Abstract
The classical non-homologous end-joining (cNHEJ) pathway is a major DNA double-strand break repair pathway in mammalian cells and is required for lymphocyte development and maturation. The DNA-dependent protein kinase (DNA-PK) is a cNHEJ factor that encompasses the Ku70-Ku80 (KU) heterodimer and the large catalytic subunit (DNA-PKcs). In mouse models, loss of DNA-PKcs (DNA-PKcs−/−) abrogates end-processing (e.g., hairpin-opening), but not end-ligation, while expression of the kinase-dead DNA-PKcs protein (DNA-PKcsKD/KD) abrogates end-ligation, suggesting a kinase-dependent structural function of DNA-PKcs during cNHEJ. Lymphocyte development is abolished in DNA-PKcs−/− and DNA-PKcsKD/KD mice due to the requirement for both hairpin-opening and end-ligation during V(D)J recombination. DNA-PKcs itself is the best-characterized substrate of DNA-PK. The S2056-cluster is the best-characterized auto-phosphorylation site on human DNA-PKcs. Here we show that radiation can induce phosphorylation of murine DNA-PKcs at the corresponding S2053. We also generated knockin mouse models with alanine- (DNA-PKcsPQR) or phospho-mimetic aspartate (DNA-PKcsSD) substitutions at the S2053 cluster. Despite moderate radiation sensitivity in the DNA-PKcsPQR/PQR fibroblasts and lymphocytes, both DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice retained normal kinase activity, and underwent efficient V(D)J recombination and class switch recombination, indicating that phosphorylation at the S2053-cluster of mouse DNA-PKcs (corresponding to S2056 of human DNA-PKcs), although important for radiation resistance, is dispensable for the end-ligation and hairpin-opening function of DNA-PK essential for lymphocyte development.
Introduction
Lymphocyte development requires ordered assembly and subsequent modifications of the antigen receptor loci through programmed DNA double-strand breaks (DSBs). While the initiation of these DNA breaks is lymphocyte specific, these physiological DSBs activate the DNA damage response and are repaired by the ubiquitously expressed classical non-homologous end-joining (cNHEJ) pathway. DNA-dependent protein kinase (DNA-PK) is a vertebrate-specific cNHEJ factor. DNA-PK holoenzyme includes the evolutionarily conserved DNA-binding Ku70 and Ku80 (KU86 in human) heterodimer (referred to as KU together) and the vertebrate-specific large catalytic subunit (DNA-PKcs). DNA-PK also belongs to the PI3 kinase related serine/threonine protein kinase (PI3KK) family, which also includes ATM and ATR kinases (1). DNA-PKcs and ATM are both activated by DNA DSBs in vivo and share many common substrates, including histone H2AX, KAP1 and DNA-PKcs itself, which contributes to their critical and redundant functions in embryonic development, lymphocyte-specific gene rearrangement, and DNA repair (2–6).
The cNHEJ pathway is one of the two best characterized DNA DSB repair pathways in mammalian cells. As its name implies, cNHEJ ligates two DNA ends together independent of sequence homology. KU initiates cNHEJ by binding to the dsDNA ends, which in turn recruits and activates DNA-PKcs (7). Among other functions, DNA-PK holoenzyme further recruits and activates another vertebrate specific cNHEJ factor, Artemis endonuclease, which processes the DNA ends (e.g., hairpin opening) (8). Finally, the end-ligation is completed by the evolutionarily conserved cNHEJ factors - Ligase4/XRCC4/XLF complex with the help of KU. Two new cNHEJ factors, PAXX, and CYREN/MRI were recently identified and interact with KU directly (9–13). While not essential for cNHEJ in otherwise wild-type cells, they are essential for end-ligation in XLF-deficient cells (9–13). Since cNHEJ functions throughout the cell cycle, it is the predominant repair pathway in non-proliferating cells (e.g., post-mitotic neurons). Complete loss of KU, Ligase4, or Xrcc4 abrogates end-ligation and leads to post-mitotic neuronal apoptosis, and in the case of Ligase4 or Xrcc4-deficiency, p53-dependent embryonic lethality (14). Loss of DNA-PKcs or Artemis abrogates hairpin opening, but not end-ligation. Accordingly, DNA-PKcs−/− and Artemis−/− mice were born of normal size at the expected ratio (15–17). But in contrast to the normal development of DNA-PKcs−/− mice, mice expressing the kinase-dead DNA-PKcs protein (DNA-PKcsKD/KD) die embryonically with severe end-ligation defects (18), indicating that the presence of catalytic inactive DNA-PKcs blocks end-ligation while lack of DNA-PKcs does not.
In developing lymphocytes, the cNHEJ pathway is exclusively required for the assembly of the antigen receptor gene products via V(D)J recombination. Recombination activating gene (RAG) initiates V(D)J recombination by generating a pair of blunt 5’-phosphorylated signal ends and a pair of covalently-sealed hairpin coding ends. The two signal ends can be ligated directly to form a signal joint. The coding end hairpin has to be opened by Artemis with the help of DNA-PKcs before the coding ends can be ligated together to form a coding joint, which encodes the variable region of the Immunoglobulin (Ig) and T cell receptor genes and is ultimately required for lymphocyte development (8). Thus, loss of DNA-PKcs (null) prevents coding end hairpin opening and causes T- and B- severe combined immunodeficiency (T-B-SCID) in animal models (15, 16). In DNA-PKcsKD/KD B cells, both signal joints and coding joints cannot be formed due to end-ligation defects (18). Mature B lymphocytes also undergo additional gene rearrangement - class switch recombination (CSR) at the Immunoglobulin heavy (IgH) chain gene locus to generate antibodies of different isotypes, and therefore different effector functions. End-ligation during CSR is also mediated by the cNHEJ pathway. In the absence of cNHEJ (e.g. XRCC4 deficiency), a subset of CSR can be completed by the alternative end-joining pathway that preferentially uses micro-homologies (MHs) at the junction (19, 20). Consistent with DNA-PKcs not being essential for end-ligation, DNA-PKcs−/− B cells have only moderate defects in CSR (21, 22), while DNA-PKcsKD/KD mature B cells carrying pre-assembled Ig heavy and light chain (IgH/IgL) display severe CSR defects like in XRCC4−/− B cells (23). Nevertheless, high throughput sequencing analyses showed that the residual CSR in both DNA-PKcs−/− and DNA-PKcsKD/KD B cells preferentially use MH at the junctions (23).
How DNA-PKcs kinase activity regulates end-ligation and end-processing is still not fully understood. Loss of KU rescues the embryonic lethality of DNA-PKcsKD/KD mice and deletion of the KU80 C-terminus that is necessary for the recruitment of DNA-PKcs partially restores end-ligation in DNA-PKcsKD/KD cells, indicating that the catalytically inactive DNA-PKcs protein physically blocks end-ligation at the DNA ends. DNA-PKcs is the best-characterized substrate of DNA-PK(24). Several DNA damage-induced phosphorylation sites have been identified on DNA-PKcs, including S2056, T2609, and S3590 (25–28). While the T2609 clusters can be phosphorylated by DNA-PKcs itself (25), ATM (29, 30), or ATR (31), DNA-PKcs specific inhibitors or a catalytically inactive DNA-PKcs mutation abolished the phosphorylation of human DNA-PKcs at the S2056 cluster, establishing S2056 as the bona fide auto-phosphorylation site in human cells (26). Since then, phosphorylation at the S2056 has been widely used as the marker for DNA-PKcs activation. In Chinese hamster ovary (CHO) cells expressing human DNA-PKcs, phosphorylation of DNA-PKcs at the S2056 and T2069 clusters alone and in combination were found to be important for radiation sensitivity, repair pathway choices and release of DNA-PKcs from DNA breaks (26, 32–35). In mouse models, alanine substitutions in the T2609 cluster (DNA-PKcs3A/3A) leads to neonatal lethality associated with p53-dependent bone marrow failure, which also abolished lymphocyte development (36, 37). But the biological consequence of S2056 phosphorylation remains unknown. In fact, whether endogenous murine DNA-PKcs can be phosphorylated upon irradiation has not been demonstrated. Here we developed a customized antibody and showed, for the first time, murine DNA-PKcs can be phosphorylated at S2053 (corresponding to S2056 in human) upon irradiation. Using mouse models with knockin alanine-substitution (DNA-PKcsPQR) or phosphomimetic mutations (DNA-PKcsSD) at the S2056 cluster, we further show that although moderately sensitive to radiation, DNA-PKcsPQR/PQR and DNA-PKcsSD/SD lymphocytes are proficient for both hairpin-opening and end-ligation necessary for development and maturation, even measured by sensitive junctional sequencing analyses. Moreover, the end-joining in DNA-PKcsPQR/PQR or DNA-PKcsSD/SD cells does not require ATM kinase activity and, by extension, compensatory phosphorylation of DNA-PKcs by ATM. Thus, in contrast to the T2609 cluster, the DNA-PKcs S2056 cluster phosphorylation is largely dispensable for cNHEJ during lymphocyte development in mice.
