia-Vicente JG, Jansson HB, Abdullah SK, Descals E, Salinas J, Lopez-Llorca LV. 2008. Fungal root endophytes from natural vegetation in Mediterranean environments with special reference to Fusarium spp. FEMS Microbiol Ecol 64:90–105. doi: 10.1111/j.1574-6941.2007.00443.x. [DOI] [PubMed] [Google Scholar]
- 61.De Boer W, Folman LB, Summerbell RC, Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Hendrickson OQ. 1991. Abundance and activity of N2-fixing bacteria in decaying wood. Can J For Res 21:1299–1304. doi: 10.1139/x91-183. [DOI] [Google Scholar]
- 63.Li CY, Massicote HB, Moore L. 1992. Nitrogen-fixing Bacillus sp. associated with Douglas-fir tuberculate ectomycorrhizae. Plant Soil 140:35–40. doi: 10.1007/BF00012804. [DOI] [Google Scholar]
- 64.Xavier LJC, Germida JJ. 2003. Bacteria associated with Glomus clarum spores influence mycorrhizal activity. Soil Biol Biochem 35:471–478. doi: 10.1016/S0038-0717(03)00003-8. [DOI] [Google Scholar]
- 65.Citterio B, Malatesta M, Battistelli S, Marcheggiani F, Baffone W, Saltarelli R, Stocchi V, Gazzanelli G. 2001. Possible involvement of Pseudomonas fluorescens and Bacillaceae in structural modifications of Tuber borchii fruit bodies. Can J Microbiol 47:264–268. doi: 10.1139/w01-005. [DOI] [PubMed] [Google Scholar]
- 66.Whipps JM. 2001. Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52:487–511. doi: 10.1093/jexbot/52.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- 67.Weller DM, Raaijmakers JM, Mcspadden Gardener BB, Thomashow LS. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol 40:309–348. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- 68.Wheatley RE. 2002. The consequences of volatile organic compound mediated bacterial and fungal interactions. Int J Gen Mol Microbiol 81:357–364. [DOI] [PubMed] [Google Scholar]
- 69.Kohlmeier S, Smits THM, Ford RM, Keel C, Harms H, Wick LY. 2005. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol 39:4640–4646. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- 70.Dziadowiec H, Pokojska U, Prusinkiewicz Z. 2004. Eco-pedological studies. PWN, Warsaw, Poland. (In Polish.) [Google Scholar]
- 71.van Reeuwijk LP. 2002. Procedures for soil analysis, 6th ed. ISRIC/FAO, Wageningen, The Netherlands. [Google Scholar]
- 72.Gołębiewski M, Deja-Sikora E, Cichosz M, Tretyn A, Wróbel B. 2014. 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb Ecol 67:635–647. doi: 10.1007/s00248-013-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thiem D, Gołebiewski M, Hulisz P, Piernik A, Hrynkiewicz K. 2018. How does salinity shape bacterial and fungal microbiomes of Alnus glutinosa roots? Front Microbiol 9:651. doi: 10.3389/fmicb.2018.00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White TJ, Bruns TD, Lee SB, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY. doi: 10.1016/B978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- 75.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, De Wit P, Sánchez-García M, Ebersberger I, de Sousa F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH. 2013. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:919. [Google Scholar]
- 80.Westcott SL, Schloss PD. 2017. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere 2:e00073-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang T, Nagy I, Mancinotti D, Otterbach SL, Bundgaard Andersen T, Motawia MS, Asp T, Geu-Flores F. 2017. Transcript profiling of a bitter variety of narrow-leafed lupin to discover alkaloid biosynthetic genes. J Exp Bot 68:5527–5537. doi: 10.1093/jxb/erx362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H, Fan W, Yu PS, Han J. 2003. Mining concept-drifting data streams using ensemble classifiers, p 226. In Proceedings of the ninth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: KDD ‘03 ACM Press, New York, NY. [Google Scholar]
- 83.Astudillo-García C, Bell JJ, Webster NS, Glasl B, Jompa J, Montoya JM, Taylor MW. 2017. Evaluating the core microbiota in complex communities: a systematic investigation. Environ Microbiol 19:1450–1462. doi: 10.1111/1462-2920.13647. [DOI] [PubMed] [Google Scholar]
- 84.ter Braak CJF, Schaffers AP. 2004. Co-correspondence analysis: a new ordination method to relate two community compositions. Ecology 85:834–846. doi: 10.1890/03-0021. [DOI] [Google Scholar]
- 85.Simpson GL. 2009. Analogue and weighted averaging methods for palaeoecology. R package analogue v0.17-0. https://www.rdocumentation.org/packages/analogue.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bacterial and fungal sequence data generated in this study using MiSeq have been deposited and are available in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA412808.