Summary
Polyploidy, or whole‐genome duplication often with hybridization, is common in eukaryotes and is thought to drive ecological and evolutionary success, especially in plants. The mechanisms of polyploid success in ecologically relevant contexts, however, remain largely unknown.
We conducted an extensive test of functional trait divergence and plasticity in conferring polyploid fitness advantage in heterogeneous environments, by growing clonal replicates of a worldwide genotype collection of six allopolyploid and five diploid wild strawberry (Fragaria) taxa in three climatically different common gardens.
Among leaf functional traits, we detected divergence in trait means but not plasticities between polyploids and diploids, suggesting that increased genomic redundancy in polyploids does not necessarily translate into greater trait plasticity in response to environmental change. Across the heterogeneous garden environments, however, polyploids exhibited fitness advantage, which was conferred by both trait means and adaptive trait plasticities, supporting a ‘jack‐and‐master’ hypothesis for polyploids.
Our findings elucidate essential ecological mechanisms underlying polyploid adaptation to heterogeneous environments, and provide an important insight into the prevalence and persistence of polyploid plants.
Keywords: adaptation, adaptive plasticity, common gardens, functional traits, polyploidy, wild strawberry
Introduction
Polyploidy (or whole‐genome duplication often with hybridization) results in heritable occurrence of more than two sets of chromosomes of the same (autopolyploidy) or disparate origins (allopolyploidy), which enlarges and diversifies an organism's genome with profound influence on phenotype and fitness (Otto & Whitton, 2000; Ramsey & Ramsey, 2014; Soltis et al., 2016). While polyploidy is common in eukaryotic lineages, some of the best‐known examples of polyploids in flowering plants include important crops (Salman‐Minkov et al., 2016) and many invasive species (te Beest et al., 2012), and the repeated and pervasive occurrence of polyploidy throughout the plant kingdom reflects its widespread adaptive significance (Van de Peer et al., 2017). Despite its evolutionary importance, the mechanisms of polyploid advantage in ecological contexts are largely unknown. A leading, yet rarely tested, hypothesis is that polyploid fitness advantage arises from altered phenotype (i.e. functional trait divergence from diploids) and/or enhanced ability to adjust phenotype (i.e. functional trait plasticity) in response to environmental change (Levin, 1983; Van de Peer et al., 2017).
Polyploidy can alter plant phenotype (Levin, 1983; Soltis et al., 2014). Phenotypic variation at the cellular level (e.g. an increase in cell size) as a result of an increase in ploidy was first recognized in early cytological studies of synthetic polyploids (reviewed in Ramsey & Ramsey, 2014). This positive correlation between genome size and cell size holds across angiosperm lineages (Masterson, 1994; Beaulieu et al., 2008), whereas for phenotype at higher levels (e.g. tissues or organs), the nucleotypic effects of genome size are shown to be weaker or absent (Knight & Beaulieu, 2008). In addition to the genome size effect, polyploidy can also diversify a plant genome by incorporating multiple copies of genes from the same or different species, which can have important implications for phenotype (Chen, 2010; Soltis et al., 2014). Comparisons of functional trait divergence between diploids and naturally occurring polyploids, in ecologically relevant contexts, have been primarily conducted in autopolyploids with intraspecific ploidal variation (Ramsey & Ramsey, 2014), and have yielded mixed, and often species‐specific, conclusions (e.g. Li et al., 1996, 2012; Maherali et al., 2009; Balao et al., 2011; Hao et al., 2013). The phenotypic consequences on functional traits of allopolyploidy – which generates diverse genetic backgrounds and the potential to express transgressive phenotypes relative to autopolyploidy (Chen, 2010) – remains unclear for the vast majority of wild allopolyploid taxa that account for half of the extant polyploids (Barker et al., 2016), with few exceptions (Buggs & Pannell, 2007; Hahn et al., 2012; Manzaneda et al., 2015; Leal‐Bertioli et al., 2017).
Polyploidy has the potential to alter functional trait plasticity (referred to as trait plasticity hereafter), owing to genomic redundancy and versatility in gene expression (Stebbins, 1971; Adams & Wendel, 2005; Leitch & Leitch, 2008; Jackson & Chen, 2010; Madlung & Wendel, 2013). Relative to diploids, polyploids can potentially employ alternative copies of duplicated genes gained from diverse and possibly adaptive genetic backgrounds to respond to novel environments (Bardil et al., 2011; Dong & Adams, 2011; Shimizu‐Inatsugi et al., 2017). Thus, it is hypothesized that polyploids can exhibit higher trait plasticity than diploids in response to varying environment. Previous work has primarily emphasized gene expression changes of polyploidy (Soltis et al., 2016), and, as a result, the questions of whether genome duplication translates into increased trait plasticity in the wild (Madlung, 2013), and how trait plasticity differs between diploids and polyploids (Buggs & Pannell, 2007; Hahn et al., 2012; Manzaneda et al., 2015), persist.
Polyploidy has been demonstrated to provide selective advantages to plants under environmental stresses and instabilities (Chao et al., 2013; Yang et al., 2014; Van de Peer et al., 2017). However, it remains controversial whether such polyploid fitness advantage occurs only in a particular environment or can be maintained consistently across environments (Ramsey, 2011; Madlung, 2013). Several competing adaptive hypotheses, based on the fitness reaction norm extended from theories of invasion (Richards et al., 2006), have been proposed. First, elevated genetic heterozygosity and gene expression versatility may enable polyploids to occupy broader ecological niches (i.e. higher ecological amplitude) than diploids. As a result of possessing such ‘general purpose’ genotypes (Baker, 1965; Stebbins, 1971), polyploids could exhibit high fitness and fitness homoeostasis (i.e. constant fitness) in heterogeneous environments (manifesting as high intercept and low slope in a fitness reaction norm; ‘jack‐of‐all‐trades’) (Richards et al., 2006). Alternatively, in the absence of fitness homoeostasis, polyploids may still maintain higher fitness than diploids across a broad range of environments. This fitness strategy (manifesting as high intercept and high slope) can be referred to as ‘jack‐and‐master’ (Richards et al., 2006). Lastly, polyploids and diploids may both be habitat specialists, exhibiting high fitness in alternative environments (i.e. ‘master‐of‐some’). While these adaptive hypotheses have been tested among invasive and native plant species (e.g. Richards et al., 2006; Davidson et al., 2011), tests with respect to polyploidy are not only limited to a few intra‐ and interspecific systems between diploids (2n = 2x) and mostly tetraploids (2n = 4x) (Petit & Thompson, 1997; Bretagnolle & Thompson, 2001; McIntyre & Strauss, 2017), but more importantly these lack the mechanisms that connect the fitness of diploids and polyploids to functional traits and trait plasticity.
