Key Points
Question
What is the accuracy of computer-aided diagnosis of melanoma and how does it translate to clinical practice?
Findings
In this meta-analysis of 70 studies, the accuracy of computer-aided diagnosis is comparable to that of human experts. However, current studies are heterogeneous and most deviate significantly from real-world scenarios and are prone to biases.
Meaning
Although computer-aided diagnosis for melanoma appears to be accurate according to the included studies, more standardized and realistic study settings are required to explore its full potential in clinical practice.
Abstract
Importance
The recent advances in the field of machine learning have raised expectations that computer-aided diagnosis will become the standard for the diagnosis of melanoma.
Objective
To critically review the current literature and compare the diagnostic accuracy of computer-aided diagnosis with that of human experts.
Data Sources
The MEDLINE, arXiv, and PubMed Central databases were searched to identify eligible studies published between January 1, 2002, and December 31, 2018.
Study Selection
Studies that reported on the accuracy of automated systems for melanoma were selected. Search terms included melanoma, diagnosis, detection, computer aided, and artificial intelligence.
Data Extraction and Synthesis
Evaluation of the risk of bias was performed using the QUADAS-2 tool, and quality assessment was based on predefined criteria. Data were analyzed from February 1 to March 10, 2019.
Main Outcomes and Measures
Summary estimates of sensitivity and specificity and summary receiver operating characteristic curves were the primary outcomes.
Results
The literature search yielded 1694 potentially eligible studies, of which 132 were included and 70 offered sufficient information for a quantitative analysis. Most studies came from the field of computer science. Prospective clinical studies were rare. Combining the results for automated systems gave a melanoma sensitivity of 0.74 (95% CI, 0.66-0.80) and a specificity of 0.84 (95% CI, 0.79-0.88). Sensitivity was lower in studies that used independent test sets than in those that did not (0.51; 95% CI, 0.34-0.69 vs 0.82; 95% CI, 0.77-0.86; P < .001); however, the specificity was similar (0.83; 95% CI, 0.71-0.91 vs 0.85; 95% CI, 0.80-0.88; P = .67). In comparison with dermatologists’ diagnosis, computer-aided diagnosis showed similar sensitivities and a 10 percentage points lower specificity, but the difference was not statistically significant. Studies were heterogeneous and substantial risk of bias was found in all but 4 of the 70 studies included in the quantitative analysis.
Conclusions and Relevance
Although the accuracy of computer-aided diagnosis for melanoma detection is comparable to that of experts, the real-world applicability of these systems is unknown and potentially limited owing to overfitting and the risk of bias of the studies at hand.
This meta-analysis evaluates the accuracy of computerized systems in the diagnosis of melanoma in patients with skin lesions.
Introduction
The rising incidence of melanoma, the benefits of early diagnosis, and the limited access to dermatologic services in some countries entailed increased efforts to develop diagnostic systems that are independent of human expertise. Most systems fall into the category of image-based, automated diagnostic systems and use either clinical or dermoscopic images. The hope is that computer-aided diagnosis (CAD) could provide decision support for physicians or could screen large numbers of images for teleconsultation services. Early studies on CAD of skin lesions relied on hand-crafted feature engineering and segmentation masks. These methods showed promising results and reached a diagnostic accuracy comparable to human ratings in experimental settings.1 The accuracy of the automated diagnostic system in real-life settings in the only prospective controlled trial to date was lower than expected.2 Recent advances in computer science3 and the introduction of convolutional neural networks and deep-learning–based approaches revolutionized the classification of medical image analysis.4 Since the last meta-analysis that was published in 2003,1 a significant number of new studies have been published on this topic, but to our knowledge, there is no study that summarizes the body of literature. The aims of this meta-analysis were to critically review the current literature on CAD for melanoma, evaluate the diagnostic accuracy in comparison with that of dermatologists, analyze the association between methodologic differences and performance measures, and explore the applicability of CAD in real-world settings.
Methods
Search Strategy and Selection Process
We searched the online databases MEDLINE, arXiv, and PubMed Central, using specific search terms for each database for articles published between January 1, 2002, and December 31, 2018, without any additional limitations, as well as the reference lists of included articles. Data were analyzed from February 1 to March 10, 2019. The key words used were melanoma and (diagnosis or detection) for MEDLINE, the word melanoma included in abstracts for arXiv, and melanoma and (diagnosis or detection) and (computer aided or artificial intelligence) for PubMed Central. Additional studies were identified by 2 of us (H.K. and P.T.).
Studies were eligible for inclusion if they investigated the accuracy of CAD systems that were or could be used in a screening setting for cutaneous melanoma. Diagnostic methods for lesions that have already been excised, methods that differentiate only between different types of malignant skin lesions, or methods processing information gained by invasive techniques were excluded. If an article discussed more than 1 diagnostic method, only the best-performing method was included.
The titles and abstracts of retrieved articles were screened by 2 of us (V.D. and H.K.). At this stage, articles were excluded if they were not published in English or German, if an abstract was unavailable, or if the content was not relevant to the research question. The full texts of articles that were not excluded during initial screening were retrieved and studied by the same readers. At that time, articles were excluded if they did not present original data or if their content was not relevant to the research question. Discrepancies regarding inclusion or exclusion of specific studies were discussed and resolved by consensus. One of us (P.T.) was available to be the decision maker in case no consensus could be reached.
Data Extraction
We used a standardized data extraction sheet to collect data from all included studies. The extracted data fields were determined in advance and included study, test, and sample characteristics, and outcome measures.
We extracted information on the selection of the study sample, characteristics of included lesions, type of diagnostic reference standard, method of automated analysis, type of classifier, preprocessing, segmentation, and extracted features, if applicable. With regard to the method of automated analysis, we differentiated between hardware-based methods, image analysis with feature extraction (computer vision), and deep learning. According to our definition, hardware-based methods use specific devices beyond simple consumer cameras or smartphones (eg, spectroscopy, multispectral imaging, or photometric stereo device). To obtain outcome measures, we extracted the raw numbers of true and false positives and true and false negatives from each study to calculate summary statistics for the diagnostic accuracy of automated diagnostic systems and, if available, of dermatologists.
