Abstract
Cell‐free microRNA (miRNA) in biofluids released by tumors in either protein or vesicle‐bound form, represent promising minimally‐invasive cancer biomarkers. However, a highly abundant non‐tumor background in human plasma and serum complicates the discovery and detection of tumor‐selective circulating miRNAs. We performed small RNA sequencing on serum and plasma RNA from Nasopharyngeal Carcinoma (NPC) patients. Collectively, Epstein Barr virus‐encoded miRNAs, more so than endogenous miRNAs, signify presence of NPC. However, RNAseq‐based EBV miRNA profiles differ between NPC patients, suggesting inter‐tumor heterogeneity or divergent secretory characteristics. We determined with sensitive qRT‐PCR assays that EBV miRNAs BART7‐3p, BART9‐3p and BART13‐3p are actively secreted by C666.1 NPC cells bound to extracellular vesicles (EVs) and soluble ribonucleoprotein complexes. Importantly, these miRNAs are expressed in all primary NPC tumor biopsies and readily detected in nasopharyngeal brushings from both early and late‐stage NPC patients. Increased levels of BART7‐3p, BART9‐3p and particularly BART13‐3p, distinguish NPC patient sera from healthy controls. Receiver operating characteristic curve analysis using sera from endemic NPC patients, other head and neck cancers and individuals with asymptomatic EBV‐infections reveals a superior diagnostic performance of EBV miRNAs over anti‐EBNA1 IgA serology and EBV‐DNA load (AUC 0.87–0.96 vs 0.86 and 0.66 respectively). The high specificity of circulating EBV‐BART13‐3p (97%) for NPC detection is in agreement with active secretion from NPC tumor cells. We conclude EV‐bound BART13‐3p in circulation is a promising, NPC‐selective, biomarker that should be considered as part of a screening strategy to identify NPC in endemic regions.
Keywords: nasopharyngeal carcinoma, Epstein–Barr virus, noninvasive diagnosis, tumor markers, BART‐microRNAs
Short abstract
What's new?
Analysis of DNA from human tumor viruses in patient blood is a non‐invasive screening method for individuals at risk for developing cancer. A drawback is over‐diagnosis as these sensitive methods also detect non‐cancer‐related infections. Here the authors show by RNA sequencing and PCR amplification that a microRNA (BART13‐3p) encoded by the Epstein–Barr Virus (EBV) is associated with circulating vesicles in patients with nasopharyngeal carcinoma, thus distinguishing between cancer and non‐cancer‐related EBV infections.
Introduction
Nasopharyngeal carcinoma (NPC) prevails in southern China, Southeast Asia, North Africa, the Middle East, and the Arctic.1 Although NPC is rare in most other parts of the world, NPC incidence is increasing in low‐risk populations such as Europe and USA due to worldwide migration.2, 3 Most patients with NPC present in the clinic with advance stage disease.1 Given the high risk of NPC in endemic regions and among certain family members,4 and the high cure rate for early‐stage of disease, early detection of NPC using a noninvasive method is considered crucial to reduce mortality from NPC.
Epstein–Barr virus (EBV) is a primary etiologic agent in the pathogenesis of undifferentiated NPC. Abnormal anti‐EBV antibody profiles, increased circulating EBV DNA levels, and a distinct EBV gene expression profile in tumor cells illustrate the active role of EBV in the carcinogenic process.5, 6, 7, 8, 9 Various noninvasive blood‐based EBV assays have been accepted for detection of circulating biomarkers for NPC, such as EBV‐specific IgA antibody testing by ELISA and PCR‐based measurement of cell‐free EBV DNA.8, 10, 11, 12, 13 In endemic regions, majority of individuals with NPC have elevated antibody titers to defined EBV antigens (EBNA1, EA, VCA) that may persist up to 10 years prior to diagnosis.4, 8, 9, 13 Although highly sensitive for NPC, these antibody assays do not meet the desired specificity as a single diagnostic marker.4 If used indiscriminately, EBV IgA serology may lead to overdiagnosis of NPC with associated psycho‐social consequences and extra costs in endemic countries.Quantitative measurement of circulating cell‐free viral loads in plasma or serum by real‐time PCR revealed higher specificity than serological test.1, 8, 11 Measurement of plasma cell‐free EBV DNA load is recognized as tumor marker for prognostic assessment of NPC patients undergoing radiotherapy and prediction of recurrent disease rather than for early detection of NPC.8, 11, 14 Although this circulating biomarker has been proposed for screening purposes15 it does not reflect intact living tumor cells and awaits validation in prospective cohorts.16 A noninvasive circulating biomarker with high diagnostic accuracy is needed for identifying high‐risk subjects prior to the onset of clinical signs of NPC. Ideally, such a marker would provide tumor‐specific information to warrant further confirmation by NP brushing or biopsy. Circulating microRNAs (miRNAs) provide insight into both the biology and clinical behavior of various types of cancer.17 EBV‐miRNAs play an important role in EBV‐driven transformation and NPC pathogenesis targeting anti‐growth and apoptotic pathways.18, 19, 20, 21, 22 However a clear consensus has not yet emerged from several studies concerning circulating EBV‐miRNAs as biomarkers for NPC detection and patient monitoring.23, 24, 25 Indeed, several studies revealed that there is a relatively high inter‐tumor variation of EBV‐miRNA expression,20 which may complicate the design a standardized noninvasive EBV‐miRNA assay for NPC detection. While RNAseq has been performed on plasma miRNA from NPC patients only endogenous miRNAs have thus far been considered.26 However, cell‐free miRNAs in cancer patients can be of both tumor and nontumor (micro‐environment) origin a proportion of which enclosed in Extracellular Vesicles (EVs).27
Here, we profiled with RNAseq serum and plasma samples of NPC patients and healthy controls with the purpose of identifying circulating EBV‐miRNAs that are consistently associated with NPC and not other diseases to reduce false positives. We validated EBV‐BART7‐3p, BART9‐3p and BART13‐3p with qRT‐PCR in total serum, serum EVs, C666.1 NPC cells, primary NPC tissues and NP brushings. Circulating BART13‐3p is a strongly EV‐associated miRNA that outperforms classic EBV‐DNA load and EBV IgA serology for identifying patients with NPC in comparison to healthy controls and individuals with various EBV‐associated diseases.
Materials and Methods
Patient selection and biological sample collection
We used archived pre‐treatment serum samples from NPC patients of Indonesian (endemic) origin with regional controls as detailed in recent publications.12, 13, 28, 29 We selected 56 NPC patients collected at the Dr. Cipto Mangunkusumo Hospital in Indonesia with available frozen biopsy or nasopharyngeal brushing, serum and whole blood samples at diagnosis (Table 1). EBV status of tumor biopsies was assessed by EBER in situ hybridization. All patients were clinically staged according to the UICC/AJCC guidelines (Supporting Information Table S1). Serum was prepared by blood clotting at RT for less than 2 h or overnight at 4C. After clotting, the liquid phase was centrifuged (1,000–2,000x g) for 10 min in a refrigerated centrifuge and the supernatant (serum) stored at −80 for further analysis. Healthy donor sera (n = 33) from Indonesian were used as controls for miRNA and DNA analysis. In addition, a total of 29 pre‐treatment serum samples from patients with various non‐NPC diseases in head and neck (Table 1) referred to VU Medical Center in Amsterdam, the Netherlands, Jeroen Bosch Hospital in the Netherlands, the Dr. Cipto Mangunkusumo Hospital, Indonesia29 (enrolled during the same period as NPC patients) were analyzed for miRNA and anti‐EBV IgA serology. All patients signed for informed consent and the study was approved by the medical ethical committees of the participating hospitals. In addition, plasma samples from 14 histologically confirmed NPC collected at the National Cancer Institute, Centro di Riferimento Oncologico, Aviano, Italy and healthy control plasma samples were used as external validation cohort.30 EDTA‐plasma was centrifuged (800‐1,200 g) in a refrigerated centrifuge for 10 min within 2 h after blood draw and the subjected to a second spin at 2500 g for 10 min. The platelet poor plasma was than stored at −80 for further analysis.
Table 1.
