Abstract
The effects of feeding behavior and diet composition, as well as their possible interactions, on daily (clock) gene expression rhythms have mainly been studied in the liver, and to a lesser degree in white adipose tissue (WAT), but hardly in other metabolic tissues such as skeletal muscle (SM) and brown adipose tissues (BAT). We therefore subjected male Wistar rats to a regular chow or free choice high-fat-high sugar (fcHFHS) diet in combination with time restricted feeding (TRF) to either the light or dark phase. In SM, all tested clock genes lost their rhythmic expression in the chow light fed group. In the fcHFHS light fed group rhythmic expression for some, but not all, clock genes was maintained, but shifted by several hours. In BAT the daily rhythmicity of clock genes was maintained for the light fed groups, but expression patterns were shifted as compared with ad libitum and dark fed groups, whilst the fcHFHS diet made the rhythmicity of clock genes become more pronounced. Most of the metabolic genes in BAT tissue tested did not show any rhythmic expression in either the chow or fcHFHS groups. In SM Pdk4 and Ucp3 were phase-shifted, but remained rhythmically expressed in the chow light fed groups. Rhythmic expression was lost for Ucp3 whilst on the fcHFHS diet during the light phase. In summary, both feeding at the wrong time of day and diet composition disturb the peripheral clocks in SM and BAT, but to different degrees and thereby result in a further desynchronization between metabolically active tissues such as SM, BAT, WAT and liver.
Keywords: Soleus muscle (SM), Brown adipose tissue (BAT), free choice High-fat High-sugar (fcHFHS), Time-restricted feeding (TRF), desynchronization
Highlights
-
•
Both timing of feeding and diet composition affect clock genes in BAT and SM.
-
•
Light phase time-restricted feeding abolishes SM clock gene rhythms.
-
•
A fcHSHS diet strengthens rhythmic expression of several clock genes in BAT and SM.
-
•
Metabolic genes PDK4 and UCP1/3 are affected by both timing of feeding and diet.
-
•
Light phase time-restricted feeding causes desynchronization of BAT and SM clocks.
1. Introduction
Many studies support the idea that both food consumption and energy metabolism are under strong influence of the biological clock (Bray and Young, 2009, Summa and Turek, 2014). It is therefore not surprising that recent epidemiological studies have found a correlation between conditions that disturb the biological clock, such as shift work, and metabolic diseases, such as obesity and type 2 diabetes mellitus (T2DM). The molecular mechanism of the biological clock is made up of a transcriptional-translational feedback loop consisting of various clock genes, such as Clock, Bmal1, Per1/2/3, Cry1/2, Rev-erbα and clock controlled genes (CCGs). CLOCK and BMAL1 are part of the core clock mechanism and form the positive limb through hetero-dimerization. The Per and Cry genes form the negative limb of the core clock mechanism and bind over the promoter regions of Bmal1 and Clock genes (Gekakis et al., 1998, Hogenesch and Hahn, 1998, Ohno et al., 2007, Yoo et al., 2005). When the PER and CRY proteins are present at sufficiently high levels in the cytoplasm they translocate back to the nucleus to inhibit their own transcription (Ramsey et al., 2007). REV-ERBα/β and RORα/β show competitive binding to promoters of Bmal1 and Clock with binding of Reverbα/β inhibiting and binding of Rorα/β promoting the transcription of Bmal1 and Clock. The transcriptional-translational feedback loop is set to revolve roughly every 24 hours (i.e., with a circadian period), but can be adjusted and synchronized by several environmental cues, so called Zeitgebers. In mammals the master, or central, clock is located in the suprachiasmatic nucleus (SCN) in the hypothalamus and is mainly synchronized by the environmental light/dark cycle. The strongest known Zeitgeber for peripheral clocks such as those in liver, white and brown adipose tissues and skeletal muscle is food or energy availability (Froy, 2010).
The biological clocks use CCGs as an output mechanism to regulate a broad range of processes, including many metabolic processes. A number of CCGs are metabolic genes that are involved in lipogenesis, fatty-acid oxidation and glucose metabolism (e.g. Pparα, Pgc-1α, Srebp1c, several glucose transporters, Fas and Lpl and many more). Strikingly, the exact effects of the biological clocks on these metabolic processes differ in a tissue-dependent manner (Marcheva et al., 2013, Eckel-Mahan and Sassone-Corsi, 2013), indicating the importance of timely orchestrated metabolic processes, both within and between different tissues. As a result, there is a need to investigate the interplay between the biological clocks and metabolic processes for the major tissue types involved in energy metabolism. Thus, most studies concentrated on the liver and, to a lesser extent, on WAT. Two peripheral tissues that until recently have often been overlooked in studies on circadian rhythms and metabolism are brown adipose tissue (BAT) and skeletal muscle (SM), despite their clear importance for whole body energy metabolism. SM is the organ with the highest overall metabolic rate (Wang et al., 2010) and is important for glucose homeostasis. SM alone is responsible for 60–80% of insulin-mediated glucose uptake (de Lange et al., 2007, Wilcox, 2005) and 80% of postprandial glucose uptake (DeFronzo et al., 1981, DeFronzo et al., 1985, Ferrannini et al., 1988, Shulman et al., 1990). SM is also responsible for a major proportion of fatty acid oxidation and the ability to oxidize this metabolic substrate is reduced in obese and T2DM patients (Berggren et al., 2008, Mensink et al., 2001). SM genes involved in carbohydrate catabolism show peak expression early in the active phase, whilst genes involved in the storage of carbohydrate substrates peak in the middle of the active phase. Conversely, genes involved in lipid metabolism peak in the middle of the inactive phase, whilst genes involved in lipogenesis and storage of lipids peak at the end of the active phase (Hodge et al., 2015). More interestingly, the 7 highest enriched gene ontology sets of mRNA found to be oscillating with a 24-h periodicity in SM, were all involved in the regulation of metabolic processes. Combined, these metabolic transcripts represented approximately 62% of the circadian transcriptome of mouse SM (Hodge et al., 2015).
BAT is a metabolically highly active tissue important for heat production. Activation of BAT for thermogenesis results in increased energy expenditure via the uncoupling protein UCP1. BAT maintains thermogenesis through oxidation of lipids and glucose and its activation results in oxidative phosphorylation as well as heat production (Bartelt et al., 2011, Cannon and Nedergaard, 2004, Mulya and Kirwan, 2016, Stanford and Goodyear, 2013). BAT has been long known to be activated by various high-calorie diets, such as high-fat and high-sucrose diets, likely through the increased UCP1 levels seen during these diets, thereby providing a potential mechanism to limit weight/fat gain (Bukowiecki et al., 1983, LeBlanc and Labrie, 1997, Mercer and Trayhurn, 1987, Rothwell and Stock, 1979).
The catabolism and storage of different substrates (i.e., carbohydrates and lipids) in metabolically active tissues is thus regulated in a time-dependent manner, which coincides with the natural daily rhythm of food intake during the active phase and resting during the inactive phase. Disturbing this biological rhythm of feeding behavior by restricting access to food to the inactive phase is a widely accepted animal model for shift-work in humans (Opperhuizen et al., 2015). Several studies, including from our own group, have investigated the effects of time-restricted feeding (TRF) (Dyar et al., 2015, Hatori et al., 2012, Oosterman et al., 2015, Opperhuizen et al., 2016, Reznick et al., 2013, Salgado-Delgado et al., 2010, Vollmers et al., 2009, Yasumoto et al., 2016, Zarrinpar et al., 2014), some of these studies even compared different diets in combination with TRF (Hatori et al., 2012, Oosterman et al., 2015, Reznick et al., 2013).
