Abstract
The σ70 family alternative σI factors and their cognate anti-σI factors are widespread in Clostridia and Bacilli and play a role in heat stress response, virulence, and polysaccharide sensing. Multiple σI/anti-σI factors exist in some lignocellulolytic clostridial species, specifically for regulation of components of a multienzyme complex, termed the cellulosome. The σI and anti-σI factors are unique, because the C-terminal domain of σI (SigIC) and the N-terminal inhibitory domain of anti-σI (RsgIN) lack homology to known proteins. Here, we report structure and interaction studies of a pair of σI and anti-σI factors, SigI1 and RsgI1, from the cellulosome-producing bacterium, Clostridium thermocellum. In contrast to other known anti-σ factors that have N-terminal helical structures, RsgIN has a β-barrel structure. Unlike other anti-σ factors that bind both σ2 and σ4 domains of the σ factors, RsgIN binds SigIC specifically. Structural analysis showed that SigIC contains a positively charged surface region that recognizes the promoter –35 region, and the synergistic interactions among multiple interfacial residues result in the specificity displayed by different σI/anti-σI pairs. We suggest that the σI/anti-σI factors represent a distinctive mode of σ/anti-σ complex formation, which provides the structural basis for understanding the molecular mechanism of the intricate σI/anti-σI system.
INTRODUCTION
Bacterial σ factors are key components of RNA polymerases (RNAPs) responsible for gene transcription. The bacterial RNAP holoenzyme includes a core enzyme consisting of five subunits (ααββ′ω) to bind the template DNA and catalyze RNA synthesis, and a dissociable σ subunit from a large number of σ factors to specifically recognize gene promoters (1). Housekeeping σ factors that are homologues of Escherichia coli σ70 are responsible for the majority of transcription in exponentially growing cells, while various alternative σ factors control specialized regulons that are activated by specific sources of stress, growth transitions, and morphological changes (2). Except for a distinct σ54 family in some species, most alternative σ factors belong to the σ70 family and have diverse sequences and functions (2). σ70 factors are classified into four groups according to sequence and structural homology, including the primary housekeeping σ factors (Group 1) and three alternative σ factors (Groups 2–4) (3). σ factors of Group 4 are also called extracytoplasmic function (ECF) σ factors, because most of them work with a co-transcribed trans-membrane anti-σ factor which senses the stimulation of external agents, although members of a small subset of ECF σ factors are linked to soluble cytoplasmic anti-σ factors (2,4). ECF σ factors are significantly divergent in sequence, with relatively large numbers in some organisms. The trans-membrane anti-σ factors generally contain an extracytoplasmic sensory domain, a transmembrane helix, and an intracellular inhibitory domain to specifically bind corresponding σ factors (4–7). Recent studies have elucidated the structural and regulatory mechanism of some ECF σ factors, and their large diversity may provide novel regulation strategies and constitute very promising tools for applied synthetic biology (8). Indeed, ECF σ factors and their promoters have been successfully applied in the design of orthogonal regulators for synthetic biology (9,10).
The alternative σI and anti-σI (i.e. SigI and RsgI) were first discovered in Bacillus subtilis as related to the heat-shock response (11) and they are found widely in Bacilli and Clostridia of Firmicutes (12). Multiple paralogous σI and anti-σI pairs have been discovered in many lignocellulolytic bacteria to regulate the components of secreted multi-enzyme complexes, termed cellulosomes, by sensing the status of environmental polysaccharides (13,14). Cellulosomes are assembled by specific modular interactions (cohesins and dockerins) between a scaffolding protein and the enzymes, and are considered the most efficient nano-machines for lignocellulose degradation in nature through the synergistic effects of their various component enzymes (15,16). A number of omics studies have revealed that the expression of cellulosomal enzymes is regulated by the type of extracellular polysaccharide substrate (17–21). The σI and anti-σI factors were found to play crucial roles in cellulosome regulation, and genomic studies have revealed that several cellulosome-producing bacteria contain 8–16 pairs of σI and anti-σI factors (13,14). Previous studies of σI and anti-σI factors in Clostridium (Ruminiclostridium) thermocellum, an anaerobic thermophilic lignocellulolytic bacterium that produces complex cellulosomes, have shown that anti-σIs contain a C-terminal module that functions as a polysaccharide-binding component for sensing different types of lignocellulosic substrates in the extracellular environment. The anti-σIs also bear a conserved cytoplasmic region responsible for binding the respective σI. Upon selective binding of the extracellular polysaccharide, the σI factor is then released from the anti-σI to activate the transcription of corresponding cellulosomal genes (22–24). It has been shown that the recognition between individual σI and anti-σI pairs is specific among the multiplicity of homologous σI and anti-σI factors, which raises the question of how such specificity is achieved (22).
The σIs were classified as Group 3 sigma factors based on phylogenetic analysis, but they are distant from other Group 3 members (2). Further sequence analysis determined that the σIs exhibit many features consistent with ECF σ factors (i.e. Group 4 sigma factors) but show distinct structural features (25). ECF σ factors generally have σ70-homologous σ2 and σ4 domains for recognition of promoter –10 and –35 regions, respectively. However, σIs have only the σ2 domain, and the C-terminal domains (SigIC) of σIs have no sequence homology to other known proteins. The anti-σIs share homologous N-terminal regions, including an N-terminal cytoplasmic domain (RsgIN), a transmembrane helix, and a periplasmic domain (RsgIP), whereas neither RsgIN nor RsgIP has sequence homology to any other known proteins. Therefore, σI and anti-σI represent a novel family of alternative σ factors, and determination of their structures is required to understand their functional mechanism. Here, we employed NMR spectroscopy, structural analysis, mutagenesis, and interaction analyses to investigate the structure and functional mechanism of these unique types of σ and anti-σ factors.
MATERIALS AND METHODS
Plasmid construction
The gene fragments encoding intracellular N-terminal domains of RsgIs, full-length SigIs, and domains of SigIs were amplified by PCR from C. thermocellum ATCC 27405 genomic DNA using relevant primers (Supplementary Table S1). The purified PCR products were ligated into the pET30a, the pET28a, or the pET28a-SMT3 (26) for different purposes. The constructs using the pET30a vector were used to express SigI1 N-terminal domain (SigI1N) and SigI1C containing a C-terminal His6-tag. The constructs using the pET28a were used to express SigI2C containing a C-terminal His6-tag. The constructs using the pET28a-SMT3 were used to express proteins containing an N-terminal His6-SMT3 tag, including RsgI1N, RsgI2N, and full-length SigI1, whereas the His6-SMT3 tag can be removed by the ULP1 protease treatment when needed. The mutants of RsgI1N and SigI1C were constructed by the QuikChange method using designed primers and appropriate templates (Supplementary Table S2).
Recombinant protein expression and purification
The recombinant plasmid pET28a-SMT3-RsgI1N and derived mutants were transformed into Escherichia coli Rosseta (DE3), and the other expression vectors were transformed into E. coli BL21 (DE3) for protein expression. The bacterial cells were cultured at 37°C, and when the absorbance at 600 nm reached ∼0.8, target protein expression was induced for ∼18 h with 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG). Cells were collected by centrifugation at 6000 rpm for 15 min.