Clinical characteristics of serum samples collected in our study
| Serum biomarkers | |||
|---|---|---|---|
| Healthy | NPC | non‐NPC malignancies (n = 29) | |
| (n = 33) | (n = 39) | ||
| Mean age (range) | 37 (20/56) | 42 (14/63) | 55 (14/85) |
| Sex (male/female) | 18/15 | 31/8 | 18/11 |
| Ethnicity | |||
| Asian | 26 | 39 | 5 |
| Caucasian | 7 | 24 | |
| Disease types | |||
| Infectious mononucleosis | 3 | ||
| Chronic active EBV | 1 | ||
| Adenoid hyperplasia | 1 | ||
| Squamous cell carcinoma | 2 | ||
| Non‐Hodgkin B cell Lymphoma | 1 | ||
| NK/T cell lymphoma | 1 | ||
| non‐NPC head and neck carcinoma | 20 | ||
| Nasopharyngeal carcinoma | 39 | ||
Cell lines
EBV positive NPC cell line C666‐1 and EBV negative cell lines NPC‐derived HONE1 were obtained and cultured as previously described and recently authenticated.31 These cell lines were used to profile intra and extracellular viral and human small noncoding RNAs for NPC microRNA biomarker discovery. Upon confluence, media were removed and cells were used for RNA extraction for intracellular miRNA profiling.19, 32 The supernatants were collected and centrifuged at 500 × g for 10 min. to remove remaining cells and re‐centrifuged at 2,000 × g for 15 min to eliminate cell debris. Pre‐cleared supernatants were further subjected to size‐exclusion chromatography (SEC) for EV enrichment.33
Sample processing and nucleic acid purification
Serum, plasma, whole blood, nasopharyngeal brushing and frozen tumor biopsy samples were collected and processed for DNA and RNA isolation according to published protocols.12, 29, 34 Extracellular vesicles (EVs) were isolated from serum and plasma by SEC Sepharose CL‐2B column (GE Healthcare) as described previously.33 TRIZOL LS (Invitrogen) was used to isolate total RNA from 250 μL total serum or plasma according to the manufacturer's instruction. Glycogen (Invitrogen, R0551) was added to increase the RNA yield. Trizol (Invitrogen) was added to (a) tumor cells, (b) tumor biopsy, (c) 250 μL of pooled SEC fractions 9 and 10 (EV fractions) and (d) 250 μL of pooled fractions 20 and 21 (HDL/protein fractions). The total extracellular RNA was precipitated and pooled from four times TRIzol‐based RNA purification of EV and HDL/protein fractions. The rest of each SEC fraction was stored at −80°C for subsequent western blot and transmission electron microscopy (TEM) as previously described.33 RNA was finally eluted in 11 μL RNAse‐free water and stored at −80°C until further use. DNA and RNA concentration and quality were assessed by NanoDrop ND‐100.
Small RNA sequencing
Sequencing libraries were generated using a maximum volume of RNA sample (7 μL) for each plasma and serum sample and an equal amount of RNA input (1,500 ng) was used for each cell line according to Illumina's TruSeq small RNA Sequencing Protocol. A 3′ adapter was ligated to the RNA template using a truncated T4 RNA ligase 2. Next, 5′ adapters were added, using T4 RNA ligase and ATP (Illumina). Single stranded cDNA was created using Superscript II Reverse Transcriptase (Invitrogen). The cDNA was then PCR amplified and PCR products were size‐selected for 145–160 bp. Quality of sequence libraries was validated using Agilent high sensitivity DNA kit (Agilent Technologies) by measuring size, purity, and concentration on an Agilent 2,100 Bioanalyzer. Sequencing was performed on Illumina HiSeq 2000 paired‐end 125‐cycle (PE125) run, using HiSeq v4 reagents (Illumina). After in silico adapter removal, 15‐125 nt fragments were mapped to the reference genome using several remote databases hosted by NCBI. Deep sequencing data was analyzed using sRNAToolbox.35
Molecular analysis
Viral and endogenous small noncoding RNA profiling was performed on 5 μL (250 to 350 ng RNA) total RNA extracted from cells, frozen biopsies, nasopharyngeal brush, serum and plasma samples with labeled primers (Life Technologies, 4,427,975). All necessary reagents for noncoding small RNA‐specific cDNA synthesis prior to multiplex stem‐loop RT‐PCR followed by real time PCR with TaqMan probes (Applied Biosystems AB7500) have been previously detailed.34 Two small noncoding RNAs (viral EBER1 and host Vault RNA) were analyzed by LightCycler stem‐loop RT‐PCR (Roche, LC480) as previously published.36 C666‐1 and HONE1 cells as EBV positive and negative control were used as calibrators. A true amplification curve with raw cycle threshold (Ct) mean value <40 (Ct SD <0.3) and was considered reliable in Taqman and LightCycler systems. As endogenous controls we used Vault RNA and miRNA16‐5p.32 A Ct value of 40 (Ct SD >0.3) for vaultRNA and a Ct value >30 (Ct SD >0.3) for miRNA16‐5p was considered as adequate levels for defining RNA quantity while hsa‐miR155 was considered as potential marker of inflammation. Relative quantification of miRNAs was calculated using the 2‐ΔΔCt approach when indicated.31 Normalization for vesicle concentration was achieved by measuring vault RNA as a surrogate as it is a highly enriched small ncRNA in EVs. EBV DNA load was measured using protocol as published before.10 All experiments were run as independent duplicates. Primers and probes used in our study are listed in the Supporting Information Table S2.
EBV EBNA1 and VCAp18 antibody assays
IgA antibody levels against the EBV EBNA1 and VCAp18 proteins was measured according to previously published protocols.4, 13, 37 A cut‐off calibrator, a blank sample, the positive and four known EBV negative controls were included in each test run. The cut‐off calibrator was run in triplicate, and the cutoff value (COV) for each ELISA plate defined by calculating the mean OD450 value +2x SD of the four negative reference sera. Results of the peptide‐based ELISA assays were reported as normalized values (i.e. the mean of the duplicate absorbance value observed for each IgA antibody, divided by the COV for each plate.
Statistical methods
Graphical analysis was plotted using Prism 6.07 software (GraphPad) and statistical evaluations were executed by using SPSS 22 and R software (version 3.2.5). All tests were two‐sided and considered statistically significant at the 0.05 level. In miRNA analysis, 2(−ΔΔCt) from each microRNA were square root transformed, standardized and then evaluated by the nonparametric Rank Products test using the R statistics. Serum EBV miRNA was presented in log2 transformed expression value, EBV DNA load was presented as copies per ml whole blood or copies per biopsy/brush and anti‐EBV antibody level was presented in mean per COV.4, 29 All values in the text and figures represent the mean or mean with SD.
The Receiver Operating Characteristic (ROC) analysis38 was performed to evaluate the diagnostic performance of each circulating EBV miRNA in differentiating patients with NPC (n = 39) from the healthy control subjects (n = 33) and patient with other diseases (n = 31) Table 1. The ΔΔCt ratio for each and every marker was transformed to the Square root scale to have less skewed‐distributed data. Next, we built univariable logistic regression models for each individual marker. The optimal cut‐off thresholds for diagnosis were set for maximum sensitivity and specificity. By analyzing serum associated EBV miRNA levels in NPC patients and healthy subjects, a diagnostic miRNA panel for NPC was built using logistic regression model. The performance of each univariable model was measured by area under the receiver operating characteristic (ROC) curve (AUC).
Pearson correlation test was used to examine the relation between expression level of EBV‐encoded miRNA and EBV DNA load in the circulation and primary tumor by using the log2 scale of quantitative EBV DNA level and the Ct value of each miRNA. The Wilcoxon matched‐pairs signed rank test was used to measure differentially expressed levels of each miRNA in serum fractions. Asterisks indicate statistical significance * p <0.05, ** p <0.005, *** p <0.001, **** p <0.0001).