Earlier we found that the combination of eating at the wrong time-of-day and diet composition (i.e., with a high-fat or high-sugar content) affects substrate metabolism on a whole body level. However, the independent contributions of TRF and diet composition could not be established in that study (Oosterman et al., 2015). Here we show the effects of different combinations of TRF and diet composition in male Wistar rats, both on a whole body level as well as in two peripheral organs: SM and BAT. We focused on these two metabolically active tissues since they are critical for glucose and lipid metabolism and they have not been investigated as thoroughly as other tissues (e.g. liver and WAT). In these tissues, we specifically targeted genes of the core clock mechanism and genes involved in glucose and lipid metabolism. The present study shows that expression patterns of the BAT and SM molecular clocks, as well as several metabolic genes, are clearly affected by changes in the daily timing of food intake as well as by diet composition.
2. Materials and methods
2.1. Animal experiments
2.1.1. Influence of diet composition and TRF
One hundred and ninety three male Wistar rats were housed under 12:12 light:dark conditions for the entire experiment, with Zeitgeber Time (ZT) 0 being the time of lights on and ZT12 the time of lights off. The animals were divided over 5 different batches. Animals were randomly assigned to either a standard chow or free-choice high-fat high-sugar (fcHFHS) diet group and to one of the TRF groups: ad libitum, Dark or Light.
The fcHFHS diet animals could freely choose between pelleted chow, a bottle of tap water, a bottle with a 30% sugar solution (Kristalsuiker; Van Gilse) and a dish with saturated fat (Ossewit Blanc de Boeuf; Vandemoortele Lipids NV). The chow diet animals had access to pelleted chow and tap water only. The ad libitum group had free access to food and water for 24 h/day, the Dark and Light TRF groups had free access to food for 10 h/day between ZT 13–23 and between ZT1-11, respectively. After 3 weeks of diet and TRF, a randomized subset of 63 animals was placed in metabolic cages for 4 days to measure the respiratory exchange ratio, locomotor activity and heat production whilst remaining on their assigned diet and TRF conditions. After 5 weeks of diet and TRF conditions animals were sacrificed at 3 hour intervals throughout a 24 hour period (at ZT 0, 3, 6, 9, 12, 15, 18, 21) and soleus muscle and BAT tissues were carefully collected, snap frozen in liquid nitrogen and stored at -80 °C until RNA isolation was performed.
2.2. Activity and respirometry
Metabolic PhenoCages (TSE systems) were used to measure several metabolic parameters, whilst animals remained on their diet and TRF conditions. Animals were individually housed in these cages. After a day of acclimatization to this new environment, the parameters for food intake, locomotor activity, respiratory exchange ratio (RER) and heat production were measured for three consecutive days (72 hours).
2.3. RNA isolation
Soleus muscle tissue was mechanically homogenized while kept on dry ice. The BAT tissue was crushed in Trizol by using the homogenizer machine. For both tissues, RNA isolation was done using the NucleoSpin RNA isolation kit (Machery-Nagel). For muscle RNA isolation, three additional washing steps with 75% ethanol were performed. RNA was eluted from the spin column using 40 μl of H2O and RNA concentration and quality of the RNA were determined using a DS-11 (DeNovix) spectrophotometer and a nanochip using Agilent 2100 Bioanalyzer (Agilent Technologies), respectively. Although RNA integrity number (RIN) values above 5 were considered acceptable, all samples had a RIN above 8.
2.4. Muscle and BAT cDNA synthesis
Two hundred ng from muscle and 350 ng from BAT isolated RNA were used as input template for cDNA synthesis. The Transcriptor First Strand cDNA synthesis kit (Roche) was used. RT-PCRs were run using an UNO-Thermoblock (Biometra).
2.5. RT-qPCR
One to nineteen (1:19) diluted cDNA was used for all qPCRs to detect muscle and BAT gene expression profiles. Expression levels of all genes were standardized by dividing over the geometric mean of three housekeeping genes: TBP, GAPDH and Cyclophilin for muscle; TBP, HPRT1 and GAPDH for BAT. RT-qPCR was performed using a LightCycler 480 (Roche). Expression levels were calculated using dedicated software for linear regression of qPCR data (LinRegPCR). All used primers are listed in Table S4. Melting curves of the RT-qPCR and fragment length of the DNA amplicons were inspected as a means of quality control.
3. Statistics
Rhythmicity of gene expression profiles was determined by the non-parametric algorithm JTK_CYCLE version 3.1 which was run under R version 3.3.1. T-tests, one-way and two-way ANOVAs, as well as Tukey’s Multiple Comparison post-hoc tests were executed by GraphPad Prism 7. All graphs were plotted by GraphPad Prism 7.
4. Results
A detailed description of the physiological and metabolic results from the metabolic cages is provided elsewhere (Oosterman et al., Submitted), below is a short description of the most important results.
4.1. Caloric intake and body weight gain
Caloric intake was not different between the TRF groups, but caloric intake was about 20% higher for the fcHFHS animals as compared to chow fed groups (two-way ANOVA: Diet p < 0.001, TRF p > 0.05, Diet*TRF p > 0.05). Similar to previous experiments animals on the fcHFHS diet consumed about 37.5% of their calories from fat, 15% from sugar and 47.5% from chow (la Fleur et al., 2007). Similarly, body weight gain after 5 weeks was not different between the TRF groups, but body weight gain was higher for the fcHFHS fed animals as compared to chow fed groups (105 g and 90 g, respectively; two-way ANOVA: Diet p < 0.001, TRF p > 0.05, Diet*TRF p > 0.05).
