Abstract
Aromatic compounds such as 4-hydroxybenzoic acid are broadly applied in industry for a myriad of applications used in everyday life. However, their industrial production currently relies heavily on fossil resources and involves environmentally unfriendly production conditions, thus creating the need for more sustainable biotechnological alternatives. In this study, synthetic biology was applied to metabolically engineer Pseudomonas taiwanensis VLB120 to produce 4-hydroxybenzoate from glucose, xylose, or glycerol as sole carbon sources. Genes encoding a 4-hydroxybenzoate production pathway were integrated into the host genome and the flux toward the central precursor tyrosine was enhanced by overexpressing genes encoding key enzymes of the shikimate pathway. The flux toward tryptophan biosynthesis was decreased by introducing a P290S point mutation in the trpE gene, and degradation pathways for 4-hydroxybenzoate, 4-hydroxyphenylpyruvate and 3-dehydroshikimate were knocked out. The resulting production strains were tailored for the utilization of glucose and glycerol through the rational modification of central carbon metabolism. In batch cultivations with a completely mineral medium, the best strain produced 1.37 mM 4-hydroxybenzoate from xylose with a C-mol yield of 8% and 3.3 mM from glucose with a C-mol yield of 19.0%. Using glycerol as a sole carbon source, the C-mol yield increased to 29.6%. To our knowledge, this is the highest yield achieved by any species in a fully mineral medium. In all, the efficient conversion of bio-based substrates into 4-hydroxybenzoate by these deeply engineered P. taiwanensis strains brings the renewable production of aromatics one step closer.
Keywords: 4-hydroxybenzoate, aromatics, metabolic engineering, Pseudomonas taiwanensis VLB120, shikimate pathway, glycerol, synthetic biology
Introduction
Aromatic compounds are broadly applied in every-day life. Being the basic substance of the paraben group, 4-hydroxybenzoate and its derivatives mainly serve as a preservative in cosmetic as well pharmaceutical products, and beyond that, it is used for the manufacturing of Vectran fibers (Beers and Ramirez, 1990; Menczel et al., 1997). Currently, 4-hydroxybenzoate is produced commercially via the Kolbe-Schmitt-reaction from potassium phenoxide and carbon dioxide (Lindsey and Jeskey, 1957). This way of production, however, harbors major drawbacks in that fossil resources are used as substrates and that the reaction takes place under harsh conditions. Due to relatively low and isomerically poor product yields (Lindsey and Jeskey, 1957), improved production strategies using toxic ionic liquids have been developed (Zhao et al., 2007; Benaskar et al., 2009; Stark et al., 2009). In total, these routes render the overall process non-sustainable and the production conditions hazardous and environmentally unfavorable. Biotechnological production of 4-hydroxybenzoate using microbial cell factories, however, would overcome these issues, since renewable substrates such as glucose, xylose, or glycerol are converted under far milder conditions. Nature offers a variety of candidates for the production of aromatics and other bulk and fine chemicals. Among those, Pseudomonads recently have gained great biotechnological interest (Sun et al., 2006; Elbahloul and Steinbüchel, 2009; Loeschcke and Thies, 2015; Tiso et al., 2016; Nikel and de Lorenzo, 2018). In comparison to other bacteria, these Gram-negative soil bacteria exhibit a high tolerance toward organic solvents (Kieboom et al., 1998), and due to their versatile metabolism, they accept a great variety of substrates (Dos Santos et al., 2004; Wierckx et al., 2015). Furthermore, sophisticated tools for genetic manipulation are available (Martínez-García and de Lorenzo, 2011; Silva-Rocha et al., 2013; Martínez-García et al., 2014; Nikel et al., 2014b; Belda et al., 2016) and cultivation conditions are undemanding. Different Pseudomonas strains have been used for the production of a range of bio-based aromatics including anthranilate, phenylalanine and derivatives, 4-coumarate, phenazines, 4-hydroxystyrene, and phenol (Wierckx et al., 2005; Nijkamp et al., 2007; Verhoef et al., 2009; Kuepper et al., 2015; Schmitz et al., 2015; Molina-Santiago et al., 2016; Wynands et al., 2018).
In microbes, 4-hydroxybenzoate can be produced in two ways. On the one hand, chorismate, which is formed via the shikimate pathway, can be converted into 4-hydroxybenzoate and pyruvate by the chorismate-pyruvate-lyase (EC 4.1.3.40, UbiC). This reaction was already exploited in Escherichia coli, Corynebacterium glutamicum, Klebsiella pneumoniae, and Pseudomonas putida KT2440 (Müller et al., 1995; Barker and Frost, 2001; Yu et al., 2016; Kallscheuer and Marienhagen, 2018; Kitade et al., 2018; Syukur Purwanto et al., 2018). However, in order to assure sufficient flux toward product formation, shutting down competing chorismate consuming pathways is often required. This often results in auxotrophic production strains which require co-feeding of aromatic amino acids or complex medium components such as yeast extract. By overexpression of an ubiC gene from E. coli, Müller et al. (1995) produced 0.12 g l−1 4-hydroxybenzoate from 18 g l−1 glucose with a strain of K. pneumoniae which was auxotrophic for all aromatic amino acids. Likewise, Barker and Frost (2001) developed an E. coli production strain deficient in biosynthesis of tyrosine, phenylalanine and tryptophan and were able to accumulate 4-hydroxybenzoate with a C-mol yield of 15.2% during fed-batch fermentation. A P. putida KT2440 biocatalyst engineered by Yu et al. (2016) achieved a 4-hydroxybenzoate titer of 1.7 g l−1 corresponding to a C-mol yield of 18.1%. However, this strain lacked the ability to natively produce tryptophan as well as phenylpyruvate, so that, as in the case for the aforementioned hosts, additional supplementation of intermediates was necessary. Recently, Kallscheuer and Marienhagen (2018) metabolically engineered a C. glutamicum strain and produced 3.3 g l−1 4-hydroxybenzoate in shake flasks, which corresponds to a C-mol yield of 12.6%. Using the same species as a platform, Kitade et al. (2018) conducted growth-arrested fermentations by which a 4-hydroxybenzoate titer of 36.6 g l−1 and a C-mol yield of 47.8% could be reached. Remarkably, this was achieved without the introduction of auxotrophies by employing a novel UbiC variant from Providencia rustigianii which is insensitive to product inhibition. However, this very high yield was achieved in a 2-step process where biomass was first grown on a rich medium, followed by 4-hydroxybenzoate production in a mineral medium using a high density (5% wet weight) of biomass from the rich medium culture. The 1st-stage culture in rich medium, and thus all biomass formation, was not accounted for in the yield calculation. Syukur Purwanto et al. (2018) produced 137.6 mM 4-hydroxybenzoate with a C-mol yield of 9.65% during fed-batch cultivations using an engineered C. glutamicum strain. Due to the deletion of the trpE gene, however, supplementation of tryptophan was required, and high 4-hydroxybenzoate titers were not reached in a fully mineral medium.
On the other hand, 4-hydroxybenzoate biosynthesis can also be accomplished via tyrosine. The first step of this pathway is deamination of tyrosine into 4-coumarate through a tyrosine ammonia-lyase (EC 4.3.1.23, TAL). Subsequently, 4-coumarate is further processed by moonlight activities of feruloyl-CoA synthetase (EC 6.2.1.34, Fcs), enoyl-CoA hydratase (EC 4.2.1.17, Ech), and vanillin dehydrogenase (EC 1.2.1.67, Vdh) to yield 4-hydroxybenzoate (Harwood and Parales, 1996; Jiménez et al., 2002). Although this approach has a lower maximum theoretical yield due to the loss of one C-atom as CO2 in the conversion of chorismate to tyrosine, it has the benefit of not requiring auxotrophies since tyrosine is the endpoint of the metabolic pathway rather than a central intermediate as in the case of chorismate. Also, chorismate was shown to be non-enzymatically rearranged to phenylpyruvate in engineered phenylalanine/tyrosine auxotrophic Saccharomyces cerevisiae strains, thus lowering its availability as a substrate for production (Winter et al., 2014). Further, both chorismate and tyrosine are key precursors for the production of many other aromatics, thus making it useful to have platform strains that efficiently channel carbon flux toward both of these metabolites. Following the route via tyrosine, Verhoef et al. (2007) accomplished 4-hydroxybenzoate formation with a C-mol yield of 19.3% during shake flask cultivations using glycerol as a sole carbon source and 11.0% with glucose with a modified strain of P. putida S12, whereas fed-batch cultivations using glycerol as a sole carbon source lead to a C-mol yield of 8.5%. Further improvement was achieved through the deletion of the hpd gene coding for the 4-hydroxyphenylpyruvate dioxygenase, resulting in a C-mol yield of 13.4% on glucose (Verhoef et al., 2010). Implementation of a xylose-degrading pathway and subsequent laboratory evolution resulted in a strain that was able to form 4-hydroxybenzoate with C-mol yield of 12.4% on xylose, 17.5% on glucose, and 19.3% on glycerol in shake flask experiments, whereas cultivation in fed-batch mode using a mixed-substrate feeding strategy with glycerol and xylose achieved a C-mol yield of 16.3% 4-hydroxybenzoate (Meijnen et al., 2011).
In this study, Pseudomonas taiwanensis VLB120 was subject to metabolic engineering in order to generate a whole-cell biocatalyst for the production of 4-hydroxybenzoate via tyrosine (Figure 1). This species is naturally capable of using xylose as a sole carbon source (Köhler et al., 2015), rendering it a superior candidate for conversion of lignocellulosic feedstock compared to other Pseudomonads. For the generation of an efficient 4-hydroxybenzoate production host, we downregulated and disrupted competing pathways, and we overexpressed key precursor-supplying genes through stable genomic integration. Primary metabolic genes were also overexpressed, tailored for the use of glucose or glycerol as sole carbon source, and the intrinsic use of xylose as a substrate for 4-hydroxybenzoate production was demonstrated.
Figure 1.
Metabolic overview of 4-hydroxybenzoate production by an engineered P. taiwanensis VLB120. Overexpressed genes are shown as green arrows, deleted genes as red arrows with a cross. The dashed red arrow symbolizes a point mutation resulting in a flux decrease. 6PGL, 6-phosphogluconate; G6P, glucose-6-phosphate; KDPG, 2-keto-3-deoxy-6-phosphogluconate; GA3P, glyceraldehyde-3-phosphate; F6P, fructose-6-phosphate; S7P, sedoheptulose-7-phosphate; E4P, erythrose-4-phosphate; P5P, pentose-5-phosphate; 1,3bPG, 1,3-bisphosphoglycerate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; Pyr, pyruvate; DAHP, 3-Deoxy-D-arabinoheptulosonate 7-phosphate; 3DQ, 3-dehydroquinate; 3DHS, 3-dehydroshikimate; S3P, shikimate-3-phosphate; 5O3PS, 5-O-(1-carboxyvinyl)-3-phosphoshikimate; 4HPP, 4-hydroxyphenylpyruvate; PP, phenylpyruvate; Aro, arogenate; HG, homogentisate; HPHP-CoA, 4-Hydroxyphenyl-β-hydroxypropionyl-CoA; Genes: pgi, glucose-6-phosphate isomerase; ppsA, phosphoenolpyruvate synthase; tktA, transketolase; aroG, DAHP synthase; tyrA, prephenate dehydrogenase; quiC, 3-dehydroshikimate dehydratase; trpE, anthranilate synthase; hpd, 4-hydroxyphenylpyruvate dioxygenase; tal, tyrosine ammonia-lyase; pal, phenylalanine ammonia-lyase; fcs, feruloyl-CoA synthetase; ech, enoyl-CoA dehydratase; vdh, vanillin dehydrogenase; pobA, p-hydroxybenzoic acid hydroxylase.
Materials and Methods
Chemicals
Unless indicated otherwise, all chemicals were purchased from Sigma Aldrich (Taufkirchen, Germany) or Carl Roth (Karlsruhe, Germany).