Materials and Methods
Generation and characterization of the mouse models
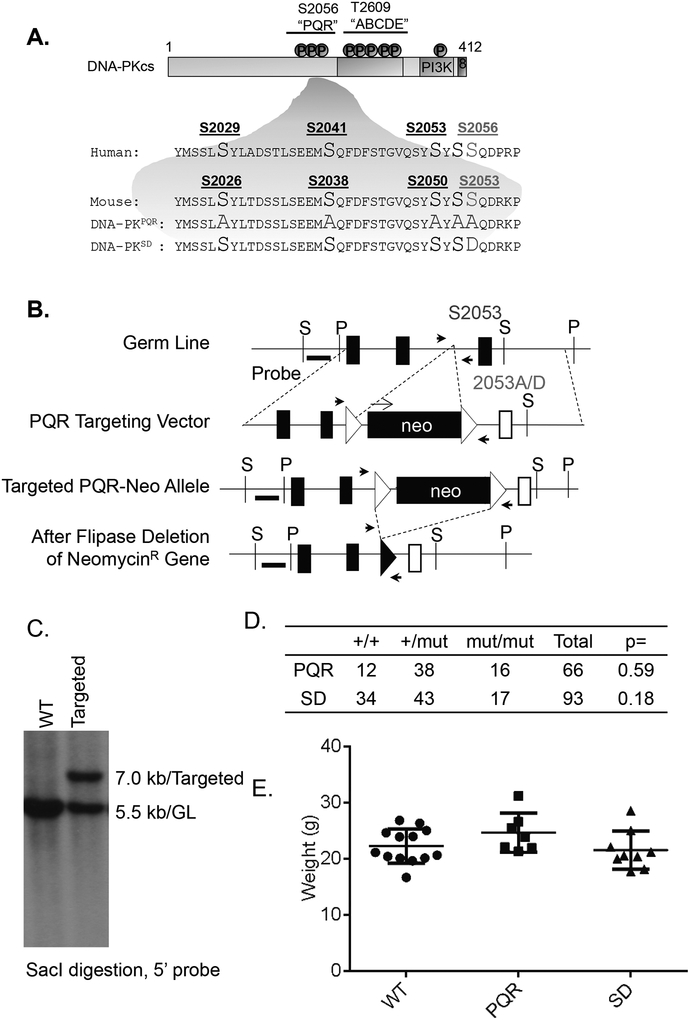
DNA-PKcs−/− and Atm−/− mice have been described previously (15, 38). The DNA-PKcs SD allele and DNA-PKcsSD/SD embryonic stem cells were also described before (18). The DNA-PKcs PQR mutation substitutes five serine residues within the S2053 (corresponding to S2056 in human) cluster with alanine using the same targeting arms as before (Fig.1 and Supplementary Fig 1A). Briefly, the pEMC based targeting construct was used to insert a neomycin resistant (NeoR) cassette in intron 45, about ~600bp upstream from the S2053A mutation site in exon 46 of the murine DNA-PKcs (Prkdc) locus. The 5’ arm is ~3.5 kb and the 3’ arm is ~4.9 kb. The sequence (~665 base pairs) with all mutations was custom synthesized by Genewiz and integrated into the 3’ arm (Fig.1A and Supplementary Fig 1A). CSL3 embryonic stem cells (129/sv background) were targeted by electroporation and screened via Southern blotting (SacI digestion, with the 5’ probe generated by PCR using primers 5’-CAG AAA TGA ACA ATT CCT CCT G-3’ and 5’-CAG TTG TAT GCA GCA CAA TGA-3’) (Fig.1C). The expected germline (GL) band is ~5.5kb and the targeted band is ~7.0 kb (with the insertion of NeoR) (Fig.1C). The correct clones were confirmed with a 3’ probe generated by PCR with primers (5’-GGT TGG TGT GAC GGA GTT TT-3’, 5’-GCT GAG GCT GCT CTT GAA CT-3’). Three out of six targeted clones were sequenced and verified to have the desired DNA-PKcs PQR mutations. Two were injected for germline transmission. The chimeras were bred into the Rosa26FLIP/FLIP mice (JAX Catalogue: 003946) to remove the NeoR cassette. Tail DNA from DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice were PCR amplified and sequenced to confirm the desired mutations (Supplementary Fig 1A and 1B). Genotyping was performed with primers (5’ TTT CCT CTC AAG TAC CAC A-3’ and 5’ AAA TTA TGT CAG GGA TTT AGA 3’) flanking the FRT left after NeoR-deletion. The genotyping primers are marked on Figure 1B as solid black arrows. The PCR product corresponding to the WT allele is 280bp. The PCR product corresponding to the PQR or SD allele is 387bp. A set of representative genotyping results are included in Supplementary Figure 1C. The DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice used in this study were all of the 129/Sv backgrounds. All animal work was conducted in a specific pathogen-free facility and all the procedures were approved by Institutional Animal Care and Use Committee (IACUC) at Columbia University Medical Center.
Figure 1. Generation of the DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mouse models.
(A) Diagram of the S2056 (S2053 in mouse) cluster of DNA-PKcs in human and mouse. (B) The targeting scheme of DNA-PKcsPQR and DNA-PKcsSD allele. Murine DNA-PKcs locus (top), targeting vectors (2nd row), targeted allele (3rd row), and the neo-deleted allele (DNA-PKcsPQR and DNA-PKcsSD, bottom). The Neo-R cassette was inserted at ~600bp upstream from the S2053D mutation site. Not all exons on the arm were marked. P=PstI site and S=SacI site. The 5’ and 3’ probes are marked as thick black lines. The exons and FRT sites are shown as solid boxes and open triangles, respectively. The genotyping primers are marked as solid black arrows. A set of representative genotyping results is shown in Supplementary Figure 1C. The map is not drawn to scale. This clone contains the Neo-R cassette, which is subsequently deleted upon breeding with the constitutive ROSAFLIP/FLIP allele (JAX 003946). The open box indicates the mutated exon (A2053-PQR) and the filled-box indicates the WT exon (S2053). (C) Southern Blot analyses of SacI-digested DNA from DNA-PKcs+/+ and DNA-PKcsPQR targeted ES cells, blotted with the 5’ probe. WT (7.0kb) and targeted (5.5 kb) clone (GL: germline). (D) Breeding frequency to obtain DNA-PKcsPQR and DNA-PKcsSD mice from heterozygous crossing. P value was calculated based on the Fisher exact test. (E) The body weight of adult DNA-PKcsPQR/PQR, DNA-PKcsSD/SD and control mice. The bar represents the average and standard error. The p-value was calculated using student’s t-test. “n.s.” = not significant.
Derivation of Murine Embryonic Fibroblasts (MEFs) and Radiation sensitivity assays.
MEFs were derived from E13.5-E14.5 embryo from timed breeding. The individual embryo was manually dissected. Cells were segregated using trypsin (GIBCO) and cultured in DMEM medium (GIBCO) supplemented with 15% fetal bovine serum. For radiation sensitivity assays, the early passage MEFs (passage 1 or 2) were plated on 96 well plates (2,000 cells/well) in quadruplication. The plates were subjected to ionizing radiation at 1, 2 or 5 Gy. The cellularity was determined by CyQUANT Direct Cell Proliferation Assay(Invitrogen) four days after irradiation. To measure radiation sensitivity in B cells, purified splenic B cells were activated for CSR (see below), irradiated within 1hr after purification in triplicates or quadruplicates, and the cell numbers were determined by an automatic cell counter (Invitrogen Countess) 4 days later. Relative survival was calculated using the cell counts of the un-irradiated group as the denominators.
The development and use of a phosphorylation-specific antibody.
