Abstract
Background
Epicutaneous immunotherapy (EPIT) is a promising method for treating food allergies. In animal models, EPIT induces sustained unresponsiveness and prevents further sensitization mediated by Tregs. Here, we elucidate the mechanisms underlying the therapeutic effect of EPIT, by characterizing the kinetics of DNA methylation changes in sorted cells from spleen and blood and by evaluating its persistence and bystander effect compared to oral immunotherapy (OIT).
Methods
BALB/c mice orally sensitized to peanut proteins (PPE) were treated by EPIT using a PPE‐patch or by PPE‐OIT. Another set of peanut‐sensitized mice treated by EPIT or OIT were sacrificed following a protocol of sensitization to OVA. DNA methylation was analyzed during immunotherapy and 8 weeks after the end of treatment in sorted cells from spleen and blood by pyrosequencing. Humoral and cellular responses were measured during and after immunotherapy.
Results
Analyses showed a significant hypermethylation of the Gata3 promoter detectable only in Th2 cells for EPIT from the 4th week and a significant hypomethylation of the Foxp3 promoter in CD62L+ Tregs, which was sustained only for EPIT. In addition, mice treated with EPIT were protected from subsequent sensitization and maintained the epigenetic signature characteristic for EPIT.
Conclusions
Our study demonstrates that EPIT leads to a unique and stable epigenetic signature in specific T‐cell compartments with downregulation of Th2 key regulators and upregulation of Treg transcription factors, likely explaining the sustainability of protection and the observed bystander effect.
Keywords: bystander effect, epicutaneous immunotherapy, food allergy, DNA methylation, Tregs
1. INTRODUCTION
Allergen‐specific immunotherapy is an attractive strategy for the treatment of food allergy.1, 2 Epicutaneous immunotherapy (EPIT) has proven efficacious in animal models 3, 4, 5, 6 and more recently in humans.7, 8, 9 The aim of EPIT is to reduce sensitivity to an allergen (desensitization) or to abolish sensitivity altogether (tolerance or sustained unresponsiveness, defined as the continued absence or reduction of sensitivity after the completion of therapy and discontinuation of repeated allergen exposure). The mechanism of immune tolerance to allergens remains largely unknown, but preliminary studies demonstrated increased levels of allergen‐specific blocking IgG antibody associated with a reduction in specific IgE levels,3, 4 reduced recruitment of inflammatory cells such as eosinophils,5 and prevention of sensitization to further allergens.6 In this study, we investigated a well‐characterized mouse model of peanut sensitization combined with EPIT to define epigenetic mechanisms underlying the induction of desensitization to peanut with EPIT. More precisely, the DNA methylation patterns were evaluated in the gene regulatory regions of four key transcription factors involved in T‐cell lineage differentiation (Gata3 (Th2), Tbx21 (Th1), Rorγ (Th17), and Foxp3 (Tregs)).
As key immune‐regulatory cells, Tregs have been shown to play a pivotal role in maintaining immune tolerance following epicutaneous immunotherapy in a mouse model.10, 11 Moreover, several studies have demonstrated that epigenetic modifications in CpG‐rich regions within the FOXP3 locus are associated with stable FOXP3 expression and cell‐suppressive functions of Tregs.12, 13, 14, 15, 16
We hypothesized that Foxp3+ Tregs might play a key role in the process of immune tolerance in both animal models and humans and thus investigated epigenetic modification at the Foxp3 locus within T cells. We also evaluated whether the decrease in IgE and/or Th2 cytokines could be associated with epigenetic alterations of the Th2 key regulator Gata3. To decipher the regulation of Treg and Th2 signaling, we engaged in epigenetic analyses of specific T‐cell subsets (Th1, Th2, CD62L+ Tregs, and CD62L− Tregs).
Here, we identify an epigenetic signature of Th2 cells and CD62L+ Tregs unique to EPIT‐treated animals that could be considered for monitoring immunotherapy.
2. MATERIALS AND METHODS
2.1. Animals
Three‐week‐old female BALB/c mice (Charles River, Lyon, France) were housed under standard animal husbandry conditions. All experiment was performed according to the European Community rules on animal care and with a positive evaluation from the Ethical Committee no 26 (2012‐041). Mice were acclimated for 1 week before starting the sensitization to peanut protein (PPE).
2.2. Induction of sensitization and epicutaneous immunotherapy
Mice were first sensitized to PPE by means of 6 intragastric gavages as previously described,3, 4, 17 with 1 mg of PPE mixed with 10 μg of Cholera Toxin (Servibio, USA). Sensitization was monitored by evaluating blood samples for the production of specific IgE (ie, 10 days after the last gavage) as detailed in the Data S1.
2.2.1. Experiment 1
In a first experiment, sensitized mice were divided into two groups, one group treated by EPIT and the other one not treated (Sham). Epicutaneous immunotherapy was performed using the Viaskin® patch (DBV Technologies, Paris, France) loaded with 100 μg of PPE and an 8‐week consecutive treatment protocol, which has previously been described (Figure 1A).3, 4 Two groups of 8 mice were sacrificed after sensitization at weeks 1, 2, 4, and 8 of immunotherapy and 8 weeks after the end of immunotherapy (8 + 8 weeks).
Figure 1.
