Abstract
MicroRNA-15a (miR-15a) and miR-16, which are transcribed from the miR-15a/miR-16-1 cluster, inhibit post-ischemic angiogenesis. MicroRNA (miRNA) binding to mRNA coding sequences (CDSs) is a newly emerging mechanism of gene expression regulation. We aimed to (1) identify new mediators of the anti-angiogenic action of miR-15a and -16, (2) develop an adenovirus (Ad)-based miR-15a/16 decoy system carrying a luciferase reporter (Luc) to both sense and inhibit miR-15a/16 activity, and (3) investigate Ad.Luc-Decoy-15a/16 therapeutic potential in a mouse limb ischemia (LI) model. LI increased miR-15a and -16 expression in mouse muscular endothelial cells (ECs). The miRNAs also increased in cultured human umbilical vein ECs (HUVECs) exposed to serum starvation, but not hypoxia. Using bioinformatic tools and luciferase activity assays, we characterized miR-15a and -16 binding to Tie2 CDS. In HUVECs, miR-15a or -16 overexpression reduced Tie2 at the protein, but not the mRNA, level. Conversely, miR-15a or -16 inhibition improved angiogenesis in a Tie2-dependent manner. Local Ad.Luc-Decoy-15a/16 delivery increased Tie2 levels in ischemic skeletal muscle and improved post-LI angiogenesis and perfusion recovery, with reduced toe necrosis. Bioluminescent imaging (in vivo imaging system [IVIS]) provided evidence that the Ad.Luc-Decoy-15a/16 system responds to miR-15a/16 increases. In conclusion, we have provided novel mechanistic evidence of the therapeutic potential of local miR-15a/16 inhibition in LI.
Keywords: microRNAs, miR-15a, miR-16a, Tie2, angiopoietins, angiogenesis, ischemia, IVIS, mouse ischemia model, miR decoy
Introduction
Peripheral artery disease affects 20% of individuals over 70 years of age worldwide1, 2 and can result in critical limb ischemia (CLI), a debilitating condition characterized by pain at rest, tissue loss, and gangrene. The current management of CLI involves revascularization of the affected limb by percutaneous angioplasty or surgical bypass. However, many CLI patients are not suitable for revascularization for reasons such as severe comorbidities or late diagnosis.
In approximately 30% of severely affected patients, minor or major amputations are inevitable.3 Novel therapeutics to locally promote angiogenesis, and hence reperfusion, would help to overcome a significant unmet clinical need.
Historically, human clinical trials have been informed by pre-clinical studies based on the delivery of single proangiogenic growth factors of the vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) families.4, 5 Such approaches have not yet yielded the desired results.6, 7 To reach the bedside of CLI patients, angiogenic therapies require further refinement, including revisiting the realm of therapeutic targets and their mechanisms of action.8
MicroRNAs (miRNAs) are small non-coding RNAs that, in their mature form, act at the post-transcriptional level by targeting multiple mRNAs.9 miRNAs are transcribed as a primary transcript (pri-miRNA) from miRNA genes or intronic regions of protein-coding genes, and they are subsequently processed to reach their mature and functional status.10 The initial and canonical model of miRNA-induced mRNA-silencing activity relies on semi-complementary binding between the miRNA seed sequence of 7 nt to miRNA-binding sequences contained in the 3′ UTR) of target mRNAs. However, it is becoming increasingly evident that miRNA binding to the 5′ UTR and coding sequences (CDSs) of mRNAs have powerful functional consequences.11 Each miRNA can target up to several hundred genes and multiple pathways, and miRNAs are known to be involved in the regulation of a plethora of cellular processes, including angiogenesis.12 Additionally, functionally active miRNAs are released from parent cells and taken up by bordering and possibly distant cells,13 where they can repress their target genes, thus providing a widespread method of gene expression regulation. Investigations based on individual miRNA experiments have identified miRNAs with either proangiogenic or anti-angiogenic effects.14, 15
miR-15 and miR-16 are organized in two different clusters, miR-15a/16-1 and miR-15b/16-2, and they are conserved between humans and mice.16 The sequences of mature miR-15a and miR-15b differ by 4 nt,16 while miR-16-1 and miR-16-2 have the same mature sequence and, hence, are commonly indicated as miR-16. miR-15a/b and -16 belong to the miR-16 family consisting of miR-15a, miR-15b, miR-16, miR-195, miR-424, and miR-497. The family is extendable to miR-103, miR-107, miR-646, and miR-503, due to homologies in the seed sequences.17 The extended miR-16 family is involved in angiogenesis regulation: miR-16 targets VEGF receptor 2 and FGF receptor 1, thus decreasing the proangiogenic activity of their ligands.18 Moreover, following LI, therapeutic angiogenesis is impaired in mice with miR-15a gene knockin.19 We previously described that miR-15a and -16 are increased in both the proangiogenic circulating cells (PACs) and the serum of CLI patients (versus healthy subjects). We also showed that serum concentrations of miR-15a and miR-16 predict the need for amputation at 1 year from revascularization in CLI subjects.20 In further support of the relevance of miR-15a and miR-16 in the CLI setting, we provided evidence that ex vivo transfection with miR-15a/16 inhibitors increases the potential of human PACs to induce therapeutic angiogenesis in an immunocompromised mouse LI model.20
Among the different ways to inhibit miRNA, the use of miRNA decoys or sponges that consist of multiple specific miRNA-binding site sequences inserted downstream of a reporter gene represents a promising approach.21, 22, 23, 24 Using a decoy for the diabetes-associated miR-503, we have already provided proof of concept that adenovirus (Ad)-mediated local delivery of a miRNA decoy can improve post-ischemic angiogenesis and blood flow recovery in mice with LI.23, 24 When delivered into cells, the binding of the targeted miRNA to the decoy sequences not only inhibits the miRNA by sequestration but also reduces the expression of the reporter gene used, often EGFP22 or luciferase.25 This construct could, therefore, be used as a sensor of the targeted miRNA quantity, as the protein activity of the reporter used is inversely correlated to the presence of the miRNA.26, 27 Additionally, more recent technological advancements allow non-invasive and precise measurement of luciferase activity in vivo in mice.28, 29 On these bases, we reasoned that Ad-mediated local delivery of a double miR-15a/16 decoy could provide therapeutic advantages by ensuring release from miRNA inhibition in a spatiotemporally defined window that is supportive of post-ischemic vascular repair.
This study was designed to mechanistically investigate the anti-angiogenic effect of miR-15a/16 and to develop an Ad.miR-15a/16 decoy to be tested for its therapeutic potential, in a mouse LI model.
Results
miR-15 and -16 Expressions in Human and Mouse Tissues
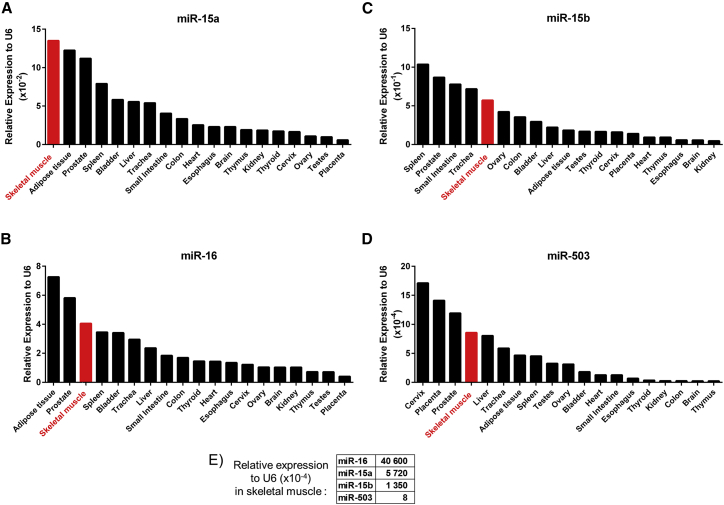
Expressions of miR-15a/b and miR-16 were assessed by qRT-PCR in 19 different human tissues. As shown in Figures 1A–1D, skeletal muscle showed the highest expression of miR-15a and was the tissue with the third highest expression of miR-16, after adipose and prostate tissues. Skeletal muscle was the fifth highest localization of miR-15b and the fourth for miR-503, which we previously found to be increased in diabetic CLI.23 The relative expressions of the four individual miRNAs in human limb muscles are reported in Figure 1E. Considering these data, we focused the study on miR-15a and miR-16.
Figure 1.
miR-15a, -15b, -16, and -503 Expressions in Human Tissues
Expressions of miR-15a (A), miR-16 (B), miR-15b (C), and miR-503 (D) in human tissues assessed by RT-PCR and relative expression of each miRNA in skeletal muscle (E). Results were normalized to small nuclear RNA U6 (U6) expression (n = 1/tissue, resulting from the pooling of three different donor samples).