Bacterial Strains and Culture Conditions
All strains used in this work are listed in Table 1. E. coli strains were cultivated in Lysogeny Broth (LB) medium (10 g l−1 N-Z-Amine, 5 g l−1 yeast extract, 5 g l−1 NaCl). When grown on plates, agar was added to a final concentration of 1.5% before autoclaving. When selection was necessary, the medium was supplemented with the respective antibiotics: 50 mg l−1 kanamycin, 10 mg l−1 gentamicin, 100 mg l−1 ampicillin, 10 mg l−1 tetracycline. Gentamicin concentration was raised to 30 mg l−1 during selection on agar plates. Shake flasks were filled with 10% of the flask volume and incubated at 37°C and 300 rpm. All Pseudomonas strains were grown on LB plates or cetrimide agar plates and selected with the respective antibiotics as described for E. coli, at a temperature of 30°C.
Table 1.
Strains used or engineered in this study.
| Strain | Characteristics | References |
|---|---|---|
| E. coli PIR1 | F−Δlac169 rpoS(Am) robA1 creC510 hsdR514 endA recA1uidA(ΔMluI)::pir-116; host for oriV(R6K) vectors in high copy number | Thermo Fisher Scientific |
| E. coli DH5α λpir | λpir lysogeny of DH5α | Glover and Hames, 1985 |
| E. coli DH5α pTNS1 | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 (rk- mk+) recA1 endA1 gyrA96 thi-1 relA1 | de Lorenzo lab |
| E. coli HB101 pRK2013 | Helper strain carrying plasmid pRK2013; F−λ−hsdS20(rB− mB−) recA13 leuB6(Am) araC14 Δ(gpt-proA)62 lacY1 galK2(Oc) xyl-5 mtl-1 thiE1 rpsL20(SmR) glnX44(AS) | Figurski and Helinski, 1979 |
| E. coli DH5α pBBFLP | Strain used for excision of kanamycin resistance cassette F−endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK−mK+), λ− |
de las Heras et al., 2008 |
| P. taiwanensis VLB120 | Wild type | Panke et al., 1998 |
|
P. taiwanensis VLB120 CL1 |
Derived from P. taiwanensis VLB120; Deficient for pobA, hpd; carrying genes ech, vdh and fcs randomly integrated via Tn5 transposon. |
This study |
|
P. taiwanensis VLB120 CL1 RtPAL |
P. taiwanensis VLB120 CL1 carrying plasmid pJT'RtPAL | This study |
|
P. taiwanensis VLB120 CL1 FjTAL |
P. taiwanensis VLB120 CL1 carrying plasmid pJT'FjTAL | This study |
|
P. taiwanensis VLB120 CL1 TcXAL |
P. taiwanensis VLB120 CL1 carrying plasmid pJT'TcXAL | This study |
|
P. taiwanensis VLB120 CL1 RsTAL |
P. taiwanensis VLB120 CL1 carrying plasmid pJT'RsTAL | This study |
|
P. taiwanensis VLB120 CL1.1 RtPAL |
Derived from P. taiwanensis VLB120 CL1; Deficient for quiC, quiC1, quiC2; carrying plasmid pJT'RtPAL |
This study |
|
P. taiwanensis VLB120 CL2 |
Derived from P. taiwanensis VLB120 CL1.1; Point mutation in trpE resulting in P290S substitution |
This study |
|
P. taiwanensis VLB120 CL2 RtPAL |
P. taiwanensis VLB120 CL2 carrying plasmid pJT'RtPAL | This study |
|
P. taiwanensis VLB120 CL2 RtPALa |
P. taiwanensis VLB120 CL2 carrying plasmid pJT'RtPALa | This study |
|
P. taiwanensis VLB120 CL2 RtPALat |
P. taiwanensis VLB120 CL2 carrying plasmid pJT'RtPALat | This study |
|
P. taiwanensis VLB120 CL2 RtPALatt |
P. taiwanensis VLB120 CL2 carrying plasmid pJT'RtPALatt | This study |
|
P. taiwanensis VLB120 CL2 BG14a RtPALat |
P. taiwanensis VLB120 CL2 carrying genes RtPAL, aroGfbr, tyrAfbr at attTn7 site under control of 14a promoter |
This study |
|
P. taiwanensis VLB120 CL2 BG14d RtPALat |
P. taiwanensis VLB120 CL2 carrying genes RtPAL, aroGfbr, tyrAfbr at attTn7 site under control of 14d promoter |
This study |
|
P. taiwanensis VLB120 CL2 BG14e RtPALat |
P. taiwanensis VLB120 CL2 carrying genes RtPAL, aroGfbr, tyrAfbr at attTn7 site under control of 14e promoter |
This study |
|
P. taiwanensis VLB120 CL3 |
P. taiwanensis VLB120 CL2 carrying genes RtPAL, aroGfbr, tyrAfbr at attTn7 site under control of 14g promoter |
This study |
|
P. taiwanensis VLB120 CL2 BG14fg RtPALat |
P. taiwanensis VLB120 CL2 carrying genes RtPAL, aroGfbr, tyrAfbr at attTn7 site under control of 14fg promoter |
This study |
|
P. taiwanensis VLB120 CL2 BG14ffg RtPALat |
P. taiwanensis VLB120 CL2 carrying genes RtPAL, aroGfbr, tyrAfbr at attTn7 site under control of 14ffg promoter |
This study |
|
P. taiwanensis VLB120 CL3.1 |
P. taiwanensis VLB120 CL3 carrying plasmid pBNT'ppsA | This study |
|
P. taiwanensis VLB120 CL3.2 |
P. taiwanensis VLB120 CL3 carrying plasmid pBNT'pgi | This study |
|
P. taiwanensis VLB120 CL3.3 |
P. taiwanensis VLB120 CL3 carrying plasmid pBNT'ppsA-pgi | This study |
|
P. taiwanensis VLB120 CL4 |
Derived from P. taiwanensis VLB120 CL4; Carrying genes RsTAL, aroGfbr, tyrAfbr at attTn7 site under control of 14f promoter |
This study |
|
P. taiwanensis VLB120 CL4.3 |
P. taiwanensis VLB120 CL4 carrying plasmid pBNT'ppsA-pgi | This study |
For production experiments in batch mode, two liquid precultures were prepared. At first, 500 ml flasks containing 50 ml LB medium were inoculated with cells from a freshly grown LB agar plate or glycerol stock and incubated overnight at 30°C and 300 rpm. The second preculture was conducted in 500 ml flasks containing 50 ml mineral salts medium (MSM) (Hartmans et al., 1989) (buffer: 11.64 g l−1 K2HPO4, 4.89 g l−1 NaH2PO4. Nitrogen source: 2 g l−1 (NH4)2SO4. Trace elements: 10 mg l−1 EDTA, 100 mg l−1 MgCl2 · 6 H2O, 2 mg l−1 ZnSO4 · 7 H2O, 1 mg l−1 CaCl2 · 2 H2O, 5 mg l−1 FeSO4 · 7 H2O, 0.2 mg l−1 Na2MoO4 · 2 H2O, 0.2 mg l−1 CuSO4 · 5 H2O, 0.4 mg l−1 CoCl2 · 6 H2O, 1 mg l−1, MnCl2 · 2 H2O. The medium contained either 20 mM glucose, 40 mM glycerol, or 24 mM xylose as sole carbon source, unless stated otherwise. When using glycerol as a sole carbon source in the main culture, the preculture contained 40 mM glycerol and 5 mM glucose to decrease the lag phase on glycerol. All MSM precultures were incubated for 16–20 h at 300 rpm and 30°C and inoculated into the main culture to a final OD600 of 0.1. For the main culture, cells from the MSM preculture were harvested via centrifugation at 5,000 rpm and 4°C for 10 min and washed twice with 0.9% NaCl. Subsequently, 24-well System Duetz plates (Duetz et al., 2000) containing 1.5 ml fresh MSM per well and 0.2 mM IPTG for induction of expression of the ferulic genes (ech, vdh, and fcs) and, if necessary, 0.1 mM salicylate for induction of pBNT plasmids, were inoculated to a final OD600 of 0.1 and incubated at 30°C and 300 rpm. Unless indicated otherwise, all strains were cultivated in biological triplicates, and the standard error of the mean was used for indication of error bars. Pulsed fed-batch cultivations using MSM with glycerol as a sole carbon source were conducted with a Biostat® A Plus system (Sartorius Stedim, Göttingen, Germany) with a maximum volume of 2 l. Initial batch phase took place in 1 l MSM containing 120 mM glycerol and twice the amount of mineral salts. Initial stirrer speed was set to 480 rpm and aeration to 1 vvm. Dissolved oxygen tension was regulated at >30% through a stirrer cascade and continuously monitored using an InPro 6800 amperometric oxygen sensor (Mettler Toledo, Columbus, USA). During the course of cultivation, this value was maintained by mixing in pure oxygen using manual control. Monitoring of the pH was carried out with a 405-DPAS-SC-K8S/225 pH electrode (Mettler Toledo, Columbus, USA) and maintained at pH 7 by titration of 4 M HCl and 2 M NH4OH. During fed-batch phase, glycerol was pulsed through a syringe to a final concentration of 200 mM each time dissolved oxygen tension increased and glycerol in the fermenter was consumed.
DNA Techniques
All primers (see Table S1) were designed either with Clone Manager Professional (Sci-Ed, Denver, USA) or NEBuilder® Assembly Tool (New England Biolabs, Ipswich, USA) and purchased from eurofins Genomics (Ebersberg, Germany). Codon-optimization for genes RsTAL (Rhodobacter sphaeroides ATCC 17025), FjTAL (Flavobacterium johnsoniae ATCC 17061), and TcXAL (Trichosporon cutaneum ATCC 90039) for P. taiwanensis VLB120 was carried out using the OPTIMIZER online tool (Puigbò et al., 2007), whereas preferences were set as follows: genetic code: eubacterial; method: guided random; manual exclusion of undesired restriction sites and rare codons exhibiting a usage of <6%. Optimized DNA fragments were ordered and purchased from Thermo Fisher Scientific (Thermo Fisher Scientific, Waltham, USA) or Integrated DNA Technologies (Coralville, USA). Inserts for all plasmids were amplified via PCR using Q5® High-Fidelity DNA polymerase (New England Biolabs, Ipswich, USA). Likewise, this was done for backbone plasmids, when cloning took place using the NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, USA). When vectors were assembled using T4 DNA Ligase (Thermo Fisher Scientific, Waltham, USA), backbone DNA was isolated from an E. coli overnight culture using Monarch® Plasmid Miniprep Kit (New England Biolabs, Ipswich, USA). Subsequently, all DNA fragments were digested with the respective restriction enzymes (New England Biolabs, Ipswich, USA) and purified with the Monarch® PCR & DNA Cleanup Kit (New England Biolabs, Ipswich, USA). Assembled and purified plasmids (Table 2) were transferred into E. coli and Pseudomonas via transformation, whereas conjugation was used for Pseudomonas as well. For E. coli, heat-shock transformation was done according to a protocol by Sambrook et al. (1989) Conjugation was performed through triparental mating using a streamlined method as outlined by Wynands et al. (2018). Selection of Pseudomonas took place on cetrimide agar plates. When pEMG and pBELK plasmids were transferred, helper strain E. coli HB101 pRK2013 was used, whereas this strain as well as DH5α pTNS1 were taken for conjugation of pBG14-based constructs.
Table 2.
