Abstract
Background:
Congenital heart defects (CHD) are a leading cause of morbidity and mortality in children and despite advanced surgical treatments, many patients progress to heart failure. Currently, transplantation is the only effective cure and is limited by donor availability and organ rejection. Recently, cell therapy has emerged as a novel method for treating pediatric heart failure with several ongoing clinical trials. However, efficacy of stem cell therapy is variable and choosing stem cells with highest reparative effects has been a challenge.
Methods:
We previously demonstrated the age-dependent reparative effects of human c-kit+ progenitor cells (hCPC) in a rat model of juvenile heart failure. Using a small subset of patient samples, computational modeling analysis showed that regression models could be made linking sequencing data to phenotypic outcomes. In the current study, we used a similar quantitative model to determine whether predictions can be made in a larger population of patients and validated the model using neonatal hCPCs. We performed RNAseq from CPCs isolated from 32 patients, including 8 neonatal samples. We tested 2 functional parameters of our model, cellular proliferation and chemotactic potential of conditioned media.
Results:
Interestingly, the observed proliferation and migration responses in each of the selected neonatal hCPC lines matched their predicted counterparts. We then performed canonical pathway analysis to determine potential mechanistic signals that regulated hCPC performance, and identified several immune response genes that correlated with performance. ELISA analysis confirmed the presence of selected cytokines in good performing hCPCs, and provided many more signals to further validate.
Conclusions:
These data show that cell behavior may be predicted using large datasets like RNAseq, and that we may be able to identify patients whose CPC exceed or underperform expectations. With systems biology approaches, interventions can be tailored to improve cell therapy, or mimic the qualities of reparative cells.
Journal Subject Terms: Basic Science Research, Cell Therapy, Computational Biology
Keywords: congenital, computer-based model, stem cell
Introduction
Congenital heart defects (CHD) are present in 8–9 of 1000 newborns (35,000 annually in the United States) and palliative surgical therapy has greatly increased their survival1. Despite improved surgical outcomes, many children still develop reduced cardiac function and go on to heart failure and transplantation. CHD are the leading cause of right-ventricular (RV) failure in pediatric populations, especially in patients with diseases like Hypoplastic Left Heart Syndrome (HLHS) and Tetralogy of Fallot. Surgical palliation puts an increased load on the RV that is balanced by hypertrophy and fibrosis, leading to reduced function. In addition, these patients also suffer great morbidity in the form of reduced exercise capacity and intellectual deficiencies. Currently, transplantation is the only cure for many of these children; however, it has limitations such as organ rejection, organ shortage, and limited transplant longevity2.
While hundreds of cellular therapy trials have been investigated in adults3, 4, the use of stem/progenitor cells to treat cardiac dysfunction in congenital patients is relatively new. The TICAP trial using autologous cardiosphere-derived cells has shown short and long-term safety with modest efficacy5, 6. Additionally, studies on cord blood-derived cells ( NCT01883076 ) and mesenchymal stem cells (MSCs, NCT03525418 ) are ongoing in children and based on positive animal studies7, 8. We have recently published that human c-kit+ progenitor cells (hCPCs) can be isolated from pediatric biopsies and can improve the failing RV in a donor age-dependent manner9. Specifically, our studies and work from other laboratories show that neonatal hCPCs show the maximum reparative effect compared with CPCs derived from infant (1–12 month) and child (>12 month) donors9–12. A clinical trial study is now underway applying neonatal hCPCs for cardiac reparative therapy in children ( NCT03406884 ).
For stem cell therapy, given the potential for patient-to-patient variability, it is critical to select cells with the highest reparative potential. An unbiased way to determine what cellular signals and cues direct functional outcomes is to use statistical modeling as part of the cue-signal-response paradigm13, 14. While linking cellular cues to functional outcomes using modeling has been well-published, our recent studies applied statistical modeling approaches on a small subset of patients and showed that regression models can link sequencing data from each cell to its phenotypic outcome9, 15, 16. Additionally, using systems biology, we showed what mechanisms and signaling pathways regulate the efficacy of CPCs and are potentially involved in the reparative ability of these cells. Statistical models using neonatal CPC mRNA showed high predictability rates for different cardiac functions including RV ejection fraction (EF), migration and proliferation9.
As data gathering has increased for biological data sets and the variables and conditions driving a biological response are being increased more than the number of observations or cell responses, reduction of the dimensionality of the problem is helpful to identify a unique solution. Capturing covariance with PCA and PLSR has distinct advantages relative to simply observing variation of each individual signal across all conditions14, 17. Principal component analysis considers the entire dataset in an unsupervised manner, and captures how the signals being measured vary in a coordinated manner. PLSR allows the user to propose a relationship, introducing hypotheses, by splitting the variables into dependent and independent variables. Then a linear relationship is identified relating these variables to each other based on biological information; this allows the solution to be related to the independent variables most connected with the dependent variables. PLSR has the added benefit of accommodating unknown coefficients and incomplete datasets because it will reduce the dimensions into principal component space from which contributing vectors are considered, instead of each individual data point for every single perturbation or condition17–19.
In the current study, the focus was to increase the predictive power of our statistical model by adding more samples to our analysis, and to validate predicted responses in each neonate cell line. We selected proliferation and migration-induction as two key indicators of cardiac reparative ability of the cells based on our prior studies and performed in vitro assays to define the functional outcome of each neonate cell line. In vitro results verified our predictive model and identified cardiac and immune response genes that are enriched in better performing CPCs. We also ran validation on selected markers and found that at least one of the outcomes could be functionally predicted using an ELISA assay. With clinical trials in cell therapy ongoing for children, the possibility of matching transcriptomic sequencing data to clinical endpoints could allow for personalized and predictive medicine.
Material and Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request. All studies were approved by the Children’s Healthcare of Atlanta and Emory Institutional Review Board (IRB00005500). All other methods are available as an online supplement.
Results
Statistical modeling analysis of different ages of pediatric cardiac progenitor cells shows a tight clustering of similar aged hCPCs
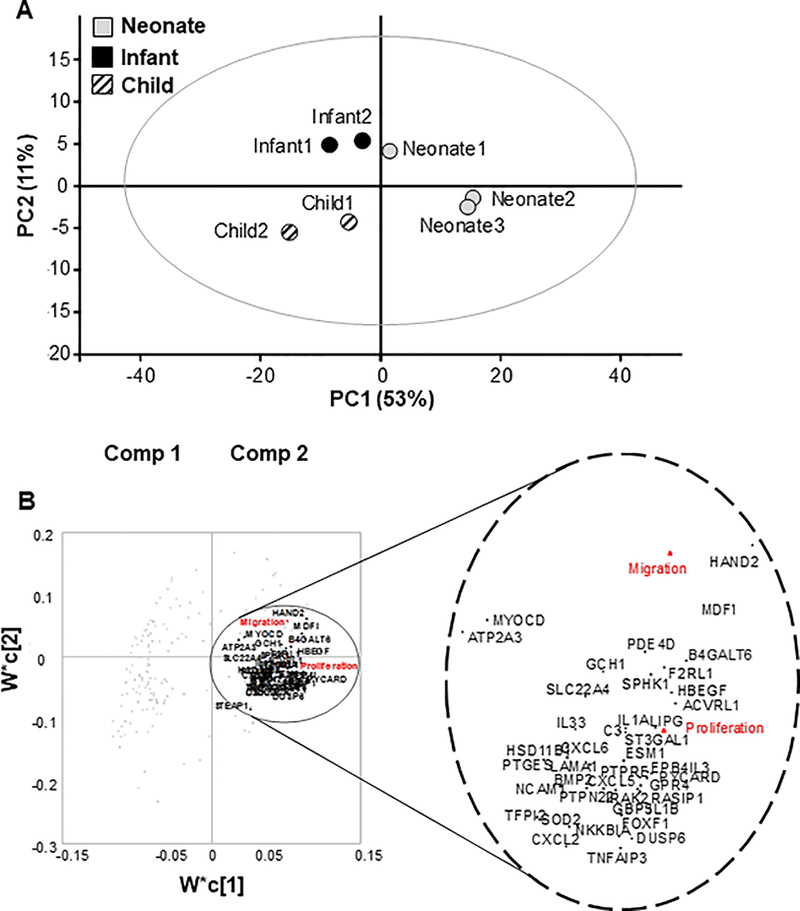
Individual patient CPCs were grouped in 3 different age groups as previously published; neonates (0–1 month), infants (1–12 months), and children (2–5 years)9. In order to build our initial model, we used mRNA sequencing data of CPCs from selected patients from our published study (that used microarray) and plotted them in principal component space. Three individual patients from the neonate patient group, and 2 individual patients from each of the infant and child groups with known cardiac reparative effects were selected. Any patients with known genetic mutations were excluded from the study; however, these patients may have unknown genetic abnormalities that have not been detected in our study. Keeping with our published study, a tight clustering of similar donor aged hCPCs was observed in the PCA plot (Figure 1A). While the neonate-1 sample locates more closely with the two infant samples, suggesting variability in rate of maturation with respect to chronological age, it nevertheless has a positive PC1 score as do the other neonate samples. Infant and child samples were found in close proximity to each other and in the same quadrant as their counterparts. The amount of variance captured by the first two components is demonstrated on the x and y axes in Figure 1A.