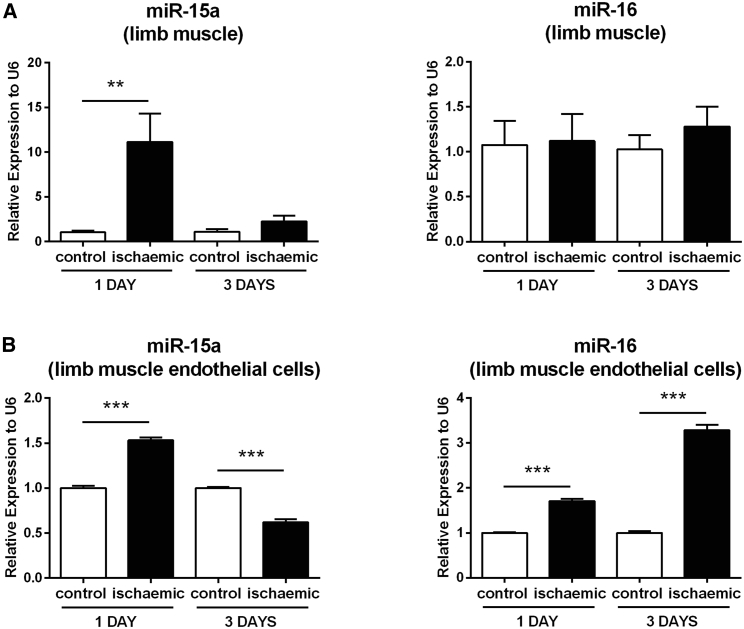
To investigate whether miR-15a and -16 expressions are regulated by LI, we harvested mouse adductor and gastrocnemius muscles at 1 and 3 days post-femoral artery ligation or sham operation, and we measured miRNA expression in the whole tissues and in muscle CD146+ microvascular cells. Ischemia-associated expressional changes in the whole muscles were limited to miR-15a (Figure 2A). However, both miRNAs were increased in the ischemic microvascular cells (Figure 2B).
Figure 2.
Expressions of miR-15a and -16 Are Differentially Modulated in Mouse Adductor Muscles and Muscle-Derived Endothelial Cells after Limb Ischemia
At 1 and 3 days after surgical induction of limb ischemia, ischemic and contralateral (control) adductor muscles were collected, and the expressions of miR-15a and -16 were assessed by RT-PCR in the total muscle tissue (A) and in muscle-derived CD146+ endothelial cells (ECs) (B). Results were normalized to the expression of small nuclear RNA U6, relative to control muscle or EC normo-perfused and expressed as mean ± SEM. n = 3 per condition. **p < 0.01 and ***p < 0.001 versus control, matched for time after ischemia.
We next moved to model the ischemic environment in vitro, by exposing cultured human umbilical vein endothelial cells (HUVECs) to either oxygen or nutrient deprivation. When combined, hypoxia (1% pO2) and serum starvation increased the expression of the prototypical hypoxamiRNA proangiogenic miR-21030 (Figure S1), used here as a positive control to validate the culture conditions. The expression of miR-16 was also increased, while miR-15a was unchanged. Taking each condition separately, hypoxia alone increased miR-210 expression, but it did not affect miR-15a/16 levels (Figures S2A–S2C). By contrast, serum deprivation for 24 h increased the expressions of both miR-15a and -16 (Figures S2D and S2E). As expected, miR-210 expression was unaffected by serum starvation (Figure S2F). As shown in Figure S3, the serum starvation-induced expressional changes were limited to the mature forms of miR-15a and -16, and neither the primary transcript (pri-miR-15a/16-1) nor the individual miRNA precursors (pre-miR-15a, pre-miR-16-1, and pre-miR-16-2) were affected.
miR-15a and -16 Target Tie2 in CDS, and Tie2 Repression Mediates the Anti-angiogenic Responses to miR-15a and -16
miR-16 has already been shown to inhibit the angiogenic potential of HUVECs.18 Using a 2D Matrigel assay, we have confirmed these data and additionally characterized that miR-15a inhibits angiogenesis in vitro (Figure S4). Next, we set out to investigate the miRNA putative target genes that could elucidate the anti-angiogenesis response. In previous studies, bioinformatic predictions assuming miRNAs targeting the 3′ UTR of mRNAs have identified VEGF-R2, FGF-R1, VEGF-A, FGF-1, FGF-2, and AKT-3 to be repressed by miR-15/16.18, 20, 31, 32 More recently, members of the miR-16 family have been shown to bind in the CDS region of their targeted mRNAs.11, 33, 34 Therefore, we expanded our search to putative miRNA-binding sites in the CDS of proangiogenic genes. A genome-wide prediction using miRcode revealed 12,269 common targets of miR-15a and -16. To identify gene ontology (GO) terms and processes to which these common targets correspond, a GO enrichment analysis was performed using the program CLUEGO. Overall 48 GO terms were significantly enriched among the common targets of miR-15a and -16. The top 15 of the GO terms for these predicted targets included blood vessel development (Figure S5A).
From this search, TEK (aka, Tie2 angiopoietin receptor) was declared an interesting candidate, because it is one of the most studied regulators of angiogenesis.35 Four angiopoietin (Ang) ligands can bind to Tie2.36 Among them, the most studied are Ang1 and Ang2. Ang1 has a clear agonistic impact on Tie2 and promotes angiogenesis and vessel stability. The type of interaction of Ang2 and Tie2 and vascular consequences are more debated and context dependent.36, 37, 38 Ang2 has been reported to increase by inflammatory and hypoxic stimuli and to promote neoangiogenesis.39 However, Ang2 can also disrupt the connections between the endothelium and perivascular cells and promote cell death and vascular regression.38, 40 We decided to focus on TEK because a closer inspection of the miRcode results revealed that there is only one binding site for miR-15a and -16 in the CDS region of TEK and no binding sites in the 3′ or 5′ UTR. This implied that the possible direct regulatory actions of miR-15a and miR-16 on TEK are exclusively mediated via miRNA binding in the CDS. In more detail, this single binding site is a 6-bp region, and the exact location is at chromosome (chr)9: 27204938–27204943 (Figure S5B). This site is highly conserved in mammals (83%) followed by primates (67%) and vertebrates (46%) (Figure S5C).
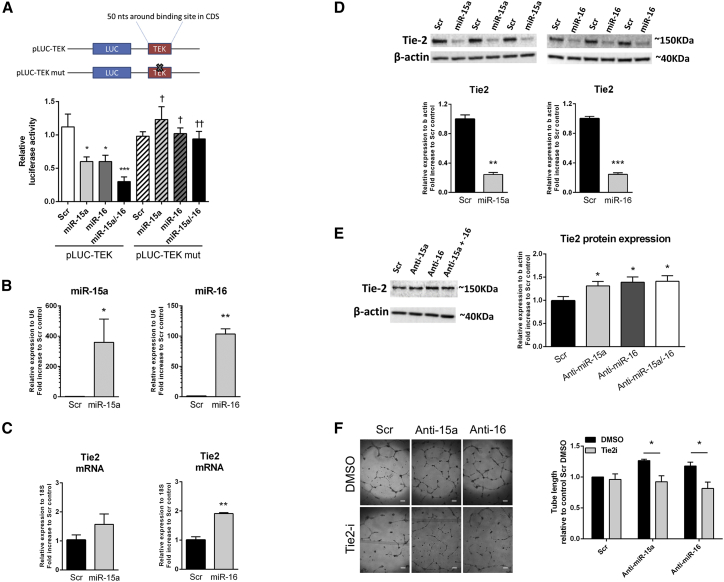
Next, to validate the direct binding of miR-15a or miR-16 to this CDS of Tie2 mRNA, we developed a luciferase reporter assay in which a sequence of 50 nt either side of the predicted binding site of miR-15a/16 on TEK (LUC-TEK), or to the same sequence but where the binding site was mutated (LUC-TEKmut), was cloned downstream of luciferase gene (Figure 3A). In HeLa cells carrying LUC-TEK, overexpression of miR-15a and/or miR-16 by pre-miRNA transfection decreased luciferase activity. This response was not observed if cells were carrying LUC-TEKmut (Figure 3A). To obtain evidence of the consequence of Tie2 targeting by miR-15/16 for angiogenesis, we next moved to HUVECs. In HUVECs transfected to overexpress either miR-15a or miR-16 (Figure 3B), Tie2 mRNA expression was increased (Figure 3C), while Tie2 protein level was decreased (Figure 3D), consistent with the notion that miRNA binding to the mRNA CDS leads to RNA translation inhibition.41 Conversely, inhibition of miR-15a and -16 using anti-miRNAs increased Tie2 protein expression (Figure 3E). This was associated with a proportional increase in Tie2 phosphorylation, indicating that Tie2 receptors released from the miRNA inhibitory block were functionally active42 (Figure S6). The importance of Tie2 in mediating the anti-angiogenic effect of either miR-15a or miR-16 was finally confirmed by using a pharmacological inhibitor of Tie2 activity.43 Tie2 inhibition prevented the proangiogenic response to inhibition of either anti-miR-15a or anti-miR-16 (Figure 3F).
Figure 3.
Tie2 Is Targeted by miR-15a and -16 through an Unusual Binding Site on Tie2 mRNA, and It Is Involved in the Proangiogenic Effect of miR-15a and -16 Inhibition in HUVECs In Vitro
(A) Luciferase activity at 48 h post-co-transfection of HEK293T cells with pre-miR-15a (miR-15a), pre-miR-16 (miR-16), a combination of the two (miR-15a/16), or miR-scrambled oligonucleotides (Scr) and plasmid containing luciferase open reading frame (pLUC), followed by a portion of 50 nt of the TEK-coding sequence surrounding the putative miRNA-binding site (pLUC-TEK). A mutated version of the miRNA-binding site was used as a control (pLUC-TEKmut). A schematic of the plasmid construct is also provided. *p < 0.05, ***p < 0.001 versus pLUC-TEK control Scr; †p < 0.05, ††p < 0.01 versus pLUC-TEK to the pre-miRNA matching condition (n = 5). (B) Human umbilical vein endothelial cells (HUVECs) were transfected with a Scramble oligo (Scr), pre-miR-15a (miR-15a), or pre-miR-16 (miR-16) (5 nM) to increase their respective mature miRNA expression. miR-15a and -16 expressions were assessed by RT-PCR and normalized to small nuclear U6 expression. Tie2 mRNA (C) or protein (D) expressions were assessed by RT-PCR and western blot and normalized to 18S and β-actin, respectively. n = 3 for each condition. (E) HUVECs were transfected with a Scramble oligo (Scr) or an anti-miRNA against miR-15a, miR-16, or the two anti-miRNAs simultaneously (total concentration of 50 nM) to inhibit their activity. Tie2 protein expression was assessed by western blot and normalized to β-actin. n = 9 for each condition. *p < 0.05 versus Scr condition. (F) Effect of inhibition of Tie2 on miR-15a/16 inhibition-induced angiogenesis was assessed in HUVECs transfected with a Scramble oligo (Scr) or an anti-miRNA against miR-15a or miR-16 (50 nM) and treated with Tie2 inhibitor (Tie2-I, 5 μM) or DMSO control. The angiogenic capacity was measured by network formation on Matrigel, calculated as total length in millimeters and expressed as a percentage of the Scr and DMSO conditions. Representative pictures are provided (scale bar, 200 μm). n = 3. *p < 0.05, **p < 0.01, ***p < 0.001. All results are expressed as mean ± SEM.