Results
To search for commonly deregulated EBV‐miRNAs in the circulation of NPC patients, we performed small RNA (Illumina) sequencing on a small panel of plasma and serum samples. The volcano plot in Figure 1 a reveals that EBV‐BART miRNAs distinguish NPC samples from healthy controls compared endogenous miRNAs (Fig. 1 a). In 3 out 5 NPC samples robust EBV‐miRNAs read counts (>10 RC) allowed us to delineate profiles. Pie‐charts reveal that the EBV‐miRNAs BART11‐3p, BART2‐5p and BART10‐3p are most frequently detected in NPC patient plasma and serum but that the profiles are not uniform (Fig. 1 b). This is consistent with the described inter‐tumor heterogeneity in EBV‐miRNA expression profiles.20, 39 BART2‐5p was detected in all EBV‐miRNA positive NPC samples consistent with a recent PCR‐based study.40 In one NPC patient there was a high relative abundance of BART13‐3p, a miRNA described before as potential NPC‐associated noninvasive marker.23 However some EBV‐miRNAs, in particular BART2‐5p and 11‐3p may not be selective for NPC, as they have been detected in the plasma of patients with non‐NPC EBV‐associated diseases.41, 42, 43 In addition, BART10‐3p was detected above cut‐off level in one control serum sample casting doubt on the NPC‐specificity for these EBV‐miRNAs as measured by RNAseq.
Figure 1.
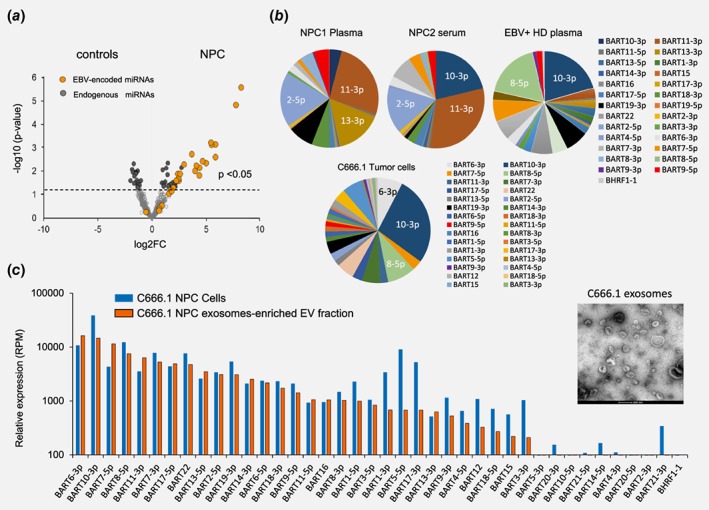
small RNAseq reveals distinct circulating EBV BART‐miRNA profiles in NPC patients. (a) Volcano plot of RNA Illumina sequence analysis of EBV miRNAs in NPC patients (n = 5) and Healthy controls (n = 3). Each dot represents the type of EBV‐encoded miRNA (orange) and endogenous miRNA (gray). (b) EBV miRNA distribution in a NPC plasma (NPC1), serum (NPC2) sample and a healthy control (CTRL1) plasma sample compared to relative expression pattern as measured in C666.1 NPC tumor cells. Data are shown as percentage read counts. (c) Relative EBV miRNA expression in NPC C666.1 tumor cells and secreted exosomes. Data are shown as reads per million miRNA reads (RPM). EM image of pure exosomes isolated from NPC C666.1 cell line. Scale bar: 200 nm.
To resolve this we performed small RNAseq on C666.1 NPC cells and secreted extracellular vesicles (EVs) that enclose tumor‐derived miRNAs and have been used for noninvasive cancer detection.27, 33 Electron Microscopy analysis revealed pure populations of EVs, the majority of which, in the size range (70‐100 nm) of endosomes‐derived exosomes (Fig. 1 b and 1 c). The EBV‐miRNA profile in C666.1 NPC cells is largely reflected by their relative presence in the exosomes showing high relative abundance of BART6‐3p, 10‐3p and 7‐5p (Figs. 1 b and 1 c and Supporting Information Fig S1A) and the EV marker vtRNA1‐1. This data clearly indicated that NPC C666.1 cells have a different overall EBV‐miRNA RNAseq profile than the NPC patient sera. Further detailed isomiR analysis indicated that BART7‐3p and BART13‐3p are highly uridylated at the 5′‐end (Supporting Information Fig. S1B). Such nontemplated addition (NTAs) may act as a possible export signal for selective incorporation of a subset of miRNAs into exosomes.44
Based on the sequencing data, BART10‐3p, BART6‐3p, BART7‐3p/−5p and BART13‐3p were selected for further validation. In addition we tested BART17‐3p/5p and BART9‐3p based on published literature.23, 24 Finally, we included multiple endogenous miRNAs (miR16, miR223 and miR155) and vault1‐1 ncRNA as a control for EV concentration.36 We detected abundant expression of EBV‐BART7‐3p, −9‐3p, 10‐3p and −13‐3p (Fig. 2 a) in the NPC C666.1 cells, secreted exosomes‐enriched EV and protein‐enriched fractions (Figs. 2 c and 2d). These markers were, as expected, absent in EBV‐negative HONE NPC cells (Fig. 2 b). Interestingly, BART6‐3p, which is highly expressed by C666.1 NPC cells as judged by RNAseq and qRT‐PCR, was virtually undetectable in the exosomes‐enriched EV‐ and protein‐enriched fractions, suggesting that not all EBV‐miRNAs have the same propensity for exosome incorporation (Supporting Information Fig. S1). Although not further detailed here, it is noteworthy that EBER1 (but not EBER2, data not shown) is abundantly present in exosomes of C666.1 cells, but not in the protein‐enriched secretory fraction.
Figure 2.
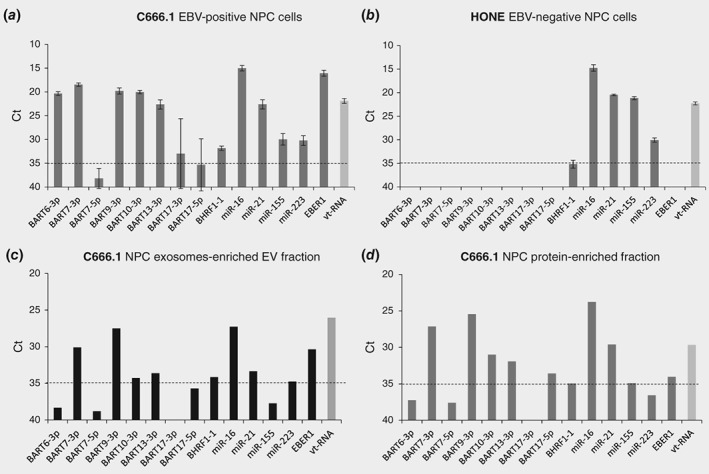
NPC C666.1 cells secrete a panel of EBV‐BARTs via exosomes and proteins. RT‐PCR analysis of NPC selective EBV miRNAs and endogenous cellular miRNAs in (a) C666.1 EBV positive NPC cells, (b) HONE‐1 EBV negative NPC cells, (c) C666.1 NPC exosomes‐enriched EV fraction and (d) C666.1 NPC protein‐enriched fraction. Data were presented as Ct values. Error bars represent the mean with SD. Dotted horizontal line represents the detection limit.
Based on these results combined, we selected BART7‐3p, −9‐3p, −10‐3p and − 13‐3p as potentially NPC‐selective, cell‐free miRNA biomarkers.
Circulating EBV BART‐miRNAs discriminate NPC patients from control groups
Given the differential intra‐ and extracellular EBV‐miRNA expression, we questioned which candidate BART‐miRNAs are most selective for detecting NPC tumors. We analyzed serum specimens obtained from 39 NPC Indonesian patients and two control serum cohorts from a mixed population of healthy individuals (n = 33) and Indonesian non‐NPC patients (n = 31) of which some (n = 7) had reported EBV‐associated disease (for details on sample characteristics see Table 1).