Figure 1.
Statistical modeling analysis of different ages of pediatric c-kit+ progenitor cells. (A). Principal component analysis (PCA). Patient c-kit+ progenitor cells (CPCs) (neonate, infant, child) were analyzed based on their mRNA expression levels. Results were plotted in principal component (PC) space. Individuals in each age group are clustered together and are localized in close proximity. (B). Partial least squares regression (PLSR) analysis of gene array. PLSR analysis of patient (neonate, infant, child) mRNAs and cardiac functions identified the most important signals from mRNA contents by calculating variable importance for projection (VIP).1 Top 300 genes with the most important signals were selected from the aforementioned PLSR analysis, and a new PLSR prediction model was trained for these patients by plotting the 300 genes in principal component space. Enlarged inset demonstrates selected cardiac and immune system related genes that are clustered with two selected cardiac functions (proliferation and migration-red font).
Furthermore, partial least squares regression (PLSR) analysis was performed to define a mathematical relationship between hCPC signals (mRNA) and cardiac reparative functions of MSC migration and CPC proliferation. PLSR analysis of patient mRNAs and cardiac functions identified the most important signals from mRNA contents by calculating importance of variable projection (VIP). VIP is calculated using a weighted sum of squares variable and is a relative value for determination of the contribution of each gene, either positively or negatively, to the associated outcome of reparative function. Genes with top 300 VIPs (exactly 256 top genes were selected, but here we refer to it as 300 genes, expanded in Table 1) were selected in an unbiased manner and a new PLSR prediction model was retrained only with these 300 genes from the patients, and the loadings plot is shown in principal component space (Figure 1C). In this study, we focused on proliferation and migration as two selected cardiac reparative effects of hCPCs. Mapping the loading plot of these genes in PC space demonstrated the contribution of many cardiac and immune response genes such as HAND2, MYOCD, as well as interleukins and cytokines, with proliferation and migration functions.
Table 1.
List of genes with corresponding VIP values
| Gene Name | VIP Value | Gene Name | VIP Value | Gene Name | VIP Value | ||
|---|---|---|---|---|---|---|---|
| ARL4C | 1.45 | FZD2 | 1.35 | PDE4D | 1.64 | ||
| ARMC9 | 1.37 | GABRE | 1.30 | PDE5A | 1.63 | ||
| ASS1 | 1.45 | GALNT6 | 1.52 | PDGFRB | 1.63 | ||
| ATP10A | 1.62 | GBGT1 | 1.46 | PDZRN3 | 1.51 | ||
| ATP2A3 | 1.31 | GBP5 | 1.41 | PHACTR1 | 1.54 | ||
| B3GNT5 | 1.50 | GCH1 | 1.38 | PIK3CD | 1.51 | ||
| B4GALT6 | 1.66 | GCNT4 | 1.70 | PML | 1.46 | ||
| BACH1 | 1.40 | GDF5 | 1.28 | PNPLA3 | 1.36 | ||
| BAIAP2L1 | 1.52 | GFPT2 | 1.35 | POPDC3 | 1.42 | ||
| BCHE | 1.76 | GK | 1.56 | POU2F2 | 1.38 | ||
| BCL11A | 1.71 | GPR4 | 1.40 | PPAP2B | 1.31 | ||
| BCL2A1 | 1.32 | GPR63 | 1.45 | PRKAA2 | 1.57 | ||
| BMP2 | 1.54 | GREB1L | 1.35 | PRKAR2B | 1.36 | ||
| BMP4 | 1.34 | HAND2 | 1.56 | PRR16 | 1.50 | ||
| BMPR1B | 1.35 | HAS2 | 1.35 | PRTFDC1 | 1.38 | ||
| BNC1 | 1.35 | HBEGF | 1.44 | PRUNE2 | 1.70 | ||
| BVES | 1.55 | HES1 | 1.32 | PTGES | 1.38 | ||
| BVES-AS1 | 1.48 | HIP1 | 1.29 | PTPN22 | 1.46 | ||
| C1QTNF1 | 1.49 | HIST1H4B | 1.29 | PTPRE | 1.40 | ||
| C3 | 1.59 | HSBP1L1 | 1.34 | PYCARD | 1.30 | ||
| CACNA1A | 1.45 | HSD11B1 | 1.35 | QPCT | 1.37 | ||
| CACNA1C | 1.37 | HTR2B | 1.64 | RAB3D | 1.53 | ||
| CACNA1H | 1.64 | ICAM1 | 1.58 | RAP1GAP2 | 1.45 | ||
| CADPS2 | 1.33 | IFI30 | 1.31 | RASA4 | 1.46 | ||
| CARD6 | 1.31 | IL18R1 | 1.38 | RASA4B | 1.58 | ||
| CCDC102B | 1.34 | IL1A | 1.47 | RASIP1 | 1.30 | ||
| CCDC148 | 1.54 | IL1B | 1.56 | RFTN2 | 1.63 | ||
| CCDC36 | 1.81 | IL23A-1 | 1.38 | RGCC | 1.51 | ||
| CDCP1 | 1.36 | IL32 | 1.49 | RGL3 | 1.50 | ||
| CEACAM19 | 1.28 | IL33 | 1.46 | RGS5 | 1.67 | ||
| CGNL1 | 1.39 | IRAK2 | 1.32 | RGS7 | 1.57 | ||
| CHD7 | 1.59 | ITGA8 | 1.29 | RGS7BP | 1.31 | ||
| CHST6 | 1.32 | ITGBL1 | 1.85 | RHBDF2 | 1.40 | ||
| CHSY3 | 1.66 | ITPKB | 1.33 | RNF144B | 1.29 | ||
| CNKSR2 | 1.35 | ITPR3 | 1.51 | RNF212 | 1.50 | ||
| COL4A5 | 1.45 | KCNC4 | 1.32 | RTKN2 | 1.35 | ||
| CPS1 | 1.62 | LAMA1 | 1.35 | SAMD12 | 1.31 | ||
| CRISPLD2 | 1.43 | LAMA4 | 1.33 | SATB1 | 1.30 | ||
| CRYAB | 1.29 | LAMA5 | 1.51 | SCN3A | 1.59 | ||
| CTH | 1.39 | LIPG | 1.59 | SCN8A-1 | 1.29 | ||
| CTHRC1 | 1.43 | LOXL3 | 1.59 | SCUBE3 | 1.45 | ||
| CXCL1 | 1.42 | LPPR4 | 1.49 | SDHAP2 | 1.35 | ||