In this study, we take advantage of the fact that polyploidy is an important mode of speciation in wild strawberries (Fragaria L.), a genus that originated around 3–8 million yr ago (Liston et al., 2014; Qiao et al., 2016) and has 22 extant species with a broad distribution in the northern hemisphere (Staudt, 1999; Liston et al., 2014). While Fragaria has two centers of species diversification (in East Asia and Europe–North America; Liston et al., 2014), we focused on diploid and polyploid Fragaria that occur in North America, South America, Europe and Northeast Asia (Fig. 1), among which repeated and independent events of allopolyploid speciation (Fig. 1) have been revealed by polyploid Fragaria genomes (Tennessen et al., 2014; Kamneva et al., 2017; Wei et al., 2017a,b; Dillenberger et al., 2018).
Figure 1.
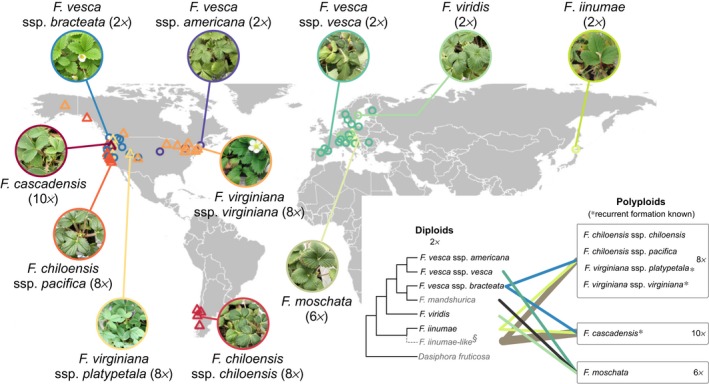
Seventy‐two source populations of diploid (circles) and polyploid (triangles) Fragaria, and their reticulate evolutionary histories (inset). The inset dendrogram represents the known evolutionary relationships among the five diploid (2x) taxa in this study (black), as well as those not in this study (grey; F. mandshurica, and an extinct F. iinumae‐like diploid§ with dashed branch), along with an outgroup taxon (Dasiphora). Among the six polyploids, the octoploid (8x) taxa are derived from the 2x F. vesca ssp. bracteata, F. iinumae and the extinct F. iinumae‐like diploid (each contributing, respectively, two, two and four sets of chromosomes to the 8x genomes, reflected by line width) (Tennessen et al., 2014; Wei et al., 2017a). The 10x F. cascadensis has two sets of chromosomes from F. vesca ssp. bracteata, two sets from F. iinumae and six sets from the F. iinumae‐like diploid (Wei et al., 2017b). The 6x F. moschata is derived from F. vesca ssp. vesca, F. viridis and F. mandshurica (Kamneva et al., 2017). Recurrent formation* of the same polyploid taxon in different populations has been previously identified (Dillenberger et al., 2018), whereas such information remains unclear for the remaining polyploid taxa.
By growing clonal replicates of a worldwide collection of Fragaria genotypes of five diploid and six allopolyploid taxa (2n = 6x–10x, primarily 8x; Fig. 1; Supporting Information Table S1) in three climatically different common gardens in Oregon, USA, we addressed the following questions: do functional traits differ between diploids and polyploids; do polyploids demonstrate higher trait plasticity than diploids in response to environmental change; is there a polyploid fitness advantage across diverse garden environments; and, if so, is the polyploid fitness advantage conferred by trait means or trait plasticities, or both?
Materials and Methods
Study system
Fragaria are perennial herbaceous plants that reproduce both sexually by seed and asexually by plantlets on stolons (Staudt, 1999). The six allopolyploid strawberries studied here are hexaploid (6x) F. moschata, octoploid (8x) F. chiloensis ssp. pacifica, F. chiloensis ssp. chiloensis, F. virginiana ssp. platypetala, F. virginiana ssp. virginiana, and decaploid (10x) F. cascadensis. The five diploid strawberries are F. vesca ssp. bracteata, F. vesca ssp. americana, F. vesca ssp. vesca, F. viridis, and F. iinumae. We defined ploidy level broadly as diploid or polyploid, owing to distinct separation between diploids and high‐order polyploids (2n ≥ 6x; Fig. S1), and the dominance of the 8x taxa and genotypes within high‐order polyploids (Table S1). Among these polyploids, the 10x and 8x taxa are derived from the 2x F. vesca ssp. bracteata, F. iinumae and an extinct F. iinumae‐like 2x taxon (Fig. 1; Tennessen et al., 2014; Wei et al., 2017a,b), and the 6x is derived from the 2x F. viridis and F. vesca ssp. vesca as well as the Asian 2x F. mandshurica (Fig. 1; Kamneva et al., 2017). Recurrent formation, representing multiple independent origins, has been observed in the 10x and 8x taxa (Dillenberger et al., 2018). The worldwide collection of Fragaria was conducted as an international collaborative effort in 2013–2014; details are available on our Wild Strawberry website (http://wildstrawberry.org/; see also Fig. S2).
Genotype and clone cultivation
In April 2015, we germinated and grew four genotypes (i.e. each from a single, open‐pollinated seed of a distinct wild plant) from each of 72 total populations across the 11 taxa (10 populations of less than four genotypes; Table S1), in a glasshouse at the University of Pittsburgh following standard protocols (Wei et al., 2017b). In September 2015, we harvested 12 plantlets (clones) from stolons of each of the 269 genotypes (24 genotypes of < 12 clones; Table S1). These clones (N = 3137) were sent to Oregon State University, kept in the dark at 16°C for 1 wk to stimulate root growth, and then transplanted to 107 cm2 conetainers (Stuewe & Sons Inc., Tangent, OR, USA) filled with Sunshine Mix #4 soil (Sun Gro Horticulture, Agawam, MA, USA). Plantlets were grown at 18°C under natural lighting in a glasshouse for 3 wk, and moved outside for 1 wk before transplanting in common gardens during autumn (28 October to 15 November 2015). At transplanting, clones had one to two leaves, most of which senesced over winter.