We found that circulating EBV miRNAs (BART7‐3p, −9‐3p, −13‐3p) were consistently detected (average Ct levels between 33 and 35) and were more abundant in the NPC patient sera than in the two control groups (Figs. 3 a– 3 c) (Mann–Whitney, p <0.0001). We concluded that presenting raw Ct values for the EBV miRNAs is reasonable as the endogenous miR155‐3p levels were highly similar (average Ct levels 27) between all groups (Fig. 3 d). This provided confidence in the RNA quality, extraction efficiency and PCR outcome of the samples. While BART10‐3p was detectable by RT‐PCR in most NPC patient serum samples, the average levels were either at or slightly below the detection limit of this assay (Ct <35–36). This may cast doubt on the robustness of this marker and was therefore excluded from further analysis (Supporting Information Fig. S2A). While cell‐free BART7‐3p and BART9‐3p were detected above detection limits in a small proportion of the healthy subjects, BART13‐3p was virtually undetectable in this group. When comparing the results with EBV‐associated, non‐NPC patients (i.e. infectious mononucleosis, chronic active EBV infection and natural killer/T cell lymphoma and non‐NPC head and neck tumors) the levels of BART7‐3p and BART9‐3p were higher than in healthy individuals in some of the patients (Figs. 3 a and 3b). In contrast, BART13‐3p was only detected in a minor proportion of this control group and mostly below the detection limit of the assay (Fig. 3 c). Interestingly, the levels of BART7‐3p and BART13‐3p appear on average are higher both in early stage (I/II) and late stage NPC compared to non‐NPC controls (Supporting Information Fig. S2B). Combined these data strongly indicate that three candidate BART miRNAs secreted by C666.1 NPC cells are robustly detected in NPC patient sera with the stemloop qRT‐PCR method. Of these, BART13‐3p appears to be most selective for NPC status in both early and late stage disease when compared to non‐NPC controls that include samples from patients with EBV‐associated disease (see Table 1).
Figure 3.
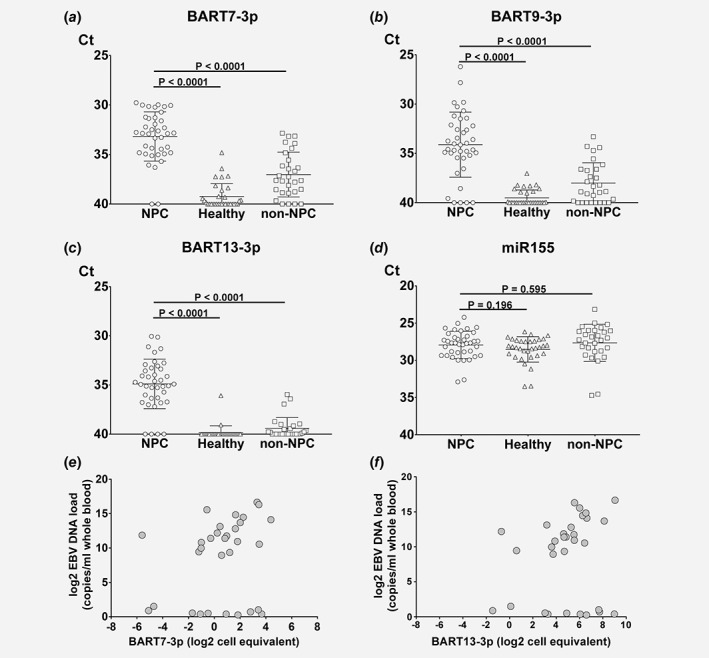
EBV BART7‐3p, BART9‐3p and BART13‐3p levels are increased in NPC patient sera compared to non‐NPC disease controls even in EBV‐DNA load negative sera. Relative EBV‐BART microRNA expression levels of (a) BART7‐3p, (b) BART9‐3p, (c) BART13‐3p and (d) endogenous miR155 control in total serum of NPC patients (circles, n = 39), healthy Individuals (triangles, n = 33) and ‘non‐NPC’ disease controls (squares, n = 29). Data are presented as Ct values, errors bars represent from the average between each group. Significant differences were analyzed by Mann–Whitney test and the p‐values are indicated above the plots. Correlation between EBV DNA copies in blood and EBV miRNA serum level (e) BART7‐3p and (f) BART13‐3p. Data are presented as a log2 scale.
To further evaluate NPC relatedness, we investigated the circulating EBV‐BART miRNA levels correlate with EBV‐DNA load in the blood of NPC patients and whether the absence of cell‐free EBV‐DNA (cfDNA) copies would mean absence of the EBV‐miRNAs. To this end we first determined the correlation between EBV‐DNA copies numbers and EBV‐miRNA expression in NPC666.1 cells as determined by qRT‐PCR (Supporting Information Fig. S3). When analyzing the correlation between EBV DNA copies in blood and the level of serum EBV‐BART miRNAs, we determined a significant positive correlation for BART7‐3p and BART13‐3p: p = 0.0007, p <0.0001 respectively (Figs. 3 e and 3f). The same correlation was found for BART9‐3p (data not shown). It is noteworthy that most NPC cases in which we failed to detect EBV‐DNA (n = 12) we clearly could detect cell‐free EBV BART‐miRNAs.
Together these results strongly suggest that BART7‐3p, BART9‐3p and BART13‐3p are released by EBV‐infected NPC tumor cells into the bloodstream detectable by qRT‐PCR, making these markers potentially useful for minimally‐invasive detection of early‐ and late stage NPC.
Evaluation of circulating versus tissue markers
To validate whether the EBV‐BART miRNA candidates are consistently expressed by NPC tumor cells in vivo, we profiled 20 NPC tissues using hypertrophic tonsil tissues (n = 11) as controls tissues. We subsequently matched the relative expression in the NPC tissues with their presence in nasopharyngeal (NP) brushings (mucosal scrapes of tumor surface, n = 20) in addition to NP brushings from a group of healthy EBV carriers (n = 11). We could confirm strongly elevated levels of BART7‐3p, −9‐3p and − 13‐3p in NPC biopsy specimens compared to tonsils (p <0.0001, Fig. 4 a). The relative expression level in the NP brushings and biopsies of patients with NPC was very similar (Fig. 4 b), suggesting that these BART miRNAs in the NP brushings primarily report NPC tumor presence and no other infections. In agreement with previous studies we show strong inter‐tumor heterogeneity of EBV‐miRNA expression levels. Even after miR16‐5p normalization levels differ up to 1,000‐fold which was most pronounced (10e5‐fold) for EBER1 expression levels. This heterogeneity was less obvious for most endogenous miRNAs (Fig. 4 a). We detected low levels of BHRF1‐1 miRNA both in tonsil tissues and normal NP brushings, indicative of low level of ongoing viral replication either in plasma cells and/or oropharyngeal epithelial cells lining. This was evident from the NPC brushings, indicative of ongoing viral replication in undifferentiated NPC tumors45 as we reported previously.12, 45
Figure 4.
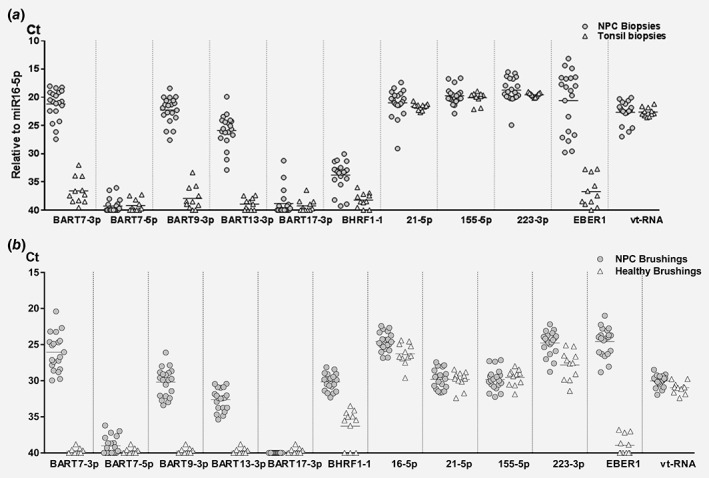
Heterogeneity of EBV‐miRNA expression profiles in primary NPC tumor tissue is reflected in NP brushings. (a) Normalized expression levels of EBV miRNAs and endogenous miRNAs in NPC biopsies (n = 20, gray dots) compared to tonsils (n = 11, white triangles). (b) Same as (a) but showed relative expression levels in NPC brushings (n = 20, gray dots) and healthy brushings (n = 11, white triangles). Data were presented as Ct values. Short horizontal lines represent the average.
