Abstract
Background
We previously discovered that 3 long non-coding RNAs (lncRNAs) NONHSAT089447, NONHSAT021545, and NONHSAT041499 were differentially expressed in the peripheral blood of patients with schizophrenia, in comparison to those in normal healthy controls. In this study, we conducted bioinformatic analysis of these 3 lncRNAs and the regulatory role of lncRNA NONHSAT089447 in the dopamine signaling pathway in patients with schizophrenia.
Material/Methods
There lncRNAs in peripheral blood mononuclear cells (PBMCs) were screened using microarray analysis. Pearson’s correlation analysis was performed to assess the levels of co-expressed mRNAs of respective lncRNAs. The Database for Annotation, Visualization and Integrated Discovery (DAVID) software was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes or Genomes (KEGG) enrichment analysis for these lncRNAs. Human neuroblastoma cell lines (SK-N-SH) were cultured and treated with dopamine or olanzapine (OLP), or transfected with siRNA targeting NONHSAT089447 or plasmid expressing NONHSAT089447. Levels of lncRNAs were detected by quantitative real-time reverse transcription polymerase chain reaction (RT-PCR). Then, mRNA and protein expression of the dopamine receptors DRD1, DRD2, DRD3, DRD4, and DRD5 were measured by RT-PCR and western blot analysis, respectively.
Results
OLP treatment significantly inhibited the expression of NONHSAT089447. Knockdown of NONHSAT089447 by siRNA decreased DRD3 and DRD5 expression, while overexpression of NONHSAT089447 significantly upregulated expression of DRD3 and DRD5. Western blot analysis confirmed that levels of NONHSAT089447 regulated downstream DRD signaling.
Conclusions
Our results revealed that the lncRNA NONHSAT089447 participated in the dopamine signaling pathway via upregulation of DRDs.
MeSH Keywords: Dopamine; Neuroblastoma; RNA, Long Noncoding; Schizophrenia
Background
Schizophrenia (SZ), characterized by a dissociation between mental events and the real world, is a mental disorder in perception, thought, emotion, and behavioral performance in social interactions [1–3], with an estimated lifetime morbidity of 1% [4]. SZ not only damages the social functioning of patients but also heavily affects families of patients and society. Currently, diagnosis of SZ is commonly considered as heavily influenced by genetics; in fact, the heredity rate for SZ is reported to be up to 70–80% [5–7]. However, an understanding of the molecular mechanisms involved in SZ remains unclear.
Long non-coding RNAs (lncRNAs) are described as mRNA-like, long RNAs that are greater than 200 nucleotides in length and lack any evident protein-coding capacity [8,9]. Although lncRNAs lack protein-coding ability, they can regulate the expression of genes at the levels of transcription, post-transcription, translation, etc. [10–12]. LncRNAs can interact and cooperate with microRNAs (miRNAs) to control genetic transcription by targeting and binding to gene promoters [13–15]. Studies have revealed that more than half of lncRNAs that have been discovered are expressed in the central nervous system and participate in important biological processes, such as cell recognition, stress response, plasticity mechanisms, and the onset and development of mental disorders [16]. It was previously reported that several lncRNAs localized to chromosomal regions associated with mental disorders [17], such as the Gomafu/MIAT/Rncr2 gene locus [18,19], which was demonstrated to be involved in cell singularization, stem cell differentiation, alternative splicing, and other cellular processes related to SZ. In recent years, research on the mechanism of control of SZ by miRNAs has greatly improved [20,21]. Meanwhile, studies investigating lncRNAs and their regulatory roles in SZ are still developing and many areas of knowledge remain unclear.
Dopamine and 5-hydroxytryptamine (serotonin) are both important neurotransmitters in the central nervous system [22,23], and we used them to stimulate cells and simulate SZ disease, while measuring expression of lncRNAs to determine their association with dopamine and serotonin. Yi et al. [24] hypothesized that lncRNAs were related to SZ susceptibility genes, such as those involved in oxidative pressure, serine-threonine kinases, peroxidases, and protein kinases, and affected nervous system function via signal transduction. Furthermore, lncRNAs can act as biomolecular scaffolds, play important roles in signal relay and intermolecular interaction, and control the specificity and dynamics of a signal [25,26]. Consequently, we hypothesized that the differential expression of lncRNAs we previously reported may be associated with the dopamine signaling pathway. Dopamine is an endogenous neurotransmitter that regulates the function of dopamine receptors (DRs). DRs are G protein-coupled receptors that can be divided into 2 families, D1 and D2. The D1 family includes the D1 receptor (DRD1) and D5 receptor (DRD5), while the D2 family includes the D2 receptor (DRD2), D3 receptor (DRD3), and D4 receptor (DRD4) [27,28]. DR function is primarily mediated by the adenylate cyclase (AC)/cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA) signaling pathway [29,30]. There is little evidence that DRs are pathogenic factors, but the dopamine signaling pathway has been a focus of research due to past findings that demonstrate certain receptor subtypes can control SZ by tightly regulating the dopamine network system [31].
In our previously published study, we identified and validated 3 lncRNAs, NONHSAT089447, NONHSAT021545, and NONHSAT041499, in peripheral blood mononuclear cells (PBMCs) that were differentially expressed between SZ patients and healthy participants, by lncRNA microarray profiling and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) [32]. In the present study, we selected NONHSAT089447 and NONHSAT041499 to further investigate their association with SZ. We used human neuroblastoma cell lines as model systems to observe the mechanism of effect of lncRNAs on the dopamine signaling pathway. This study might provide new insight into the regulation and molecular mechanism of SZ, which might be beneficially applied to the early diagnosis and treatment of SZ.
Material and Methods
Participants
A total of 160 participants, 40 patients with SZ, 40 patients with major depressive disorder (MDD), 40 patients with generalized anxiety disorder (GAD), and 40 healthy adults, were enrolled in the study between September 2013 and May 2015 at the No. 102 Hospital of the Chinese People’s Liberation Army (Changzhou, China). Inclusion criteria for patients comprised: 1) all patients fulfilled criteria as defined by the American Diagnostic and Statistical Manual of Mental Disorders (DSM-IV-TR); and 2) all patients were in the absence of antidepressant or antipsychotic medication within at least 3 months. Exclusion criteria for patients included: 1) serious organic disease, effective disorder, mental disorder or personality disorder from sequential medicine, alcohol abuse, and other substance abuse; and 2) any modified and regular electro convulsion therapy (MECT) treatment within 6 months or long-term use of antipsychotic medication. All healthy participants had no history of severe traumatic events and did not receive a blood transfusion within a month before blood sampling. This study was approved by the Medical Ethics Board of the No. 102 Hospital of the Chinese People’s Liberation Army.
