Abstract
There are hundreds of essential genes in multidrug resistant bacterial genomes, but only a few of their products are exploited as antibacterial targets. An example is the electron flavoprotein (ETF) which is required for growth and viability in Burkholderia cenocepacia. Here, we evaluated ETF as an antibiotic target for Burkholderia cepacia complex (Bcc). Depletion of the bacterial ETF during infection of Caenorhabditis elegans significantly extended survival of the nematodes, proving that ETF is essential for survival of B. cenocepacia in this host model. In spite of the arrest in respiration in ETF mutants, the inhibition of etf expression did not increase the formation of persister cells, when treated with high doses of ciprofloxacin or meropenem. To test if etf’translation could be inhibited by RNA interference, antisense oligonucleotides that target the etfBA operon were synthesized. One antisense oligonucleotide was effective in inhibiting etfB translation in vitro but not in vivo, highlighting the challenge of reduced membrane permeability for the design of drugs against B. cenocepacia.
This work contributes to the validation of ETF of B. cenocepacia as a target for antibacterial therapy and demonstrates the utility of a C. elegans liquid killing assay to validate gene essentiality in an in vivo infection model.
Keywords: Burkholderia cenocepacia, cystic fibrosis, electron transfer flavoprotein (ETF), essential genes, antibacterial targets, persister cells, antisense oligonucleotides, C. elegans
Burkholderia cepacia complex (Bcc) is a group of Gram-negative bacteria that causes a substantial detriment in pulmonary function in patients with the genetic disease cystic fibrosis (CF) (Mahenthiralingam et al. 2005). In 20% of the cases, Bcc infections result in sepsis and death. Further, Bcc bacteria can survive in the presence of disinfectants (Kim et al. 2015, Ahn et al. 2016) causing occasional outbreaks in hospitals worldwide (Lee et al. 2013). Unfortunately, there are very limited options to treat Bcc infections as these bacteria are intrinsically resistant to most antibiotics (Avgeri et al. 2009). Thus, novel antibiotics are needed to treat Bcc infections.
The targets of effective antibiotics are typically the products of ‘essential genes’, which are those coding for components required for bacteria to grow in rich medium (Arigoni et al. 1998, Akerley et al. 2002). Bacterial genomes encode hundreds of essential genes; however, only a few of those gene products are currently used as targets of antibiotics (Fields et al. 2016). Among the challenges for validating such essential products as novel antibiotic targets is the possibility that essential genes may code for functions that are essential in laboratory conditions, which differ from the host environment (Brinster et al. 2009). Indeed, certain metabolic pathways have been ruled out as drug targets due to the ability of bacteria to bypass the essential requirement by utilizing bioproducts from the host (Brinster et al. 2009). Another challenge for validating an essential product as an antibacterial target is whether targeting an essential process can increase persister cell formation (Lewis 2007, Kwan et al. 2013). Persistence is a phenotypic state of dormancy experienced by up to 1% of the bacterial population that allows surviving under high concentrations of antibiotic (Lewis 2007). Persistence has been strongly linked to the recalcitrant CF infections and failure of treatment (Mulcahy et al. 2010). Previous studies have demonstrated that a decrease in ATP intracellular levels leads to an increase in persister cell formation in Staphylococcus aureus (Conlon et al. 2016). On the other hand, loss of respiration in Escherichia. coli showed a ~1000 fold reduction in the number of persisters in stationary phase (Orman and Brynildsen 2015).