To understand whether the expression of miR-15a and -16 and Tie2 are correlated in ischemic limb muscles and their microvascular cells, we measured Tie2 expression in similar conditions by RT-PCR and by immunohistochemistry. While miR-15a and -16 were increased by ischemia (Figures 2A and 2B), Tie2 protein level was unchanged in ischemic capillaries (Figures S7A–S7H), suggesting that additional molecular mechanisms are responsible for Tie2 expressional regulation. Ang1 expression was not changed in muscles at 1 and 3 days, post-ischemia. By contrast, Ang2 increased at 3 days post-ischemia (Figures S7A and S7B).
Ad.Luc-miR-15a/16 Decoy Can Be Used to Sense Expressional Changes in miR-15a and -16
We next prepared Luc-Decoy in the form of a miRNA decoy system linked to a luciferase reporter gene (Figure S8A), with the intention to both sense changes in endogenous miR-15a and -16 levels and inhibit the functionality of miR-15a/16 in cultured cells and in murine ischemic limb muscles. The capacity of this tool to sequester the endogenous miRNA, thus inhibiting miRNA activity, was already proven in our previous publications on miR-503.23, 24 Here, we additionally evaluated whether Ad.Luc-miR-15a/16 Decoy was useful in tracking expressional changes in miR-15a and -16 expression in vitro and in vivo. We used HeLa cells transfected with either pre-miR-15a or pre-miR-16, and infected with either Ad.Luc-Decoy or Ad.Luc to validate that increases in intracellular miR-15a and -16 reduce luciferase activity detected in an in vivo imaging system (IVIS) apparatus (Figure S8B). Importantly, we could confirm that Ad.Luc-Decoy was selectively sensing changes in miR-15a and -16 in comparison with other miR-16 family members, such as miR-15b and miR-503 (Figure S8C).
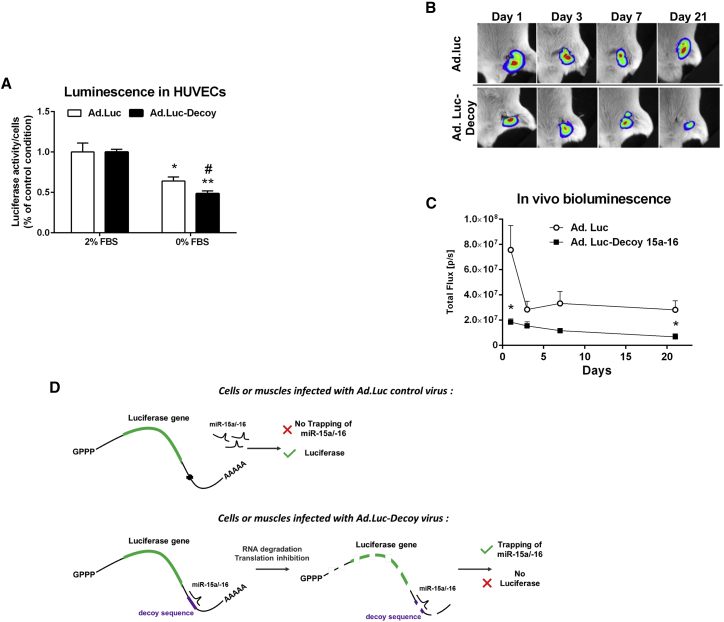
We next investigated whether the Ad.Luc-Decoy system could be used to sense increases in endogenous miR-15a and -16 expressions. To this aim, HUVECs were infected with Ad.Luc-Decoy or Ad.Luc and submitted to serum withdrawal. After 24 h, we found a decrease in luciferase activity in cells infected with Ad.Luc-Decoy in comparison to cells infected with Ad.Luc control (Figure 4A). We then evaluated the effect of Ad.Luc-Decoy injection into murine muscles after the induction of LI in vivo. At 1 day post-LI, in keeping with the increased miR-15a and -16 expressions in total ischemic muscle (see Figure 2A), the luciferase activity was reduced in mice injected with Ad.Luc-Decoy compared with mice injected with the control virus (Figures 4B and 4C). A reduction in luciferase activity was also observed at day 21, when miR-16 was found upregulated in ischemic muscle (Figure S9). Figure 4D summarizes the modality of action and potential of Ad.Luc-Decoy.
Figure 4.
Adenovirus Carrying a Decoy Sequence for miR-15a and -16 (Ad.Luc-Decoy) Sensor Activity and Inhibition Efficiency In Vivo
(A) Luciferase activity measured on an IVIS of HUVECs infected with Ad.Luc control or Ad.Luc-Decoy virus cultured under complete (2% FBS) or serum-deprived (0% FBS) media for 24 h. Results are expressed per cell and relative to Ad.-matching, 2% FBS condition. Results are presented as the mean of n = 4–5 per condition ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 versus Ad.-matching, 2% FBS condition; #p < 0.05 versus Ad.-Luc, 0% FBS condition, Student’s t test. (B and C) After limb ischemia, bioluminescence activity (expressed as photons per second per centimeter squared per steradian (P⋅s−1⋅cm−2⋅sr−1) was measured and analyzed in the ischemic legs using the Xenogen IVIS. D-Luciferin was subcutaneously injected into the ischemic limb, and the consequent bioluminescence from the reaction with the luciferase in either Ad.Luc- or Ad.Luc-Decoy-injected legs was measured from day 1 to day 21 after injection of the viruses. Representative images are presented. Data are shown as mean ± SEM (n ≥ 10 per condition). *p < 0.05 versus time-matched Ad.Luc condition using Student’s t test. (D) Schematic explanation of the mechanisms of miR-15a and -16 inhibition, trapping, and sensing in cells expressing the Luc control construct, lacking complementary-decoy sequence for miR-15a/16 or the Luc-Decoy construct.
Ad.Luc-Decoy Improves Post-ischemic Blood Flow Recovery and Therapeutic Angiogenesis
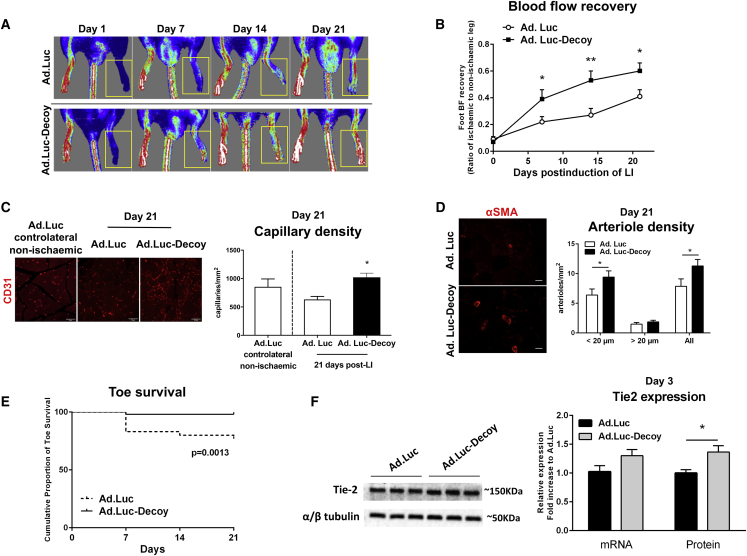
We finally provided evidence of the therapeutic potential of Ad.Luc-Decoy in the mouse LI model. When compared with Ad.Luc, Ad.Luc-Decoy improved post-ischemic blood flow recovery (Figures 5A and 5B) and increased the reparative angiogenesis response in the ischemic muscles (Figures 5C and 5D). The latter was demonstrated by a 35% increase in capillary density (CD31+ capillaries) and 47% increase in the density of small (lumen diameter <20 μm) arterioles (Figures 5C and 5D) in the ischemic muscles. A similar post-ischemic increase in capillary density was found using Isolectin B4 as a marker for ECs (Ad.Luc versus Ad.Luc-Decoy at 21 days post-ischemia: 557 ± 69 versus 812 ± 97 capillaries/mm2 (mean ± SEM).
Figure 5.