For Western blotting, the cells were lysed with SDS-containing total protein lysis buffer (150mM NaCl, 10mM Tris-HCl (pH7.4), 0.1% SDS, 0.1% Triton-100, 1% Deoxylcholate and 5mM EDTA) and sonicated to disrupt genomic DNA. The protein concentration was measured with the DC protein assay reagent (BioRad). Approximately 100μg total protein (unless otherwise indicated) were electrophoresed on a 6% polyacrylamide gel, transferred to PVDF membrane (with 10% methanol) and blotted for indicated antibodies. The phosphorylation-specific antibody (polyclonal rabbit) recognizing murine DNA-PKcs (Cat No. BL15933) at the S2053 clusters was developed in collaboration with the Bethyl Labortary Inc. using Bethyl’s standard phosphorylation-specific antibody pipeline and was used at 1:500 dilution.
Lymphocyte development and class switch recombination analyses.
Single-cell suspensions were prepared from thymus, bone marrow, spleen, and lymph node from young adult mice. Splenocytes were treated with red blood cell lysis buffer (Lonza ACK Lysis Buffer) for 1–2 minutes at room temperature. Approximately 1 × 105 cells were stained using fluorescence-conjugated antibodies and analyzed by flow cytometry (13, 18). The following antibody cocktails were used for B cells (FITC anti-mouse CD43, Biolegend, 553270; PE Goat anti-mouse IgM, Southern Biotech, 1020–09; PE-Cyanines5 anti-Hu/Mo CD45R (B220), eBioScience 15-0452-83; and APC anti-mouse TER119, Biolegend 116212) and T cells (PE rat anti-mouse CD4, Biolegend 557308; FITC anti-mouse CD8a, Biolegend 100706; PE/Cy5 anti-mouse CD3e, eBioscience 15-0031-83; and APC anti-mouse TCRβ, BD Pharmingen 553174) analyses, respectively. We used the following gate strategies in order when applicable: 1) the dead cells and debris were excluded based on high side scatter and low forward scatter; 2) for bone marrow and spleen (with significant and sometimes variable erythrocytes after red blood cell lysis), the Ter119+ population (mostly erythrocytes) was gated out; 3) the remaining cells were analyzed for B or T cell specific markers as shown in Figures 3 and 4. The endogenous V(D)J recombination junctions were analyzed as previously described (13). DNA from purified splenic B cells were amplified via nesting PCR, first with JH_DQ52: 5’- TGA TAG GCA CCC AAG TAC ACT A −3’ and JH4 Int: 5’- CCT CTC CAG TTT CGG CTG AAT CC-3’ and then with the same JH_DQ52 primer and JH4E: 5’- AGG CTC TGA GAT CCC TAG ACA G −3’. The ~536bp amplicon corresponding to the coding joint between DQ52-JH4 was isolated and cloned into pGEM-T (Promega) and sequenced.
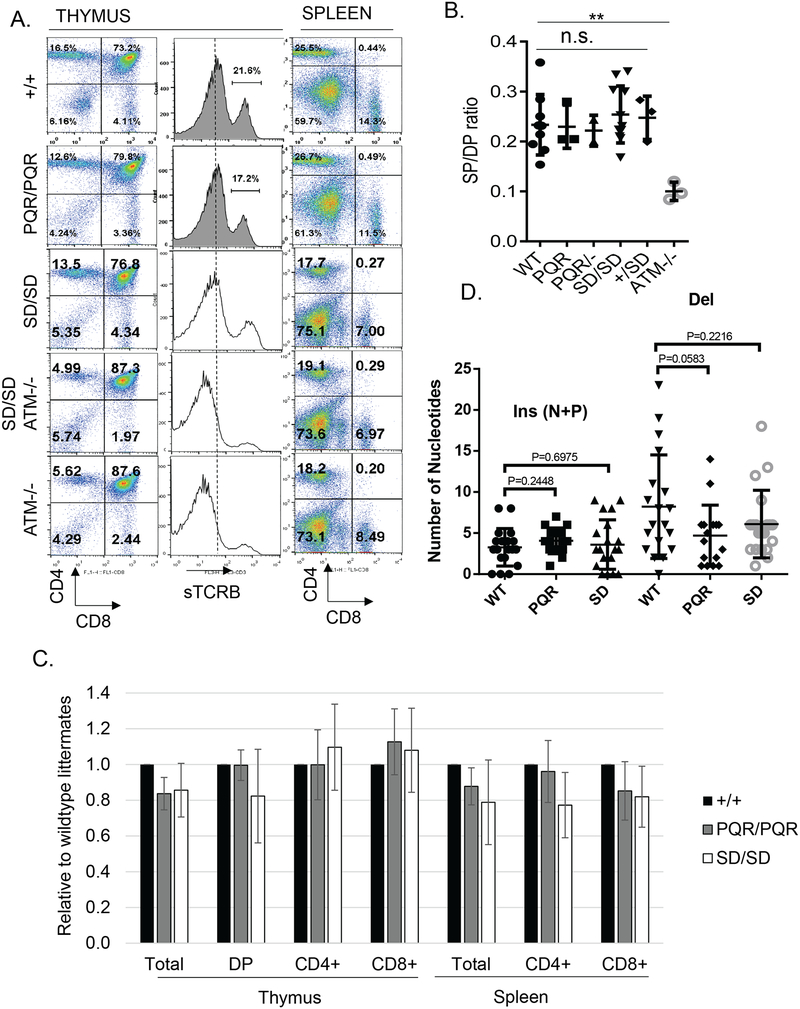
Figure 3. Normal T lymphocyte development in DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice.
(A) Representative flow cytometry analyses of T cell development in the thymus and spleen of DNA-PKcs+/+, DNA-PKcsPQR/PQR, DNA-PKcsSD/SD as well as DNA-PKcsSD/SDATM−/− and ATM−/− mice. (B) The ratio between the sum of CD4+ or CD8+ single T cells and CD4+CD8+ double positive immature T cells in the thymus from mice of different genotypes. The bars represent the average and standard derivation of n>=3 mice of each genotype. Student t-test was used to calculate the p-value. There is no significant difference between WT and all DNA-PKcs mutants. (C) The relative cell number of the different T cell population in the thymus and spleen. The data represent the cellularity as the percentage of the age-matched WT controls analyzed at the same time. More than three biological repeats were performed for each genotype. The DNA-PKcsPQR/- mice are compound heterozygous for the PQR (PQR) and the null (–) alleles. The bar represents the average and standard error. n.s. = not significant for student’s t-test. (D) Comparison of the number of nucleotide deletions and insertions (including both N- and P- elements) in de novo coding joints formed in splenic B cells from DNA-PKcs+/+, DNA-PKcsPQR/PQR, and DNA-PKcsSD/SD mice. Each dot represents a unique joint (n=19 for DNA-PKcs+/+, n=16 for DNA-PKcsPQR/PQR and n=20 for DNA-PKcsSD/SD). The junction sequences are presented in Supplementary Table 1, 2 and 3 (p >0.1 for both insertion and deletion, and for both genotypes).
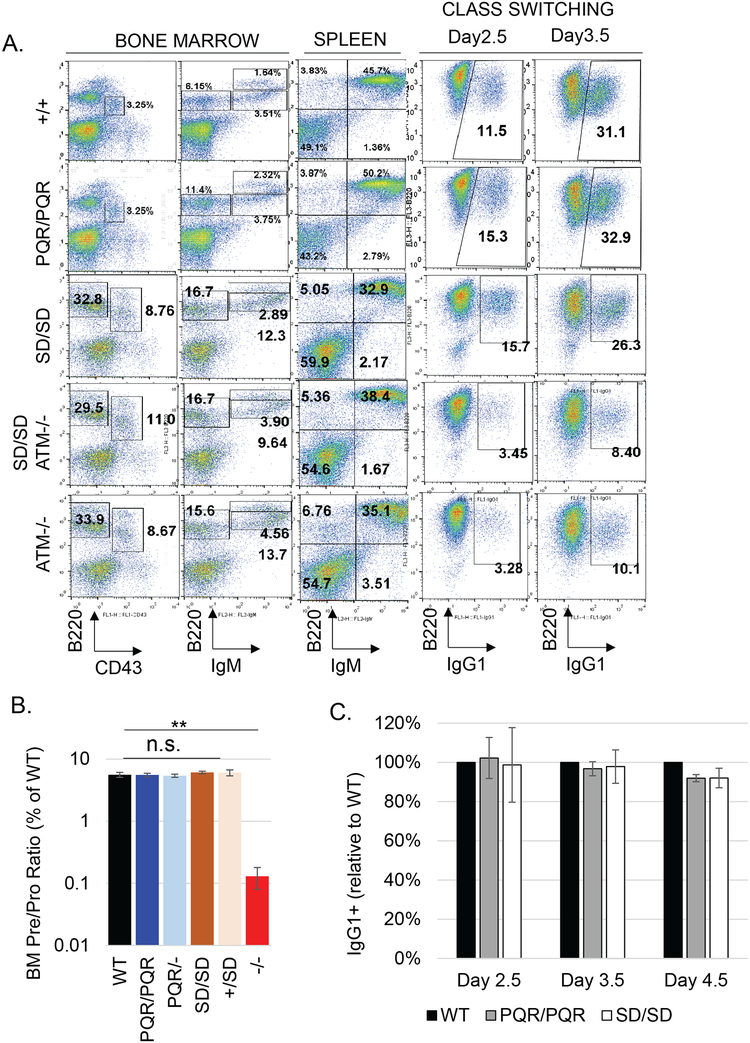
Figure 4. Normal B lymphocyte development and class switch recombination in DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice.