All the cell pellets were resuspended in binding buffers of 20 mM Tris, 500 mM NaCl, 30 mM imidazole at pH 8.0 and lysed by high-pressure homogenization (for SigI1C) or ultrasonication (for other SigI or RsgI proteins). All the proteins were first purified by the Ni-chelating affinity chromatography using a Histrap column with the elution buffer containing 20 mM Tris, 500 mM NaCl, 500 mM imidazole at pH 8.0. The second step for the protein purification was optimized by considering the different properties for various target proteins. The second step for the proteins RsgI1N and RsgI2N was a ULP1 protease treatment, and the proteins were then passed through a Histrap column to remove the cleaved SMT3 tag. The target proteins were further purified using ion exchange chromatography with a HiTrap SP-FF column (for RsgI1N) or a HiTrap Q-FF column (for RsgI2N). The binding buffers were 20 mM Bis–Tris at pH 7.0 for RsgI1N and 20 mM Tris–HCl at pH 7.5 for RsgI2N. The proteins were eluted by adding 1 M NaCl into the corresponding binding buffers. The second purification step of the proteins SigI1C, SigI1N and SigI2C was gel filtration with a HiLoad 16/600 Superdex 75 column with buffers of 20 mM Tris, 150 mM NaCl at pH 8.0 (for SigI1C) or 7.5 (for SigI2C) and 20 mM Bis–Tris, 150 mM NaCl at pH 6.8 (for SigI1N,). The complex of SigI1C–RsgI1N was obtained by mixing the purified RsgI1N and SigI1C and was further purified by gel filtration with a HiLoad 16/600 Superdex 75 column with buffers of 20 mM Tris, 150 mM NaCl at pH 8.0. The purification procedures of RsgI1N and SigI1C mutants were the same as those of the wild-type proteins. During purification, all protein solutions and buffers were kept on ice. The final purity of proteins was detected by SDS-PAGE. The samples were exchanged by dialysis or ultrafiltration with appropriate buffers for subsequent NMR and SPR experiments.
The 15N- and 13C-labeled proteins for NMR experiments were obtained by cell cultivation using M9 minimal medium containing 15N-NH4Cl and 13C-glucose as sole nitrogen and carbon sources, respectively. The labeled proteins were purified using the same procedures as the unlabeled proteins.
NMR spectroscopy and structural calculations
Protein samples for NMR experiments were dissolved in various optimized buffers containing 90% H2O, 10% D2O and 0.02% (w/v) sodium 2,2-dimethylsilapentane-5-sulfonate (DSS). The buffer for RsgI1N was 20 mM Bis–Tris, 50 mM NaCl, 2 mM EDTA at pH 6.2; the buffer for the RsgI1N–SigI1C complex was 20 mM Bis–Tris, 50 mM NaCl, 2 mM EDTA at pH 6.5. All NMR experiments were performed at 298 K on a Bruker Avance III 600 MHz NMR spectrometer using a z-gradient triple resonance cryoprobe, except that the NOESY spectra of the RsgI1N–SigI1C complex were acquired on a Bruker Avance III 850 MHz NMR spectrometer. NMR data for chemical shift assignments include 2D 1H–15N HSQC, 2D 1H–13C HSQC, 3D 1H–13C–15N HNCACB, CBCA(CO)NH, HNCA, HNCO, HN(CA)CO, HBHA(CBCA)(CO)NH, HBHA(CBCA)NH, H(C)CH-TOCSY, (H)CCH-TOCSY, H(C)CH-COSY and (H)CCH-COSY. The NOESY spectra for distance restraints of structure calculation include 1H–15N NOESY-HSQC and 1H–13C NOESY-HSQC. The mixing time for NOESY experiments was 200 ms for RsgI1N and 120 ms for the RsgI1N–SigI1C complex. All the spectra were processed using NMRPipe (27) and analyzed using NMRViewJ (28). The backbone chemical shift assignments were obtained using the program MARS (29) with manual verification. The side chain assignments were obtained manually in NMRViewJ (28).
The initial structures were calculated using the program CYANA (30) and refined using SANE (31) and CNS (32) with explicit water refinement protocol implemented in RECOORDScripts (33). The dihedral angle restraints of backbone φ, ψ, and side chain χ1 obtained by the program TALOS-N (34) were used in the structure calculation. The hydrogen bond restraints according to the secondary structural elements were also used in the late stage of structure refinements. The final structures were validated by PROCHECK_NMR (35) and WHAT_CHECK (36). MOLMOL (37) and PyMol (http://www.pymol.org/) were used for visual structure validation and structure figure plotting.
The search for structure homology was performed using the Dali server (38) and SSM server (39). The structural alignments were obtained using the SSM server (39).
Homology modeling
The structure models of the RsgIxN–SigIxC (x = 2–8) complexes were obtained using the program Modeller (40). The NMR structure of the RsgI1N–SigI1C complex was used as the template, and the sequence alignments were obtained by ClustalX (41).
Surface plasmon resonance (SPR) experiments
SPR experiments were performed on Biacore T100 (GE healthcare) with Series S Sensor Chip NTA (GE healthcare). The buffer containing 20 mM Tris–HCl, 50 mM NaCl, 0.05% Triton X-100 at pH 7.5 was used in the experiments. The standard Single Cycle Kinetics (SCK) (42) protocol was used in the SPR experiments and analysis. Each experiment was repeated three times.
Expression and purification of RNA polymerase from C. thermocellum
The gene of the β′ subunit of RNA polymerase (RNAP) was amplified by PCR from C. thermocellum DSM 1313 genomic DNA. The primers contained an additional coding region for adding 10 histidines at the C-terminus of the β′ subunit. The amplified DNA fragment was inserted into a pHK plasmid (43) for protein expression in C. thermocellum DSM 1313. The plasmid pHK-β′ was transformed into the C. thermocellum DSM 1313. The correct transformants were amplified into 3 L of GS-2 media for fermentation. The cells were harvested by centrifugation at 10 200 g for 30 min and lysed by ultrasonication. The RNAP core enzyme complex (ααββ′ω) was first purified by affinity chromatography with a Histrap column using the elution buffer of 20 mM Tris–HCl, 500 mM NaCl, 500 mM imidazole, at pH 8.0. The complex was further purified by heparin affinity chromatography using a HiTrap Heparin HP column. The binding buffer was 20 mM Tris–HCl, 100 mM NaCl, 5% (v/v) glycerol, pH 8.0, and the elution buffer was 20 mM Tris–HCl, 1 M NaCl, 5% (v/v) glycerol, pH 8.0. The α, β, β′ and ω subunits in the purified RNAP core enzyme complex were identified by SDS-PAGE and HPLC-Q-TOF high resolution mass spectrometry.
The holoenzyme of RNAP was prepared by adding purified SigI1 into a solution containing the core enzyme. The full length SigI1 was expressed with an His6-SMT3 tag and purified by Ni-chelating affinity chromatography. After ULP1 protease treatment, the mixture of SigI1 and His6-SMT3 was excessively added into the solution of the RNAP core enzyme. The protein mixture was then applied onto a HiLoad 26/600 Superdex 200 column with a buffer containing 20 mM Tris–HCl, 150 mM NaCl at pH 8.0. The RNAP holoenzyme (ααββ′ω-SigI1), the excess SigI1 and His6-SMT3 eluted at different elution volumes. In the competition experiment, the purified RsgI1N was added to the holoenzyme with a molar ratio of 1.3:1, and the mixture was applied onto the HiLoad 26/600 Superdex 200 column. Different components were collected and detected by SDS-PAGE.