Inhibition of miR-15a/16 Improves Blood Flow (BF) Recovery, Post-ischemic Neovascularization, and Toe Survival following Limb Ischemia
Limb ischemia was induced in mice by femoral artery ligation, and Ad.Luc or Ad.Luc-Decoy was injected into the ischemic adductor muscle immediately after surgery. (A and B) Line graph shows the time course of post-ischemic BF recovery (calculated as the ratio between the BF of the ischemic foot and the contralateral foot) in mice injected with Ad.Luc or Ad.Luc-Decoy. Representative color laser Doppler images taken at days 0, 7, 14, and 21 post-ischemia are provided. Yellow frames show the ischemic foot analyzed. Data are presented as mean ± SEM (n ≥ 10 per group). *p < 0.05, **p < 0.01 versus time-matched Ad.Luc condition. (C) Immunohistological analysis and representative microphotographs showing the capillary density in mice injected with Ad.Luc or Ad.Luc-Decoy 21 days after LI and in contralateral (control) muscle of mice injected with Ad.Luc (n = 5 per group), assessed by immunolabelling of CD31 as a marker of endothelial cells. Scale bar, 50 μm. (D) Immunohistological analysis and representative microphotographs showing arteriolar density in mice injected with Ad.Luc or Ad.Luc-Decoy (n = 11 or 8 mice per group, respectively) after limb ischemia, assessed by immunolabelling of α-smooth muscle actin (αSMA) as a marker of vascular smooth muscle cells. Arterioles were quantified according to vessel diameter (<20 μm or ≥20 μm). Data are presented as mean + SEM. Scale bar, 20 μm. (E) Cumulative proportion of toe survival over 21 days of follow-up after ischemia in mice injected with Ad.Luc or Ad.Luc-Decoy (n ≥ 10 mice per group). p = 0.0013, log rank test. (F) Tie2 expression in ischemic adductor muscles, 3 days after limb ischemia surgery and injection with either Ad.Luc or Ad.Luc-Decoy virus, was assessed at mRNA and protein levels by RT-PCR and western blot, respectively. Results were normalized to GAPDH for mRNA analysis (n = 6) or to α/β-tubulin for protein analysis (n = 9) and presented as mean ± SEM. *p < 0.05 versus Ad.Luc group.
Using toe necrosis as a marker of ischemia severity,44 we showed that Ad.Luc-Decoy improved toe survival (Figure 5E). Finally, in line with in vitro data reported in Figures 3C and 3D, Ad.Luc-Decoy increased Tie2 protein expression (Figure 5F), and it did not change the expressions of Tie2 ligands Ang1 and Ang2 (Figure S10). The mRNA expressions of VEGF-A, VEGF- R2, FGF-2, and FGF-1, which were all described to be bound in their 3′ UTRs by miR-15a or -16, were also unchanged (Figure S11).
Discussion
This study has characterized the expressional regulation of miR-15a and -16 in ischemic ECs, and it identified that the anti-angiogenic actions of these miRNAs is in part mediated by the repression of Tie2. Additionally, we have utilized a miRNA decoy approach to sense and inhibit the activity of endogenous miR-15a and -16 in cultured cells and mouse ischemic muscles. In vivo work has provided evidence of the therapeutic potential of Ad.Luc-Decoy in CLI.
miR-15a and -16 were previously described to be expressed in various solid and blood cancers.45, 46 Additionally, the group of van Rooij47, 48 has studied miR-15 in the ischemic heart, showing that it induces cell-cycle arrest in neonatal cardiomyocytes and contributes to ischemic damage of the heart in adult rodent and swine models. Early in this study, we found that, under basal conditions and comparing several human organs and tissues, miR-16 and especially miR-15a are more highly expressed in the skeletal muscle compared to the heart.
To the best of our knowledge, the consequences of endogenous changes in miR-15a and -16 in LI, and their possible impact on post-ischemic vascular responses, have never been directly investigated. However, we previously reported increased serum levels of miR-15a and -16 in diabetic CLI patients, where circulating miR-15a and -16 levels were able to predict clinical outcome (amputation) at 12 months post-revascularization.20 In this new study, we have been able to show that LI increases miR-15a and -16 in the muscular microvascular cells and that the ischemia-induced increases in these endogenous miRNAs impair post-ischemic vascular repair, worsening blood flow recovery and toe survival.
It should be noted that, under our ethics license, we are not allowed to study more severe forms of LI outcome, and we must humanely euthanize our research animals if these occur. The angiogenic response observed in mice after local delivery of Ad.Luc-Decoy is in line with our previous work on human PACs, which were shown to carry higher miR-15a and -16 levels when obtained from CLI patients. Moreover, PACs improved their proangiogenic profile when ex vivo simultaneously transfected with inhibitors for the two miRNAs before their efficacy testing in immunosuppressed mice with LI.20 Similar to the LI setting, miR-15a is elevated in the ischemic heart.48 In contrast, hypoxic cancer tissues expressed less miR-15a/miR-16 than non-hypoxic ones.49 miR-15/16 function as a tumor suppressor by targeting BCL2.45 They also inhibit cancer angiogenesis by targeting FGF-2.49 Therefore, in cancer, miR-15/16 expression deregulation might contribute to explaining why hypoxia, a common micro-environmental feature of rapidly growing tumors, is associated with worse clinical outcomes.50 According to our data, in ischemic non-cancerous ECs, the regulation of miR-15a and -16 expression is hypoxia independent and triggered by nutrient deprivation. We have also clarified that increases in mature miR-15a and -16 are not associated with equivalent changes in primary or precursor miRNA expression and, hence, are possibly due to either slower miRNA decay or accelerated maturation of the pre-miRNAs to mature miRNAs.
This study has newly identified Tie2 as a target of miR-15a and -16. The angiopoietin Tie2 receptor is mainly expressed in the vascular endothelium and involved in embryonic and post-natal blood vessel development.51, 52 Interestingly, we found that Ang2 was upregulated in ischemic muscles, while Ang1 was unchanged. Tie2 levels were also unchanged by ischemia.
The modality of action of Ang2 on Tie2 is still debated, and it has been suggested to function as a context-dependent agonist or antagonist42 and to be associated with inflammatory responses.53 Direct intramuscular injection of plasmid DNA encoding Ang1, but not Ang2, was shown to augment revascularization in a rabbit ischemic hindlimb model.54 Increasing the Ang1/Tie2 signaling in ischemic limb muscles could be beneficial for reparative angiogenesis.
In fact, Tie2 disruption was shown to impair post-ischemic regeneration in mice with LI.55 This is in line with our data showing that an Ad.Luc-Decoy-induced increase in limb Tie2 is associated with improved post-ischemic reparative angiogenesis and blood flow recovery. We have clarified that miR-15a/16 bind to the CDS of Tie2. A few studies have started to report efficient targeting of miRNAs on CDS. The canonical mechanism of action of a miRNA involves the stabilization of mature miRNA by Argonaute-2 (Ago2) proteins, functioning to direct the RISC (RNA-induced silencing complex) to target mRNAs. According to this model, binding between the miRNA seed sequence to one or more semi-complementary regions within the mRNA 3′ UTR induces post-transcriptional inhibition, manifesting as reduced mRNA and protein levels.11 It is progressively emerging that this model is probably insufficient to explain miRNA actions and that, in some cases, the most predominant Ago2:miR-binding sites may be found within the CDS.11
It has already been shown that miR-103 and -107, which belong to the wider miR-16 family, do make ample use of CDS targeting.34 The miRNA target sites in the CDS are under evolutionary selection.41 Sequence conservation has usually been taken as evidence of functional importance.56, 57 In this context, it is of interest that the miR-15a/16-binding sites to CDS targets are conserved between species (human and macaque58). miR-15a/b and -16 contain the AGCAGC motif at the 5′ end. Using computational analyses, Hausser et al.41 found that miRNAs with this motif location have more complementary sites in the CDS compared with 3′ UTR. Such miRNAs also have in common the capacity to regulate the cell cycle.41 Hausser et al.41 additionally suggested that CDS binding is more potent in inhibiting mRNA translation, as compared to mRNA degradation. This is consistent with our finding that pre-miR-15a or -16 transfection of HUVECs reduced protein and increased mRNA expression for Tie2.
In our study we used a homology-of-sequence strategy to identify miR-15a and -16 mRNA targets. miRNA-mRNA interactions are largely based on sequence homology, but they are also dependent on additional factors. As an example, mRNA 3D configuration can affect the accessibility of the RISC-binding site.59 Increasing evidence shows that binding of a miRNA to its mRNA targets can be cell type and environment dependent60 and that this can be relevant for anti-miRNA cardiovascular therapies.61 This was already exemplified by Eding et al.,61 who found that, upon in vivo inhibition of the cardiomyocyte-specific miR-208a, the quality and quantity of direct targets of miR-208a that are expressionally modulated depend on the presence and type of cardiac stresses, such as hypertension or myocardial infarction. The anti-miR-208 treatment in healthy and diseased hearts revealed 3 subsets of targets: (1) health-specific targets, (2) disease-specific targets, and (3) targets common to health and disease. In line with that, we have also noted a context-dependent target regulation after the inhibition of miRNA-15a and -16. In fact, Chamorro-Jorganes et al.18 showed that miR-16 regulates VEGFR2 in human ECs cultured under normal conditions. In our model of LI, miR-15a/16 inhibition using the Ad.Luc-Decoy virus did not change the expression of VEGF-R2 or other previously described targets of miR-15a or -16, such as VEGF-A, FGF-1, and FGF-2.18, 20, 32
We have shown that viral vector-mediated local delivery of a miRNA decoy increases the density of small arterioles, which contain vascular smooth muscle cells (VSMCs) and play an important role in the resistance of blood flow in skeletal muscles.62, 63 The data presented here are consistent with our previous report that miR-15a decreased human VSMC migration and proliferation in vitro,20 biological process involved in arteriole growth.64
Our study also showed the potential of using a bioluminescence IVIS apparatus to non-invasively detect changes in endogenous miRNA expression in wild-type animals. Conventional techniques to measure miRNA expression usually consist of RT-PCR, microarray analyses, and northern blots. Even though these techniques are accurate, they require lysed cells or tissues that represent a further constraint for time course studies, especially in vivo.65 A major strength versus genome editing of small animal models, which is otherwise possible, is that, once optimized, it can be translated for use in large animal models and human patients. In future studies, the prototypical Ad.Luc-Decoy can be refined to increase precision of measurements of different miRNA types, and it can be adapted for use with different viral vectors, with different tropism and length of transgene expression.