4.2. Respiratory exchange ratio (RER)
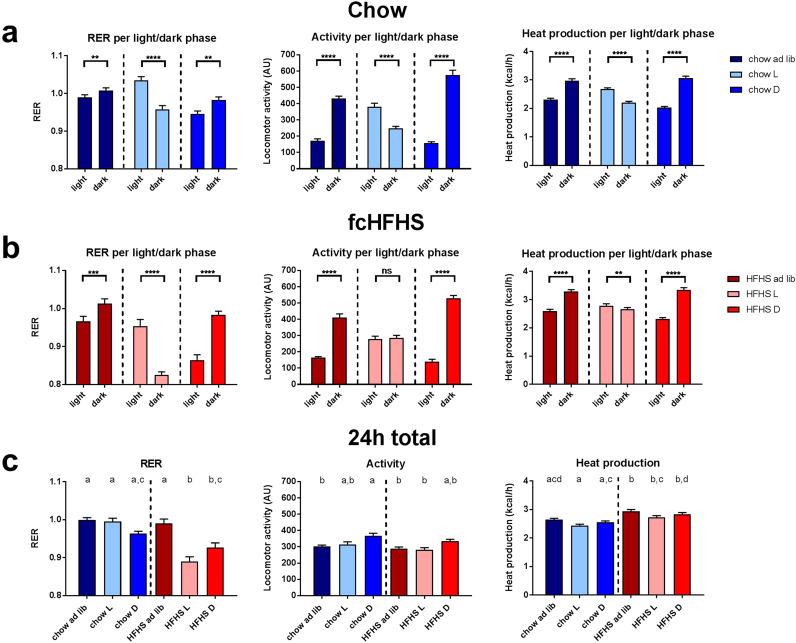
Animals fed ad libitum showed a clear day/night rhythm in their RER, with highest levels found during the active phase for both the chow and fcHFHS fed groups (Fig. 1a & b). TRF to either the dark or light phase greatly increased the amplitude of the RER for both diet groups. The RER for groups restricted to feeding during the light phase was strikingly anti-phasic as compared to both the ad libitum and dark fed groups. The L/D difference in RER of all groups clearly follows the daily feeding pattern, with highest RER levels being reached during the feeding period, independent of the time of day (Fig. 1a & b). Analysis of the average RER values per 24 hour period (Fig. 1c) revealed significant differences between the different diet and TRF conditions (two-way ANOVA: Diet p < 0.001, TRF p < 0.001, Diet*TRF p < 0.001). Closer inspection of the individual diet and TRF combinations revealed that the different chow fed groups did not significantly differ in their average 24 h RER, but that the RER of both the light and dark fed groups on the fcHFHS diet was significantly lower than that of the ad libitum group on a fcHFHS diet as well as that of the chow ad libitum and light fed groups (one-way ANOVA, p < 0.0001) (Fig. 1c). Aside from this, the RER of the light fed fcHFHS group was also significantly lower than the RER of the dark fed chow group (p < 0.0001).
Fig. 1.
Analysis of the metabolic parameters RER (left), locomotor activity (middle) and heat production (right) of the animals inside the metabolic cages during TRF. Whilst in the metabolic cages animals remained on their assigned diet composition and TRF conditions. (a) Difference within metabolic parameters between light and dark phase for the chow fed groups. (b) Difference within metabolic parameters between light and dark phase for the fcHFHS fed groups. (c) Average 24 hour values of the metabolic parameters for all diet composition and TRF groups. Data are depicted as means ± SEM. ns = non significant, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001, n = 10–11 per group. Identical letters indicate similar mean values, Tukey’s Multiple Comparison post-hoc test was performed to correct for multiple testing. Locomotor activity is presented as arbitrary units (AU). ad lib = ad libitum fed animals, L = light fed animals, D = dark fed animals.
4.3. Locomotor activity
Animals fed ad libitum showed a clear day/night rhythm in their locomotor activity with most activity occurring during the dark phase (71% of total activity; Fig. 1a & b). During TRF in the dark phase this L/D difference in locomotor activity is strengthened due to the increased activity during the dark phase (79% of total activity; Fig. 1a & b). Diet composition does not seem to affect the locomotor activity for ad libitum and dark fed groups (71% and 79% of total activity during the dark phase, respectively). TRF to the light phase, however, does alter the daily pattern of locomotor activity. Animals fed chow during the light phase showed an inverted activity pattern, with most locomotor activity during the light phase (61% of total activity), i.e., in their feeding period (Fig. 1a). Interestingly, light fed animals on a fcHFHS diet lost the day/night rhythm in locomotor activity and showed equal activity during the light (49% activity) and dark period (51% activity; Fig. 1b). Analysis of the total locomotor activity per 24 hour period (Fig. 1c) revealed significant differences between the different diet and TRF conditions (two-way ANOVA: Diet p < 0.018, TRF p < 0.0005, Diet*TRF p = 0.721). Total locomotor activity for the combination of chow diet and TRF to the dark phase was significantly higher compared to the chow ad libitum, fcHFHS ad libitum and fcHFHS light fed groups, but no other diet or TRF combination differed (one-way ANOVA, p = 0.0002).
4.4. Heat production
Similar to locomotor activity, heat production was highest during the feeding phase for all groups, including the light fed groups (Fig. 1a & b). Akin to the locomotor activity data the difference between the light and dark period in heat production was largest in the dark fed animals (Fig. 1a & b). Two-way ANOVA showed significant effects of both diet composition and TRF on mean heat production per 24 hours (two-way ANOVA: Diet p < 0.0005, TRF p = 0.001, Diet*TRF p = 0.985) (Fig. 1c). Specifically, heat production was lowest in the chow light group and differed significantly from all fcHFHS groups, chow ad libitum differed from fcHFHS ad libitum, and chow dark differed from fcHFHS ad libitum and fcHFHS dark. Interestingly, this result seems to be caused primarily by diet composition and not by TRF, since the three fcHFHS groups did not differ from each other, nor did the 3 chow groups differ from each other, contrasting the results from the two-way ANOVA (Fig. 1c).
4.5. Clock gene expression in soleus muscle and BAT
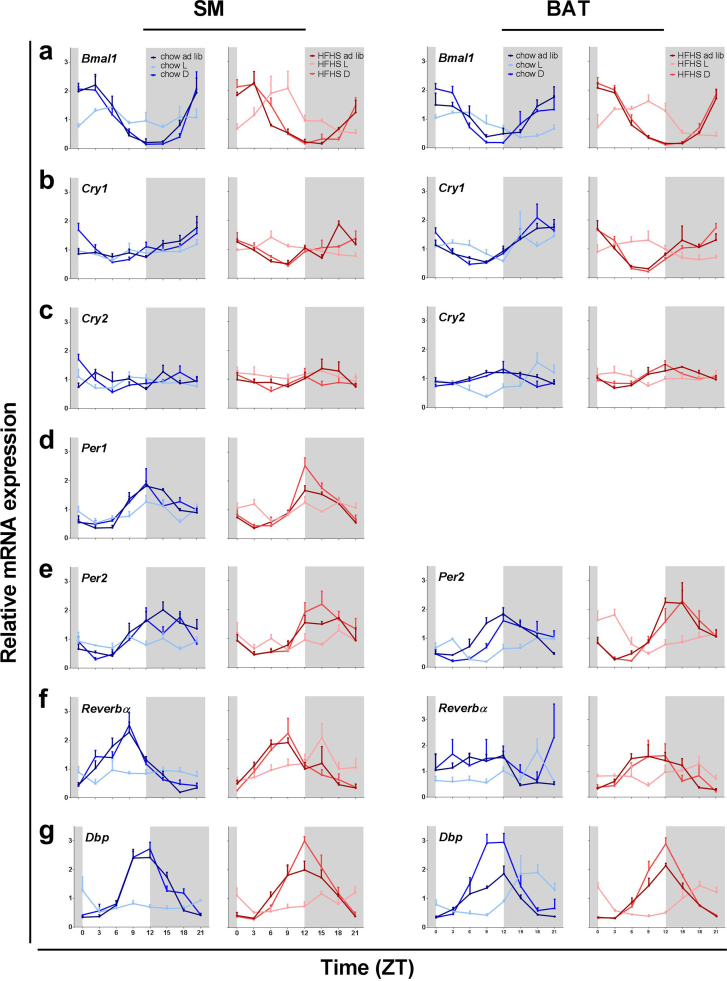
Gene expression analysis using qPCR and JTK_CYCLE analysis confirmed rhythmicity of six of the seven clock genes investigated in both soleus muscle (Bmal1, Cry1, Per1, Per2, Dbp and Rev-erbα) (Fig. 2, Table 1) and BAT (Bmal1, Cry1, Cry2, Per2, Dbp and Rev-erbα) (Fig. 2, Table 2). TRF to the dark phase did not alter the expression patterns of these core clock genes in either tissue type, although it did induce slight phase-shifts for some of the clock genes (Table 1, Table 2). These results were similar for animals on a chow and a fcHFHS diet, although in BAT the fcHFHS diet enhanced the rhythmicity of some of the core clock genes (Fig. 2a,b,c,e & f). The amplitude of the expression rhythm tended to be enlarged for Bmal1, Per2, Cry1, Cry2, Rev-erbα, although this never reached significance (Table S3).