Common gardens
Three common gardens were located in Oregon, USA (Fig. 2a): cool/coastal ‘Newport’ (44.62046°N, 124.04410°W; altitude, 5 m), temperate/valley ‘Corvallis’ (44.56107°N, 123.28911°W; 70 m) and arid/montane ‘Bend’ (44.08895°N, 121.26192°W; 1063 m), each differing in temperature, precipitation and soil (Fig. 2c; Table S2). At each location, we established four raised wooden beds (18 × 1.5 m; Fig. 2b), filled with soil derived from local sources (Methods S1; Table S2).
Figure 2.
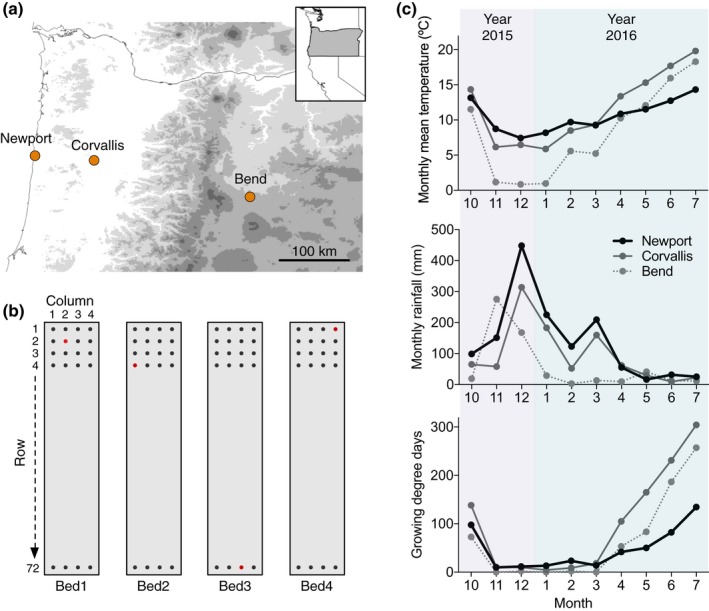
The location, design and climate of common gardens. (a) Three common gardens were located in Oregon, USA, including the coastal garden at Newport, the valley garden at Corvallis and the montane garden at Bend. (b) We established four raised wooden beds at each garden location. Each bed (18 × 1.5 m) can host 72 × 4 plants, indicated by the dots. For each genotype, the four clones (red dots) were assigned to the four beds, and the position within each bed was chosen randomly. (c) The monthly mean temperature, rainfall and growing degree days (i.e. the cumulative heat > 10°C) were obtained (see Supporting Information Methods S1) for the three common gardens, during the course of the field experiment from October 2015 to mid‐July 2016.
Plants were arranged in a complete randomized block design with c. 25 cm spacing, and one clone per genotype was randomly assigned a position in one of the four beds (blocks) at each garden location (Fig. 2b). For the 24 genotypes with < 12 clones, we distributed available clones evenly across garden locations, but within each garden we prioritized filling beds 1 and 2 to have at least two complete blocks each location. Empty positions (N = 319) were filled with nonexperimental clones, which were cultivated in the same manner as the others, to maintain even plant spacing and density. Throughout the course of the experiment (October 2015–July 2016), plants received only natural precipitation at Newport and Corvallis, which reached a total of 138.5 and 95.5 cm, respectively (Fig. 2c); however, at Bend (natural precipitation of 58.2 cm), plants were given supplemental water totaling 14.2 cm during the months of near‐zero rainfall (February–April 2016; Fig. 2c). All beds were protected from large herbivores using polypropylene mesh (1.6 × 1.6 cm) netting. Beds at Bend received straw cover (November 2015–February 2016) to minimize winter freeze damage to plant crowns.
Given the amelioration of freezing and drought stress at Bend, we considered Corvallis the most favorable environment, and Newport the least favorable based on growing degree days (i.e. the cumulative heat > 10°C; Fig. 2c).
Functional traits and fitness components
We assessed a suite of leaf functional traits that capture essential plant ecophysiological processes (Table 1) in May 2016 on experimental plants in beds 1 and 2 of each garden (N = 1429). We counted the number of leaves of individual plants, and collected the largest, fully expanded leaf over a 14 d period for all selected beds to measure leaf area and seven functional traits as described in Methods S1. Among these traits, vein density and trichome density were measured only at Corvallis and Bend (N = 950), as the collected leaves from plants at Newport were too small for these additional measurements (see Methods S1). Leaf nitrogen content and carbon isotope discrimination were obtained for a subset of randomly chosen genotypes per population at individual gardens (N = 210).
Table 1.
Key variables of the common garden experiment
| Description (unit) | Functiona | |
|---|---|---|
| Leaf functional traits | ||
| Specific leaf area (SLA) | Light‐capturing leaf area per unit dry mass (mm2 mg−1) | SLA reflects the thickness and/or dry mass content of leaf tissue. High SLA permits high leaf carbon gain. |
| Stomatal density (SD) | Abaxial stomata per unit leaf area (mm−2) | SD regulates CO2 intake and water transpiration, reflecting the tradeoff between gas conductance and epidermal construction cost. |
| Stomatal length (SL) | Abaxial guard cell length (μm) | SL regulates CO2 intake and water transpiration. SL correlates negatively with SD. |
| Vein density (VLA) | Total minor vein lengths per unit leaf area (mm mm−2) | VLA supports leaf hydraulic conductance. Low VLA, however, reduces construction cost. |
| Trichome density (TD) | Total abaxial and adaxial trichomes per unit leaf area (mm−2) | TD influences the ability of plants to prevent water loss. |
| Nitrogen content (N mass) | Leaf nitrogen per unit dry mass (%) | N mass, required for photosynthetic proteins, supports leaf photosynthetic potential. |
| Carbon isotope discrimination (Δ13C) | Amount of isotope discrimination against 13C relative to 12C during photosynthesis (‰) | Δ13C reflects photosynthetic water‐use efficiency, integrated over the life span of a leaf. Low Δ13C indicates high water‐use efficiency. |
| Plant fitness components | ||
| Survival | Presence (1) or absence (0) of a plant | It is used to estimate genotypic survival rate here. |
| Plant size | Leaf number × leaf area (dm2) | It reflects plant growth since transplanting. |
| Stolon mass | Dry mass of stolons (g) | It reflects asexual reproduction. |
We scored plant survival in May 2016 on plants in all four beds each garden. For plants in beds 1 and 2 of each garden, we estimated plant size as the product of leaf number and the area of the largest leaf (Table 1). For reproduction, as most plants did not flower in 2016, we focused on asexual reproduction (stolon mass). All experimental plants survived to the time (7–14 July 2016) when we harvested stolons, which were dried at 65°C for 1 wk before weighing.