Plasmids used and constructed in this work.
| Plasmid | Characteristics | References |
|---|---|---|
| pJT'mcs | AmpR GmR, expression vector, under control of Ptac promoter | Nijkamp et al., 2007 |
| pTacPAL | AmpR GmR, expression vector, harboring RtPAL under control of Ptac promoter | Nijkamp et al., 2007 |
| pBELK | Mini-Tn5 delivery vector; tnpA oriV(R6Kγ) oriT(RK2) lacIQ Ptrc bla FRT-aphA-FRT, AmpR KmR | Nikel and de Lorenzo, 2013 |
| pBELK ferulic s | Mini-Tn5 delivery vector harboring genes ech, vdh and fcs from P. putida S12 | This study |
| pJT'RsTAL | pJT expression vector harboring RsTAL | This study |
| pJT'FjTAL | pJT expression vector harboring FjTAL | This study |
| pJT'TcXAL | pJT expression vector harboring TcXAL | This study |
| pJT RtPAL | pJT expression vector harboring RtPAL | This study |
| pBWatt | pSEVA234 expression vector harboring genes aroGfbr, tyrAfbr, tktA; KmR, ori pBBR1, laqIq-Ptrc | This study |
| pJT'RtPALa | pJT expression vector harboring gene aroGfbr | This study |
| pJT'RtPALat | pJT expression vector harboring gene aroGfbr, tyrAfbr | This study |
| pJT'RtPALatt | pJT expression vector harboring gene aroGfbr, tyrAfbr, tktA | This study |
| pBG14a | Tn7 delivery vector; KmR GmR, ori R6K, Tn7L, and Tn7R flanks, BCD2–msfgfp fusion, promoter no. 28 | Zobel et al., 2015 |
| pBG14d | Tn7 delivery vector; KmR GmR, ori R6K, Tn7L, and Tn7R flanks, BCD2–msfgfp fusion, promoter no. 51 | Zobel et al., 2015 |
| pBG14e | Tn7 delivery vector; KmR GmR, ori R6K, Tn7L, and Tn7R flanks, BCD2–msfgfp fusion, promoter no. 17 | Zobel et al., 2015 |
| pBG14f | Tn7 delivery vector; KmR GmR, ori R6K, Tn7L, and Tn7R flanks, BCD2–msfgfp fusion, promoter no. 25 | Zobel et al., 2015 |
| pBG14g | Tn7 delivery vector; KmR GmR, ori R6K, Tn7L, and Tn7R flanks, BCD2–msfgfp fusion, promoter no. 42 | Zobel et al., 2015 |
| pBG14fg | Tn7 delivery vector; KmR GmR, ori R6K, Tn7L, and Tn7R flanks, BCD2–msfgfp fusion, hybrid promoter composed of No. 25 & No. 42 | Köbbing et al. in preparation |
| pBG14ffg | Tn7 delivery vector; KmR GmR, ori R6K, Tn7L, and Tn7R flanks, BCD2–msfgfp fusion, hybrid promoter 2 x No. 25 & No. 42 | Köbbing et al. in preparation |
| pBG14a RtPALat | Tn7 delivery vector harboring genes RtPAL, aroGfbr, tyrAfbr, promoter no. 28 | This study |
| pBG14d RtPALat | Tn7 delivery vector harboring genes RtPAL, aroGfbr, tyrAfbr, promoter no. 51 | This study |
| pBG14e RtPALat | Tn7 delivery vector harboring genes RtPAL, aroGfbr, tyrAfbr, promoter no. 17 | This study |
| pBG14g RtPALat | Tn7 delivery vector harboring genes RtPAL, aroGfbr, tyrAfbr, promoter no. 42 | This study |
| pBG14fg RtPALat | Tn7 delivery vector harboring genes RtPAL, aroGfbr, tyrAfbr, hybrid promoter no. 25 & no. 42 | This study |
| pBG14ffg RtPALat | Tn7 delivery vector harboring genes RtPAL, aroGfbr, tyrAfbr, hybrid promoter 2 × no. 25 & no. 42 | This study |
| pBG14f RsTALat | Tn7 delivery vector harboring genes RsTAL, aroGfbr, tyrAfbr, promoter no. 25 | This study |
| pRK2013 | Helper plasmid for conjugational transfer; KmR, oriV(RK2/ColE1), mob+ tra+ |
Figurski and Helinski, 1979 |
| pTNS1 | Helper plasmid; AmpR, ori R6K, TnSABC+D operon | Choi et al., 2005 |
| pSW-2 | GmR, oriRK2, xylS, Pm → I-sceI (transcriptional fusion of I-sceI to Pm) | Martínez-García and de Lorenzo, 2011 |
| pBBFLP | Helper plasmid for excision of antibiotic markers, oriV(pBBR1), oriT(RK2) RK2 mob+ λPR::FLP λ(cI857) sacB tet, TetR | de las Heras et al., 2008 |
| pEMG pobA | Knockout vector carrying TS1 and TS2 flanking regions of p-hydroxybenzoic acid hydroxylase (pobA) | Wynands et al., 2018 |
| pEMG hpd | Knockout vector carrying TS1 and TS2 flanking regions of 4-hydroxyphenylpyruvate dioxygenase (hpd) | Wynands et al., 2018 |
| pEMG quiC | Knockout vector carrying TS1 and TS2 flanking regions of dehydroshikimate dehydratase (quiC) | Wynands et al., 2018 |
| pEMG quiC1 | Knockout vector carrying TS1 and TS2 flanking regions of dehydroshikimate dehydratase (quiC1) | Wynands et al., 2018 |
| pEMGu quiC2 | Knockout vector carrying TS1 and TS2 flanking regions dehydroshikimate dehydratase (quiC2) | Wynands et al., 2018 |
| pEMGu trpEP290S | Knockout vector carrying TS1 and TS2 flanking regions of the codon for the P290S substitution of trpE gene | Wynands et al., 2018 |
| pBNT'mcs | Expression vector harboring salicylate-inducible nagR/PNagAa promoter, KmR | Verhoef et al., 2010 |
| pBNT'ppsA | pBNT expression vector harboring gene encoding phosphoenolpyruvate synthase (ppsA) | This study |
| pBNT'pgi | pBNT expression vector harboring gene encoding glucose-6-phosphate isomerase (pgi) | This study |
| pBNT'ppsA-pgi | pBNT expression vector harboring gene encoding phosphoenolpyruvate synthase (ppsA), and glucose-6-phosphate isomerase (pgi) | This study |
Gene knockouts were carried out according to a protocol developed by Martínez-García and de Lorenzo (2011). The pEMG suicide vectors containing TS1 and TS2 flanking regions of the gene to be eliminated were conjugated into the respective recipient strain, whereas successful integration was verified via colony PCR. Due to high ampicillin resistance of P. taiwanensis VLB120, plasmid pSW-2 encoding enzyme I-SceI was used instead of pSW-I. Induction of expression by addition of 3-methylbenzoate was not required. Knockouts were confirmed by colony PCR and DNA sequencing. Genes ech, vdh, and fcs, were amplified from the genome of P. putida S12 (Hartmans et al., 1989) and randomly integrated into the genome of P. taiwanensis VLB120 via the pBELK Tn5 mini-transposon system developed by Nikel and de Lorenzo (2013). Integration sites were identified through arbitrary-primed PCR as described by Martínez-García et al. (2014) using the Q5® DNA polymerase and subsequent DNA sequencing and BLAST analysis against the genome of P. taiwanensis VLB120. In order to excise the kanamycin resistance cassette, plasmid pBBFLP harboring a flippase gene was transformed into selected clones and loss of resistance was confirmed via selection of Kms clones on LB agar plates. Stable integration of key genes and modulated expression was achieved by using the Tn7-based calibrated promoter system developed by Zobel et al. (2015).
Cloning procedures were routinely verified via colony PCR using OneTaq® DNA polymerase (New England Biolabs, Ipswich, USA). Colonies were picked from transformation plates and lysed in 30 μl PEG200 (pH 12) for 5–10 min (Chomczynski and Rymaszewski, 2006).
Analytical Methods
Optical densities of cell cultures were measured at a wavelength of 600 nm using an Ultrospec 10 spectrophotometer (GE Healthcare, Chicago, USA). Cell dry weight was determined by multiplying OD600 values by the empirical factor 0.505. Aromatics were analyzed by HPLC. Samples taken during cultivations were centrifuged at 17,000 g for 2 min and the supernatant was filtered using syringe filters with a pore size of 0.2 μm. After addition of methanol (Th. Geyer, Renningen, Germany) in a 1:1 ratio, samples were stored at 4°C overnight in order to precipitate any salts or proteins. After another centrifugation at 13,000 rpm for 2 min, the supernatant was taken for analysis. HPLC was performed using a System Gold 168 diode array detector (Beckman Coulter, Brea, USA) and an ISAspher 100-5 C18 BDS reversed phase HPLC column (ISERA, Düren, Germany) at 30°C and a flow rate of 0.8 ml min. Elution took place with a gradient starting at 95% of 0.1% (v/v) TFA and 5% methanol for 2 min, followed by gradual increase to 100% methanol over 18 min. After 2 min at 100% methanol, initial ratios were reached again within 2 min. UV detection of aromatics was conducted at a wavelength of 260 nm. Glycerol concentrations were determined using the Glycerol GK Assay Kit (Megazyme, Bray, Ireland). Glucose concentrations were determined via HPLC using an Aminex Ion Exclusion HPX-87H column (Bio-Rad, Hercules, USA) and a Smartline RI detector 2300 (Knauer, Berlin, Germany), whereas isocratic elution took place in 5 M H2SO4 at 1.2 ml/min and 70°C. For calculation of production rates, CDW was estimated by multiplying OD values with the empirical factor 0.505. The increase of 4-hydroxybenzoate concentration between two time points was then divided by average biomass of those time points and the period of time. Growth data of the 4-hydroxybenzoate pulse experiment carried out in the Growth Profiler® (Enzyscreen, Heemstede, Netherlands) were normalized by subtracting the initial offset values of each curve compared to that of the cultivation with no 4-hydroxybenzoate pulse, thereby setting all initial points to the same value.
Results and Discussion
Establishing 4-Hydroxybenzoate Production in P. taiwanensis
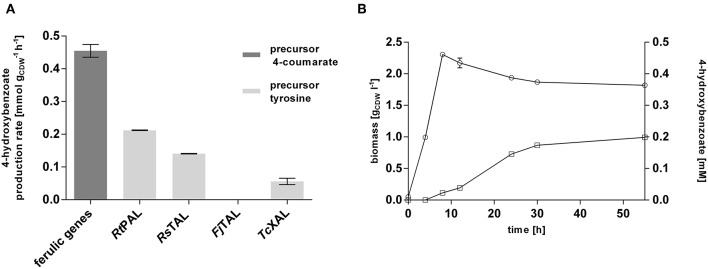
One important requirement for an efficient biocatalyst to be functional is that degradation of the product itself, as well as that of potential precursors, is prevented. Wierckx et al. (2008) and Verhoef et al. (2010) showed that producers of aromatic compounds had upregulated metabolic pathways for the degradation of 4-hydroxybenzoate, protocatechuate, and tyrosine. Therefore, the pobA gene (PVLB_11545) responsible for the conversion of 4-hydroxybenzoate into protocatechuate, and the hpd gene (PVLB_11760), encoding the 4-hydroxyphenylpyruvate dioxygenase, were knocked out, rendering the cells unable to use 4-hydroxybenzoate and tyrosine as a sole carbon sources (Wynands et al., 2018). In doing so, the strain P. taiwanensis VLB120 ΔpobAΔhpd was constructed. Unlike many Pseudomonads, the genome of P. taiwanensis VLB120 does not contain genes encoding a ferulic acid degradation pathway, and indeed this strain is unable to grow on ferulate. Since this pathway is necessary to convert 4-coumarate into 4-hydroxybenzoate, the operon containing ferulic genes fcs, ech, and vdh was amplified from the genome of P. putida S12 and cloned into the pBELK transposon vector (Nikel and de Lorenzo, 2013) under the control of the IPTG-inducible Ptrc promoter. Subsequently, this operon was randomly integrated into the genome of P. taiwanensis VLB120 ΔpobAΔhpd. Three clones were randomly picked and cultivated in MSM containing 20 mM glucose and 3 mM 4-coumarate. Under these conditions, all three strains produced 4-hydroxybenzoate at a rate of 0.45 mmol h−1, proving the functionality of the pathway encoded by the ferulic operon. Another variant of the ferulic operon containing the additional genes encoding feruloyl-CoA dehydrogenase (fcd) and β-ketothiolase (aat) was also tested, but the addition of these two genes did not affect 4-hydroxybenzoate production rates from 4-coumarate, and they were therefore omitted in further strain engineering (data not shown). The integration sites of the operon were identified by arbitrary PCR and DNA sequencing as described in Martínez-García et al. (2014). The transposons had integrated 1,049 bp downstream from the start codon of the gene encoding the plug domain of a TonB-dependent receptor (PVLB_16205, clone 1), 242 bp downstream from the start codon of a radical SAM protein (PVLB_20680, clone 2), and 1,696 bp downstream from the start codon of a gene encoding a sensory box protein (PVLB_25350, clone 3). A map depicting the integration sites can be found in Figure S1. Since no detectable differences regarding growth or production could be observed among the different clones, a negative effect of disruption of these genes on production performance can be excluded. All further experiments were carried out with clone 1, which hereafter is named P. taiwanensis VLB120 CL1.