Assessment of SZ
SZ patients were assessed by 3 psychiatrists who were collectively trained to administer a standardized procedure using the Positive and Negative Symptom Scale (PANSS). The PANSS [33] is comprised of rating parameters on 30 symptoms divided between 3 scales (positive, negative, and general psychopathology) from 1 to 7, with a higher grade indicating higher psychotic severity. The reliability and validity of PANSS was naturalized and verified as applicable to Chinese people.
Quantitative RT-PCR analysis
Complementary DNA (cDNA) was synthesized using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, CA, USA). Real-time PCR was performed using the 7900HT Real-Time PCR System (Applied Biosystems). Each sample was run in triplicate for analysis. Data were collected using the SDS 2.4 software (Applied Biosystems). After normalizing to the expression level of RNU48 (control), expression levels of the lncRNAs were calculated using the formula, 2−ΔCt.
Gene ontology (GO) and Kyoto Encyclopedia of Genes or Genomes (KEGG) pathway enrichment analysis
We used the Database for Annotation, Visualization and Integrated Discovery (DAVID) software (http://david.abcc.ncifcrf.gov/) to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes or Genomes (KEGG) pathway enrichment analysis of the 3 mRNAs co-expressed with the lncRNAs of interest. We set the threshold of statistical significance as P value less than 0.1, as readjusted by the Fisher’s exact test.
Cell culture and transfection
SK-H-SH cells (Chinese Academy of Sciences, Beijing, China) were maintained in Dulbecco’s modified Eagle medium (DMEM) with high glucose (GIBCO, CA, USA) and 10% fetal bovine serum (GIBCO). Cells (2×105) were cultured per well of a 6-well plate or 35 mm dish at 37°C and 5% CO2. For cell transfection, cells at 70–80% confluence were transfected with RNA using Lipofectamine 2000 reagent. In brief, plasmid RNA (1–2 μg) and Lipofectamine 2000 reagent were each diluted in Opti-MEM (50 μL) at room temperature for 5 minutes, respectively, and then mixed gently at room temperature for 15–45 minutes. Cells were washed with Opti-MEM. Fresh medium was added to cells, followed by the mixture of DNA and Lipofectamine 2000 reagent. Cells were incubated at 37°C. Medium was replaced 6 hours after transfection, and transfection efficiency was observed 24 hours after transfection.
Western blot analysis
Cells were pelleted by centrifugation and washed with cold phosphate-buffered saline (PBS) (1 mL). The cell pellet was lysed with cell lysis buffer, containing 1 mM phenylmethylsulfonyl fluoride (PMSF) added fresh, and incubated on ice for 20–30 minutes. After centrifugation at 12 000 rpm for 5 minutes at 4°C, supernatant was aspirated and stored at −70°C prior to use. Protein (10 μg) was quantified and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto polyvinylidene fluoride or polyvinylidene difluoride (PVDF) film and blocked in 5% bovine serum albumin at room temperature for 1 hour. Primary and secondary antibodies were added sequentially.
Statistical analysis
All data were processed by SPSS (v 17.0. Mev4.6) software, and cluster analysis was conducted on lncRNA expression levels from the gene microarray profile. Independent sample t-test was used to test the differences of expression levels of 10 lncRNAs between groups. All statistical tests were 2-tailed, and P<0.05 was considered statistically significant.
Results
We previously identified that 3 lncRNAs, NONHSAT089447, NONHSAT021545, and NONHSAT041499, were upregulated in SZ patients in comparison to levels in normal healthy people, using microarray analysis and qRT-PCR validation [32]. In this study, we selected to further analyze these lncRNAs.
LncRNAs co-expressed with protein-coding genes
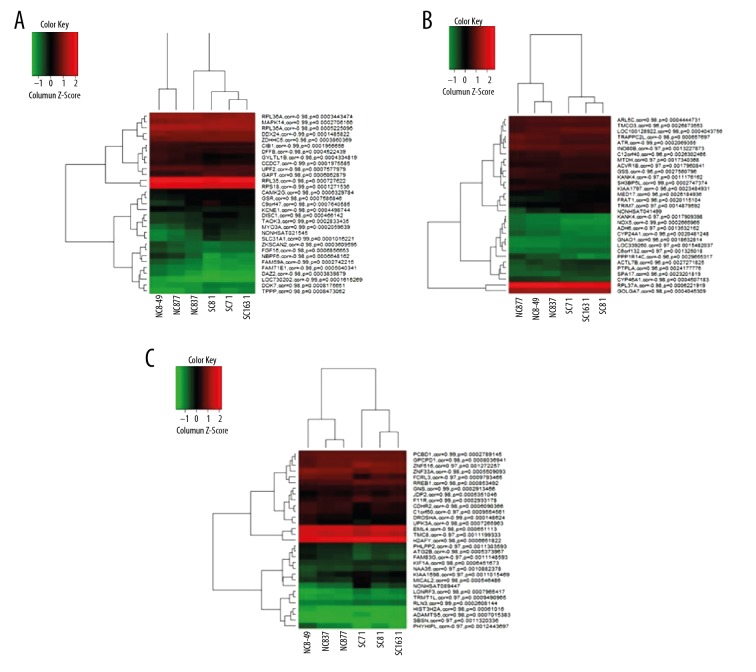
We identified 1773 mRNAs that were co-expressed with NONHSAT021545, 638 mRNAs co-expressed with NONHSAT041499, and 1602 mRNAs co-expressed with NONHSAT089447. Figure 1A–1C illustrate the 30 most relevant mRNAs and their expression levels associated with the 3 lncRNAs, by unsupervised hierarchical clustering analysis. Overall, there were 89 mRNAs that were identified to co-express simultaneously with the 3 lncRNAs used in this study (Supplementary Table 1).