Finally, this study has provided the first evidence that local inhibition of miR-15a/16 is effective at improving post-ischemic vascular repair in limbs. This finding has translational implications, and it significantly expands on what has already been reported by Hullinger et al.48 in heart ischemia models, where locked nucleic acid (LNA)-modified anti-miRNA was employed to target miR-15, reducing infarct size. Hullinger et al.48 focused exclusively on the heart and did not investigate the vascular response to miRNA inhibition. Clinical trials based on LNA have already started in patients with different forms of lymphoma and leukemia (https://www.cancer.gov/about-cancer/treatment/clinical-trials/intervention/lna-based-antimir-155-mrg-106). Additionally, gene therapy clinical trials to test proangiogenic therapies have been already authorized by regulatory authorities.66 Therefore, our proof of concept that miR-15a/16 inhibition holds therapeutic potential in the setting of LI has the potential to be progressed through the translational pipeline via at least two alternatives (LNA or viral vector-mediated Decoy transfer) to reach the bedside of CLI patients.
Materials and Methods
Human RNA Tissue Bank Real-Time qPCR Analysis
RNA from a human tissue bank (resulting from the pooling of three samples from different patients) was obtained from Ambion, Applied Biosystems. Reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, 4366596), and real-time PCR was performed in triplicate using the TaqMan universal master mix with the LightCycler480 qPCR detection system (Roche, 04707516001).
Expression was normalized to the small nucleolar RNA U6 (snRU6). Primer identification numbers are indicated in Table S1. Relative miRNA expression was defined from threshold cycle (Ct) values, calculated using the 2(−ΔΔCt) method.
Mouse LI Model
The experiments involving mice were performed in accordance with the Guide for the Care and Use of Laboratory Animals prepared by the Institute of Laboratory Animal Resources and with the prior approval of the UK Home Office and the University of Bristol ethics committee.
Sample size was determined using a power calculation with blood flow at 21 days post-ischemia, measured using laser Doppler, as the primary outcome. Specifically, for an 80% power at a 5% significance level, to detect a 50% increase in ischemic:contralateral limb blood flow ratio (see below) in the test group (from a mean of 0.4 to 0.6, based on previous experience), at least 12 mice were required in each group. Based on our own pilot data on the use of in vivo bioluminescence analyses, this number of mice would give us a 90% power to detect a 3-fold difference in bioluminescence activity.
Left LI was induced in 10- to 12-week-old CD1 male mice (Charles River Laboratories, Margate, UK). Mice were anesthetized (tribromoethanol, 880 mmol/kg intraperitoneally [i.p.], Sigma-Aldrich, T48402), and the left femoral artery was ligated with a 6-0 silk suture at two points, followed by electrocoagulation of the ligated segment, leaving the femoral vein and nerve untouched.67 Analgesia (Vetergesic, 0.02 mL/30 g animals) was administrated after surgery. Mice were excluded and humanely euthanized where they developed signs or symptoms of severe LI, as per the conditions of our ethics license. Any data accrued for excluded mice were still analyzed up to the point of exclusion.
In vivo experiments were performed by three investigators (S.S., L.H., and A.C.T.). Where experiments involved the randomization of mice into one of two groups, the operator and outcome assessor were blinded as to the group allocation. For evaluation of miR-15a/16 expression in ischemic muscles, mice were sacrificed at 1 and 3 days after surgery. Ischemic and contralateral adductor muscles were harvested, and RNA extraction was performed on either the total muscle or muscle CD146+ microvascular cells extracted following a previously described protocol.68
RNA Extraction from Mouse Tissue and Cells and Real-Time qPCR Analysis
Total RNA was extracted from adductor muscle tissues and mouse cells using miRNeasy Mini 343 Kit (QIAGEN, 217004), according to the manufacturer’s instructions. RT-PCR was performed in triplicate as described above.
Cell Culture
HUVECs (Lonza, Slough, UK, CC-2519) were cultured between passages 2 and 6 at 37°C with 5% CO2 in EBM-2 EC basal medium (Lonza, CC-3156), with the addition of the SingleQuot Kit (EGM-2 medium, Lonza, CC-4176) containing 2% fetal bovine serum (FBS, included in the kit). For hypoxia experiments, cells were exposed to 1% pO2 for 24 and 48 h, whereas for the normoxia control condition cells were exposed to 21% pO2. For the serum deprivation condition experiments, cells were cultured in EGM-2 medium without FBS for 24 and 48 h, whereas in the control condition cells were exposed to complete medium. HeLa cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco, 15140122). Cells were incubated in a humidified atmosphere at 37°C with 5% CO2.
Bioinformatic Prediction of New miR-15a and -16 Targets
The program miRcode (http://www.mircode.org/index.php) was used to predict common genome-wide targets of miR-15a and miR-16 by looking for their binding sites in the 3′ UTR, 5′ UTR, and CDS regions of human genes. The predicted common targets of the two were then subjected to a GO analysis to identify enriched terms and processes. GO analysis was performed using CLUEGO,69 with significant terms considered at a Bonferroni-corrected p value of ≤0.05.
Luciferase Assay for the Validation of miR-15a and -16 Binding to Tie2 mRNA
The binding sequence for the miR-16 family on Tie2 CDS was identified using miRcode software. The binding site is in position chr9: 27204938–27204943. At 450 bp to either side of the binding site was cloned in pMIR-Reporter (Life Technologies). Primers used for the cloning were as follows: Tie2 CDS, forward 5′-ATAGTGGTTAGGTGGCAGGG-3′ and reverse 5′-CTGCCTGTACTTGGACTTGC-3′. Primers for 3′ UTR mutation were as follows: Tie2 CDS, forward 5′-AGGGGGGAAGAATAATAAATTAGCCATCCTTGG-3′ and reverse 5′-CCAAGGATGGCTAATTTATTATTCTTCCCCCCT-3′.
Luciferase assays were performed as previously described.24 Briefly, luciferase constructs were transfected into HEK293T cells together with miR-15 and/or miR-16 mimics or p-SV-β-gal control vector. Cells were cultured for 48 h and assayed with the Luciferase and β-Galactosidase Reporter Assay Systems (Promega, E1500 and E2000, respectively). Luciferase values were normalized to protein concentration and β-galactosidase activity.
Tie2 Expression Regulation by miR-15a and -16 in HUVECs
HUVECs were plated on a 6-multiwell plate at a density of 2.0 × 105 cells per well in 2 mL EGM-2 and grown for 24 h until 70%–80% confluent. Cells were then washed once with PBS and maintained in OPTIMEM (Invitrogen, 31985062) for 0.5 h prior to transfection with pre-miRNAs (hsa-miR-15a-5p, PM10235; hsa-miR-16-5p, PM10339, Ambion, 5 nM), anti-miRNAs (MH10339 and MH10235, Ambion, total 50 nM), or a scramble (Scr) control (Cy3 dye-labeled Pre-miR Negative Control, AM17120 or Anti-miR Negative Control AM17011, Ambion) using Lipofectamine RNAiMAX (Invitrogen, 13778150), according to the manufacturer’s protocol. At 2 days after transfection, cells were lysed either in ice-cold Pierce radioimmunoprecipitation assay (RIPA) buffer (Thermoscientific, 89900) completed with cOmplete Protease Inhibitor Cocktail (Roche, 11697498001) and phosphatase inhibitor PhosSTOP (Roche, 4906845001) for protein extraction or with QIAzol for RNA extraction (miRNeasy Mini Kit, 217004).
For reverse transcription, cDNA was synthesized from 500 ng total RNA using the QuantiTect RT kit (QIAGEN, Manchester, UK, 205314), as per the manufacturer’s instructions, including a step to remove genomic DNA. All cDNA was stored at −20°C. qPCR was performed using Power SYBR (Life Technologies, Paisley, UK, 4368708) with a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific), according to the manufacturer’s instructions. Primers used to measure the expression of human Tie2 are indicated in Table S2. Data were normalized to 18S as an endogenous housekeeping gene, and relative expression n was calculated using the 2−(ΔΔCT) method. To extract protein, lysates were incubated at 4°C on a rotator. Samples were centrifuged at 14,000 × g for 15 min at 4°C, and the supernatant fractions were used for western blot. Protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225). Detection of proteins by western blot analyses was done using 10 μg protein extract on SDS-polyacrylamide gels.
Proteins were transferred to nitrocellulose membranes and probed with the following antibodies: Goat anti-human Tie2 (R&D Systems, AF313, 1:1,000), Rabbit anti-human/mouse phospho-Tie2 (Y992) (R&D Systems, AF2720, 1:3,000), and mouse and human β-actin (Sigma, A5441; 1:50,000). Secondary antibodies used were enhanced chemiluminescence (ECL) mouse immunoglobulin G (IgG) (GE Healthcare, NA931; 1:5,000), mouse anti-goat- horseradish peroxidase (HRP) (Santa Cruz Biotechnology, sc-2354, 1:2,000), and donkey anti-rabbit-HRP (Santa Cruz Biotechnology, sc-2313, 1:2,000). Membranes were stripped between imaging of phospho-Tie2 and total Tie2.