(A) Representative flow cytometry analyses of the development in the bone marrow and spleen and in vitro class switch recombination of B cells from DNA-PKcs+/+, DNA-PKcsPQR/PQR, DNA-PKcsSD/SD as well as DNA-PKcsSD/SDATM−/− and ATM−/− mice. (B) Quantification of bone marrow PreB (B220+IgM-CD43-) vs ProB (B220+IgM-CD43+) B cell ratio in the bone marrow. More than three biological repeats were performed for each genotype. (C) The quantification of CSR kinetics in purified B cells activated in vitro. The bar represents the average and standard error. The p-value was calculated using student’s t-test. n.s. = not significant, and **<0.01.
For the CSR assay, CD43-splenocytes were purified with magnetic beads (MACS, Miltenyi Biotec) and cultured (5 × 105 cells ml−1) in RPMI (GIBCO) supplemented with 10% FBS and 2 μg ml−1 anti-CD40 (BD Bioscience) plus 20 ng ml−1 of IL-4 (R&D). Cells were maintained daily at 1 × 106 cells ml−1 and collected for flow cytometry with directly conjugated antibodies (FITC anti-IgG1, BD Pharmingen, and PE Cy5 anti-B220, eBioscience) after gating out dead cells and debris. Flow cytometry was performed on a FACSCalibur flow cytometer (BD Bioscience) and data were processed using the FlowJo V10 software package. B cell radiation sensitivity is detailed in the radiation sensitivity section above.
High throughput genomic translocation sequence (HTGTS) of CSR junctions
HTGTS was performed as described (23, 39). Briefly, genomic DNA was collected from activated B cells after 4 days, sonicated (Diagenode Bioruptor), and amplified with an Sμ specific biotinylated primer (5’/5BiosG/CAGACCTGGGAATGTATGGT3’) and nested primer (5’CACACAAAGACTCTGGACCTC3’). AflII was used to remove germline (non-arranged) sequences. Since all mice are of pure 129 background, the IgH switch region (from JH4 to the last Cα exon, chr12 114, 494, 415–114, 666, 816) of the C57/BL6 based mm9 was replaced by the corresponding region in the AJ851868.3 129 IgH sequence (1415966–1592715) to generate the mm9sr (switch region replacement) genome and the sequences analyses were performed as detailed previously (39, 40). The best-path searching algorithm (related to YAHA (41)) was used to identify optimal sequence alignments from Bowtie2-reported top alignments (alignment score > 50). The reads were filtered to exclude mis-priming, germline (unmodified), sequential joints, and duplications. To plot all the S-region junctions, including those in the repeats but unequivocally mapped to an individual switch region, we combined the ones filtered by a mappability filter but unequivocally mapped to S regions with ‘good’ reads passing both the mappability (both de-duplicated) filters (39). MHs are defined as regions of 100% homology between the bait and prey-break site. Insertions are defined as regions containing nucleotides that map to neither the bait nor prey-break site. Blunt junctions are considered to have no MHs or insertions. The HTGTS data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE117628 and GSE129895).
Results
DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice
The human DNA-PKcs S2056 phosphorylation cluster contains two SQ sites (human 2041/2056, corresponding to murine S2038/2053) and three other Serine residues (S2029, S2053, and S2055 in human, corresponding to S2026, S2050, and S2052 in murine DNA-PKcs) (Fig. 1A) (24, 33, 34). To interrogate the impact of DNA-PKcs S2053 cluster phosphorylation, we generated a mouse model with alanine substitutions at all 5 potential phosphorylation targets (all serine residues) within the S2053 cluster (DNA-PKcsPQR – corresponding to human S2056) (Fig.1A, 1B and 1C). The desired mutations were confirmed by sequencing the tail DNA from DNA-PKcsPQR/PQR mice and the cDNA from DNA-PKcs+/PQR mice (Supplementary Fig. 1A). The equal presentation of wild type and mutated bases in the cDNA derived from DNA-PKcs+/PQR cells suggests that the DNA-PKcs PQR allele is expressed normally. In parallel, we also injected the previously generated ES cells with a phospho-mimetic mutation at the best-characterized S2056 (S2053D in mouse) (DNA-PKcsSD) (18) for germline transmission (Fig.1A and Supplementary Fig 1B). Complete loss of the end-ligation function of the cNHEJ pathway (e.g., in Ligase4−/−, Xrcc4−/− or DNA-PKcsKD/KD) leads to severe post-mitotic neuronal apoptosis and Tp53-dependent late embryonic lethality in mice (18). In this regard, both DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice were born at Mendelian ratios (Fig.1D), are of normal size (Fig.1E and Supplementary Fig. 1D) and are fertile in both males and females, suggesting that phosphorylation at the S2056 cluster is not essential for embryonic development and, by extension, the end-ligation required in post-mitotic neurons.
Irradiation dependent phosphorylation of S2053 in murine DNA-PKcs
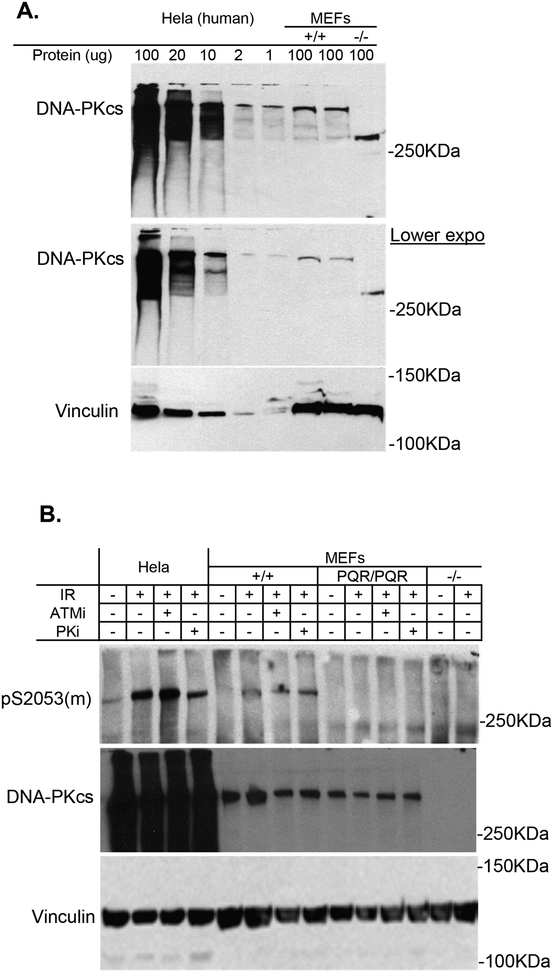
Despite the sequence similarity (Fig. 1A), the commercial antibodies developed against S2056 phosphorylated human DNA-PKcs was not able to recognize the corresponding phosphorylation (S2053) in murine DNA-PKcs (Supplementary Fig. 1E). This cannot be solely explained by the nearly 20 fold lower abundance of DNA-PKcs protein in mouse cells (MEFs) relative to human counterparts (Hela) (Fig. 2A and Supplementary Fig. 1E), since commercial antibodies against human DNA-PKcs T2609 phosphorylation can effectively recognize corresponding phosphorylation in murine cells (Supplementary Fig. 1E). We noted that there is a proline to arginine difference 3 amino acids, after the conserved S2053 (2056 in human DNA-PKcs), which could potentially affect antibody recognition. Another possibility is that the percentage of DNA-PKcs phosphorylated at S2053 might be much lower than that at T2609, which could also further compromise detection. We reasoned that a phosphorylation-specific antibody developed against pS2053 of murine DNA-PKcs might be able to recognize both human and murine DNA-PKcs phosphorylation. In collaboration with the Bethyl Laboratories Inc., we developed a novel antibody using phosphorylated murine DNA-PKcs peptide (corresponding to pS2053) as an immunogen. Our result shows that irradiation can induce phosphorylation of murine DNA-PKcs at S2053. Importantly, radiation-induced S2053 phosphorylation is not detectable in DNA-PKcs null cells and is abolished in DNA-PKcsPQR/PQR cells, supporting the specificity of the antibody (Fig. 2B). As predicted, this antibody can effectively detect phosphorylation of human DNA-PKcs at S2056, which is significantly reduced in the presence of a DNA-PKcs-specific inhibitor (NU7441) (Fig. 2B). Notably, radiation-induced phosphorylation of murine DNA-PKcs at S2053 was not significantly decreased by either ATM-specific inhibitor (KU95533) or DNA-PKcs inhibitor (NU7441) alone (Fig. 2B), suggesting potential redundancy. Nevertheless, the results, for the first time, demonstrate that endogenous murine DNA-PKcs can be phosphorylated at S2053 upon irradiation and confirm that the DNA-PKcsPQR allele leads to the expression of DNA-PKcs that cannot be phosphorylated at S2053.
