Abstract
Recent studies of human respiratory secretions using culture-independent techniques have found a surprisingly diverse array of microbes. Interactions among these community members can profoundly impact microbial survival, persistence and antibiotic susceptibility and, consequently, disease progression. Studies of polymicrobial interactions in the human microbiota have shown that the taxonomic and structural compositions, and resulting behaviours, of microbial communities differ substantially from those of the individual constituent species and in ways of clinical importance. These studies primarily involved oral and gastrointestinal microbiomes. While the field of polymicrobial respiratory disease is relatively young, early findings suggest that respiratory tract microbiota members also compete and cooperate in ways that may influence disease outcomes. Ongoing efforts therefore focus on how these findings can inform more ‘enlightened’, rational approaches to combat respiratory infections. Among the most common respiratory diseases involving polymicrobial infections are cystic fibrosis (CF), non-CF bronchiectasis, COPD and ventilator-associated pneumonia. While respiratory microbiota can be diverse, two of the most common and best-studied members are Staphylococcus aureus and Pseudomonas aeruginosa, which exhibit a range of competitive and cooperative interactions. Here, we review the state of research on pulmonary coinfection with these pathogens, including their prevalence, combined and independent associations with patient outcomes, and mechanisms of those interactions that could influence lung health. Because P. aeruginosa–S. aureus coinfection is common and well studied in CF, this disease serves as the paradigm for our discussions on these two organisms and inform our recommendations for future studies of polymicrobial interactions in pulmonary disease.
Keywords: bacterial infection, cystic fibrosis, paediatric lung disaese, respiratory infection
Introduction
Antonie van Leeuwenhoek noted during his first microscopic observations in the 16th century that micro-organisms rarely live in isolation.1 However, only during the past few decades have researchers began to examine how interactions between microbes might influence their human hosts. This recent interest in the clinical consequences of polymicrobial interactions is largely fuelled by the broader application of deep-sequencing technologies to human microbiota, identifying highly abundant and diverse communities throughout the body. The resulting enhanced insight into the composition of host-associated microbiota has spurred investigation into how the behaviours of microbial communities differ from those of their individual constituents, and how polymicrobial community interactions influence disease progression and treatment efficacy. Investigation of microbial community interactions in dental and gut communities reveal the following key lessons which may inform our discussions of respiratory communities: (1) a ‘healthy’ microbial community may defend against invading pathogens2; (2) the presence of certain keystone species and spatial organisation drive community development, composition and functional behaviours3; and (3) community members interact/collaborate metabolically in ways that are difficult to predict by studying individual species.4
The investigation of polymicrobial infections in the respiratory tract has been dominated by studies of the genetic disease cystic fibrosis (CF). The airways of people with CF are infected with an abundant, dynamic and diverse array of micro-organisms throughout the course of their lives, offering investigators a unique opportunity to interrogate microbial interactions and adaptations. The CF respiratory pathogens commonly identified by culture include Pseudomonas aeruginosa, Staphylococcus aureus, Stenotrophomonas maltophilia and multiple species of Achromobacter, Haemophilus and Burkholderia cepacia complex.5 Of these, P. aeruginosa and S. aureus are the most frequently cultured CF pathogens, and their co-isolation has been observed since Dorothy Andersen’s initial descriptions of the disease.6 As CF is among the best-studied chronic respiratory infection, the vast majority of studies of polymicrobial airway infections have focused on P. aeruginosa and S. aureus. These studies, most of which were performed in vitro or in animal models, have revealed a range of clinically important interactions that may promote microbial growth and survival, enhance antimicrobial tolerance and/or alter virulence factor production. Understanding the mechanisms of these interactions and their clinical relevance, if any, is essential to rationally guide current therapies and to develop improved treatments for respiratory infections.
In addition to CF, polymicrobial infections commonly occur with other chronic airway diseases, including non-CF bronchiectasis, COPD and ventilator-associated pneumonia. Table 1 summarises published clinical studies on respiratory microbial communities and reveals that while P. aeruginosa and S. aureus coinfections are common in chronic airway diseases, their interactions during these infections remain relatively unexamined. In addition to sharing similar microbial communities, these chronic airway diseases have many pathophysiological similarities, including reduced airway clearance, heightened yet unproductive inflammatory response and persistent infections often recalcitrant to treatment.7 By integrating the knowledge gained from the study of microbial interactions in these airway diseases, we may identify general principles regarding how microbes interact during airway infections. Moreover, we can apply the resulting knowledge to emerging areas, including infections beyond the lung and commensal microbiota.
Table 1.
P. aeruginosa and S. aureus infections in pulmonary disease
| Study | Number | Country | Age (years) | Specimen | Analysis | P. aeruginosa (%) | S. aureus (%) | Coinfection (%) | Outcome analysed? |
| Cystic fibrosis* | |||||||||
| 35 | n=283 | Germany | 0–57 (md 7) | Various† | Culture | NA | 59 | 29 | Yes |
| 36 | n=354 | USA | 6–32.8 (mean 15.7) | Various | Culture | 30‡ | 11.9§ | 11.0¶ | Yes |
| 40 | n=234 | USA | 25.4±10.9 | Various | Culture | 26 | 32 | 31 | Yes |
| 37 | n=84 | Canada | >18 | Sputum | Culture | 60 | 24 | 12 | Yes |
| 39 | n=100 | USA | 0.4–16.9 (mean 9.1) | Various | Culture | 46 | 88 | 40? | No |
| 41 | n=419 | France | 23.1±9.8 (mean±SD) | Sputum | Culture | 67 | 72 | 60 | Yes |
| 43 | n=111 | USA | <6 | BAL | Culture | 53 | 24 | 18 | Yes |
| 44 | n=81 | USA | ≤13 | OP | Culture | 19 | 33 | 8.6 | Yes |
| Non-CF bronchiectasis | |||||||||
| 86 | n=150 | Saudi Arabia | 7.3±4.1 (mean±SD) | NP, sputum | Culture | 16 | 7 | NA | No |
| 87 | n=93 | UK | 1.6–18.8 (md 7.2) | Various | Culture | 6 | 8 | NA | No |
| 88 | n=111 | Turkey | 1–17.5 (md 7.4) | Sputum | Culture | 11 | 17 | NA | No |
| 89 | n=136 | UK | 3–18 (md 12.1) | Various | Culture | 11 | 4 | NA | No |
| 90 | n=92 | Ireland | 1.5–13 (md 6.4) | Sputum, BAL | Culture | 9 | 15 | NA | No |
| 91 | n=104 | Australia | 0.4–12.9 (md 2.4) | BAL | Culture | 0 | 3 | 0 | No |
| 92 | n=113 | Australia | 0.4–12.9 (md 2.4) | BAL | Culture | 2 | 5 | 0 | Yes |
| 93 | n=2596 | Various (Europe) | 57–74 (md 67) | Various | Culture | 15 | 6 | NA | Yes, not coinfection |
| Ventilator-associated pneumonia | |||||||||
| 94 | n=12 | UK | 23–70 (md 43.5) | BAL | 16S sequencing | 350 000 OTU† | 150 000 OTU | NA | No |
| 95 | n=43 | USA | 65.9±16.8 | BAL | Culture | 40 | 28 | NA | No |
| 96 | n=36 | India | 18–78 | ETA | Culture | 3.77 | 9.43 | 0 | No |
| 97 | n=20 | UK | 20–79 | ETT | 16S DGGE | 20 | 30 | 5 | No |
| 98‡ | n=107 | UK | >18 (mean 54) | Dental, BAL, NAL | Culture PCR | 23, 29, 4 | 43, 37, 14 | 10, NA, NA | No |
*For CF, only studies reporting coinfection are included.
