Abstract
Summary
Ancient whole-genome duplications (WGDs) have been uncovered in almost all major lineages of life on Earth and the search for traces or remnants of such events has become standard practice in most genome analyses. This is especially true for plants, where ancient WGDs are abundant. Common approaches to find evidence for ancient WGDs include the construction of KS distributions and the analysis of intragenomic colinearity. Despite the increased interest in WGDs and the acknowledgment of their evolutionary importance, user-friendly and comprehensive tools for their analysis are lacking. Here, we present an easy to use command-line tool for KS distribution construction named wgd. The wgd suite provides commonly used KS and colinearity analysis workflows together with tools for modeling and visualization, rendering these analyses accessible to genomics researchers in a convenient manner.
Availability and implementation
wgd is free and open source software implemented in Python and is available at https://github.com/arzwa/wgd.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
In this era of whole-genome sequencing, many ancient whole-genome duplication (WGD) events have been uncovered across the eukaryotic tree of life (Van de Peer et al., 2017). One of the main approaches for revealing ancient WGDs using genomic data is the construction of whole paranome KS distributions (e.g. Blanc and Wolfe, 2004; Cui et al., 2006; Lynch and Conery, 2000; Vanneste et al., 2013), where KS is the synonymous distance or the estimated number of synonymous substitutions per synonymous site. Under the assumption of neutral evolution at synonymous sites, the synonymous distance between two coding sequences serves as a proxy for the divergence time of two sequences. Under a model of continuous small-scale gene duplication (SSD) and loss of duplicated copies not under selection, a whole paranome KS distribution is expected to show an exponential decay of the number of retained duplicates in function of age (Blanc and Wolfe, 2004; Lynch and Conery, 2000). Against this background of SSDs, large-scale duplication events, such as WGDs, are visible as peaks in the number of retained duplicates at a particular age.
Several issues compromise the use of KS distributions for WGD inference, and these were extensively addressed in Vanneste et al. (2013). When high-quality genome assemblies are available, gene colinearity (often called synteny) based analyses may further aid in unveiling WGDs or large segmental duplications (Van de Peer, 2004). WGDs are expected to leave large blocks with high intragenomic colinearity, and paralogs located in such colinear segments (anchor pairs) can therefore be traced back more reliably to a particular event, enabling their use for downstream analyses such as molecular dating (Vanneste et al., 2014) or functional analysis.
While these methods have been used frequently in genomics research, no comprehensive and user-friendly software is available to perform these analyses, and researchers have often resorted to custom pipelines. Here, we fill this gap with an integrated suite for KS and colinearity based analysis of ancient WGDs. We briefly discuss the methods implemented here, but refer to the documentation and Supplementary Material for more information.
2 Materials and methods
2.1 Gene family delineation
Delineation of paralogous gene families and one-to-one orthologs starts from all-versus-all BLASTp similarity searches or precomputed BLAST results and is performed using ‘wgd mcl’. For whole paranome delineation, MCL (van Dongen, 2000) is then used to cluster sequences in paralogous gene families. One-to-one orthologs are determined using the commonly employed reciprocal best hit strategy.
2.2 KS distribution construction
A KS distribution for a set of paralogous families or one-to-one orthologs can be constructed using the ‘wgd ksd’ subcommand, and we closely follow the approach used by Vanneste et al. (2013). We refrain from a full description of the methodology here and refer to the Supplementary Material instead.
2.3 Colinearity analyses
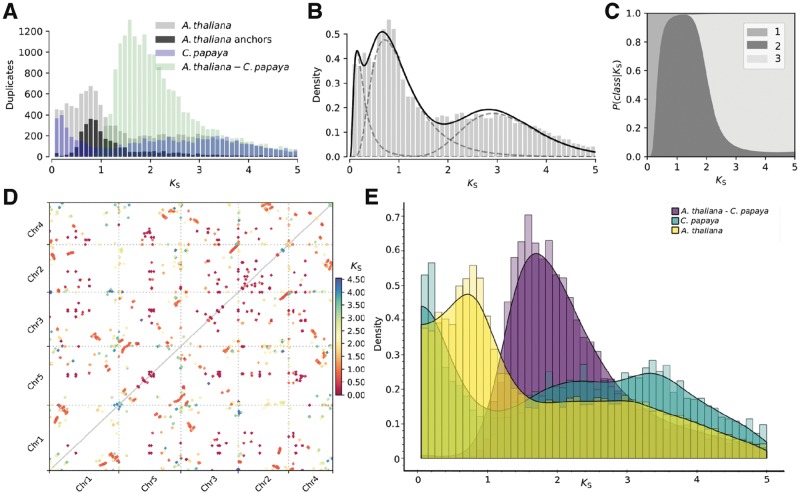
When high-quality structural genome annotations are available, the ‘wgd syn’ tool allows the identification of intragenomic colinear blocks and their corresponding anchor pairs using I-ADHoRe 3.0 (Proost et al., 2012). Whole-genome syntenic dotplots are generated, and if a KS distribution is provided, KS-colored dotplots and anchor pair KS distributions are generated (Fig. 1).