Methodologic Analysis
We assessed applicability and risk of bias according to the modified version of the QUADAS-2 tool,5 which we adapted for our specific purpose with regard to sample selection, index test, reference standard, flow, and timing. The studies were also evaluated using the good-quality criteria suggested by Rosado et al,1 which, if followed, should ensure transferability of the results to a real-world setting. These criteria, however, were not applicable to studies from the field of computer science.
Statistical Analysis
For the quantitative meta-analysis, we used the results of studies that either presented absolute numbers for true and false positives and true and false negatives or offered sufficient information to calculate these numbers for the detection of melanoma vs benign lesions. If the same group of authors published more than 1 study with overlapping sets of lesions, only 1 publication was included in the statistical analysis. If studies directly compared CAD with dermatologists, the corresponding sensitivities and specificities of dermatologists were extracted in the same way.
The R Statistics6 package mada7 and the SAS8 Macro MetaDAS9 were used for analyses. A coupled forest plot of sensitivity and specificity was created using RevMan, version 5.3.10 Summary receiver operating characteristic (ROC) curves and mean estimates of sensitivity and specificity and the corresponding 95% CIs were calculated by the bivariate model of Reitsma et al.11 Heterogeneity and the presence of outliers were visually checked and the presence of between-study variance was tested.12 A bivariate meta-regression with potential covariables was modeled to reduce any heterogeneity noted between the studies. For all studies, the use of independent test sets, of proprietary or public test sets, and the method of analysis (computer vision, deep learning, or hardware based) were available and investigated. In sensitivity analyses, potential outliers were excluded to assess their association with the results. All tests were based on a 2-sided significance level of P = .05.
Results
General Study Characteristics
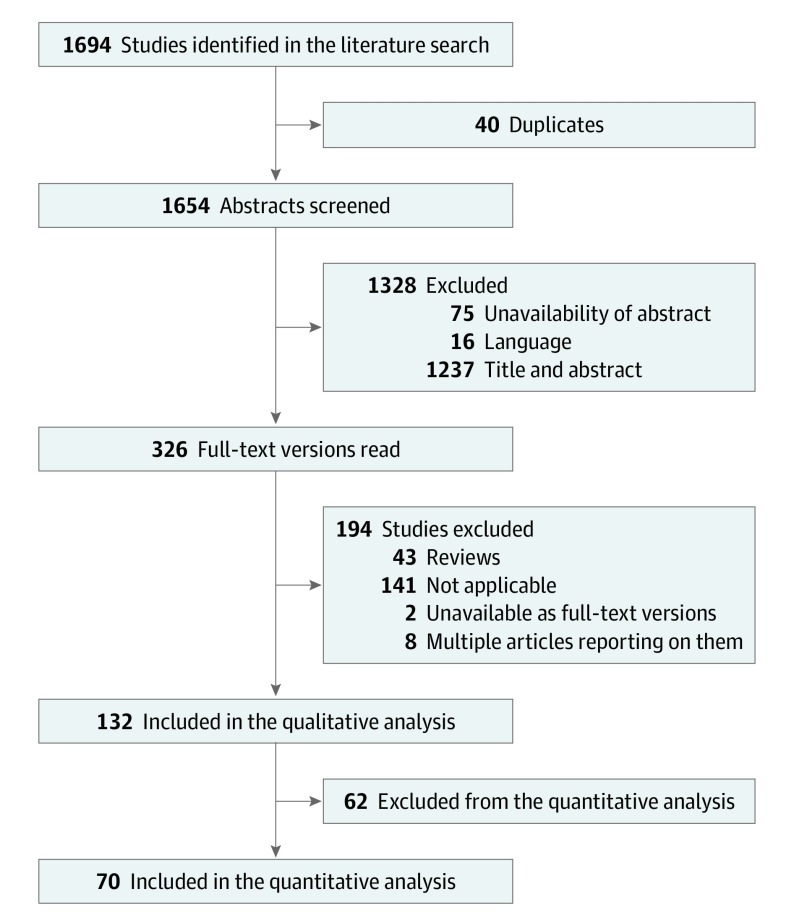
We identified 1694 potentially eligible articles, of which 132 were included in the qualitative analysis and 70 provided sufficient data for a quantitative meta-analysis (Figure 1, Figure 2, and Figure 3).2,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81 We attributed 105 articles to the field of computer science and 27 to the field of medicine. The methods used were computer vision (n = 58), deep learning (n = 55), and hardware based (n = 19) . Artificial neural networks and support vector machines were the most commonly applied machine-learning techniques for classification (eTable 1 in the Supplement).
Figure 1. Study Selection Process.
Selection of studies according to inclusion and exclusion criteria at different stages of the meta-analysis.
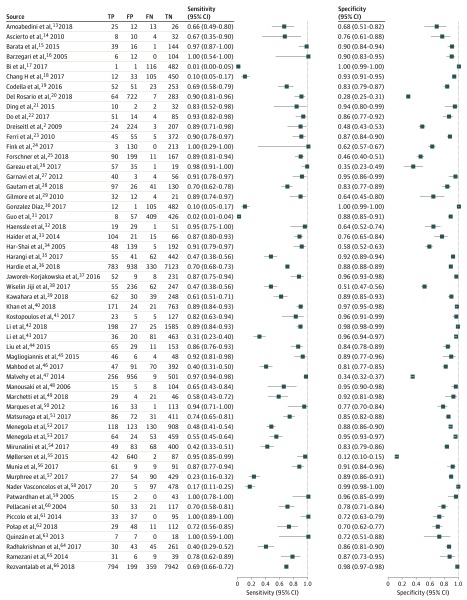
Figure 2. Sensitivity and Specificity of 55 of 70 Included Studies.
FN indicates false negative; FP, false positive; TN, true negative; and TP, true positive.