| CXCL2 | 1.51 | LRRC32 | 1.64 | SECTM1 | 1.40 | ||
| CXCL5 | 1.34 | MAGI1 | 1.39 | SEMA5A | 1.29 | ||
| CXCL6 | 1.51 | MAN1A1 | 1.58 | SEMA6D | 1.32 | ||
| CXXC5 | 1.74 | MAP2 | 1.54 | SGCD | 1.56 | ||
| CYP1B1 | 1.51 | MASP1 | 1.40 | SH3D21 | 1.36 | ||
| DAAM2 | 1.39 | MBNL3 | 1.28 | SHROOM4 | 1.33 | ||
| DHRS3 | 1.34 | MDFI | 1.38 | SLC1A1 | 1.44 | ||
| DNAH11 | 1.73 | MEF2C | 1.32 | SLC1A3 | 1.32 | ||
| DNER | 1.32 | MEST | 1.46 | SLC22A4 | 1.44 | ||
| DNM1 | 1.32 | MICA | 1.28 | SLC23A3 | 1.34 | ||
| DOK5 | 1.33 | MOXD1 | 1.40 | SLC24A3 | 1.73 | ||
| DOK6 | 1.53 | MRVI1 | 1.54 | SLC39A8 | 1.44 | ||
| DSP | 1.33 | MUC1 | 1.35 | SLC4A4 | 1.54 | ||
| DUSP6 | 1.40 | MUC20 | 1.29 | SLC7A2 | 1.46 | ||
| EBF1 | 1.34 | MYCT1 | 1.29 | SNED1 | 1.29 | ||
| ECM2 | 1.40 | MYLIP | 1.75 | SOD2 | 1.49 | ||
| EDA | 1.38 | MYLK | 1.42 | SPEG | 1.41 | ||
| EDA2R | 1.35 | MYOCD | 1.40 | SPHK1 | 1.54 | ||
| EHD4 | 1.40 | NALCN | 1.42 | SQRDL | 1.35 | ||
| EHF | 1.30 | NAMPT-1 | 1.54 | ST3GAL1 | 1.49 | ||
| EMB | 1.59 | NCAM1 | 1.68 | ST6GALNAC3 | 1.53 | ||
| EMCN | 1.52 | NDUFA4L2 | 1.48 | STEAP1 | 1.31 | ||
| EPB41L3 | 1.35 | NEK10 | 1.47 | TBX15 | 1.50 | ||
| ESM1 | 1.48 | NFASC | 1.65 | TDRP | 1.37 | ||
| F2RL1 | 1.41 | NFATC2IP | 1.40 | TFPI2 | 1.42 | ||
| F3 | 1.57 | NFKBIA | 1.39 | TGM2 | 1.45 | ||
| FAM162B | 1.56 | NOVA1 | 1.36 | TIE1 | 1.44 | ||
| FBN2 | 1.68 | NOVA2 | 1.34 | TIFA | 1.40 | ||
| FGD4 | 1.42 | NOX4 | 1.37 | TINAGL1 | 1.39 | ||
| FGF7 | 1.30 | NR2F2 | 1.70 | TMEM130 | 1.31 | ||
| FMN2 | 1.62 | NRCAM | 1.45 | TMEM132A | 1.62 | ||
| FMNL1 | 1.31 | OSCAR | 1.54 | TMEM132B | 1.32 | ||
| FMOD | 1.46 | OSR1 | 1.53 | TMEM154 | 1.48 | ||
| FOXD1 | 1.31 | P4HA3 | 1.56 | TMOD2 | 1.55 | ||
| FOXF1 | 1.32 | PABPC4L | 1.44 | TNFAIP3 | 1.38 | ||
| FST | 1.47 | PCSK5 | 1.66 | TP53I11 | 1.52 | ||
| FUOM | 1.40 | PDE1A | 1.64 |
Predictability measurements of CPC outcomes for proliferation and migration show a high predictability rate which remains relatively high even after reducing the number of the genes
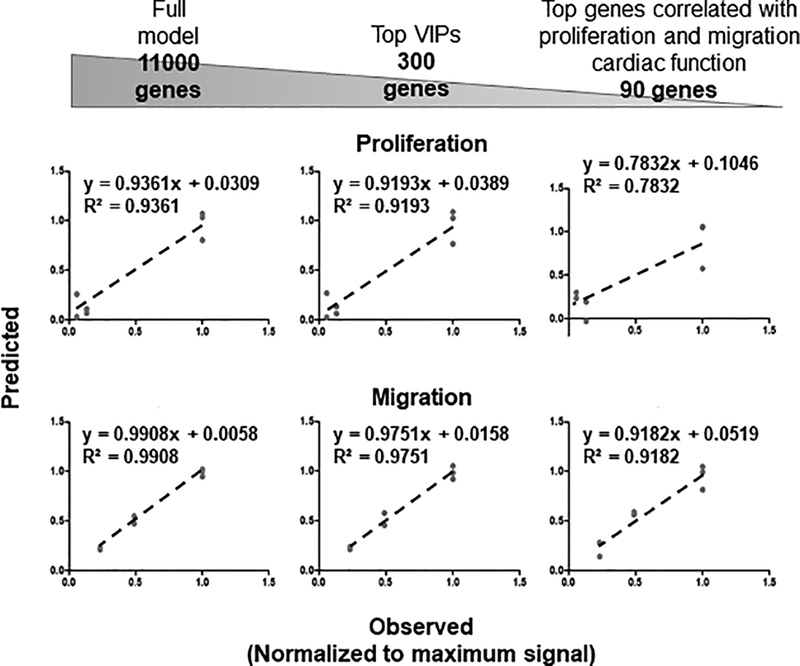
To determine the power of our model to use mRNA levels as inputs and predict cardiac reparative response outputs, we ran several regression analyses (Figure 2) using our bootstrapping method. We calculated predictability for both proliferation and migration responses of hCPCs using the complete set of signals (11,000 genes), then compared the calculated, predicted number to that of the actual number determined experimentally. Training the model with 11,000 genes provided 94% predictability for proliferation and 99% for migration. We have shown that we could reduce the number of signals, keeping only those that project the greatest on one or more of the PCs, and still retain high predictability for measured responses13,15. From 11,000 genes, we reduced the model to the top 300 genes with the highest variable importance of projections (VIPs), and retained predictability (92% and 98% for proliferation and migration responses respectively), only slightly lower than when the whole set of signals was used (Figure 2). To further reduce the model, we picked mRNA signals that were highly correlated with both proliferation and migration responses, as opposed to those that projected greater for one or the other outcomes, and retrained the PLSR model with these signals (comprehensive list in Supplementary Table 1). To select highly correlated genes with both proliferation and migration responses, we arbitrarily chose top 150 genes correlated with either proliferation or migration responses. Ninety one out of 150 genes were overlapping between the two sets. Predictability of proliferation and migration responses was slightly reduced in the model with 91 genes (78% and 92% respectively). These results suggested that even with a reduced number of mRNA signals we still predict measured responses with a model including just the 300 top mRNA signals differentially expressed with respect to the functional traits. This gene set was regarded as optimal for further statistical modeling and systems biology analyses.
Figure 2.
Predictability measurements of cardiac reparative functions. Predictability of selected cardiac functions was calculated by the PLSR model trained with all signals (11,000 genes), top 300 VIP signals, and top 91 genes correlated with both proliferation and migration responses. Reduction of the 10,000+ genes to 300 genes does not significantly reduce the high predictability rate measured with all signals. Observed proliferation and migration responses are normalized to the maximum signal.