Figure 1.
Heatmaps generated for (A) NONHSAT021545, (B) NONHSAT041499, and (C) NONHSAT089447.
GO enrichment analysis of genes co-expressed with the lncRNAs
GO enrichment analysis was performed on the list of mRNAs that co-expressed with the lncRNAs to determine associated biological processes. The results indicated that these associated genes were involved in signal transduction, multicellular organismal development, cell cycle, cell proliferation, differentiation, apoptosis, and metabolism. Several notable biological processes included pallium development, axon guidance and extension, synaptic transmission, learning, and memory (Table 1), which play significant roles in brain development and function, and potentially, the pathophysiology of SZ.
Table 1.
List of gene ontology (GO) terms from enrichment analysis associated with central neural systems.
| GO No. | Biological process items | P value | FDR |
|---|---|---|---|
| 0051402 | Neuron apoptosis | 0.004 | 7.240738 |
| 0046578 | Regulation of Ras protein signal transduction | 0.004 | 7.854287 |
| 0046579 | Positive regulation of Ras protein signal transduction | 0.005 | 9.803511 |
| 0032496 | Response to lipopolysaccharide | 0.008 | 14.27691 |
| 0007254 | JNK cascade | 0.011 | 18.9682 |
| 0070302 | Regulation of stress-activated protein kinase signaling pathway | 0.011 | 19.06017 |
| 0007611 | Learning or memory | 0.012 | 20.08509 |
| 0046328 | Regulation of JNK cascade | 0.013 | 21.97039 |
| 0050803 | Regulation of synapse structure and activity | 0.020 | 30.90909 |
| 0007265 | Ras protein signal transduction | 0.020 | 31.94657 |
| 0031098 | Stress-activated protein kinase signaling pathway | 0.021 | 32.6278 |
| 0007610 | Behavior | 0.032 | 46.07315 |
| 0031175 | Neuron projection development | 0.039 | 52.61498 |
| 0007264 | Small GTPase mediated signal transduction | 0.042 | 55.32477 |
| 0043523 | Regulation of neuron apoptosis | 0.042 | 55.33304 |
| 0048812 | Neuron projection morphogenesis | 0.051 | 62.58074 |
| 0007612 | Learning | 0.057 | 66.421 |
| 0021819 | Layer formation in the cerebral cortex | 0.060 | 68.9358 |
| 0007409 | Axonogenesis | 0.064 | 71.271 |
| 0007219 | Notch signaling pathway | 0.079 | 78.71841 |
| 0007605 | Sensory perception of sound | 0.081 | 79.65405 |
| 0050807 | Regulation of synapse organization | 0.082 | 79.83228 |
| 0000187 | Activation of MAPK activity | 0.098 | 85.58835 |
| 0043525 | Positive regulation of neuron apoptosis | 0.099 | 85.81547 |
KEGG signal pathway enrichment analysis of genes co-expressed with the lncRNAs
Similarly, KEGG pathway enrichment analysis revealed the significant enrichment of 10 pathways related to neuronal brain function, such as the ribosome signaling pathway, lysosome signaling pathway, gonadotropin-releasing hormone (GnRH) signaling pathway, mammalian target of rapamycin (mTOR) signaling pathway, insulin signaling pathway, and apoptotic signaling pathway (Figure 2).
Figure 2.
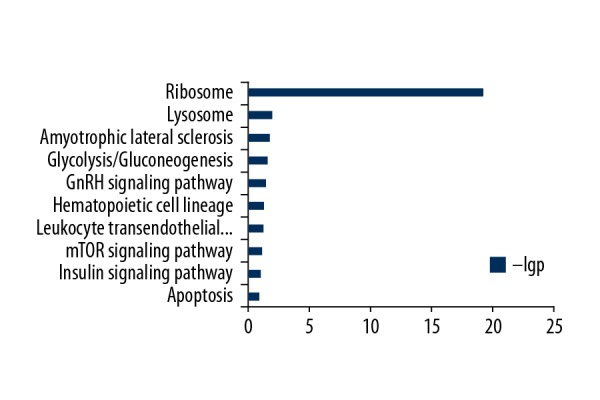
Kyoto Encyclopedia of Genes or Genomes (KEGG) pathway analysis of mRNA-coding genes co-expressed with lncRNAs.
Association analysis between the lncRNAs and transcription factors
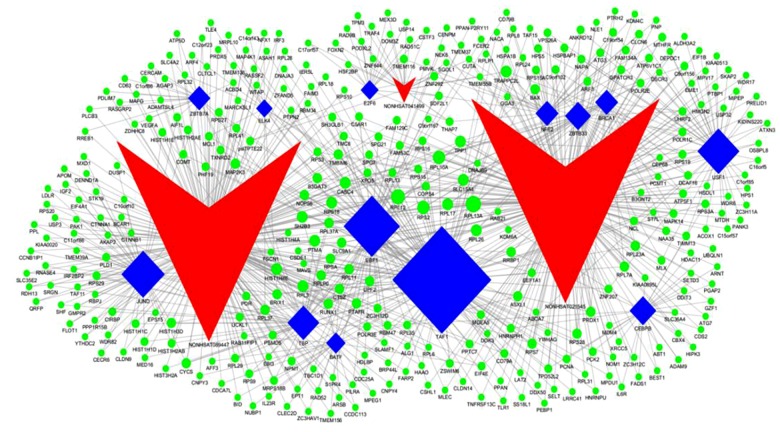
We identified 7 transcription factors that were significantly associated with NONHSAT021545, namely TATA-binding protein-associated factor 1 (TAF1), upstream transcription factor 1 (USF1), CCAAT/enhancer-binding protein beta (CEBPB), breast cancer factor 1 (BRCA1), early B-cell factor 1 (EBF1), zinc finger and BTB domain-containing protein 33 (ZBTB33), and nuclear factor erythroid 2 (NFE2); 2 transcription factors that were significantly associated with NONHSAT041499, namely E2F transcription factor 6 (E2F6) and NFE2; and 7 transcription factors that were significantly associated with NONHSAT089447, namely JunD, TATA-binding protein (TBP), TAF1, basic leucine zipper transcription factor (BATF), ETS domain-containing protein Elk-4, EBF1, and zinc finger and BTB domain-containing protein 7A (ZBTB7A). Figure 3 illustrates a topographical map of the lncRNAs, associated transcriptional factors, and their target genes.