This strain was subsequently equipped with different ammonia-lyases which catalyze the conversion of tyrosine into 4-coumarate, including a native pal from Rhodosporidium toruloides (RtPAL; Verhoef et al., 2007) and codon-optimized versions of a tal from Rhodobacter sphaeroides (RsTAL; Xue et al., 2007), from Flavobacterium johnsoniae (FjTAL), and a xal from Trichosporon cutaneum (TcXAL) (Jendresen et al., 2015), cloned into plasmid pJT'mcs (Nijkamp et al., 2007) under the control of the constitutive Ptac promoter. This setup of genomic integration of the ferulic operon and plasmid-based expression of PAL/TAL-encoding genes was chosen because the ammonia-lyase is generally the rate-limiting step (Verhoef et al., 2007; Jendresen et al., 2015), and expression from a multicopy plasmid was expected to increase its activity. Co-feeding experiments with 20 mM glucose and 3 mM tyrosine confirmed the ammonia-lyase reaction as the rate-limiting step of the pathway. Among the four tested lyases, RtPAL enabled the highest 4-hydroxybenzoate production rate (0.20 mmol h−1), reaching 44% of the conversion velocity of the downstream pathway encoded by the ferulic operon (Figure 2A). The second best performance was exhibited by the strain harboring the RsTAL construct with a rate of 0.14 mmol h−1. When cultivated with 20 mM glucose in mineral medium without precursor supplementation (Figure 2B), the strain harboring the ferulic operon and the RtPAL construct produced 0.20 mM 4-hydroxybenzoate, corresponding to a C-mol yield of 1.2% with a maximum production rate of 0.004 mmol h−1, which is only 2% of the rate achieved with additional feed of tyrosine. However, considerable accumulation of 4-hydroxybenzoate could only be detected after cells had reached stationary phase, suggesting that at least some of the pathway genes are catabolite-repressed. Indeed, the recognition sequence motif (5′-AANAANAA-3′) characteristic for catabolite-repressed expression (Moreno et al., 2009b, 2015) could be found 37 base pairs downstream from the start codon of fcs, encoding the first enzymatic step in 4-coumarate conversion, and 20 bp upstream from the start codon of ech, which is the first gene downstream from the promoter. These findings are in accordance to those described by Verhoef et al. (2010). However, the latter site was deleted during the cloning into the pBELK vector. Upon binding of the Crc global regulator protein in conjunction with the Hfq chaperone to this motif on the mRNA level, translation is prevented (Wolff et al., 1991; MacGregor et al., 1996; Hester et al., 2000a,b; Morales et al., 2004; Ruiz-Manzano et al., 2005; Moreno et al., 2009a; Hernández-Arranz et al., 2016). Knockout of crc or hfq in order to override catabolite-repression, however, did not beneficially influence 4-hydroxybenzoate production (Figure S2). In summary, an efficient 4-hydroxybenzoate production pathway from tyrosine could be implemented in P. taiwanensis VLB120 CL1, in which the selection of an efficient ammonia-lyase was a key enabling factor. The fact that feeding of precursors such as tyrosine results in a greatly increased 4-hydroxybenzoate production rate suggests that, as expected, the de novo formation of tyrosine is a bottleneck for efficient 4-hydroxybenzoate production from glucose or glycerol.
Figure 2.
Evaluation of the 4-hydroxybenzoate production pathway in P. taiwanensis VLB120 CL1 in co-feeding experiments. (A) Comparison of the conversion rates of the two different pathway components (encoded by the ferulic operon and the ammonia-lyase genes). Cells were grown in biological triplicates in MSM with 5 mM glucose in System Duetz plates at 30°C and 300 rpm. After ~6 h, 3 mM 4-coumarate (ferulic operon) or tyrosine (PAL and TAL genes), and another 20 mM glucose were added. Highest measured rates are indicated. (B) 4-hydroxybenzoate production (squares) and growth (circles) of P. taiwanensis VLB120 CL1 (containing RtPAL). Cells were cultivated in triplicates in MSM supplemented with 20 mM glucose as a sole carbon source at 30°C and 300 rpm in System Duetz plates. Expression of ferulic genes was induced with 0.2 mM IPTG. Error bars indicate standard error of the mean.
Enhancing Supply of Tyrosine
As in the case of pobA and hpd, upregulation of 3-dehydroshikimate (3DHS) dehydratase (quiC1) (Peek et al., 2017) was observed in the phenol producing strain P. putida S12 TPL3 (Wierckx et al., 2008), converting 3DHS, one of the upper metabolites of the shikimate pathway, into protocatechuate. Recently, investigations on quinate metabolism in P. taiwanensis VLB120 revealed the presence of a total of three genes encoding 3DHS dehydratase (Wynands et al., 2018). Therefore, genes quiC (PVLB_18200), quiC1 (PVLB_10935), and quiC2 (PVLB_13075) were knocked out in P. taiwanensis VLB120 CL1 in order to prevent the degradation of aromatic pathway intermediates, thus generating VLB120 CL1.1. Besides tyrosine, tryptophan is also formed via the shikimate pathway. In P. putida S12 TPL3, genome analysis revealed a mutation in the gene encoding the anthranilate synthase (trpE), resulting in a P290S amino acid exchange (Wierckx et al., 2008). This mutation substantially increased the carbon flux toward tyrosine in a P. taiwanensis-based phenol overproducer (Wynands et al., 2018). Therefore, the same point mutation as described in Wynands et al. (2018) resulting in a P290S substitution was introduced in P. taiwanensis VLB120 CL1.1, thereby generating P. taiwanensis VLB120 CL2. These genetic modifications together with the pJT'RtPAL construct lead to an almost 2-fold increase in 4-hydroxybenzoate titer for P. taiwanensis VLB120 CL1.1 and 6.9-fold for P. taiwanensis VLB120 CL2 compared to P. taiwanensis VLB120 CL1, corresponding to C-mol yields of 2.4 and 8.1%, respectively (Figure 3). The large increase in production obtained with CL2 confirms the importance of the trpEP290S mutation. The difference between CL1 and CL1.1 indicates that indeed a substantial portion of the carbon flux into the shikimate pathway is diverted into the 3DHS degradation pathway, thereby creating a futile cycle. These strains also produced 0.10 ± 0.01 mM (CL1.1) and 0.30 ± 0.01 mM (CL2) of trans-cinnamate due to loose substrate specificity of the used RtPAL enzyme which is capable of converting both tyrosine and phenylalanine.
Figure 3.
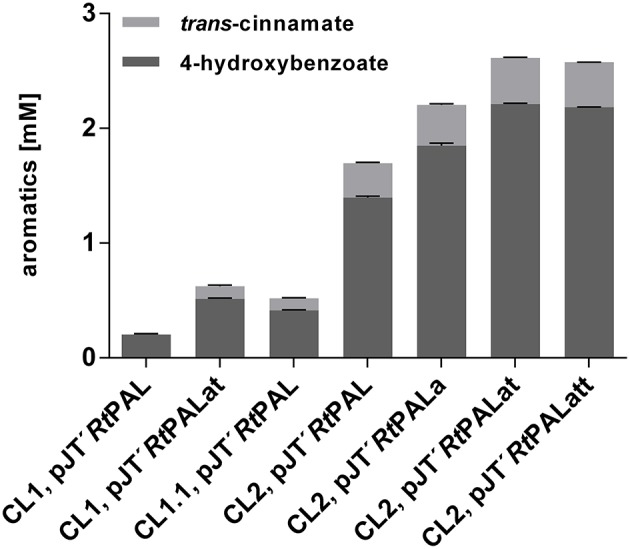
Comparison of aromatics titers of P. taiwanensis VLB120 CL1, P. taiwanensis CL1.1, and P. taiwanensis CL2 with overexpression of different bottleneck genes. All strains were cultivated for 96 h in biological triplicates in MSM containing 20 mM glucose as a sole carbon source in System Duetz plates at 30°C and 300 rpm, expression of ferulic genes was induced with 0.2 mM IPTG. Error bars indicate standard error of the mean.
Former studies in E. coli revealed genes aroG, tyrA, and tktA as key steps in increasing tyrosine supply (Kikuchi et al., 1997; Li et al., 1999; Lütke-Eversloh and Stephanopoulos, 2007; Kim et al., 2014). Therefore, along with the gene encoding RtPAL, feedback-resistant versions of aroG and tyrA (aroGfbr, with a D146N substitution, and tyrAfbr, with A354V and M53I substitutions) as well as tktA, from E. coli, were codon-optimized and cloned into the pJT'mcs vector. In the experiment shown in Figure 3, synergistic effects of these genes on 4-hydroxybenzoate production were investigated in MSM with 20 mM glucose as sole carbon source. When using the pJT'RtPALat construct (overexpression of RtPAL, aroGfbr, and tyrAfbr) in VLB120 CL1, the 4-hydroxybenzoate titer was raised to 0.51mM with a C-mol yield of 3%. Strain P. taiwanensis VLB120 CL2 harboring the same pJT'RtPALat plasmid produced 2.21 ± 0.01 mM 4-hydroxybenzoate with a C-mol yield of 12.9%, confirming that a major contribution to enhanced production is made by downregulation and elimination of competing pathways (quiC, quiC1, quiC2, and trpEP290S), but also showing that the additional expression of both aroGfbr and tyrAfbr significantly enhances production. Compared to these strains, the inclusion of aroGfbr alone shows an intermediate phenotype. The additional overexpression of the tktA gene (pJT'RtPALatt) did not further increase the product concentration. The highest trans-cinnamate concentration of 0.40 ± 0.01 mM was found for P. taiwanensis VLB120 CL2 harboring pJT'RtPALat, making up 15.9% of the total aromatics produced.
Stable Genomic Integration of 4-Hydroxybenzoate Production Modules
The use of plasmid-based expression systems in a bioprocess has the disadvantage that selective pressure has to be maintained throughout the entire cultivation, thus leading to increased burden (Mi et al., 2016). Furthermore, plasmids inherently vary in their copy number from cell to cell, which results in greater variability and instability (Gao et al., 2014; Jahn et al., 2014). To address these issues, a Tn7 transposon system with a calibrated promoter library (Zobel et al., 2015) was used in order to stably integrate one copy of RtPAL, aroGfbr, and tyrAfbr into the genome of P. taiwanensis VLB120 CL2. Different promoter strengths were tested in order to investigate optimal constitutive expression levels (Figure 4). In addition, the 14fg and 14ffg variants were included, which are stacked promoters with activities higher than 14g (Sebastian Köbbing, RWTH Aachen University, personal communication). The highest final 4-hydroxybenzoate concentration was achieved with the 14g construct, which reached a titer equal to that of the plasmid-based system. Unexpectedly, the strongest investigated promoter 14ffg resulted in a tyrosine titer of 2.4 mM, but no accumulation of 4-hydroxybenzoate or trans-cinnamate. Since this promoter exhibits 80% more activity than the BG14g promoter, one explanation for this may be that expression levels of RtPAL were too high, leading to the selection of mutated constructs. The strain harboring the P14g RtPALat construct exhibited the best performance and is hereafter designated P. taiwanensis VLB120 CL3.
Figure 4.