Figure 2. DNA-PKcsPQR allele abolished irradiation induced phosphorylation at S2053 of murine DNA-PKcs.
(A) Western blotting analyses show that the abundance of DNA-PKcs in human cells (Hela) is between 10–50 fold higher than that of murine embryonic fibroblast. Anti-DNA-PKcs (1:200, Invitrogen). (B) The phosphorylation-specific antibody developed for S2053 of murine DNA-PKcs can reliably detect irradiation (20 Gy, after 1hr) induced DNA-PKcs phosphorylation at S2053 in murine DNA-PKcs and S2056 in human DNA-PKcs.
Normal lymphocyte development in DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice
The cNHEJ pathway is exclusively required for the somatic assembly of the Ig and TCR gene products via V(D)J recombination. So next, we examined lymphocyte development in DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice. The weight and total cellularity of the lymphoid organs (thymus and spleens) of DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice were indistinguishable from age-matched control DNA-PKcs+/+ littermates (Supplementary Fig 2A and 2B). Flow cytometry analyses showed that the number and frequency of T cell progenitors and mature T cells are indistinguishable in DNA-PKcsPQR/PQR, DNA-PKcsSD/SD, and littermate control DNA-PKcs+/+ mice (Fig. 3A and 3C, Supplementary Fig. 2A and 2B). Sequential rearrangements of the TCRα locus in CD4+CD8+ double positive (DP) thymocytes are coupled with both positive and negative selections. Relatively moderate V(D)J recombination defects in ATM- or 53BP1-deficient mice result in reduced surface TCRβ and its co-receptor CD3 levels on the DP thymocytes (Fig. 3A) (42, 43) and a significant reduction of the mature single positive (SP) vs immature DP T cells ratio (Fig. 3B). Yet, surface expression of TCRβ/CD3 in DP cells and the SP/DP ratio are unaffected in DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice (Fig. 3A and 3B). Likewise, the frequency of immature pro-B (CD43+B220+IgM−), pre-B (CD43−B220+IgM−), newly generated naïve B (IgM+B220low), and re-circulating B (IgM+B220hi) cells in the bone marrow from DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice are also comparable to those of DNA-PKcs+/+ mice (Fig. 4A). Successful V(D)J recombination at the IgH locus is required for the transition from pro-B to pre-B cells. Thus, DNA-PKcs−/− mice with hairpin opening defects, demonstrate a severe blockade in pro-B to pre-B cell transition, evidenced by a drop of the pre-B/pro-B ratio in the bone marrow (Fig. 4B). In contrast, the ratios of bone marrow-derived pre-B/pro-B cells are comparable in DNA-PKcsPQR/PQR, DNA-PKcsSD/SD and the DNA-PKcs+/+mice (Fig. 4B). Furthermore, lymphocyte development and number are also normal in DNA-PKcsPQR/- mice with compound heterozygosity of the PQR allele and a true null allele (Figure 3C, 4B and supplementary 2A), suggesting even expression of ~50% of DNA-PKcs-PQR polypeptide is sufficient to support lymphocyte development in vivo. Finally, sequence analyses of the non-selective DQ52 to JH4 junctions from B cells derived from DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice did not find excess nucleotide insertion or deletion (Fig. 3D and Supplementary Table 1–2). Together, these data indicate that phosphorylation at the S2053 (S2056 in human) cluster is not required for the end-ligation and hairpin opening that are necessary for V(D)J recombination and early lymphocyte development.
End-ligation during Class Switch Recombination is also unaffected in DNA-PKcsPQR/PQR and DNA-PKcsSD/SD B cells
Mature B cells undergo CSR at the IgH locus to generate antibodies with different effector functions. During CSR, a DSB in the upstream μ switch region (Sμ) is joined to another DSB in a downstream switch region (e.g., Sγ1). The end-ligation during CSR is also mediated by the cNHEJ pathway. Consistent with significant ligation defects, DNA-PKcsKD/KD mature B cells have severe CSR defects, like Xrcc4−/− B cells (23). In cNHEJ hypomorphic XLF-deficient B cells, chromosomal V(D)J recombination is not significantly affected, but CSR is reduced by nearly 50% (44), suggesting CSR might be more sensitive to mild cNHEJ deficiency. So, we analyzed CSR of purified splenic B cells drived from DNA-PKcsPQR/PQR, DNA-PKcsSD/SD mice, and control DNA-PKcs+/+ littermates. In vitro stimulation with anti-CD40 and interleukin 4 (IL-4) induced robust CSR to IgG1, and expression of surface IgG1 in ~30% of DNA-PKcs+/+, DNA-PKcsPQR/PQR, and DNA-PKcsSD/SD B cells by day 4 (Fig. 4A and 5C and Supplementary Fig.2C), suggesting normal CSR. Moreover, the kinetics of CSR to IgG1 is also not affected in DNA-PKcsPQR/PQR and DNA-PKcsSD/SD B cells (Fig. 4C).
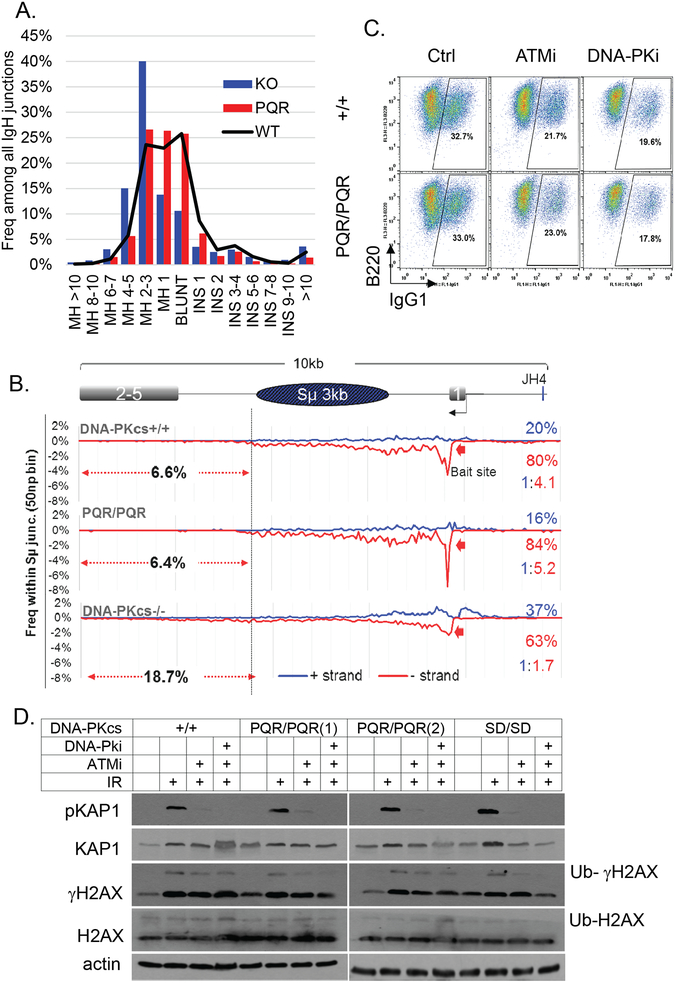
Figure 5. Normal Class Switch Recombination junction and DNA-PKcs kinase activity in DNA-PKcsPQR/PQR mice.