Climatic niche distance
Plant functional traits and fitness can be influenced by climatic differences between source populations and experimental gardens (Rehfeldt et al., 1999), or the ‘climatic niche distance’ (CND). To estimate CND, we extracted the 19 bioclimatic variables (current conditions, 1970–2000) at 30 arcsec resolution (or 2.5 arcmin resolution for west coast populations of North America), as well as altitude estimates, from worldclim v2.0 (Fick & Hijmans, 2017) for the 72 source populations and the three garden locations. We conducted a principal component analysis (PCA) of these 20 variables using prcomp in R v3.3.3 (R Core Team, 2017). The first five PCs, accounting for 94.2% of the variation (Fig. S3), were used to calculate the Euclidean CND between each source population and each garden using the R package pdist (Wong, 2013). Owing to the lack of soil data from source Fragaria populations, our estimates of CND did not include soil variables.
General linear mixed models
We addressed each of the four questions in the Introduction using linear mixed models (LMMs) with the package lme4 (Bates et al., 2015). While the response variables and predictors (fixed effects) of LMMs were specific to each question, all LMMs accounted for evolutionary dependence among populations and taxa using nested random effects (i.e. populations nested in taxa and taxa in ploidy level, ploidy/taxon/population), which outperformed phylogenetic LMMs based on Fragaria plastid tree (Fig. S4) that simplifies the reticulate relationships (Fig. 1) between diploid and polyploid taxa (see Methods S1; Figs S5, S6). Moreover, for all LMMs, response variables were power‐transformed using the Box–Cox method in the package car (Fox & Weisberg, 2011) to improve normality, and the absence of multicollinearity among predictors was confirmed using the variance inflation factor. We evaluated the statistical significance (by type III sums of squares) of predictors and their least‐squares means in LMMs using the package lmertest (Kuznetsova et al., 2017). The variance explained by predictors and random effects of LMMs was assessed using the package mumin (Bartoń, 2017).
To evaluate whether diploids and polyploids differ in functional traits (the first question), the response variables of LMMs considered genotypic values of each functional trait (i.e. the average of two clones) at each garden. The predictors included central leaflet width + CND + garden + ploidy + ploidy : garden + ploidy : CND. We incorporated central leaflet widths, which were similar among diploid and polyploid taxa (Fig. S7), to account for functional trait variation potentially attributable to leaf characteristics (e.g. expansion, vigor), despite using a standardized leaf collection protocol as described in the section ‘Functional traits and fitness components’.
To evaluate whether polyploids express higher trait plasticity than diploids (the second question), we estimated plasticity for each trait and genotype using relative distance plasticity index (RDPI) and phenotypic plasticity index (PI; Valladares et al., 2006). For traits that were only measured at two gardens (vein density and trichome density), plasticity was calculated as trait distance (in absolute value) of the same genotype between the two environments, divided by the mean (for RDPI) or by the maximum (for PI) of the two genotypic trait values. For traits measured at all three gardens, RDPI and PI were calculated as the mean of the three pairwise distances. The response variables of LMMs considered genotypic plasticity of each trait, and the predictors included CND mean (i.e. genotypic CND averaged across gardens) + ploidy + ploidy : CND mean.
To evaluate whether polyploids have higher fitness than diploids (the third question), we estimated genotypic fitness of these perennial plants at each garden using a composite fitness index as the multiplicative product of genotypic survival rate, growth (plant size) and asexual reproduction (stolon mass). The genotypic survival rate was calculated as the proportion of clones that survived to May 2016 in all four beds per garden. The genotypic plant size and stolon mass were the average of the two clones measured per garden. As many plants produced zero stolons at Newport, we adjusted the genotypic stolon mass at each garden by adding 0.01 g. Our estimate of fitness represented a relatively equal contribution from each of the three components, given their similar scales across gardens (Fig. S8). Fitness (with the power transformation parameter λ = 0.1) was taken as the response variable, and the predictors of the LMM included CND + garden + ploidy + ploidy : garden + ploidy : CND.
To determine whether plant fitness over all garden environments is associated with trait means or plasticities (the fourth question), we used genotypic average fitness (across gardens) as the response variable (power transformation, λ = 0.1) in LMMs. The predictors of each LMM included CND mean + trait plasticity + trait mean (i.e. genotypic trait averaged across gardens) + ploidy : trait plasticity + ploidy : trait mean, for each functional trait. Correlations between trait plasticity and trait mean were weak (Table S3). LMMs with trait plasticities of RDPI and PI were performed separately, but as they yielded similar patterns, we only reported the results based on RDPI. To compare the magnitude of the respective effects of trait mean and trait plasticity on average fitness and to assess how they differ between ploidy levels, we estimated the effect sizes (i.e. standardized coefficients, β′) of the predictors using the package sjplot (Lüdecke, 2017).
Results
Do functional traits differ between diploids and polyploids?
Diploid and polyploid Fragaria differed in most leaf functional traits (Fig. 3; Table S4), either consistently across environments (e.g. stomatal length and vein density) or in certain environments (e.g. specific leaf area, stomatal density and nitrogen content).
Figure 3.
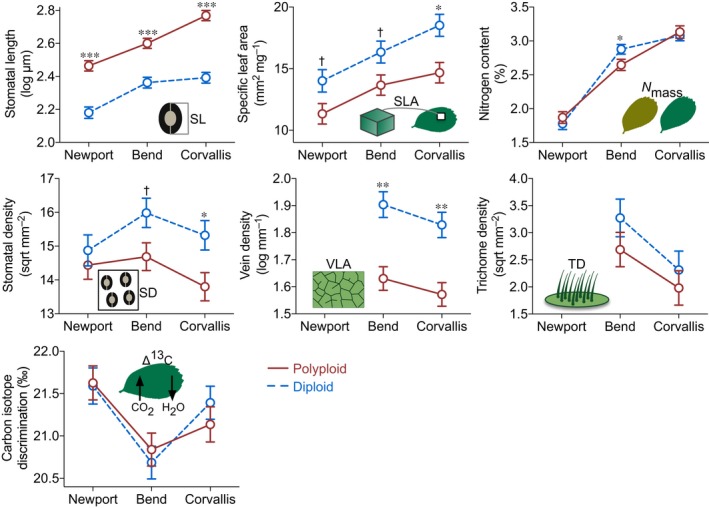
Diploid and polyploid Fragaria differ in leaf functional traits. The least‐squares mean ±1 SE of each trait are plotted for diploids (dashed lines) and polyploids (solid lines) at each garden location, estimated from general linear mixed models where the response variables were power‐transformed if necessary (see the Materials and Methods section; sqrt, square root; log, natural logarithm). The x‐axis is arranged from the least favorable, cool/coastal garden at Newport to the most favorable, temperate/valley garden at Corvallis. SL, stomatal length; SLA, specific leaf area; N mass, nitrogen content; SD, stomatal density; VLA, vein density; TD, trichome density; Δ13C, carbon isotope discrimination. VLA and TD were not available for plants at Newport. Significance levels: ***, P < 0.001; **, P < 0.01; *, P < 0.05; †, P = 0.053.