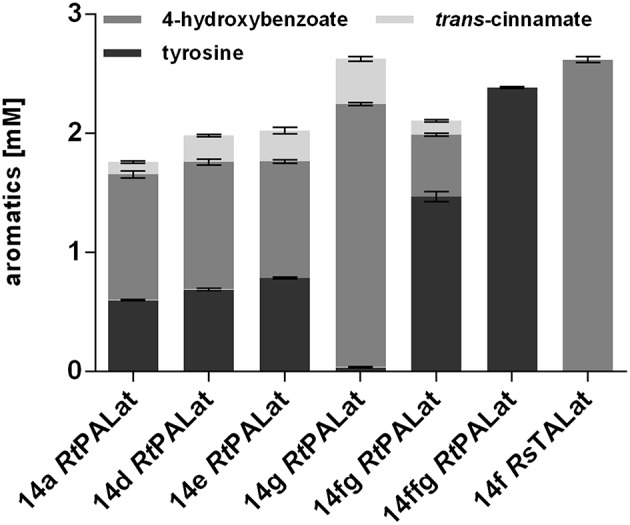
Modulation of expression of genomically integrated RtPAL, RsTAL, aroGfbr, and tyrAfbr. Increasing promoter strengths are listed from left to right. Cells were cultivated in biological triplicates in MSM containing 20 mM glucose as a sole carbon source at 30°C and 300 rpm in System Duetz plates, expression of ferulic genes was induced with 0.2 mM IPTG. Titers of final samples (96 h) are shown. Error bars indicate standard error of the mean.
Although the RtPAL enzyme enabled the highest production rate of 4-hydroxybenzoate in co-feeding experiments, one major disadvantage of this enzyme is its relaxed substrate specificity, causing it to also convert phenylalanine into trans-cinnamate. Thus, not only is a fraction of valuable precursor converted into a byproduct, it also hampers the downstream purification of 4-hydroxybenzoate. In an attempt to circumvent this issue, RtPAL was replaced by a codon-optimized gene encoding the tyrosine ammonia-lyase from Rhodobacter sphaeroides (RsTAL) (Xue et al., 2007). Although this TAL supports a lower 4-hydroxybenzoate production rate (Figure 2), it has a much higher relative affinity for tyrosine, thus potentially generating less byproduct. The gene was cloned along with aroGfbr and tyrAfbr into the pBG14f transposon vector and integrated at the attTn7 site into the genome of P. taiwanensis VLB120 CL2, thereby generating P. taiwanensis VLB120 CL4. The construct containing the 14f promoter was chosen because cloning attempts using stronger promoters failed. After 96 h of cultivation in MSM containing 20 mM glucose, a 4-hydroxybenzoate titer of 2.62 ± 0.03 mM was reached, with no detectable formation of trans-cinnamate. The increase of 4-hydroxybenzoate corresponds to the amount of trans-cinnamate produced by the same strain using RtPAL, indicating that in this case the RsTAL enzyme efficiently redirected the flux toward the main product, likely aided by the phenylalanine hydroxylase PhhAB which converts phenylalanine into tyrosine (Arias-Barrau et al., 2004).
Enhanced 4-Hydroxybenzoate Production Through Overexpression of ppsA and pgi
Overall, the biosynthesis of one mole of tyrosine requires one mole of erythrose-4-phosphate (E4P) and two moles of phosphoenolpyruvate (PEP). In E. coli, glucose is taken up through a phosphotransferase system which converts PEP into pyruvate. This system poses a severe drain on PEP, and enhanced production of tyrosine could be achieved by overexpression of the ppsA gene encoding phosphoenolpyruvate synthase A, which re-converts pyruvate into PEP (Yi et al., 2002; Lütke-Eversloh and Stephanopoulos, 2007; Juminaga et al., 2012). Although Pseudomonads do not take up glucose via a phosphotransferase system (Romano et al., 1970), they mainly metabolize glucose via the Entner-Doudoroff pathway (Nikel et al., 2015), which yields one PEP and one pyruvate per glucose molecule. The Embden-Meyerhof-Parnas pathway employed by E. coli yields two PEP per mole of glucose, and thus both organisms theoretically yield the same net production of one PEP and one pyruvate per mole of transported glucose through their major metabolic pathway. In order to increase the PEP precursor supply, we overexpressed ppsA in the 4-hydroxybenzoate producing strains. The gene was amplified from the genome of P. taiwanensis VLB120 and cloned into the pBNT'mcs expression vector (Verhoef et al., 2010) under the control of the salicylate-inducible NagR/PnagAa-promoter system. The second precursor E4P is formed by transketolase (tktA), the overexpression of which did not increase 4-hydroxybenzoate production (Figure 3). In Pseudomonads, the flux through the pentose phosphate pathway (PPP) is generally low (Fuhrer et al., 2005; Wierckx et al., 2009; Nikel et al., 2015), and enhancement of the flux through the PPP through evolution on xylose also increased 4-hydroxybenzoate production in an engineered P. putida S12 (Meijnen et al., 2011), making it likely that this pathway poses a bottleneck in the engineered P. taiwanensis strains as well. Given that overexpression of tktA did not improve production, phosphoglucose isomerase (pgi) was chosen as upstream target, converting glucose-6-phosphate into fructose-6-phosphate.
To investigate whether the overexpression of these two genes has a beneficial effect on 4-hydroxybenzoate formation, they were cloned separately and together into the pBNT'mcs plasmid and transformed into P. taiwanensis VLB120 CL3, thereby generating P. taiwanensis VLB120 CL3.1 overexpressing ppsA, P. taiwanensis VLB120 CL3.2 overexpressing pgi and P. taiwanensis VLB120 CL3.3 overexpressing both genes. The strains were cultivated in mineral medium containing 20 mM glucose as a sole carbon source (Figure 5). Overexpression of the individual genes did not result in an increase of 4-hydroxybenzoate titer. However, a synergistic effect of both genes could be observed, by which a 4-hydroxybenzoate concentration of 2.62 ± 0.07 mM was produced from 20 mM glucose, corresponding to a C-mol yield of 15.3%, which is an increase of 18.6% compared to the equivalent strain without ppsA-pgi overexpression. As the best performance of 4-hydroxybenzoate production was achieved upon combined overexpression of both ppsA and pgi, the RsTAL-harboring strain P. taiwanensis VLB120 CL4 was equipped with plasmid pBNT'ppsA-pgi and hereafter named P. taiwanensis VLB120 CL4.3. Under the same conditions as strain P. taiwanensis VLB120 CL3.3, this strain produced 3.26 ± 0.05 mM 4-hydroxybenzoate with a C-mol yield of 19.0%. Compared to the reference strain, overexpression of ppsA and/or pgi had a negative effect on growth. On the one hand, this may be due to the additional plasmid-borne metabolic load. However, since lowest biomass values can be observed in strains harboring ppsA-pgi constructs, it can be assumed that poorer growth may be due to reduced availability of PEP and E4P which is now channeled into the shikimate pathway and toward 4-hydroxybenzoate. In addition, PpsA phosphorylates pyruvate through the conversion of ATP into AMP (Berman and Cohn, 1970), and may thus constitute an ATP-wasting futile cycle in conjunction with the reverse pyruvate kinase reaction (Hädicke et al., 2015).
Figure 5.
Overexpression of ppsA and pgi in P. taiwanensis VLB120 CL3 and CL4. Cells were cultivated in biological triplicates in MSM with 20 mM glucose as a sole carbon source at 30°C and 300 rpm in System Duetz plates. Expression of ppsA and pgi was induced with 0.1 mM salicylate and expression of the ferulic genes with 0.2 mM IPTG. Concentrations of aromatics (A) and biomass (B) measured after 96 h. Error bars indicate standard error of the mean.
Production of 4-Hydroxybenzoate Is Enhanced on Glycerol, but Not on Xylose
Former studies showed that glycerol represents a promising alternative to glucose as a renewable biotechnological feedstock (Murarka et al., 2008; Yang et al., 2012; West, 2013; Zambanini et al., 2017; Wynands et al., 2018). Being a byproduct formed during biodiesel production, it is a cheap substrate, and another advantage over glucose is that it can be applied in higher concentrations for Pseudomonas batch cultivations, since no gluconate is formed which acidifies the medium. To evaluate the performance of P. taiwanensis VLB120 CL3 and P. taiwanensis VLB120 CL3.3 on this substrate, they were cultivated in MSM containing glycerol as sole carbon source. When grown on 40 mM glycerol (Figure 6A), a final titer of 4.72 ± 0.01 mM 4-hydroxybenzoate was achieved, corresponding to a C-mol yield of 27.5%. A maximum production rate of 0.57 mmol h−1 was reached after 16 h, which is a 1.8-fold increase compared to the same strains on glucose, and a 2.6-fold increase compared to the P. putida S12 strain of Verhoef et al. (2007) under similar conditions. The overexpression of ppsA-pgi in P. taiwanensis VLB120 CL3.3 further increased the titer to 5.1 ± 0.1 mM of 4-hydroxybenzoate with a C-mol yield of 29.6% (Figure 6B). Considering formation of trans-cinnamate as a byproduct of RtPAL, P. taiwanensis VLB120 CL3 and P. taiwanensis VLB120 CL3.3 reached a total aromatics C-mol yield of 32.3% and 35.1%, respectively.
Figure 6.
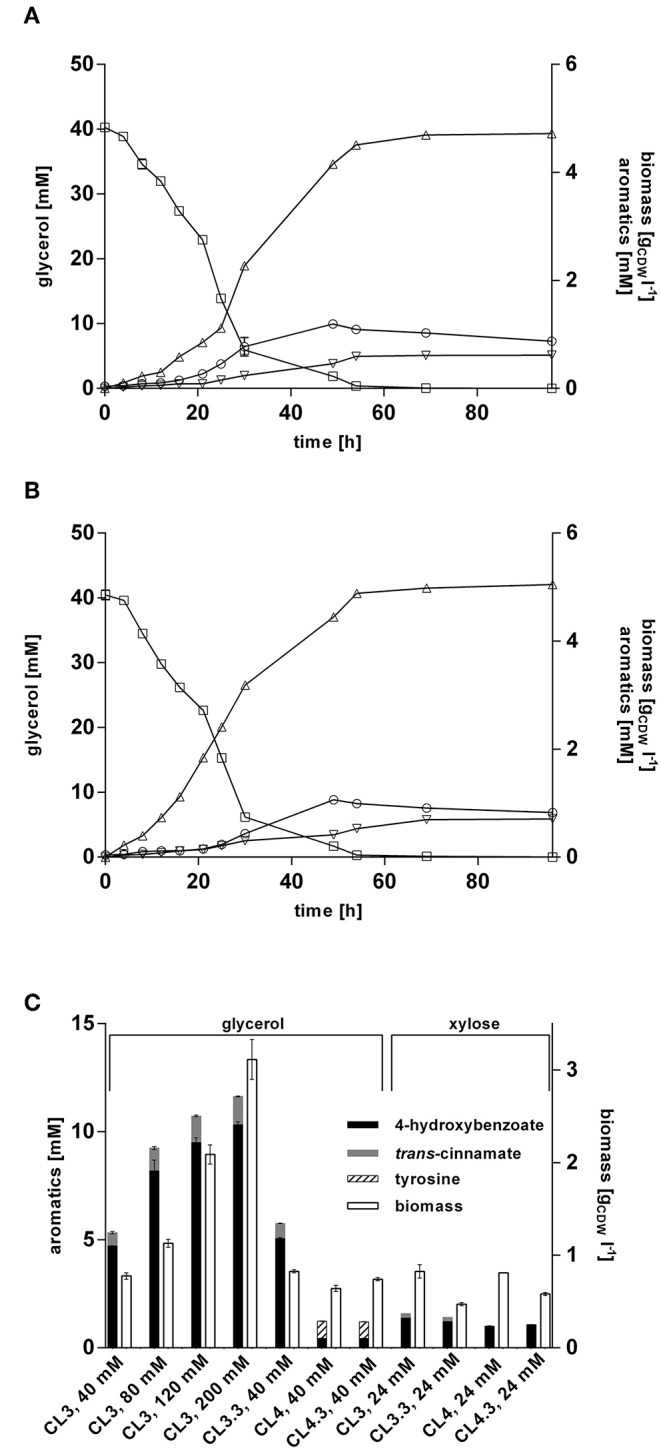
4-hydroxybenzoate production from glycerol and xylose by engineered P. taiwanensis VLB120 strains. Cells were grown in biological triplicates in MSM containing glycerol or 24 mM xylose as a sole carbon source in 24-well System Duetz plates at 30°C and 300 rpm. Expression of ferulic genes was induced with 0.2 mM IPTG. Cultivations of (A) strain CL3 and (B) CL3.3 Values of biomass (circles), glycerol (squares), 4-hydroxybenzoate (triangles), and trans-cinnamate (inverted triangles) are shown. (C) Aromatics concentrations and biomass after 96 h. The used substrate concentrations are indicated on the X-axis. For P. taiwanensis VLB120 CL3.3 and CL4.3, expression of ppsA-pgi was induced with 0.1 mM salicylate. Error bars indicate standard error of the mean.