Fig. 2.
Effect of diet composition and TRF on expression profiles of clock genes (a-f) and clock controlled gene Dbp (g) in SM and BAT tissues. Expression profiles are presented as means ± SEM. Tissues were collected at 8 different time points across 24 hours. Shaded areas represent the dark phase.
Table 1.
Effects of diet and timing of food intake on daily rhythms in clock and metabolic gene expression in SM. Data was analyzed by JTK Cycle. The acrophase (in ZT) is only given for genes that are rhythmically expressed (p < 0.05). NR = non-rhythmic.
|
JTK cycle analysis for Muscle |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes Muscle |
CHOW |
HFHS |
|||||||||||
| Clock | Acrophase |
P-value |
Acrophase |
P-value |
|||||||||
| Ad lib | D | L | Ad lib | D | L | Ad lib | D | L | Ad lib | D | L | ||
| Bmal1 | 1.5 | 3 | NR | 0.001 | 0.001 | 1 | 3 | 3 | 9 | 0.001 | 0.001 | 0.001 | |
| Per1 | 13.5 | 15 | NR | 0.001 | 0.001 | 0.420 | 15 | 15 | NR | 0.001 | 0.001 | 0.170 | |
| Per2 | 16.5 | 16.5 | NR | 0.001 | 0.001 | 1 | 16.5 | 16.5 | NR | 0.003 | 0.001 | 1 | |
| Rev-erbα | 9 | 9 | NR | 0.001 | 0.001 | 1 | 9 | 9 | 15 | 0.001 | 0.001 | 0.001 | |
| Cry1 | 21 | 22.5 | NR | 0.005 | 0.001 | 0.380 | 19.5 | 22.5 | 7.5 | 0.001 | 0.005 | 0.001 | |
| Cry2 | NR | NR | NR | 1 | 0.110 | 1 | NR | NR | NR | 1 | 1 | 1 | |
| DBP | 12 | 13.5 | NR | 0.001 | 0.001 | 1 | 13.5 | 13.5 | NR | 0.001 | 0.001 | 0.078 | |
| Metabolic | |||||||||||||
| Srebp-1c | NR | 0 | 10.5 | 0.230 | 0.006 | 0.004 | NR | NR | NR | 0.290 | 0.078 | 0.078 | |
| Glut4 | NR | NR | NR | 1 | 0.140 | 1 | NR | NR | NR | 1 | 0.380 | 1 | |
| Ucp3 | 4.5 | 4.5 | 21 | 0.001 | 0.001 | 0.001 | 4.5 | 6 | NR | 0.001 | 0.001 | 0.450 | |
| PDK4 | 4.5 | 6 | 21 | 0.001 | 0.001 | 0.001 | 4.5 | 6 | 22.5 | 0.016 | 0.001 | 0.035 | |
| Pgc1α | NR | NR | NR | 1 | 1 | 0.38 | NR | NR | NR | 0.890 | 0.230 | 1 | |
| Pparα | NR | NR | NR | 1 | 0.980 | 1 | NR | NR | NR | 1 | 1 | 1 | |
| Fas | NR | 19.5 | NR | 0.210 | 0.049 | 0.300 | NR | NR | NR | 0.24 | 0.083 | 0.086 | |
Table 2.
Effects of diet and timing of food intake on daily rhythms in clock and metabolic gene expression in BAT. Data was analyzed by JTK Cycle. The acrophase (in ZT) is only given for genes that are rhythmically expressed (p < 0.05). NR = non-rhythmic.
|
JTK cycle analysis for BAT |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes BAT |
CHOW |
HFHS |
||||||||||
| Clock | Acrophase |
P-value |
Acrophase |
P-value |
||||||||
| Ad lib | D | L | Ad lib | D | L | Ad lib | D | L | Ad lib | D | L | |
| Bmal1 | 22.5 | 0 | 4.5 | 0.001 | 0.001 | 0.001 | 1.5 | 1.5 | 9 | 0.001 | 0.001 | 0.001 |
| Per2 | 13.5 | 16.5 | 21 | 0.001 | 0.001 | 0.001 | 16.5 | 16.5 | 0 | 0.001 | 0.001 | 0.001 |
| Rev-erbα | 9 | NR | NR | 0.001 | 1 | 0.092 | 10.5 | 10.5 | NR | 0.001 | 0.001 | 0.133 |
| Cry1 | 21 | 21 | 0 | 0.001 | 0.001 | 0.005 | 21 | 22.5 | 7.5 | 0.001 | 0.001 | 0.005 |
| Cry2 | 12 | 12 | 21 | 0.001 | 0.034 | 0.001 | 16.5 | 13.5 | NR | 0.001 | 0.001 | 1 |
| DBP | 12 | 12 | 19.5 | 0.001 | 0.001 | 0.001 | 13.5 | 13.5 | 21 | 0.001 | 0.001 | 0.001 |
| Metabolic | ||||||||||||
| Srebp-1c | 10.5 | NR | NR | 0.024 | 1 | 0.071 | NR | 13.5 | NR | 0.090 | 0.013 | 1 |
| Glut4 | NR | NR | NR | 1 | 0.432 | 0.189 | NR | 3 | NR | 1 | 0.007 | 1 |
| Ucp1 | NR | NR | NR | 1 | 1 | 1 | NR | 7.5 | NR | 0.736 | 0.023 | 0.736 |
| Pgc1α | 9 | 10.5 | NR | 0.001 | 0.017 | 0.117 | NR | NR | NR | 1 | 0.083 | 0.844 |
| Pparα | NR | 10.5 | NR | 1 | 0.016 | 1 | NR | 12 | NR | 0.429 | 0.004 | 0.376 |
| Fas | NR | NR | NR | 0.950 | 1 | 1 | NR | NR | NR | 1 | 1 | 1 |
| LPL | NR | NR | 0 | 0.264 | 0.07 | 0.002 | NR | NR | NR | 1 | 1 | 0.488 |
| HSL | 9 | NR | 22.5 | 0.036 | 0.253 | 0.001 | NR | NR | NR | 1 | 1 | 0.736 |
In contrast, TRF to the light phase whilst on a chow diet completely abolished the rhythmicity of all 6 rhythmic clock genes in the soleus muscle. Contrasting, in BAT the clock genes still displayed rhythmicity, although with a somewhat altered pattern of expression as compared to that of the ad libitum and dark fed animals. When on a fcHFHS diet, rhythmicity for several core clock genes in the soleus muscle was rescued from the dampening effect of a TRF to the light phase, as seen in animals on a chow diet. In BAT, TRF to the light phase showed similar effects in the fcHFHS and chow groups.