Electrophoretic mobility shift assay
The electrophoretic mobility shift assay was performed using the native polyacrylamide gel electrophoresis. A 12% polyacrylamide gel was prepared with buffer containing 5× Tris-borate-EDTA (TBE) buffer at pH 8.3. The promoter –35 region DNAs (5′-ATCGATAATATACACAAAAA-3′ of SigI1 and 5′-TTATTGGTATCCCCCGAAAA-3′ of SigI2) were synthesized and annealed. Samples contained 500 ng promoter DNA and different molar ratios of SigI1C (or SigI1C mutants). The electrophoresis was performed using a buffer containing 0.5× TBE buffer at pH 8.3. The polyacrylamide gel was dyed with ethidium bromide and detected with ultraviolet transilluminator.
RESULTS
RsgI1N shows a β-barrel structure which is unique among all anti-sigma factors
The N-terminal domains of RsgI factors in C. thermocellum generally contain 50–60 residues and have 12–40% sequence identity to each other (Supplementary Figure S1 and Table S3). Among the nine RsgIs from C. thermocellum, RsgI1 contains a CBM3-type C-terminal domain, which was demonstrated to recognize cellulose (22). The promoter sequence recognized by the cognate SigI1 has been analyzed, and SigI1 can recognize the promoter of major cellulosomal components, including Cel48S and Cel8A (22,25). Therefore, we chose RsgI1 and SigI1 to study the structure and interaction of σI and anti-σI factors. RsgI1N showed good solubility and well-dispersed peaks in the 1H–15N HSQC NMR spectrum (Supplementary Figure S2), suggesting that it has a well-folded structure and is suitable for structure determination by NMR spectroscopy. The NMR structure of RsgI1N was finally determined to high quality as indicated by the statistics in Supplementary Table S4.
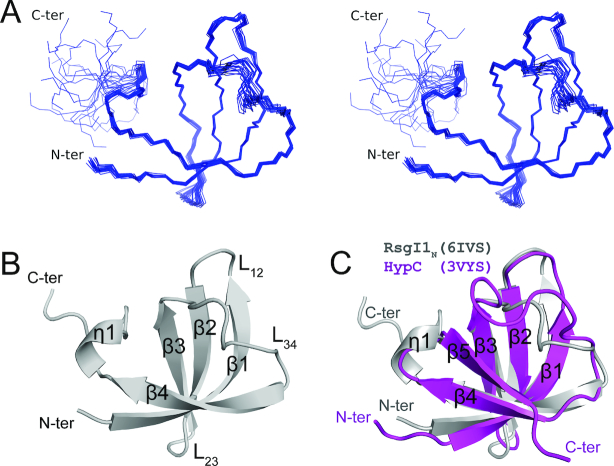
The structure of RsgI1N contains four anti-parallel β-strands and a short C-terminal 310 helix, which exhibits a β-barrel structure (Figure 1A and B). The central portion of the barrel comprises the hydrophobic core, formed by I7, I10, A15, V17, L25, I27, M33, V35, V39, F41 and I46, which are largely conserved in all RsgIs. Hydrophobic residues, including L4, I6, M13, V16, L18, F24 and I26, are exposed on the surface and are not conserved in all RsgIs. RsgI1N is the inhibitory domain of RsgI1, because RsgI1N specifically binds to SigI1 and inhibits the transcriptional activity of the RNAP-SigI1 complex (22). Interestingly, the β-barrel structure of RsgI1N is completely different from those of known anti-sigma factors, which have α-helical structures as the inhibitory domains, according to previous reports (44–46). Therefore, the RsgIs represent a unique family of anti-sigma factors.
Figure 1.
NMR structures of RsgI1N. (A) Stereo view of the backbone ensemble of 20 RsgI1N structures. (B) Ribbon representation of the RsgI1N structure. Secondary structure elements are labeled on the structure. (C) Superposition of RsgI1N (gray) and its structurally homologous protein, metallochaperone HypC (PDB 3VYS, magenta) from Thermococcus kodakarensis.
Despite the lack of sequence homology to known protein structures as determined by Blast search of the Protein Data Bank (PDB), we continued to examine potential structural homology of RsgI1N in the PDB, using the Dali and SSM servers. Both servers identified a large number of proteins with significant structural similarity. The structure with the highest Dali Z score is a metallochaperone HypC (PDB 3vyt:A, Z-score 6.1, RMSD 1.7 Å) from Thermococcus kodakarensis (47) (Figure 1C). Most of the structurally similar proteins have OB (oligonucleotide/oligosaccharide binding)-fold domains. Classical OB-fold domains consist of five β-strands that form a closed β-barrel and an extra α-helix between β3 and β4 (48,49). The OB-fold proteins also show great structural diversity with additional secondary structural elements or lack of either the β5 or the α-helix (50). RsgI1N represented a simplified OB-fold which lacks both β5 and the α-helix. OB-fold proteins have high functional diversity (49) and no OB-fold protein has previously been found among the anti-σ factors, so the structural similarity with OB-fold proteins does not tell us the functional mechanism of RsgI1N.
SigI1 binds to RsgI1N through its C-terminal domain
To gain insight into the inhibitory mechanism of RsgIN towards SigI, we first investigated the interaction of SigI1 and RsgI1N. The purification of full-length SigI1 was difficult because SigI1 is prone to precipitation. SigI contains an N-terminal domain (SigI1N), which is homologous to the σ2 domain of σ70 factors, and a putative C-terminal domain (SigI1C), which has no sequence homology to other proteins and is proposed to be functionally similar to the σ4 domain of σ70 factors. Because the structures of both the σ2 and σ4 domains of σ70 have been determined (51,52), we tried to purify SigI1N and SigI1C separately and found that each domain was more stable than the full-length protein. Although both domains are soluble and could be successfully purified, their 1H–15N HSQC NMR spectra showed poor spectral quality, indicating that they are aggregated under the conditions for solution NMR (i.e. concentrations in the μM to mM range). In NMR titration experiments the 1H–15N HSQC spectra of SigI1N and RsgI1N showed slight, gradual changes, which suggests a weak interaction between them (Supplementary Figure S3). However, the 1H–15N HSQC spectra of RsgI1N and SigI1C showed dramatic changes during the titration and no further change in the spectrum was observed when the ratio was over 1:1, indicating strong and equimolar binding of SigI1C and RsgI1N.