To search for potential antibacterial targets, we previously built a conditional growth mutant library of the clinically relevant strain B. cenocepacia K56–2 (Table 1), in which essential genes were placed under the control of a rhamnose-inducible promoter (Bloodworth et al. 2013). In one of these mutants, rhamnose-dependent downregulation of an electron transfer flavoprotein (ETF), coded for by etfBA caused the strongest defect in cell viability and abolition of aerobic respiration (Bloodworth et al. 2015). Analysis of the mechanisms that resulted in loss of cell viability showed a pleiotropic effect that included abolition of aerobic respiration and changes in cell morphology. The essentiality of the B. cenocepacia ETF was surprising as ETF has a non-essential role as an electron acceptor for dehydrogenases involved in fatty acid degradation in other organisms (Matsuoka et al. 2007, Fujita et al. 2007). Identification of essential genes in other bacteria also show that ETF is essential in relevant human pathogens such as Acinetobacter baylyi (Berardinis et al. 2008), Mycobacterium tuberculosis (Griffin et al. 2011), Porphyromonas gingivalis (Klein et al. 2012), Burkholderia pseudomallei (Moule et al. 2014), and Pseudomonas aeruginosa (Lee et al. 2015). These findings prompted us to investigate whether the essential ETF could be considered as an antibacterial target. Although the reasons for ETF essentiality are unknown in Bcc, many ETFs are components of the fatty acid metabolism pathway (Fujita et al. 2007). Therefore, it is possible that fatty acids produced by the host could bypass the bacterial requirements for survival under ETF depletion. In addition, because of the known function of ETF as an electron transporter, ATP depletion caused by downregulation of ETF could cause an increase in the formation of persister cells, as observed in S. aureus (Conlon et al. 2016). Both aspects would rule out ETF as a promising antibacterial target.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains | Relevant phenotype | Reference |
|---|---|---|
| E. coli OP50 | Nonpathogenic strain for feeding C. elegans | Caenorhabditis Genetic Center (CGC), University of Minnesota, Minneapolis, USA |
| B. cenocepacia J2315 | (LMG18863), ET12 linage, CF isolate | (Mahenthiralingam et al. 2002) |
| B. cenocepacia K56–2 | ||
| B. cenocepacia RSF34 | K56–2, ΔhldA, LPS defective | |
| B. cenocepacia CGetf | K56–2; Tpr *, rhamnose-dependent etfBA expression | (Bloodworth et al. 2015) |
| Plasmid | ||
| pSCrhaB2 | oripBBRl rhaR rhaS PrhaB Tpr *mob+ | (Cardona and Valvano 2005) |
Tpr, trimethoprim resistant.
In this work, we used the Caenorhabditis elegans host infection model (Desalermos et al. 2011, Kim et al. 2017), to determine that worms infected with an ETF knockdown mutant overcome the infection when ETF is downregulated. We also demonstrate that ETF depletion, which causes lack of aerobic respiration, does not increase the formation of persister cells. In an attempt to look for venues to inhibit expression of etfBA in wild type cells as a mean of treatment, we designed antisense oligonucleotides that effectively interfered with expression in vitro.
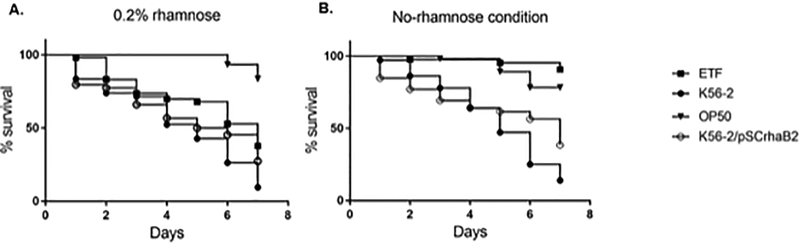
We first hypothesized that a C. elegans liquid killing assay (Selin et al. 2015), where infected worms are suspended in liquid medium with or without the addition of rhamnose would allow us to modulate the bacterial ETF expression inside the host. If this was the case, ETF depletion should cause a decrease in bacterial intestinal growth increasing the survival of the worms. Briefly, L4 worms grown on E. coli OP50 lawns were transferred to plates containing lawns of B. cenocepacia K56–2 WT or the B. cenocepacia knockdown mutant of ETF (CGetf) (Table 1) grown in nematode growth medium (NGM) or NGM supplemented with trimethoprim 100 μg/mL and 0.2% rhamnose, respectively. The plates were incubated at 25°C for 16 hours, to allow bacterial