4.6. Metabolic gene expression in soleus muscle and BAT
4.6.1. Soleus muscle
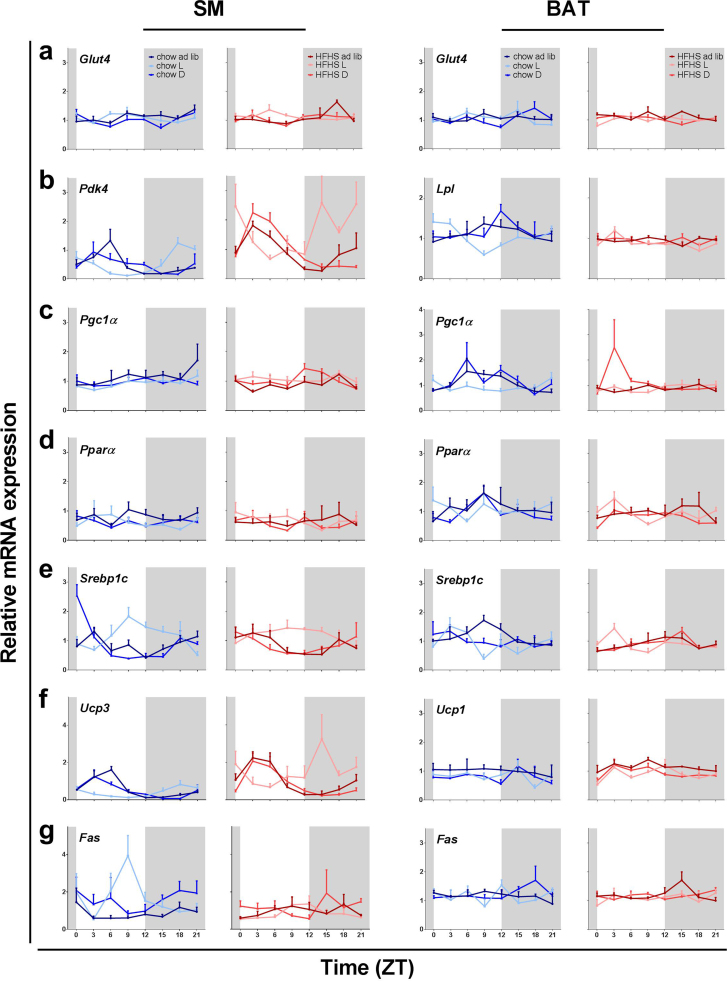
Most of the studied metabolic genes in muscle do not show rhythmicity under ad libitum conditions whether fed with chow or fcHFHS diet, except for substrate switch pyruvate dehydrogenase kinase (Pdk4) and the most abundant uncoupling protein in skeletal muscle uncoupling protein 3 (Ucp3) (p < 0.001 for both genes) Fatty acid synthase (Fas) was only rhythmically expressed in the dark fed animals on a chow diet (p = 0.049). When fed during the light phase, both Pdk4, and Ucp3 show phase shifts of 7.5 hours as compared to the dark and ad libitum groups, with the exception of Ucp3 for the fcHFHS group that was fed during the light period which was not rhythmically expressed (p = 0.45). Interestingly, Srebp1c expression became rhythmic when animals on a chow diet were subjected to TRF to either dark or light phase (Fig. 3e), whilst the ad libitum group did not display rhythmic expression for this gene (p < 0.005 for both dark and light phase TRF; acrophase at ZT = 0 and ZT = 10.5, respectively). None of the fcHFHS groups displayed significant rhythmic expression of Srebp1c. The insulin sensitive glucose transporter Glut4 and the transcription factors Pgc-1α and Pparα were not rhythmically expressed in any of the groups (Fig. 3a, c & d). Both Pdk4 and Ucp3 showed a main effect of diet composition for all three TRF groups, due to the higher expression levels in the fcHFHS groups as compared to the chow groups (two way ANOVA, Table S1). Additionally, Pgc-1α had a main effect of diet composition for the ad libitum fed groups, with higher levels for the chow fed group, and for Glut4 an interaction between diet composition and time was found (p = 0.011 and p = 0.006 respectively, two Way ANOVA, Table S1). For the light fed groups there was also a main effect of diet composition for Fas expression, with higher levels for the chow fed group (two way ANOVA, Table S1).
Fig. 3.
Effect of diet composition and TRF on expression profiles of genes involved in glucose and lipid metabolism (a-g) in SM and BAT tissues. Expression profiles are presented as means ± SEM. Tissues were collected at 8 different time points across 24 hours. Shaded areas represent the dark phase.
4.6.2. Brown adipose tissue
Similar to the soleus muscle most of the studied metabolic genes in BAT did not show rhythmicity in both chow and fcHFHS under ad libitum feeding conditions. In contrast to soleus muscle Hsl (Fig not shown), Srebp1c and Pgc-1α expression in BAT showed rhythmicity under ad libitum conditions (p = 0.036, p = 0.024 and p = 0.001, respectively) (Figs. 3e & 3c respectively, Table 2). Srebp1c lost rhythmicity in both light and dark chow fed condition while Pgc-1α lost rhythmicity in the chow light fed group (Fig. 3c). Under TRF to either dark or light phase a few genes gained rhythmicity, such as Pparα in both chow and fcHFHS dark fed groups (p = 0.016 and p = 0.004). Similarly Lpl and Hsl (p = 0.002 and p = 0.001) both show rhythmicity in the light fed chow group, where the acrophase of Hsl is shifted by almost 12 hours. Additionally some genes upon dark feeding with the fcHFHS diet gained rhythmicity, such as Srebp1c, Glut4 and Ucp1 (p = 0.013, p = 0.007 and p = 0.023, respectively). A significant effect of diet composition as well as a significant interaction between diet composition and time were found for Pgc-1α in the ad libitum fed groups (Diet p = 0.028 and Diet*Time p = 0.011 two way ANOVA Table S2) and for Pparα dark fed groups (Diet p = 0.016 and Diet*Time p < 0.001, two way ANOVA, Table S2). Glut4 showed a significant interaction between diet composition and time for the dark fed groups (p < 0.001, two way ANOVA Table S2).
5. Discussion
Many recent studies have investigated the effects of disturbed rhythms on energy metabolism by focusing on (clock) gene expression in liver and WAT (Hatori et al., 2012, Reznick et al., 2013, Salgado-Delgado et al., 2010). Here we investigated the effects of different TRF paradigms as well as diet composition on (clock) gene expression rhythms in the soleus SM and BAT, two tissue types also important for energy metabolism but often overlooked. Both TRF and diet composition affected daily rhythms in energy metabolism, at the whole body level as well as at the tissue level (SM and BAT). Daily clock gene expression patterns in BAT and SM tissue were strongly affected by TRF and to a lesser extent by diet composition, with clock gene rhythms in SM being completely abolished by daytime TRF whilst shifted in BAT.