Furthermore, we investigated whether RsgI1N can inhibit holoenzyme formation of SigI1 and RNAP. The β′ subunit of C. thermocellum RNAP was overexpressed in C. thermocellum using a plasmid containing the β′ subunit gene with an additional C-terminal His10-tag. The RNAP was successfully purified from the recombinant C. thermocellum, and the bands of RNAP subunits on SDS-PAGE gels were verified by mass spectrometry (Supplementary Figure S4). SigI1, expressed and purified in E. coli, was then added to the RNAP, and the holoenzyme would be purified by gel filtration if SigI1 and RNAP can form a complex. SigI1 was eluted together with RNAP, thus indicating formation of the holoenzyme (Figure 2A and C). However, when RsgI1N was added into the solution containing the holoenzyme, the SigI1 band was significantly weakened in the eluted RNAP fraction and appeared in a separate fraction with RsgI1N (Figure 2B and C). These results indicate that RsgI1N inhibits SigI1 by preventing it from interacting with RNAP, and the binding affinity between RsgI1N and SigI1 is much higher than that between SigI1 and the RNAP core enzyme.
Figure 2.
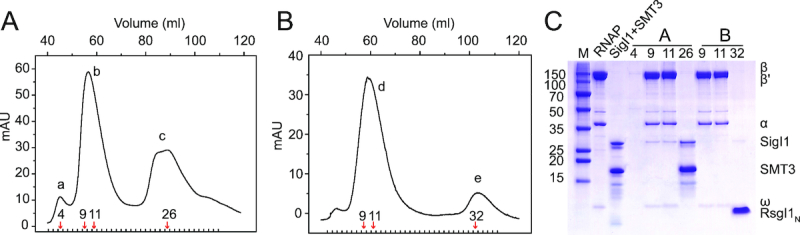
RsgI1N prevents SigI1 from binding to RNA polymerase. (A) Purification of the RNAP-SigI1 complex. The chromatography of the mixture of full-length recombinant SigI1 (containing a SMT3 tag) purified from E. coli and RNAP from C. thermocellum was performed using a Superdex200 gel filtration column. The fractions of peak b represent the RNAP-SigI1 complex. (B) Gel filtration chromatography of the RNAP–SigI1 complex (peak b in Figure A) after addition of RsgI1N. (C) SDS-PAGE of samples after the gel filtration steps. Lane M is the molecular weight marker; lane RNAP is the purified RNAP from C. thermocellum; lane SigI1 + SMT3 is the purified SigI1 and SMT3 after ULP1 protease treatment; other lanes are labeled according to the eluted fraction numbers indicated in Panels A and B by red arrows.
The structure of the SigI1C–RsgI1N complex represents a novel structural type of alternative σ/anti-σ complex
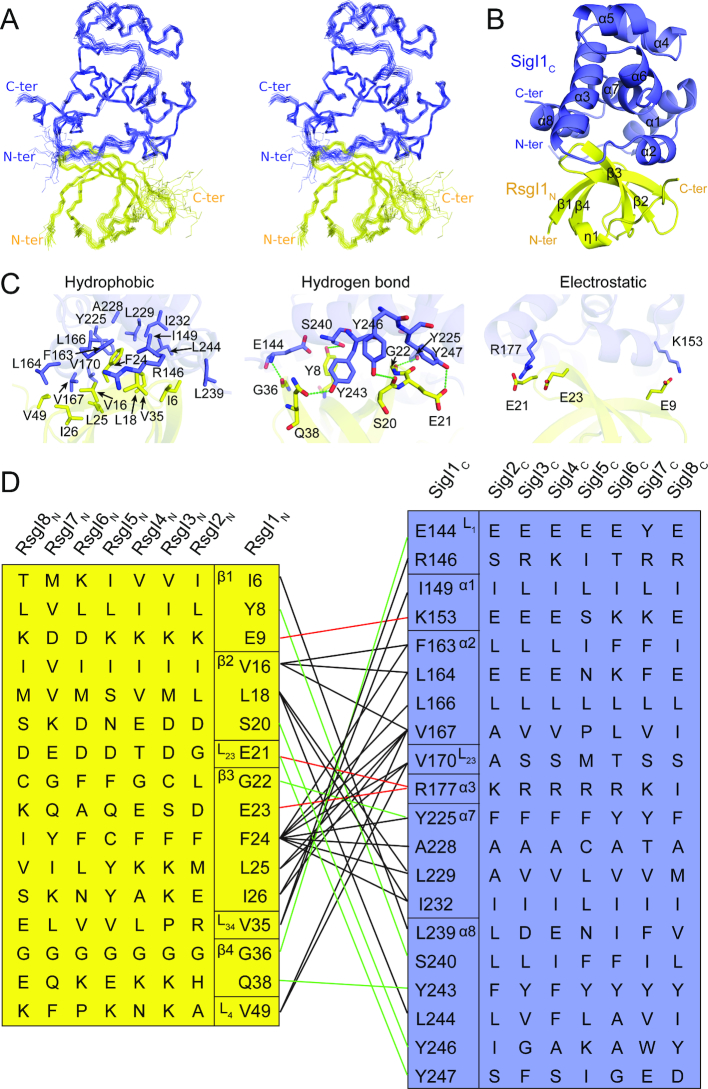
The NMR titration experiments showed the complex of SigI1C and RsgI1N has a well-dispersed 1H–15N HSQC spectrum and is suitable for NMR structure determination (Supplementary Figure S3 and S5). High-quality structures of the SigI1C–RsgI1N complex were determined using NMR (Figure 3A), and the final structural statistics of the structures are shown in Supplementary Table S4. In the structure of the SigI1C–RsgI1N complex, RsgI1N forms a simplified OB-fold structure almost identical to the structure of the free RsgI1N. A slight difference in the C-terminal region including the 310 helix and the flexible tail of RsgI1N was observed, which is likely caused by the hydrophobic interaction between V49 of RsgI1N and V167-L168 of SigI1C (Supplementary Figure S6). SigI1C is composed of eight α-helices, and the interacting surface includes the outer surfaces of all four β-strands of RsgI1N and helices α1, α2, α3, α7 and α8 of SigI1C, with 1132 ± 56 Å2 buried surface area (Figure 3B). These α-helices of SigI1C are stacked mainly involving hydrophobic interactions, while the packing between SigI1C and RsgI1N involves hydrophobic, hydrogen-bonding, and electrostatic interactions (Figure 3C and D).
Figure 3.
The structure of the SigI1C–RsgI1N complex and the interaction between SigI1C and RsgI1N. (A) Stereo view of the backbone ensemble of 20 SigI1C-RsgI1N complex structures. SigI1C is colored in blue and RsgI1N is in yellow. (B) Ribbon representation of the overall SigI1C–RsgI1N complex. (C) The interaction between SigI1C and RsgI1N in the structure. Key interaction residues from SigI1C (blue) and RsgI1N (yellow) are shown as sticks and are labeled. (D) The interaction network in the SigI1C–RsgI1N complex. Residues at the corresponding positions in other SigIs and RsgIs are also shown. The black, red and green lines represent the hydrophobic, electrostatic, and hydrogen-bonding interactions, respectively.