(A) HTGTS analyses of CSR junctions show the micro-homology (MH) and insertion (INS) usage among all IgH junctions recovered from DNA-PKcsPQR/PQR (n=1559), DNA-PKcs+/+ (n=10465), and control DNA-PKcs−/− (n=717) mice. (B) The distribution of the Sμ-Sμ joins within the 10Kb region include the core Sμ region. The percentage shows the frequency of junctions that fall downstream of the core Sμ region. The red (in the telomere to centromere orientation) and the blue (in the centromere to telomere orientation) numbers on the right show the percentage and ratio of junctions that fall in given orientations, respectively. Since the IgH locus resides in the telomere to centromere orientation, the normal deletional joints fall in the telomere to centromere orientation (red). (C) (D) Western blotting of ATM and DNA-PKcs substrate phosphorylation upon IR (10 Gy) with or without treatment with ATM inhibitor (KU55933, 10μM) and DNA-PK kinase inhibitor (NU7441, 10μM). Primary antibodies were used at the following dilutions: anti-H2AX (1:1000, Millipore), anti-gH2AX (1:1000, Millipore), anti-KAP1 (1:1000, Cell Signaling), anti-phospho-KAP1 (S824) (1:1000, Bethyl Laboratories), and anti-actin (1:5000, Sigma).
In the absence of cNHEJ (e.g., Xrcc4-deficiency), a subset of CSR can be completed by the alternative-end-joining pathways that preferentially use MH at the junction (19, 20). We used HTGTS (40) to analyze thousands of CSR junctions between the 5’ of Sμ and its joining partner genome-wide, including both internal deletion (Sμ-Sμ) and potentially productive switching (Sμ-Sγ1). In a recent study (23), using HTGTS, we uncovered a prominent shift to MH, extensive erosion within the switch region, and a relative increase in inversions (marked by red: blue ratio) in DNA-PKcs−/− B cells with only moderate CSR defects (21, 22) (Fig. 5A). So we performed HTGTS analyses of CSR junctions from DNA-PKcsPQR/PQR and control DNA-PKcs−/− B cells. While the results confirmed S region erosion and increased MH usage in DNA-PKcs−/− B cells, no change in MH usage (Fig. 5A) or Sμ junction distribution (Fig. 5B) was noted in DNA-PKcsPQR/PQR B cells. Together, these results indicate that phosphorylation at the S2056 cluster, unlike expression of the kinase-dead DNA-PKcs or loss of DNA-PKcs, does not affect cNHEJ during V(D)J recombination or CSR.
S2053 cluster phosphorylation does not affect DNA-PK kinase activity or DNA damage response in activated B cells
DNA end-ligation in murine B cells with alanine substitutions at the T2609 clusters (DNA-PKcs3A/3A) is hypersensitive to ATM kinase inhibition, suggesting ATM might phosphorylate other substrates, including other sites on DNA-PKcs to promote end-ligation in DNA-PKcs3A/3A cells (45). So next, we determined whether the S2056 mutation affects DNA-PKcs kinase activity and whether the end-ligation in DNA-PKcsPQR/PQR cells depends on compensatory phosphorylation by ATM kinase. Ionizing radiation (IR) induced phosphorylation of KAP1 and H2AX, two substrates shared by DNA-PK and ATM (4–6), is not different in DNA-PKcsPQR/PQR and DNA-PKcsSD/SD B cells, compared to DNA-PKcs+/+ controls, both in the presence or absence of ATM kinase inhibitor (KU55933) (Fig. 5D). This is consistent with in vitro findings (26) and indicates that S2056 phosphorylation does not affect DNA-PK kinase activity. ATM and DNA-PK have critical redundant functions during embryonic development (2, 3) and CSR (6) because of their overlapping substrates. Consistent with normal kinase activity in DNA-PKcsSD/SD mice and the lack of dependence on ATM kinase activity, DNA-PKcsSD/SDAtm−/− mice were born at the expected ratio (Supplementary Figure 2D). DNA-PK kinase inhibitor (NU7441, 10μM), ATM kinase inhibitor (KU55933, 7.5μM) or ATM deletion did not further reduce the CSR in DNA-PKcsPQR/PQR or DNA-PKcsSD/SD B cells beyond what was observed in DNA-PKcs+/+ B cells (Fig. 4A and Supplementary Figure 2C). Thus, the S2056 phosphorylation does not affect DNA-PK kinase activity or its sensitivity to ATM inhibition.
S2053 cluster affects the radiation sensitivity of both murine fibroblasts and activated B cells.
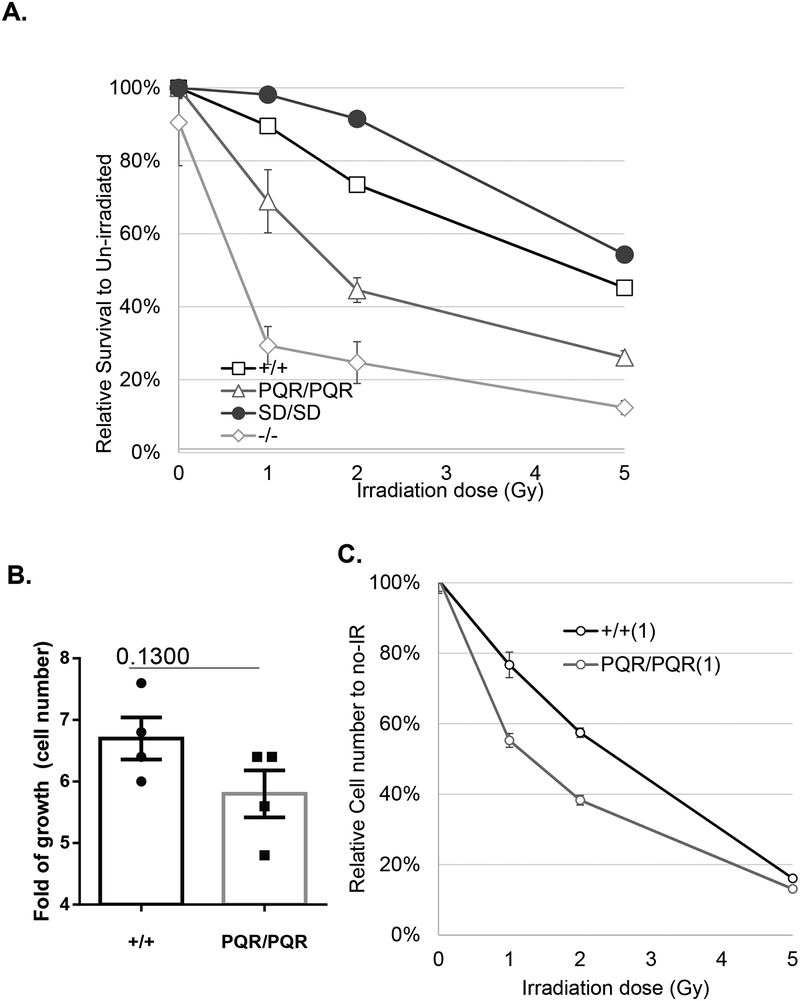
S2056 cluster phosphorylation was implicated in IR sensitivity in human cells (26). Next, we derived primary MEFs from both DNA-PKcsPQR/PQR and DNA-PKcsSD/SD mice. While DNA-PKcs−/− MEFs are sensitive to IR as previously reported, DNA-PKcsPQR/PQR MEFs are only moderately sensitive to IR, and DNA-PKcsSD/SD MEFs even display a mild resistance to IR (Fig. 6A). To determine whether S2053 phosphorylation has cell type-dependent roles, which might explain the lack of lymphocyte development defects in DNA-PKcsPQR/PQR mice despite the mild IR sensitivity in MEFs, we measured IR sensitivity in DNA-PKcsPQR/PQR B cells activated for CSR. Activated DNA-PKcsPQR/PQR B cells proliferate well upon activation (Fig. 6B), consistent with normal development of DNA-PKcsPQR/PQR mice and normal CSR of DNA-PKcsPQR/PQR B cells. Yet, in two independent experiments, DNA-PKcsPQR/PQR B cells displayed moderate, yet consistent IR sensitivity (Fig. 6C and Supplementary Figure 2E). Thus, we conclude that phosphorylation of murine DNA-PKcs at the S2053 cluster promotes radiation resistance in both MEFs and B cells. Given the normal lymphocyte development, the results suggest that S2056 phosphorylation might be necessary for the repair of a subset of complex DNA lesions generated upon radiation or perhaps S2056 phosphorylation plays a role in the response to acute pathological insult, but not to physiological DNA strand breaks during lymphocyte development.