5.1. Altered feeding behavior leads to desynchrony within and between peripheral clocks, whilst diet composition mainly affects whole body metabolism
The effects of TRF and diet composition on mRNA expression patterns were different between SM and BAT, clearly indicating that these tissues are differently regulated by the same Zeitgebers, feeding behavior and diet composition in this case. These results are in line with previous TRF experiments in rats, as it was shown that TRF has different effects on clock gene rhythms in muscle compared to those in liver (Opperhuizen et al., 2016, Reznick et al., 2013). In the liver, daytime TRF shifts most clock genes by approximately 12 hours (Damiola et al., 2000, Salgado-Delgado et al., 2013, Yamajuku et al., 2009). Clock gene expression rhythms in muscle on the other hand are mostly obliterated by daytime TRF (Opperhuizen et al., 2016, Reznick et al., 2013). Here we confirm the disruptive effects of daytime TRF on muscle clock gene rhythms. On the other hand, clock gene expression rhythms in BAT remained rhythmic (although with a ~12 h shift) upon daytime TRF as was also shown in mice (Hatori et al., 2012, Zvonic et al., 2006), further adding to the notion that different tissue types are regulated differently by the same Zeitgeber.
Clock gene rhythms in BAT became more pronounced with the fcHFHS diet, which correlates to the larger L/D difference in RER seen in the fcHFHS-fed groups. Consuming the fcHFHS diet also seemed to strengthen the rhythm of several clock genes in SM, in a gene- and TRF-dependent manner. For example, in the light fed group on a fcHFHS diet, Bmal1, Cry1 and Rev-erbα remained rhythmic in contrast to those in the chow fed group. This finding implicates that not all components of the molecular clock are regulated similarly within the same tissue.
5.2. Different Zeitgebers in skeletal muscle tissue
The above data show that metabolic genes in muscle react to changes in feeding behavior to a similar extent as metabolic genes in BAT, but that clock genes in SM clearly are differently affected as compared to BAT. These data again indicated that metabolic genes seem to be controlled more by behavior and hormonal rhythms than the local tissue clock (Su et al., 2016). It seems plausible that different Zeitgebers affect clock gene expression in a tissue-specific manner. Locomotor activity and exercise have previously been shown to be important Zeitgebers for SM (Dyar et al., 2015). In our study TRF to the light/inactive phase not only changed the timing of food intake, but also caused clear changes in locomotor activity patterns. Therefore, clock gene rhythms in SM could be adjusted less by feeding behavior and putatively more by energy use, e.g., locomotor activity. Of note, although locomotor activity was highest during the light phase, the day/night difference was dampened in the light fed group. This might explain why in SM, unlike in tissues such as liver and BAT, the clock gene rhythms were not inverted by TRF to the light phase. Additionally, entrainment by the SCN or the endogenous peripheral oscillators present in virtually all cells might be differently regulated in different tissues (Yamazaki et al., 2000).
Previous experiments by our group have found results consistent with the present study. In an experimental set-up similar to the present study, but with chow fed animals only, a similar loss of rhythmicity of core clock genes in SM was found for animals on a TRF regimen (Opperhuizen et al., 2016). Notably, the muscle examined in Opperhuizen et al. was a different muscle (a mixture of hind leg muscles as opposed to our isolated soleus muscle), indicating that the effects found here are likely not muscle-type specific. A similar result was found in mice in which expression of core clock genes in two different muscle types were directly compared. Expression patterns of core clock genes in the fast tibialis anterior and slow soleus muscle were found to be essentially identical (Dyar et al., 2015). Dyar et al. describe that in both fast tibialis anterior and slow soleus muscles, TRF to the inactive phase shifted the expression peak phase of core genes Bmal1, Per1, and Per2 by around 12 h in mice. On the other hand, they report that denervation of the hind limb by sciatic nerve lesions caused relatively minor changes in the expression patterns of most core clock genes, showing that clock gene rhythms are not solely affected by muscle activity. In another study in mice gastrocnemius muscle TRF to the light phase eliminated the rhythm in Per2 expression, but not in other genes, although the amplitude of expression of several clock genes was dampened and only small shifts in acrophase were found (3.46 ± 1.41 hours compared to dark fed animals) (Bray et al., 2013). In their study locomotor activity was found to be mainly nocturnal, in contrast to our experiment. It might well be possible that this persistence of a nocturnal activity pattern acts as a mechanism of retained rhythmic expression in most SM clock genes. Unfortunately, locomotor activity was not reported in the experiments by Dyar et al.
Another explanation for the differing results between our study and both Dyar et al. and Bray et al. might be that the duration of the TRF treatment was shorter in those studies, lasting for either 9 days (Bray et al.) or 2 weeks (Dyar et al.) versus 5 weeks in our study. If the arrhythmicity in clock gene expression establishes only after a more prolonged period of TRF, this could also explain the discrepancy between the Dyar and Bray studies. Additionally, in our experimental protocol, the TRF animals only had access to food for 10 hours per day, whilst in both Bray et al. and Dyar et al. the animals had access to food for 12 hours a day. Duration of the fasting period is important as most improvements in metabolic health in rodent studies are seen when food access is limited to 8–12 hours a day (Chaix et al., 2014, Longo and Panda, 2016). Another explanation of the differing results might be a difference between species. In another rat study, chow TRF to the light phase for 3 weeks also resulted in a diminished amplitude of the core clock genes Bmal1 (rhythm lost) and Dbp (still rhythmic) in SM (Reznick et al., 2013). Opposing our activity results, in this study rats maintained a clear nocturnal pattern of activity, albeit with a slight dampening in the normal difference between the light and dark phase, which might explain the differing results in changes in Dbp expression. Interestingly, in the Reznick et al. (2013) study, rats on a HF diet were also subjected to TRF to the light phase. Whilst on a HF diet, the effects of TRF to the light phase on SM core clock expression were less pronounced than whilst on a chow diet. This suggests that increased lipid metabolism resulting from a HF diet in muscles attenuates or rescues the effects of activity at the wrong time of day on the muscle molecular clock and is in line with our results using the fcHFHS diet.
Future experiments that can distinguish between the effects of feeding behavior and locomotor activity are needed to reveal the separate contributions of feeding and locomotor activity to the muscle peripheral clock. For example, experiments including a forced mild exercise paradigm could provide more insight into these matters.
5.3. Clinical relevance of diet composition and TRF interventions
Total food intake and timing of food intake in relation to TRF and shiftwork have been extensively studied, both in humans and animal models.
Several lines of evidence suggest that reinforcing behavioral rhythms, such as through rigid schedules of sleep, exercise or TRF, has beneficial effects on longevity and several parameters of health, including lowered risk of obesity and T2DM (e.g. (Manoogian and Panda, 2016). The exact mechanism of these effects remains to be elucidated, but a prominent role for the circadian clock system can be expected. With respect to the present TRF experiment, it is interesting to note that in both the chow and fcHFHS fed animals, TRF to the dark phase resulted in a more pronounced difference between daytime and nighttime locomotor activity, indicating a less fragmented rhythm. Similar results were seen for RER and heat production, where TRF to the dark phase caused a more pronounced difference between daytime and nighttime RER and heat production, both whilst on the chow and fcHFHS diet. It therefore is tempting to speculate that the beneficial effects that are seen during TRF in animal models result from a more clear distinction between rest and active phase on several physiological parameters such as feeding behavior, metabolism and activity and that this beneficial effect of TRF could also protect against the consequences of an “unhealthy” hypercaloric diet. However, the effects seen could also result from the prolonged (14 hours) periods of fasting, which is also associated with enforcement of stronger behavioral rhythmicity, such as a clearer distinction between the resting and active phase (Manoogian and Panda, 2016). During this prolonged fasting, the body possibly uses up most of its glucose reserves and starts catabolizing more lipids as energy source. Something similar has been shown in mice where TRF during the active phase reduced the fasting glucose level when fed a high caloric diet as well as increased lipid oxidation, proportional with the fasting duration (Chaix et al., 2014). This is also in line with our experiments where the overall 24 hour RER level did not differ between chow groups, whether rats were fed during the light or dark phase or ad libitum. However, in the TRF groups on the fcHFHS diet, the prolonged period of fasting in combination with an increased intake of lipids resulted in an overall lower 24 hour RER. This indicates that regardless of the timing of fasting, fasting has a beneficial effect on overall oxidation of lipids whilst on a high-fat high-sugar diet.