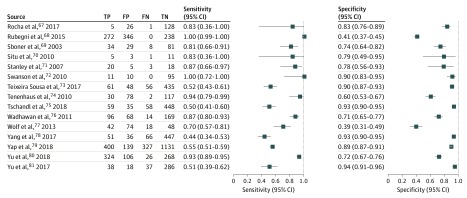
Figure 3. Sensitivity and Specificity of 15 of 70 Included Studies.
FN indicates false negative; FP, false positive; TN, true negative; and TP, true positive.
Fifty studies included only melanocytic lesions, while nonmelanocytic lesions were included in 67 studies. Fifteen studies did not specify whether nonmelanocytic lesions were included (eTable 1 in the Supplement). Twenty-two studies reported melanoma thickness and 28 studies noted the inclusion of in situ melanomas. The median thickness of invasive melanomas ranged from 0.2 to 1.5 mm. Publicly available images were used in 76 studies, while 56 studies used proprietary data sets. Most studies (n = 119) did not select lesions randomly and 13 studies used consecutively collected samples.
Quality Assessment
According to the QUADAS-2 tool,5 13 studies showed moderate applicability, and the concerns about the applicability of the remaining studies was judged as low (eTable 2 in the Supplement). The concerns about the risk of bias were judged as high in at least 1 category in all but 4 studies, and 58 studies presented a high risk of bias in at least 2 categories (eTable 2 and eFigure in the Supplement). The quality assessment of the 27 studies from the medical field, using the quality criteria proposed by Rosado et al,1 showed that between 1 and 7 of 9 quality criteria were met (eFigure in the Supplement). The general characteristics of the 70 studies that were included in the quantitative analysis are shown in eTable 1 in the Supplement.
Diagnostic Accuracy
Based on the 70 studies that were included in the quantitative analysis, the summary estimate for the melanoma sensitivity of CAD systems was 0.74 (95% CI, 0.66-0.80) and the specificity was 0.84 (95% CI, 0.79-0.88) (Table). The sensitivity was significantly lower for the 45 studies that used independent test sets (0.51; 95% CI, 0.34-0.69 vs 0.82; 95% CI, 0.77-0.86; P < .001). The summary estimates for the corresponding specificities were similar (0.83; 95% CI, 0.71-0.91 vs 0.85; 95% CI, 0.80-0.88; P = .67).
Table. Summary Estimates for Sensitivity and Specificity.
| Variable | Summary Estimate (95% CI) | P Value | |
|---|---|---|---|
| Univariate | Multiplea | ||
| CAD Overall (n = 70) | |||
| Sensitivity | 0.74 (0.66-0.80) | [Reference] | [Reference] |
| Specificity | 0.84 (0.79-0.88) | [Reference] | [Reference] |
| Independent Test Set (n = 25) | |||
| Sensitivity | 0.51 (0.34-0.69) | [Reference] | [Reference] |
| Specificity | 0.83 (0.71-0.91) | [Reference] | [Reference] |
| Nonindependent Test Set (n = 45) | |||
| Sensitivity | 0.82 (0.77-0.86) | <.001b | .002b |
| Specificity | 0.85 (0.80-0.88) | .67b | .006b |
| Public Test Set Source (n = 37) | |||
| Sensitivity | 0.57 (0.44-0.68) | [Reference] | [Reference] |
| Specificity | 0.91 (0.88-0.94) | [Reference] | [Reference] |
| Proprietary Test Set Source (n = 33) | |||
| Sensitivity | 0.87 (0.82-0.91) | <.001c | .003c |
| Specificity | 0.72 (0.63-0.79) | <.001c | <.001c |
| Computer Vision (n = 35) | |||
| Sensitivity | 0.85 (0.80-0.88) | [Reference] | [Reference] |
| Specificity | 0.77 (0.69-0.84) | [Reference] | [Reference] |
| Deep Learning (n = 26) | |||
| Sensitivity | 0.44 (0.30-0.59) | <.001d | <.001d |
| Specificity | 0.92 (0.89-0.95) | <.001d | <.001d |
| Hardware-Based (n = 9) | |||
| Sensitivity | 0.86 (0.77-0.92) | .71d | .71d |
| <.001e | .008e | ||
| Specificity | 0.70 (0.54-0.82) | .47d | .28d |
| <.001e | .17e | ||
| Dermatologists (n = 14) | |||
| Sensitivity | 0.88 (0.79-0.93) | NA | NA |
| Specificity | 0.78 (0.76-0.79) | NA | NA |
| Corresponding CADs (n = 14) | |||
| Sensitivity | 0.89 (0.87-0.91) | .50f | NA |
| Specificity | 0.68 (0.60-0.77) | .052f | NA |
Abbreviations: CAD, computer-aided diagnosis; NA, not applicable.
Multiple P value refers to significance testing with a multiple bivariate meta-regression model with variables test-set independence, test-set source, and CAD method included.
Compared with independent test set.
Compared with public test set source.
Compared with computer vision.
Compared with deep learning.
Compared with dermatologists.
The 33 studies that used proprietary test sets had a significantly higher sensitivity than the 37 studies that used publicly available test sets (0.87; 95% CI, 0.82-0.91 vs 0.57; 95% CI, 0.44-0.68; P < .001); however, the specificity was significantly lower (0.72; 95% CI, 0.63-0.79 vs 0.91; 95% CI, 0.88-0.94; P < .001).
Computer-aided diagnosis systems using deep learning achieved a sensitivity of 0.44 (95% CI, 0.30-0.59; P < .001) and a specificity of 0.92 (95% CI, 0.89-0.95; P < .001) and behaved significantly differently from the other 2 methods. The 35 studies using computer vision achieved a sensitivity of 0.85 (95% CI, 0.80-0.88; P < .001) and a specificity of 0.77 (95% CI, 0.69-0.84; P < .001), and 9 studies using hardware-based methods reached a sensitivity of 0.86 (95% CI, 0.77- 0.92; P < .001) and a specificity of 0.70 (95% CI, 0.54-0.82; P = .001). Studies based on computer vision and hardware-based methods were not significantly different with respect to sensitivity and specificity.