Neonatal hCPCs shows the highest predictability rate for functional outcomes compared to other pediatric hCPCs
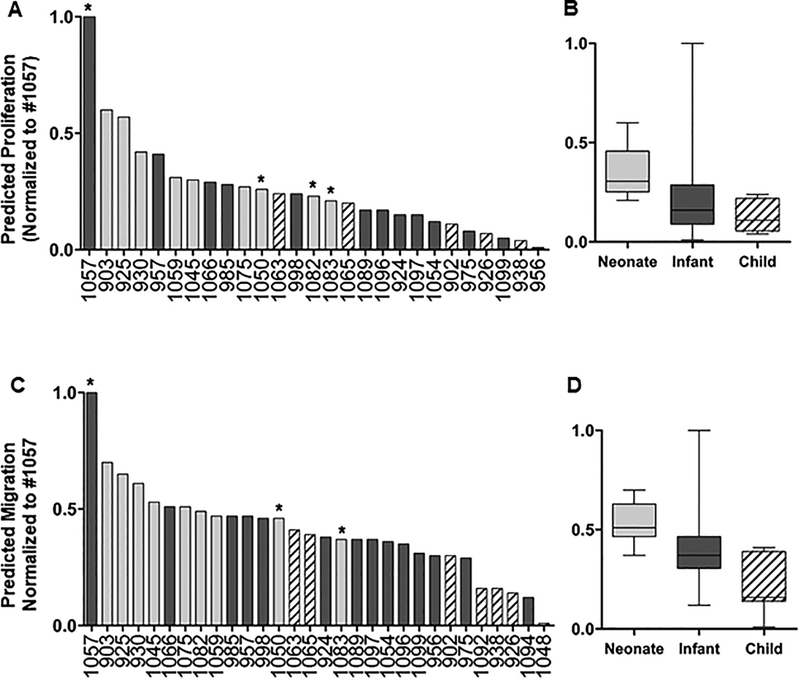
Our recent publication demonstrated that there was a donor age-dependent effect of hCPC therapy following injury9. To determine whether this could be predicted on a larger scale, we used the model generated with the top 300 mRNA signals, and analyzed the data of 32 pediatric patients with this model and predicted functional outcomes of these cells. For each output, the response was normalized to the highest predicted function in the group, patient #1057 for both proliferation and migration responses. Of these samples, 27 patients for proliferation and 30 patients for migration had measurable predictions and are shown in Figure 3. As expected, neonatal CPCs showed the highest predicted response (7 of the top 10 lines) for both proliferation (Figure 3A and 3B) and migration (Figure 3C and 3D). Additionally, consistent with previous findings and based on our prediction analysis9,10, child CPCs showed the least predicted outcomes with infants being between the two. While our analysis confirmed the previous finding that neonatal CPCs have the highest reparative effect, we observed patient-to-patient variability with regard to proliferation and migration outcomes in each age group that are highlighted with asterisks. Our results also showed that proliferation and migration functional outcomes are highly correlated (R2=0.93) and relative reparative effect of proliferation and migration functions are similar in each patient (Supplementary Figure S1).
Figure 3.
Prediction analysis of selected hCPCs. Top 300 RNA-seq data from pediatric CPCs were used in our predictive model and proliferation (A,B) and migration (C,D) responses for each patient were determined. Many of the neonates (gray bars) cluster together and reside close to the top with the most functional improvement. Most child CPCs (striped bars) cluster together near the bottom with the least functional improvement and infants (black bars) cluster together and reside intermediately between neonate and child patients. There are outliers in each age group that reside outside of their cluster (such as neonates #1050 and #1083, and infant #1057 labeled with asterisks. Each sample is normalized by the highest scoring individual for each function (Infant #1057).
In vitro model validation in neonate patients verifies variability between different neonatal cell lines
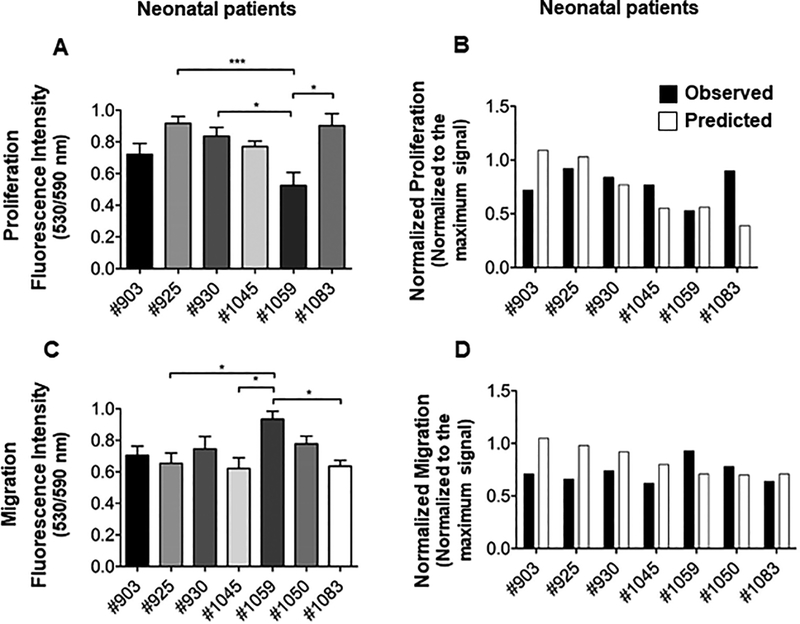
To test patient variability observed in our model and to examine if predicted responses match the observed ones in each patient, we performed in vitro experiments to test proliferation and chemotaxis capacity of seven different neonatal hCPC lines. Neonatal patients were chosen due to our ongoing clinical trial. Our results confirmed patient variability between different neonatal cell lines, with patient #1059 having significantly lower proliferation response than several within the cohort (Figure 4A). When plotted with predicted values, our observed results matched for many of the samples with 78% accuracy for proliferation (Figure 4B).
Figure 4.
Proliferation and migration analysis and validation of the predictive model. (A). Proliferation analysis of neonate CPCs in vitro. Cell proliferation assay was performed on 10,000 cultured CPCs of each neonate using Click iT-Edu microplate assay and the absorbance was calculated after 24 hours of incubation. Some neonate CPCs showed a higher proliferation rate compare to the others. Absorbance was normalized to the maximum signal. N=5 replicates for each cell line; ANOVA followed by Tukey test, P<0.05. (B). Comparison of in vitro proliferation responses and the predicted responses. Observed proliferation response for each neonate patient was plotted against its predicted response. Proliferation assay was not performed with neonatal #1050 cell lines. This cell line was lost after collecting the conditioned media for migration assay. (C). Migration analysis of neonate CPCs in vitro. Migration assay was performed by applying conditioned media obtained from each neonate CPC on human mesenchymal stem cells (MSCs) in a Boyden chamber. Migrated cells were labeled with CMRA and cell migration was quantified after 24 hours by measuring CMRA fluorescence intensity. Absorbance was normalized to the maximum signal. N=4 replicates for each cell line; One-way ANOVA followed by Tukey test, P<0.05. (D). Comparison of in vitro migration responses and the predicted responses. Observed migration response for each neonate patient was plotted against its predicted response.
Furthermore, we measured the chemotactic potential of media from the same neonate cell lines using MSCs in a modified Boyden chamber. MSCs subjected to 24 hour quiescence were treated with conditioned media and allowed to migrate to the other side of the chamber for 24 hours. Similar to proliferation rates, our results verified significant patient variability for migration capacity of neonatal hCPC media (Figure 4C). While patient #1059 was a moderate performing line for proliferation, media from these cells demonstrated significantly higher MSC migration. Again, the observed migration response for each neonate patient closely matched the predicted responses with 80% accuracy (Figure 4D). Taken together these results confirm the patient variability within the neonatal age group and validate our predictive model as closely matching observed functional outcomes.