6. Conclusion
The interactions found between diet composition and TRF indicate that for rats it matters what they eat and when they eat it. Moreover, these interactions show that the combination of what is eaten and when it is eaten can both attenuate or worsen the effects seen by either diet composition or TRF. This is especially true in skeletal muscle but is not excluded for BAT. Together these data provide further evidence for the occurrence of desynchronization between metabolic tissues as a result of TRF in the light period. Additionally, since the molecular clocks in BAT and SM are differently affected, potentially different mechanisms could be regulating these peripheral clocks. Locomotor activity and (prolonged) fasting are two putative candidates that deserve further studies.
Declaration of interest
Conflicts of interest: none.
Acknowledgements
We acknowledge Unga A. Unmehopa and Bernadine Snell for their assistance on the quality control of RNA isolation and RT-qPCR. PdG was supported by a ZonMW TOP grant (#91214047). SS was supported by “NeuroTime” Erasmus Mundus Program. JEO was supported by an Academic Medical Center Ph.D. scholarship.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.nbscr.2017.09.002.
Appendix A. Supplementary material
Supplementary material
.
References
- Bartelt A., Bruns O.T., Reimer R., Hohenberg H., Ittrich H., Peldschus K., Kaul M.G., Tromsdorf U.I., Weller H., Waurisch C., Eychmuller A., Gordts P.L., Rinninger F., Bruegelmann K., Freund B., Nielsen P., Merkel M., Heeren J. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- Berggren J.R., Boyle K.E., Chapman W.H., Houmard J.A. Skeletal muscle lipid oxidation and obesity: influence of weight loss and exercise. Am. J. Physiol. Endocrinol. Metab. 2008;294:E726–E732. doi: 10.1152/ajpendo.00354.2007. [DOI] [PubMed] [Google Scholar]
- Bray M.S., Young M.E. The role of cell-specific circadian clocks in metabolism and disease. Obes. Rev. 2009;10(Suppl 2):6–13. doi: 10.1111/j.1467-789X.2009.00684.x. [DOI] [PubMed] [Google Scholar]
- Bray M.S., Ratcliffe W.F., Grenett M.H., Brewer R.A., Gamble K.L., Young M.E. Quantitative analysis of light-phase restricted feeding reveals metabolic dyssynchrony in mice. Int. J. Obes. (Lond.) 2013;37:843–852. doi: 10.1038/ijo.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowiecki L.J., Lupien J., Follea N., Jahjah L. Effects of sucrose, caffeine, and cola beverages on obesity, cold resistance, and adipose tissue cellularity. Am. J. Physiol. 1983;244:R500–R507. doi: 10.1152/ajpregu.1983.244.4.R500. [DOI] [PubMed] [Google Scholar]
- Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Chaix A., Zarrinpar A., Miu P., Panda S. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20:991–1005. doi: 10.1016/j.cmet.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFronzo R.A., Simonson D., Ferrannini E., Barrett E. Insulin resistance: a universal finding in diabetic states. Bull. Schweiz Akad. Med Wiss. 1981:223–238. [PubMed] [Google Scholar]
- DeFronzo R.A., Gunnarsson R., Bjorkman O., Olsson M., Wahren J. Effects of insulin on peripheral and splanchnic glucose metabolism in noninsulin-dependent (type II) diabetes mellitus. J Clin. Investig. 1985;76:149–155. doi: 10.1172/JCI111938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyar K.A., Ciciliot S., Tagliazucchi G.M., Pallafacchina G., Tothova J., Argentini C., Agatea L., Abraham R., Ahdesmaki M., Forcato M., Bicciato S., Schiaffino S., Blaauw B. The calcineurin-NFAT pathway controls activity-dependent circadian gene expression in slow skeletal muscle. Mol. Metab. 2015;4:823–833. doi: 10.1016/j.molmet.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K., Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol. Rev. 2013;93:107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrannini E., Simonson D.C., Katz L.D., Reichard G., Jr., Bevilacqua S., Barrett E.J., Olsson M., DeFronzo R.A. The disposal of an oral glucose load in patients with non-insulin-dependent diabetes. Metabolism. 1988;37:79–85. doi: 10.1016/0026-0495(88)90033-9. [DOI] [PubMed] [Google Scholar]
- la Fleur S.E., Vanderschuren L.J., Luijendijk M.C., Kloeze B.M., Tiesjema B., Adan R.A. A reciprocal interaction between food-motivated behavior and diet-induced obesity. Int. J. Obes. (Lond.) 2007;31:1286–1294. doi: 10.1038/sj.ijo.0803570. [DOI] [PubMed] [Google Scholar]
- Froy O. Metabolism and circadian rhythms--implications for obesity. Endocr. Rev. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- Gekakis N., Staknis D., Nguyen H.B., Davis F.C., Wilsbacher L.D., King D.P., Takahashi J.S., Weitz C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E.A., Gill S., Leblanc M., Chaix A., Joens M., Fitzpatrick J.A., Ellisman M.H., Panda S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge B.A., Wen Y., Riley L.A., Zhang X., England J.H., Harfmann B.D., Schroder E.A., Esser K.A. The endogenous molecular clock orchestrates the temporal separation of substrate metabolism in skeletal muscle. Skelet. Muscle. 2015;5:17. doi: 10.1186/s13395-015-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch H., Hahn F.F. Primary vascular neoplasms of lymph nodes in the dog. Vet. Pathol. 1998;35:74–76. doi: 10.1177/030098589803500109. [DOI] [PubMed] [Google Scholar]
- de Lange P., Moreno M., Silvestri E., Lombardi A., Goglia F., Lanni A. Fuel economy in food-deprived skeletal muscle: signaling pathways and regulatory mechanisms. FASEB J. 2007;21:3431–3441. doi: 10.1096/fj.07-8527rev. [DOI] [PubMed] [Google Scholar]
- LeBlanc J., Labrie A. A possible role for palatability of the food in diet-induced thermogenesis. Int. J. Obes. Relat. Metab. Disord. 1997;21:1100–1103. doi: 10.1038/sj.ijo.0800520. [DOI] [PubMed] [Google Scholar]
- Longo V.D., Panda S. Fasting, Circadian Rhythms, and Time-Restricted Feeding in Healthy Lifespan. Cell Metab. 2016;23:1048–1059. doi: 10.1016/j.cmet.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoogian E.N., Panda S. Circadian clock, nutrient quality, and eating pattern tune diurnal rhythms in the mitochondrial proteome. Proc. Natl. Acad. Sci. USA. 2016;113:3127–3129. doi: 10.1073/pnas.1601786113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B., Ramsey K.M., Peek C.B., Affinati A., Maury E., Bass J. Circadian clocks and metabolism. Handb. Exp. Pharmacol. 2013:127–155. doi: 10.1007/978-3-642-25950-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensink M., Blaak E.E., van Baak M.A., Wagenmakers A.J., Saris W.H. Plasma free Fatty Acid uptake and oxidation are already diminished in subjects at high risk for developing type 2 diabetes. Diabetes. 2001;50:2548–2554. doi: 10.2337/diabetes.50.11.2548. [DOI] [PubMed] [Google Scholar]
- Mercer S.W., Trayhurn P. Effect of high fat diets on energy balance and thermogenesis in brown adipose tissue of lean and genetically obese ob/ob mice. J. Nutr. 1987;117:2147–2153. doi: 10.1093/jn/117.12.2147. [DOI] [PubMed] [Google Scholar]
- Mulya A., Kirwan J.P. Brown and Beige adipose tissue: therapy for obesity and Its comorbidities? Endocrinol. Metab. Clin. North Am. 2016;45:605–621. doi: 10.1016/j.ecl.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno T., Onishi Y., Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 2007;35:648–655. doi: 10.1093/nar/gkl868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterman, J.E., Koekkoek, L.L., Foppen, E., Unmehopa, U., Eggels, L., Verheij, J., Fliers, E., la Fleur, S.E. and Kalsbeek, A. Effects of daily timing of saturated fat and liquid sugar intake on the development of obesity and the rhythmic expression of hepatic clock and metabolic genes, (Submitted for publication).