Increasing the initial glycerol concentration also increased product titers. This did, however, affect production efficiency, as C-mol yields of 23.9, 20.0, and 16.0% were reached using 80, 120, and 200 mM glycerol, respectively (Figure 6C). The relatively low yield achieved with 200 mM glycerol is likely, at least in part, caused by the fact that such a substrate concentration will lead to a nitrogen limitation under the conditions tested. Still, it is remarkable that 4-hydroxybenzoate production is more than twice as efficient from glycerol as from an equivalent concentration of glucose. For P. putida KT2440, it was shown that growth on glycerol leads to metabolic and regulatory rearrangements within the cell (Nikel et al., 2014a). Genes belonging to the Entner-Doudoroff pathway as well as stress-related genes are downregulated and enhanced expression of genes of the glyoxylate shunt as well as pgi was detected. In concert, these actions may result in increased availability of PEP and E4P which, together with the lower growth rate, could lead to a more efficient channeling of these central metabolites into the shikimate pathway.
Production of 4-hydroxybenzoate from glycerol was also investigated for strain P. taiwanensis VLB120 CL4 and P. taiwanensis VLB120 CL4.3. Unexpectedly, these strains only reached a final 4-hydroxybenzoate concentration of 0.44 ± 0.01 mM without and 0.43 ± 0.03 mM with ppsA-pgi overexpression, which is only 9.4% of the 4-hydroxybenzoate concentrations produced by P. taiwanensis VLB120 CL3. Additionally, for the strain P. taiwanensis VLB120 CL4, a residual concentration of 0.81 ± 0.01 mM tyrosine and 0.78 ± 0.01 mM for strain P. taiwanensis VLB120 CL4.3 was measured in the supernatant. These low titers may be caused by the lack of conversion of phenylalanine into trans-cinnamate, which could lead to an increased intracellular phenylalanine concentration, triggering feedback-inhibition mechanisms in the upstream pathway. This may be exacerbated by a reduced expression of phhAB, which is 20–30-fold less strongly induced on glycerol than on glucose in the 4-hydroxybenzoate producing P. putida S12palB1 (Verhoef et al., 2010). Indeed, the culture supernatant of the RsTAL strains turned brownish during cultivation, hinting toward the accumulation of shikimate pathway intermediates, as described by Wynands et al. (2018).
Lignocellulosic biomass contains up to 25% pentose sugars such as xylose (Lee, 1997), making it a relevant alternative substrate especially on lignocellulosic hydrolysates. In contrast to e.g., C. glutamicum and P. putida, P. taiwanensis VLB120 is natively capable of assimilating xylose via the Weimberg pathway (Köhler et al., 2015). To exploit this trait for 4-hydroxybenzoate biosynthesis, production performance of strains P. taiwanensis VLB120 CL3 harboring RtPAL and the ppsA-pgi overexpressing variant CL3.3, as well as their RsTAL-harboring equivalents P. taiwanensis VLB120 CL 4 and CL4.3, was investigated in MSM containing 24 mM of xylose (Figure 6C). Strain CL3 produced 1.37 mM and strain CL3.3 (ppsA-pgi overexpression) 1.21 mM 4-hydroxybenzoate, which is 38% and 54%, respectively, less than on the equivalent amount of glucose. The counterparts CL4 and CL4.3. containing RsTAL. reached similar titers of 1.0 mM and 1.1 mM. During its multi-step assimilation, xylose is converted to α-ketoglutarate and thus enters the primary metabolism at the level of the TCA cycle (Weimberg, 1961). This sets it apart from glucose and glycerol in that PEP and E4P as precursors for 4-hydroxybenzoate formation are formed by gluconeogenic reactions at a higher energetic expense compared to glycolysis. This is likely one explanation for the lower production performance. In accordance with Köhler et al. (2015), growth on xylose also resulted in decreased formation of biomass compared to glucose, with 0.83 gCDW l−1 for strain CL3, 0.47 gCDW l−1 for CL3.3, 0.8 gCDW l−1 for CL4, and 0.58 gCDW l−1 for strain CL4.3. In summary, the feasibility of 4-hydroxybenzoate production upon intrinsic use of xylose as sustainable carbon source could be demonstrated. However, the fact that neither the avoidance of trans-cinnamate formation through the use of RsTAL, nor ppsA-pgi overexpression could considerably enhance 4-hydroxybenzoate titers, underlines that, as in the case of glucose and glycerol, production has to be tailored to the respective substrate. To this end, the insertion of a non-oxidative xylose metabolic pathway, likely coupled to adaptive laboratory evolution, may be a promising strategy (Meijnen et al., 2008, 2011).
Production of 4-Hydroxybenzoate From Glycerol Via Pulsed Fed-Batch Cultivation
In order to increase final product titers beyond those enabled by simple shake-flask cultivations, strain P. taiwanensis VLB120 CL3.3 was cultivated in 1 l MSM with glycerol as sole carbon source in controlled bioreactors under pulsed fed-batch conditions (Figure 7). After an initial batch phase on 120 mM glycerol, the cultures were pulsed multiple times with 200 mM glycerol when an increase in the dissolved oxygen tension was observed, indicating a depletion of the carbon source. The reactors were titrated with ammonium hydroxide to maintain a neutral pH and to avoid a nitrogen limitation at higher cell densities. With this system, a final 4-hydroxybenzoate titer of 72.0 ± 0.96 mM (9.9 g l−1) was reached, and 11.2 ± 0.37 mM (1.7 g l−1) trans-cinnamate was produced, whereas no tyrosine was detected in the culture supernatant. After 85 h of cultivation, both growth and production ceased immediately, so that the last pulse of glycerol had no more effect on biomass and product formation and appeared to be only consumed for homeostasis. Excluding this final pulse, a 4-hydroxybenzoate C-mol yield of 19.2% and total aromatics C-mol yield of 23.0% were reached, with maximum production rate of 0.49 mmol h −1.
Figure 7.

Glycerol pulsed fed-batch cultivation of P. taiwanensis VLB120 CL3.3. Cultivation took place in MSM containing an initial glycerol concentration of 120 mM as a sole carbon source. Expression of the ferulic operon and ppsA-pgi was induced by addition of 0.2 mM IPTG and 0.1 mM salicylate. Two biological replicates were cultivated. (A) Mean values of biomass (circles), 4-hydroxybenzoate (squares), trans-cinnamate (triangles) are shown. (B) Dissolved oxygen tension (no symbols) and glycerol (triangles). Dissolved oxygen tension values from one replicate are shown. Error bars indicate the deviation from the mean.
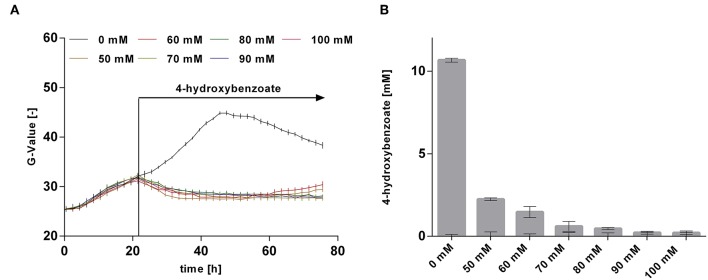
The rather abrupt cessation of growth and product formation is likely due to product toxicity, product inhibition, or both. In order to test this, batch cultures of P. taiwanensis VLB120 CL3.3 on MSM with 120 mM glycerol were pulsed with different concentrations of 4-hydroxybenzoate in the exponential growth phase after 23 h (Figure 8). At this point, the culture had already produced 3.7 ± 0.11 mM 4-hydroxybenzoate. Growth was monitored using a Growth Profiler® and 4-hydroxybenzoate concentrations were measured immediately after the pulse and at the end of the experiment. Compared to the control (no addition) which continued growing as expected and produced 14.4 ± 0.04 mM 4-hydroxybenzoate, a strong inhibition of growth was already observed at the lowest added concentration of 50 mM 4-hydroxybenzoate. Upon addition of 50 and 60 mM 4-hydroxybenzoate, a significant additional 2.2 ± 0.08 mM, respectively, 1.5 ± 0.33 mM 4-hydroxybenzoate were produced, which amounts to total titers of 5.9 ± 0.19 mM and 5.2 ± 0.33 mM, respectively. No significant increase in concentration was measured after a pulse of 70 mM 4-hydroxybenzoate and higher concentrations. These results are in good accordance with the final titer of 72 mM reached in the pulsed fed-batch cultivation, and indicate that both product toxicity and product inhibition are severe at this concentration of 4-hydroxybenzoate, and that product toxicity likely is the predominant factor. In comparison, an impressive tolerance of C. glutamicum toward up to 300 mM 4-hydroxybenzoate was shown by Kitade et al. (2018). During growth-arrested production, the engineered C. glutamicum strains produced 4-hydroxybenzoate with a remarkable titer of 37 g l−1 and a yield of 41% (mol/mol). This was achieved in a two-step-process comprising formation of biomass in a rich medium and subsequent high-cell density production in minimal medium. The P. taiwanensis VLB120 strains developed in this study, however, were subject to 4-hydroxybenzoate production using a fully mineral medium combined with cell growth, which hence was included in all yield calculations. The two-stage process reported by Kitade et al. (2018) could provide a promising approach to reduce product toxicity for Pseudomonas, given that the cells only need to survive, rather than grow. Alternatively, in situ product removal could be applied to circumvent the accumulation of inhibiting concentrations of 4-hydroxybenzoate, and thereby increase production performance of P. taiwanensis VLB120 hosts. Regarding this approach, strategies such as Calcium salt precipitation (Zambanini et al., 2016), co-crystallization (Urbanus et al., 2010) as well as reactive extraction (Schügerl and Hubbuch, 2005; Kreyenschulte et al., 2018) have been proven to be applicable for organic acids.
Figure 8.
Growth and production upon a 4-hydroxybenzoate pulse. P. taiwanensis VLB120 CL3.3 was cultivated in 24-well System Duetz plates in a Growth Profiler in MSM containing 120 mM glycerol as sole carbon source. Each well was filled with a culture volume of 1,350 μl. At an OD600 of ~1, different concentrations of 4-hydroxybenzoate were applied as indicated, using 150 μl of a 10x stock solution for each concentration adjusted to pH 7, whereas water was added to the wells with no 4-hydroxybenzoate pulse. Growth (A) was monitored by image analysis and concentrations were measured immediately after addition and at the end of experiment. (B) Difference between 4-hydroxybenzoate concentrations just after the pulse and after 96 h. Values of triplicate samples are shown, error bars indicate standard error of the mean.
Conclusion
This study describes the rational metabolic engineering of a P. taiwanensis VLB120 strain for high-yield microbial catalysis of glucose or glycerol into 4-hydroxybenzoate via the central metabolite L-tyrosine. This was achieved in a completely minimal medium, without the use of auxotrophies to force metabolic flux toward the product of interest. The formation of trans-cinnamate as a byproduct could be avoided through the use of the tyrosine-specific RsTAL, but this only enabled efficient 4-hydroxybenzoate production from glucose, and not from glycerol. Large differences in product to substrate yields were achieved on these two carbon sources, with glycerol being the preferred substrate with C-mol yields up to 29.6% (= 0.19 g/g) as long as the less specific RtPAL was used. To the best of our knowledge, this is the highest reported 4-hydroxybenzoate yield on a fully mineral medium. The unexpected interplay between the up- and downstream pathways of tyrosine should be further investigated, which would likely yield valuable insights into the underlying mechanisms of the fundamental synthetic biology concept of chassis and modules. Production of 4-hydroxybenzoate from xylose was also demonstrated, although in this case the relatively low performance compared to glucose or glycerol indicates that further improvement of central precursor supply is needed. In all, this work provides a significant advance in the efficiency of aromatics production in non-pathogenic Pseudomonads, the fundamentals of which can be applied to enable the sustainable production of 4-hydroxybenzoate. In addition, the development of the solvent-tolerant P. taiwanensis as aromatics platform can be further exploited for the biosynthesis of a wide range of other more toxic chemicals.