Figure 3.
lncRNA/transcription factor/target gene network system. Red denotes lncRNAs, blue denotes transcription factors, green denotes miRNAs, and lines represent the existence of a regulatory relationship.
Expression of the 3 lncRNAs in MDD and GAD patients
As shown in Table 2, ΔCt values for the 3 lncRNAs were significantly lower in SZ patients than in healthy controls, and MDD and GAD patients (P<0.001), indicating that the expression of the lncRNAs was significantly higher in SZ patients. Therefore, we observed upregulation of these 3 lncRNAs, which might differentiate SZ patients from healthy controls, and MDD and GAD patients.
Table 2.
The qRT-PCR (ΔCt value) analysis of lncRNA expression in SZ, MDD, and GAD patients and healthy normal controls.
| Probes | NC | SZ | GAD | MDD |
|---|---|---|---|---|
| NONHSAT089447 | 6.28±5.37 | 1.24±3.27# | 8.3±2.08* | 8.28±1.94 |
| NONHSAT021545 | 6.48±4.5 | 1.75±3.26# | 8.64±1.91* | 7.46±3.09 |
| NONHSAT041499 | 6.7±4.45 | 2.81±3.44# | 9.2±1.55** | 8.43±1.66 |
P<0.05;
P<0.01, versus NC;
P<0.001 versus GAD/MDD.
qRT-PCR – quantitative real-time reverse transcription polymerase chain reaction; SZ – schizophrenia; MDD – major depressive disorder; GAD – generalized anxiety disorder; NC – normal control.
Effect of dopamine or serotonin on lncRNA expression in SK-N-SH cells
Previously, we demonstrated that antipsychotic drug treatment significantly decreased both symptomatology and total scores of SZ patients, and the levels of the 2 lncRNAs, NONHSAT041499 and NONHSAT089447, in PMBCs of SZ patients were also significantly reduced [32]. Therefore, the 2 lncRNAs, NONHSAT041499 and NONHSAT089447, were selected for further analysis.
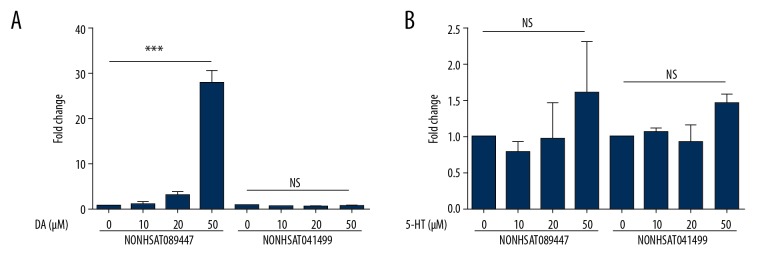
SK-N-SH cells cultured in 6-well plates were treated with either dopamine or serotonin, at varying concentrations, for 24 hours. Expression of the lncRNA NONHSAT089447 dramatically increased when cells were treated with 50 μM dopamine (Figure 4A). However, addition of serotonin did not affect the expression of NONHSAT041499 or NONHSAT089447 (Figure 4B).
Figure 4.
(A, B) The effect of dopamine/serotonin on the expression of 2 lncRNAs: NONHSAT089447 and NONHSAT041499.
Olanzapine (OLP) inhibited expression of NONHSAT089447 induced by dopamine
SK-N-SH cells were cultured in 6-well plates with or without the addition of dopamine, followed by the addition of olanzapine (OLP), obtained from the National Institutes for Food and Drug Control (serial number: 100948-200801, ID: V4V2-69GU), at increasing concentrations (0 μM, 10 μM, and 50 μM) for 24 hours. Dopamine-induced NONHSAT089447 expression was inhibited by subsequent OLP treatment (Figure 5).
Figure 5.
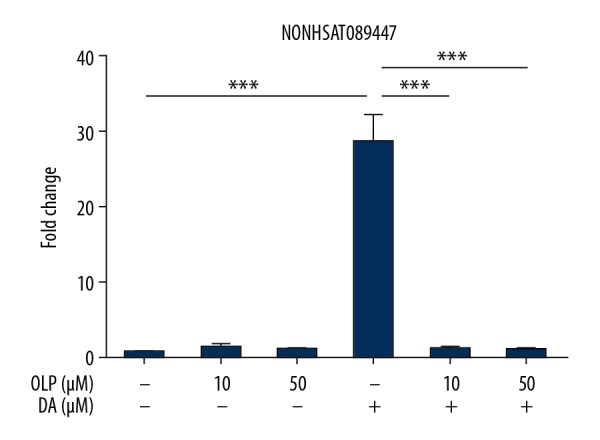
The effect of olanzapine (OLP) on the expression of NONHSAT089447 induced by dopamine treatment.
SiRNA knockdown of lncRNAs NONHSAT041499 and NONHSAT089447
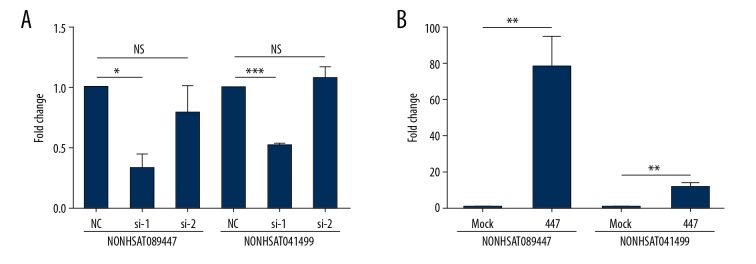
SK-N-SH cells were cultured in 6-well plates for 24 hours prior to transfecting with siRNAs that targeted the lncRNAs NONHSAT041499 and NONHSAT089447. Transfected cells were incubated for 48 hours prior to measurement of lncRNA expression. Knockdown with siRNA 1 significantly reduced expression levels of NONHSAT089447 compared to those in untreated cells (P<0.05). Interestingly, expression levels of NONHSAT041499 significantly increased (P<0.001) in contrast. Knockdown with siRNA 2 somewhat decreased expression levels of NONHSAT089447 compared to those in untreated cells, while increasing NONHSAT041499 expression levels (Figure 6A).