Transcriptomic analysis of c-kit+ progenitor cell shows the presence of immune response related genes among differentiated genes
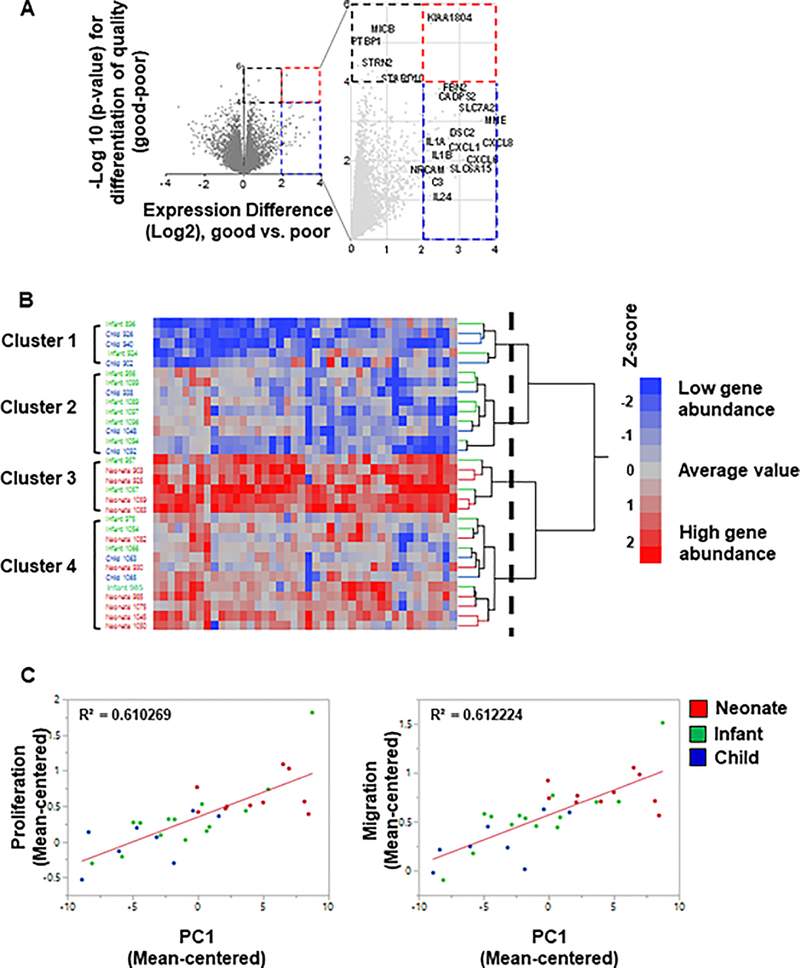
To investigate potential reparative signals in cells, a volcano plot of significance (negative log of the p-value) against difference in abundance (log2 scale) between “GOOD” and “POOR” performing patients was plotted (Figure 5A, expanded in Table 2). “GOOD” and “POOR” performing cells were characterized by their predicted functional improvements in our model (see Supplementary Table 2 for categories)8. Cells significantly increasing tricuspid annular plane systolic excursion (TAPSE) and RV ejection fraction, improving angiogenesis, and significantly reducing wall thickness and fibrosis in rats transplanted with c-kit+ cells were categorized as “GOOD” performing cells (at least within 10% of healthy values in all categories). Chemotactic and proliferative capacity of “GOOD” performing c-kit+ cells were also significantly higher than the “POOR” performing cells. At least thirty genes were upregulated at least 4-fold in “GOOD” vs “POOR” hCPCs (Figure 5A red and blue dashed lines; expanded in Supplementary Table 3). All of these are significant at unadjusted p<0.05 (NLP>1.3), but given the small sample size for the comparison do not reach experiment-wide significance, so should be regarded as candidate biomarkers that may not be individually diagnostic in each cell line. In addition, there were several genes that were significantly increased (black dashed line; p<0.0001, NLP>4) but showed only modest differential expression (Expanded in Supplementary Table 3). For our analysis, we focused on the 42 differentially expressed genes whose expression was either upregulated at least four-fold or significantly increased (P<0.0001; 10% FDR), or both (genes included in blue, black, and red dashed squares respectively in Figure 5A, all raw data included in online supplement and available at https://www.davislab.org/supplementary-data-from-publication). Many immune response related genes including interleukins (IL-1α, IL-1β, and IL24) and cytokines (CXCL6 and CXCL8) are present among the differentially expressed genes. Furthermore, we performed two-way hierarchical clustering analysis of the 42 differentially expressed genes between different age groups to indicate clustering of same patients in same groups. We applied Ward’s method and indicated strong but as expected imperfect separation of patients by age (Figure 5B). The two-way hierarchical clustering analysis showed that all patients (neonate, infant, child) are separated into 4 groups, of which neonates are grouped in either cluster 3 or 4. Neonates #903 and #925 that are among the top “GOOD” samples, along with neonates# 1059 and 1083 reside in cluster 3. The rest of neonate CPCs cluster within group 4 (Figure 5B), and it is noteworthy that neonates in the bottom half of cluster 4 also tend to be up-regulated as observed for cluster 3 containing the other neonates.
Figure 5.
Transcriptomic analysis of hCPC and potential effectors. (A) Volcano plot of GOOD vs POOR patient (neonate, infant, child) comparison. A volcano plot of significance (negative log of the p-value) against difference in abundance (log2 scale) of genes was plotted between good and poor c-kit+ progenitor cells. Dashed blue square represents genes that are upregulated in good vs poor CPCs but not statistically significant. Signals residing in the red dashed square are genes that are significantly upregulated in good vs poor CPCs (p-value <0.0001). Genes are indicated by gray circles. Many immune response related genes including interleukins and cytokines are present among differentiated genes. (B) Two-way hierarchical cluster analysis of differentially expressed CPC genes using Ward’s method. Two-way hierarchical analysis was performed on the same 42 genes between neonates (red labels), infants (green labels), and children (blue labels). The dashed vertical line indicates the cutoff on the dendrogram used to define the four clusters. (C) Correlated genes with proliferation and migration functions. The differentially upregulated genes showed high correlation with both proliferation and migration functions (running a regression analysis; R2=0.6 for both functions).
Table 2.
List of differentiated genes in good vs. poor c-kit+ progenitor cells with overlapping genes from original model identified.
| Upregulated genes in good vs. poor patients | Upregulated genes in good vs. poor patients | Overlapping upregulated genes (good vs. poor patients) with Agarwal et al9 top 300 VIPs |
|---|---|---|
| C3 | KCNK1 | C3 |
| CADPS2 | KIAA1804 | CADPS2 |
| CARD11 | LAMP3 | CXCL1 |
| CSF2 | LOC100505622 | CXCL2 |
| CXCL1 | MDFI | CXCL6 |
| CXCL2 | MICB | EPB41L3 |
| CXCL6 | MME | FBN2 |
| CXCL8 | NRCAM | GPR4 |
| DSC2 | PDZK1IP1 | HSD11B1 |
| EPB41L3 | POU2F2 | IL1A |
| FAM163A | PTBP1 | IL1B |
| FBN2 | RBM10 | MDFI |
| GPR4 | RPL14 | NRCAM |
| HIF3A | S1PR1 | POU2F2 |
| HPSE | SLC3D2 | SLC7A2 |
| HSD11B1 | SLC6A15 | TFPI2 |
| IGF2BP2 | SLC7A2 | TIE1 |
| IL1A | STARD10 | |
| IL1B | STRN4 | |
| IL24 | TFPI2 | |
| IRAK3 | TIE1 |
Finally, to show whether these highly differentiated genes (42 genes) are correlated with cardiac regenerative functions, we ran a regression analysis of proliferation and migration responses with the most variable genes (PC1) for all pediatric hCPCs (Figure 5C) and showed that both responses are highly correlated with these genes (R2 = 0.61 for both proliferation and migration). Collectively, expression of these genes is more predictive of cell function than any single transcript. We expanded our analysis and identified individual genes that are highly correlated with proliferation and migration cardiac functions (Comprehensive list of genes in Table 3). Genes playing roles in cardiac and/or immune response mechanisms are highlighted in bold. Functional analysis of the highly differentiated genes using Ingenuity Pathways Analysis demonstrated that these genes are involved in pathways such as cardiovascular system development and function, immune response pathways and networks, as well as cellular growth/proliferation (comprehensive list in Supplementary Table 4). Moreover, there were 17 overlapping genes between the 42 highly upregulated genes of “GOOD” vs “POOR” patients and the 300 top genes from the original list of genes. Interestingly, the canonical pathways that these 17 genes are involved in highly match with the pathways involved in our model (Supplementary Table 5).