Thus EBV‐BART miRNAs in brushes is a valid option for in situ NPC tumor confirmation after liquid biopsy (cell‐free DNA and circulating miRNA) analysis.
NPC‐associated ebv‐miR‐BART13 is released into circulation via extracellular vesicles
Circulating EBV‐DNA likely represents pieces of the viral genome released form dying EBV‐infected cells. We reasoned that at least a proportion of circulating EBV‐BART miRNAs in NPC patient sera are actively secreted by living tumor cells via EVs as suggested from our in vitro experiments (Fig. 1 c). To investigate this, we performed SEC‐based fractionation on serum samples46 of 18 patients with NPC, enriching fractions for EVs and separating these from smaller, soluble protein complexes. A representative transmission electron microscope (TEM) image confirmed purification of serum EVs in fractions 9 and 10 (Fig. 5 a), which were as expected highly enriched in the EV‐bound small Vault1.1 ncRNA (Fig. 5 b). We extracted RNA from these serum fractions and performed qRT‐PCR. While EBV BART7‐3p is readily detected in both EV and protein‐enriched fractions of NPC patients (Fig. 5 c), only BART13‐3p (Fig. 5 d) was significantly enriched in EVs from NPC patients when compared to the protein fraction (=0.007). Endogenous cell‐free miR‐155 was also strongly EV‐bound in both NPC patients and healthy control serum, yet there was no difference in the levels of NPC EVs and healthy (Fig. 5 e) suggesting a non‐NPC origin of these EVs.
Figure 5.
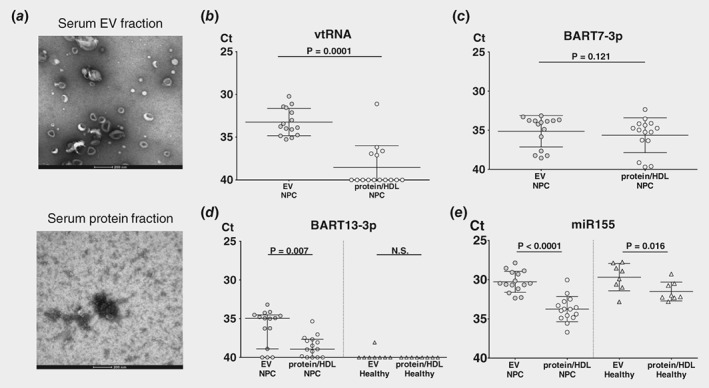
EBV BART13‐3p is selectively enriched in serum EVs from NPC patients. Sera from 18 NPC patients underwent Size Exclusion Chromatography. (SEC) to get fractionated into EVs and protein followed by stem‐loop RT and TaqMan. miRNA assays. (a) Transmission electron microscope (TEM) image of serum from NPC patient in EV fraction and protein fraction. Scale bar: 200 nm. miRNA expression levels of (b) endogenous vtRNA, (c) BART7‐3p, (d and e) BART13‐3p and miR155 respectively were measured by qRT‐PCR in extracellular vesicles (EVs) fractions and protein fraction of 18 NPC patients and 8 healthy control sera. The raw data is adjusted by volume input and presented as Ct values. Errors bars represent the averages of EV and protein fractions. Significance of differences were analyzed by Mann–Whitney test and the corresponding p‐values are indicated above the plots.
These findings, together with the in vitro data (Fig. 1 c) suggest that NPC cells actively secrete EV‐bound BART13‐3p, which is relatively stable in circulation with hitherto unknown functional consequences for EV‐target cells.
Diagnostic performance of EBV BART miRNAs compared to serology and EBV DNA load
Finally we calculated the diagnostic potential of each individual miRNA using receiver operator characteristics (ROC) using 39 NPC, 33 healthy and 29 non‐NPC serum samples. To diminish the influence of inter sample variability for this diagnostic analysis we normalized miRNA levels with Vault1‐1 ncRNA using the ΔΔCt method (Supporting Information Fig. S4A‐D). We choose Vault1‐1 since (i) it was always detected in each sample (ii) has no known relation with NPC (iii) Ct levels are in a similar range as the miRNA targets (iv) variation appears small (v) the PCR efficiency was similar as to the miRNAs and (vi) its levels are strongly EV‐associated (Figs. 2 c and 5 b).
Compared to healthy donors, the ROC curve of individual viral miRNAs revealed AUC's of 0.90, 0.87, and 0.91 for BART7‐3p, 9‐3p, and 13‐3p, respectively (Fig. 6 a). Next we used this information to determine the optimal cut‐off point for each of the EBV BART miRNAs individually to reach maximal sensitivity and specificity as identified by the R3.2.5 software. This approach shows that all miRNAs can provide a relatively high sensitivity and specificity (BART7‐3p, 85% and 90%; BART9‐3p, 97% and 82% and BART‐13‐3p 74% and 97% respectively) with circulating BART13‐3p clearly being the most strongly NPC‐associated miRNA compared to healthy individuals with an of AUC 0.91. In contrast to the circulating EBV‐miRNAs, endogenous cell‐free miR‐155 showed essentially no diagnostic potential for NPC when compared to healthy EBV carriers (data not shown). Moreover, a combination of BART13‐3p with BART7‐3p yielded an AUC of 0.93 (Fig. 6 a).
Figure 6.
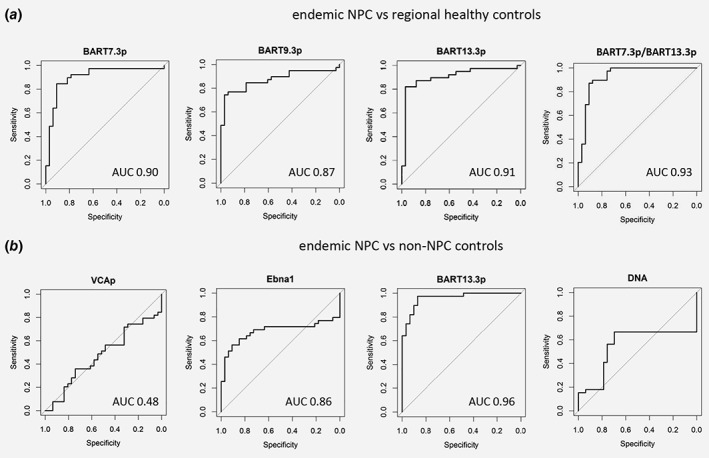
Serum BART‐miRNAs notably BART13‐3p normalized to vtRNA differentiate NPC sera from healthy and non‐NPC disease control sera. ROC curves of: (a) EBV miRNAs BART7‐3p, BART9‐3p, BART13‐3p and BART7‐3p/BART13‐3p levels measured by qRT‐PCR in the sera of NPC patients and compared to healthy controls. (b) IgA‐VCAp18, IgA‐EBNA1, BART13‐3p and EBV‐DNA levels were measured in the sera from NPC patients and compared to non‐NPC controls.
Next we compared the performance of PCR‐based EBV‐miRNAs with ELISA‐based EBV‐EBNA1‐IgA and EBV DNA‐load measurements from NPC patients and non‐NPC patients with persistent EBV infection and head and neck cancer. Consistent with previous studies,4 IgA anti‐EBNA1 ELISA yielded an AUC of 0.86, which was superior over IgA‐VCAp18 (AUC, 0.48) and EBV‐DNA (AUC 0.66) (Fig. 6 b). While the sensitivity of EBNA1 IgA ELISA was 92%, the specificity was 71%, thus performing less accurate than serum BART13‐3p levels (97% and 87%), which again appeared the best marker to identify true NPC patients with an AUC of 0.96. Finally, we evaluated whether our candidate EBV‐miRNAs can also be indicative of NPC using plasma from nonendemic (Italian) NPC patient cohort and healthy control plasma (see Table 2). We calculated that a combination of BART7‐3p and BAR13‐3p achieved an AUC of 0.88 in this independent sample set (Supporting Information Fig. S4E).
Table 2.
Clinical characteristics of plasma samples collected in our study
| Plasma | ||
|---|---|---|
| External group | ||
| Healthy | Italian NPC | |
| (n = 14) | (n = 14) | |
| Mean age (range) | 30 (22–62) | 44 (30–70) |
| Sex (male/female) | 1/13 | 13/1 |
| Ethnicity | ||
| Asian | 2 | |
| Caucasian | 12 | 14 |
Thus measuring the levels of circulating EBV miRNA, in particular serum EV‐bound BART13‐3p can be used to identify endemic and nonendemic NPC patients.