intestinal colonization (Law et al. 2008). Worms were then washed from the plate with M9 buffer, and resuspended in liquid killing medium (80% M9 and 20% NGMII). Twenty-30 worms per well, using 4 wells per condition, were spotted in a 96-well plate containing liquid killing medium with or without addition of rhamnose. We then tracked worm survival during 7 days by visually counting dead and live worms for each well and the average was calculated for each condition. Fig. 1 illustrates a representative survival curve of three biological replicates. When the worms were kept in liquid medium supplemented with 0.2% rhamnose (Fig. 1A) C. elegans infected with either WT or CGetf bacterial strains showed similar survival rates, less than 50% individuals survived after four days. This result indicates that rhamnose reached the nematode’s gut and induced ETF expression, allowing bacterial proliferation in the gut. Infection in these conditions was comparable to that of the wild type strain. Conversely, in the absence of rhamnose, the individuals infected with CGetf recovered from the infection with a survival rate close to 100% (Fig. 1B). These results suggest that B. cenocepacia CGetf cells are not able to bypass the ETF depletion in the C. elegans host model by metabolizing the contents of the intestinal lumen or components of the intestinal cells.
Figure 1.
Liquid killing assays. C. elegans was allowed to feed for 16 hours on B. cenocepacia K56–2, E. coli OP50 (non-pathogenic control), or the B. cenocepacia CGetf mutant (ETF) in the presence of 0.2% rhamnose to mimic WT levels of etf expression. Twenty to 30 worms were added to 96-wells plates containing 100 μl of liquid killing medium with (A) or without (B) the presence of 0.2% rhamnose. Four wells were inoculated per condition. The number of worms was counted every 24 hours for 7 days and plotted as survival curves using GraphPad Prism. K56–2 carrying the plasmid pSCrhaB2, which confers resistant to trimethoprim, was included as a control.
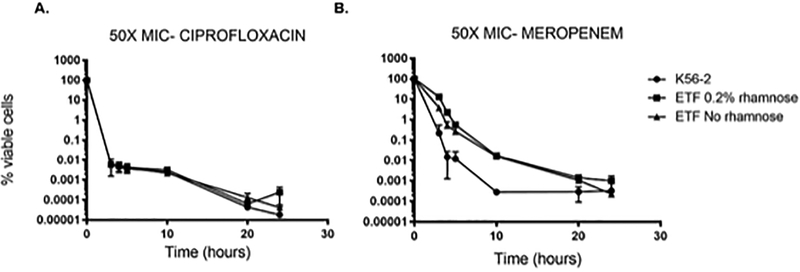
To determine the effect of ETF inhibition on persister cell formation, we quantified the cell viability of the CGetfmutant after treatment with 50X the minimal inhibitory concentration (MIC) of ciprofloxacin or meropenem. Briefly, B. cenocepacia WT and CGetf were cultured in LB broth in the presence of 0.2% rhamnose at 37°C for 16 hours. Cells were collected and washed, inoculated to an OD600 of 0.05 in LB broth with or without addition of 0.2% rhamnose, and incubated in a shaker at 37°C until late exponential phase. The cells were then standardized to an OD600 of 0.7 in the same media with the addition of 100 μg/mL ciprofloxacin or 800 μg/mL meropenem. 100 μl aliquots were removed at time 0 (before treatment), 3, 4, 5, 20 and 24 hours’ post-treatment, washed with fresh LB to remove residual antibiotic, and plated onto LB agar plates (WT) or LB agar plates containing 100 μg/mL trimethoprim and 0.2%rhamnose (CGetf). Plates were incubated at 37°C for estimation of colony forming units. Fig. 2 shows that after three hours of treatment with ciprofloxacin (Fig. 2A) or meropenem (Fig. 2B), the viability of the WT strain dropped to approximately 0.01%, which is typical of the formation of a persister population. Cells depleted of ETF showed the same killing profile as the WT strain under ciprofloxacin treatment with a survival lower than 0.0001% after 24 h of treatment. Meropenem treatment seemed to have a delayed killing effect in CG etf cells, reaching the WT killing rate after 24 h post treatment. This effect was not due to ETF depletion because the delayed killing was also observed in ETF-replete cells (CGetf plus rhamnose). While the reasons for the delayed killing of the CGetf cells in the presence of meropenem are unknown in both cases, we were able to demonstrate that the inhibition of ETF does not increase the formation of persister cells, despite its effect on aerobic respiration.