Polyploids possessed larger stomata (gauged by stomatal length; Table 1) than diploids in all environments (F = 37.56, df = 1, P < 0.001; Table S4). In the favorable environment at Corvallis, polyploids produced not only larger stomata (post hoc contrast of least‐squares means (LS means), t = 8.48, P < 0.001), but also fewer stomata per unit leaf area (t = −2.57, P = 0.028) than diploids, which may lower the epidermal construction cost of stomata per unit area for gas exchange (de Boer et al., 2016) in polyploids. The general tradeoff between stomatal length and density seen across vascular plants (Franks & Beerling, 2009) was, nevertheless, decoupled in the stressful environment at Newport (Fig. 3); reduced stomatal length was not accompanied by increased stomatal density, for polyploids and especially diploids, which could negatively affect photosynthetic potential (Tanaka et al., 2013). Polyploids and diploids also differed in leaf vein density across environments (F = 4.74, df = 1, P = 0.037; Fig. 3; Table S4), with polyploids producing lower minor vein length per unit leaf area (i.e. lower hydraulic construction cost; Sack & Scoffoni, 2013).
Although the main effect of ploidy level across gardens did not influence specific leaf area (F = 3.10, df = 1, P = 0.100; Table S4) and nitrogen content (F = 0.57, df = 1, P = 0.453), polyploids produced foliage with significantly smaller specific leaf area compared with diploids at Corvallis (LS means contrast, t = −3.18, P = 0.011), and significantly lower nitrogen content at Bend (t = −2.12, P = 0.040). By contrast, polyploids and diploids were similar in leaf traits that influence water loss (trichome density, F = 0.96, df = 1, P = 0.346) and water‐use efficiency (carbon isotope discrimination, F = 1.46, df = 1, P = 0.233) in all environments (Fig. 3; Table S4).
Do polyploids demonstrate higher trait plasticity than diploids in response to environmental change?
Fragaria genotypes expressed plasticity for the measured traits in response to different environments (Fig. 3), as demonstrated by the significant main effect of garden on each trait (all P < 0.001; Table S4). Quantifying plasticity using RDPI and PI yielded similar patterns in degrees of plasticity among traits: carbon isotope discrimination had the lowest plasticity (mean RDPI = 0.02; PI = 0.05); stomatal length, stomatal density, specific leaf area and vein density exhibited fivefold higher plasticity (RDPI = 0.10, 0.13, 0.12 and 0.10, respectively; PI = 0.18, 0.26, 0.21 and 0.22, respectively); nitrogen content and trichome density had the highest (10‐fold) plasticity (RDPI = 0.21 and 0.36, and PI = 0.32 and 0.61, respectively). Yet, plasticity of these traits showed low correlation (Table S5), suggesting limited plasticity integration (Pigliucci, 2003) among traits. LMMs indicated that polyploids and diploids had similar degrees of plasticity for all seven traits (all P > 0.05 for RDPI and PI; Table S6).
Is there a polyploid fitness advantage across diverse garden environments?
The main effect of ploidy level influenced plant fitness (F = 20.02, df = 1, P < 0.001; Fig. 4; Table S7), after accounting for the significant negative effect of climatic niche distance (F = 71.54, df = 1, P < 0.001), as revealed by the LMM (R 2 of fixed effects = 0.61; Table S7). Polyploids had significantly higher fitness than diploids at Corvallis (LS means contrast, t = 3.86, P = 0.002) and Bend (t = 3.02, P = 0.011), and marginally higher at Newport (t = 1.97, P = 0.072), a pattern that refutes the ‘master‐of‐some’ strategy for polyploids or diploids. Fitness changed dramatically for both polyploids and diploids across the three gardens (garden effect, F = 563, df = 2, P < 0.001; Table S7), contradicting fitness homoeostasis of the ‘jack‐of‐all‐trades’ hypothesis but instead supporting the ‘jack‐and‐master’ hypothesis for polyploids.
Figure 4.
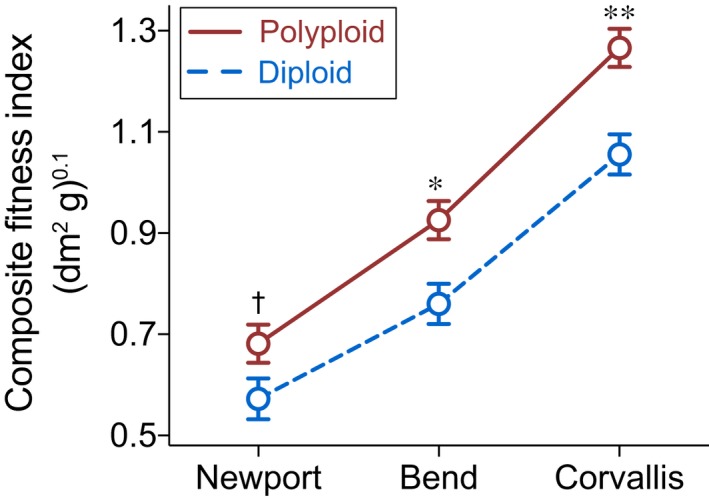
Polyploid Fragaria exhibit higher fitness compared with diploids. The composite fitness index, which was the product of genotypic survival rate, growth (plant size) and asexual reproduction (stolon mass), was transformed (with a power parameter λ = 0.1) in the general linear mixed model. The least‐squares means ±1 SE are plotted for diploids (dashed line) and polyploids (solid line) at each garden location. Significance levels: **, P < 0.01; *, P < 0.05; †, P = 0.072.
Is the polyploid fitness advantage associated with trait means or trait plasticities?