Figure 6.
The effect of (A) siRNA knockdown and (B) overexpression by plasmid RNAs on the expression levels of 2 lncRNAs in SK-N-SH cells.
Overexpression of the lncRNAs NONHSAT041499 and NONHSAT089447
SK-N-SH cells were cultured in 6-well plates for 24 hours prior to transfection with plasmid RNAs for the lncRNAs NONHSAT041499 and NONHSAT089447, respectively. Transfected cells were incubated for 24 hours. As shown in Figure 6B, expression levels of the lncRNAs NONHSAT041499 and NONHSAT089447 significantly increased in transfected cells (P<0.01).
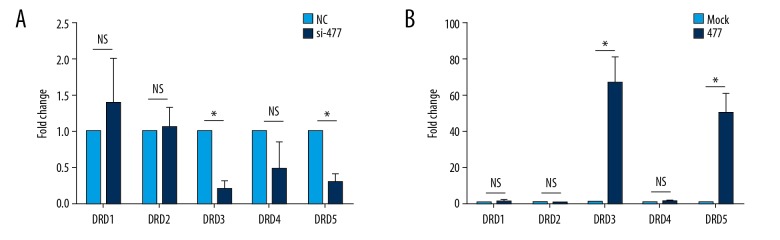
Effect of siRNA knockdown on expression of dopamine receptors (DRDs)
SK-N-SH cells were cultured in 6-well plates for 24 hours prior to transfection with siRNAs. Transfected cells were incubated for an additional 24 hours. siRNA knockdown significantly upregulated expression of DRD1 and DRD2, in comparison to untreated cells (Figure 7A). In addition, expression of DRD3, DRD4, and DRD5 decreased, but only DRD4 and DRD5 expression decreased significantly (P<0.05).
Figure 7.
The effect of (A) siRNA knockdown and (B) overexpression by plasmid RNAs on the mRNA expression levels of dopamine receptors (DRDs) in SK-N-SH cells.
Effect of overexpression of lncRNAs on expression of DRDs
SK-N-SH cells were cultured in 6-well plates for 24 hours, and then transfected and incubated with siRNAs for 48 hours. Following siRNA knockdown, cells were overexpressed with plasmid RNAs for NONHSAT041499 and NONHSAT089447. Expression of DRD1, DRD2, and DRD4 in rescued cells did not exhibit any significant differences compared to those in untreated cells. However, expression of DRD3, DRD4, and DRD5 increased significantly when NONHSAT041499 and NONHSAT089447 expression was rescued (P<0.05) (Figure 7B).
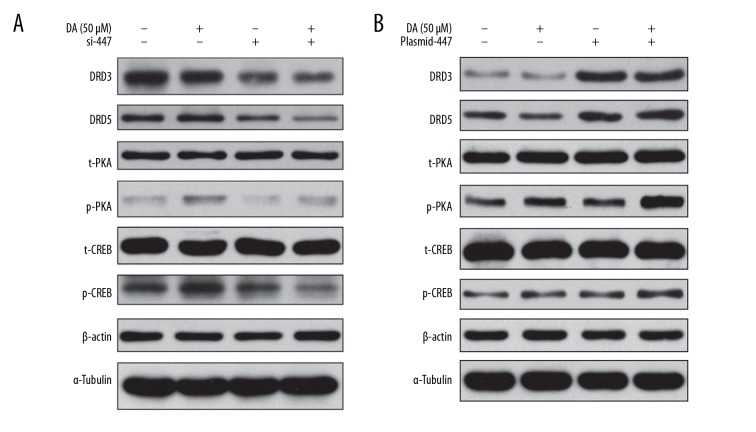
Effect of siRNA knockdown on downstream signaling of DRDs
SK-N-SH cells were cultured in 6-well plates for 24 hours prior to transfection with siRNAs. Transfected cells were incubated for an additional 24 hours, followed by addition of dopamine for 15 minutes. siRNA knockdown downregulated expression levels of DRD3 and DRD5, and phosphorylation levels of PKA and cAMP response element-binding protein (CREB) (Figure 8A).
Figure 8.
The effect of (A) siRNA knockdown and (B) overexpression by plasmid RNAs on the protein expression levels of downstream dopamine receptors (DRDs) signaling in SK-N-SH cells, with or without the interference of NONHSAT089447. Results are representative of experiments conducted in triplicate.
Effect of overexpression of lncRNAs on downstream signaling of DRDs
SK-N-SH cells were cultured in 6-well plates for 24 hours, and then overexpressed with plasmid RNAs for the lncRNAs NONHSAT041499 and NONHSAT089447 for 24 hours. Cells were stimulated with dopamine for 15 minutes. Overexpression of lncRNA NONHSAT089447 significantly downregulated expression levels of DRD3 and DRD5, and phosphorylation levels of PKA and CREB (Figure 8B).
Discussion
Currently, the diagnosis of SZ is solely based on qualitative analysis of patient symptoms [34,35] due to a lack of specific biological or molecular markers. LncRNAs can regulate the expression of genes at various molecular levels, including epigenetic, transcriptional, and post-transcriptional levels [11,12]. Previously, we screened and identified 3 lncRNAs, NONHSAT089447, NONHSAT021545, and NONHSAT041499, that were significantly upregulated in SZ patients compared to healthy controls [32]. In the present study, we showed that these lncRNAs were significantly upregulated in SZ patients compared to MDD and GAD patients. These observations suggested that upregulation of these lncRNAs might be specifically characteristic of SZ. Investigation of these lncRNAs and their regulatory role in SZ might provide potential diagnostic value.
To further explore the role of these lncRNAs in the pathogenesis of SZ, bioinformatic analysis of these lncRNAs was conducted. We identified relevant mRNAs that associated with the lncRNAs NONHSAT089447, NONHSAT021545, and NONHSAT041499 by Pearson’s correlation analysis, which totaled hundreds to thousands. This suggested that individual lncRNA regulated multiple target genes, participating in a wide range of biological processes. Particularly, a total of 89 mRNAs were co-expressed with all 3 aberrant lncRNAs, indicating that interaction between these lncRNAs and multiple miRNAs might be involved in the pathogenesis of SZ.