Figure 2.
Persister cells formation of the CGetf mutant strain. The percentage of viable cells (y-axis) was calculated after treatment with 100 μg/mL ciprofloxacin (A) or 800 μg/mL meropenem (B) of exponentially growing cells of, wild type K56–2 (black circles) and CGetf (ETF) in the presence (black squares) or absence (black triangles) of 0.2% rhamnose. X-axis indicates time in hours. Time 0 indicates the initial % of viable cells before treatment. An aliquot of each culture was taken at 3, 4, 5, 20 and 24 hours’ post addition of antibiotic, washed with LB and plated for colony counting. The graphs include standard deviation (SD) of three biological replicates.
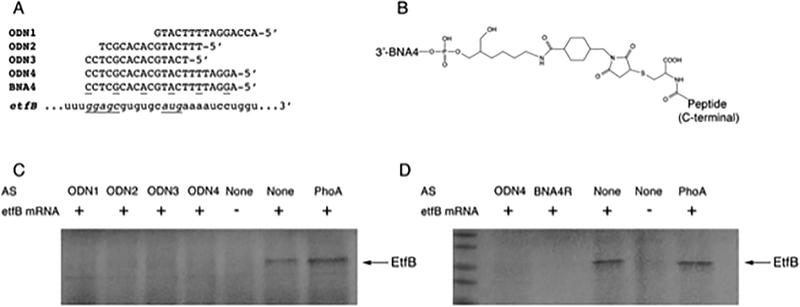
A limitation of the use of ETF as a drug target is that the human mitochondrial electron transport chain contains an ETF homolog (Roberts et al. 1996). Inhibition of human ETF could cause metabolic disorders (Frerman and Goodman 2013), which emphasizes the need of selective inhibition of the bacterial ETF. Antisense therapy is a powerful tool for development of new drugs that inhibit the translation of a target mRNA (Kole et al. 2012). The main advantage of this technology is target specificity, which minimizes side effects on commensal bacteria and host (Sully and Geller 2016). Antisense oligonucleotides can inhibit gene expression through different mechanisms such as steric hindrance, and RNase H- or RNase P-induced degradation of the mRNA (Rasmussen et al. 2007). In this case, the antisense sequence was designed to target the initiation of translation region by steric hindrance. To test the potential of ETF as a target for antisense inhibition, we synthesized four antisense oligodeoxynucleotides (ODNs) complementary to the 5’end of the etfB coding gene (See ODN sequences in Fig. 3A) and evaluated the effect of these ODNs on etfBA expression in an in vitro expression system (Lopez et al. 2015).
Figure 3.