For both diploids and polyploids, average fitness was influenced by mean values for four of the seven functional traits (i.e. stomatal length, specific leaf area, vein density and trichome density; grey colour, Fig. 5a; Table S8), with the magnitude of effect sizes often differing between ploidy levels (Fig. 5a). The trait mean of stomatal length had a significant positive effect on average fitness (Fig. 5a), indicating that plants having larger stomata were associated with higher fitness, and the magnitude of this positive effect was similar between polyploids (β′ = 0.28, P < 0.001) and diploids (β′ = 0.28, P < 0.01). The trait mean of specific leaf area also positively influenced average fitness (Fig. 5a), but the magnitude was stronger in polyploids (β′ = 0.27, P = 0.015) than in diploids (β′ = 0.16, P = 0.074). While plants producing foliage of higher vein density and trichome density were associated with lower fitness (Fig. 5a), these negative effects were especially strong in diploids (β′ = −0.25, P < 0.001, and β′ = −0.30, P < 0.001, respectively) relative to polyploids (β′ = −0.08, P = 0.40, and β′ = −0.10, P = 0.42, respectively).
Figure 5.
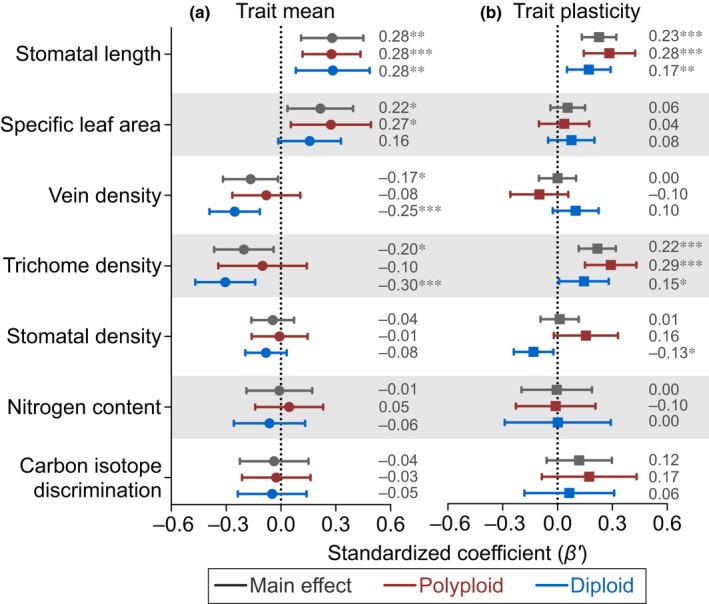
Trait means (a) and trait plasticities (b) predict average fitness of diploids and polyploids across the heterogeneous garden environments. Standardized regression coefficients (β′) of the main (grey) and ploidy‐specific (red, polyploid; blue, diploid) effects of trait means and plasticities on fitness are presented from general linear mixed models fitted separately for each functional trait. The average estimates of standardized coefficients are denoted by the symbols and the values to the right of the error bars (95% confidence intervals). Significance levels: ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Trait plasticities had significant positive effects on average fitness for only two of the seven traits (i.e. stomatal length and trichome density; Fig. 5b; Table S8), and the effect sizes for these adaptive plasticities were nearly twofold higher in polyploids (β′ = 0.28 and 0.29, respectively) than in diploids (0.17 and 0.15, respectively). It is also notable that plasticity in stomatal density was maladaptive for diploids (β′ = −0.13, P = 0.019) but marginally adaptive for polyploids (β′ = 0.16, P = 0.084), despite the overall neutral effect on average fitness (Fig. 5b).
Discussion
Using a worldwide genotype collection of Fragaria grown in three different climatic regions (cool/coastal, temperate/valley, arid/montane), we derive important insights into the mechanisms underlying polyploid adaptation to heterogeneous environments. Our results indicate divergence between allopolyploids and diploids in several leaf functional traits. Although different functional traits display varying degrees of plasticity, trait plasticity is of similar magnitude between diploids and allopolyploids, suggesting that increased genomic redundancy does not necessarily translate into greater trait plasticity in polyploids, as is often predicted (Stebbins, 1971; Levin, 1983). More importantly, this is the first study, to our knowledge, to explicitly link functional traits and plasticity to fitness differences between wild polyploids and diploids in natural environments. We find that both trait mean and trait plasticity contribute to higher allopolyploid fitness, and provide support for the ‘jack‐and‐master’ hypothesis for allopolyploid advantage over diploids in the genus Fragaria.
Similar trait plasticity between diploids and polyploids
Our findings of similar trait plasticity between diploids and allopolyploids contradict the long‐held idea (Stebbins, 1971; Levin, 1983) that greater trait plasticity in polyploids enables them to occupy broader ecological niches by expressing suitable phenotypes across a wider range of environments than is the case with diploids. Despite rich theory (Ramsey & Ramsey, 2014), there have been few empirical evaluations of trait plasticity and polyploidy, and none as extensive as our study in terms of the geographic, genetic and phylogenetic diversity of the source material. A glasshouse experiment of trait plasticity in response to water variation (Manzaneda et al., 2015) revealed similar plasticity between annual allotetraploid Brachypodium hybridum and its diploid progenitor B. distachyon in stomatal conductance and carbon isotope discrimination, although the diploid exhibited higher plasticity in photosynthetic rate. In response to nutrient variation (Sánchez Vilas & Pannell, 2017), similar plasticity in specific leaf area was found in glasshouse conditions between autotetraploid and allohexaploid cytotypes of the annual Mercurialis annua. For perennial allotetraploid and diploid Centaurea stoebe (Hahn et al., 2012), similar plasticity between ploidy levels was observed in all measured functional traits in response to water and nutrient variation in garden settings, and only a few traits exhibited higher plasticity in the polyploid in response to garden sites for one of two measuring occasions. These case studies, along with ours, draw the general picture of comparable degrees of plasticity in functional traits between diploids and polyploids. This pattern appears consistent across diverse plant genera, life‐history strategies and environments, suggesting that it may well be the rule rather than the exception, at least for herbaceous polyploid plants.