GO enrichment analysis of the mRNAs that co-expressed with the lncRNAs implicated a variety of important biological processes, including those involved in the central nervous system such as neuronal apoptosis and regulation, Ras signal transduction, lipopolysaccharide reaction, c-Jun N-terminal kinase (JNK) signaling and regulation, stress-related protein kinase signaling and regulation, learning and memory, regulation of synaptic structure and activity, and the formation, development, cortical layering, and axonogenesis of neural projection. These results might provide additional insight into the mechanism of pathogenesis of SZ.
Next, we investigated the relationship between dopamine/serotonin and lncRNA expression. Dopamine and/or 5-hydroxytryptamine (serotonin) was added to cultured human neuroblastoma cell lines (SK-N-SH) to simulate the nerve cells of SZ patients. Cells stimulated by both dopamine and serotonin exhibited an exponentially greater induction of lncRNA expression, compared to that in cells treated with serotonin alone, indicating that dopamine-treated cells simulated SZ nerve cells better than serotonin-treated cells. Upon stimulation with dopamine, the expression level of NONHSAT089447 increased exponentially compared to that of NONHSAT041499, suggesting that NONHSAT089447 might be more likely to play a role in SZ. Our results confirmed that increased NONHSAT089447 expression can be induced by dopamine treatment. Therefore, NONHSAT089447 expression might be tightly associated with the dopamine signaling pathway.
OLP, an antagonist of dopamine [36], was used to verify whether NONHSAT089447 expression was induced by dopamine. OLP was added at varying concentrations to cells pre-treated with dopamine. We observed that NONHSAT089447 expression was downregulated by OLP treatment in a dose-dependent manner. Since OLP is a dopamine antagonist, these results validated that NONHSAT089447 expression was, in fact, induced by dopamine.
To determine the relationship between the lncRNAs and the DRD signaling pathway, we transfected siRNAs or overexpressed plasmid RNAs into PBMCs and monitored alterations in signal transduction. Expression levels of the lncRNAs in SK-H-SN cells significantly decreased with siRNA knockdown. Overexpression of the lncRNAs by transfecting plasmid RNAs significantly increased expression levels of both lncRNAs in cells. However, relative to one another, NONHSAT041499 expression remained lower than NONHSAT089447 expression. Therefore, we concluded that NONHSAT089447 might be a more representative biomarker of SZ.
We demonstrated the relationship between lncRNA expression and the DRD signaling pathway by analyzing the regulation of DRD1, DRD2, DRD3, DRD4, and DRD5 in cells treated for siRNA knockdown of NONHSAT089447. When NONHSAT089447 expression was rescued in cells by transfecting with plasmid RNA, DRD1, DRD3, and DRD5 expression levels were upregulated, with DRD3 and DRD5 upregulation being statistically significant. From these results, we speculated that DRD3 and DRD5 might be involved in the mechanism of pathogenesis of SZ and might be possible targets for treatment. Some studies have reported that DRD3 plays an important role in mental diseases, including SZ [37,38], regulating the composition and release of dopamine, and regulating dopamine’s neuronal function [39,40]. Thus, there is evidence that it might be a significant molecular target in SZ. With extensive linkage disequilibrium approach, Domínguez et al. compared 260 SZ patients with 354 healthy people from a homogeneous population, which revealed significant differences in frequencies of the DRD3 haplotypes at the 3’half of the gene region [37,41]. Furthermore, a plethora of pre-clinical studies have demonstrated that pharmacological treatment with dopamine receptor antagonists or knockout of DRD3 remedies the negative symptoms and improves the cognitive function of animal models [42]. Moreover, there are reports that DRD5 is predominantly distributed in the limbic system, and due to the close relationship between the limbic system and mental activities, these findings suggest that DRD5 might be associated with mental disorders [43,44]. A study involving northern Han Chinese by Zhao Y et al. has proven that DRD5 might be the contributor for the genetic susceptibility to SZ in Han Chinese [45]. What’s more, Williams et al. [46] have observed a significant changes on the allele of the gene coding for DRD5 in SZ patients compared with healthy controls. Our studies give more evidence supporting the possibility that DRD3 and DRD5 were associated with SZ.
Our study demonstrated that the dopamine receptors DRD3 and DRD5, and their downstream signals were activated by NONHSAT089447 expression. Consequently, DRD downstream signaling was suppressed when NONHSAT089447 expression was suppressed. Therefore, highly expressed NONHSAT089447 might activate dopamine receptors and their downstream signaling pathways; more specifically, there might be a tight association between NONHSAT089447 and the downstream signaling of DRD3 and DRD5.
Conclusions
We demonstrated that the differential expression of the 3 lncRNAs NONHSAT089447, NONHSAT021545, and NONHSAT041499, might be characteristic of SZ, suggesting novel molecular targets for study and application in the diagnosis and treatment of SZ. We conducted bioinformatic analysis of these lncRNAs and determined that NONHSAT089447 might play a regulatory role and activate the DRD signaling pathway.
In this study, OLP was selected to be consistent with the previous study, in which we observed the altered expressions of lncRNAs in patients with SZ using second-generation antipsychotic drugs. So, the deficiency of this study was that we used OLP which acts as antagonist at both 5HT2A and D2 receptors. It would be a better practice to choose a pure DR receptor antagonists.
Ethics approval
The procedures of this study were approved by the Independent Ethics Committee (IEC) of No. 102 Hospital of Chinese People’s Liberation Army.
Supplementary Table
Supplementary Table 1.