- Oosterman J.E., Foppen E., van der Spek R., Fliers E., Kalsbeek A., la Fleur S.E. Timing of fat and liquid sugar intake alters substrate oxidation and food efficiency in male Wistar rats. Chronobiol. Int. 2015;32:289–298. doi: 10.3109/07420528.2014.971177. [DOI] [PubMed] [Google Scholar]
- Opperhuizen A.L., van Kerkhof L.W., Proper K.I., Rodenburg W., Kalsbeek A. Rodent models to study the metabolic effects of shiftwork in humans. Front. Pharmacol. 2015;6:50. doi: 10.3389/fphar.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperhuizen A.L., Wang D., Foppen E., Jansen R., Boudzovitch-Surovtseva O., de Vries J., Fliers E., Kalsbeek A. Feeding during the resting phase causes profound changes in physiology and desynchronization between liver and muscle rhythms of rats. Eur. J. Neurosci. 2016;44:2795–2806. doi: 10.1111/ejn.13377. [DOI] [PubMed] [Google Scholar]
- Ramsey K.M., Marcheva B., Kohsaka A., Bass J. The clockwork of metabolism. Annu Rev. Nutr. 2007;27:219–240. doi: 10.1146/annurev.nutr.27.061406.093546. [DOI] [PubMed] [Google Scholar]
- Reznick J., Preston E., Wilks D.L., Beale S.M., Turner N., Cooney G.J. Altered feeding differentially regulates circadian rhythms and energy metabolism in liver and muscle of rats. Biochim. Biophys. Acta. 2013;1832:228–238. doi: 10.1016/j.bbadis.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Rothwell N.J., Stock M.J. A role for brown adipose tissue in diet-induced thermogenesis. Nature. 1979;281:31–35. doi: 10.1038/281031a0. [DOI] [PubMed] [Google Scholar]
- Salgado-Delgado R., Angeles-Castellanos M., Saderi N., Buijs R.M., Escobar C. Food intake during the normal activity phase prevents obesity and circadian desynchrony in a rat model of night work. Endocrinology. 2010;151:1019–1029. doi: 10.1210/en.2009-0864. [DOI] [PubMed] [Google Scholar]
- Salgado-Delgado R.C., Saderi N., Basualdo Mdel C., Guerrero-Vargas N.N., Escobar C., Buijs R.M. Shift work or food intake during the rest phase promotes metabolic disruption and desynchrony of liver genes in male rats. PLoS One. 2013;8:e60052. doi: 10.1371/journal.pone.0060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman G.I., Cline G., Schumann W.C., Chandramouli V., Kumaran K., Landau B.R. Quantitative comparison of pathways of hepatic glycogen repletion in fed and fasted humans. Am. J. Physiol. 1990;259:E335–E341. doi: 10.1152/ajpendo.1990.259.3.E335. [DOI] [PubMed] [Google Scholar]
- Stanford K.I., Goodyear L.J. The therapeutic potential of brown adipose tissue. Hepatobiliary Surg. Nutr. 2013;2:286–287. doi: 10.3978/j.issn.2304-3881.2013.09.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Foppen E., Zhang Z., Fliers E., Kalsbeek A. Effects of 6-meals-a-day feeding and 6-meals-a-day feeding combined with adrenalectomy on daily gene expression rhythms in rat epididymal white adipose tissue. Genes Cells. 2016;21:6–24. doi: 10.1111/gtc.12315. [DOI] [PubMed] [Google Scholar]
- Summa K.C., Turek F.W. Chronobiology and obesity: interactions between circadian rhythms and energy regulation. Adv. Nutr. 2014;5:312S–319SS. doi: 10.3945/an.113.005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C., Gill S., DiTacchio L., Pulivarthy S.R., Le H.D., Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ying Z., Bosy-Westphal A., Zhang J., Schautz B., Later W., Heymsfield S.B., Muller M.J. Specific metabolic rates of major organs and tissues across adulthood: evaluation by mechanistic model of resting energy expenditure. Am. J. Clin. Nutr. 2010;92:1369–1377. doi: 10.3945/ajcn.2010.29885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox G. Insulin and insulin resistance. Clin. Biochem Rev. 2005;26:19–39. [PMC free article] [PubMed] [Google Scholar]
- Yamajuku D., Okubo S., Haruma T., Inagaki T., Okuda Y., Kojima T., Noutomi K., Hashimoto S., Oda H. Regular feeding plays an important role in cholesterol homeostasis through the liver circadian clock. Circ. Res. 2009;105:545–548. doi: 10.1161/CIRCRESAHA.109.199034. [DOI] [PubMed] [Google Scholar]
- Yamazaki S., Numano R., Abe M., Hida A., Takahashi R., Ueda M., Block G.D., Sakaki Y., Menaker M., Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yasumoto Y., Hashimoto C., Nakao R., Yamazaki H., Hiroyama H., Nemoto T., Yamamoto S., Sakurai M., Oike H., Wada N., Yoshida-Noro C., Oishi K. Short-term feeding at the wrong time is sufficient to desynchronize peripheral clocks and induce obesity with hyperphagia, physical inactivity and metabolic disorders in mice. Metabolism. 2016;65:714–727. doi: 10.1016/j.metabol.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Yoo S.H., Ko C.H., Lowrey P.L., Buhr E.D., Song E.J., Chang S., Yoo O.J., Yamazaki S., Lee C., Takahashi J.S. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. USA. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A., Chaix A., Yooseph S., Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonic S., Ptitsyn A.A., Conrad S.A., Scott L.K., Floyd Z.E., Kilroy G., Wu X., Goh B.C., Mynatt R.L., Gimble J.M. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material