Figure 6. DNA-PKcsPQR/PQR cells are moderately sensitive to radiation.
(A) The sensitivity to ionizing radiation (1, 2, 5 Gy) of DNA-PKcs+/+, DNA-PKcs−/−, DNA-PKcsPQR/PQR, and DNA-PKcsSD/SD primary MEFs. The relative survival represents the relative cellularity at the given dose as a proportion of the un-irradiated sample of the same genotype. The data represent the average and standard error of at least 5 independent wells per genotype per dose. The radiation sensitivity of DNA-PKcsPQR/PQR MEFs is statistically significantly different from both DNA-PKcs+/+ and DNA-PKcs−/− MEFs at all three doses tested (p<0.001). (B) Relative fold of growth (by cell counts) for DNA-PKcs+/+ and DNA-PKcsPQR/PQR splenic B cells during class switch recombination (4 days). The bars mark the means and standard errors. The unpaired student’s t-test p-value is 0.13. (C) The sensitivity to ionizing radiation (1, 2, 5 Gy) of DNA-PKcs+/+ and DNA-PKcsPQR/PQR B cells activated for CSR. Two independent pairs of DNA-PKcs+/+ and DNA-PKcsPQR/PQR B cells were assayed in two independent experiments (another is shown in Supplementary Figure 2E). DNA-PKcsPQR/PQR B cells are significantly more sensitive to radiation than the DNA-PKcs+/+ controls at all three radiation doses (p<0.05).
Discussion
Here we developed a phosphorylation-specific antibody and showed that murine DNA-PKcs can be phosphorylated at the corresponding S2053 site upon radiation. Notably, in contrast to the phosphorylation of S2056 in human DNA-PKcs, radiation-induced phosphorylation of murine DNA-PKcs at S2053 is not abolished by either DNA-PKcs or ATM specific inhibitors, suggesting potential redundancy. Nevertheless, analyses of the DNA-PKcsPQR/PQR, DNA-PKcsPQR/- (presumably ~50% PQR protein) and DNA-PKcsSD/SD mice confirmed the role of S2053 phosphorylation in radiation resistance of both fibroblasts and B cells. Yet unexpectedly, the in vivo analyses of lymphocyte development clearly indicate that S2053 phosphorylation is dispensable for end-ligation and the hairpin-opening required during both V(D)J recombination and Ig CSR. In this context, DNA-PKcsPQR/PQR MEFs are much less sensitive to IR than DNA-PKcs−/− MEFs, suggesting the S2053 phosphorylation is only required for a subset of DNA-PKcs dependent repair. In contrast to the relatively clean DNA ends generated during lymphocyte development, radiation generates complex DNA lesions with potential base modifications, which might require additional DNA-PKcs function. Alternatively, the acute activation of DNA-PKcs upon radiation might require more robust DNA-PKcs activity than physiological gene rearrangement. Nevertheless, careful analyses of V(D)J and CSR junctions show that S2056 phosphorylation does not affect the quality or the quantity of the junctions. Collectively, these findings are consistent with the phosphorylation of DNA-PKcs at the S2056 cluster being largely dispensable for physiological DSB repair during lymphocyte development and suggest a potential substrate-specific role of S2056 phosphorylation during DNA repair.
This apparent discrepancy between the strong phenotype in the kinase-dead model (18) vs. the lack of detectable phenotype in the S2056 auto-phosphorylation site mutant is not unique to DNA-PKcs. Similar findings were also noted for ATM kinase (46–49), forcing us to consider other phosphorylation sites or phosphorylation-independent effects of catalysis on DNA-PKcs and ATM. In this context, the mouse model with alanine substitutions at another DNA-PKcs phosphorylation cluster (T2609) (DNA-PKcs3A/3A) succumbed to bone marrow failure shortly after birth (36). Although DNA-PKcs3A/3A B cells are proficient at chromosomal V(D)J recombination, cNHEJ in DNA-PKcs3A/3A B cells is hypersensitive to ATM inhibition (45), suggesting that ATM kinase might phosphorylate other targets, or other sites of DNA-PKcs to promote cNHEJ. Yet, ATM inhibition or ATM deletion does NOT further impair end-ligation required for lymphocyte development or CSR in DNA-PKcsPQR/PQR and DNA-PKcsSD/SD lymphocytes, suggesting that at least the cNHEJ function in DNA-PKcsPQR/PQR cells cannot be explained by compensatory phosphorylation via ATM. Although we did not formally exclude the contribution of ATR kinase, ATR kinase activity is essential for cell and embryonic viability and ATR is usually activated by RPA coated ssDNA, not DSBs (50). In addition, the catalysis itself might trigger conformational changes of DNA-PKcs independent of any specific auto-phosphorylation sites. Early biochemical studies suggested that binding of purified DNA-PKcs to DNA is affected by ATP hydrolysis, but not the resulted phosphorylation (51). Such catalysis dependent allosteric changes have been reported for PARP1, which is also activated upon DNA binding (52). The published structure of purified DNA-PKcs adopted a few different conformations (32, 53), but how and which reflects the in vivo structure of DNA-PKcs remain elusive.
In summary, our findings unequivocally demonstrate an important distinction between auto-phosphorylation and catalysis itself, indicating that auto-phosphorylation at S2056, although a good marker for DNA-PK activation, is largely dispensable for cNHEJ and DNA damage responses required for lymphocyte development. Although end-ligation in DNA-PKcsPQR/PQR cells and mice are not hypersensitive to ATM loss, the moderate radiation sensitivity of DNA-PKcsPQR/PQR cells suggest the possibility that the apparently efficient end-ligation in DNA-PKcsPQR/PQR cells might be vulnerable to other disturbances, including DNA-PKcs mediated phosphorylation of other sites on DNA-PKcs and the loss of other non-essential cNHEJ factors (e.g., XLF, PAXX or MRI). This possibility should be tested in future experiments.
Supplementary Material
Key points.
Radiation induces S2053 phosphorylation of murine DNA-PKcs.
S2053 phosphorylation is dispensible for V(D)J and class switch recombination.
Acknowledgment
We thank Dr. Eric Mcintush and the Bethyl Laboratories Inc. for helping us develop the phosphorylation-specific antibody against murine DNA-PKcs. We thank Dr. Chyuan-Sheng (Victor) Lin and Ms. Li Chen for technical assistance and advice on the generation of the new mouse models. We thank other members of the Zha lab for helpful discussions and technical advice. We apologize to colleagues, whose work could not be cited due to space limitations and was covered by reviews instead.
Grant Support
This work is in part supported by NIH/NCI 5R01CA158073, 5R01CA184187 and R01CA226852. SZ is the recipient of the Leukemia Lymphoma Society Scholar Award. WJ was supported by NIH/NCI T32-CA09503. JC was supported by NIH/NCI F31CA183504–01A1 and CTSA/NIH TL1 TR000082. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30CA013696 to Herbert Irving Comprehensive Cancer Center (HICCC) of Columbia University.