Inhibition of etf expression by antisense oligodeoxynucleotides (ODNs). A) Sequences of the ODNs, BNA, and mRNA target regions used in this study. Upper case, lower case, and underlined letters represent deoxyribonucleotide, ribonucleotide, and BNA residues, respectively. The ribosome binding site and start codon are shown as underlined italics. B) Chemical structure of the BNA4 linked to penetrating peptides (RXR)4XB or (RFF)3RXB. C and D) In vitro inhibition of translation of mRNA etfB by ODNs using the E. coli S30 Cell Free Extract System. Additions to the reaction mixtures are shown on top. AS, antisense ODN added. PhoA, antisense targeting the region encompassing the start codon of thephoA gene. The arrow shows the location of the EtfB protein. BNA4R, BNA4-(RXR)4XB conjugate. ODNs were purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa). (RXR)4XB-Cys-SMCC-C6 amino-2’,4’-BNANC-DNA (R, arginine; X, 6-aminohexanoic acid; B, β-alanine) and (RFF)3RXB-Cys-SMCC-C6 amino-2’−4’-BNANC-DNA (BNA4-(RXR)4XB and BNA4-(RFF)3RXB, respectively) were purchased from Bio-Synthesis Inc., Lewisville, Texas.
In vitro translation of etfB was carried out by adding a 941 bp PCR fragment including ribosome binding site (RBS) and coding sequence of the etfB gene to the E. coli S30 Cell Free Extract System for Circular DNA kit (Promega). The amino acid mixture (minus methionine) plus 10 μCi of radiolabeled [35S] methionine (Perkin-Elmer) were added to the reaction to facilitate subsequent detection. 6.6 μM of the different ODNs were added for translation inhibition when indicated. An unrelated ODN complementary to the alkaline phosphatase phoA was included as a control. The products were visualized by autoradiography in 18% SDS-PAGE after treatment with En3hance (Perkin Elmer). ETF in vitro synthesis produced the expected 26.6 KDa protein band that corresponds to the EtfB subunit (Fig. 3C). When the protein synthesis was carried out in the presence of any of the four ODNs tested, no protein band was observed in the gel. All the four ODNs were able to inhibit the production of ETF protein in vitro. However, practical utilization of antisense oligonucleotides as drugs require them to resist the action of the ubiquitous nucleases and to penetrate the cell membrane to reach the bacterial cytosol. The former problem can be solved by utilizing nucleotide analogs so that the oligonucleotides composed of units of one analog or different analogs are resistant to most nucleases but still form duplexes with the target mRNA (Rasmussen et al, 20017). We have shown before hybrid oligomers that are nuclease-resistant (Soler Bistue et al. 2009) and good inhibitors of translation by steric hindrance (Lopez et al. 2015). Furthermore, the antisense compounds must penetrate the cell wall, reach the cytosol, and be active inside the cells. Conjugation of antisense analogs to permeabilizer peptides has been proposed as a solution to this problem in several bacteria (Boisguerin et al., 2015). Therefore, one of the ODNs, ODN4, was selected for further analysis and an isosequential oligonucleotide analog containing 2’,4’-bridged nucleic acid (BNA) and deoxynucleotide residues with the configuration A+GGAT+TTTC+ATGC+ACAC+GCTC+C, where the “+” before the base indicates that it is BNA residue, was covalently bound to the cell penetrating peptides (RXR)4XB or (RFF)3RXB (purchased from Biosynthesis Inc) (Greenberg et al. 2010). These compounds should be nuclease resistant and capable of reaching the cytosol where they should interact with the mRNA and inhibit translation. Fig. 3D shows that BNA4- (RXR)4XB was active in the cell-free in vitro system and inhibited etfB translation at the same level as the original ODN4. This result demonstrated that etfB expression can be turned off by