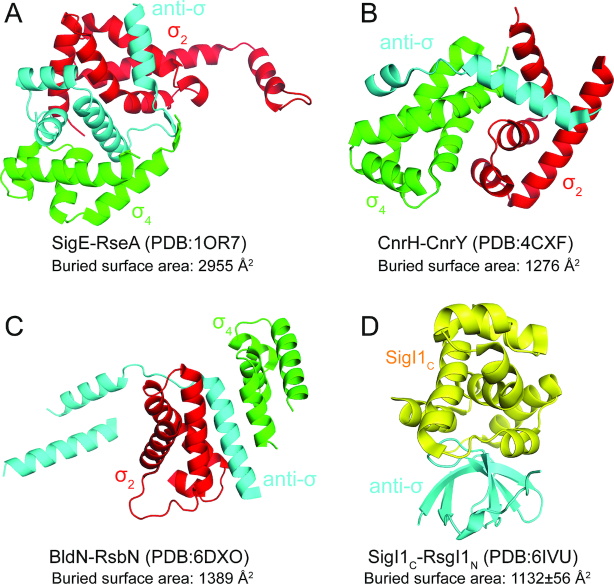
The structure of SigI1C is clearly distinct from the σ4 domain of other σ70 factors (Figure 4). The structure of SigI1C resembles a compact globular protein, whereas the σ4 domain is more extended and composed of four helices which form two helix-turn-helix (HTH) motifs to bind the -35 region of the promoter DNA (44,52). Previously known anti-σ factors bind both the σ2 and the σ4 domains of ECF sigma factors by forming either a σ2/anti-σ/σ4 sandwich structure or an anti-σ factor-embracing a compact σ2/σ4 structure (Figure 4A–C) (8,46,53). In contrast, the binding interface of SigI1C and RsgI1N is located at one side of the globular SigI1C molecule with a large buried surface area (Figure 4D). Therefore, the structure of the SigI1C-RsgI1N complex represents a novel structural type of alternative σ/anti-σ complex, completely distinct, according to the structural characteristics, from the three known classes of these complexes (3,46,53).
Figure 4.
The structure of the SigI1C-RsgI1N complex is distinct from known structures of ECF anti-σ factors. SigE-RseA, CnrH-CnrY and BldN-RsbN are the σ/anti-σ factors from Mycobacterium tuberculosis, Rhodobacter sphaeroides and Streptomyces venezuelae, respectively. The σ2 domains, σ4 domains, anti-σ factors, and SigIC are in red, green, cyan and yellow, respectively.
Structure analysis reveals the promoter binding region on SigI1C
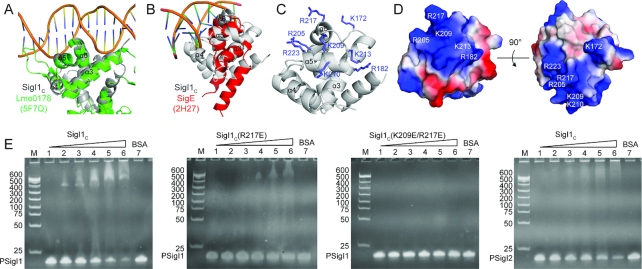
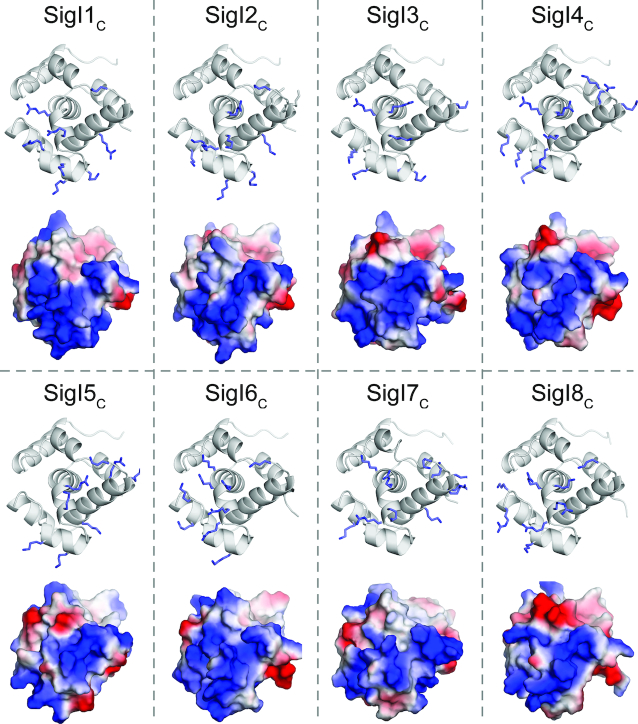
Because SigI1C has no sequence homology to other proteins in the PDB, we used the Dali and SSM servers to identify proteins with structural similarity. Both servers identified many nucleic acid binding proteins, some of which are transcriptional regulators containing helix-turn-helix (HTH) motifs. For example, the protein Lmo0178 (PDB 5F7Q, Dali Z score 4.2, RMSD 3.0 Å) is a transcriptional repressor, which recognizes the operator of its operon by binding to the major and minor groove of DNA using its HTH domain and an additional loop, respectively (54). It is known that the basic core HTH domain generally contains three-helix bundles which recognize the major groove of the target DNA region (55,56). Interestingly, the α-helices (α3, α5, and α6) of SigI1C shared high homology with the HTH domain of Lmo0178 (Figure 5A), thus suggesting that SigI1C may play a potential role in DNA binding. Analysis of the corresponding α-helices of SigI1C revealed that many positively charged residues are distributed on these three helices (Figure 5C and D). However, sequence alignment of the eight SigICs in C. thermocellum indicates that most of these residues are not conserved (Supplementary Figure S1). Nevertheless, on every modelled SigIC structure, these regions contain basic residues at different positions which allow these regions to form positively charged surfaces (Figure 6). Therefore, we speculate that the three helices of SigIC contain the putative DNA binding region for specific recognition of the promoter -35 regions of the target genes.
Figure 5.
Structural comparison of SigI1C with transcriptional factors and σ4 domain reveals the promoter binding site of SigI1C. (A) Structural superposition of SigI1C (gray) and a transcription repressor–DNA complex (PDB 5F7Q, green) from Listeria monocytogenes. (B) Structural superposition of SigI1C (gray) and SigE (PDB 2H27, red) in complex with the –35 region promoter DNA from E. coli. (C) Positively charged residues on the proposed DNA binding sites of SigI1C. (D) Electrostatic surface of SigI1C. The positively and negatively charged surfaces are colored in blue and red, respectively. The residues involved in the proposed promoter-binding region are labeled. (E) EMSAs of SigI1C and -35 region promoter DNA. The promoter from SigI1 (PSigI1) was used in the assays of SigI1C and its single (R217E) and double (K209E/R217E) mutants (first three panels). The promoter from SigI2 (PSigI2) was also used in an assay of SigI1C (last panel). Lanes 1–6 are the samples with the DNA:protein molar ratio of 1:0, 1:0.5, 1:1, 1:2, 1:4, 1:8. Lane 7 is a sample containing the promoter DNA and BSA with molar ratio of 1:8.
Figure 6.
Electrostatic surfaces of SigIC structures. The structures of SigI2C-SigI8C were obtained by homology modeling using the SigI1C structure as the template. Positively charged residues on the proposed DNA binding sites are shown as sticks. Electrostatic surfaces are shown in blue and red for positively and negatively charged surfaces, respectively.