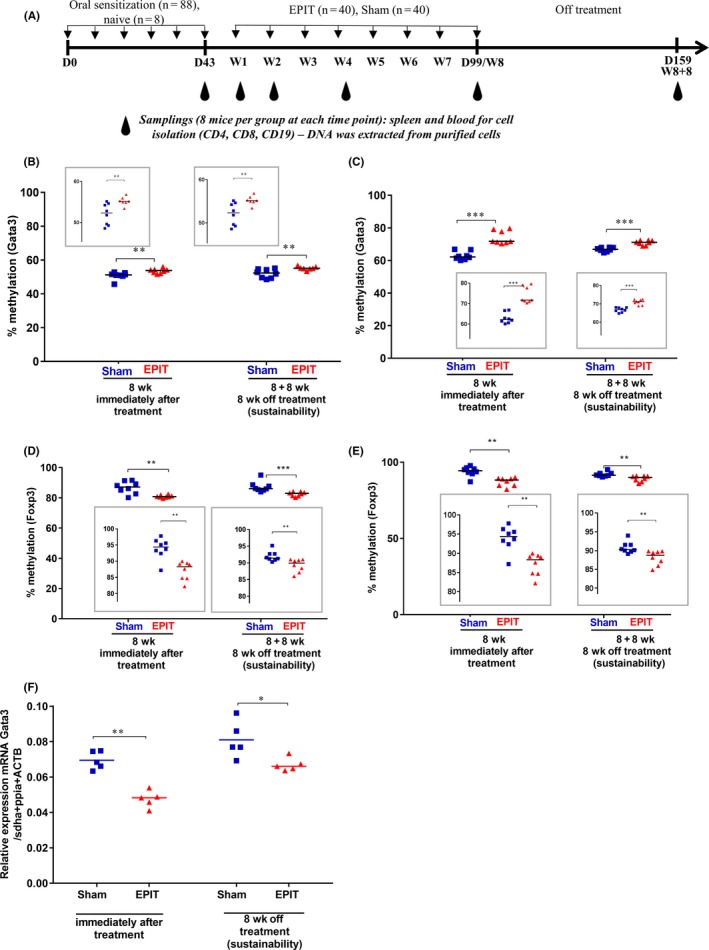
Epicutaneous immunotherapy (EPIT) induces DNA hypermethylation of Gata3 and hypomethylation of Foxp3 in CD4+ cells purified from spleen and blood during EPIT, which is accompanied by a reduction of Gata3 mRNA expression in CD4+ cells purified from the spleen. A, Experimental design for the methylation analysis of DNA isolated from CD4+ T cells, CD8+ T cells, and CD19+ B cells (of spleen and blood) occurring during EPIT for sensitized mice epicutaneously treated using a patch loaded with peanut protein extract (EPIT) or a placebo (Sham). Analysis of the methylation levels of the Gata3 promoter in CD4+ cells isolated from (B) spleen and (C) whole blood at week 8 (8 wk) of EPIT and 8 weeks after the end of EPIT (8 + 8 wk). Analysis of the methylation levels of Foxp3 in CD4+ cells isolated from (D) spleen and (E) whole blood at week 8 (8 wk) of EPIT and 8 weeks after the end of EPIT (8 + 8 wk). (F) Measurement of the expression level of Gata3 mRNA by RT‐qPCR. Results are expressed as individual data and median. Differences between groups were analyzed by a Kruskall‐Wallis test followed by Dunn's multiple comparison test. *P < .05, **P < .01 and ***P < .001
2.2.2. Experiment 2
In a second experiment, sensitized mice treated as indicated in experiment 1 were sacrificed at the end of the sensitization procedure, at the end of EPIT and 8 weeks after the end of EPIT for harvesting spleens for cell extraction and sorting of Th1, Th2, CD62L+ Tregs, and CD62L− Tregs (detailed procedure in the Data S1).
2.2.3. Experiment 3
In a third experiment, sensitized mice were divided into three groups, one group treated by EPIT, the second one by oral immunotherapy (OIT), and the third one not treated (Sham). Epicutaneous immunotherapy was performed as described above, and OIT was performed following the protocol published by Diozeghy et al11 adapted from Leonard et al18 consisting of the administration of 1 mg of PPE the first week, 2 mg the second week, then 5 mg the 5 following weeks. Mice were sacrificed as previously described, with groups dedicated to specific time points. In addition, 3 groups (EPIT, OIT, and Sham) were exposed to a procedure of sensitization to ovalbumin (OVA) after completing the immunotherapy phase as previously described 3, 19 and then were sacrificed 10 days after the end of sensitization to OVA.
For the two experiments, blood and spleens were collected after sacrifice to stimulate in vitro splenocytes and/or purify cell populations using magnetic beads isolation kits (Miltenyi Biotec, Paris, France).
2.2.4. Experiment 4
In a fourth experiment, we applied a previously described procedure to isolate Tregs from milk‐sensitized mice treated by EPIT.6 More precisely, a group of mice were first sensitized to milk and then epicutaneously treated with milk EPIT or placebo (Sham) patch. After sacrifice, spleens were harvested to isolate 2 subsets of Tregs (at least 95% purity): CD62L+ expressing cells or not (detailed procedure in Data S1). Tregs were then transferred to nonsensitized mice before initiating sensitization to peanuts.6 A group of mice, which did not receive Tregs and were sensitized to peanuts, served as positive controls. Mice were challenged intravenously, and thirty minutes after the challenge, body temperature was measured as well as mouse mast cell protease‐1 (mMCP1) in plasma.6 This experiment was reproduced 2 times.
2.3. Evaluation of the methylation level of transcription factors in cells isolated from spleen and blood
Amplification products were designed to regions known to exhibit T‐cell lineage‐specific differential DNA methylation including the Treg‐specific demethylated region in Foxp3,15 the CpG island in the first exon of Rorc,20, 21 the CpG island in the first intron of Tbx21,22 and the promoter‐associated CpG island of Gata3.6 Details on the pyrosequencing analysis are given in the Data S1.23
2.4. Measurement of Gata3 mRNA expression
mRNA expression of Gata3 was analyzed by qPCR using the following amplification primers: Gata3 5′‐GAGGAGGAACGCTAATGG‐3′ and 5′‐TTTCGATTTGCTAGACATCTTC‐3′ and normalized to the geometric mean of the expression of three reference genes (Sdha, Actb, and Ppia; detailed procedure in Data S1).24
2.5. Statistical analysis
Differences between groups were analyzed by a Mann‐Whitney, ANOVA, or Kruskall‐Wallis test followed by post hoc analysis with Dunn's or Tukey's multiple comparisons test. The GraphPad Prism Software 6.0 (San Diego, CA, USA) was used for statistical analysis. Results are expressed as individuals and/or median with range. A value of P < .05 after correction for multiple testing was considered significant.
3. RESULTS
3.1. Epicutaneous immunotherapy modifies the level of DNA methylation for Th2 and Treg transcription factors
In a first experiment, sensitized mice were divided into two groups, one group treated by EPIT and the other one not treated (Sham) (Figure 1A). The level of DNA methylation at the Gata3 promoter in spleen and blood CD4+ cells was comparable in mice peanut sensitized and in naive mice (Figure S1 a and b). During EPIT, the level of methylation increased significantly from the 4th week of treatment in CD4+ cells from spleen of peanut‐sensitized mice (P < .05, Figure S1a) and blood (P < .01, Figure S1b). The methylation level observed at the end of EPIT (week 8) (Figure 1B and C) was sustained 8 weeks after the end of EPIT (P < .01 in spleen and P < .001 in blood). The Gata3 hypermethylation was accompanied by a decrease in Gata3 mRNA expression observed at the end (P < .01 vs Sham) and 8 weeks after the end of EPIT (P < .05 vs Sham) (Figure 1F). No change in the DNA methylation was seen at the Gata3 promoter in CD19+ and CD8+ cells from spleen and blood (Figure S2a‐d).
In the Treg‐specific demethylated regions (TSDR) of Foxp3, methylation in CD4+ cells in spleen and blood was comparable in mice sensitized to peanut and in naive mice (Figure S1c and d). The level of methylation for the TSDR of Foxp3 significantly decreased during EPIT, from the 4th week of treatment (Figure S1c‐d, P < .001 and P < .01) until the end (Figure 1D and E, P < .01 for both). The methylation decrease was sustained 8 weeks after the end of treatment in spleen (P < .001) and blood (P < .01) (Figure 1D and E). The increase in the expression of Foxp3 has previously been published.5, 10 No change was observed in the other cells (CD8+ and CD19+) isolated from spleen and blood as expected (Figure S2e‐h).