Author Contributions
All authors saw and approved the manuscript. All authors contributed significantly to the work. NW conceived the project. NW, CL, and LB designed experiments and analyzed results. CL and NW wrote the manuscript with the help of LB, BW, and MO. CL performed experiments, supported by BW, MO, JB, and PM.
Conflict of Interest Statement
NW is employed by the Institute for Bio-und Geosciences (IBG-1): Biotechnology, Forschungszentrum Jülich GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sebastian Köbbing for providing plasmids pBG14fg and pBG14ffg.
Footnotes
Funding. This project was funded by the German Research Foundation through the Emmy Noether program (WI 4255/1-1). LB also acknowledges funding by the Cluster of Excellence The Fuel Science Center–Adaptive Conversion Systems for Renewable Energy and Carbon Sources, which is funded by the Excellence Initiative of the German federal and state governments to promote science and research at German universities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2019.00130/full#supplementary-material
References
- Arias-Barrau E., Olivera E. R., Luengo J. M., Fernández C., Galán B., García J. L., et al. (2004). The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 186, 5062–5077. 10.1128/JB.186.15.5062-5077.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. L., Frost J. W. (2001). Microbial synthesis of p-hydroxybenzoic acid from glucose. Biotechnol. Bioeng. 76, 376–390. 10.1002/bit.10160 [DOI] [PubMed] [Google Scholar]
- Beers D. E., Ramirez J. E. (1990). Vectran high-performance fibre. J. Text. Inst. 81, 561–574. 10.1080/00405009008658729 [DOI] [Google Scholar]
- Belda E., van Heck R. G., José Lopez-Sanchez M., Cruveiller S., Barbe V., Fraser C., et al. (2016). The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 18, 3403–3424. 10.1111/1462-2920.13230 [DOI] [PubMed] [Google Scholar]
- Benaskar F., Hessel V., Krtschil U., Lo P. (2009). Intensification of the capillary-based Kolbe-Schmitt synthesis from resorcinol by reactive ionic liquids, microwave heating, or a combination thereof. Org. Process Res. Dev. 13, 970–982. 10.1021/op9000803 [DOI] [Google Scholar]
- Berman K. M., Cohn M. (1970). Phosphoenolpyruvate synthetase Partial reactions studied with adenosine triphosphate analogues and the inorganic phosphate-h2 18O exchange reaction. J. Biol. Chem. 245, 5319–5325. [PubMed] [Google Scholar]
- Choi K. H., Gaynor J. B., White K. G., Lopez C., Bosio C. M., Karkhoff-Schweizer R. R., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2, 443–448. 10.1038/nmeth765 [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Rymaszewski M. (2006). Alkaline polyethylene glycol-based method for direct PCR from bacteria, eukaryotic tissue samples, and whole blood. Biotechniques 40, 454–458. 10.2144/000112149 [DOI] [PubMed] [Google Scholar]
- de las Heras A., Carreño C. A., de Lorenzo V. (2008). Stable implantation of orthogonal sensor circuits in Gram-negative bacteria for environmental release. Environ. Microbiol. 10, 3305–3316. 10.1111/j.1462-2920.2008.01722.x [DOI] [PubMed] [Google Scholar]
- Dos Santos V. A., Heim S., Moore E. R., Strätz M., Timmis K. N. (2004). Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6, 1264–1286. 10.1111/j.1462-2920.2004.00734.x [DOI] [PubMed] [Google Scholar]
- Duetz W. A., Rüedi L., Hermann R., O'Connor K., Büchs J., Witholt B. (2000). Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl. Environ. Microbiol. 66, 2641–2646. 10.1128/AEM.66.6.2641-2646.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbahloul Y., Steinbüchel A. (2009). Large-scale production of poly(3-hydroxyoctanoic acid) by Pseudomonas putida GPo1 and a simplified downstream process. Appl. Environ. Microbiol. 75, 643–651. 10.1128/AEM.01869-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer T., Fischer E., Sauer U. (2005). Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187, 1581–1590. 10.1128/JB.187.5.1581-1590.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Liu C., Ding Y., Sun C., Zhang R., Xian M. (2014). Development of genetically stable Escherichia coli strains for poly (3-hydroxypropionate) production. PLoS ONE 9:e97845. 10.1371/journal.pone.0097845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. M., Hames B. D. (1985). DNA Cloning: A Practical Approach. Oxford; Washington, DC: IRL press Oxford. [Google Scholar]
- Hädicke O., Bettenbrock K., Klamt S. (2015). Enforced ATP futile cycling increases specific productivity and yield of anaerobic lactate production in Escherichia coli. Biotechnol. Bioeng. 112, 2195–2199. 10.1002/bit.25623 [DOI] [PubMed] [Google Scholar]
- Hartmans S., Smits J. P., van der Werf M. J., Volkering F., de Bont J. A. (1989). Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl. Environ. Microbiol. 55, 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S., Parales R. E. (1996). The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50, 553–590. 10.1146/annurev.micro.50.1.553 [DOI] [PubMed] [Google Scholar]
- Hernández-Arranz S., Sánchez-Hevia D., Rojo F., Moreno R. (2016). Effect of Crc and Hfq proteins on the transcription, processing, and stability of the Pseudomonas putida CrcZ sRNA. RNA 22, 1–16. 10.1261/rna.058313.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester K. L., Lehman J., Najar F., Song L., Roe B. A., Macgregor C. H., et al. (2000a). Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 182, 1144–1149. 10.1128/JB.182.4.1144-1149.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester K. L., Madhusudhan K. T., Sokatch J. R. (2000b). Catabolite repression control by Crc in 2xYT medium is mediated by posttranscriptional regulation of bkdR expression in Pseudomonas putida. J. Bacteriol. 182, 1150–1153. 10.1128/JB.182.4.1150-1153.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn M., Vorpahl C., Türkowsky D., Lindmeyer M., Bühler B., Harms H., et al. (2014). Accurate determination of plasmid copy number of flow-sorted cells using droplet digital PCR. Anal. Chem. 86, 5969–5976. 10.1021/ac501118v [DOI] [PubMed] [Google Scholar]
- Jendresen C. B., Stahlhut S. G., Li M., Gaspar P., Siedler S., Förster J., et al. (2015). Highly active and specific tyrosine ammonia-lyases from diverse origins enable enhanced production of aromatic compounds in bacteria and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 81, 4458–4476. 10.1128/AEM.00405-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I., Miñambres B., García J. L., Díaz E. (2002). Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4, 824–841. 10.1046/j.1462-2920.2002.00370.x [DOI] [PubMed] [Google Scholar]
- Juminaga D., Baidoo E. E., Redding-Johanson A. M., Batth T. S., Burd H., Mukhopadhyay A., et al. (2012). Modular engineering of L-tyrosine production in Escherichia coli. Appl. Environ. Microbiol. 78, 89–98. 10.1128/AEM.06017-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallscheuer N., Marienhagen J. (2018). Corynebacterium glutamicum as platform for the production of hydroxybenzoic acids. Microb. Cell Fact. 17, 1–13. 10.1186/s12934-018-0923-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieboom J., Dennis J. J., de Bont J. A. M., Zylstra G. J. (1998). Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 273, 85–91. 10.1074/jbc.273.1.85 [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Tsujimoto K., Kurahashi O. (1997). Mutational analysis of the feedback sites of phenylalanine-sensitive 3- deoxy-d-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. Appl. Environ. Microbiol. 63, 761–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Park H., Na D., Lee S. Y. (2014). Metabolic engineering of Escherichia coli for the production of phenol from glucose. Biotechnol. J. 9, 621–629. 10.1002/biot.201300263 [DOI] [PubMed] [Google Scholar]
- Kitade Y., Hashimoto R., Suda M., Hiraga K., Inui M. (2018). Production of 4-hydroxybenzoic acid by an aerobic growth-arrested bioprocess using metabolically engineered Corynebacterium glutamicum. Appl. Environ. Microbiol. 84:e02587-17. 10.1128/AEM.02587-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler K. A. K., Blank L. M., Frick O., Schmid A. (2015). D-Xylose assimilation via the Weimberg pathway by solvent-tolerant Pseudomonas taiwanensis VLB120. Environ. Microbiol. 17, 156–170. 10.1111/1462-2920.12537 [DOI] [PubMed] [Google Scholar]
- Kreyenschulte D., Heyman B., Eggert A., Maßmann T., Kalvelage C., Kossack R., et al. (2018). In situ reactive extraction of itaconic acid during fermentation of Aspergillus terreus. Biochem. Eng. J. 135, 133–141. 10.1016/j.bej.2018.04.014 [DOI] [Google Scholar]
- Kuepper J., Dickler J., Biggel M., Behnken S., Jäger G., Wierckx N., et al. (2015). Metabolic engineering of Pseudomonas putida KT2440 to produce anthranilate from glucose. Front. Microbiol. 6:1310. 10.3389/fmicb.2015.01310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. (1997). Biological conversion of lignocellulosic biomass to ethanol. J. Biotechnol. 56, 1–24. 10.1016/S0168-1656(97)00073-4 [DOI] [PubMed] [Google Scholar]
- Li K., Mikola M. R., Draths K. M., Worden R. M., Frost J. W. (1999). Fed-batch fermentor synthesis of 3-dehydroshikimic acid using recombinant Escherichia coli. Biotechnol. Bioeng. 64, 61–73. [DOI] [PubMed] [Google Scholar]
- Lindsey A. S., Jeskey H. (1957). The Kolbe-Schmitt reaction. Chem. Rev. 57, 583–620. 10.1021/cr50016a001 [DOI] [Google Scholar]
- Loeschcke A., Thies S. (2015). Pseudomonas putida—a versatile host for the production of natural products. Appl. Microbiol. Biotechnol. 99, 6197–6214. 10.1007/s00253-015-6745-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütke-Eversloh T., Stephanopoulos G. (2007). L-tyrosine production by deregulated strains of Escherichia coli. Appl. Microbiol. Biotechnol. 75, 103–110. 10.1007/s00253-006-0792-9 [DOI] [PubMed] [Google Scholar]
- MacGregor C. H., Arora S. K., Hager P. W., Dail M. B., Phibbs P. V. (1996). The nucleotide sequence of the Pseudomonas aeruginosa pyrE-crc-rph region and the purification of the crc gene product. J. Bacteriol. 178, 5627–5635. 10.1128/jb.178.19.5627-5635.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García E., Aparicio T., de Lorenzo V., Nikel P. I. (2014). New transposon tools tailored for metabolic engineering of Gram-negative microbial cell factories. Front. Bioeng. Biotechnol. 2, 1–13. 10.3389/fbioe.2014.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García E., de Lorenzo V. (2011). Engineering multiple genomic deletions in Gram-negative bacteria: Analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 13, 2702–2716. 10.1111/j.1462-2920.2011.02538.x [DOI] [PubMed] [Google Scholar]
- Meijnen J. P., de Winde J. H., Ruijssenaars H. J. (2008). Engineering Pseudomonas putida S12 for efficient utilization of D-xylose and L-arabinose. Appl. Environ. Microbiol. 74, 5031–5037. 10.1128/AEM.00924-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijnen J. P., Verhoef S., Briedjlal A. A., de Winde J. H., Ruijssenaars H. J. (2011). Improved p-hydroxybenzoate production by engineered Pseudomonas putida S12 by using a mixed-substrate feeding strategy. Appl. Microbiol. Biotechnol. 90, 885–893. 10.1007/s00253-011-3089-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menczel J. D., Collins G. L., Saw S. K. (1997). Thermal analysis of Vectran fibers and films. J. Therm. Anal. 49, 201–208. 10.1007/BF01987440 [DOI] [Google Scholar]