Matrigel Assay of HUVECs Transfected with miR-15a and miR-16 Inhibitors and Tie2 Inhibition
HUVECs were transfected using Lipofectamine RNAiMAX (Invitrogen, 13778150) and anti-miRNA negative control or anti-miR-15a or miR-16 inhibitors, following the protocol described above. The cells were used for Matrigel assay at 48 h after transfection. On the day of the Matrigel assay, HUVECs were plated onto duplicate wells containing 70 μL Reduced Growth Factors Matrigel (BD Biosciences, 356231), on a 96-well plate (104 cells/well) in complete medium with or without 5 μM Tie2 inhibitor (ab141270, Abcam) or the equivalent volume of the inhibitor diluent (DMSO, final concentration 0.05%). The experiment continued for 24 h before imaging each well at 5×. Network formation was quantified in randomly captured microscopic fields by calculating the length of cellular network using Image-Pro plus software.
Ad.Luc-Decoy
The fragment of firefly luciferase-decoy 15-16 (Luc-Decoy) under the control of the cytomegalovirus (CMV) promoter (2,662 bp) was excised from TW-3′ UTR LUC DEC15-CYAN70 with ClaI and EcoRV, blunt ended using mung bean nuclease and ligated into pDC511 shuttle vector (Microbix Biosystems, Canada). Replication-deficient E1-E3-deleted adenovirus was generated by recombination in 293 cells.71 The DNA construct consists of the CMV promotor followed by a luciferase gene and tandem sequences complementary to miR-15a and also 80% complementary to miR-16 (decoy sequences), separated by an 18-bp unrelated spacer.70
Preparation of HeLa Cells and HUVECs for In Vitro Bioluminescence Analyses Using the IVIS System
HeLa cells were seeded on a T75 flask at a density of 8 × 105 cells/flask in 10 mL complete DMEM and grown 24 h until 70%–80% confluent. Cells were then infected overnight with Ad.Luc-Decoy virus at 10 MOI in DMEM without FBS. To transfect the cells with miRNA precursors, HeLa cells were plated in duplicate on a 6-well plate at a density of 2.5 × 105 cells/well in 2 mL DMEM 10% FBS and grown for 24 h until 70%–80% confluent. Cells were then maintained in OPTIMEM medium (Invitrogen, 31985062) for 30 min prior to transfection with 25 nM pre-miR-15a, pre-miR-16, or Scr. For luciferase activity measurement, after transfection, HeLa cells were plated in a 96-well plate at a density of 1 × 103, 2 × 103, 4 × 103, 6 × 103, 8 × 103, 1 × 104, 2 × 104, and 3 × 104 cells/well in duplicates. After 12 h, the medium was removed and replaced by 100 μL pre-warmed PBS containing 150 μL/mL D-Luciferin (Caliper LifeSciences, 122796). Luciferase activity was measured 1 min after the addition of the D-Luciferin using an IVIS. The average of n = 2 per condition plated in duplicate was then expressed in units of maximum photons per second per centimeter squared per steradian (P·s−1·cm−2·sr−1).
HUVECs were seeded on a 12-well plate at a density of 1 × 105 cells/well in 1 mL complete EGM-2 medium and grown until 70%–80% confluent. Cells were then infected overnight with adenoviruses Ad.Luc-Decoy or Ad.Luc at 100 MOI in EGM-2 complete medium. The following day, the cells were washed with PBS and the medium was replaced with normal complete or serum-free EGM-2 medium for 24 h. After luciferase activity measurement (performed as in HeLa cells), HUVECs were then trypsinized and counted. The average of n = 5 per condition was then expressed in units of maximum photons per second per centimeter squared per steradian (P·s−1·cm−2·sr−1) per cell.
Preparation of Mice for In Vivo Bioluminescence Analyses Using the IVIS System
The sample size for in vivo work was based on power calculations. Mice were randomized in two treatment groups (n = 14 mice/group) and underwent LI induction.
Immediately after the procedure, Ad.Luc-Decoy or Ad.Luc (109 plaque-forming units per mouse in a total volume of 30 μL injected into 3 equidistant sites) was delivered into the ischemic adductor muscle. The luciferase activity in the ischemic muscles was evaluated at 1, 3, 7, and 21 days post-surgery using the Xenogen In Vivo Imaging System. Following the exclusion criterion stated above, the day 21 analysis included 12 mice in the Ad.Luc-Decoy group and 10 in the Ad.Luc group. Briefly, after anesthesia induction (tribromoethanol, 880 mmol/kg i.p., Sigma-Aldrich, T48402), the mouse ischemic leg was injected subcutaneously with the reporter probe D-luciferin (Caliper LifeScience, 122796) at 150 mg/kg body weight.
After 10 min, animals were imaged. Bioluminescence was quantified and expressed in units of maximum photons per second per centimeter squared per steradian (P·s−1·cm−2·sr−1), as described previously.72
Efficacy Testing the Therapeutic Potential of Ad.Luc-Decoy
The mice used for the IVIS analyses were tested for post-ischemic blood flow recovery, therapeutic angiogenesis, and clinical outcome in terms of toe necrosis. The superficial blood flow (BF) of the ischemic and contralateral foot was sequentially analyzed, starting immediately after surgery and at 7, 14, and 21 days, using a high-resolution laser color Doppler imaging system (Moor LDI2, Moor Instruments, Devon, UK). The ratio between BF in the ischemic and contralateral foot was calculated and used as an index of percentage BF recovery. At 21 days post-ischemia, mice were perfusion fixed under terminal anesthesia, and adductors were harvested and paraffin embedded for histological and immunohistochemical analyses, as previously described.67 Toe necrosis occurrence, defined as darkening of the toe followed by loss of the tissue, was evaluated along the LI protocol. The cumulative proportion of toe survival at 7, 14, and 21 days after the induction of LI was calculated, excluding mice that showed symptoms of femoral nerve damage.
Histology and Immunohistochemistry on Muscles
The 4-μm-thick adductor muscle slices were stained using Rabbit anti-mouse/human CD31 (Abcam, ab28364, 1:50) and Donkey anti-rabbit Alexa-647 (Jackson ImmunoResearch Laboratories, 711-605-152, 1:200) to detect ECs and with a Cy3-conjugated rabbit polyclonal anti-α-smooth muscle actin antibody (Sigma, C6198, 1:400) to stain VSMCs, present in arterioles. The slides were then mounted using a mounting medium (Fluoromount G, South Biotech, 0100-01) containing DAPI to stain nuclei. The number of capillary and arterioles per millimeter squared was counted in 10 randomly selected high-power fields (magnification 40×) using a Zeiss inverted fluorescent microscope. The lumen diameter of α-smooth muscle actin-positive arterioles was measured using ImageJ software. Tie2 staining in non-ischemic and ischemic muscles was performed using a mouse anti-mouse/human Tie2 antibody (Abcam, ab24859 [Cl. 16], 1:50) and a goat anti-mouse Alexa-568 (Abcam, ab175473, 1:200) on cryosections. At least 5 fields of 40× magnification using Zeiss AxioObserver inverted fluorescent microscope were used to analyze the percentage of Tie2+/CD31+ capillaries. Intensity of staining of Tie2 within the Tie2+/CD31+ capillaries was analyzed using Fiji software.
Tie2 Expression in Ischemic Skeletal Muscle Injected with Ad.Luc and Ad.Luc-Decoy Virus
Animals received surgery of LI and were injected into the adductor muscle with 109 plaque-forming unit (PFU)/animal with either Ad.Luc or Ad.Luc-Decoy. At 3 days after surgery, animals were euthanized and ischemic muscles were collected and snap frozen. Tissues were mechanically disrupted either in Qiazol (QIAGEN) for RNA extraction or in RIPA buffer (Thermo Scientific, 89900) completed with cOmplete protease inhibitor cocktail and phosphatase inhibitor PhosSTOP for protein extraction, as described above. Total RNA from adductor muscle tissues was extracted using miRNeasy Mini extraction Kit (QIAGEN, 217004), according to the manufacturer’s instructions. For tissues collected at 21 days post-LI, total RNA from formalin-fixed and paraffin-embedded tissue sections was isolated using deparaffinization solution and miRNeasy FFPE Kit (QIAGEN 217504), according to the manufacturer’s protocol (deparaffinization solution 19093).
For mRNA analysis, reverse transcription was performed using the QuantiTect Reverse Transcription kit (QIAGEN, 205314), according to the manufacturer’s protocols. cDNA was amplified by real-time qPCR. cDNA (equivalent to 1 μg total RNA) was incubated in triplicate with gene-specific primers for mouse Angiopoietin-1, Angiopoetin-2, FGF-2, FGF-1, VEGF-A, VEGF-R2, Tie2, GAPDH, and 18S (sequences presented in Table S2). To extract protein, lysates were incubated at 4°C on a rotator. Samples were centrifuged at 14,000 × g for 15 min at 4°C, and the supernatant fractions were used for western blot. Protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225). Detection of proteins by western blot analyses was done using 30 μg protein extract on SDS-polyacrylamide gels for Tie2 and 120 μg for VEGF-R2. Proteins were transferred to nitrocellulose membranes and probed with the following antibodies: goat anti-mouse/rat Tie2 (R&D Systems, AF762, 1:1,000), rabbit anti-human/mouse VEGF-R2 (Abcam, ab39256), and rabbit anti-mouse α/β-Tubulin (R&D Systems, 2148S, 1:2,000). Secondary antibodies used were mouse anti-goat-HRP (Santa Cruz Biotechnology, sc-2354, 1:2,000) and donkey anti-rabbit-HRP (Santa Cruz Biotechnology, sc-2313, 1:2,000).