No change was observed for the Rorγ and Tbx21 promoters whatever the source (spleen or blood) or the cell type (CD4+, CD8+ or CD19+) (Figure S3 for RORγ and Figure S4 for Tbx21).
3.2. Foxp3 demethylation and Gata3 hypermethylation correlate with biological effects induced by EPIT
Specific antibodies (sIgE, sIgG1, and sIgG2a) were monitored after sensitization and every 2 weeks during the 8 weeks of EPIT and after 8 weeks without EPIT (Figure 2A‐C). During the 8 weeks of EPIT, sIgE slowly decreased and there was a significant difference between EPIT and Sham (P < .05) at the end of the treatment (Figure 2A). This significant difference was maintained after the end of the treatment by EPIT. There was no modification during and after EPIT for sIgG1 (Figure 2B), while sIgG2a significantly increased in EPIT‐treated mice compared with Sham (P < .05) from the 2nd week until the end of immunotherapy (Figure 2C). No changes were observed in Sham‐treated mice. The significant increase in sIgG2a observed for EPIT was sustained 8 weeks after the end of immunotherapy (P < .01).
Figure 2.
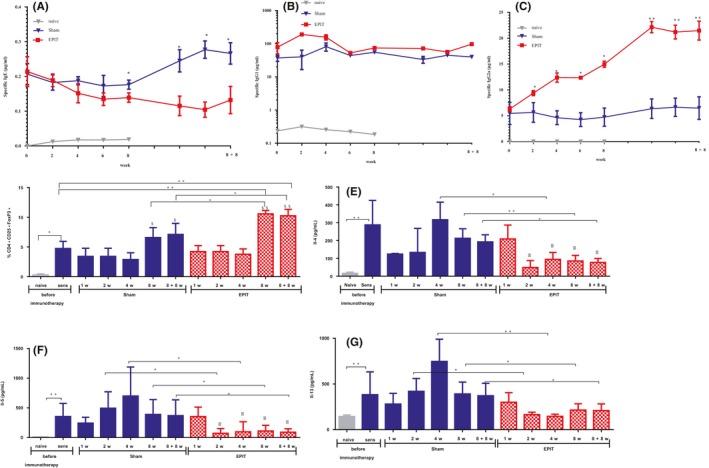
Epicutaneous immunotherapy (EPIT) decreases the Th2 profile in serological and cellular responses (spleen) to peanut protein extract. (A‐C) Sera were harvested after sensitization to peanut protein (PPE) (W0) and during EPIT (W2, W4, W6, and W8) and after the end of EPIT, in the Sham, EPIT, and naive groups until 8 weeks off treatment (W8 + 8), to measure, respectively IgE, IgG1, and IgG2a reactive to PPE. (D) The percentage of CD4+ CD25+Foxp3+ cells was evaluated after harvesting of splenocytes. (E‐G) Measurement of cytokines secreted by splenocytes after 3 d of reactivation. Results are expressed as mean ± SD in μg/mL or in percentage of total splenocytes for Tregs and in pg/mL for cytokines. Differences between groups were analyzed by a Kruskall‐Wallis test followed by Dunn's multiple comparison test, and differences between time points for the same treatment groups were analyzed by ANOVA followed by Tukey's multiple comparisons test. *P < .05, **P < .01, $ P < .05 Sham 8 wk or Sham 8 + 8 wk vs Sham 1 wk, 2 wk, 4 wk; $$ P < .01 EPIT 8 wk or EPIT 8 + 8 wk vs EPIT 1 wk, 2 wk, 4 wk; Φ P < .05 EPIT 1 wk vs EPIT 2 wk, 4 wk, 8 wk, 8 + 8wk
Moreover, the proportion of CD4+CD25+Foxp3+ Tregs was significantly enhanced when analyzed directly after harvesting splenocytes in peanut‐sensitized mice compared with negative control/naïve mice and further increased by EPIT compared with Sham after 8 weeks of immunotherapy (P < .05, Figure 2D). This induction remained significant compared with Sham after the end of the treatment of EPIT (P < .05, Figure 2D). No significant increase in Tregs compared with sensitized mice was obtained for Sham whatever the time point analyzed.
In parallel, Th2 cytokines were measured in the supernatants of in vitro reactivated splenocytes. Splenocytes from EPIT‐treated mice produced significantly less Il4, Il5, and Il13 (at least P < .05), compared with Sham, from the 2nd week of treatment until the end of treatment (Figure 2E‐G) and remained significantly lower 8 weeks after the end of EPIT (P < .05, Figure 2E‐G). We evaluated also expression of Ifn‐γ and Il10, but did not observe any change after EPIT in comparison with Sham (data not shown).
3.3. Foxp3 demethylation and Gata3 hypermethylation occur in different cell populations, respectively CD62L+ Tregs and Th2 cells
Tregs and Th2 cells are involved in allergic sensitization to food, and we monitored whether those changes reflect an altered distribution of T‐cell subsets or a change in the epigenetic profile of a particular T‐cell subpopulation. After peanut sensitization and 8 weeks of peanut EPIT, the proportions of Th1, Th2, and Foxp3+ Treg cells were analyzed by cell sorting (Figure 3A and D). The proportion of Th1 and Th2 cells was not modified by EPIT (Figure 3B‐C). As previously shown in independent experiments,11 Foxp3+ Tregs increased during EPIT to a similar extent in CD62L+ and CD62L− subsets (Figure 3E‐G). The methylation level for Gata3 did not vary in Th1, CD62L+, and CD62L− Tregs (Figure 4B, F and H). In contrast, the methylation level of Gata3 increased significantly following EPIT in Th2 cells (P < .05; Figure 4D) and was maintained 8 weeks after the end of EPIT (P < .05; Figure 4D). Of note, Th2 cells were the only analyzed cell subtype in which Gata3 was not methylated, Foxp3 demethylation did not occur in Th1 (Figure 4C), Th2 (Figure 4E), which were found nearly completely methylated, or CD62L− Tregs (Figure 4I). A significant decrease in methylation level in Foxp3 was only seen in CD62L+ Tregs after EPIT (P < .05 vs Sham; Figure 4G) and was maintained 8 weeks after the end of EPIT (P < .001 vs Sham; Figure 4G).
Figure 3.