†Cough swab, sputum or bronchoalveolar lavage (BAL).
‡Chronic infection >50% positive cultures.
§Chronic MRSA (≥3 previous cultures positive).
¶Chronic coinfection with P. aeruginosa and MRSA.
CF, cystic fibrosis; DGGE, denaturing gradient gel electrophoresis; ETA, endotracheal aspiration; ETT, endotracheal tube; md, median; MRSA, methicillin-resistant S. aureus; NA, not analysed; NAL, non-directed alveolar lavage; NP, nasopharyngeal; OP, oropharyngeal swabs; OTT, operational taxonomic units.
Because of the importance of P. aeruginosa and S. aureus as a binary model system for studying polymicrobial interactions in human disease, here we review the current state of this field and discuss how published analyses may be used as a paradigm to inform further studies in similar chronic airway diseases. Several excellent reviews have recently been published regarding the in vitro interactions between P. aeruginosa and S. aureus.8 9 We will focus on what is known (and not known) about the clinical relevance of these findings, in order to examine how current treatments might be improved and to identify important gaps in the literature for future study.
Epidemiology of P. aeruginosa and S. aureus infections in cystic fibrosis: compete or coexist?
S. aureus is the micro-organism cultured most frequently, and often the earliest, from CF respiratory samples. In 2016, more than 70% of people with CF in the USA were culture positive for S. aureus, and approximately 30% of individuals between the ages of 10 and 30 years were positive for methicillin-resistant S. aureus (MRSA).5 P. aeruginosa, on the other hand, is cultured only intermittently from young patients with CF, but it predominates later in life, with approximately 60% of patients infected by age 30.5 The general inverse patterns of infection by P. aeruginosa and S. aureus has led to speculation that competition between these organisms prevents their coexistence during infection or that the conditions favouring the persistence of these organisms in the airway are present at different phases of CF pulmonary infection.
Reciprocal interspecies exclusion/inclusion during respiratory infection may influence infection patterns
Speculation that S. aureus may prevent or delay the acquisition of initial P. aeruginosa infections is based, in part, on studies indicating that paediatric patients undergoing prophylactic anti-Staphylococcus treatment may be at a higher risk for P. aeruginosa infections.10–12 The clinical impact of these results is controversial, however, resulting in different recommendations internationally regarding antibiotic usage for paediatric S. aureus infections.13 14 While a recent longitudinal observational study of children with CF receiving prophylactic flucloxacillin in the UK confirmed an association between prophylaxis and earlier P. aeruginosa detection, it remains unknown if this was due to reduction in S. aureus infection.10 A recent study also sought insight into this question by using US CF Foundation Patient Registry Database (CFFPRD, 28 942 patients, age 6 or older) data to ask how identification of each of the most prevalent CF pathogens influences the presence and future acquisition of another pathogen.15 In cross-sectional analyses, P. aeruginosa detection was negatively associated with concurrent detection of MSSA, B. cepacia complex, S. maltophilia and A. xylosoxidans (OR ~0.5) and positively associated with Aspergillus species (OR ~1.5). A. xylosoxidans exhibited a negative association with concurrent B. cepacia (OR ~0.7), while in contrast, S. maltophilia was positively associated with Aspergillus species (OR >2.5), suggesting these coinfections are common. Positive associations between Aspergillus and P. aeruginosa or S. maltophilia suggest these organisms either competitively interact during infection or that the physiochemical environment of the airway is not concurrently suitable for both pathogens. The authors then examined if the presence of one organism was a predictor for the presence of another in the subsequent year. Detection of MSSA, B. cepacia complex, S. maltophilia and A. xylosoxidans were all negatively associated with P. aeruginosa detection in the following year. These data support the hypothesis that P. aeruginosa infections may be delayed, either directly or indirectly, by the presence of other CF pathogens.
On the other hand, some observations support the contrary hypothesis that early S. aureus colonisation promotes P. aeruginosa acquisition. For example, a longitudinal analysis of the CFFPRD identified S. aureus detection as a risk factor for earlier acquisition of P. aeruginosa.16 In the laboratory, Cigana and colleagues aimed to establish a murine model mimicking infection patterns observed in patients with CF by infecting mice with S. aureus embedded in agar beads, followed by infection with agar-embedded P. aeruginosa.17 While pre-infection with S. aureus did not alter the total burden of P. aeruginosa recovered from the lung, P. aeruginosa clearance and mouse mortality were reduced, which the authors cited as evidence for enhanced transition to chronic P. aeruginosa infection. Histological examination of coinfection revealed S. aureus formed abscess-like lesions in the lung parenchyma, whereas P. aeruginosa was found primarily in the bronchial lumen, suggesting S. aureus influences P. aeruginosa colonisation indirectly in this model. How these observations relate to infections in patients with CF is unclear, given these mice were wild-type and that formation of abscesses by S. aureus is not generally observed during infection in the human CF airway. Further studies are required to determine if and how early S. aureus infections might influence the acquisition of P. aeruginosa during respiratory disease.