Joint Co-expressed Coding Genes for 3 lncRNAs.
| EPHA3 | IL6R | EPHA4 | PPTC7 | PRELID1 | ANKRD36 | CT62 | MGEA5 |
| ZNF155 | SEL1L3 | MTRF1L | LOC153811 | REXO1L1 | TRERF1 | EIF4G3 | RASGRP3 |
| SH2D1A | FZD1 | LSM1 | TRPS1 | RD3 | RNF220 | ZNF550 | FAM196B |
| CD79A | IFT46 | SSB | GCNT2 | COL19A1 | PAK1 | RPL26 | IL1RAP |
| ARID5B | PCDHB4 | CTNNA1 | ASCC3 | BSDC1 | TRMT5 | ITGA1 | OXCT1 |
| KIAA0485 | DIAPH3 | psiTPTE22 | FLT3 | SIGLEC5 | UPF2 | MRPL9 | RPL35 |
| LOC100507650 | EYA1 | ROS1 | LEUTX | DOK7 | RIMS3 | MGC20647 | ANO1 |
| MAPK14 | GJB3 | MEX3D | TMEM164 | SLC9A1 | DISC1 | CCR2 | KCNE1 |
| LHFPL1 | GYLTL1B | CDCA7L | PPP2CB | RPL36A | OR2L3 | FCRL1 | LBH |
| HMGN2 | WDFY3 | LOC100505857 | HNRNPA1L2 | KCNJ15 | RPL37A | GPATCH3 | FAM129C |
| MCF2 | PET117 | MYL10 | PIK3C2B | RAB21 | FAM190B | RPS19 | FMO5 |
| AUTS2 |
Footnotes
Source of support: No. 102 Hospital of Chinese People’s Liberation Army
Conflict of interests
None.
References
- 1.Zhang LY, Cheng HS. Clinical psychology. 4th ed. People’s Military Medical Press of Beijing; Beijing: 2015. [Google Scholar]
- 2.Jetha MK, Zheng X, Goldberg JO, et al. Shyness and emotional face processing in schizophrenia: an ERP study. Biol Psychol. 2013;94(3):562–74. doi: 10.1016/j.biopsycho.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Kudumija Slijepcevic M, Jovanovic N. EPA-1103 – personality across the aggressive spectrum – a cross-sectional study of 358 males with paranoid schizophrenia. European Psychiatry. 2014;29(1):12014. [Google Scholar]
- 4.Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005;2(5):e141. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenwood TA, Braff DL, Light GA, et al. Initial heritability analyses of endophenotypic measures for schizophrenia: The consortium on the genetics of schizophrenia. Arch Gen Psychiatry. 2007;64(11):1242–50. doi: 10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyajyan AS, Atshemyan SA, Zakharyan RV. [Association of schizophrenia with variations in genes encoding transcription factors]. Mol Biol (Mosk) 2015;49(6):977–83. doi: 10.7868/S0026898415060038. [in Russian] [DOI] [PubMed] [Google Scholar]
- 7.Gogos JA, Gerber DJ. Schizophrenia susceptibility genes: Emergence of positional candidates and future directions. Trends Pharmacol Sci. 2006;27(4):226–33. doi: 10.1016/j.tips.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 9.Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29(8):461–68. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Caley DP, Pink RC, Trujillano D, Carter DR. Long noncoding RNAs, chromatin, and development. ScientificWorldJournal. 2010;10:90–102. doi: 10.1100/tsw.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152(6):1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Wu Z, Fu X, Han W. Long noncoding RNAs: Insights from biological features and functions to diseases. Med Res Rev. 2013;33(3):517–53. doi: 10.1002/med.21254. [DOI] [PubMed] [Google Scholar]
- 13.Faghihi MA, Zhang M, Huang J, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11(5):R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martianov I, Ramadass A, Serra BA, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 15.Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer. Hum Mol Genet. 2010;19(R2):R152–61. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent JB, Petek E, Thevarkunnel S, et al. The RAY1/ST7 tumor-suppressor locus on chromosome 7q31 represents a complex multi-transcript system. Genomics. 2002;80(3):283–94. doi: 10.1006/geno.2002.6835. [DOI] [PubMed] [Google Scholar]
- 18.Popkie AP, Zeidner LC, Albrecht AM, et al. Phosphatidylinositol 3-kinase (PI3K) signaling via glycogen synthase kinase-3 (Gsk-3) regulates DNA methylation of imprinted loci. J Biol Chem. 2010;285(53):41337–47. doi: 10.1074/jbc.M110.170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishizuka A, Hasegawa Y, Ishida K, et al. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Genes Cells. 2014;19(9):704–21. doi: 10.1111/gtc.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun XY, Song H, Zhao L, et al. [A control study on altered microRNA expression in peripheral blood mononuclear cells from patients with schizophrenia]. Chinese Journal of Behavioral Medicine and Brain Science. 2013;22(12):1095–98. [in Chinese] [Google Scholar]
- 21.Song HT, Sun XY, Zhang L, et al. A preliminary analysis of association between the down-regulation of microRNA-181b expression and symptomatology improvement in schizophrenia patients before and after antipsychotic treatment. J Psychiatr Res. 2014;54:134–40. doi: 10.1016/j.jpsychires.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Creese I, Burt DR, Snyder SH. Dopamine receptor binding predicts clinical and pharmacological potencies of antischizophrenic drugs. Science. 1976;192(4238):481–83. doi: 10.1126/science.3854. [DOI] [PubMed] [Google Scholar]
- 23.Seeman P. Dopamine receptors and the dopamine hypothesis of schizophrenia. Synapse. 1987;1(2):133–52. doi: 10.1002/syn.890010203. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Luo ZH, Yang H. [Biological function of lncRNA and its effect on nervous system disease]. Journal of International Neurology and Neurosurgery. 2013;40(5):484–88. [in Chinese] [Google Scholar]
- 25.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6(5):539–43. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valadkhan S, Gunawardane LS. LncRNA-mediated regulation of the interferon response. Virus Res. 2016;212:127–36. doi: 10.1016/j.virusres.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gingrich JA, Caron MG. Recent advances in the molecular biology of dopamine receptors. Annu Rev Neurosci. 1993;16:299–321. doi: 10.1146/annurev.ne.16.030193.001503. [DOI] [PubMed] [Google Scholar]