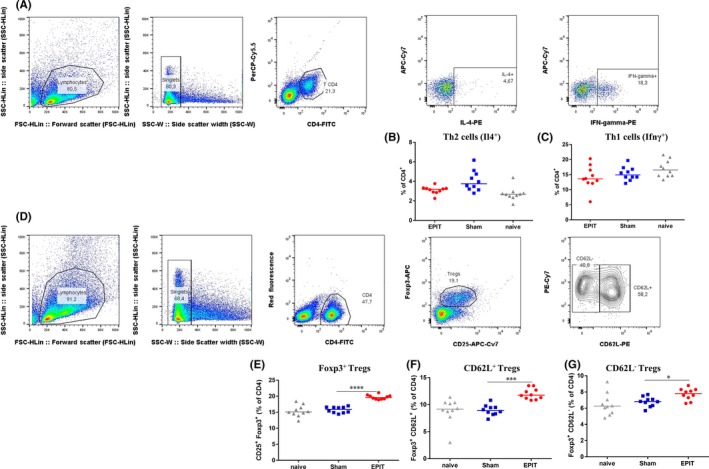
Epicutaneous immunotherapy (EPIT) does not modify the proportions of Th1 and Th2 cells, but increases Foxp3+ Tregs, CD62L+, and CD62L− Tregs obtained from spleen. Gating strategy (A and D) and analysis of the proportions of Th2 (B), Th1 (C) after a short in vitro stimulation with PMA‐ionomycin, and Foxp3+ Tregs (E), Foxp3+ CD62L+ Tregs (F), and Foxp3+ CD62L− Tregs (G) ex vivo after EPIT. For CD4+, IL4+, and IFNγ+ gates, the y‐axis was arbitrarily defined with a fluorochrome different from those used in the gating strategy and without any specific antibody (PerCP‐Cy5.5 or APR‐Cy7). Results are expressed as mean ± SD in percentage of total CD4+ T cells. Differences between groups were analyzed by a Mann‐Whitney test. *P < .05, ***P < .001
Figure 4.
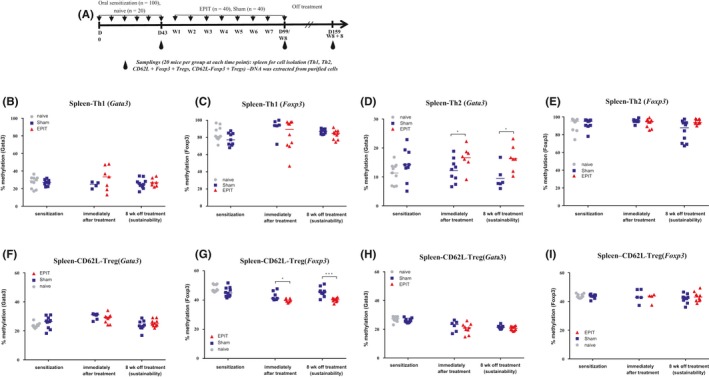
Hypermethylation of Gata3 is restricted to Th2 cells and hypomethylation to Foxp3 in CD62L+ Tregs at the end of epicutaneous immunotherapy (EPIT) and 8 weeks after the end of EPIT. (A) Experimental design for the methylation analysis in T‐cell compartments (Th1, Th2, and Tregs). Analysis of the methylation levels of the Gata3 promoter and the Foxp3 Treg‐specific demethylated regions (TSDR) in Th1 (B and C) and Th2 cells (D and E), CD62L+ Tregs (F and G), and CD62L− Tregs (H and I) isolated from spleen at week 8 (immediately after treatment) or 8 weeks after the end of EPIT (sustainability). Results are expressed as individual data, and median. Differences between groups were analyzed by a Kruskall‐Wallis test followed by Dunn's multiple comparison test. ns nonsignificant, *P < .05, **P < .01
3.4. The epigenetic signature observed with EPIT is unique compared with OIT
In an independent experiment, we assessed the methylation changes in spleen and blood CD4+ T cells isolated from peanut‐sensitized mice in EPIT, Sham and in a third group receiving OIT. The efficiency of OIT was verified through the measurement of specific IgE and IgG2a, yielding similar results than those obtained in Dioszeghy et al11 (data not shown). This confirmed that the degree of methylation of the Gata3 promoter increased significantly during EPIT in CD4+ T cells from spleen (P < .05 at the end of treatment, Figure 5B) and blood (P < .01 at the end of treatment, Figure 5C), whereas the DNA methylation level of the Foxp3 TSDR decreased significantly during EPIT in spleen (P < .01 at the end of treatment, Figure 5D) and in blood (P < .01 at the end of treatment, Figure 5E). For EPIT, epigenetic changes were sustained after the end of the treatment (8 + 8 week, Figure 5B‐E).
Figure 5.
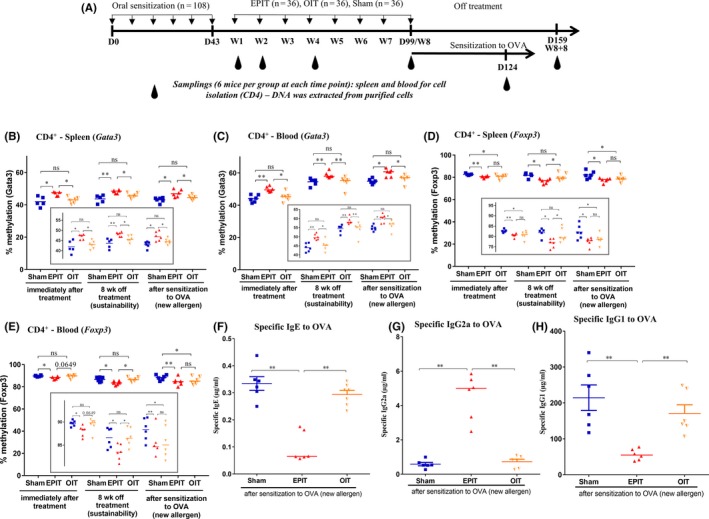
Epicutaneous immunotherapy (EPIT) leads to a unique DNA methylation profile in CD4+ cells (spleen and blood) compared with oral immunotherapy (OIT) in peanut‐sensitized mice, which is sustained after sensitization to ovalbumin (OVA) (new allergen) and correlates with serological response to OVA. (A) Experimental design. Analysis of the methylation levels of the Gata3 promoter in CD4+ cells isolated from (B) spleen and (C) whole blood at week 8 (8 wk) of EPIT or OIT, 8 weeks after the end of immunotherapies (8 + 8 wk) and after sensitization to OVA. Analysis of the methylation levels of Foxp3 in CD4+ cells isolated from (D) spleen and (E) whole blood at week 8 (8 wk) of EPIT or OIT, 8 weeks after the end of immunotherapies (8 + 8 wk) and after sensitization to OVA. (F‐H) Sera were harvested after EPIT to peanut protein (PPE) and sensitization to OVA for the EPIT, OIT, and Sham groups, to measure, respectively, IgE, IgG1, and IgG2a reactive to OVA. For methylation analyses, differences between groups were analyzed by a Kruskall‐Wallis test followed by Dunn's multiple comparison test. For sera, differences between groups were analyzed by a Mann‐Whitney test. Results are expressed as individual data and median. ns nonsignificant, *P < .05, **P < .01
Oral immunotherapy did not modify the methylation level of the Gata3 promoter in spleen or blood compared with Sham (Figure 5B and C). Furthermore, the level of DNA methylation remained unaltered compared with Sham 8 weeks after the end of treatment (8 + 8 week). In the Foxp3 TSDR, a methylation decrease was observed in the spleen (P < .05) together with a trend for a decrease in the blood (P = .0649) (Figure 5D and E). Those modifications were not sustained after the end of OIT treatment (8 + 8 week) in both spleen and blood. In agreement with the first experiment, there was no modification in the DNA methylation levels for the Tbet and Rorγ promoters in spleen or blood whatever the immunotherapy treatment.