Does P. aeruginosa competitively exclude S. aureus? At late stages of CF infection, P. aeruginosa is most often the predominant organism.18 It has been hypothesised that this pattern is due, in part, to the ability of P. aeruginosa to outcompete other organisms. Additionally, the prevalence of S. aureus infections later in life has been rising in recent years, a trend often attributed to the widespread adoption of aggressive eradication strategies for P. aeruginosa.5 However, as with S. aureus, establishing a direct link between interspecies competition and observed clinical trends is challenging. In the laboratory, in vitro studies demonstrate early P. aeruginosa isolates competitively exclude S. aureus during coculture,19–23 supporting the hypothesis that P. aeruginosa infection decreases the likelihood of S. aureus infection. These hypotheses are supported by several in vitro studies that demonstrate P. aeruginosa can inhibit the growth or reduce the viability of S. aureus through multiple mechanisms, including sequestration of iron, production of extracellular proteases and surfactants, and inhibition of S. aureus respiration via production of redox-active secondary metabolites, including 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO) and the phenazine molecule pyocyanin.20 23–32 As discussed below, these interactions are in turn impacted by P. aeruginosa adaptive genetic changes during chronic infection, further complicating this model of interaction.33 34 The mechanisms of these interactions have been extensively reviewed elsewhere and we refer the reader to excellent reviews by Hotterbeek et al 8 and Nguyen et al.9
One third of patients with CF are coinfected with P. aeruginosa and S. aureus
However, some clinical observations are at odds with these models of interspecies competition; for example, concurrent isolation of these organisms is common in CF respiratory disease and also in other respiratory diseases and in chronic wound infections. Table 1 presents a summary of clinical studies reporting coinfection with P. aeruginosa and S. aureus. For CF, coinfection frequencies ranging from 8.6% to 60% of patients were reported, with an average of 28.3% (eight independent studies, 1432 patients total). The highest rates of coinfection have been observed for patients in their mid-twenties, but substantial frequencies have also been reported in patients ranging from 8 to 55 years.35–37
Identifying consistent and clinically meaningful definitions for ‘infection’ for individual organisms is challenging for chronic diseases, a problem that is further amplified when applied to polymicrobial infections. Thus, the challenges encountered using epidemiological data to discern functional relationships is not surprising. For CF, monoinfections are most frequently defined using modifications of the ‘Leeds Criteria’ for P. aeruginosa. The categories provided by this definition include never having a positive culture, free of a positive culture in the past 12 months, intermittent infection (fewer than 50% of cultures in the past year positive) and chronic infection (more than 50% of cultures in the past year positive).38 For polymicrobial infection, certain studies considered patients coinfected when they were intermittently or chronically infected with both organisms in the past year.36 37 39 While useful for identifying patients with possible or likely coinfection, these criteria do not require a single respiratory sample to be culture positive for both organisms. Other studies include only patients who were culture positive for both organisms from the same respiratory samples during any point in the study period or at specific time points.40–44 This definition decreases the rate of false positives, but without examining multiple samples, it may exclude patients due to sampling errors. Properly defining coinfection may also be influenced by the sampling methods used, particularly in young children, who often do not expectorate. Some studies indicate that both sputum and oropharyngeal swab cultures can, in some contexts and for specific organisms, reflect lower airway microbiology identified by bronchoalveolar lavage (BAL)45 or direct sampling of mucous plugs by protected brush (PB) sampling,46 while other studies found poor such correlations. For example, Seidler and colleagues found that while the positive predictive value of both sputum and throat swabs for identifying P. aeruginosa was 100%, in comparison with BAL, the positive predictive values of these two sample types for S. aureus were only 57% and 41%, respectively.45 Thus, the rates of S. aureus monoinfection and coinfection may be underestimated using these samples. Conversely, the negative predictive values of sputum and throat swabs for P. aeruginosa were 60% and 50%, respectively, while those for S. aureus were both 100%. These results, and those of other studies,47 underscore the importance of considering sample type when describing prevalence of specific organisms in patients with CF. Additionally, little is known about how well an individual sputum sample appropriately reflects the airway community diversity of a patient’s entire airway tree. For example, serial sputum samples from the same patient could conceivably originate from different anatomical locations. However, at least one study is reassuring in this regard: by evaluating communities in different areas of the airway by PB and BAL sampling, CF individuals with mild to moderate lung disease, Hogan and colleagues found highly similar communities among all sampled sites for eight of nine subjects.46 For a more detailed discussion on the influence of CF specimen collection on microbial community composition determination, we refer the reader to a recent review.48
The relative abundance of each organism is also likely to influence the presence and/or the magnitude of potentially relevant microbial interactions. Several recent studies have used culture-independent methods to quantitatively and longitudinally examine community dynamics of the CF airway microbiome during changes in clinical disease state, antimicrobial therapy and restoration of CF transmembrane conductance regulator (CFTR) activity (please see the following recent reviews on this topic49 50). However, while studies of patients with CF have rigorously examined the co-occurrence (simultaneous presence and absence) of organisms in sputum,51 the examination of how varying levels of specific microbial species influence the presence or amount of another species in these studies is limited. One study outside of CF performing such an analysis included 60 adult patients with bronchiectasis and determined that when the abundance of P. aeruginosa was high, that of H. influenzae was low, and vice versa, suggesting a competitive relationship between the two species.52 Further studies of this type may help to more effectively define relationships among respiratory pathogens.
Influence of P. aeruginosa and S. aureus on patient outcomes
Coinfection may correlate with poor outcomes for patients with CF
Progressive obstructive pulmonary disease with bronchiectasis is the primary cause of morbidity and mortality in patients with CF.5 Chronic infection with P. aeruginosa has been associated, in multiple studies, with worse outcomes, including survival, lung function, pulmonary exacerbations and nutritional status.53–55 However, the individual influence of S. aureus infections on outcomes in CF is less clear. While data suggest S. aureus infections are associated with increased lower airway inflammation and risk for bronchiectasis, primarily in paediatric studies,42 43 56 others have found an association between S. aureus colonisation and milder disease, particularly in adult patients.37 57 For coinfected patients, accumulating evidence from multiple independent studies suggests an association between coinfection and lower lung function, increased numbers of pulmonary exacerbations, intravenous antibiotics used per year, rates of CF-related diabetes and risk of death, compared with patients who were infected with P. aeruginosa or S. aureus only (other pathogens may be present).36 40 41 44 Patients who are coinfected with P. aeruginosa and MRSA are consistently reported to have the worst outcomes among patients with either of these two species.36 41 While in each study, coinfected patients (P. aeruginosa with S. aureus, either MSSA or MRSA) have worse outcomes compared with patients monoinfected with S. aureus (MSSA or MRSA), not every study performed thus far has observed worse outcomes compared with P. aeruginosa alone.35 37 While the latter study consisted of only a small group of patients, the reasons for the differences in observations in this study and that by Schwerdt and colleagues remain unclear; they may result from differing demographics, treatment strategies or definitions of ‘coinfection’.
Putative interspecies interactions influencing patient health
The correlations observed in some studies between coinfection with P. aeruginosa and S. aureus and poor patient outcomes raise questions about why these patients might do so poorly, and whether interspecies interactions drive clinical observations. In vitro studies have identified several possible mechanisms, which are illustrated in figure 1 and described in detail below.
Figure 1.
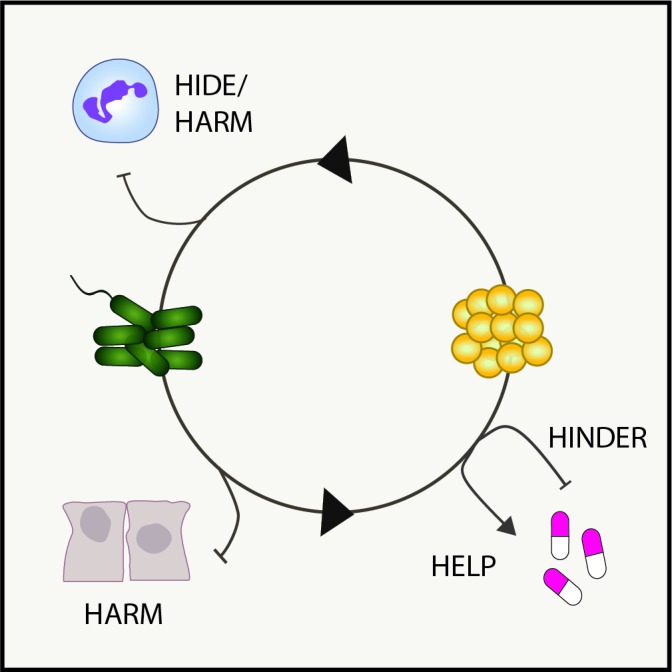
Model of interspecies interactions in pulmonary disease. Shown are examples of how known bacterial interactions during polymicrobial infections can alter host innate immunity (hide or harm), antibiotic susceptibility (help or hinder) or damage the lung microenvironment (harm).