Statistical Analysis
Results are presented as mean ± SEM. Tests for statistical significance were conducted using GraphPad Prism (version 6; GraphPad, San Diego, CA).
Comparisons between two groups were performed using Student’s t test, with the exception of the analysis of the capillary and arteriole densities for which a Welch’s unequal variances t test was used. For comparison among more than two groups, ANOVA followed by a Tukey post hoc test was used. Differences in necrosis outcomes were assessed using the log rank test. Exceptions are indicated in the captions. A p value of <0.05 was interpreted to be statistically significant.
Author Contributions
Participated in research design: M.B., A.C., and C.E. Conducted experiments: M.B., S.S., M.A., P.D., A.C-J., W.S., G.S-N., A-C.T., L.H., and S.M. Contributed new reagents or analytic tools: G.S-N., P.M., and E.P. Performed data analysis: M.B., S.S., M.A., A.C-J., A.C., and C.E. Wrote or contributed to the writing of the manuscript: M.B. and C.E.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
Authors would like to acknowledge the Facility for Imaging by Light Microscopy (FILM) at Imperial College London, which is in part supported by funding from the Wellcome Trust (grant 104931/Z/14/Z) and BBSRC (grant BB/L015129/1); we are grateful to Mr. Stephen Rothery (laboratory manager and senior technician at FILM) for his help with fluorescence imaging and analysis. We also acknowledge some technical help from Dr. Micol Marchetti (University of Bristol). This study was funded through the following (to C.E.): British Heart Foundation grants (BHF) research programme grant “microRNAs from cardiac surgery to basic science – and back?” (RG/15/5/31446), and BHF Chair in Cardiovascular Science (CH/15/1/31199); BHF PhD studentship for S.S. (FS/10/61/28566); and Leducq Foundation transatlantic grant on microRNAs in vascular disease (miRVAD).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.002.
Supplemental Information
References
- 1.Annex B.H. Therapeutic angiogenesis for critical limb ischaemia. Nat. Rev. Cardiol. 2013;10:387–396. doi: 10.1038/nrcardio.2013.70. [DOI] [PubMed] [Google Scholar]
- 2.Robless P., Mikhailidis D.P., Stansby G.P. Cilostazol for peripheral arterial disease. Cochrane Database Syst. Rev. 2007;(1):CD003748. doi: 10.1002/14651858.CD003748.pub2. [DOI] [PubMed] [Google Scholar]
- 3.Adam D.J., Beard J.D., Cleveland T., Bell J., Bradbury A.W., Forbes J.F., Fowkes F.G., Gillepsie I., Ruckley C.V., Raab G., Storkey H., BASIL trial participants Bypass versus angioplasty in severe ischaemia of the leg (BASIL): multicentre, randomised controlled trial. Lancet. 2005;366:1925–1934. doi: 10.1016/S0140-6736(05)67704-5. [DOI] [PubMed] [Google Scholar]
- 4.Gowdak L.H., Poliakova L., Wang X., Kovesdi I., Fishbein K.W., Zacheo A., Palumbo R., Straino S., Emanueli C., Marrocco-Trischitta M. Adenovirus-mediated VEGF(121) gene transfer stimulates angiogenesis in normoperfused skeletal muscle and preserves tissue perfusion after induction of ischemia. Circulation. 2000;102:565–571. doi: 10.1161/01.cir.102.5.565. [DOI] [PubMed] [Google Scholar]
- 5.Caron A., Michelet S., Caron A., Sordello S., Ivanov M.A., Delaère P., Branellec D., Schwartz B., Emmanuel F. Human FGF-1 gene transfer promotes the formation of collateral vessels and arterioles in ischemic muscles of hypercholesterolemic hamsters. J. Gene Med. 2004;6:1033–1045. doi: 10.1002/jgm.594. [DOI] [PubMed] [Google Scholar]
- 6.Muona K., Mäkinen K., Hedman M., Manninen H., Ylä-Herttuala S. 10-year safety follow-up in patients with local VEGF gene transfer to ischemic lower limb. Gene Ther. 2012;19:392–395. doi: 10.1038/gt.2011.109. [DOI] [PubMed] [Google Scholar]
- 7.Zachary I., Morgan R.D. Therapeutic angiogenesis for cardiovascular disease: biological context, challenges, prospects. Heart. 2011;97:181–189. doi: 10.1136/hrt.2009.180414. [DOI] [PubMed] [Google Scholar]
- 8.Carmeliet P., Conway E.M. Growing better blood vessels. Nat. Biotechnol. 2001;19:1019–1020. doi: 10.1038/nbt1101-1019. [DOI] [PubMed] [Google Scholar]
- 9.Mendell J.T. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 10.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 11.Matkovich S.J., Boudreau R.L. When Knowing “Enough” May Still Not Be Enough. Circ. Res. 2018;123:412–414. doi: 10.1161/CIRCRESAHA.118.313387. [DOI] [PubMed] [Google Scholar]
- 12.Yang W.J., Yang D.D., Na S., Sandusky G.E., Zhang Q., Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J. Biol. Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 13.Caporali A., Miscianinov V., Saif J., Emanueli C. MicroRNA transport in cardiovascular complication of diabetes. Biochim. Biophys. Acta. 2016;1861:2111–2120. doi: 10.1016/j.bbalip.2016.01.010. 12 Pt B. [DOI] [PubMed] [Google Scholar]
- 14.Saif J., Emanueli C. miRNAs in post-ischaemic angiogenesis and vascular remodelling. Biochem. Soc. Trans. 2014;42:1629–1636. doi: 10.1042/BST20140263. [DOI] [PubMed] [Google Scholar]
- 15.Bonauer A., Carmona G., Iwasaki M., Mione M., Koyanagi M., Fischer A., Burchfield J., Fox H., Doebele C., Ohtani K. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 16.Yue J., Tigyi G. Conservation of miR-15a/16-1 and miR-15b/16-2 clusters. Mamm. Genome. 2010;21:88–94. doi: 10.1007/s00335-009-9240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporali A., Emanueli C. MicroRNA-503 and the extended microRNA-16 family in angiogenesis. Trends Cardiovasc. Med. 2011;21:162–166. doi: 10.1016/j.tcm.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chamorro-Jorganes A., Araldi E., Penalva L.O., Sandhu D., Fernández-Hernando C., Suárez Y. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler. Thromb. Vasc. Biol. 2011;31:2595–2606. doi: 10.1161/ATVBAHA.111.236521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin K.J., Olsen K., Hamblin M., Zhang J., Schwendeman S.P., Chen Y.E. Vascular endothelial cell-specific microRNA-15a inhibits angiogenesis in hindlimb ischemia. J. Biol. Chem. 2012;287:27055–27064. doi: 10.1074/jbc.M112.364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spinetti G., Fortunato O., Caporali A., Shantikumar S., Marchetti M., Meloni M., Descamps B., Floris I., Sangalli E., Vono R. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ. Res. 2013;112:335–346. doi: 10.1161/CIRCRESAHA.111.300418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 22.Ebert M.S., Sharp P.A. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporali A., Meloni M., Völlenkle C., Bonci D., Sala-Newby G.B., Addis R., Spinetti G., Losa S., Masson R., Baker A.H. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123:282–291. doi: 10.1161/CIRCULATIONAHA.110.952325. [DOI] [PubMed] [Google Scholar]
- 24.Caporali A., Meloni M., Nailor A., Mitić T., Shantikumar S., Riu F., Sala-Newby G.B., Rose L., Besnier M., Katare R. p75(NTR)-dependent activation of NF-κB regulates microRNA-503 transcription and pericyte-endothelial crosstalk in diabetes after limb ischaemia. Nat. Commun. 2015;6:8024. doi: 10.1038/ncomms9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horie T., Ono K., Nishi H., Iwanaga Y., Nagao K., Kinoshita M., Kuwabara Y., Takanabe R., Hasegawa K., Kita T., Kimura T. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem. Biophys. Res. Commun. 2009;389:315–320. doi: 10.1016/j.bbrc.2009.08.136. [DOI] [PubMed] [Google Scholar]
- 26.Xie J., Ameres S.L., Friedline R., Hung J.H., Zhang Y., Xie Q., Zhong L., Su Q., He R., Li M. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A.D., Tung N., Israelow B., Evans M.J., Sachidanandam R., Brown B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J.C., Sundaresan G., Iyer M., Gambhir S.S. Noninvasive optical imaging of firefly luciferase reporter gene expression in skeletal muscles of living mice. Mol. Ther. 2001;4:297–306. doi: 10.1006/mthe.2001.0460. [DOI] [PubMed] [Google Scholar]