3.5. The epigenetic signature is maintained after sensitization to a new allergen (OVA) and correlates with the absence of sensitization to OVA (nonallergen‐specific effect)
The DNA methylation patterns were assessed in spleen and blood CD4+ T cells from peanut‐sensitized mice treated by EPIT, Sham or OIT and subsequently submitted to a protocol of sensitization to OVA. During this protocol, the methylation level of the Gata3 promoter remained significantly increased in EPIT mice, compared to Sham, in the spleen (P < .05, Figure 5B) and blood (P < .05, Figure 5C). In parallel, the methylation degree of the Foxp3 TSDR in EPIT‐treated mice remained significantly decreased, compared to Sham, in spleen (P < .05, Figure 5D) and blood (P < .01, Figure 5C). Oral immunotherapy did not modify the Gata3 promoter methylation in spleen or blood (Figure 5B‐C), compared to Sham. On the contrary, during the OIT protocol of sensitization to OVA, the Foxp3 TSDR methylation level significantly decreased, compared to Sham, in spleen (P < .05, Figure 5D) and blood (P < .05, Figure 5E).
Specific antibodies (sIgE, sIgG1, and sIgG2a) were monitored after the sensitization to OVA as new allergen (Figure 5F‐H). sIgE and sIgG1 levels were significantly enhanced for Sham and OIT compared with EPIT (P < .01), whereas sIgG2a was significantly increased for EPIT compared with Sham and OIT (P < .01).
3.6. Foxp3 DNA demethylation on CD62L+ Tregs is associated with prevention from further sensitization conferred by EPIT (nonallergen‐specific effect)
Given the link between CD62L+ Treg cell numbers, Foxp3 expression, the desensitization process, and long‐term protection, we determined whether protection could be transferred by CD62L+ and/or CD62L− Tregs. The latter were isolated from milk‐EPIT‐treated mice and injected to naïve mice before peanut sensitization and intravenous challenge. Serum levels of peanut‐specific IgE increased significantly in the positive control and the CD62L− Treg groups (P < .01; Figure 6B), compared to the negative control group. No increase in peanut‐specific IgG2a was observed for these groups (Figure 6C). Mice with CD62L+ Tregs transfer before peanut sensitization did not develop peanut‐specific IgE, compared to the positive control group (P < .01, Figure 6B), but significantly increased peanut‐specific IgG2a (P < .001; Figure 6C). The positive control and CD62L− Treg groups developed anaphylaxis during peanut challenge marked by a drop in temperature (−6°C, P < .001 vs negative control group and −5°C, P < .01 negative control group, respectively) (Figure 6D) and increase in plasma mMCP1 (P < .001 and P < .01 vs negative control group) (Figure 6E). Mice with CD62L+ Treg transfer before peanut sensitization were protected from anaphylaxis compared with the positive control group (P < .001 for temperature drop and P < .01 for plasma mMCP1, Figure 6D and E).
Figure 6.
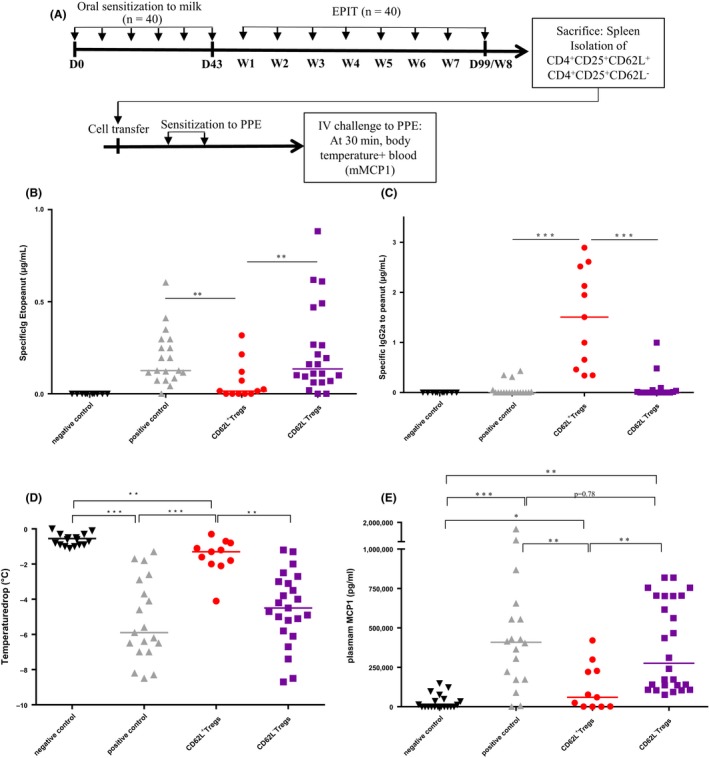
Demethylated CD62L+ Tregs confer protection against sensitization to a new allergen (peanut). (A) Experimental design for the evaluation of the bystander effect conferred by CD62L+ Tregs and CD62L− Tregs after epicutaneous immunotherapy (EPIT). (B) and (C) serological response to sensitization to peanut expressed in μg/mL; (D) and (E) anaphylaxis was evaluated by measuring body temperature and the level of mMCP1 in plasma 30 min after intravenous injection of peanut solution in negative control, positive control, CD62L+ Treg EPIT, and CD62L− Treg EPIT groups. Differences between groups were analyzed by a Kruskall‐Wallis test followed by Dunn's multiple comparison test. Individual data and medians are plotted *P < .05, **P < .01, ***P < .001
4. DISCUSSION
Epicutaneous immunotherapy is a promising treatment option for food allergy, based on animal models3, 4, 5 and clinical trials.7, 8, 9 A comprehensive analysis of epigenetic modifications induced by EPIT and their role in the regulation of the immune response is currently lacking. The present study, in a model of peanut‐sensitized mice, shows that the induction of a unique epigenetic signature by EPIT might account for both the sustainable effect of EPIT and its ability to prevent sensitization to further allergens, as previously shown.6
In this study, we tested whether the methylation of key transcription factors (Gata3, Tbet, Rorγ, and Foxp3) is involved in the efficacy of EPIT, its sustainability, and the bystander effect.6 Mice sensitized to peanuts have been widely used to prove the biological and clinical efficacy of EPIT.3, 4, 5, 6 Changes in DNA methylation were measured in cells sorted from spleen and blood, more precisely T cells (CD4+ and CD8+) and B cells (CD19+), and then more specifically in Th1, Th2, CD62L+ Tregs, and CD62L− Tregs, likely the key cell populations for EPIT, based on current knowledge.10 Sorting cell populations is crucial: DNA methylation differences may relate to individual variations or be diluted or masked if the cell population of interest is a minor component of PBMCs (reviewed in25), such as for Tregs.26, 27 Genome‐wide studies in autoimmune diseases have highlighted the diverging DNA methylation patterns in disease‐relevant blood cell populations,28, 29 and the advantage of detecting larger (and more robust) methylation differences when working with cell‐sorted populations compared with whole blood.30 In this study, sorted cells were not antigen‐specific, which could explain some differences compared with published data.16