Discussion
EBV‐encoded miRNAs are suggested to have a functional role in the pathogenesis NPC.18, 19, 47 Many studies have demonstrated that circulating miRNAs represent promising minimally‐invasive biomarkers for cancer detection but selectivity for tumor cell origin is not always confirmed.27, 48 Using multiple in vitro and in vivo strategies we tested and validated BART7‐3p, 9‐3p, and 13‐3p as promising liquid biopsy targets. EV‐bound BART13‐3p appeared the best performing NPC‐selective marker in diagnostic serum analysis as compared to existing methods (DNA‐load and IgA EBV serology) using healthy‐, non‐NPC cancer‐ and EBV‐associated disease as controls (Fig. 6).
To date, noninvasive EBV serology tests confirmed by endoscopic assessment and pathological examination of tissue biopsies is the standard approach for screening and diagnosis of NPC.1, 4, 9, 13, 37 Elevated IgA antibodies that are indicative of aberrant Epstein–Barr virus (EBV) activity are noninvasive tools to identify high‐risk individuals and IgA titers against EBV antigens are helpful in stratifying NPC risk.4 However a screening test based on serology alone would lead to unacceptable over diagnosis and treatment and thus requires improvement. Next to serology, determining the cell free EBV‐DNA load has been extensively evaluated for noninvasive NPC diagnosis.10, 12 However, given that the origin of EBV‐DNA in the blood is unclear, diagnosis of primary NPC relying solely on the detection of (fragmented) EBV DNA in circulation may still not be specific enough, or practical, for large scale screening.15, 16, 49 This is especially true for poorer regions of the world where high‐throughput sequencing‐based assays remain prohibitively expensive.
To discern what approach would be optimal for early NPC detection we contend that practical and affordable methods need to be compared directly. We found in 2 separate NPC cohorts (endemic, nonendemic) that EBNA1‐specific IgA serology has a 92% sensitivity to identify NPC, while specificity reached 77%, consistent with prior studies in large cohorts.4, 37 Importantly, measuring BART13‐3p miRNA levels is serum outperforms the diagnostic accuracy of IgA serology (AUC 0.96 vs. 0.86) (Fig. 6 a). Apart from relatively easy and absolute detection techniques, EBV miRNAs are promising biomarkers for early NPC detection due to their biological role in NPC pathogenesis with a tight tissue‐, cell‐ and time‐specific expression. However there is currently no consensus on which circulating miRNA panel is most optimal for NPC population screening.17, 23, 26, 40 We confirmed that EBV miRNAs are promising markers in more‐invasive NP brushings or swabs, directly reflecting the local tumor load and disease process.9, 12, 28, 50, 51 Our results showed indeed that BARTs were both highly expressed in NP brushings and tumor biopsies (Figs. 4 a and 4b). Although repeated brush sampling is technically feasible, this is an invasive and bothersome procedure that may be used to follow up inconclusive circulating miRNA results to rule out false negatives. Our data suggest that the NP‐brushing procedure provides molecular evidence (miRNA and intact tumor derived EBV DNA) on the presence of actual NPC tumor cells at the site of tumor initiation and serves as a valid confirmation tool for NPC.12, 50, 51
Could the circulating EBV miRNA approach be improved and do they have a biological role as suggested from our prior findings?52 We determined by size‐exclusion chromatography that at least two different populations of circulating EBV miRNAs exist, extracellular vesicle EVs and ribonucleoprotein (RNP) complexes (Figs. 1, 2, 5). Recently BART2‐5p was identified as a potential early stage NPC serum marker,40 although BART2‐5p serum levels may also be elevated in non‐NPC diseases43 hence our focus on different EBV‐miRNAs. Recent evidence suggest that serum miRNAs in cancer patients can be both tumor‐cell derived or from stroma cells that are either EV‐bound and/or RNP‐associated and possibly functionally related to clinical response to therapy.27 Selective packaging of cell free miRNAs into vesicle carriers is probably related to the specific biological functions of the secreted miRNAs as we found previously for EBV‐miRNAs.52 Our findings show that BART13‐3p is strongly EV‐associated, possibly explaining its superior diagnostic performance over the other candidates. Curiously, candidate markers that were detected by RNAseq in the serum of NPC patients such as BART6‐3p and BART10‐3p were deemed less attractive when their secretion was analyzed from NPC cells in vitro (Fig. 2). Tailored design of primers and probes for BART6‐3p isomiRs for example could improve the detection of the secreted form of this miRNA.53
Despite the small sample size, we found that RNAseq‐based detection of circulating EBV‐miRNAs is feasible and discriminates NPC patients from controls. Although not all potential markers found by RNAseq could be subsequently validated by qRT‐PCR (probably due to the semi‐quantitative nature of this technique), technological improvements such as inclusion of unique‐molecular identifiers (UMIs) might increase accuracy. Nevertheless as group, EBV‐miRNAs are most selective for NPC detection when compared to endogenous serum miRNAs (Fig. 1). Despite technical limitations the qRT‐PCR results for EBV‐miRNAs in total serum of endemic (Indonesian NPC samples) are in strong agreement with an independent study performed by Zhang and colleagues who also found that BART7‐3p and BART13‐3p levels are significantly increased in NPC patients compared to healthy controls, that decrease upon treatment.23 In our study, we compared these markers and BART9‐3p with a relevant control group existing of noncancer patients with EBV associated diseases and non‐NPC head and neck malignancies, a crucial comparison that is needed to assess true NPC‐specificity (Table 1, Fig. 3). One important limitation of our study design is the relatively low numbers of early stage disease samples in our cohorts (Table 1) precluding firm predictions whether EBV miRNAs are superior over serology in detecting early stage NPC disease. In addition, we determined EBV‐DNA load in whole blood for comparison purposes, whereas a novel sequencing based technique of cell‐free EBV‐DNA plasma shows high sensitivity and specificity (AUC 0.93)16 similar to BART13‐3p with qRT‐PCR (AUC 0.96) found in our study.
Since the sensitivity and specificity for NPC detection as measured by ELISA in our cohorts, corresponded with prior published large‐scale studies,37 the outcome of our comparative analysis and prior studies,23, 40 warrants a large scale validation of EBV‐miRNAs in a prospective study that should include (EV‐bound) BART13‐3p, preferably in combination with cell‐free EBV‐DNA fragment detection.16 It is likely that a combined method will ultimately yield the desired sensitivity and specificity necessary for population‐based screening.
Supporting information
Figure S1. small RNAseq highlights high concordance between C666.1 NPC cells and exosomes
(A) and (B) EBV miRNA distribution in NPC C666.1 cells and exosomes. (A) Data are shown as percentage read counts. (B) IsomiRs with nucleotide additions (NTA‐A and NTA‐U) as described previously by Koppers‐Lalic et al.44 Data are shown as frequency distribution in percentage.
Figure S2A. EBV BART10‐3p levels are low in NPC sera compared to other BARTs
EBV BART miRNA expression level of (A) BART10‐3p in NPC patients (n = 21), Healthy Individuals (n = 20) and non‐NPC various disease controls (n = 26). Endogenous miRNA expression level of (B) miR16 and (C) vtRNA in NPC patients (n = 39), Healthy Individuals (n = 33) and non‐NPC various disease controls (n = 31). Data were presented as Ct values, errors bars represent from the average between each group. Significant differences were analyzed by Mann–Whitney test and the p‐values are indicated. Circles represent individual NPC patients, whereas healthy controls are triangles and non‐NPC controls are signified as squares.
Figure S2B. BART9‐3p does not distinguish stage I/II NPC from non‐NPC
Relative EBV miRNA expression levels of (A) BART7‐3p, (B) BART9‐3p and (C) BART13‐3p in NPC patients stage I/II (n = 8), NPC patients stage II/IV (n = 31) and non‐NPC various disease controls (n = 31). (D, E and F) Same groups as in (A, B and C), but showed expression levels of endogenous miRNAs (D) miR155), (E) miR16 and (F) vtRNA. Data were presented as Ct values, errors bars represent from the average between each group. Significant differences were analyzed by Mann–Whitney test and the p‐values are indicated above the plots. Circles represent individual NPC patients stage I/II, whereas NPC patients stage III/IV are triangles and non‐NPC controls are squares.