Abbreviations
- cNHEJ
classical non-homologous end-joining
- CSR
class switch recombination
- DNA-PK
DNA-dependent protein kinase
- DSB
double-strand break
- HTGTS
High throughput genomic translocation sequence
- IR
Ionizing radiation
- MEF
Murine Embryonic Fibroblast
- MH
micro-homology
References
- 1.Shiloh Y 2003. ATM and related protein kinases: safeguarding genome integrity. Nat.Rev.Cancer 3:155–168. [DOI] [PubMed] [Google Scholar]
- 2.Gurley KE, Kemp CJ 2001. Synthetic lethality between mutation in Atm and DNA-PK(cs) during murine embryogenesis. Curr.Biol. 11:191–194. [DOI] [PubMed] [Google Scholar]
- 3.Sekiguchi J, Ferguson DO, Chen HT, Yang EM, Earle J, Frank K, Whitlow S, Gu Y, Xu Y, Nussenzweig A, Alt FW 2001. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc.Natl.Acad.Sci.U.S.A 98:3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gapud EJ, Dorsett Y, Yin B, Callen E, Bredemeyer A, Mahowald GK, Omi KQ, Walker LM, Bednarski JJ, McKinnon PJ, Bassing CH, Nussenzweig A, Sleckman BP 2011. Ataxia telangiectasia mutated (Atm) and DNA-PKcs kinases have overlapping activities during chromosomal signal joint formation. Proc Natl Acad Sci U S A 108:2022–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zha S, Jiang W, Fujiwara Y, Patel H, Goff PH, Brush JW, Dubois RL, Alt FW 2011. Ataxia telangiectasia-mutated protein and DNA-dependent protein kinase have complementary V(D)J recombination functions. Proc Natl Acad Sci U S A 108:2028–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, Nussenzweig M 2009. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell 34:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falck J, Coates J, Jackson SP 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434:605–611. [DOI] [PubMed] [Google Scholar]
- 8.Ma Y, Pannicke U, Schwarz K, Lieber MR 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781–794. [DOI] [PubMed] [Google Scholar]
- 9.Hung PJ, Johnson B, Chen BR, Byrum AK, Bredemeyer AL, Yewdell WT, Johnson TE, Lee BJ, Deivasigamani S, Hindi I, Amatya P, Gross ML, Paull TT, Pisapia DJ, Chaudhuri J, Petrini JJH, Mosammaparast N, Amarasinghe GK, Zha S, Tyler JK, Sleckman BP 2018. MRI Is a DNA Damage Response Adaptor during Classical Non-homologous End Joining. Mol Cell 71:332–342 e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, Travers J, Wu Q, Draviam VM, Robinson CV, Blundell TL, Jackson SP 2015. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347:185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing M, Yang M, Huo W, Feng F, Wei L, Jiang W, Ning S, Yan Z, Li W, Wang Q, Hou M, Dong C, Guo R, Gao G, Ji J, Zha S, Lan L, Liang H, Xu D 2015. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nature communications 6:6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar V, Alt FW, Frock RL 2016. PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1-arrested progenitor B-cell line. Proc Natl Acad Sci U S A 113:10619–10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Shao Z, Jiang W, Lee BJ, Zha S 2017. PAXX promotes KU accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice. Nature communications 8:13816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieber MR 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver DT, Alt FW 1998. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 9:367–376. [DOI] [PubMed] [Google Scholar]
- 16.Taccioli GE, Amatucci AG, Beamish HJ, Gell D, Xiang XH, Torres Arzayus MI, Priestley A, Jackson SP, Marshak RA, Jeggo PA, Herrera VL 1998. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 9:355–366. [DOI] [PubMed] [Google Scholar]
- 17.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, Alt FW 2002. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol.Cell 10:1379–1390. [DOI] [PubMed] [Google Scholar]
- 18.Jiang W, Crowe JL, Liu X, Nakajima S, Wang Y, Li C, Lee BJ, Dubois RL, Liu C, Yu X, Lan L, Zha S 2015. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol Cell 58:172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis J, Nussenzweig A, Nussenzweig M, Alt FW 2010. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med 207:417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482. [DOI] [PubMed] [Google Scholar]
- 21.Bosma GC, Kim J, Urich T, Fath DM, Cotticelli MG, Ruetsch NR, Radic MZ, Bosma MJ 2002. DNA-dependent protein kinase activity is not required for immunoglobulin class switching. J.Exp.Med. 196:1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manis JP, Dudley D, Kaylor L, Alt FW 2002. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity. 16:607–617. [DOI] [PubMed] [Google Scholar]
- 23.Crowe JL, Shao Z, Wang XS, Wei PC, Jiang W, Lee BJ, Estes VM, Alt FW, Zha S 2018. Kinase-dependent structural role of DNA-PKcs during immunoglobulin class switch recombination. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP 2004. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res 32:4351–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev 16:2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ 2005. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem 280:14709–14715. [DOI] [PubMed] [Google Scholar]
- 27.Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, Ye R, Morrice N, Lees-Miller SP, Meek K 2007. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol Cell Biol 27:1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, Bazett-Jones DP, Lees-Miller SP 2002. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry 41:12706–12714. [DOI] [PubMed] [Google Scholar]
- 29.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ 2007. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem 282:6582–6587. [DOI] [PubMed] [Google Scholar]
- 30.Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, Meek K, Alessi DR, Lees-Miller SP 2002. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem J 368:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yajima H, Lee KJ, Chen BP 2006. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol 26:7520–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K 2003. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol 23:5836–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K 2005. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol 25:10842–10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP 2007. trans Autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol 27:3881–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ 2007. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 177:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen HT, Wong N, Bunting S, Lin YF, Li M, Lee KJ, Story M, Gapud E, Sleckman BP, Nussenzweig A, Zhang CC, Chen DJ, Chen BP 2011. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol 193:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S, Matsunaga S, Lin YF, Sishc B, Shang Z, Sui J, Shih HY, Zhao Y, Foreman O, Story MD, Chen DJ, Chen BP 2016. Spontaneous tumor development in bone marrow-rescued DNA-PKcs(3A/3A) mice due to dysfunction of telomere leading strand deprotection. Oncogene 35:3909–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zha S, Sekiguchi J, Brush JW, Bassing CH, Alt FW 2008. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc.Natl.Acad.Sci.U.S.A. 105:9302–9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong J, Panchakshari RA, Zhang T, Zhang Y, Hu J, Volpi SA, Meyers RM, Ho YJ, Du Z, Robbiani DF, Meng F, Gostissa M, Nussenzweig MC, Manis JP, Alt FW 2015. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature 525:134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, Frock RL 2016. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat Protoc 11:853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faust GG, Hall IM 2012. YAHA: fast and flexible long-read alignment with optimal breakpoint detection. Bioinformatics 28:2417–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borghesani PR, Alt FW, Bottaro A, Davidson L, Aksoy S, Rathbun GA, Roberts TM, Swat W, Segal RA, Gu Y 2000. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc.Natl.Acad.Sci.U.S.A 97:3336–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, Sleckman BP, Nussenzweig A 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456:529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S 2008. Lymphocyte-Specific Compensation for XLF/Cernunnos End-Joining Functions in V(D)J Recombination. Mol.Cell. 31:631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee BS, Gapud EJ, Zhang S, Dorsett Y, Bredemeyer A, George R, Callen E, Daniel JA, Osipovich O, Oltz EM, Bassing CH, Nussenzweig A, Lees-Miller S, Hammel M, Chen BP, Sleckman BP 2013. Functional Intersection of ATM and DNA-PKcs in Coding End Joining During V(D)J Recombination. Mol Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto K, Wang Y, Jiang W, Liu X, Dubois RL, Lin CS, Ludwig T, Bakkenist CJ, Zha S 2012. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J Cell Biol 198:305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A 2006. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. [DOI] [PubMed] [Google Scholar]
- 48.Daniel JA, Pellegrini M, Lee JH, Paull TT, Feigenbaum L, Nussenzweig A 2008. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J.Cell Biol. 183:777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniel JA, Pellegrini M, Lee BS, Guo Z, Filsuf D, Belkina NV, You Z, Paull TT, Sleckman BP, Feigenbaum L, Nussenzweig A 2012. Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol 198:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menolfi D, Jiang W, Lee BJ, Moiseeva T, Shao Z, Estes V, Frattini MG, Bakkenist CJ, Zha S 2018. Kinase-dead ATR differs from ATR loss by limiting the dynamic exchange of ATR and RPA. Nature communications 9:5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jovanovic M, Dynan WS 2006. Terminal DNA structure and ATP influence binding parameters of the DNA-dependent protein kinase at an early step prior to DNA synapsis. Nucleic Acids Res 34:1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langelier MF, Zandarashvili L, Aguiar PM, Black BE, Pascal JM 2018. NAD(+) analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nature communications 9:844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobbs TA, Tainer JA, Lees-Miller SP 2010. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 9:1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.