The hypermethylation of the Gata3 promoter and hypomethylation of the Foxp3 TSDR induced by EPIT occur only in CD4+ T cells and correlate with previous findings of biological parameters. Gata3 is a key transcription factor in immune regulation,31 for example, Th2 differentiation and function32 and in the regulation of Treg cell function by binding to the regulatory regions of the Foxp3 locus. The methylation of a CpG‐rich island of Gata3 decreases its expression, as confirmed in the present study, directly decreasing allergy‐skewing cytokines (Il4, Il5 and Il13) secretion. Although numerous disease‐relevant cell populations express Gata3 (eg, eosinophils, mast cells, innate lymphoid cells, and other T‐cell subsets), sorting CD4+ Th1, Th2, and Treg cell subsets delineated the effect only to the Th2 lineage. This change was not observed in CD4+ T cells isolated after OIT. A growing body of evidence suggests that DNA methylation in combination with other epigenetic modifications such as histone modifications is critical for the development of Th2 immunity and allergic disease.33, 34 The promoter of Gata3 is marked by both activating and repressing histone modifications, a bivalent state also reported after Th2 lineage determination,35 which commonly goes along with a larger plasticity compared with DNA methylation at Th1/Th2 regulatory regions.36 Interestingly, the proportion of Th2 cells in spleen was not modified by EPIT, whereas their methylation level was enhanced, suggesting a change of the phenotype of the Th2 population upon EPIT similar to what was recently observed for eosinophils in asthma and which probably correspond to a different activation state.37
Our data suggest that changes at the Gata3 locus may dramatically influence cellular responses, including T‐cell cytokine secretion and B‐cell production (decrease of specific IgE). At the clinical level, DNA methylation changes in genes with direct relevance to Th2 immunity and asthma are associated with allergic asthma in innercity children38 as well as the protective farm environment.39 Binding sites of GATA3 are found in the promoter region of Il5 and Il13.40 Therefore, the modulation of Gata3 could allow the simultaneous abrogation of the expression of a number of inflammatory cytokines and the decline of the excessive Th2 lineage specification.41, 42
FOXP3 is a specific marker of Treg cells and serves as a lineage specification transcriptional factor of Treg cells. Both mice and human mutations of FOXP3 result in a complex syndrome of dysregulation and enteropathy.43, 44 DNA methylation of promoter and gene regulatory elements of Foxp3 were shown to influence the development of regulatory T cells.14, 45 The sustained expression of FOXP3 is critical for maintaining regulatory function,45 and a demethylated pattern of the TSDR is a prerequisite for stable Foxp3 expression and their suppressive phenotype.15, 46, 47 The TSDR is the region ensuring both a persistent expression and the suppressive functions, through a positive feedback mechanism, during which FOXP3 binds to its own gene. Our experiment suggests that EPIT decreased the methylation level of the Foxp3 TSDR, which is linked to the induction of Tregs observed during EPIT in a mouse model.10 A similar demethylation of the same region has previously been reported after OIT in a small human cohort.16 Surprisingly, hypomethylation of the Foxp3 TSDR was specifically identified in only CD62L+ Tregs, shown to be specifically induced by EPIT compared with other forms of immunotherapy.11, 48 CD62L+ Tregs have been associated with a more suppressive phenotype49 and prevented severe tissue damage to the colon and protected recipients from lethal GVHD,50 suggesting a broad range of action.
The ultimate goal for specific immunotherapy of food allergy is a long‐term effect. Discordant results have been published about the sustained unresponsiveness following the termination of OIT (reviewed in51). Recently, it has been shown that 3 (of 23) patients with sustained unresponsiveness for 3 months had persistent hypomethylation of FOXP3.16 Similarly, successful dual sublingual immunotherapy (to timothy grass and dust mite) with possible long‐term tolerance was supported by epigenetic modifications of the FOXP3 promoter and TSDR in memory regulatory T cells.12 Here, we show the sustainability of epigenetic modifications at Foxp3, but also at Gata3. Our data on the hypomethylation of the Foxp3 TSDR provide one explanation for the long‐lasting production and suppressive function of EPIT‐induced Tregs obtained by our group in similar mouse experiments.10 Interestingly, the alteration of the methylation status—hypermethylation of the Gata3 promoter and hypomethylation of the Foxp3 TSDR—persisted beyond 2 months and even appeared stable despite exposure of animals to a protocol of further sensitization with 2 injections of OVA mixed with Alum. In addition, the large preventive action of EPIT against further sensitization—the bystander effect—observed in the mouse model6 might be supported by persistent DNA methylation changes. Previously, we have shown that the transfer of EPIT‐induced Tregs, and not Tregs from sensitized, but nontreated mice, conferred a protection against further sensitization to a different allergen, independently of the antigen sequence used. Here, the role of CD62L+ Tregs in this bystander effect is supported by adoptive transfer of this cell population generated during milk‐EPIT in naive mice prior to initiating a protocol of sensitization to peanut. The protective effect against further sensitization could be clearly attributed to CD62L+ Tregs. As the antigen sequence was not responsible for the effect as previously demonstrated,6 we hypothesize that their higher suppressive effect could be at least partly conferred by the hypomethylation of Foxp3, observed in CD62L+ Tregs isolated after peanut EPIT. A more detailed investigation on CD62L+ Tregs isolated after milk‐EPIT is required to confirm this hypothesis. These data suggest that CD62L+ Tregs induced by EPIT can prevent sensitization to new allergens, and our findings are consistent with previous results on this Treg population.50, 52, 53
Finally, this work suggests a potential biomarker to identify patients that have responded to EPIT. Martino et al54 suggested that DNA methylation biomarkers could be a novel diagnostic test in patients with food allergy. In our study, DNA methylation of two key genes (Gata3 and Foxp3) was modulated by EPIT and could be measured in the spleen and in blood from the 4th week of EPIT onward, without analyzing antigen‐specific cells.