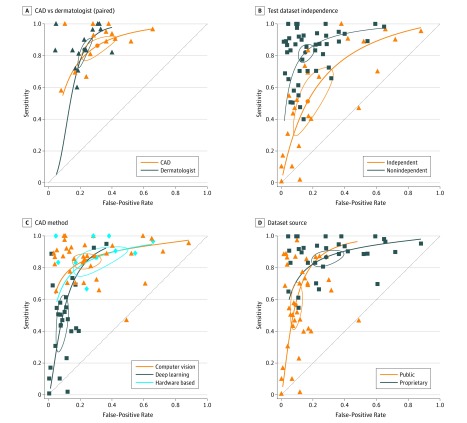
A multiple bivariate meta-regression model showed that sensitivity and specificity depended significantly on the test set characteristics, the test set source, and the method of analysis. Sensitivity was significantly lower for independent (0.51; 95% CI, 0.34-0.69) versus nonindependent test sets (0.82; 95% CI, 0.77-0.86; P = .002). Specificity was significantly higher for deep learning (0.92; 95% CI, 0.89-0.95) than for computer vision (0.77; 95% CI, 0.69-0.84; P < .001) or hardware-based methods (0.70; 95% CI, 0.54-0.82; P < .001) (Table). Analyses were repeated with studies of Wolf et al77 and Wiselin Jiji38 excluded because their distance from the summary ROC indicated potential outliers; however, their influence on the outcome was minor (eTable 3 in the Supplement). Although the bivariate meta-regression reduced heterogeneity in the meta-analysis, a significant between-study heterogeneity remained in all subgroups (Figure 4).
Figure 4. Summary Receiver Operating Characteristic (ROC) Curves.
Bivariate summary ROC curves comparing computer-aided diagnosis (CAD) and dermatologists for the detection of melanoma vs benign lesions in studies when both methods are available (A). Bivariate summary ROC curves comparing studies on automated systems for the detection of melanoma vs benign lesions using independent and nonindependent test sets (B), different CAD methods (C), and public or proprietary test data sets (D).
A subset of 14 studies compared CAD with dermatologists, who reached a sensitivity of 0.88 (95% CI, 0.79-0.93) and a specificity of 0.78 (95% CI, 0.76-0.79). Dermatologists and CAD attained a similar sensitivity (sensitivity of CAD: 0.89; 95% CI, 0.87-0.91; P = .496); specificity, however, was 10 percentage points lower for CAD, although not statistically significant (0.68; 95% CI, 0.60-0.77; P = .052).
Discussion
Computer-aided diagnosis of melanoma is an instructive example of the current mismatch between expectations and the actual outcome of machine-learning approaches for accurate predictions and diagnoses in health care. Despite numerous breakthrough studies that demonstrate expert-level accuracy of CAD for melanoma, existing devices or applications are not widely used. A potential reason for this mismatch may be that the results of the studies conducted in this field cannot be transferred directly to clinical practice. We performed this meta-analysis with the aim to better characterize the studies at hand and identify factors that explain the mismatch between expectations and reality.
Most studies on CAD came from the field of computer science, whereas clinical studies were sparse. The studies from the field of computer science typically focused on technical issues, such as preprocessing of images, image augmentation, segmentation, feature extraction and architecture, and fine tuning of the classification algorithm. These studies usually did not address typical limitations of diagnostic studies, such as the potential ambiguity of pathologic reports, the complexity of clinical decision making in the presence of uncertainty, and the types of biases involved in such studies. Clinical information, such as age, anatomic site, and history of melanoma, was rarely used, although it may significantly improve the accuracy for melanoma detection.82
Computer-aided diagnosis studies were highly heterogeneous and at high risk for bias. Half of the studies, and practically all studies coming from the field of computer science, were conducted in an experimental setting and used images from publicly available databases. Most used convenience samples or, at best, retrospectively collected consecutive samples. These data sets are usually prone to selection and verification bias. Overfitting is an inherent problem of machine learning resulting in lack of generalizability, especially if the training set and the test set are different from the group of lesions encountered in clinical practice. It is not surprising that studies that used independent test sets reached a lower sensitivity than the remaining studies. The fact that specificity is not affected by overfitting may be explained by class imbalance. Most data sets used for training and testing are imbalanced and contain more nevi than melanomas. Because overfitting is more likely if the sample size is small, the sensitivity for melanoma is more likely affected by overfitting than the specificity.
Ideally, CAD should be trained and tested in the setting of its intended use. The clinical setting may vary from general population screening to surveillance of high-risk patients with multiple nevi and a personal history of melanoma. Most clinical studies were conducted in specialized referral centers with high melanoma prevalence. The systems were not tested in the general population or as screening tools.
Dermoscopy was most widely used for classification. Dermoscopic images can be obtained with different devices, including smartphones, which makes them widely available. Although dermoscopy is regarded as the state of the art in vivo technique for the diagnosis of melanoma, most prospective clinical studies used other methods, such as spectroscopy or multispectral images, which require exclusive hardware. Prospective, controlled clinical studies of automated systems of dermoscopic images or conventional close-ups are currently missing.