- 29.Riegler J., Gillich A., Shen Q., Gold J.D., Wu J.C. Cardiac tissue slice transplantation as a model to assess tissue-engineered graft thickness, survival, and function. Circulation. 2014;130(11, Suppl 1):S77–S86. doi: 10.1161/CIRCULATIONAHA.113.007920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greco S., Gaetano C., Martelli F. HypoxamiR regulation and function in ischemic cardiovascular diseases. Antioxid. Redox Signal. 2014;21:1202–1219. doi: 10.1089/ars.2013.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Q., Ren X., Chen J., Li Y., Tang X., Wen X., Yang X., Zhang J., Wang Y., Ma J., Liu N. miR-16 targets fibroblast growth factor 2 to inhibit NPC cell proliferation and invasion via PI3K/AKT and MAPK signaling pathways. Oncotarget. 2016;7:3047–3058. doi: 10.18632/oncotarget.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schelch K., Kirschner M.B., Williams M., Cheng Y.Y., van Zandwijk N., Grusch M., Reid G. A link between the fibroblast growth factor axis and the miR-16 family reveals potential new treatment combinations in mesothelioma. Mol. Oncol. 2018;12:58–73. doi: 10.1002/1878-0261.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quann K., Jing Y., Rigoutsos I. Post-transcriptional regulation of BRCA1 through its coding sequence by the miR-15/107 group of miRNAs. Front. Genet. 2015;6:242. doi: 10.3389/fgene.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson P.T., Wang W.X., Mao G., Wilfred B.R., Xie K., Jennings M.H., Gao Z., Wang X. Specific sequence determinants of miR-15/107 microRNA gene group targets. Nucleic Acids Res. 2011;39:8163–8172. doi: 10.1093/nar/gkr532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suri C., Jones P.F., Patan S., Bartunkova S., Maisonpierre P.C., Davis S., Sato T.N., Yancopoulos G.D. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 36.Jeltsch M., Leppänen V.M., Saharinen P., Alitalo K. Receptor tyrosine kinase-mediated angiogenesis. Cold Spring Harb. Perspect. Biol. 2013;5:5. doi: 10.1101/cshperspect.a009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biel N.M., Siemann D.W. Targeting the Angiopoietin-2/Tie-2 axis in conjunction with VEGF signal interference. Cancer Lett. 2016;380:525–533. doi: 10.1016/j.canlet.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerald D., Chintharlapalli S., Augustin H.G., Benjamin L.E. Angiopoietin-2: an attractive target for improved antiangiogenic tumor therapy. Cancer Res. 2013;73:1649–1657. doi: 10.1158/0008-5472.CAN-12-4697. [DOI] [PubMed] [Google Scholar]
- 39.Eklund L., Kangas J., Saharinen P. Angiopoietin-Tie signalling in the cardiovascular and lymphatic systems. Clin. Sci. (Lond.) 2017;131:87–103. doi: 10.1042/CS20160129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fagiani E., Christofori G. Angiopoietins in angiogenesis. Cancer Lett. 2013;328:18–26. doi: 10.1016/j.canlet.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 41.Hausser J., Syed A.P., Bilen B., Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23:604–615. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leppänen V.M., Saharinen P., Alitalo K. Structural basis of Tie2 activation and Tie2/Tie1 heterodimerization. Proc. Natl. Acad. Sci. USA. 2017;114:4376–4381. doi: 10.1073/pnas.1616166114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasenstein J.R., Kasmerchak K., Buehler D., Hafez G.R., Cleary K., Moody J.S., Kozak K.R. Efficacy of Tie2 receptor antagonism in angiosarcoma. Neoplasia. 2012;14:131–140. doi: 10.1593/neo.111770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varu V.N., Hogg M.E., Kibbe M.R. Critical limb ischemia. J. Vasc. Surg. 2010;51:230–241. doi: 10.1016/j.jvs.2009.08.073. [DOI] [PubMed] [Google Scholar]
- 45.Pekarsky Y., Balatti V., Croce C.M. BCL2 and miR-15/16: from gene discovery to treatment. Cell Death Differ. 2018;25:21–26. doi: 10.1038/cdd.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aqeilan R.I., Calin G.A., Croce C.M. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 47.Porrello E.R., Mahmoud A.I., Simpson E., Johnson B.A., Grinsfelder D., Canseco D., Mammen P.P., Rothermel B.A., Olson E.N., Sadek H.A. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. USA. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hullinger T.G., Montgomery R.L., Seto A.G., Dickinson B.A., Semus H.M., Lynch J.M., Dalby C.M., Robinson K., Stack C., Latimer P.A. Inhibition of miR-15 protects against cardiac ischemic injury. Circ. Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue G., Yan H.L., Zhang Y., Hao L.Q., Zhu X.T., Mei Q., Sun S.H. c-Myc-mediated repression of miR-15-16 in hypoxia is induced by increased HIF-2α and promotes tumor angiogenesis and metastasis by upregulating FGF2. Oncogene. 2015;34:1393–1406. doi: 10.1038/onc.2014.82. [DOI] [PubMed] [Google Scholar]
- 50.Rey S., Schito L., Wouters B.G., Eliasof S., Kerbel R.S. Targeting Hypoxia-Inducible Factors for Antiangiogenic Cancer Therapy. Trends Cancer. 2017;3:529–541. doi: 10.1016/j.trecan.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Augustin H.G., Koh G.Y., Thurston G., Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat. Rev. Mol. Cell Biol. 2009;10:165–177. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 52.Peters K.G., Kontos C.D., Lin P.C., Wong A.L., Rao P., Huang L., Dewhirst M.W., Sankar S. Functional significance of Tie2 signaling in the adult vasculature. Recent Prog. Horm. Res. 2004;59:51–71. doi: 10.1210/rp.59.1.51. [DOI] [PubMed] [Google Scholar]
- 53.Thurston G., Daly C. The complex role of angiopoietin-2 in the angiopoietin-tie signaling pathway. Cold Spring Harb. Perspect. Med. 2012;2:a006550. doi: 10.1101/cshperspect.a006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shyu K.G., Manor O., Magner M., Yancopoulos G.D., Isner J.M. Direct intramuscular injection of plasmid DNA encoding angiopoietin-1 but not angiopoietin-2 augments revascularization in the rabbit ischemic hindlimb. Circulation. 1998;98:2081–2087. doi: 10.1161/01.cir.98.19.2081. [DOI] [PubMed] [Google Scholar]
- 55.Lekas M., Lekas P., Mei S.H., Deng Y., Dumont D.J., Stewart D.J. Tie2-dependent neovascularization of the ischemic hindlimb is mediated by angiopoietin-2. PLoS ONE. 2012;7:e43568. doi: 10.1371/journal.pone.0043568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 57.Xu J., Zhang R., Shen Y., Liu G., Lu X., Wu C.I. The evolution of evolvability in microRNA target sites in vertebrates. Genome Res. 2013;23:1810–1816. doi: 10.1101/gr.148916.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guojing L., Zhang R., Xu J., Wu C.I., Lu X. Functional Conservation of Both CDS- and 3′-UTR-Located MicroRNA Binding Sites between Species. Mol. Biol. Evol. 2015;32:3276. doi: 10.1093/molbev/msv192. [DOI] [PubMed] [Google Scholar]
- 59.Cloonan N. Re-thinking miRNA-mRNA interactions: intertwining issues confound target discovery. BioEssays. 2015;37:379–388. doi: 10.1002/bies.201400191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erhard F., Haas J., Lieber D., Malterer G., Jaskiewicz L., Zavolan M., Dölken L., Zimmer R. Widespread context dependency of microRNA-mediated regulation. Genome Res. 2014;24:906–919. doi: 10.1101/gr.166702.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eding J.E., Demkes C.J., Lynch J.M., Seto A.G., Montgomery R.L., Semus H.M., Jackson A.L., Isabelle M., Chimenti S., van Rooij E. The Efficacy of Cardiac Anti-miR-208a Therapy Is Stress Dependent. Mol. Ther. 2017;25:694–704. doi: 10.1016/j.ymthe.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gödény I., Pollesello P., Edes I., Papp Z., Bagi Z. Levosimendan and its metabolite OR-1896 elicit KATP channel-dependent dilation in resistance arteries in vivo. Pharmacol. Rep. 2013;65:1304–1310. doi: 10.1016/s1734-1140(13)71488-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anversa P., Capasso J.M. Loss of intermediate-sized coronary arteries and capillary proliferation after left ventricular failure in rats. Am. J. Physiol. 1991;260:H1552–H1560. doi: 10.1152/ajpheart.1991.260.5.H1552. [DOI] [PubMed] [Google Scholar]
- 64.Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat. Med. 2000;6:389–395. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- 65.Hernandez R., Orbay H., Cai W. Molecular imaging strategies for in vivo tracking of microRNAs: a comprehensive review. Curr. Med. Chem. 2013;20:3594–3603. doi: 10.2174/0929867311320290005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ylä-Herttuala S., Bridges C., Katz M.G., Korpisalo P. Angiogenic gene therapy in cardiovascular diseases: dream or vision? Eur. Heart J. 2017;38:1365–1371. doi: 10.1093/eurheartj/ehw547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emanueli C., Salis M.B., Stacca T., Gaspa L., Chao J., Chao L., Piana A., Madeddu P. Rescue of impaired angiogenesis in spontaneously hypertensive rats by intramuscular human tissue kallikrein gene transfer. Hypertension. 2001;38:136–141. doi: 10.1161/01.hyp.38.1.136. [DOI] [PubMed] [Google Scholar]
- 68.Mitić T., Caporali A., Floris I., Meloni M., Marchetti M., Urrutia R., Angelini G.D., Emanueli C. EZH2 modulates angiogenesis in vitro and in a mouse model of limb ischemia. Mol. Ther. 2015;23:32–42. doi: 10.1038/mt.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonci D., Coppola V., Musumeci M., Addario A., Giuffrida R., Memeo L., D’Urso L., Pagliuca A., Biffoni M., Labbaye C. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 71.Sala-Newby G.B., Freeman N.V., Curto M.A., Newby A.C. Metabolic and functional consequences of cytosolic 5′-nucleotidase-IA overexpression in neonatal rat cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H991–H998. doi: 10.1152/ajpheart.00053.2003. [DOI] [PubMed] [Google Scholar]
- 72.Cao F., Lin S., Xie X., Ray P., Patel M., Zhang X., Drukker M., Dylla S.J., Connolly A.J., Chen X. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–1014. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.