Table 3.
List of top genes correlated with proliferation and migration (from 42 upregulated genes in good vs. poor c-kit+ progenitor cells). Bolded genes are associated with cardiac and/or immune response mechanisms. Twenty-seven genes are shared between proliferation and migration.
| Selected top correlated genes with proliferation | Correlation (r2) | Selected top correlated genes with migration | Correlation (r2) |
|---|---|---|---|
| S1PR1 | 0.62 | S1PR1 | 0.59 |
| POU2F2 | 0.59 | POU2F2 | 0.56 |
| HPSE | 0.54 | IL1A | 0.55 |
| KCNK1 | 0.52 | IL1B | 0.51 |
| SLC35D2 | 0.52 | SLC35D2 | 0.51 |
| CXCL1 | 0.51 | CXCL1 | 0.50 |
| CXCL8 | 0.51 | CXCL8 | 0.50 |
| IL1B | 0.51 | HPSE | 0.49 |
| GPR4 | 0.48 | KCNK1 | 0.48 |
| IL1A | 0.48 | KIAA1804 | 0.48 |
| LOC100505622 | 0.48 | NRCAM | 0.48 |
| MME | 0.48 | TIE1 | 0.47 |
| CXCL6 | 0.47 | CXCL6 | 0.46 |
| HIF3A | 0.46 | HIF3A | 0.43 |
| NRCAM | 0.43 | LOC100505622 | 0.43 |
| TFPI2 | 0.42 | GPR4 | 0.42 |
| TIE1 | 0.42 | PTBP1 | 0.41 |
| KIAA1804 | 0.41 | STARD10 | 0.41 |
| STARD10 | 0.40 | MME | 0.39 |
| C3 | 0.37 | SLC7A2 | 0.39 |
| CSF2 | 0.37 | IGF2BP2 | 0.37 |
| FAM163A | 0.37 | C3 | 0.36 |
| IL24 | 0.36 | MICB | 0.36 |
| PTBP1 | 0.35 | CADPS2 | 0.35 |
| SLC6A15 | 0.35 | CXCL2 | 0.35 |
| SLC7A2 | 0.35 | EPB41L3 | 0.35 |
| CADPS2 | 0.32 | FBN2 | 0.35 |
| CXCL2 | 0.32 | CSF2 | 0.34 |
| HSD11B1 | 0.32 | IRAK3 | 0.33 |
| EPB41L3 | 0.31 | LAMP3 | 0.32 |
| FBN2 | 0.31 | CARD11 | 0.30 |
| IGF2BP2 | 0.31 |
Protein validation of model demonstrates CXCL6 as a mediator of hCPC function
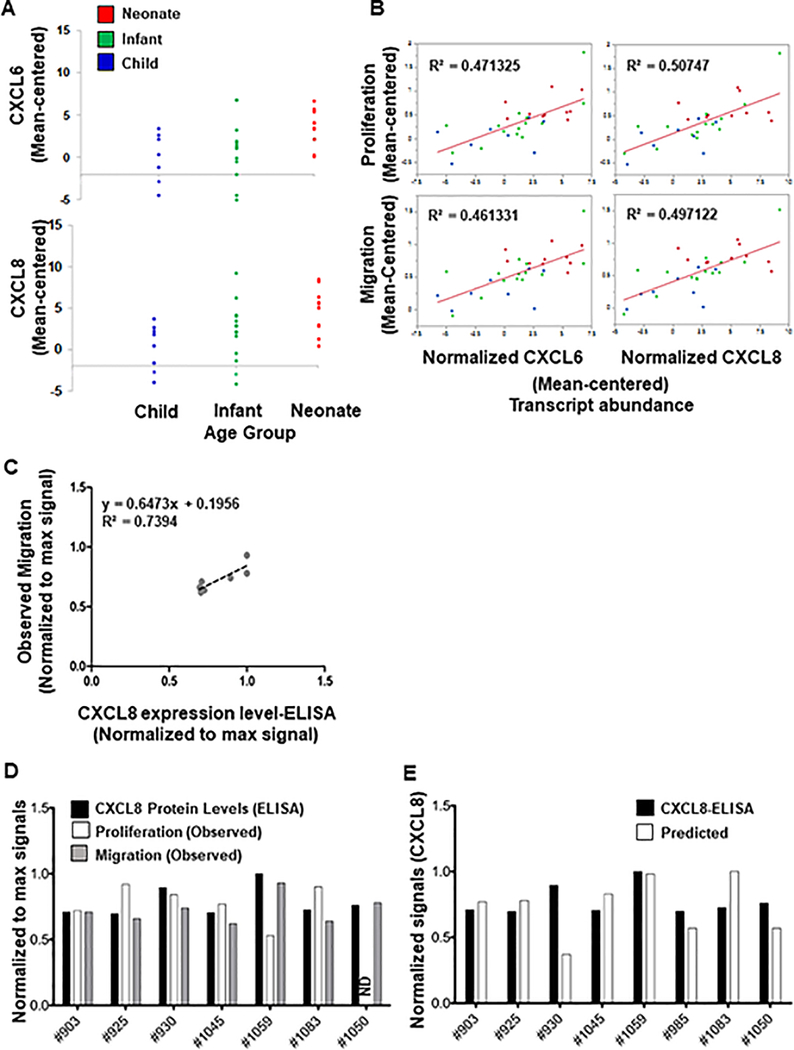
Based on our data from Figure 5A, we selected several differentially expressed genes for protein validation. As shown in Figure 6A, RNA for both CXCL6 and CXCL8 were enriched in newborns as compared to both infant and child samples. Additionally, there was a high correlation between both of these molecules and either proliferation or migration (Figure 6B). We also examined IL-1α, IL-1β, and IL-24, which had high correlations, but were unable to detect levels in several patients by ELISA (correlations shown in Supplementary Figure S2). We examined both CXCL6 (Supplementary Figure S3) and CXCL8 (Figure 6C and Supplementary Figure S3) by ELISA and correlated normalized protein values to migration and proliferation. While there was no correlation between CXCL6 protein levels and either function, there was a strong correlation (R2 = 0.74) between CXCL8 and migration. Figure 6D shows individual data for normalized CXCL8 levels, migration, and proliferation, showing a strong relationship between CXCL8 and migration with >80% accuracy. Finally, using our RNAseq statistical model, we were able to successfully predict CXCL8 protein levels based on expression data with 82% accuracy (Figure 6E).
Figure 6.
Confirmation of CXCL6 as a mediator of hCPC function. (A) High expression levels of CXCL6 and CXCL8 in “GOOD” vs “POOR” pediatric patients. Statistical modeling demonstrated high expression levels of CXCL6 and CXCL8 mRNA in GOOD pediatric CPCs. Patients above threshold (dotted lines) are GOOD samples with significantly high expression of CXCL6 (top panel) and CXCL8 (bottom panel). (B) Correlation of CXCL6 and CXCL8 mRNAs with proliferation and migration functions in pediatric patients. CXCL6 and CXCL8 mRNA expression showed relatively high positive correlation with both proliferation and migration functions. Signals are the mean TMM values from edgeR. They have centered with respect to the overall mean gene expression of each sample (A and B). (C) Regression analysis of observed migration responses with protein expression levels of CXCL8 measured by ELISA in neonatal patients. Observed proliferation or migration responses for each neonate patient were plotted against CXCL8 protein expression levels. Observed migration responses in neonatal patients showed high correlation with CXCL8 protein expression levels (R2=0.74). (D). Comparison of in vitro proliferation and migration responses and protein expression levels of CXCL8 measured by ELISA. The abundance of CXCL8 protein levels (black bars) matched the migration induction potential of neonatal CPCs (gray bars). Expression level of CXCL8 protein did not fully match with the proliferation ability of the cells (white bars). Proliferation assay was not performed with neonatal #1050 cell lines. This cell line was lost after collecting the conditioned media for migration assay. (E) Comparison of the abundance of CXCL8 proteins measured with ELISA and predicted expression levels of CXCL8 mRNA in neonatal CPCs. Predicted richness of CXCL8 mRNA (white bars) and the observed protein level of CXCL8 expressed in neonatal CPCs (black bars) matched very closely and showed an average accuracy of 82%. ND=not detected. CXCL8 protein expression (ELISA) and observed proliferation and migration responses are normalized to the maximum signal.