Previous studies have revealed that the σ70 family sigma factors have a σ4 domain which also belongs to the HTH group of structures and the σ4 domains recognize the major groove of DNA with several similar conserved sites (57,58). We tried to align the structure of SigI1C with a known σ4 domain structure (PDB 2H27) (57) using the SSM server, and found they could also be aligned at the DNA-binding region of the σ4 domain with low scores (SSM Q-score 0.0698, RMSD 2.975) (Figure 5B). Helices α3, α5, and α6 of SigI1C are indeed similar to the helices of σE in the major groove of the DNA, but loops L23 and L45 of SigI1C would clash with the DNA in this binding mode. Therefore, if this region is the –35 promotor-binding region of SigI, it must either have a different binding mode or it undergoes additional conformational changes when it binds RNAP to form the active holoenzyme.
To further validate the proposed promoter binding region of SigI1C, electrophoretic mobility shift assays (EMSAs) were performed to detect the binding of SigI1C and its mutants with the SigI1 promoter –35 region DNA (Figure 5E). The results show that the wild-type SigI1C can bind well to the promoter DNA, while the single mutant on the proposed binding region significantly weakens the binding and the double mutation can abolish the binding completely. Therefore, the positively charged region on SigI1C is responsible for the promoter binding. The region is on the opposite side of the RsgI-binding surface, which is consistent with the proposal that RsgI inhibits the activity of SigI by blocking RNAP binding instead of promoter binding.
Mutagenesis analysis reveals the structural basis for the specific σI/anti-σI interactions
The structure of the SigI1C–RsgI1N complex indicates that the large interfacial surfaces and consequent interactions are involved in the formation of the complex. We constructed several RsgI1N variants with mutated interfacial residues to examine the importance of these residues in the interaction. The correct folding of RsgI1N mutants was confirmed by NMR experiments (Supplementary Figure S7). Surface plasmon resonance (SPR) experiments were performed to check the affinity of SigI1C and RsgI1N mutants (Table 1). Wild-type RsgI1N showed very strong binding to SigI1C, the equilibrium dissociation constants for which reached values of 10−11 M. The results showed that most of the single mutations of interfacial residues weakened the interaction between the two proteins, such as E9G, Y8L and V16K, which reduced the electrostatic, hydrogen bonding, and hydrophobic interactions, respectively. However, none of these mutations fully abolished complex formation. When all of the negatively charged residues (E9, E21 and E23) were mutated simultaneously to positively charged lysine, the interaction decreased dramatically and binding was not detectable by SPR. Therefore, multiple interactions contribute synergistically to the strong SigI–RsgI interaction. Additionally, the configuration of hydrophobic residues is also important since the mutants V16I, L18I and F24I showed weakened interactions.
Table 1.
Equilibrium dissociation constants for interaction between SigI1C and wild-type (WT) or mutants of RsgI1N
| RsgI1N | K D (M) |
|---|---|
| WT | 1.1 ± 0.4 × 10–11 |
| Y8L | 8.4 ± 0.9 × 10–11 |
| Y8I | 4.5 ± 0.5 × 10–10 |
| E9K | 9.7 ± 2.7 × 10–11 |
| E9G | 7.1 ± 1.4 × 10–11 |
| V16I | 3.6 ± 0.7 × 10–10 |
| V16K | 2.4 ± 0.9 × 10–9 |
| L18I | 6.7 ± 0.2 × 10–10 |
| L18F | 3.1 ± 0.9 × 10–12 |
| E21K | 1.6 ± 0.1 × 10–11 |
| E23K | 3.6 ± 1.0 × 10–10 |
| F24I | 1.9 ± 0.5 × 10–10 |
| V35K | 6.5 ± 1.6 × 10–12 |
| V35I | 2.3 ± 1.0 × 10–11 |
| Y8L-E9K | 1.6 ± 0.6 × 10–10 |
| Y8L-V35R | 4.0 ± 0.7 × 10–11 |
| E9K-E21K | 2.0 ± 0.3 × 10–11 |
| E9K-E23K | 6.4 ± 0.9 × 10-9 |
| Y8L-E9K-V35R | 9.3 ± 0.8 × 10-9 |
| E9K-E21K-E23K | Not detected |
Although different SigI–RsgI pairs share significant homology (Supplementary Table S3), the structure of the SigI1C–RsgI1N complex and the sequence alignments indicate that the interfacial residues are not well conserved (Figure 3D, Supplementary Figure S1). At least eight residues are different among the 16 and 20 interfacial residues of RsgI and SigI, respectively, and only G36 of RsgI1 and L166 of SigI1 are completely conserved in the eight SigI–RsgI pairs (Figure 3D, Supplementary Table S5). This phenomenon is consistent with the specific recognition of multiple SigI–RsgI pairs.
To further understand the structural basis for the specific recognition, we performed structural and mutagenesis analyses, using two pairs of σI/anti-σI factors, SigI1/RsgI1 and SigI2/RsgI2, as examples. By comparing the interfacial interactions in the structure of the SigI1C–RsgI1N complex and the structural model of SigI2C-RsgI2N (Supplementary Table S6), we selected residues Y8, E9 and V35 of RsgI1N which form hydrogen bonding, electrostatic, and hydrophobic interactions with SigI1C, respectively, and the corresponding residues L9, K10 and R36 of RsgI2N which form hydrophobic, reversed electrostatic, and new electrostatic interactions with SigI2C, respectively. Therefore, these residues may play roles in the specific recognition for SigI1/RsgI1 and SigI2/RsgI2 pairs. Because SigI2C showed non-specific binding to the SPR chip and was unstable during the SPR experiments, we used NMR titration experiments to check the interactions by adding wild-type RsgI1N, the Y8L, Y8L-E9K or Y8L-E9K-V35R mutants, or wild-type RsgI2N into a solution of 15N-labeled SigI1C or SigI2C (Supplementary Figure S8). The results indicate that these mutations increasingly weaken the interaction with SigI1C but enhance interaction with SigI2C. The triple mutant of RsgI1N, however, still failed to abolish the interaction with SigI1C completely, and its interaction with SigI2C was not as strong as wild-type RsgI2N, indicating that multiple interfacial residues contribute synergistically to the specificity of the two pairs of σI/anti-σI factors.
DISCUSSION
SigI and RsgI are distinctive pairs of alternative σ/anti-σ factors. In this study, we presented the structures and recognition mechanisms of the key domains of SigI and RsgI, which were discovered to be notably different from all other known σ/anti-σ factors. The results reveal a novel β-barrel inhibitory domain structure for RsgI and a distinct 8-helical structure for SigIC, which differs from the well-known σ4 domain of σ70 factors. The 3D structure of the SigI1C–RsgI1N complex revealed the structural basis of the specific recognition between multiple pairs of σI/anti-σI factors. Previous studies have shown that the –35 element is important for the specific recognition of promoters by different SigIs (25), and our analysis of the SigI1C structure revealed the promoter binding site of SigIC for –35 region recognition. The low sequence homology of the interaction regions for either RsgI binding or promoter recognition provides the functional specificity of each σI-anti-σI pair.