Interactions influence production of microbial virulence factors and other exoproducts
Both P. aeruginosa and S. aureus elaborate an array of secreted products that have been shown in a variety of models to enable bacterial cells to evade killing by innate immune effectors, acquire essential nutrients, adhere to surfaces and other cells, and disseminate to new sites. While evidence is limited regarding the clinical relevance of many of these exoproducts, a number of laboratory studies have shown that the interspecies interactions between these two pathogens often alter virulence factor production by one or both species, potentially influencing pathogenesis, persistence and/or antibiotic susceptibility.
For S. aureus, cocultivation with P. aeruginosa can alter the expression of at least three potentially important virulence factors: Panton-Valentine Leukocidin (PVL, also called LukSF), alpha-haemolysin (Hla) and S. aureus protein A (SpA), as demonstrated in vitro or during coinfection in a partial thickness porcine burn wound model.58 PVL and Hla have potent abilities to lyse host cells such as neutrophils, resulting in tissue necrosis. SpA, which exists in both secreted and membrane-bound forms, has been shown to influence P. aeruginosa behaviours thought to be required for colonisation and survival during CF airway infection. Secreted SpA from S. aureus binds to molecules on the P. aeruginosa cell surface, including the polysaccharide Psl (Pseudomonas polysaccharide locus) and a motility appendage, type IV pilus (T4P). These interactions result in both inhibition of biofilm formation and protection from neutrophil phagocytosis.59 Among these interactions, perhaps the strongest evidence for clinical relevance is for SpA: a recent study suggests that this interspecies interaction may protect P. aeruginosa from eradication by antibiotics in young patients with CF.60 These studies highlight the potential for P. aeruginosa to modulate virulence behaviours and other clinically relevant functions of S. aureus; however, much remains to be studied in this young field.
P. aeruginosa also has the potential to produce an extensive arsenal of virulence determinants, which may be influenced by the presence of S. aureus during coinfection. Many of the virulence determinants previously discussed, which may provide P. aeruginosa a competitive advantage during growth with S. aureus, are also capable of inflicting significant collateral damage to the airway. For example, P. aeruginosa has been shown during in vitro cocultivation with S. aureus to increase expression of genes required for production of the Pseudomonas quinolone signal, which regulates expression of P. aeruginosa virulence factors including pyocyanin and elastase.22 27 Pyocyanin is a secreted redox-active pigment that promotes virulence by interfering with several cellular functions in host cells, including electron transport, cellular respiration, energy metabolism, gene expression and innate immune mechanisms.61 Elastase is a secreted protease with broad substrate specificity that can damage diverse host molecules, including elastin, collagens types I and IV, laminin, immunoglobulins A and G, complement components and αl anti-proteinase inhibitor.62 Additional P. aeruginosa-produced anti-staphylococcal factors capable of inflicting significant damage to the respiratory environment include HQNO (discussed above), the siderophores pyoverdine and pyochelin, and rhamnolipids.63 However, whether expression of these factors is increased during coinfection with S. aureus, and if they are responsible for associated differences in clinical outcomes, are currently unknown.
Polymicrobial interactions influence antimicrobial tolerance
Further insult to the patient may occur as S. aureus responds and/or adapts to the presence of P. aeruginosa. At levels otherwise sublethal to S. aureus, many P. aeruginosa exoproducts have also been shown to influence S. aureus survival in the presence of clinically relevant antibiotics. For example, inhibition of electron transport by P. aeruginosa-produced HQNO and phenazines can enhance tolerance to frontline S. aureus antimicrobials, including vancomycin, tobramycin and ciprofloxacin.23 64 65 Rhamnolipids, on the other hand, increase uptake and sensitivity of S. aureus to tobramycin.66 Production of these exoproducts varies among clinical isolates from patients with CF and under different environmental conditions; thus, the susceptibility of S. aureus to treatment may vary among patients depending on the composition of their microbial communities.67 68 Moreover, S. aureus may also genetically adapt to prolonged exposure to P. aeruginosa respiration-inhibiting exoproducts through formation of small-colony-variant (SCV) mutants. SCVs exhibit defects in electron transport, which simultaneously confer slow growth on most laboratory media, reduced killing by P. aeruginosa compared with wild-type parent strains20 23 and resistance to many antibiotics.69 SCVs are found commonly in patients with CF70 and are associated with an increased decline in lung function.39
Polymicrobial interactions may influence host immunity
The diseased CF airway lumen is characterised by excessive, sustained and ineffective neutrophil-dominated inflammatory response, which correlates closely with disease progression.71 Specifically, studies have shown dysregulation of pro-inflammatory mediators, such as IL-1772 and pro-resolution factors.73 Thus, one mechanism where interspecies interactions may influence pulmonary health is by altering host immune responses. Findings from cocultivation experiments with host cells in vitro have demonstrated that one species can interfere with the inflammatory response to another; for example, S. aureus has been observed to suppress IL-8 production by airway epithelial cells exposed to P. aeruginosa exoproducts74; similarly, S. aureus exoproducts have been shown to inhibit neutrophil phagocytosis of P. aeruginosa.59 On the other hand, a study of coinfection in a porcine burn wound model revealed increased inflammation and delayed healing at the site of coinfection, compared with monoinfected wounds.75 Clinically, children participating in the AREST-CF study who were culture positive for both P. aeruginosa and S. aureus were found to have significantly higher BAL markers of inflammation than children with neither species detected, with a non-significant trend towards higher inflammation in coinfected compared with monoinfected children.42 Sagel and colleagues’ investigation of the relationship between coinfection and inflammation found coinfection to be significantly correlated with increased markers of inflammation in BAL fluid compared with monoinfection with either pathogen.43 However, no significant interactions between P. aeruginosa and S. aureus were observed in this model, suggesting the effects on the inflammatory response were additive, not synergistic. Consideration of these collective clinical, animal model and in vitro findings highlights the difficulty of defining the net proinflammatory effects of this two-species coinfection in a complex system with diverse and variable host cell types, bacterial phenotypes and tissue physicochemical conditions.
Mechanisms of coexistence
How do P. aeruginosa and S. aureus coexist in the CF airway despite their great potential for competition? Putative factors promoting coexistence during infection have been identified on both the bacterial and the host side and are described in detail below.