The restriction to melanocytic lesions was also a limitation found in most of the studies of this meta-analysis. If nonmelanocytic lesions were included, it was usually by chance. The restriction to melanocytic lesions limits the applicability of such systems in clinical practice. In a population of individuals with extensive chronic sun damage, a significant portion of pigmented lesions that are excised or biopsied for diagnostic reasons are nonmelanocytic. A system that is trained to differentiate melanoma from nevi will not be suitable in a setting in which a significant portion of lesions are seborrheic keratosis, solar lentigines, basal cell carcinomas, actinic keratoses, or Bowen disease. Such a system would need preselection of melanocytic lesions by experts, but if experts are needed to handle the system it would defy its own purpose. The lack of generalizability and the problem of out-of-distribution lesions, such as rare or unknown disease categories, is a limitation that is not addressed by current studies. In a recent study, an otherwise accurate CAD missed amelanotic melanomas, most probably because they were underrepresented in the training set.83
When compared directly, CAD differentiated melanoma from nevi with similar sensitivity to dermatologists but with lower specificity. This difference, however, was not statistically significant and can also be attributed to a threshold effect. The optimal threshold and the tradeoff between sensitivity and specificity is a problem. Although metrics exist that take into account the consequences of diagnostic decisions, they are rarely used in the realm of machine learning, which has also been criticized recently in an editorial by Shah et al.84 As shown in Figure 2, most automated diagnostic systems used thresholds that balanced sensitivity and specificity and avoided extreme values. This selection of thresholds makes sense clinically because if the sensitivity is maximized at an expense of an intolerable low specificity, the system would be useless in clinical practice.
Limitations
This meta-analysis has limitations. Because of the heterogeneity of the studies, the summary estimates of the quantitative part have to be interpreted with caution and in light of the methodologic quality of the studies. Two studies in particular occurred as visual outliers in summary ROC space. Wiselin Jiji et al38 used a support-vector machine with significantly lower performance compared with other competitors of the International Symposium on Biomedical Imaging 2017 challenge, suggesting that implementation may have been suboptimal. Wolf et al77 used a smartphone-based approach on a retrospective convenience sample without specific details on technical implementation.
Conclusions
It is likely that parts of the mismatch between promising experimental results and limited usefulness in reality can be attributed to issues beyond accuracy. Dermatologists, who are regarded as the experts in the field of melanoma diagnosis, probably benefit the least and feel threatened the most. There is a fear that less-skilled physicians or even nonmedical personnel will use such systems to deliver a service that should be restricted to dermatologists. One could argue that not all dermatologists are experts in dermoscopy and that even experts could benefit from computer assistance when related to repetitive tasks, such as comparing sequential images. Therefore, a successful CAD would most probably enhance and support dermatologists rather than replace them. It is currently unclear in which setting and for which task CADs are most useful, but if the setting and the tasks are unclear, the systems cannot be trained and tested sufficiently. If the systems are used in a setting in which they are not accepted or for a task they have not been trained for, they will be wasted, even if the technology is exciting and accurate.
eTable 1. General Characteristics of All Included Studies (n=132)
eTable 2. Assessment of Bias Risk and Applicability Concerns of the Included Studies (n=132) Using the QUADAS-2 Tool
eTable 3. Sensitivity, Specificity and Covariable Effects Calculated With Two Identified Outliers (Wolf 2013 and Jiji 2017) Excluded
eFigure. Risk of Bias, Applicability Concerns, and Methodologic Quality
eReferences
References
- 1.Rosado B, Menzies S, Harbauer A, et al. Accuracy of computer diagnosis of melanoma: a quantitative meta-analysis. Arch Dermatol. 2003;139(3):361-367. doi: 10.1001/archderm.139.3.361 [DOI] [PubMed] [Google Scholar]
- 2.Dreiseitl S, Binder M, Hable K, Kittler H. Computer versus human diagnosis of melanoma: evaluation of the feasibility of an automated diagnostic system in a prospective clinical trial. Melanoma Res. 2009;19(3):180-184. doi: 10.1097/CMR.0b013e32832a1e41 [DOI] [PubMed] [Google Scholar]
- 3.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst. 2012;25(2):1097-1105. [Google Scholar]
- 4.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115-118. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whiting PF, Rutjes AW, Westwood ME, et al. ; QUADAS-2 Group . QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529-536. doi: 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 6.The R Project for Statistical Computing. https://www.R-project.org/. Accessed May 11, 2019.
- 7.mada Meta-analysis of diagnostic accuracy. https://cran.r-project.org/web/packages/mada/index.html. Accessed May 11, 2019.
- 8.SAS Institute Inc SAS Version 9.4. Chapel Hill, NC: SAS Institute Inc; 2016. [Google Scholar]
- 9.Takwoingi Y, Deeks JJ MetaDAS: A SAS macro for meta-analysis of diagnostic accuracy studies. Version 1.3. https://methods.cochrane.org/sdt/software-meta-analysis-dta-studies. Accessed May 11, 2019.
- 10.Cochrane Community Review manager (RevMan). version 5.3. https://community.cochrane.org/help/tools-and-software/revman-5/revman-5-download. Accessed May 11, 2019.
- 11.Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58(10):982-990. doi: 10.1016/j.jclinepi.2005.02.022 [DOI] [PubMed] [Google Scholar]
- 12.Bossuyt P, Davenport C, Deeks JJ, Hyde C, Leeflang M, Scholten R Interpreting results and drawing conclusions. In: Deeks JJ, Bossuyt PM, Gatsonis C, eds. Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 0.9. The Cochrane Collaboration. http://srdta.cochrane.org/. Accessed May 11, 2019.
- 13.Amoabedini A, Farsani MS, Saberkari H, Aminian E. Employing the local radon transform for melanoma segmentation in dermoscopic images. J Med Signals Sens. 2018;8(3):184-194. doi: 10.4103/jmss.JMSS_40_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ascierto PA, Palla M, Ayala F, et al. The role of spectrophotometry in the diagnosis of melanoma. BMC Dermatol. 2010;10:5. doi: 10.1186/1471-5945-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barata C, Emre Celebi M, Marques JS. Melanoma detection algorithm based on feature fusion. Conf Proc IEEE Eng Med Biol Soc. 2015;2015:2653-2656. doi: 10.1109/embc.2015.7318937 [DOI] [PubMed] [Google Scholar]
- 16.Barzegari M, Ghaninezhad H, Mansoori P, Taheri A, Naraghi ZS, Asgari M. Computer-aided dermoscopy for diagnosis of melanoma. BMC Dermatol. 2005;5(1):8. doi: 10.1186/1471-5945-5-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi L, Kim J, Ahn E, Feng DDF Automatic skin lesion analysis using large-scale dermoscopy images and deep residual networks. https://arxiv.org/abs/1703.04197. Published March 17, 2017. Accessed May 11, 2019.