Discussion
Stem cell therapy has been widely tested in the context of adult coronary heart diseases using a variety of stem and progenitor cells. Despite this, very few studies have been performed in the pediatric population. An initial clinical trial of stem cell therapy in a small cohort of children with HLHS (TICAP) demonstrated an improvement in cardiac function in children undergoing intracoronary infusion of cardiosphere-derived cells5, 6. With that improvement, a follow-up trial (PERSEUS) is planned as a Phase II study. In addition, clinical trials based on cord blood-derived cells and mesenchymal stem cells, as well as another clinical trial approved for c-kit+ progenitor cells, are underway7, 8. While the adult data have been mixed, it is clear that there is much patient-to-patient variability that may lead to confounding results in planned pediatric trials with small numbers of patients.
It is essential to select stem cells with highest cardiac reparative ability for therapy, therefore, our lab previously compared CPCs from donors of varying ages to determine optimal characteristics of cells. Our data demonstrated that both cells and exosomes from newborn patients had the highest ability to repair the damaged myocardium9, 15. This was keeping with several published studies showing that cells from newborn donors were optimal12, 20, 21. We then performed statistical modeling analysis on a small number of samples to determine potential mechanisms in an unbiased manner, as well as create predictive regression models relating mRNA and microRNA levels to select outputs. PLSR analysis identified the 300 top VIP mRNAs and a predictive model was regenerated using these 300 signals. We have generated our predictive model based on a slightly small sample size. It should be noted that our samples were obtained from children with various congenital heart diseases who were undergoing heart surgeries, therefore our sample size was limited. Additionally, the model in this study is based on our previously published model which contained a similar sample size.9 However, now that we have validated the current model, we can begin to develop larger scale models with obtaining more clinical samples.
In the current study, we selected the same top 300 genes from Agarwal et al.9 and retrained our predictive model with an expanded set of pediatric patients. We used proliferation and migration as responses due to the ease of testing and the high involvement of these pathways in our mechanistic models. Furthermore, several published studies demonstrate the importance of implanted cell proliferation and stem cell migration in healing the damaged myocardium22–24. Our data show that the prediction power is comparable between the full model (11,000 genes) and the model with 300 genes (all above 90% predictability). However, when we further reduced the number of signals to 91 top genes that were highly correlated with both proliferation and migration responses, prediction power was decreased for both responses especially for the proliferation response (78% vs 92% for 91 signals). Similarly, the prediction power in the model generated with 42 differentially expressed genes was further reduced for both proliferation and migration responses (66% and 71% respectively, data not shown). Therefore, we selected the refined model generated with 300 genes for further analysis which had the predictability rates of 92% and 98% for proliferation and migration respectively. In previous published data, Gray et al. were able to obtain a high predictability with only 11 signals which was about 3% of their original data of 337 signals16. However it should be noted that in our study, the refined model with 300 genes was 2.7% of the 11,000 signals of the full model. Therefore, similar to the previous study, we were able to narrow down the number of our initial signals to the same extent and still train a model with high predictability power. This is critical for clinical studies in which it may not be feasible to perform RNAseq on all samples, but rather to select a focused list of candidate biomarkers that may predict efficacy as well as the full set, but better than any individual biomarker.
With smaller patient numbers, it is difficult to isolate and study variability, which we were able to do with our expanded bank of nearly 40 samples. We fit the transcriptomic data to our regression models and made predictions regarding proliferation and ability to induce MSC migration. We were able to confirm that there was variability across several newborn samples, and that we could predict outcomes a priori with high accuracy. It is worth noting that the observed variability across samples (including infant #1057 which showed the highest proliferation and migration improvements across all age groups) could be the result of biological, technical, or experimental problems, which are not unusual in this kind of studies. While migration and proliferation are two important functions, they are not the only indicators of cardiac repair. Future studies will need to be performed to determine whether these two outcomes correlate with functional recovery, and potentially inform the use of these in vitro assays as surrogates for efficacy. In addition, we only examined newborn samples in this study. We chose this population as all the current clinical trials are centered on HLHS and newborn cells and understanding which cells may not have efficacy is critically important. Despite this, future work will expand confirmation to the older pediatric population.
To further investigate which specific genes play a role in improving the functionality of CPCs, we examined the outliers in an unbiased way. Our full model consisted of around 11000 genes. Volcano plot of significance against difference in abundance was created for these genes and we showed that 42 genes are upregulated in “GOOD” vs “POOR” patients, characterized by their predicted functional improvement. We selected these parameters to include as many genes as possible based on prior studies25. We excluded downregulated genes from this part of the study and focused only on the upregulated genes. Studying downregulated genes in vivo due to cell therapy is extremely hard and in order to be more clinically relevant, something must be more easily measurable. It may be possible that our list is too relaxed or stringent, but we were sensitive to excluding false negatives. Published data have shown the association of many of these genes with their cardiac repair potential (Bolded genes in Table 3)26–28. Our original model includes 300 top genes and about 17 genes from this list matched with the 42 highly upregulated genes in the updated model, which makes about 6% of the total 300 genes (Table 2). Similar to 42 genes, these 17 genes play important roles in cardiac and immune response pathways that overlap between the two sets of genes. These results suggest that with narrowing down our model to only a few genes (17 genes), we still capture similar cardiac and immune response pathways (Supplementary Figure S1), which implies that we could potentially determine reparative response from a small number of genes, similar to studies looking at serum biomarkers for injury and/or recovery29, 30.
The unbiased statistical analysis of our dataset showed CXCL6 and CXCL8 cytokines among the highly upregulated genes in “GOOD” patients. The role of innate and adaptive immune mechanisms in deriving regenerative responses have been identified after cardiac injuries25. These two genes were also highly correlated with proliferation and migration responses. To validate our analysis and determine the levels of these cytokines in “GOOD” patients, we tested the abundance of these secreted proteins in the conditioned media collected from neonatal CPCs and observed that the amount of secreted CXCL8 in the media is positively correlated with the migration-inducing potency of these cells. There was no correlation of CXCL8 with proliferation, for example, despite the high expression levels of CXCL8 protein present in the conditioned media of neonate #1059 CPCs, the proliferative rate of these cells was lower. CXCL8 has been shown to induce migration of MSCs directly, and thus it potentially serves as validation of the model31, 32. We found that protein levels of CXCL6 did not correlate with either output measured. CXCL6 increases proliferation and migration in other cell types33; however, our data does not determine that there is no role of CXCL6 in CPC function, rather that there was no correlation. In fact, recently CXCL6 was shown to be important in the CPC secretome, but mainly with regard to angiogenesis34. As we gather data from more patients, we may be able to test additional outputs as well. Moreover, we only tested a small subset of genes and proteins in our predictive model, and testing was done under basal cell culture conditions. As the microenvironment of the injured pediatric heart is likely different, these studies should be repeated under more pathological conditions such as hypoxia and hypertrophic signaling. In vivo testing of all cell lines is both labor and cost intensive, thus finding correlative in vitro readouts will help support testing of more signals.