There are several potential explanations for the lack of differentiation in trait plasticity between ploidy levels. First, polyploidy‐induced versatility in gene and the resultant trait expression (e.g. Gaeta et al., 2007) may quickly diminish during the course of polyploid formation, as a result of gene loss or silencing of duplicated copies (Wendel et al., 2018), particularly for genes involved in essential biological processes such as photosynthesis (De Smet et al., 2013). Second, even given gene retention in polyploids, it is possible that only one copy responds to a specific aspect of the abiotic environment, such as in allotetraploid cotton (Liu & Adams, 2007), where one copy of the alcohol dehydrogenase gene responds to cold stress and the other to water treatment owing to subfunctionalization of gene duplicates. Third, similar trait plasticity between ploidy levels may arise from biased gene expression towards one of the subgenomes in allopolyploids. Such subgenome expression bias has been seen in both synthetic and wild polyploids (Jackson & Chen, 2010; Grover et al., 2012), such as Brassica rapa (Cheng et al., 2016) and Mimulus peregrinus (Edger et al., 2017). Thus, linking gene expression of polyploids and diploids to trait plasticity will be critical in disentangling the mechanisms underlying similar trait plasticity between ploidy levels, as well as resolving when plasticity in gene expression (Adams & Wendel, 2005; Leitch & Leitch, 2008) is – or is not – correlated with trait plasticity.
Polyploid fitness advantage and its ecological mechanisms
Among the heterogeneous environments provided by our climatic gardens, allopolyploid Fragaria displayed the ‘jack‐and‐master’ strategy, showing higher fitness in each environment, and overall higher average fitness than diploids. Such polyploid fitness advantage has also been detected in the autotetraploids Arrhenatherum elatius (Petit & Thompson, 1997) and Dactylis glomerata (Bretagnolle & Thompson, 2001), and the allotetraploid Centaurea stoebe (Hahn et al., 2012). In a Claytonia complex (two 2x, one 4x and two 6x cytotypes) growing in California, one 6x cytotype possessed higher biomass than the others consistently across elevational gardens, albeit not for polyploid cytotypes as a whole (McIntyre & Strauss, 2017). In our study, although sexual fitness (e.g. flower number and/or fruit production) was not measured, owing to extremely low incidence of flowering across gardens (c. 20 plants out of 3137 total), sexual reproduction scales with clonal reproduction in long‐lived, perennial Fragaria (Ashman, 2005), unlike in annual plants. Thus, we expect that polyploid advantage also holds when considering sexual fitness in Fragaria.
Here allopolyploids performed as habitat generalists relative to diploids; yet, as they did not exhibit fitness homeostasis across climatic gardens, the ‘jack‐of‐all‐trades’ hypothesis must be rejected. Such plasticity in fitness (i.e. enhanced fitness in response to a favorable environment) is ubiquitous in both polyploids and diploids (Petit & Thompson, 1997; Bretagnolle & Thompson, 2001; Buggs & Pannell, 2007; Hahn et al., 2012; Sánchez Vilas & Pannell, 2017). Although these previous studies and ours often support the ‘jack‐and‐master’ hypothesis for polyploids (but see Buggs & Pannell, 2007), we cannot rule out the possibility that some diploid Fragaria may exhibit the ‘master‐of‐some’ strategy in environments beyond the climatic variation captured by this study (e.g. locations with higher dry season precipitation or greater seasonality; Fig. S3), albeit our gardens are contained within the climatic niches of Fragaria species, and niche distances were taken into account. Thus, generalizing about the adaptive strategies of polyploids and diploids will require not only genetically and geographically broad sampling of taxa as we have here, but also more diverse field environments.
Our study is the first to explicitly connect fitness differences between wild polyploids and diploids in natural environments to functional traits and trait plasticity. While trait plasticity contributes to fitness of diploid and allotetraploid Centaurea stoebe (Hahn et al., 2012), our results revealed not only the importance of trait plasticity for fitness, but also stronger consequences of adaptive plasticity in allopolyploids than in diploids. Relative to trait plasticity, we found that functional trait divergence between allopolyploids and diploids, owing to genomic changes in size and structure (Levin, 1983; Balao et al., 2011), probably plays a more important role in determining fitness differences between ploidy levels, as more traits predict fitness in terms of trait means than plasticities. Also significant is the fact that allopolyploids benefit from stronger positive fitness effects and weaker negative fitness effects of their functional traits, perhaps because their trait means are closer to optima than is the case with diploids in the experimental habitats. One should note, however, that these conclusions rest on the assumption that the statistical covariance between traits and fitness reflect causal relationships that would need to be verified with experiments manipulating the predicted causal traits and/or environments (Kingsolver et al., 2012).
In conclusion, the broad phylogenetic, genetic and geographic scope of this study provides the most robust evaluation to date of adaptive hypotheses for fitness advantage of wild polyploids in changing environments, and elucidates functional trait divergence and adaptive plasticity as the underlying ecological mechanisms. We emphasize that our findings are based on naturally occurring diploids and allopolyploids; as such, they reflect the ‘effective’ adaptions of allopolyploidy, resulting from the cumulative effects of allopolyploid formation via genome duplication and hybridization, and allopolyploidy‐enabled establishment and divergence. Thus the generalizability of our findings to autopolyploidy (i.e. genome duplication without hybridization) remains to be determined. Such comparisons among diploids, autopolyploids and allopolyploids will ultimately be valuable for informing the respective roles of genome duplication and hybridization on polyploid adaptation. Here in light of allopolyploid fitness advantage, the coexistence of allopolyploid and diploid Fragaria in parts of their ranges (Fig. 1) may reflect the collective roles of ecological adaptation to abiotic environment as addressed here as well as other mechanisms (e.g. demographic history, dispersal, and biotic interactions). Future research on linking the biotic adaptation of polyploidy to functional traits and trait plasticity is necessary. Nevertheless, our results add significantly to the understudied ecological adaptations of polyploids, especially allopolyploids (Ramsey & Ramsey, 2014), and offer important insights into the causes of evolutionary success of repeated and pervasive occurrence of polyploids (Van de Peer et al., 2017).
Author contributions
T‐LA and AL designed the research, and all authors contributed to refinement of the design. RC managed garden establishment. All authors collected data. NW performed data analyses. NW wrote the manuscript, and all authors contributed substantially to revisions.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Distinct separation between diploid and high‐order polyploid Fragaria, with stomatal length as an example.
Fig. S2 Collection map of Fragaria.
Fig. S3 Climatic niche distances of 72 source Fragaria populations to the common gardens.
Fig. S4 Maximum likelihood (ML) plastid phylogeny of Fragaria.
Fig. S5 Model comparisons for evolutionary dependence control, with stomatal length as an example.
Fig. S6 Model comparisons for evolutionary dependence control, with fitness as an example.
Fig. S7 Central leaflet width was similar between diploid and polyploid Fragaria taxa.
Fig. S8 Similar scales of three fitness components.
Methods S1 Additional details of Materials and Methods.
Table S1 Genotypes and populations of diploid and polyploid Fragaria.