- 18.Chang H. Skin cancer reorganization and classification with deep neural network. https://arxiv.org/abs/1703.00534. Published March 1, 2017. Accessed May 11, 2019.
- 19.Codella N, Nguyen QB, Pankanti S, et al. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Develop. 2016;64(4). doi: 10.1147/jrd.2017.2708299 [DOI] [Google Scholar]
- 20.Del Rosario F, Farahi JM, Drendel J, et al. Performance of a computer-aided digital dermoscopic image analyzer for melanoma detection in 1,076 pigmented skin lesion biopsies. J Am Acad Dermatol. 2018;78(5):927-934.e6. doi: 10.1016/j.jaad.2017.01.049 [DOI] [PubMed] [Google Scholar]
- 21.Ding Y, John NW, Smith L, Sun J, Smith M. Combination of 3D skin surface texture features and 2D ABCD features for improved melanoma diagnosis. Med Biol Eng Comput. 2015;53(10):961-974. doi: 10.1007/s11517-015-1281-z [DOI] [PubMed] [Google Scholar]
- 22.Do T, Hoang T, Pomponiu V, et al. Accessible melanoma detection using smartphones and mobile image analysis. https://arxiv.org/abs/1711.09553. Published November 27, 2017. Accessed May 11, 2019.
- 23.Ferri M, Stanganelli I. Size functions for the morphological analysis of melanocytic lesions. Int J Biomed Imaging. 2010;2010:621357. doi: 10.1155/2010/621357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fink C, Jaeger C, Jaeger K, Haenssle HA. Diagnostic performance of the MelaFind device in a real-life clinical setting. J Dtsch Dermatol Ges. 2017;15(4):414-419. [DOI] [PubMed] [Google Scholar]
- 25.Forschner A, Keim U, Hofmann M, et al. Diagnostic accuracy of dermatofluoroscopy in cutaneous melanoma detection: results of a prospective multicentre clinical study in 476 pigmented lesions. Br J Dermatol. 2018;179(2):478-485. [DOI] [PubMed] [Google Scholar]
- 26.Gareau DS, Correa da Rosa J, Yagerman S, et al. Digital imaging biomarkers feed machine learning for melanoma screening. Exp Dermatol. 2017;26(7):615-618. doi: 10.1111/exd.13250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garnavi R, Aldeen M, Bailey J. Computer-aided diagnosis of melanoma using border and wavelet-based texture analysis. IEEE Trans Inf Technol Biomed. 2012;16(6):1239-1252. doi: 10.1109/TITB.2012.2212282 [DOI] [PubMed] [Google Scholar]
- 28.Gautam D, Ahmed M, Meena YK, Ul Haq A. Machine learning-based diagnosis of melanoma using macro images. Int J Numer Method Biomed Eng. 2018;34(5):e2953. doi: 10.1002/cnm.2953 [DOI] [PubMed] [Google Scholar]
- 29.Gilmore S, Hofmann-Wellenhof R, Soyer HP. A support vector machine for decision support in melanoma recognition. Exp Dermatol. 2010;19(9):830-835. doi: 10.1111/j.1600-0625.2010.01112.x [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez Diaz I. Incorporating the knowledge of dermatologists to convolutional neural networks for the diagnosis of skin lesions. https://arxiv.org/abs/1703.01976. Published March 6, 2017. Accessed May 11, 2019. [DOI] [PubMed]
- 31.Guo S, Luo Y, Song Y Random forests and VGG-NET: an algorithm for the ISIC 2017 skin lesion classification challenge. https://arxiv.org/abs/1703.05148. Published March 15, 2017.
- 32.Haenssle HA, Fink C, Schneiderbauer R, et al. ; Reader study level-I and level-II Groups . Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29(8):1836-1842. doi: 10.1093/annonc/mdy166 [DOI] [PubMed] [Google Scholar]
- 33.Haider S, Cho D, Amelard R, Wong A, Clausi DA. Enhanced classification of malignant melanoma lesions via the integration of physiological features from dermatological photographs. Conf Proc IEEE Eng Med Biol Soc. 2014;2014:6455-6458. [DOI] [PubMed] [Google Scholar]
- 34.Har-Shai Y, Glickman YA, Siller G, et al. Electrical impedance scanning for melanoma diagnosis: a validation study. Plast Reconstr Surg. 2005;116(3):782-790. doi: 10.1097/01.prs.0000176258.52201.22 [DOI] [PubMed] [Google Scholar]
- 35.Harangi B. Skin lesion detection based on an ensemble of deep convolutional neural network. https://arxiv.org/abs/1705.03360. Published May 7, 2017. Accessed May 11, 2019.
- 36.Hardie R, Ali R, De Silva M, Kebede T Skin lesion segmentation and classification for ISIC 2018 using traditional classifiers with hand-crafted features. https://arxiv.org/abs/1807.07001. Published July 18, 2018. Accessed May 11, 2019.
- 37.Jaworek-Korjakowska J, Kłeczek P. Automatic classification of specific melanocytic lesions using artificial intelligence. Biomed Res Int. 2016;2016:8934242. doi: 10.1155/2016/8934242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiselin Jiji G, Johnson Duraj Raj P An extensive technique to detect and analyze melanoma: a challenge at the International Symposium on Biomedical Imaging (ISBI) 2017. https://arxiv.org/ftp/arxiv/papers/1702/1702.08717.pdf. Accessed May 11, 2019.