In summary, our data represents a novel advance in pediatric cell therapy that can be expanded in the future. The use of transcriptomic data to create predictive models based on preclinical studies may help inform ongoing clinical trials. The idea of using personalized, precision medicine to determine potential stem cell efficacy, as well as potential mechanisms, is attractive from a therapeutic standpoint. Identification of important signals may inform FDA-required potency assays and release criteria. Further, as some of the ongoing trials are allogeneic, finding the optimal donor profile could greatly improve outcomes35. With limited number of genes, a more efficient therapeutic strategy can be applied in cardiac cell therapy by rapid identification of optimal donors with strategies such as quantitative PCR or ELISA. While our approach certainly has caveats, such as inconsistencies between RNA profile and protein content of the cells and difficulties in discerning whether these signals are causative mechanisms or merely biomarkers, the unbiased and quantitative selection of cues, signals, and responses not only allows for better understanding of hCPC therapy, but the potential to extend this to other cell types and similar outcomes for other diseases.
Supplementary Material
Sources of Funding:
This work was supported by grant #2245 from the Marcus Foundation, funds from the Betkowski Family Fund, funding from Alliance Data, and HL145644 to MED. The Yerkes Non-Human Primate Genomics Core is supported in part by NIH P51OD011132. UA was supported by T32HL007745
Footnotes
Disclosures: None
Reference:
- 1.Benjamin EJ, et al. Heart Disease and Stroke Statistics-2019 Update: A Report From the American Heart Association. Circulation. 2019:CIR0000000000000659. [DOI] [PubMed] [Google Scholar]
- 2.Feinstein JA, et al. Hypoplastic left heart syndrome: current considerations and expectations. J Am Coll Cardiol. 2012;59:S1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolli R, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–57. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Makkar RR, et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishigami S, et al. Intracoronary autologous cardiac progenitor cell transfer in patients with hypoplastic left heart syndrome: the TICAP prospective phase 1 controlled trial. Circ Res. 2015;116:653–64. [DOI] [PubMed] [Google Scholar]
- 6.Tarui S, et al. Transcoronary infusion of cardiac progenitor cells in hypoplastic left heart syndrome: Three-year follow-up of the Transcoronary Infusion of Cardiac Progenitor Cells in Patients With Single-Ventricle Physiology (TICAP) trial. J Thorac Cardiovasc Surg. 2015;150:1198–1207, 1208 e1–2. [DOI] [PubMed] [Google Scholar]
- 7.Burkhart HM, et al. Regenerative therapy for hypoplastic left heart syndrome: first report of intraoperative intramyocardial injection of autologous umbilical-cord blood-derived cells. J Thorac Cardiovasc Surg. 2015;149:e35–7. [DOI] [PubMed] [Google Scholar]
- 8.Kaushal S, et al. Study design and rationale for ELPIS: A phase I/IIb randomized pilot study of allogeneic human mesenchymal stem cell injection in patients with hypoplastic left heart syndrome. Am Heart J. 2017;192:48–56. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal U, et al. Age-Dependent Effect of Pediatric Cardiac Progenitor Cells After Juvenile Heart Failure. Stem Cells Transl Med. 2016;5:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boopathy AV, et al. The modulation of cardiac progenitor cell function by hydrogel-dependent Notch1 activation. Biomaterials. 2014;35:8103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra R, et al. Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation. 2011;123:364–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson DL, et al. A strong regenerative ability of cardiac stem cells derived from neonatal hearts. Circulation. 2012;126:S46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HD, et al. Signaling network state predicts twist-mediated effects on breast cell migration across diverse growth factor contexts. Mol Cell Proteomics. 2011;10:M111 008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platt MO, et al. Multipathway kinase signatures of multipotent stromal cells are predictive for osteogenic differentiation: tissue-specific stem cells. Stem Cells. 2009;27:2804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal U, et al. Experimental, Systems, and Computational Approaches to Understanding the MicroRNA-Mediated Reparative Potential of Cardiac Progenitor Cell-Derived Exosomes From Pediatric Patients. Circ Res. 2017;120:701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray WD, et al. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ Res. 2015;116:255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janes KA, et al. Cue-signal-response analysis of TNF-induced apoptosis by partial least squares regression of dynamic multivariate data. J Comput Biol. 2004;11:544–61. [DOI] [PubMed] [Google Scholar]
- 18.Janes KA and Lauffenburger DA. A biological approach to computational models of proteomic networks. Curr Opin Chem Biol. 2006;10:73–80. [DOI] [PubMed] [Google Scholar]
- 19.Miller-Jensen K, et al. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–8. [DOI] [PubMed] [Google Scholar]
- 20.Capogrossi MC. Cardiac stem cells fail with aging: a new mechanism for the age-dependent decline in cardiac function. Circ Res. 2004;94:411–3. [DOI] [PubMed] [Google Scholar]
- 21.Ye J, et al. Aging Impairs the Proliferative Capacity of Cardiospheres, Cardiac Progenitor Cells and Cardiac Fibroblasts: Implications for Cell Therapy. J Clin Med. 2013;2:103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen B, et al. Co-expression of Akt1 and Wnt11 promotes the proliferation and cardiac differentiation of mesenchymal stem cells and attenuates hypoxia/reoxygenation-induced cardiomyocyte apoptosis. Biomed Pharmacother. 2018;108:508–514. [DOI] [PubMed] [Google Scholar]
- 23.Loffredo FS, et al. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell. 2011;8:389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh A, et al. Mesenchymal stem cells in cardiac regeneration: a detailed progress report of the last 6 years (2010–2015). Stem Cell Res Ther. 2016;7:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson G and Weir B. The quantitative genetics of transcription. Trends Genet. 2005;21:616–23. [DOI] [PubMed] [Google Scholar]
- 26.Epelman S, et al. Role of innate and adaptive immune mechanisms in cardiac injury and repair. Nat Rev Immunol. 2015;15:117–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiu K, et al. A heart-brain-kidney network controls adaptation to cardiac stress through tissue macrophage activation. Nat Med. 2017;23:611–622. [DOI] [PubMed] [Google Scholar]
- 28.Yang T, et al. TGF-beta/Smad3 pathway enhances the cardio-protection of S1R/SIPR1 in in vitro ischemia-reperfusion myocardial cell model. Exp Ther Med. 2018;16:178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackie AR, et al. Sonic hedgehog-modified human CD34+ cells preserve cardiac function after acute myocardial infarction. Circ Res. 2012;111:312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, et al. The potential role of exosomes in the diagnosis and therapy of ischemic diseases. Cytotherapy. 2018;20:1204–1219. [DOI] [PubMed] [Google Scholar]
- 31.Bayo J, et al. IL-8, GRO and MCP-1 produced by hepatocellular carcinoma microenvironment determine the migratory capacity of human bone marrow-derived mesenchymal stromal cells without affecting tumor aggressiveness. Oncotarget. 2017;8:80235–80248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stich S, et al. Human periosteum-derived progenitor cells express distinct chemokine receptors and migrate upon stimulation with CCL2, CCL25, CXCL8, CXCL12, and CXCL13. Eur J Cell Biol. 2008;87:365–76. [DOI] [PubMed] [Google Scholar]
- 33.Ma JC, et al. Fibroblast-derived CXCL12/SDF-1alpha promotes CXCL6 secretion and co-operatively enhances metastatic potential through the PI3K/Akt/mTOR pathway in colon cancer. World J Gastroenterol. 2017;23:5167–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toran JL, et al. CXCL6 is an important paracrine factor in the pro-angiogenic human cardiac progenitor-like cell secretome. Sci Rep. 2017;7:12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakravarty T, et al. ALLogeneic Heart STem Cells to Achieve Myocardial Regeneration (ALLSTAR) Trial: Rationale and Design. Cell Transplant. 2017;26:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.