Microbial adaptation
P. aeruginosa is notorious for its ability to adapt both phenotypically and genetically to changing conditions during chronic infection, and such adaptations have recently been demonstrated to enhance coexistence with S. aureus. Whether these adaptions occur in response to S. aureus or other unidentified selective pressures is unknown, but it is clear that CF-adapted P. aeruginosa isolates are more capable of peaceful coexistence in vitro with S. aureus. Independent groups have investigated the competitive behaviour of isogenic CF P. aeruginosa isolates collected during different evolutionary stages of infection and found that while laboratory strains and early P. aeruginosa CF isolates generally outcompete S. aureus in vitro, late CF isolates exhibit a more commensal-like behaviour with S. aureus during coculture.21 76 Frydenlund Michelsen and colleagues determined that late isolates exhibit an altered metabolic profile compared with early isolates, where decreased production of 4-hydroxy-2-alkylquinoline by the P. aeruginosa late isolates allowed for S. aureus survival.65 We recently observed similar commensal-like behaviour in P. aeruginosa isolates from coinfected patients with CF compared with isolates from patients culture negative for S. aureus. Isolates with reduced antagonism exhibited a phenotype characteristic of late CF isolates—overproduction of the exopolysaccharide alginate (referred to as mucoidy)—that consequently reduced the production of P. aeruginosa quinolone signal, siderophores and rhamnolipids, each of which are required for full P. aeruginosa virulence towards S. aureus.19 Tognon and colleagues also evolved P. aeruginosa in vitro in the presence of S. aureus and identified mutations in P. aeruginosa genes involved in the production of O-antigen; however, the role of this modification in S. aureus interactions remains unclear.77 S. aureus may also adapt to the exoproducts of P. aeruginosa in a fashion that promotes coexistence. As discussed above, S. aureus SCVs can emerge after prolonged cocultivation that are resistant to these P. aeruginosa exoproducts, enabling these two organisms to grow together in vitro.23 32 While some studies have identified other adaptive changes in S. aureus CF isolates that could conceivably facilitate coinfection, such as in metabolism and iron scavenging,78 adaptation of S. aureus remains relatively unexamined compared with P. aeruginosa.
Host factors
Coexistence may also be facilitated by certain physiochemical features of the CF airway. It has been hypothesised that host molecules may reduce P. aeruginosa toxicity towards S. aureus, or alternatively that spatial constraints may prevent the proximity necessary for these interactions. For example, following the observation that laboratory P. aeruginosa strains readily kill S. aureus in standard laboratory medium yet coexist in a chronic wound model, Smith and colleagues determined that physiological concentrations of host albumin can inhibit the antimicrobial activity of P. aeruginosa and promote coexistence.79 Additionally, Wakeman and colleagues demonstrated that exposure to a host protein that sequesters metal ions (calprotectin) can reduce P. aeruginosa production of anti-staphylococcal factors and promote interspecies coexistence.80 The authors identified bacteria with morphological characteristics of P. aeruginosa and S. aureus in an explanted CF lung containing calprotectin, suggesting that mixed communities may reside in specific lung niches more suitable for coexistence. Further efforts have been made to examine the spatial organisation of polymicrobial communities ex vivo by visualising bacterial aggregates with fluorescence in situ hybridisation in CF sputum81 and a murine wound model.82 While these studies did not specifically focus on the relative distribution of P. aeruginosa and S. aureus, these two species were observed to reside in these specimens in aggregates of variable size, in close proximity to each other and to other bacterial species; however, limited mixing between aggregates of different species was observed. Spatial segregation of species may limit and/or be driven by interspecies competition. Further studies are required to examine the spatial organisation of Pseudomonas and Staphylococcus specifically in airway secretions and to determine the functional consequences of such organisation.
Conclusions and recommendations
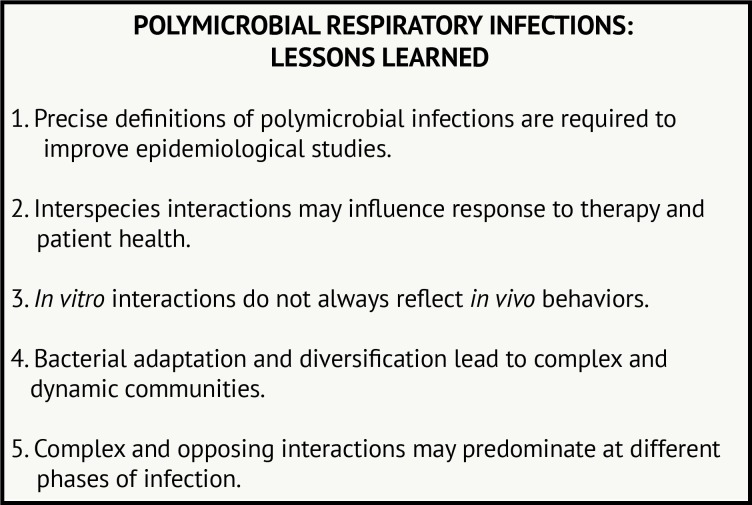
With the rise in culture-independent techniques, enormous amounts of sequence data can be acquired to examine the composition of microbial communities. Advanced culture methods are also improving microbial detection. We are now faced with the task of determining how knowing ‘who is present’ can inform how community members interact and influence outcomes. Studies from P. aeruginosa and S. aureus in patients with CF have revealed microbial physiology is altered in the presence of another species in ways that may influence patient health. These studies also demonstrate the challenge of identifying a causal link between interactions and patient outcomes. Figure 2 outlines some of the primary lessons we have learnt from studying these interactions from CF and other clinical scenarios.
Figure 2.
Polymicrobial respiratory infections: lessons learned.
In many airway infections, microbial residents can be either chronic or transient. Therefore, as in CF, one must rigorously define what is meant by ‘coinfection’. Does the identification of two or more organisms at a single time point signify coinfection or do these organisms need to be identified in multiple samples over time? It is likely that even transient interactions between species during infection contribute to community dynamics, but the contribution may be different from those of more stable community members. Crucial in this determination, however, is the manner of sampling and quantification of community member populations to ensure accurate measurements of the community composition. Several excellent reviews have addressed this important and evolving topic.48 83 84 Future studies investigating polymicrobial infections in respiratory disease should carefully consider these parameters during study design.
Several studies to date demonstrate microbial interactions can influence bacterial susceptibility to clinically relevant antibiotics; much of this research focuses on the tolerance to antibiotics by S. aureus, induced by redox-active P. aeruginosa products. However, all of these observations have been made in vitro; it is currently unknown if these interactions influence S. aureus metabolism or growth rate in the CF airway, in aggregates or other lifestyles where growth rate is already predicted to be significantly reduced. However, convergent observations from independent groups, in a variety of model systems (including biofilms), provide compelling evidence that these observations will likely be important for a range of respiratory conditions.
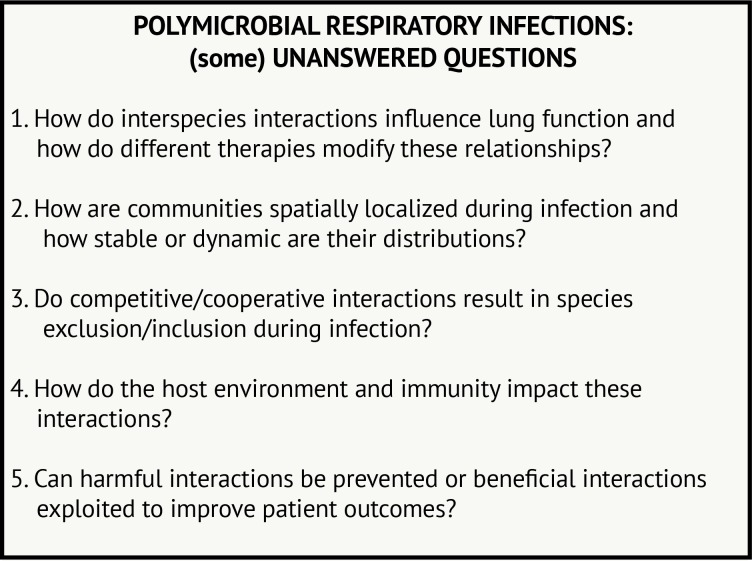
Many questions remain to be addressed regarding polymicrobial interactions in airway infections, including in CF and beyond (figure 3). Many of these questions are quite challenging to answer from epidemiological or laboratory studies alone. In CF, monitoring how respiratory microbial communities change with antibiotics or, perhaps more importantly, with newly available therapies that treat the underlying protein defect for many patients with CF85 may be particularly informative. Further development of animal or tissue culture models where rates of infection can be monitored in response to changing community members or therapeutic intervention may also lend complementary insight. Moreover, further investigation into microbial dynamics in other airway diseases will inform which interactions are broadly relevant and perhaps less influenced by environmental conditions. Thus, we propose that the lessons we have learnt from the study of P. aeruginosa–S. aureus interaction in CF airway disease can be used to study microbial interactions in a broad range of diseases, which can iteratively inform and progress the study of diverse lung infections.