short antisense oligonucleotide analogs covalently bound to permeabilizing peptides. However, neither BNA4-(RXR)4XB nor BNA4-(RFF)3RXB inhibited bacterial growth when added to cultures of the WT strains K56–2 and J2315 at a concentration of up to 20 μM (Table 2). Both strains belong to the ET12 epidemic clone but B. cenocepacia J2315 lacks O antigen (Ortega et al. 2005). In another experiment, both compounds were tested on the mutant strain B. cenocepacia RSF34, in which a modification of the LPS core increases susceptibility to different antimicrobial peptides (Loutet et al. 2006). No difference in growth inhibition was observed in the presence or absence of BNA4- (RXR)4XB or BNA4-(RFF)3RXB (Table 2). Taken together these results indicate that the antisense compounds show robust inhibition of expression of ETF in vitro but failed to inhibit growth in vivo despite the modifications to confer stability and ability to penetrate bacterial cells. The most probable causes for the lack of activity in cellulo are that the peptide conjugates did not reach the cytoplasm of B. cenocepacia or they failed to exert the inhibitory activity once inside the cells. The penetrating peptide (RFF)3RXB used in this study has been previously conjugated to a phosphorodiamidate morpholino oligomer (PMO) designed to target the acpP gene. This peptide-conjugated PMO was effective in inhibiting B. cenocepacia J2315 (Greenberg et al. 2010). While the reasons for the disagreement with previous results are not known, one possible scenario is that the penetrating properties of (RFF)3RXB are compromised when the peptide is conjugated to a BNA/DNA mixmer. Further studies modifying the chemical nature of the oligomers as well as the utilization of other cell penetrating peptides may clarify the causes of the lack of activity of the compounds used in this work and provide with active compounds capable of inhibition of growth of B. cenocepacia. These studies will also permit to start defining peptides and analogs that are appropriate for each particular bacterial species.
Table 2.
Minimum Inhibitory Concentrations (MICs) of peptide conjugated BNA4 of different B. cenocepacia strains.
| Strain | BNA4-(RXR)4XB | BNA4-(RFF)3RXB | Trimethoprim* | Polymyxin B** |
|---|---|---|---|---|
| B. cenocepacia J2315 | >20 μM | >20 μM | 8 μg/mL | >512 μg/mL |
| B. cenocepacia K56–2 | >20 μM | >20 μM | 8 μg/mL | >512 μg/mL |
| B. cenocepacia RSF34 | >20 μM | >20 μM | 8 μg/mL | 8 μg/mL |
Trimethoprim is known to inhibit B. cenocepacia growth and it was included as a control,
Polymyxin B was used to test the sensitivity of strain RSF34 to antimicrobial peptides.
In conclusion, we demonstrate that an electron transfer flavoprotein (ETF) of B. cenocepacia, previously shown to be essential in vitro, is also essential for survival in the C. elegans model of infection. Depletion of ETF does not increase formation of persister cells, and its mRNA can be successfully inhibited in vitro by antisense technology. Thus, ETF holds promise as a new antibacterial target. We also describe that the C. elegans infection model performed in liquid medium allows modulation of bacterial essential gene expression in vivo when essential genes are under the control of the rhamnose-inducible promoter. This assay can be applied to study the effect of any essential gene inhibition during infection conditions. Confirmation of the essentiality of ETF in other animal models of infection is necessary to validate ETF as an antibacterial target.
ACKNOWLEDGEMENTS
This work was supported by a research grant from Cystic Fibrosis Canada (CFC) to S.T.C. and by a Public Health Service grant 2R15AI047115–04 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health to M.E.T. We thank Dr. Miguel A. Valvano for providing strain RSF34.