Suppl Figure 3C. EBV‐miRNA (Ct‐value) vs EBV DNA copy numbers in C666.1 cells
Correlation between EBV miRNA level (A) BART7‐3p, (B) BART9‐3p and (C) BART13‐3p and EBV DNA copy numbers in NPC C666.1 cells. Data of EBV miRNA levels were presented as Ct values and EBV DNA copy numbers were presented as a log2 scale.
Suppl Figure 4. vtRNA‐normalized BART miRNA levels differentiate NPC sera from controls
(A) Endogenous vtRNA expression level in NPC patients (n = 39), Healthy Individuals (n = 33) and non‐NPC various disease controls (n = 31). Data were presented as Ct values. Normalized EBV miRNA expression levels of (B) BART13‐3p, (C) BART7‐3p and (D) BART9‐3p in the same groups as in (A).
Data are presented as a log2 scale. Circles represent individual NPC patients, whereas healthy controls are triangles and non‐NPC controls squares.
Table S1. The tumor stage of NPC cohort
Table S2. A complete list of primers and probes for detection of EBV DNA, small noncoded RNA and microRNA used in the present study
Acknowledgements
We thank Prof. Dr. Ruud Brakenhoff and Arjen Brink at Dept. Head and Neck Surgery, VU University Medical Center, Amsterdam for serum samples from untreated patients with non‐NPC head and neck tumors.
Conflict of interest: DM Pegtel is CSO of ExBiome B.V. The other authors declared no potential conflicts of interest.
References
- 1. Chua MLK, Wee JTS, Hui EP, et al. Nasopharyngeal carcinoma. Lancet 2016;387:1012–24. [DOI] [PubMed] [Google Scholar]
- 2. Arnold M, Wildeman MA, Visser O, et al. Lower mortality from nasopharyngeal cancer in The Netherlands since 1970 with differential incidence trends in histopathology. Oral Oncol 2013;49:237–43. [DOI] [PubMed] [Google Scholar]
- 3. Wang Y, Zhang Y, Ma S. Racial differences in nasopharyngeal carcinoma in the United States. Cancer Epidemiol 2013;37:793–802. 10.1016/j.canep.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coghill AE, Hsu W‐L, Pfeiffer RM, et al. Epstein–Barr virus serology as a potential screening marker for nasopharyngeal carcinoma among high‐risk individuals from multiplex families in Taiwan. Cancer Epidemiol Biomarkers Prev 2014;23:1213 LP–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tao Q, Chan ATC. Nasopharyngeal carcinoma: molecular pathogenesis and therapeutic developments. Expert Rev Mol Med 2007;9:1–24. [DOI] [PubMed] [Google Scholar]
- 6. de Oliveira DE, Ballon G, Cesarman E. NF‐kappaB signaling modulation by EBV and KSHV. Trends Microbiol 2010. Jun;18:248–57. [DOI] [PubMed] [Google Scholar]
- 7. Young LS, Yap LF, Murray PG. Epstein–Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 2016;16:789–802. [DOI] [PubMed] [Google Scholar]
- 8. Hongxin F, John N, Daniel C, et al. Laboratory markers of tumor burden in nasopharyngeal carcinoma: a comparison of viral load and serologic tests for Epstein‐Barr virus. Int J Cancer 2004;112:1036–41. [DOI] [PubMed] [Google Scholar]
- 9. Stevens SJ, Verkuijlen SA, Hariwiyanto B, et al. Noninvasive diagnosis of nasopharyngeal carcinoma: nasopharyngeal brushings reveal high Epstein‐Barr virus DNA load and carcinoma‐specific viral BARF1 mRNA. Int J Cancer 2006;119:608–14. [DOI] [PubMed] [Google Scholar]
- 10. Stevens SJ, Vervoort MB, van den Brule AJ, et al. Monitoring of epstein‐barr virus DNA load in peripheral blood by quantitative competitive PCR. J Clin Microbiol 1999;37:2852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan KCA. Plasma Epstein‐Barr virus DNA as a biomarker for nasopharyngeal carcinoma. Chin J Cancer 2014;33:598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramayanti O, Juwana H, Verkuijlen SAMW, et al. Epstein‐Barr virus mRNA profiles and viral DNA methylation status in nasopharyngeal brushings from nasopharyngeal carcinoma patients reflect tumor origin. Int J Cancer 2017;140:149–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fachiroh J, Schouten T, Hariwiyanto B, et al. Molecular diversity of Epstein‐Barr virus IgG and IgA antibody responses in nasopharyngeal carcinoma: a comparison of Indonesian, Chinese, and European subjects. J Infect Dis 2004;190:53–62. [DOI] [PubMed] [Google Scholar]
- 14. Peng H, Guo R, Chen L, et al. Prognostic impact of plasma Epstein‐Barr virus DNA in patients with nasopharyngeal carcinoma treated using intensity‐modulated radiation therapy. Sci Rep 2016;6:22000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan KCA, Woo JKS, King A, et al. Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med 2017;377:513–22. [DOI] [PubMed] [Google Scholar]
- 16. Lam WKJ, Jiang P, Chan KCA, et al. Sequencing‐based counting and size profiling of plasma Epstein–Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc Natl Acad Sci USA 2018;115:E5115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012;13:358–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riley KJ, Rabinowitz GS, Yario TA, et al. EBV and human microRNAs co‐target oncogenic and apoptotic viral and human genes during latency. EMBO J 2012;31:2207 LP–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qiu J, Cosmopoulos K, Pegtel M, et al. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog 2011;7:e1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barth S, Meister G, Grässer FA. EBV‐encoded miRNAs. Biochim Biophys Acta ‐ Gene Regul Mech 2011;1809:631–40. [DOI] [PubMed] [Google Scholar]
- 21. Tycowski KT, Guo YE, Lee N, et al. Viral noncoding RNAs : more surprises. Genes Dev 2015;29:567–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feederle R, Linnstaedt SD, Bannert H, et al. A viral microRNA cluster strongly potentiates the transforming properties of a human Herpesvirus. PLoS Pathog 2011;7:e1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gaohong Z, Jingfeng Z, Shaojun L, et al. Circulating Epstein–Barr virus microRNAs miR‐BART7 and miR‐BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer 2014;136:E301–12. [DOI] [PubMed] [Google Scholar]
- 24. Gourzones C, Gelin A, Bombik I, et al. Extra‐cellular release and blood diffusion of BART viral micro‐RNAs produced by EBV‐infected nasopharyngeal carcinoma cells. Virol J 2010;7:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirai N, Wakisaka N, Kondo S, et al. Potential interest in circulating miR‐BART17‐5p as a post‐treatment biomarker for prediction of recurrence in Epstein‐Barr virus‐related nasopharyngeal carcinoma. PLoS One 2016;11:e0163609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang H‐Y, Yan L‐X, Shao Q, et al. Profiling plasma MicroRNA in nasopharyngeal carcinoma with deep sequencing. Clin Chem 2014;60:773 LP–782. [DOI] [PubMed] [Google Scholar]
- 27. Li H, Liu J, Chen J, et al. A serum microRNA signature predicts trastuzumab benefit in HER2‐positive metastatic breast cancer patients. Nat Commun 2018;9:1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stoker SD, Wildeman MA, Novalic Z, et al. Can Epstein‐Barr virus DNA load in nasopharyngeal brushings or whole blood predict recurrent nasopharyngeal carcinoma in a non‐endemic region? A prospective nationwide study of the Dutch head and neck oncology cooperative group. Eur Arch Oto‐Rhino‐Laryngol 2016;273:1557–67. [DOI] [PubMed] [Google Scholar]
- 29. Adham M, Kurniawan AN, Muhtadi AI, et al. Nasopharyngeal carcinoma in Indonesia: epidemiology, incidence, signs, and symptoms at presentation. Chin J Cancer 2012;31:185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Elisa P, Laura C, Dal ML, et al. Undifferentiated nasopharyngeal carcinoma from a nonendemic area: protective role of HLA allele products presenting conserved EBV epitopes. Int J Cancer 2009;125:1358–64. [DOI] [PubMed] [Google Scholar]