Although we identified strong associations between DNA methylation changes at the Gata3 promoter and in the TSDR of Foxp3 with the efficacy and sustainability of EPIT, additional studies in ongoing clinical trials are required to confirm that similar epigenetic changes occur during EPIT in allergic patients and their relationship to adequate immune responses.
Finally, this methylation pattern is specifically observed following EPIT and not OIT in peanut‐sensitized mice, supporting differences in the mechanisms of actions between the two immunotherapies.7, 11
Taken together, we demonstrate in a model of peanut‐sensitized mice that epigenetic regulation of Th2 cells and CD62L+ Tregs induced by EPIT is strongly involved in its desensitization process, marked by Th2 repression and Treg enhancement, in the sustainability of those mechanisms and in the prevention of new sensitizations.
CONFLICT OF INTERESTS
The authors declare competing interests. LM, VDi, CP, ML, and VDh are DBV Technologies employees. HS is a part‐time employee serving as Chief Scientific Officer (60%) and a part‐time faculty member of the Icahn School of Medicine at Mount Sinai (40%—Professor of Pediatrics). CD received honoraria and/or compensation in regard to the study, as investigator, coordinator or expert, in relation to the time spent on the study. JT had conference and travel fees covered by DBV Technologies.
AUTHOR CONTRIBUTIONS
LM and JT jointly conceived the project and interpreted the results. VDi developed analytical tools. CP, VDh, and ML carried out the mouse experiments, cell sorting, mRNA extraction, ELISA, and cell culture. FB, KB, LL, and CDa performed pyrosequencing and the data transfer. HS commented on the manuscript at all stages. LM, JT, and CDu wrote the manuscript.
Supporting information
ACKNOWLEDGMENTS
The authors thank Valérie Domergue‐Dupont and her staff for taking care of mice during all the experiments conducted in the animal facility of the Faculty of Pharmacy (Châtenay‐Malabry, France).
Mondoulet L, Dioszeghy V, Busato F, et al. Gata3 hypermethylation and Foxp3 hypomethylation are associated with sustained protection and bystander effect following epicutaneous immunotherapy in peanut‐sensitized mice. Allergy. 2019;74:152–164. 10.1111/all.13479
Contributor Information
L. Mondoulet, Email: lucie.mondoulet@dbv-technologies.com.
J. Tost, Email: tost@cng.fr.
REFERENCES
- 1. Akdis CA. Therapies for allergic inflammation. Refining strategies to induce tolerance. Nat Med. 2012;18:736‐749. [DOI] [PubMed] [Google Scholar]
- 2. Akdis CA, Akdis M. Advances in allergen immunotherapy. Aiming for complete tolerance to allergens. Sci Transl Med. 2015;7:280ps6. [DOI] [PubMed] [Google Scholar]
- 3. Mondoulet L, Dioszeghy V, Ligouis M, Dhelft V, Dupont C, Benhamou P‐H. Epicutaneous immunotherapy on intact skin using a new delivery system in a murine model of allergy. Clin Exp Allergy. 2010;40:659‐667. [DOI] [PubMed] [Google Scholar]
- 4. Mondoulet L, Dioszeghy V, Vanoirbeek JA, Nemery B, Dupont C, Benhamou P‐H. Epicutaneous immunotherapy using a new epicutaneous delivery system in mice sensitized to peanuts. Int Arch Allergy Immunol. 2011;154:299‐309. [DOI] [PubMed] [Google Scholar]
- 5. Mondoulet L, Dioszeghy V, Larcher T, et al. Epicutaneous immunotherapy (EPIT) blocks the allergic esophago‐gastro‐enteropathy induced by sustained oral exposure to peanuts in sensitized mice. PLoS One. 2012;7:e31967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mondoulet L, Dioszeghy V, Puteaux E, et al. Specific epicutaneous immunotherapy prevents sensitization to new allergens in a murine model. J Allergy Clin Immunol. 2015;135:1546‐1557. [DOI] [PubMed] [Google Scholar]
- 7. Jones SM, Burks AW, Dupont C. State of the art on food allergen immunotherapy. Oral, sublingual, and epicutaneous. J Allergy Clin Immunol. 2014;133:318‐323. [DOI] [PubMed] [Google Scholar]
- 8. Sampson HA, Shreffler WG, Yang WH, et al. Effect of varying doses of epicutaneous immunotherapy vs placebo on reaction to peanut protein exposure among patients with peanut sensitivity. A randomized clinical trial. JAMA. 2017;318:1798‐1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones SM, Sicherer SH, Burks AW, et al. Epicutaneous immunotherapy for the treatment of peanut allergy in children and young adults. J Allergy Clin Immunol. 2017;139:1242‐1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dioszeghy V, Mondoulet L, Dhelft V, et al. The regulatory T cells induction by epicutaneous immunotherapy is sustained and mediates long‐term protection from eosinophilic disorders in peanut‐sensitized mice. Clin Exp Allergy. 2014;44:867‐881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dioszeghy V, Mondoulet L, Puteaux E, et al. Differences in phenotype, homing properties and suppressive activities of regulatory T cells induced by epicutaneous, oral or sublingual immunotherapy in mice sensitized to peanut. Cell Mol Immunol. 2016;13:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swamy RS, Reshamwala N, Hunter T, et al. Epigenetic modifications and improved regulatory T‐cell function in subjects undergoing dual sublingual immunotherapy. J Allergy Clin Immunol. 2012;130:215‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lal G, Zhang N, van der Touw W, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huehn J, Polansky JK, Hamann A. Epigenetic control of FOXP3 expression. The key to a stable regulatory T‐cell lineage. Nat Rev Immunol. 2009;9:83‐89. [DOI] [PubMed] [Google Scholar]
- 15. Floess S, Freyer J, Siewert C, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Syed A, Garcia MA, Lyu S‐C, et al. Peanut oral immunotherapy results in increased antigen‐induced regulatory T‐cell function and hypomethylation of forkhead box protein 3 (FOXP3). J Allergy Clin Immunol. 2014;133:500‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adel‐Patient K, Bernard H, Ah‐Leung S, Créminon C, Wal J‐M. Peanut‐ and cow's milk‐specific IgE, Th2 cells and local anaphylactic reaction are induced in Balb/c mice orally sensitized with cholera toxin. Allergy. 2005;60:658‐664. [DOI] [PubMed] [Google Scholar]
- 18. Leonard SA, Martos G, Wang W, Nowak‐Węgrzyn A, Berin MC. Oral immunotherapy induces local protective mechanisms in the gastrointestinal mucosa. J Allergy Clin Immunol. 2012;129:1579‐1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dioszeghy V, Mondoulet L, Dhelft V, et al. Epicutaneous immunotherapy results in rapid allergen uptake by dendritic cells through intact skin and downregulates the allergen‐specific response in sensitized mice. J Immunol. 2011;186:5629‐5637. [DOI] [PubMed] [Google Scholar]