REFERENCES
- Ahn Y, Kim JM, Kweon O, Kim SJ, Jones RC, Woodling K, Costa G.G. da, LiPuma JJ, Hussong D, Marasa BS, and Cerniglia CE 2016. Intrinsic Resistance of Burkholderia cepacia Complex to Benzalkonium Chloride. mBio 7(6): 10.1128/mBio.01716-16. doi:e01716–16 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Rubin EJ, N. V, Amaya K, Judson N, and Mekalanos JJ 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci U A 99(2): 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni F, Talabot F, Peitsch M, Edgerton MD, Meldrum E, Allet E, Fish R, Jamotte T, Curchod ML, and Loferer H 1998. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol 16(9): 851–856. [DOI] [PubMed] [Google Scholar]
- Avgeri SG, Matthaiou DK, Dimopoulos G, Grammatikos AP, and Falagas ME 2009. Therapeutic options for Burkholderia cepacia infections beyond co-trimoxazole: a systematic review of the clinical evidence. Int. J. Antimicrob. Agents 33(5): 394–404. doi: 10.1016/j.ijantimicag.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Berardinis V. de, Vallenet D, Castelli V, Besnard M, Pinet A, Cruaud C, Samair S, Lechaplais C, Gyapay G, Richez C, Durot M, Kreimeyer A, Fevre FL, Schachter V, Pezo V, Doring V, Scarpelli C, Medigue C, Cohen GN, Marliere P, Salanoubat M, and Weissenbach J 2008. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol 4: 174–189. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodworth RA, Gislason AS, and Cardona ST 2013. Burkholderia cenocepacia conditional growth mutant library created by random promoter replacement of essential genes. MicrobiologyOpen 2(2): 243–258. doi: 10.1002/mbo3.71; 10.1002/mbo3.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodworth RA, Zlitni S, Brown ED, and Cardona ST 2015. An electron transfer flavoprotein (ETF) is essential for viability and its depletion causes a rod-to-sphere change in Burkholderia cenocepacia. Microbiol. Read. Engl 161: 1909–1920. doi: 10.1099/mic.0.000156 [doi]. [DOI] [PubMed] [Google Scholar]
- Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, and Poyart C 2009. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458(7234): 83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- Cardona ST, and Valvano MA 2005. An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia. Plasmid 54(3): 219–228. doi: 10.1016/j.plasmid.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, and Lewis K 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol 1(Journal Article): 16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- Desalermos A, Muhammed M, Glavis-Bloom J, and Mylonakis E 2011. Using C. elegans for antimicrobial drug discovery. Expert Opin. Drug Discov 6(6): 645–652. doi: 10.1517/17460441.2011.573781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields FR, Lee SW, and McConnell MJ 2016. Using bacterial genomes and essential genes for the development of new antibiotics. Biochem. Pharmacol doi:S0006-2952(16)30463-4 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerman FE, and Goodman SI 2013. Defects of electron transfer flavoprotein and electron transfer flavoprotein-ubiquinone oxidoreductase: glutaric acidemia type II. In 2nd edition Edited by Scriver CR. McGraw-Hill, New York: pp. 2357–2365. [Google Scholar]
- Fujita Y, Matsuoka H, and Hirooka K 2007. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol 66(4): 829–839. doi: 10.1111/j.1365-2958.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- Greenberg DE, Marshall-Batty KR, Brinster LR, Zarember KA, Shaw PA, Mellbye BL, Iversen PL, Holland SM, and Geller BL 2010. Antisense phosphorodiamidate morpholino oligomers targeted to an essential gene inhibit Burkholderia cepacia complex. J. Infect. Dis 201(12): 1822–1830. doi: 10.1086/652807; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, and Sassetti CM 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7(9): e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Ahn Y, LiPuma JJ, Hussong D, and Cerniglia CE 2015. Survival and susceptibility of Burkholderia cepacia complex in chlorhexidine gluconate and benzalkonium chloride. J. Ind. Microbiol. Biotechnol 42(6): 905–913. doi: 10.1007/s10295-015-1605-x. [DOI] [PubMed] [Google Scholar]
- Kim W, Hendricks GL, Lee K, and Mylonakis E 2017. An update on the use of C. elegans for preclinical drug discovery: screening and identifying anti-infective drugs. Expert Opin. Drug Discov: 1–9. doi: 10.1080/17460441.2017.1319358. [DOI] [PubMed] [Google Scholar]
- Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, and Hu LT 2012. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13(1): 578–595. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R, Krainer AR, and Altman S 2012. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Discov 11(2): 125–140. doi: 10.1038/nrd3625; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan BW, Valenta JA, Benedik MJ, and Wood TK 2013. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother 57(3): 1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RJ, Hamlin JNR, Sivro A, McCorrister SJ, Cardama GA, and Cardona ST 2008. A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J Bacteriol 190(21): 7209–7218. doi: 10.1128/JB.00481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Han SW, Kim G, Song DY, Lee JC, and Kwon KT 2013. An outbreak of Burkholderia cenocepacia associated with contaminated chlorhexidine solutions prepared in the hospital. Am. J. Infect. Control doi: 10.1016/j.ajic.2013.01.024; 10.1016/j.ajic.2013.01.024. [DOI] [PubMed] [Google Scholar]
- Lee SA, Gallagher LA, Thongdee M, Staudinger BJ, Lippman S, Singh PK, and Manoil C 2015. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A 112(16): 5189–5194. doi: 10.1073/pnas.1422186112 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K 2007. Persister cells, dormancy and infectious disease. Nat. Rev 5(1): 48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Lopez C, Arivett BA, Actis LA, and Tolmasky ME 2015. Inhibition of AAC(6’)-Ib-mediated resistance to amikacin in Acinetobacter baumannii by an antisense peptide-conjugated 2’,4’-bridged nucleic acid-NC-DNA hybrid oligomer. Antimicrob. Agents Chemother 59(9): 5798–5803. doi: 10.1128/AAC.01304-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet SA, Flannagan RS, Kooi C, Sokol PA, and Valvano MA 2006. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J. Bacteriol 188(6): 2073–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E, Baldwin A, and Vandamme P 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J.Med.Microbiol 51(7): 533–538. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Urban TA, and Goldberg JB 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev 3(2): 144–156. [DOI] [PubMed] [Google Scholar]
- Matsuoka H, Hirooka K, and Fujita Y 2007. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J. Biol. Chem 282(8): 5180–5194. doi: 10.1074/jbc.M606831200. [DOI] [PubMed] [Google Scholar]
- Moule MG, Hemsley CM, Seet Q, Guerra-Assunção JA, Lim J, Sarkar-Tyson M, Clark TG, Tan PBO, Titball RW, Cuccui J, and Wren BW 2014. Genome-wide saturation mutagenesis of Burkholderia pseudomallei K96243 predicts essential genes and novel targets for antimicrobial development. mBio 5(1): e00926–00913. doi: 10.1128/mBio.00926-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy LR, Burns JL, Lory S, and Lewis K 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol 192(23): 6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman MA, and Brynildsen MP 2015. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat. Commun 6(Journal Article): 7983. doi: 10.1038/ncomms8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega X, Hunt TA, Loutet S, Vinion-Dubiel AD, Datta A, Choudhury B, Goldberg JB, Carlson R, and Valvano MA 2005. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol 187(4): 1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen LCV, Sperling-Petersen HU, and Mortensen KK 2007. Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb. Cell Factories 6: 24. doi: 10.1186/1475-2859-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DL, Frerman FE, and Kim JJ 1996. Three-dimensional structure of human electron transfer flavoprotein to 2.1-A resolution. Proc. Natl. Acad. Sci. U. S. A 93(25): 14355–14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selin C, Stietz MS, Blanchard JE, Gehrke SS, Bernard S, Hall DG, Brown ED, and Cardona ST 2015. A Pipeline for Screening Small Molecules with Growth Inhibitory Activity against Burkholderia cenocepacia. PloS One 10(6): e0128587. doi: 10.1371/journal.pone.0128587 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler Bistue AJ, Martin FA, Vozza N, Ha H, Joaquin JC, Zorreguieta A, and Tolmasky ME 2009. Inhibition of aac(6’)-Ib-mediated amikacin resistance by nuclease-resistant external guide sequences in bacteria. Proc. Natl. Acad. Sci. U. S. A 106(32): 13230–13235. doi: 10.1073/pnas.0906529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sully EK, and Geller BL 2016. Antisense antimicrobial therapeutics. Curr. Opin. Microbiol 33(Journal Article): 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]