- 31. Hui KF, Cheung AKL, Choi CK, et al. Inhibition of class I histone deacetylases by romidepsin potently induces Epstein‐Barr virus lytic cycle and mediates enhanced cell death with ganciclovir. Int J Cancer 2016;138:125–36. [DOI] [PubMed] [Google Scholar]
- 32. Cosmopoulos K, Pegtel M, Hawkins J, et al. Comprehensive profiling of Epstein‐Barr virus MicroRNAs in nasopharyngeal carcinoma. J Virol 2008;83:2357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van Eijndhoven MAJ, Zijlstra JM, et al. Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients. JCI Insight 2016;1:e89631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Verweij FJFJ, van Eijndhoven MAJMAJ, Middeldorp J, et al. Analysis of viral microRNA exchange via exosomes in vitro and in vivo. Methods Mol Biol 2013;1024:53–68. [DOI] [PubMed] [Google Scholar]
- 35. Rueda A, Barturen G, Lebrón R, et al. sRNAtoolbox: an integrated collection of small RNA research tools. Nucleic Acids Res 2015;43:W467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baglio SRR, van Eijndhoven MAJMAJ, Koppers‐Lalic D, et al. Sensing of latent EBV infection through exosomal transfer of 5′pppRNA. Proc Natl Acad Sci USA 2016;2015:201518130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yue L, Qihong H, Wanli L, et al. Establishment of VCA and EBNA1 IgA‐based combination by enzyme‐linked immunosorbent assay as preferred screening method for nasopharyngeal carcinoma: a two‐stage design with a preliminary performance study and a mass screening in southern China. Int J Cancer 2011;131:406–16. [DOI] [PubMed] [Google Scholar]
- 38. Xu YP, C . Statistical modeling of MicroRNA expression with human cancers. J Biometrics Biostat 2015;6:240. [Google Scholar]
- 39. Chen SJ, Chen GH, Chen YH, et al. Characterization of Epstein‐Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One 2010;5:e12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen J, Jinna C, Shanghang X, et al. Evaluation of circulating EBV microRNA BART2‐5p in facilitating early detection and screening of nasopharyngeal carcinoma. Int J Cancer 2018;143:3209–17. [DOI] [PubMed] [Google Scholar]
- 41. Bergallo M, Gambarino S, Pinon M, et al. EBV‐encoded microRNAs profile evaluation in pediatric liver transplant recipients. J Clin Virol 2017;91:36–41. [DOI] [PubMed] [Google Scholar]
- 42. Kawano Y, Kawada J, Ito Y. Epstein‐Barr virus MicroRNAs in plasma as potential biomarkers for chronic infections: reply to Makarewicz et al. J Infect Dis 2014;209:1298–300. [DOI] [PubMed] [Google Scholar]
- 43. Yuki K, Kan K, Toshihiro N, et al. Circulating Epstein‐Barr virus–encoded micro‐RNAs as potential biomarkers for nasal natural killer/T‐cell lymphoma. Hematol Oncol 2016;35:655–63. [DOI] [PubMed] [Google Scholar]
- 44. Koppers‐Lalic D, Hackenberg M, Bijnsdorp IVIV, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 2014;8:1649–58. [DOI] [PubMed] [Google Scholar]
- 45. Pegtel DMM, Subramanian A, Meritt D, et al. IFN‐alpha‐stimulated genes and Epstein‐Barr virus gene expression distinguish WHO type II and III nasopharyngeal carcinomas. Cancer Res 2007;67:474–81. [DOI] [PubMed] [Google Scholar]
- 46. Boing AN, Van Der PE, Grootemaat AE, et al. Single‐step isolation of extracellular vesicles by size‐exclusion chromatography. J Extracell vesicles 2014;1:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cai X, Schafer A, Lu S, et al. Epstein‐Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 2006;2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwarzenbach H, Hoon DS, Pantel K. Cell‐free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426–37. [DOI] [PubMed] [Google Scholar]
- 49. Ming‐Fang J, Qi‐Hong H, Xia Y, et al. Evaluation of plasma Epstein‐Barr virus DNA load to distinguish nasopharyngeal carcinoma patients from healthy high‐risk populations in southern China. Cancer 2014;120:1353–60. [DOI] [PubMed] [Google Scholar]
- 50. Ng RHW, Ngan R, Wei WI, et al. Trans‐Oral brush biopsies and quantitative PCR for EBV DNA detection and screening of nasopharyngeal carcinoma. Otolaryngol Neck Surg 2014;150:602–9. [DOI] [PubMed] [Google Scholar]
- 51. Xiao‐Hui Z, Li‐Xia L, Xi‐Zhao L, et al. Quantification of Epstein–Barr virus DNA load in nasopharyngeal brushing samples in the diagnosis of nasopharyngeal carcinoma in southern China. Cancer Sci 2015;106:1196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pegtel DMM, Cosmopoulos K, Thorley‐Lawson D, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA 2010;107:6328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koppers‐Lalic D, Hackenberg M, Menezes RD, et al. Noninvasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget 2016;7:22566–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. small RNAseq highlights high concordance between C666.1 NPC cells and exosomes
(A) and (B) EBV miRNA distribution in NPC C666.1 cells and exosomes. (A) Data are shown as percentage read counts. (B) IsomiRs with nucleotide additions (NTA‐A and NTA‐U) as described previously by Koppers‐Lalic et al.44 Data are shown as frequency distribution in percentage.
Figure S2A. EBV BART10‐3p levels are low in NPC sera compared to other BARTs
EBV BART miRNA expression level of (A) BART10‐3p in NPC patients (n = 21), Healthy Individuals (n = 20) and non‐NPC various disease controls (n = 26). Endogenous miRNA expression level of (B) miR16 and (C) vtRNA in NPC patients (n = 39), Healthy Individuals (n = 33) and non‐NPC various disease controls (n = 31). Data were presented as Ct values, errors bars represent from the average between each group. Significant differences were analyzed by Mann–Whitney test and the p‐values are indicated. Circles represent individual NPC patients, whereas healthy controls are triangles and non‐NPC controls are signified as squares.
Figure S2B. BART9‐3p does not distinguish stage I/II NPC from non‐NPC
Relative EBV miRNA expression levels of (A) BART7‐3p, (B) BART9‐3p and (C) BART13‐3p in NPC patients stage I/II (n = 8), NPC patients stage II/IV (n = 31) and non‐NPC various disease controls (n = 31). (D, E and F) Same groups as in (A, B and C), but showed expression levels of endogenous miRNAs (D) miR155), (E) miR16 and (F) vtRNA. Data were presented as Ct values, errors bars represent from the average between each group. Significant differences were analyzed by Mann–Whitney test and the p‐values are indicated above the plots. Circles represent individual NPC patients stage I/II, whereas NPC patients stage III/IV are triangles and non‐NPC controls are squares.
Suppl Figure 3C. EBV‐miRNA (Ct‐value) vs EBV DNA copy numbers in C666.1 cells
Correlation between EBV miRNA level (A) BART7‐3p, (B) BART9‐3p and (C) BART13‐3p and EBV DNA copy numbers in NPC C666.1 cells. Data of EBV miRNA levels were presented as Ct values and EBV DNA copy numbers were presented as a log2 scale.
Suppl Figure 4. vtRNA‐normalized BART miRNA levels differentiate NPC sera from controls
(A) Endogenous vtRNA expression level in NPC patients (n = 39), Healthy Individuals (n = 33) and non‐NPC various disease controls (n = 31). Data were presented as Ct values. Normalized EBV miRNA expression levels of (B) BART13‐3p, (C) BART7‐3p and (D) BART9‐3p in the same groups as in (A).
Data are presented as a log2 scale. Circles represent individual NPC patients, whereas healthy controls are triangles and non‐NPC controls squares.
Table S1. The tumor stage of NPC cohort
Table S2. A complete list of primers and probes for detection of EBV DNA, small noncoded RNA and microRNA used in the present study