Figure 3.
Polymicrobial respiratory infections: (some) unanswered questions.
Footnotes
Contributors: DHL and LRH contributed to the literature review, writing and editing of the manuscript.
Funding: This work was supported by funding from The Cystic Fibrosis Foundation to DHL (LIMOLI18F5) and the National Institutes of Health to LRH (K24HL141669).
Competing interests: None declared.
Provenance and peer review: Commissioned; externally peer reviewed.
Patient consent for publication: Not required.
References
- 1. van Leeuwenhoeck MA. Observations, communicated to the publisher by Mr. Antony van Leewenhoeck, in a Dutch letter of the 9th Octob. 1676. here English’d: concerning little animals by him observed in rain-well-sea- and snow water; as also in water wherein pepper had lain infused. Phil Trans 1677;12:821–31. [Google Scholar]
- 2. Desai MS, Seekatz AM, Koropatkin NM, et al. . A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016;167:1339–53. 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kolenbrander PE, Palmer RJ, Periasamy S, et al. . Oral multispecies biofilm development and the key role of cell–cell distance. Nat Rev Microbiol 2010;8:471–80. 10.1038/nrmicro2381 [DOI] [PubMed] [Google Scholar]
- 4. Andoh A. Physiological role of gut microbiota for maintaining human health. Digestion 2016;93:176–81. 10.1159/000444066 [DOI] [PubMed] [Google Scholar]
- 5. Cystic Fibrosis Foundation. Patient Registry 2016 Annual Data Report. Bethesda, Maryland: Cystic Fibrosis Foundation, 2017:1–94. [Google Scholar]
- 6. di Sant’Agnese PEA, Anderson DH. Celiac syndrome; chemotherapy in infections of the respiratory tract associated with cystic fibrosis of the pancreas; observations with penicillin and drugs of the sulfonamide group, with special reference to penicillin aerosol. Am J Dis Child 1946;72:17–61. [PubMed] [Google Scholar]
- 7. Zhou-Suckow Z, Duerr J, Hagner M, et al. . Airway mucus, inflammation and remodeling: emerging links in the pathogenesis of chronic lung diseases. Cell Tissue Res 2017;367:537–50. 10.1007/s00441-016-2562-z [DOI] [PubMed] [Google Scholar]
- 8. Hotterbeek A, Kumar-Singh S, Goossens H, et al. . In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp . Front Cell Infect Microbiol 2017;7:175 10.3389/fcimb.2017.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nguyen AT, Oglesby-Sherrouse AG. Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infections. Appl Microbiol Biotechnol 2016;100:6141–8. 10.1007/s00253-016-7596-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hurley MN, Fogarty A, McKeever TM, et al. . Early respiratory bacterial detection and antistaphylococcal antibiotic prophylaxis in young children with cystic fibrosis. Ann Am Thorac Soc 2018;15:42–8. 10.1513/AnnalsATS.201705-376OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smyth AR, Rosenfeld M. Prophylactic anti-staphylococcal antibiotics for cystic fibrosis. Cochrane Database Syst Rev 2017;4:CD001912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ratjen F, Comes G, Paul K, et al. . Effect of continuous antistaphylococcal therapy on the rate of P. aeruginosa acquisition in patients with cystic fibrosis. Pediatr Pulmonol 2001;31:13–16. [DOI] [PubMed] [Google Scholar]
- 13. Cystic Fibrosis Trust. Standards for the clinical care of children and adults with cystic fibrosis in the UK: 2nd edn, 2011. [Google Scholar]
- 14. Mogayzel PJ, Naureckas ET, Robinson KA, et al. . Cystic fibrosis pulmonary guidelines. Chronic medications for maintenance of lung health. Am J Respir Crit Care Med 2013;187:680–9. [DOI] [PubMed] [Google Scholar]
- 15. Granchelli AM, Adler FR, Keogh RH, et al. . Microbial interactions in the cystic fibrosis airway. J Clin Microbiol 2018;56:299 10.1128/JCM.00354-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosenfeld M, Emerson J, McNamara S, et al. . Risk factors for age at initial Pseudomonas acquisition in the cystic fibrosis epic observational cohort. J Cyst Fibros 2012;11:446–53. 10.1016/j.jcf.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cigana C, Bianconi I, Baldan R, et al. . Staphylococcus aureus impacts Pseudomonas aeruginosa chronic respiratory disease in murine models. J Infect Dis 2018;217:933–42. 10.1093/infdis/jix621 [DOI] [PubMed] [Google Scholar]
- 18. Goddard AF, Staudinger BJ, Dowd SE, et al. . Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci U S A 2012;109:13769–74. 10.1073/pnas.1107435109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Limoli DH, Whitfield GB, Kitao T, et al. . Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. MBio 2017;8 10.1128/mBio.00186-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filkins LM, Graber JA, Olson DG, et al. . Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol 2015;197:2252–64. 10.1128/JB.00059-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baldan R, Cigana C, Testa F, et al. . Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS One 2014;9:e89614 10.1371/journal.pone.0089614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korgaonkar AK, Whiteley M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol 2011;193:909–17. 10.1128/JB.01175-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoffman LR, Deziel E, D’Argenio DA, et al. . Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa . Proc Natl Acad Sci USA 2006;103:19890–5. 10.1073/pnas.0606756104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Machan ZA, Taylor GW, Pitt TL, et al. . 2-Heptyl-4-hydroxyquinoline N -oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa . J Antimicrob Chemother 1992;30:615–23. 10.1093/jac/30.5.615 [DOI] [PubMed] [Google Scholar]
- 25. Mashburn LM, Jett AM, Akins DR, et al. . Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 2005;187:554–66. 10.1128/JB.187.2.554-566.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nguyen AT, Jones JW, Ruge MA, et al. . Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa . J Bacteriol 2015;197:2265–75. 10.1128/JB.00072-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Korgaonkar A, Trivedi U, Rumbaugh KP, et al. . Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA 2013;110:1059–64. 10.1073/pnas.1214550110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haba E, Pinazo A, Jauregui O, et al. . Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol Bioeng 2003;81:316–22. 10.1002/bit.10474 [DOI] [PubMed] [Google Scholar]
- 29. Bharali P, Saikia JP, Ray A, et al. . Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: a novel chemotaxis and antibacterial agent. Colloids Surf B Biointerfaces 2013;103:502–9. 10.1016/j.colsurfb.2012.10.064 [DOI] [PubMed] [Google Scholar]