- 39.Kawahara J, Daneshvar S, Argenziano G, Hamarneh G. 7-Point checklist and skin lesion classification using multi-task multi-modal neural nets. IEEE J Biomed Health Inform. 2018;23(2):538-546. doi: 10.1109/JBHI.2018.2824327 [DOI] [PubMed] [Google Scholar]
- 40.Khan MA, Akram T, Sharif M, et al. An implementation of normal distribution based segmentation and entropy controlled features selection for skin lesion detection and classification. BMC Cancer. 2018;18(1):638. doi: 10.1186/s12885-018-4465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostopoulos SA, Asvestas PA, Kalatzis IK, et al. Adaptable pattern recognition system for discriminating Melanocytic Nevi from Malignant Melanomas using plain photography images from different image databases. Int J Med Inform. 2017;105:1-10. doi: 10.1016/j.ijmedinf.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 42.Li K, Li E Skin lesion analysis towards melanoma detection via end-to-end deep learning of convolutional neural networks. https://arxiv.org/abs/1807.08332. Published July 22, 2018. Accessed May 11, 2019.
- 43.Li Y, Shen L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors (Basel). 2018;18(2):556. doi: 10.3390/s18020556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z, Zerubia J. Skin image illumination modeling and chromophore identification for melanoma diagnosis. Phys Med Biol. 2015;60(9):3415-3431. doi: 10.1088/0031-9155/60/9/3415 [DOI] [PubMed] [Google Scholar]
- 45.Maglogiannis I, Delibasis KK. Enhancing classification accuracy utilizing globules and dots features in digital dermoscopy. Comput Methods Programs Biomed. 2015;118(2):124-133. doi: 10.1016/j.cmpb.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 46.Mahbod A, Ecker R, Ellinger I Skin lesion classification using hybrid deep neural networks. https://arxiv.org/abs/1702.08434. Published April 25, 2019.
- 47.Malvehy J, Hauschild A, Curiel-Lewandrowski C, et al. Clinical performance of the Nevisense system in cutaneous melanoma detection: an international, multicentre, prospective and blinded clinical trial on efficacy and safety. Br J Dermatol. 2014;171(5):1099-1107. doi: 10.1111/bjd.13121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manousaki AG, Manios AG, Tsompanaki EI, et al. A simple digital image processing system to aid in melanoma diagnosis in an everyday melanocytic skin lesion unit: a preliminary report. Int J Dermatol. 2006;45(4):402-410. doi: 10.1111/j.1365-4632.2006.02726.x [DOI] [PubMed] [Google Scholar]
- 49.Marchetti MA, Codella NCF, Dusza SW, et al. ; International Skin Imaging Collaboration . Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J Am Acad Dermatol. 2018;78(2):270-277.e1. doi: 10.1016/j.jaad.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marques JS, Barata C, Mendonça T. On the role of texture and color in the classification of dermoscopy images. Conf Proc IEEE Eng Med Biol Soc. 2012;2012:4402-4405. doi: 10.1109/embc.2012.6346942 [DOI] [PubMed] [Google Scholar]
- 51.Matsunaga K, Hamada A, Minagawa A, Koga H Image classification of melanoma, nevus and seborrheic keratosis by deep neural network ensemble. https://arxiv.org/abs/1703.03108. Published March 9, 2017. Accessed May 11, 2019.
- 52.Menegola A, Fornaciali M, Pires R, Vasques Bittencourt F, Avila S, Valle E Knowledge transfer for melanoma screening with deep learning. https://arxiv.org/abs/1703.07479. Published March 22, 2017. Accessed May 11, 2019.
- 53.Menegola A, Tavares J, Fornaciali M, Li L, Avila S, Valle E RECOD titans at ISIC challenge 2017. https://arxiv.org/abs/1703.04819. Published March 14, 2017.
- 54.Mirunalini P, Chandrabose A, Gokul V, Jaisakthi S Deep learning for skin lesion classification. https://arxiv.org/abs/1703.04364. Published March 13, 12017.
- 55.Møllersen K, Kirchesch H, Zortea M, Schopf TR, Hindberg K, Godtliebsen F. Computer-aided decision support for melanoma detection applied on melanocytic and nonmelanocytic skin lesions: a comparison of two systems based on automatic analysis of dermoscopic images. Biomed Res Int. 2015;2015:579282. doi: 10.1155/2015/579282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munia TTK, Alam MN, Neubert J, Fazel-Rezai R. Automatic diagnosis of melanoma using linear and nonlinear features from digital image. Conf Proc IEEE Eng Med Biol Soc. 2017;2017:4281-4284. [DOI] [PubMed] [Google Scholar]
- 57.Murphree DH, Ngufor C Transfer learning for melanoma detection: participation in ISIC 2017 skin lesion classification challenge. https://arxiv.org/abs/1703.05235. 2017.
- 58.Nader Vasconcelos C, Nader Vasconcelos B Convolutional neural network committees for melanoma classification with classical and expert knowledge based image transforms data augmentation. https://arxiv.org/abs/1702.07025. Published March 15, 2017. Accessed May 11, 2019.
- 59.Patwardhan SV, Dai S, Dhawan AP. Multi-spectral image analysis and classification of melanoma using fuzzy membership based partitions. Comput Med Imaging Graph. 2005;29(4):287-296. doi: 10.1016/j.compmedimag.2004.11.001 [DOI] [PubMed] [Google Scholar]
- 60.Pellacani G, Grana C, Cucchiara R, Seidenari S. Automated extraction and description of dark areas in surface microscopy melanocytic lesion images. Dermatology. 2004;208(1):21-26. doi: 10.1159/000075041 [DOI] [PubMed] [Google Scholar]
- 61.Piccolo D, Crisman G, Schoinas S, Altamura D, Peris K. Computer-automated ABCD versus dermatologists with different degrees of experience in dermoscopy. Eur J Dermatol. 2014;24(4):477-481. [DOI] [PubMed] [Google Scholar]
- 62.Połap D, Winnicka A, Serwata K, Kęsik K, Woźniak M. An intelligent system for monitoring skin diseases. Sensors (Basel). 2018;18(8):2552. doi: 10.3390/s18082552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinzán I, Sotoca JM, Latorre-Carmona P, Pla F, García-Sevilla P, Boldó E. Band selection in spectral imaging for non-invasive melanoma diagnosis. Biomed Opt Express. 2013;4(4):514-519. doi: 10.1364/BOE.4.000514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radhakrishnan A, Durham C, Soylemezoglu A, Uhler C Patchnet: interpretable neural networks for image classification. https://arxiv.org/abs/1705.08078. Published November 29, 2018.