- 20. Schmidl C, Hansmann L, Andreesen R, Edinger M, Hoffmann P, Rehli M. Epigenetic reprogramming of the RORC locus during in vitro expansion is a distinctive feature of human memory but not naïve Treg. Eur J Immunol. 2011;41:1491‐1498. [DOI] [PubMed] [Google Scholar]
- 21. Mazzoni A, Santarlasci V, Maggi L, et al. Demethylation of the RORC2 and IL17A in human CD4+ T lymphocytes defines Th17 origin of nonclassic Th1 cells. J Immunol. 2015;194:3116‐3126. [DOI] [PubMed] [Google Scholar]
- 22. Ivascu C, Wasserkort R, Lesche R, et al. DNA methylation profiling of transcription factor genes in normal lymphocyte development and lymphomas. Int J Biochem Cell Biol. 2007;39:1523‐1538. [DOI] [PubMed] [Google Scholar]
- 23. Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265‐2275. [DOI] [PubMed] [Google Scholar]
- 24. Pfaffl MW. A‐Z of quantitative PCR. Quantification strategies in real‐time PCR. La Jolla, CA, USA: S.A. Bustin, 2004. [Google Scholar]
- 25. Zhang M, Huang B. The multi‐differentiation potential of peripheral blood mononuclear cells. Stem Cell Res Ther. 2012;3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Asano M, Toda M, Sakaguchi N, Sakaguchi S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J Exp Med. 1996;184:387‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self‐tolerance maintained by activated T cells expressing IL‐2 receptor alpha‐chains (CD25). Breakdown of a single mechanism of self‐tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151‐1164. [PubMed] [Google Scholar]
- 28. Absher DM, Li X, Waite LL, et al. Genome‐wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T‐cell populations. PLoS Genet. 2013;9:e1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miceli‐Richard C, Wang‐Renault S‐F, Boudaoud S, et al. Overlap between differentially methylated DNA regions in blood B lymphocytes and genetic at‐risk loci in primary Sjögren's syndrome. Ann Rheum Dis. 2016;75:933‐940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Imgenberg‐Kreuz J, Sandling JK, Almlöf JC, et al. Genome‐wide DNA methylation analysis in multiple tissues in primary Sjögren's syndrome reveals regulatory effects at interferon‐induced genes. Ann Rheum Dis. 2016;75:2029‐2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wan YY. GATA3. A master of many trades in immune regulation. Trends Immunol. 2014;35:233‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tindemans I, Serafini N, Di Santo JP, Hendriks RW. GATA‐3 function in innate and adaptive immunity. Immunity. 2014;41:191‐206. [DOI] [PubMed] [Google Scholar]
- 33. Bégin P, Nadeau KC. Epigenetic regulation of asthma and allergic disease. Allergy Asthma Clin Immunol. 2014;10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Potaczek DP, Harb H, Michel S, Alhamwe BA, Renz H, Tost J. Epigenetics and allergy. From basic mechanisms to clinical applications. Epigenomics. 2017;9:539‐571. [DOI] [PubMed] [Google Scholar]
- 35. Wei G, Wei L, Zhu J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brand S, Teich R, Dicke T, et al. Epigenetic regulation in murine offspring as a novel mechanism for transmaternal asthma protection induced by microbes. J Allergy Clin Immunol. 2011;128:618‐625. [DOI] [PubMed] [Google Scholar]
- 37. Xu C‐J, Söderhäll C, Bustamante M, et al. DNA methylation in childhood asthma. An epigenome‐wide meta‐analysis. Lancet Respir Med. 2018;6:379–388. [DOI] [PubMed] [Google Scholar]
- 38. Yang IV, Pedersen BS, Liu A, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015;136:69‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Michel S, Busato F, Genuneit J, et al. Farm exposure and time trends in early childhood may influence DNA methylation in genes related to asthma and allergy. Allergy. 2013;68:355‐364. [DOI] [PubMed] [Google Scholar]
- 40. Yamashita M, Ukai‐Tadenuma M, Kimura M, et al. Identification of a conserved GATA3 response element upstream proximal from the interleukin‐13 gene locus. J Biol Chem. 2002;277:42399‐42408. [DOI] [PubMed] [Google Scholar]
- 41. Zhu J, Min B, Hu‐Li J, et al. Conditional deletion of Gata3 shows its essential function in T(H)1‐T(H)2 responses. Nat Immunol. 2004;5:1157‐1165. [DOI] [PubMed] [Google Scholar]
- 42. Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA‐3 for the function of regulatory T cells. Immunity. 2011;35:337‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kwon HK, Chen HM, Mathis D, et al. FoxP3 scanning mutagenesis reveals functional variegation and mild mutations with atypical autoimmune phenotypes. Proc Natl Acad Sci. 2018;115:E253‐E262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bennett CL, Christie J, Ramsdell F, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X‐linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20‐21. [DOI] [PubMed] [Google Scholar]
- 45. Williams LM, Rudensky AY. Maintenance of the Foxp3‐dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277‐284. [DOI] [PubMed] [Google Scholar]
- 46. Janson PCJ, Winerdal ME, Marits P, Thörn M, Ohlsson R, Winqvist O. FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS One. 2008;3:e1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toker A, Engelbert D, Garg G, et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J Immunol. 2013;190:3180‐3188. [DOI] [PubMed] [Google Scholar]
- 48. Dioszeghy V, Benlhassan‐Chahour K, Delache B, et al. Changes in soluble factor‐mediated CD8+ cell‐derived antiviral activity in cynomolgus macaques infected with simian immunodeficiency virus SIVmac251: relationship to biological markers of progression. J Virol. 2006;80:236‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alyanakian M‐A, You S, Damotte D, et al. Diversity of regulatory CD4+ T cells controlling distinct organ‐specific autoimmune diseases. Proc Natl Acad Sci USA. 2003;100:15806‐15811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ermann J, Hoffmann P, Edinger M, et al. Only the CD62L+ subpopulation of CD4+CD25+ regulatory T cells protects from lethal acute GVHD. Blood. 2005;105:2220‐2226. [DOI] [PubMed] [Google Scholar]
- 51. Yee CSK, Rachid R. The heterogeneity of oral immunotherapy clinical trials. Implications and future directions. Curr Allergy Asthma Rep. 2016;16:25. [DOI] [PubMed] [Google Scholar]
- 52. Bynoe MS, Bonorino P, Viret C. Control of experimental autoimmune encephalomyelitis by CD4+ suppressor T cells. Peripheral versus in situ immunoregulation. J Neuroimmunol. 2007;191:61‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carman CV, Martinelli R. T lymphocyte–endothelial interactions. Emerging understanding of trafficking and antigen‐specific immunity. Front Immunol. 2015;6:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martino D, Dang T, Sexton‐Oates A, et al. Blood DNA methylation biomarkers predict clinical reactivity in food‐sensitized infants. J Allergy Clin Immunol. 2015;135:1319‐1328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