Fig. 1.
Illustration of the various tools and visualizations in wgd. (A) Arabidopsis thaliana and Carica papaya paranome KS distributions overlayed with the KS distribution of anchor pairs for A. thaliana and KS distribution of one-to-one orthologs of C. papaya and A. thaliana. (B) Mixture of three log-normal distributions fitted to the KS distribution of A. thaliana, using the Variational Bayes algorithm with γ = 10−3. (C) Plot showing the probability to belong to a particular component of the mixture shown in (B) in function of KS. These probabilities can be used to define component-wise paralogs for further downstream analyses. (D) KS-colored dotplot for A. thaliana, showing colinear blocks identified by I-ADHoRe, colored by their median KS value. (E) Interactive histogram visualization (user interface not shown, see Supplementary Fig. S1), showing the whole paranome KS distributions using histograms and kernel density estimates for A. thaliana and C. papaya together with the KS distribution of one-to-one orthologs in these species. We refer to the Supplementary Material for detailed methods
2.4 Kernel density estimation and mixture modeling
Downstream analyses of KS distributions have often consisted in fitting statistical models and visualizing these. We provide tools (‘wgd kde’) for fitting kernel density estimates (KDEs). Importantly, we apply a correction for boundary effects, which are often neglected but may lead to artificial peaks in low KS regions. As peaks derived from WGDs are expected to be approximately log-normally distributed, Gaussian mixture models (GMMs) have also been used frequently to analyze KS distributions. We provide tools (‘wgd mix’) for fitting mixtures of log-normal components using different inference algorithms, implemented using the scikit-learn python library (Pedregosa et al., 2011). Common approaches to determine the optimal number of components are provided, using the Akaike or Bayesian information criterion, however we would like to warn prospective users to carefully interpret ‘significant’ components, as these GMMs may strongly overfit the empirical distribution (Tiley et al., 2018).
2.5 Interactive visualization
Lastly, we provide tools for (interactive) visualization of histograms and KDEs in ‘wgd viz’ (Fig. 1). These tools allow visualization of multiple KS distributions for comparative purposes as well as modification of key visualization parameters such as the histogram bin-width or the KDE bandwidth. We encourage researchers to modify and explore the influence of these to guide careful analysis of the distributions and to prevent misinterpretations of KDE or histogram artifacts as biologically interesting features.
3 Conclusion
We provide, to our knowledge, the first comprehensive toolshed for KS and colinearity based analysis of WGDs in an easy to use and freely available package named wgd. We hope that, besides being a useful tool for researchers, it will also aid in preventing common pitfalls and misinterpretations when analyzing putative WGDs in genomic data.
Funding
This work was supported by the European Union Seventh Framework Programme (FP7/2007-2013) under European Research Council Advanced Grant Agreement 322739—DOUBLEUP [to Y.V.d.P]; and a PhD Fellowship of the Research Foundation—Flanders (FWO) [to A.Z.].
Conflict of Interest: none declared.
Supplementary Material
References
- Blanc G., Wolfe K.H. (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell, 16, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., et al. (2006) Widespread genome duplications throughout the history of flowering plants. Genome Res., 16, 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J.S. (2000) The evolutionary fate and consequences of duplicate genes. Science, 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Pedregosa F., et al. (2011) Scikit-learn: machine learning in Python. J. Mach. Learn. Res., 12, 2825–2830. [Google Scholar]
- Proost S., et al. (2012) i-ADHoRe 3.0: fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res., 40, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiley G.P., et al. (2018) Assessing the performance of Ks plots for detecting ancient whole genome duplications. Genome Biol. Evol., 10, 2882–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y. (2004) Computational approaches to unveiling ancient genome duplications. Nat. Rev. Genet., 5, 752–763. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., et al. (2017) The evolutionary significance of polyploidy. Nat. Rev. Genet., 18, 411–424. [DOI] [PubMed] [Google Scholar]
- van Dongen S. (2000) Graph Clustering by Flow Simulation. PhD Thesis, University of Utrecht, Utrecht, The Netherlands. [Google Scholar]
- Vanneste K., et al. (2013) Inference of genome duplications from age distributions revisited. Mol. Biol. Evol., 30, 177–190. [DOI] [PubMed] [Google Scholar]
- Vanneste K., et al. (2014) Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res., 24, 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.