- 65.Ramezani M, Karimian A, Moallem P. Automatic detection of malignant melanoma using macroscopic images. J Med Signals Sens. 2014;4(4):281-290. [PMC free article] [PubMed] [Google Scholar]
- 66.Rezvantalab A, Safigholi H, Karimijeshni S Dermatologist level dermoscopy skin cancer classification using different deep learning convolutional neural networks algorithms. https://arxiv.org/abs/1810.10348. Published October 21, 2018. Accessed May 11, 2019.
- 67.Rocha L, Menzies SW, Lo S, et al. Analysis of an electrical impedance spectroscopy system in short-term digital dermoscopy imaging of melanocytic lesions. Br J Dermatol. 2017;177(5):1432-1438. doi: 10.1111/bjd.15595 [DOI] [PubMed] [Google Scholar]
- 68.Rubegni P, Feci L, Nami N, et al. Computer-assisted melanoma diagnosis: a new integrated system. Melanoma Res. 2015;25(6):537-542. doi: 10.1097/CMR.0000000000000209 [DOI] [PubMed] [Google Scholar]
- 69.Sboner A, Eccher C, Blanzieri E, et al. A multiple classifier system for early melanoma diagnosis. Artif Intell Med. 2003;27(1):29-44. doi: 10.1016/S0933-3657(02)00087-8 [DOI] [PubMed] [Google Scholar]
- 70.Situ N, Wadhawan T, Yuan X, Zouridakis G. Modeling spatial relation in skin lesion images by the graph walk kernel. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:6130-6133. doi: 10.1109/iembs.2010.5627798 [DOI] [PubMed] [Google Scholar]
- 71.Stanley RJ, Stoecker WV, Moss RH. A relative color approach to color discrimination for malignant melanoma detection in dermoscopy images. Skin Res Technol. 2007;13(1):62-72. doi: 10.1111/j.1600-0846.2007.00192.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swanson DL, Laman SD, Biryulina M, et al. Optical transfer diagnosis of pigmented lesions. Dermatol Surg. 2010;36(12):1979-1986. doi: 10.1111/j.1524-4725.2010.01808.x [DOI] [PubMed] [Google Scholar]
- 73.Teixeira Sousa R, Vasconcellos de Moraes L Araguaia medical vision lab at ISIC 2017 skin lesion classification challenge. https://arxiv.org/abs/1703.00856. Published March 2, 2017. Accessed May 11, 2019.
- 74.Tenenhaus A, Nkengne A, Horn JF, Serruys C, Giron A, Fertil B. Detection of melanoma from dermoscopic images of naevi acquired under uncontrolled conditions. Skin Res Technol. 2010;16(1):85-97. doi: 10.1111/j.1600-0846.2009.00385.x [DOI] [PubMed] [Google Scholar]
- 75.Tschandl P, Argenziano G, Razmara M, Yap J. Diagnostic accuracy of content-based dermatoscopic image retrieval with deep classification features. [published online September 12, 2018]. Br J Dermatol. 2018. doi: 10.1111/bjd.17189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wadhawan T, Situ N, Rui H, Lancaster K, Yuan X, Zouridakis G. Implementation of the 7-point checklist for melanoma detection on smart handheld devices. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:3180-3183. doi: 10.1109/iembs.2011.6090866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolf JA, Moreau JF, Akilov O, et al. Diagnostic inaccuracy of smartphone applications for melanoma detection. JAMA Dermatol. 2013;149(4):422-426. doi: 10.1001/jamadermatol.2013.2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang X, Zeng Z, Yeo SY, Tan C, Tey HL, Su Y A novel multi-task deep learning model for skin lesion segmentation and classification. https://arxiv.org/abs/1703.01025. Published March 3, 2017. Accessed May 11, 2019.
- 79.Yap J, Yolland W, Tschandl P. Multimodal skin lesion classification using deep learning. Exp Dermatol. 2018;27(11):1261-1267. doi: 10.1111/exd.13777 [DOI] [PubMed] [Google Scholar]
- 80.Yu C, Yang S, Kim W, et al. Acral melanoma detection using a convolutional neural network for dermoscopy images. PLoS One. 2018;13(3):e0193321. doi: 10.1371/journal.pone.0193321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu L, Chen H, Dou Q, Qin J, Heng PA. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging. 2017;36(4):994-1004. doi: 10.1109/TMI.2016.2642839 [DOI] [PubMed] [Google Scholar]
- 82.Binder M, Kittler H, Dreiseitl S, Ganster H, Wolff K, Pehamberger H. Computer-aided epiluminescence microscopy of pigmented skin lesions: the value of clinical data for the classification process. Melanoma Res. 2000;10(6):556-561. doi: 10.1097/00008390-200012000-00007 [DOI] [PubMed] [Google Scholar]
- 83.Tschandl P, Rosendahl C, Akay BN, et al. Expert-level diagnosis of nonpigmented skin cancer by combined convolutional neural networks. JAMA Dermatol. 2019;155(1):58-65. doi: 10.1001/jamadermatol.2018.4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shah ND, Steyerberg EW, Kent DM. Big data and predictive analytics: recalibrating expectations. JAMA. 2018;320(1):27-28. doi: 10.1001/jama.2018.5602 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. General Characteristics of All Included Studies (n=132)
eTable 2. Assessment of Bias Risk and Applicability Concerns of the Included Studies (n=132) Using the QUADAS-2 Tool
eTable 3. Sensitivity, Specificity and Covariable Effects Calculated With Two Identified Outliers (Wolf 2013 and Jiji 2017) Excluded
eFigure. Risk of Bias, Applicability Concerns, and Methodologic Quality
eReferences