- 28.Zawilska JB. [Dopamine receptors – structure, characterization and function]. Postepy Hig Med Dosw. 2003;57(3):293–322. [in Polish] [PubMed] [Google Scholar]
- 29.Neve KA, Seamans JK, Trantham-Davidson H. Dopamine receptor signaling. J Recept Signal Transduct Res. 2004;24(3):165–205. doi: 10.1081/rrs-200029981. [DOI] [PubMed] [Google Scholar]
- 30.Teicher MH, Glod CA, Surrey J, Swett C. Early childhood abuse and limbic system ratings in adult psychiatric outpatients. J Neuropsychiatry Clin Neurosci. 1993;5(3):301–6. doi: 10.1176/jnp.5.3.301. [DOI] [PubMed] [Google Scholar]
- 31.Laruelle M, Abi-Dargham A, van Dyck CH, et al. Single photon emission computerized tomography imaging of amphetamine-induced dopamine release in drug-free schizophrenic subjects. Proc Natl Acad Sci USA. 1996;93(17):9235–40. doi: 10.1073/pnas.93.17.9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S, Sun X, Niu W, et al. Aberrant expression of long non-coding RNAs in schizophrenia patients. Med Sci Monit. 2016;22:3340–51. doi: 10.12659/MSM.896927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13(2):261–76. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 34.Mikolajewski AJ, Scheeringa MS, Weems CF. Evaluating diagnostic and statistical manual of mental disorders, fifth edition posttraumatic stress disorder diagnostic criteria in older children and adolescents. J Child Adolesc Psychopharmacol. 2017;27(4):374–82. doi: 10.1089/cap.2016.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vats P, Juneja M, Mishra D. Diagnostic accuracy of international epidemiology network (INCLEN) diagnostic tool for autism spectrum disorder (INDT-ASD) in comparison with diagnostic and statistical manual of mental disorders-5 (DSM-5) Indian Pediatr. 2018;55(6):482–8. [PubMed] [Google Scholar]
- 36.Constantinos Z, Eleni P, Goulas A, Iakovidou-Kritsi Z. Olanzapine: Evaluation of the in vivo cytogenetic effect. Hum Psychopharmacol. 2015;30(3):189–92. doi: 10.1002/hup.2471. [DOI] [PubMed] [Google Scholar]
- 37.Domínguez E, Loza MI, Padín F, et al. Extensive linkage disequilibrium mapping at HTR2A and DRD3 for schizophrenia susceptibility genes in the Galician population. Schizophr Res. 2007;90(1–3):123–29. doi: 10.1016/j.schres.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 38.Zhang F, Fan H, Xu Y, et al. Converging evidence implicates the dopamine D3 receptor gene in vulnerability to schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(5):613–19. doi: 10.1002/ajmg.b.31203. [DOI] [PubMed] [Google Scholar]
- 39.Murray AM, Ryoo HL, Gurevich E, Joyce JN. Localization of dopamine D3 receptors to mesolimbic and D2 receptors to mesostriatal regions of human forebrain. Proc Natl Acad Sci USA. 1994;91(23):11271–75. doi: 10.1073/pnas.91.23.11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Missale C, Nash SR, Robinson SW, et al. Dopamine receptors: From structure to function. Physiol Rev. 1998;78(1):189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- 41.Mitelman SA, Buchsbaum MS, Christian BT, et al. Dopamine receptor density and white mater integrity: 18F-fallypride positron emission tomography and diffusion tensor imaging study in healthy and schizophrenia subjects. Brain Imaging Behav. 2018 doi: 10.1007/s11682-018-0012-0. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 42.Inada T, Sugita T, Dobashi I, et al. Dopamine D3 receptor gene polymorphism and the psychiatric symptoms seen in first-break schizophrenic patients. Psychiatr Genet. 1995;5(3):113–16. doi: 10.1097/00041444-199505030-00003. [DOI] [PubMed] [Google Scholar]
- 43.Beischlag TV, Nam D, Ulpian C, et al. A polymorphic dinucleotide repeat in the human dopamine D5 receptor gene promoter. Neurosci Lett. 1996;5(3):173–76. doi: 10.1016/0304-3940(96)12416-2. [DOI] [PubMed] [Google Scholar]
- 44.Hoenicka J, Aragüés M, Ponce G, et al. From dopaminergic genes to psychiatric disorders. Neurotox Res. 2007;11(1):61–72. doi: 10.1007/BF03033483. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Ding M, Pang H, et al. Relationship between genetic polymorphisms in the DRD5 gene and paranoid schizophrenia in northern Han Chinese. Genet Mol Res. 2014;13(1):1609–18. doi: 10.4238/2014.March.12.13. [DOI] [PubMed] [Google Scholar]
- 46.Williams NM, Cardno AG, Murphy KC, et al. Association between schizophrenia and a microsatellite polymorphism at the dopamine D5 receptor gene. Psychiatr Genet. 1997;7(2):83–85. doi: 10.1097/00041444-199722000-00005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Joint Co-expressed Coding Genes for 3 lncRNAs.
| EPHA3 | IL6R | EPHA4 | PPTC7 | PRELID1 | ANKRD36 | CT62 | MGEA5 |
| ZNF155 | SEL1L3 | MTRF1L | LOC153811 | REXO1L1 | TRERF1 | EIF4G3 | RASGRP3 |
| SH2D1A | FZD1 | LSM1 | TRPS1 | RD3 | RNF220 | ZNF550 | FAM196B |
| CD79A | IFT46 | SSB | GCNT2 | COL19A1 | PAK1 | RPL26 | IL1RAP |
| ARID5B | PCDHB4 | CTNNA1 | ASCC3 | BSDC1 | TRMT5 | ITGA1 | OXCT1 |
| KIAA0485 | DIAPH3 | psiTPTE22 | FLT3 | SIGLEC5 | UPF2 | MRPL9 | RPL35 |
| LOC100507650 | EYA1 | ROS1 | LEUTX | DOK7 | RIMS3 | MGC20647 | ANO1 |
| MAPK14 | GJB3 | MEX3D | TMEM164 | SLC9A1 | DISC1 | CCR2 | KCNE1 |
| LHFPL1 | GYLTL1B | CDCA7L | PPP2CB | RPL36A | OR2L3 | FCRL1 | LBH |
| HMGN2 | WDFY3 | LOC100505857 | HNRNPA1L2 | KCNJ15 | RPL37A | GPATCH3 | FAM129C |
| MCF2 | PET117 | MYL10 | PIK3C2B | RAB21 | FAM190B | RPS19 | FMO5 |
| AUTS2 |