- Mi J., Sydow A., Schempp F., Becher D., Schewe H., Schrader J., et al. (2016). Investigation of plasmid-induced growth defect in Pseudomonas putida. J. Biotechnol. 231, 167–173. 10.1016/j.jbiotec.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Molina-Santiago C., Cordero B. F., Daddaoua A., Udaondo Z., Manzano J., Valdivia M., et al. (2016). Pseudomonas putida as a platform for the synthesis of aromatic compounds. Biotechnol. Bioeng. 162, 1535–1543. 10.1099/mic.0.000333 [DOI] [PubMed] [Google Scholar]
- Morales G., Linares J. F., Beloso A., Albar J. P., Martínez J. L., Rojo F. (2004). The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 186, 1337–1344. 10.1128/JB.186.5.1337-1344.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R., Hernández-Arranz S., La Rosa R., Yuste L., Madhushani A., Shingler V., et al. (2015). The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ. Microbiol. 17, 105–118. 10.1111/1462-2920.12499 [DOI] [PubMed] [Google Scholar]
- Moreno R., Martínez-Gomariz M., Yuste L., Gil C., Rojo F. (2009a). The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics 9, 2910–2928. 10.1002/pmic.200800918 [DOI] [PubMed] [Google Scholar]
- Moreno R., Marzi S., Romby P., Rojo F. (2009b). The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic Acids Res. 37, 7678–7690. 10.1093/nar/gkp825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Wagener A., Schmidt K., Leistner E. (1995). Microbial production of specifically ring-13C-labelled 4-hydroxybenzoic acid. Appl. Microbiol. Biotechnol. 43, 985–988. 10.1007/BF00166913 [DOI] [PubMed] [Google Scholar]
- Murarka A., Dharmadi Y., Yazdani S. S., Gonzalez R. (2008). Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl. Environ. Microbiol. 74, 1124–1135. 10.1128/AEM.02192-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijkamp K., Westerhof R. G., Ballerstedt H., de Bont J. A., Wery J. (2007). Optimization of the solvent-tolerant Pseudomonas putida S12 as host for the production of p-coumarate from glucose. Appl. Microbiol. Biotechnol. 74, 617–624. 10.1007/s00253-006-0703-0 [DOI] [PubMed] [Google Scholar]
- Nikel P. I., Chavarría M., Fuhrer T., Sauer U., de Lorenzo V. (2015). Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 290, 25920–25932. 10.1074/jbc.M115.687749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel P. I., de Lorenzo V. (2013). Implantation of unmarked regulatory and metabolic modules in Gram-negative bacteria with specialised mini-transposon delivery vectors. J. Biotechnol. 163, 143–154. 10.1016/j.jbiotec.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Nikel P. I., de Lorenzo V. (2018). Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. Metab. Eng. 50, 142–155. 10.1016/j.ymben.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Nikel P. I., Kim J., de Lorenzo V. (2014a). Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT2440. Environ. Microbiol. 16, 239–254. 10.1111/1462-2920.12224 [DOI] [PubMed] [Google Scholar]
- Nikel P. I., Martínez-García E., de Lorenzo V. (2014b). Biotechnological domestication of Pseudomonads using synthetic biology. Nat. Rev. Microbiol. 12, 368–379. 10.1038/nrmicro3253 [DOI] [PubMed] [Google Scholar]
- Panke S., Witholt B., Schmid A., Wubbolts M. G. (1998). Towards a biocatalyst for (S)-styrene oxide production: characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl. Environ. Microbiol 64, 2032–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek J., Roman J., Moran G. R., Christendat D. (2017). Structurally diverse dehydroshikimate dehydratase variants participate in microbial quinate catabolism. Mol. Microbiol. 103, 39–54. 10.1111/mmi.13542 [DOI] [PubMed] [Google Scholar]
- Puigbò P., Guzmán E., Romeu A., Garcia-Vallvé S. (2007). OPTIMIZER: A web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 35, 126–131. 10.1093/nar/gkm219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A. H., Eberhard S. J., Dingle S. L., Mcdowell T. D. (1970). Distribution of the phosphoenolpyruvate: glucose phosphotransferase system in bacteria. J. Bacteriol. 104, 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Manzano A., Yuste L., Rojo F. (2005). Levels and activity of the Pseudomonas putida global regulatory protein Crc vary according to growth conditions. J. Bacteriol. 187, 3678–3686. 10.1128/JB.187.11.3678-3686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. New York, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schmitz S., Nies S., Wierckx N., Blank L. M., Rosenbaum M. A. (2015). Engineering mediator-based electroactivity in the obligate aerobic bacterium Pseudomonas putida KT2440. Front. Microbiol. 6:284. 10.3389/fmicb.2015.00284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schügerl K., Hubbuch J. (2005). Integrated bioprocesses. Curr. Opin. Microbiol. 8, 294–300. 10.1016/j.mib.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R., Martínez-García E., Calles B., Chavarría M., Arce-Rodríguez A., de las Heras A., et al. (2013). The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–D675. 10.1093/nar/gks1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Huebschmann S., Sellin M., Kralisch D., Trotzki R., Ondruschka B. (2009). Microwave-assisted Kolbe-Schmitt synthesis using ionic liquids or dimcarb as reactive solvents. Chem. Eng. Technol. 32, 1730–1738. 10.1002/ceat.200900331 [DOI] [Google Scholar]
- Sun Z., Ramsay J. A., Guay M., Ramsay B. A. (2006). Automated feeding strategies for high-cell-density fed-batch cultivation of Pseudomonas putida KT2440. Appl. Microbiol. Biotechnol. 71, 423–431. 10.1007/s00253-005-0191-7 [DOI] [PubMed] [Google Scholar]
- Syukur Purwanto H., Kang M.-S., Ferrer L., Han S.-S., Lee J.-Y., Kim H.-S., et al. (2018). Rational engineering of the shikimate and related pathways in Corynebacterium glutamicum for 4-hydroxybenzoate production. J. Biotechnol. 282, 92–100. 10.1016/j.jbiotec.2018.07.016 [DOI] [PubMed] [Google Scholar]
- Tiso T., Sabelhaus P., Behrens B., Wittgens A., Rosenau F., Hayen H., et al. (2016). Creating metabolic demand as an engineering strategy in Pseudomonas putida – rhamnolipid synthesis as an example. Metab. Eng. Commun. 3, 234–244. 10.1016/j.meteno.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus J., Roelands C. P. M., Verdoes D., Jansens P. J., ter Horst J. H. (2010). Co-crystallization as a separation technology: controlling product concentrations by co-crystals. Cryst. Growth Des. 10, 1171–1179. 10.1021/cg9010778 [DOI] [Google Scholar]
- Verhoef S., Ballerstedt H., Volkers R. J. M., de Winde J. H., Ruijssenaars H. J. (2010). Comparative transcriptomics and proteomics of p-hydroxybenzoate producing Pseudomonas putida S12: novel responses and implications for strain improvement. Appl. Microbiol. Biotechnol. 87, 679–690. 10.1007/s00253-010-2626-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef S., Ruijssenaars H. J., de Bont J. A., Wery J. (2007). Bioproduction of p-hydroxybenzoate from renewable feedstock by solvent-tolerant Pseudomonas putida S12. J. Biotechnol. 132, 49–56. 10.1016/j.jbiotec.2007.08.031 [DOI] [PubMed] [Google Scholar]
- Verhoef S., Wierckx N., Westerhof R. G., de Winde J. H., Ruijssenaars H. J. (2009). Bioproduction of p-hydroxystyrene from glucose by the solvent-tolerant bacterium Pseudomonas putida S12 in a two-phase water-decanol fermentation. Appl. Environ. Microbiol. 75, 931–936. 10.1128/AEM.02186-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimberg R. (1961). Pentose oxidation by Pseudomonas fragi. J. Biol. Chem 236, 629–635. [PubMed] [Google Scholar]
- West T. P. (2013). Citric acid production by Candida species grown on a soy-based crude glycerol. Prep. Biochem. Biotechnol. 43, 601–611. 10.1080/10826068.2012.762929 [DOI] [PubMed] [Google Scholar]
- Wierckx N., Prieto M. A., Pomposiello P., de Lorenzo V., O'Connor K., Blank L. M. (2015). Plastic waste as a novel substrate for industrial biotechnology. Microb. Biotechnol. 8, 900–903. 10.1111/1751-7915.12312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierckx N., Ruijssenaars H. J., de Winde J. H., Schmid A., Blank L. M. (2009). Metabolic flux analysis of a phenol producing mutant of Pseudomonas putida S12: verification and complementation of hypotheses derived from transcriptomics. J. Biotechnol. 143, 124–129. 10.1016/j.jbiotec.2009.06.023 [DOI] [PubMed] [Google Scholar]
- Wierckx N. J., Ballerstedt H., de Bont J. A., de Winde J. H., Ruijssenaars H. J., Wery J. (2008). Transcriptome analysis of a phenol-producing Pseudomonas putida S12 construct: genetic and physiological basis for improved production. J. Bacteriol. 190, 2822–2830. 10.1128/JB.01379-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierckx N. J., Ballerstedt H., de Bont J. A., Wery J. (2005). Engineering of solvent-tolerant Pseudomonas putida S12 for bioproduction of phenol from glucose. Appl. Environ. Microbiol. 71, 8221–8227. 10.1128/AEM.71.12.8221-8227.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G., Averesch N. J., Nunez-Bernal D., Krömer J. O. (2014). In vivo instability of chorismate causes substrate loss during fermentative production of aromatics. Yeast 31, 333–341. 10.1002/yea.3025 [DOI] [PubMed] [Google Scholar]
- Wolff J. A., MacGregor C. H., Eisenberg R. C., Phibbs P. V. (1991). Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J. Bacteriol. 173, 4700–4706. 10.1128/jb.173.15.4700-4706.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynands B., Lenzen C., Otto M., Koch F., Blank L. M., Wierckx N. (2018). Metabolic engineering of Pseudomonas taiwanensis VLB120 with minimal genomic modifications for high-yield phenol production. Metab. Eng. 47, 121–133. 10.1016/j.ymben.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Xue Z., McCluskey M., Cantera K., Sariaslani F. S., Huang L. (2007). Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from the photosynthetic bacterium Rhodobacter sphaeroides. J. Ind. Microbiol. Biotechnol. 34, 599–604. 10.1007/s10295-007-0229-1 [DOI] [PubMed] [Google Scholar]
- Yang F., Hanna M. A., Sun R. (2012). Value-added uses for crude glycerol–a byproduct of biodiesel production. Biotechnol. Biofuels 5, 1–10. 10.1186/1754-6834-5-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., Li K., Draths K. M., Frost J. W. (2002). Modulation of phosphoenolpyruvate synthase expression increases shikimate pathway product yields in E. coli. Biotechnol. Prog. 18, 1141–1148. 10.1021/bp020101w [DOI] [PubMed] [Google Scholar]
- Yu S., Plan M. R., Winter G., Krömer J. O. (2016). Metabolic engineering of Pseudomonas putida KT2440 for the production of para-hydroxybenzoic acid. Front. Bioeng. Biotechnol. 4:90 10.3389/fbioe.2016.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambanini T., Hosseinpour Tehrani H., Geiser E., Sonntag C. K., Buescher J. M., Meurer G., et al. (2017). Metabolic engineering of Ustilago trichophora TZ1 for improved malic acid production. Metab. Eng. Commun. 4, 12–21. 10.1016/j.meteno.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambanini T., Kleineberg W., Sarikaya E., Buescher J. M., Meurer G., Wierckx N., et al. (2016). Enhanced malic acid production from glycerol with high-cell density Ustilago trichophora TZ1 cultivations. Biotechnol. Biofuels 9:135. 10.1186/s13068-016-0553-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Liao Y., Zhang Z. (2007). Toxicity of ionic liquids. Clean 35, 42–48. 10.1002/clen.200600015 [DOI] [Google Scholar]
- Zobel S., Benedetti I., Eisenbach L., de Lorenzo V., Wierckx N., Blank L. M. (2015). Tn7-based device for calibrated heterologous gene expression in Pseudomonas putida. ACS Synth. Biol. 4, 1341–1351. 10.1021/acssynbio.5b00058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.