Analysis of the interactions between SigI1C, RsgI1N and RNAP revealed that RsgI blocks holoenzyme formation of RNAP and SigI. This suggests the presence of overlapping binding surfaces on SigI for interaction with RNAP or RsgI, and that the SigIC domain is important for RNAP binding. The different SigIs presumably share a conserved RNAP-binding surface to form holoenzymes with RNAP. However, the RsgI-binding surfaces of SigIs showed largely non-conserved residues for specific recognition of their cognate RsgIs. One possible explanation is that SigIC may undergo significant conformational changes to expose highly conserved regions upon RNAP binding. Future structure determination of the RNAP holoenzyme is thus needed to address this issue.
The 3D structure of the SigI1C–RsgI1N complex presented in this paper also provides the structural basis for analysis of the multiple SigI and RsgI factors in other bacterial species. Studies on the structure and specificity of σI, anti-σI, and the cognitive promoters will enhance our understanding of the molecular mechanism of these intricate systems. Furthermore, the large number of σI/anti-σI pairs in the different species provides an abundant library of regulatory components. Recently, the exquisite specificity of ECF σ factors has been successfully used in the design of orthogonal genetic switches and regulators in synthetic genetic circuits (9,10). The σI/anti-σI systems with alternative specificities are also promising components for the development of novel genetic circuits. Understanding the fine structural and molecular details of the various σI-anti-σI systems from different sources can provide a future basis for advanced regulatory design in synthetic biology.
DATA AVAILABILITY
The structures and the chemical shift assignments have been deposited into Protein Data Bank and the BioMagResBank under accession numbers 6IVS and 36220 for RsgI1N and 6IVU and 36221 for the RsgI1N–SigI1C complex, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Fei Li from Protein Material group in QIBEBT for useful discussion of the SPR experiments. We thank Professor Sarah Perrett (Institute of Biophysics, Chinese Academy of Sciences) for critically reading the manuscript.
Notes
Present address: Lizett Ortiz de Ora, Department of Chemistry, University of California, Irvine, California, USA.
Present address: Iván Muñoz-Gutiérrez, Outreach Research Training and Minority Science Programs, School of Biological Sciences, University of California, Irvine, California, USA.
Present address: Yifei Li, Vonsun Pharmatech (Suzhou) Co., Ltd., Room 213, Building A4, No 218 Xinghu Street, Suzhou industrial park, Suzhou, Jiangsu, 215000, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31670735, 31661143023 to Y.F., 31570029 to Y.-J.L., 31470210 to Q.C.]; ‘Transformational Technologies for Clean Energy and Demonstration’, Strategic Priority Research Program of the Chinese Academy of Sciences [XDA21060201 to Q.C.]; Shandong Provincial Natural Science Foundation [ZR2016CB09 to C.C.]; a joint research grant from the Israel Science Foundation (ISF) [2566/16 to E.A.B.]–National Natural Science Foundation of China (NSFC) [31661143023 to Y.F]; Israel Science Foundation (ISF) [1349 to E.A.B.]. E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry. Funding for open access charge: National Natural Science Foundation of China [31670735, 31661143023 to Y.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Murakami K.S. Structural biology of bacterial RNA polymerase. Biomolecules. 2015; 5:848–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gruber T.M., Gross C.A.. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003; 57:441–466. [DOI] [PubMed] [Google Scholar]
- 3. Paget M.S. Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules. 2015; 5:1245–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mascher T. Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Curr. Opin. Microbiol. 2013; 16:148–155. [DOI] [PubMed] [Google Scholar]
- 5. Helmann J.D. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002; 46:47–110. [DOI] [PubMed] [Google Scholar]
- 6. Brooks B.E., Buchanan S.K.. Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochim. Biophys. Acta. 2008; 1778:1930–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ho T.D., Ellermeier C.D.. Extra cytoplasmic function σ factor activation. Curr. Opin. Microbiol. 2012; 15:182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Campagne S., Allain F.H.T., Vorholt J.A.. Extra cytoplasmic function sigma factors, recent structural insights into promoter recognition and regulation. Curr. Opin. Struct. Biol. 2015; 30:71–78. [DOI] [PubMed] [Google Scholar]
- 9. Zong Y.Q., Zhang H.Q.M., Lyu C., Ji X.Y., Hou J.R., Guo X., Ouyang Q., Lou C.B.. Insulated transcriptional elements enable precise design of genetic circuits. Nat. Commun. 2017; 8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pinto D., Vecchione S., Wu H., Mauri M., Mascher T., Fritz G.. Engineering orthogonal synthetic timer circuits based on extracytoplasmic function σ factors. Nucleic Acids Res. 2018; 46:7450–7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Asai K., Ootsuji T., Obata K., Matsumoto T., Fujita Y., Sadaie Y.. Regulatory role of RsgI in sigI expression in Bacillus subtilis. Microbiology. 2007; 153:92–101. [DOI] [PubMed] [Google Scholar]
- 12. Ramaniuk O., Převorovský M., Pospíšil J., Vitovská D., Kofroňová O., Benada O., Schwarz M., Šanderová H., Hnilicová J., Krásný L.. σI from Bacillus subtilis: Impact on gene expression and characterization of σI-dependent transcription that requires new types of promoters with extended -35 and -10 elements. J. Bacteriol. 2018; 200:e00251-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Izquierdo J.A., Goodwin L., Davenport K.W., Teshima H., Bruce D., Detter C., Tapia R., Han S.S., Land M., Hauser L. et al.. Complete genome sequence of Clostridium clariflavum DSM 19732. Stand. Genomic Sci. 2012; 6:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muñoz-Gutiérrez I., Ortiz, de Ora L., Grinberg I.R., Garty Y., Bayer E.A., Shoham Y., Lamed R., Borovok I.. Decoding biomass-sensing regulons of Clostridium thermocellum alternative sigma-I factors in a heterologous Bacillus subtilis host system. PLoS One. 2016; 11:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bayer E.A., Chanzy H., Lamed R., Shoham Y.. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 1998; 8:548–557. [DOI] [PubMed] [Google Scholar]
- 16. Smith S.P., Bayer E.A.. Insights into cellulosome assembly and dynamics: from dissection to reconstruction of the supramolecular enzyme complex. Curr. Opin. Struct. Biol. 2013; 23:686–694. [DOI] [PubMed] [Google Scholar]
- 17. Stevenson D.M., Weimer P.J.. Expression of 17 genes in Clostridium thermocellum ATCC 27405 during fermentation of cellulose or cellobiose in continuous culture. Appl. Environ. Microbiol. 2005; 71:4672–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gold N.D., Martin V.J.J.. Global view of the Clostridium thermocellum cellulosome revealed by quantitative proteomic analysis. J. Bacteriol. 2007; 189:6787–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zverlov V.V., Schwarz W.H.. Bacterial cellulose hydrolysis in anaerobic environmental subsystems - Clostridium thermocellum and Clostridium stercorarium, thermophilic plant-fiber degraders. Ann. N. Y. Acad. Sci. 2008; 1125:298–307. [DOI] [PubMed] [Google Scholar]