- 30. Deziel E, Lepine F, Milot S, et al. . Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA 2004;101:1339–44. 10.1073/pnas.0307694100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hazan R, Que YA, Maura D, et al. . Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Current Biology 2016;26:195–206. 10.1016/j.cub.2015.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Biswas L, Biswas R, Schlag M, et al. . Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa . Appl Environ Microbiol 2009;75:6910–2. 10.1128/AEM.01211-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith EE, Buckley DG, Wu Z, et al. . Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA 2006;103:8487–92. 10.1073/pnas.0602138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffman LR, Kulasekara HD, Emerson J, et al. . Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros 2009;8:66–70. 10.1016/j.jcf.2008.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwerdt M, Neumann C, Schwartbeck B, et al. . Staphylococcus aureus in the airways of cystic fibrosis patients—a retrospective long-term study. Int J Med Microbiol 2018;308:631–9. 10.1016/j.ijmm.2018.02.003 [DOI] [PubMed] [Google Scholar]
- 36. Maliniak ML, Stecenko AA, McCarty NA. A longitudinal analysis of chronic MRSA and Pseudomonas aeruginosa co-infection in cystic fibrosis: a single-center study. J Cyst Fibros 2016;15:350–6. 10.1016/j.jcf.2015.10.014 [DOI] [PubMed] [Google Scholar]
- 37. Ahlgren HG, Benedetti A, Landry JS, et al. . Clinical outcomes associated with Staphylococcus aureus and Pseudomonas aeruginosa airway infections in adult cystic fibrosis patients. BMC Pulm Med 2015;15:918 10.1186/s12890-015-0062-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Twr L, Brownlee KG, Conway SP, et al. . Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. J Cyst Fibros 2003;2:29–34. [DOI] [PubMed] [Google Scholar]
- 39. Wolter DJ, Emerson JC, McNamara S, et al. . Staphylococcus aureus small-colony variants are independently associated with worse lung disease in children with cystic fibrosis. Clin Infect Dis 2013;57:384–91. 10.1093/cid/cit270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Limoli DH, Yang J, Khansaheb MK, et al. . Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur J Clin Microbiol Infect Dis 2016;35:947–53. 10.1007/s10096-016-2621-0 [DOI] [PubMed] [Google Scholar]
- 41. Hubert D, Réglier-Poupet H, Sermet-Gaudelus I, et al. . Association between Staphylococcus aureus alone or combined with Pseudomonas aeruginosa and the clinical condition of patients with cystic fibrosis. J Cyst Fibros 2013;12:497–503. 10.1016/j.jcf.2012.12.003 [DOI] [PubMed] [Google Scholar]
- 42. Gangell C, Gard S, Douglas T, et al. . Inflammatory responses to individual microorganisms in the lungs of children with cystic fibrosis. Clin Infect Dis 2011;53:425–32. 10.1093/cid/cir399 [DOI] [PubMed] [Google Scholar]
- 43. Sagel SD, Gibson RL, Emerson J, et al. . Impact of Pseudomonas and Staphylococcus infection on inflammation and clinical status in young children with cystic fibrosis. J Pediatr 2009;154:183–8. 10.1016/j.jpeds.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hudson VL, Wielinski CL, Regelmann WE. Prognostic implications of initial oropharyngeal bacterial flora in patients with cystic fibrosis diagnosed before the age of two years. J Pediatr 1993;122:854–60. 10.1016/S0022-3476(09)90007-5 [DOI] [PubMed] [Google Scholar]
- 45. Seidler D, Griffin M, Nymon A, et al. . Throat swabs and sputum culture as predictors of P. aeruginosa or S. aureus lung colonization in adult cystic fibrosis patients. PLoS One 2016;11:e0164232 10.1371/journal.pone.0164232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hogan DA, Willger SD, Dolben EL, et al. . Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS One 2016;11:e0149998 10.1371/journal.pone.0149998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rosenfeld M, Emerson J, Accurso F, et al. . Diagnostic accuracy of oropharyngeal cultures in infants and young children with cystic fibrosis. Pediatr Pulmonol 1999;28:321–8. [DOI] [PubMed] [Google Scholar]
- 48. O’Toole GA. CF Airway microbiome: overturning the old, opening the way for the new. J Bacteriol 2017;17:JB.00561-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Héry-Arnaud G, Boutin S, Cuthbertson L, et al. . The lung and gut microbiome: what has to be taken into consideration for cystic fibrosis? J Cyst Fibros 2019;18:13–21. [DOI] [PubMed] [Google Scholar]
- 50. Caverly LJ, LiPuma JJ. Cystic fibrosis respiratory microbiota: unraveling complexity to inform clinical practice. Expert Rev Respir Med 2018;12:857–65. 10.1080/17476348.2018.1513331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quinn RA, Whiteson K, Lim YW, et al. . Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microbiomes 2016;2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rogers GB, Carroll MP, Serisier DJ, et al. . Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 2004;42:5176–83. 10.1128/JCM.42.11.5176-5183.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Harun SN, Wainwright C, Klein K, et al. . A systematic review of studies examining the rate of lung function decline in patients with cystic fibrosis. Paediatr Respir Rev 2016;20:55–66. 10.1016/j.prrv.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 54. Emerson J, Rosenfeld M, McNamara S, et al. . Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol 2002;34:91–100. 10.1002/ppul.10127 [DOI] [PubMed] [Google Scholar]
- 55. Kosorok MR, Zeng L, West SEH, et al. . Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol 2001;32:277–87. 10.1002/ppul.2009.abs [DOI] [PubMed] [Google Scholar]
- 56. Caudri D, Turkovic L, Ng J, et al. . The association between Staphylococcus aureus and subsequent bronchiectasis in children with cystic fibrosis. J Cyst Fibros 2018;17:462–9. 10.1016/j.jcf.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 57. Junge S, Görlich D, den Reijer M, et al. . Factors associated with worse lung function in cystic fibrosis patients with persistent Staphylococcus aureus . PLoS One 2016;11:e0166220 10.1371/journal.pone.0166220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pastar I, Nusbaum AG, Gil J, et al. . Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE 2013;8:e56846 10.1371/journal.pone.0056846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Armbruster CR, Wolter DJ, Mishra M, et al. . Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa . MBio 2016;7:277 10.1128/mBio.00538-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beaudoin T, Yau YCW, Stapleton PJ, et al. . Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes 2017;3:25 10.1038/s41522-017-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rada B, Leto TL. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol 2013;21:73–81. 10.1016/j.tim.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wretlind B, Pavlovskis OR. Pseudomonas aeruginosa elastase and its role in Pseudomonas infections. Rev Infect Dis 1983;5(Suppl 5):S998–1004. 10.1093/clinids/5.Supplement_5.S998 [DOI] [PubMed] [Google Scholar]
- 63. Gellatly SL, Hancock REW. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 2013;67:159–73. 10.1111/2049-632X.12033 [DOI] [PubMed] [Google Scholar]
- 64. Orazi G, O’Toole GA. Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. MBio 2017;8:e00873–17. 10.1128/mBio.00873-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Frydenlund Michelsen C, Hossein Khademi SM, Krogh Johansen H, et al. . Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction. Isme J 2016;10:1323–36. 10.1038/ismej.2015.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Radlinski L, Rowe SE, Kartchner LB, et al. . Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus . PLoS Biol 2017;15:e2003981 10.1371/journal.pbio.2003981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dekimpe V, Deziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009;155:712–23. 10.1099/mic.0.022764-0 [DOI] [PubMed] [Google Scholar]