- 20. Raman B., Pan C., Hurst G.B., Rodriguez M., McKeown C.K., Lankford P.K., Samatova N.F., Mielenz J.R.. Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One. 2009; 4:e5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raman B., McKeown C.K., Rodriguez M., Brown S.D., Mielenz J.R.. Transcriptomic analysis of Clostridium thermocellum ATCC 27405 cellulose fermentation. BMC Microbiol. 2011; 11:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nataf Y., Bahari L., Kahel-Raifer H., Borovok I., Lamed R., Bayer E.A., Sonenshein A.L., Shoham Y.. Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18646–18651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kahel-Raifer H., Jindou S., Bahari L., Nataf Y., Shoham Y., Bayer E.A., Borovok I., Lamed R.. The unique set of putative membrane-associated anti-σ factors in Clostridium thermocellum suggests a novel extracellular carbohydrate-sensing mechanism involved in gene regulation. FEMS Microbiol. Lett. 2010; 308:84–93. [DOI] [PubMed] [Google Scholar]
- 24. Yaniv O., Fichman G., Borovok I., Shoham Y., Bayer E.A., Lamed R., Shimon L.J.W., Frolow F.. Fine-structural variance of family 3 carbohydrate-binding modules as extracellular biomass-sensing components of Clostridium thermocellum anti-σI factors. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2014; 70:522–534. [DOI] [PubMed] [Google Scholar]
- 25. Ortiz de Ora L., Lamed R., Liu Y.J., Xu J., Cui Q., Feng Y.G., Shoham Y., Bayer E.A., Muñoz-Gutiérrez I.. Regulation of biomass degradation by alternative σ factors in cellulolytic clostridia. Sci. Rep. 2018; 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu H.W., Gong W.B., Yao X.Z., Wang J.F., Perrett S., Feng Y.G.. Evolutionarily conserved binding of translationally controlled tumor protein to eukaryotic elongation factor 1B. J. Biol. Chem. 2015; 290:8694–8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 28. Johnson B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004; 278:313–352. [DOI] [PubMed] [Google Scholar]
- 29. Jung Y.S., Zweckstetter M.. Mars - robust automatic backbone assignment of proteins. J. Biomol. NMR. 2004; 30:11–23. [DOI] [PubMed] [Google Scholar]
- 30. Herrmann T., Güntert P., Wüthrich K.. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002; 319:209–227. [DOI] [PubMed] [Google Scholar]
- 31. Duggan B.M., Legge G.B., Dyson H.J., Wright P.E.. SANE (Structure assisted NOE evaluation): An automated model-based approach for NOE assignment. J. Biomol. NMR. 2001; 19:321–329. [DOI] [PubMed] [Google Scholar]
- 32. Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. et al.. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D-Biol. Crystallogr. 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 33. Nederveen A.J., Doreleijers J.F., Vranken W., Miller Z., Spronk C.A.E.M., Nabuurs S.B., Güntert P., Livny M., Markley J.L., Nilges M. et al.. RECOORD: a recalculated coordinate database of 500+proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005; 59:662–672. [DOI] [PubMed] [Google Scholar]
- 34. Shen Y., Bax A.. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol. Biol. 2015; 1260:17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M.. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996; 8:477–486. [DOI] [PubMed] [Google Scholar]
- 36. Hooft R.W.W., Vriend G., Sander C., Abola E.E.. Errors in protein structures. Nature. 1996; 381:272–272. [DOI] [PubMed] [Google Scholar]
- 37. Koradi R., Billeter M., Wüthrich K.. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996; 14:51–55. [DOI] [PubMed] [Google Scholar]
- 38. Holm L., Rosenström P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krissinel E., Henrick K.. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2004; 60:2256–2268. [DOI] [PubMed] [Google Scholar]
- 40. Šali A., Blundell T.L.. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993; 234:779–815. [DOI] [PubMed] [Google Scholar]
- 41. Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. et al.. Clustal W and clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 42. Frostell A., Vinterbäck L., Sjöbom H.. Protein-ligand interactions using SPR systems. Methods Mol. Biol. 2013; 1008:139–165. [DOI] [PubMed] [Google Scholar]
- 43. Mohr G., Hong W., Zhang J., Cui G.Z., Yang Y.F., Cui Q., Liu Y.J., Lambowitz A.M.. A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum. PLoS One. 2013; 8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Campbell E.A., Tupy J.L., Gruber T.M., Wang S., Sharp M.M., Gross C.A., Darst S.A.. Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-σ RseA. Mol. Cell. 2003; 11:1067–1078. [DOI] [PubMed] [Google Scholar]
- 45. Maillard A.P., Girard E., Ziani W., Petit-Härtlein I., Kahn R., Covès J.. The crystal structure of the anti-σ factor CnrY in complex with the σ factor CnrH shows a new structural class of anti-σ factors targeting extracytoplasmic function σ factors. J. Mol. Biol. 2014; 426:2313–2327. [DOI] [PubMed] [Google Scholar]
- 46. Schumacher M.A., Bush M.J., Bibb M.J., Ramos-León F., Chandra G., Zeng W., Buttner M.J.. The crystal structure of the RsbN-σBldN complex from Streptomyces venezuelae defines a new structural class of anti-σ factor. Nucleic Acids Res. 2018; 46:7467–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Watanabe S., Matsumi R., Atomi H., Imanaka T., Miki K.. Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation. Structure. 2012; 20:2124–2137. [DOI] [PubMed] [Google Scholar]
- 48. Murzin A.G. OB(Oligonucleotide/Oligosaccharide Binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993; 12:861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 2002; 12:794–801. [DOI] [PubMed] [Google Scholar]
- 50. Guardino K.M., Sheftic S.R., Slattery R.E., Alexandrescu A.T.. Relative stabilities of conserved and non-conserved structures in the OB-fold superfamily. Int. J. Mol. Sci. 2009; 10:2412–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Malhotra A., Severinova E., Darst S.A.. Crystal structure of a σ70 subunit fragment from E. coli RNA polymerase. Cell. 1996; 87:127–136. [DOI] [PubMed] [Google Scholar]
- 52. Patikoglou G.A., Westblade L.F., Campbell E.A., Lamour V., Lane W.J., Darst S.A.. Crystal structure of the Escherichia coli regulator of σ70, Rsd, in complex with σ70 domain 4. J. Mol. Biol. 2007; 372:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sineva E., Savkina M., Ades S.E.. Themes and variations in gene regulation by extracytoplasmic function (ECF) sigma factors. Curr. Opin. Microbiol. 2017; 36:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Light S.H., Cahoon L.A., Halavaty A.S., Freitag N.E., Anderson W.F.. Structure to function of an α-glucan metabolic pathway that promotes Listeria monocytogenes pathogenesis. Nat. Microbiol. 2017; 2:16202. [DOI] [PubMed] [Google Scholar]
- 55. Wintjens R., Rooman M.. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 1996; 262:294–313. [DOI] [PubMed] [Google Scholar]
- 56. Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M.. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev. 2005; 29:231–262. [DOI] [PubMed] [Google Scholar]
- 57. Lane W.J., Darst S.A.. The structural basis for promoter -35 element recognition by the group IV σ factors. PLoS Biol. 2006; 4:1491–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Staroń A., Sofia H.J., Dietrich S., Ulrich L.E., Liesegang H., Mascher T.. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) σ factor protein family. Mol. Microbiol. 2009; 74:557–581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures and the chemical shift assignments have been deposited into Protein Data Bank and the BioMagResBank under accession numbers 6IVS and 36220 for RsgI1N and 6IVU and 36221 for the RsgI1N–SigI1C complex, respectively.