- 68. Feltner JB, Wolter DJ, Pope CE, et al. . LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa . MBio 2016;7:e01513–6. 10.1128/mBio.01513-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Proctor RA, von Eiff C, Kahl BC, et al. . Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 2006;4:295–305. 10.1038/nrmicro1384 [DOI] [PubMed] [Google Scholar]
- 70. Besier S, Smaczny C, von Mallinckrodt C, et al. . Prevalence and clinical significance of Staphylococcus aureus small-colony variants in cystic fibrosis lung disease. J Clin Microbiol 2007;45:168–72. 10.1128/JCM.01510-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Roesch EA, Nichols DP, Chmiel JF. Inflammation in cystic fibrosis: an update. Pediatr Pulmonol 2018;53:S30–50. 10.1002/ppul.24129 [DOI] [PubMed] [Google Scholar]
- 72. McAllister F, Henry A, Kreindler JL, et al. . Role of IL-17A, IL-17F, and the IL-17 receptor in regulating growth-related oncogene-alpha and granulocyte colony-stimulating factor in bronchial epithelium: implications for airway inflammation in cystic fibrosis. J Immunol 2005;175:404–12. 10.4049/jimmunol.175.1.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Serhan CN. Treating inflammation and infection in the 21st century: new hints from decoding resolution mediators and mechanisms. FASEB J 2017;31:1273–88. 10.1096/fj.201601222R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chekabab SM, Silverman RJ, Lafayette SL, et al. . Staphylococcus aureus inhibits IL-8 responses induced by Pseudomonas aeruginosa in airway epithelial cells. PLoS One 2015;10:e0137753 10.1371/journal.pone.0137753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chaney SB, Ganesh K, Mathew-Steiner S, et al. . Histopathological comparisons of Staphylococcus aureus and Pseudomonas aeruginosa experimental infected porcine burn wounds . Wound Repair Regen 2017;25:541–9. 10.1111/wrr.12527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Michelsen CF, Christensen A-MJ, Bojer MS, et al. . Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage. J Bacteriol 2014;196:3903–11. 10.1128/JB.02006-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tognon M, Köhler T, Gdaniec BG, et al. . Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa . Isme J 2017;11:2233–43. 10.1038/ismej.2017.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Treffon J, Block D, Moche M, et al. . Adaptation of Staphylococcus aureus to airway environments in patients with cystic fibrosis by upregulation of superoxide dismutase M and iron-scavenging proteins. J Infect Dis 2018;217:1453–61. 10.1093/infdis/jiy012 [DOI] [PubMed] [Google Scholar]
- 79. Smith AC, Rice A, Sutton B, et al. . Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect Immun 2017;85:e00116-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wakeman CA, Moore JL, Noto MJ, et al. . The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat Commun 2016;7:11951 10.1038/ncomms11951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rudkjøbing VB, Thomsen TR, Alhede M, et al. . The microorganisms in chronically infected end-stage and non-end-stage cystic fibrosis patients. FEMS Immunol Med Microbiol 2012;65:236–44. 10.1111/j.1574-695X.2011.00925.x [DOI] [PubMed] [Google Scholar]
- 82. Dalton T, Dowd SE, Wolcott RD, et al. . An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 2011;6:e27317 10.1371/journal.pone.0027317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Paul L. Is bronchoscopy an obsolete tool in cystic fibrosis? The role of bronchoscopy in cystic fibrosis and its clinical use. J Thorac Dis 2017;9:S1139–45. 10.21037/jtd.2017.06.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res 2012;160:258–66. 10.1016/j.trsl.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schmidt BZ, Haaf JB, Leal T, et al. . Cystic fibrosis transmembrane conductance regulator modulators in cystic fibrosis: current perspectives. Clin Pharmacol 2016;8:127–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Banjar HH. Clinical profile of Saudi children with bronchiectasis. Indian J Pediatr 2007;74:149–52. 10.1007/s12098-007-0008-z [DOI] [PubMed] [Google Scholar]
- 87. Eastham KM, et al. . The need to redefine non-cystic fibrosis bronchiectasis in childhood. Thorax 2004;59:324–7. 10.1136/thx.2003.011577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Karadag B, Karakoc F, Ersu R, et al. . Non-cystic-fibrosis bronchiectasis in children: a persisting problem in developing countries. Respiration 2005;72:233–8. 10.1159/000085362 [DOI] [PubMed] [Google Scholar]
- 89. Am L, Sonnappa S, Lex C, et al. . Non-CF bronchiectasis: does knowing the aetiology lead to changes in management? Eur Respir J 2005;26:8–14. [DOI] [PubMed] [Google Scholar]
- 90. Zaid AA, Elnazir B, Greally P. A decade of non-cystic fibrosis bronchiectasis 1996–2006. Ir Med J 2010;103:77–9. [PubMed] [Google Scholar]
- 91. Hare KM, Grimwood K, Leach AJ, et al. . Respiratory bacterial pathogens in the nasopharynx and lower airways of Australian indigenous children with bronchiectasis. J Pediatr 2010;157:1001–5. 10.1016/j.jpeds.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 92. Kapur N, Grimwood K, Masters IB, et al. . Lower airway microbiology and cellularity in children with newly diagnosed non-CF bronchiectasis. Pediatr Pulmonol 2012;47:300–7. 10.1002/ppul.21550 [DOI] [PubMed] [Google Scholar]
- 93. Chalmers JD, Aliberti S, Filonenko A, et al. . Characterization of the “frequent exacerbator phenotype” in bronchiectasis. Am J Respir Crit Care Med 2018;197:1410–20. 10.1164/rccm.201711-2202OC [DOI] [PubMed] [Google Scholar]
- 94. Marino PJ, Wise MP, Smith A, et al. . Community analysis of dental plaque and endotracheal tube biofilms from mechanically ventilated patients. J Crit Care 2017;39:149–55. 10.1016/j.jcrc.2017.02.020 [DOI] [PubMed] [Google Scholar]
- 95. Behnia M, Logan SC, Fallen L, et al. . Nosocomial and ventilator-associated pneumonia in a community hospital intensive care unit: a retrospective review and analysis. BMC Res Notes 2014;7:232 10.1186/1756-0500-7-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Thakuria B, Singh P, Agrawal S, et al. . Profile of infective microorganisms causing ventilator-associated pneumonia: a clinical study from resource limited intensive care unit. J Anaesthesiol Clin Pharmacol 2013;29:361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cairns S, Thomas JG, Hooper SJ, et al. . Molecular analysis of microbial communities in endotracheal tube biofilms. PLoS ONE 2011;6:e14759 10.1371/journal.pone.0014759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sands KM, Wilson MJ, Lewis MAO, et al. . Respiratory pathogen colonization of dental plaque, the lower airways, and endotracheal tube biofilms during mechanical ventilation. J Crit Care 2017;37:30–7. 10.1016/j.jcrc.2016.07.019 [DOI] [PubMed] [Google Scholar]