Table S2 Soil properties of the three common gardens.
Table S3 Pairwise correlations between trait means and trait plasticities.
Table S4 Differences in leaf functional traits between diploids and polyploids.
Table S5 Pairwise correlations between trait plasticities and between functional traits.
Table S6 Differences in trait plasticity between diploids and polyploids.
Table S7 Differences in fitness between diploids and polyploids.
Table S8 Relationships between average fitness and trait means and trait plasticities.
Acknowledgements
We are grateful to all the collectors of wild strawberry genotypes. We thank K. Schuller, T. Jennings, C. Poklemba, P. Krabacher, M. Surplus, A. Kessler and L. Longway for assistance with genotype and clone cultivation, common garden establishment and management, and fitness measurements; A. Freundlich, E. Jiang, C. Rumrill, R. Babu, I. Alagar, S. Barratt‐Boyes and Z. Du for assistance with leaf collection and functional trait measurements; C. Muir for help with impression technique of leaf stomata; and M.S. Dillenberger for providing plastid sequence alignments. We acknowledge the University of Pittsburgh glasshouses, Oregon State University glasshouses and Hatfield Marine Science Center, and the USDA Forest Service Region 6 Bend Seed Extractory for logistic support. We also thank the members of the Ashman, Liston, and Cronn laboratories for discussion, and the Editor and four anonymous reviewers for comments. This work was supported by the National Science Foundation (DEB 1241006 to T‐LA; DEB 1241217 to AL).
Contributor Information
Na Wei, Email: na.wei@pitt.edu.
Tia‐Lynn Ashman, Email: tia1@pitt.edu.
References
- Adams KL, Wendel JF. 2005. Novel patterns of gene expression in polyploid plants. Trends in Genetics 21: 539–543. [DOI] [PubMed] [Google Scholar]
- Ashman T‐L. 2005. The limits on sexual dimorphism in vegetative traits in a gynodioecious plant. The American Naturalist 166: S5–S16. [DOI] [PubMed] [Google Scholar]
- Baker HG. 1965. Characteristics and modes of origin of weeds In: Baker HG, Stebbins LG, eds. The genetics of colonizing species. New York, NY, USA: Academic Press, 147–172. [Google Scholar]
- Balao F, Herrera J, Talavera S. 2011. Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: a multivariate morphological approach. New Phytologist 192: 256–265. [DOI] [PubMed] [Google Scholar]
- Bardil A, de Almeida JD, Combes MC, Lashermes P, Bertrand B. 2011. Genomic expression dominance in the natural allopolyploid Coffea arabica is massively affected by growth temperature. New Phytologist 192: 760–774. [DOI] [PubMed] [Google Scholar]
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA. 2016. On the relative abundance of autopolyploids and allopolyploids. New Phytologist 210: 391–398. [DOI] [PubMed] [Google Scholar]
- Bartoń K. 2017. MuMIn: multi‐model inference. R package version 1.40.40. [WWW document] URL https://CRAN.R-project.org/package=MuMIn.
- Bates D, Machler M, Bolker BM, Walker SC. 2015. Fitting linear mixed‐effects models using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA. 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist 179: 975–986. [DOI] [PubMed] [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesova M, Pysek P. 2012. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany 109: 19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer HJ, Price CA, Wagner‐Cremer F, Dekker SC, Franks PJ, Veneklaas EJ. 2016. Optimal allocation of leaf epidermal area for gas exchange. New Phytologist 210: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretagnolle F, Thompson JD. 2001. Phenotypic plasticity in sympatric diploid and autotetraploid Dactylis glomerata . International Journal of Plant Sciences 162: 309–316. [Google Scholar]
- Buggs RJA, Pannell JR. 2007. Ecological differentiation and diploid superiority across a moving ploidy contact zone. Evolution 61: 125–140. [DOI] [PubMed] [Google Scholar]
- Chao D‐Y, Dilkes B, Luo H, Douglas A, Yakubova E, Lahner B, Salt DE. 2013. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis . Science 341: 658–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. 2010. Molecular mechanisms of polyploidy and hybrid vigor. Trends in Plant Science 15: 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Sun C, Wu J, Schnable J, Woodhouse MR, Liang J, Cai C, Freeling M, Wang X. 2016. Epigenetic regulation of subgenome dominance following whole genome triplication in Brassica rapa . New Phytologist 211: 288–299. [DOI] [PubMed] [Google Scholar]
- Davidson AM, Jennions M, Nicotra AB. 2011. Do invasive species show higher phenotypic plasticity than native species and if so, is it adaptive? A meta‐analysis. Ecology Letters 14: 419–431. [DOI] [PubMed] [Google Scholar]
- De Smet R, Adams KL, Vandepoele K, Van Montagu MCE, Maere S, Van de Peer Y. 2013. Convergent gene loss following gene and genome duplications creates single‐copy families in flowering plants. Proceedings of the National Academy of Sciences, USA 110: 2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillenberger MS, Wei N, Tennessen JA, Ashman T‐L, Liston A. 2018. Plastid genomes reveal recurrent formation of allopolyploid Fragaria . American Journal of Botany 105: 1–13. [DOI] [PubMed] [Google Scholar]
- Dong S, Adams KL. 2011. Differential contributions to the transcriptome of duplicated genes in response to abiotic stresses in natural and synthetic polyploids. New Phytologist 190: 1045–1057. [DOI] [PubMed] [Google Scholar]
- Edger PP, Smith RD, McKain MR, Cooley AM, Vallejo‐Marin M, Yuan Y‐W, Bewick AJ, Ji L, Platts AE, Bowman MJ et al 2017. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140‐year‐old naturally established neo‐allopolyploid monkeyflower. Plant Cell 29: 2150–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehleringer JR, Björkman O. 1978. Pubescence and leaf spectral characteristics in a desert shrub, Encelia farinosa . Oecologia 36: 151–162. [DOI] [PubMed] [Google Scholar]
- Farquhar GD, Richards RA. 1984. Isotopic composition of plant carbon correlates with water‐use efficiency of wheat genotypes. Australian Journal of Plant Physiology 11: 539–552. [Google Scholar]
- Fick SE, Hijmans RJ. 2017. WorldClim 2: new 1‐km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37: 4302–4315. [Google Scholar]
- Fox J, Weisberg S. 2011. An R companion to applied regression. Thousand Oaks, CA, USA: SAGE. [Google Scholar]
- Franks PJ, Beerling DJ. 2009. Maximum leaf conductance driven by CO2 effects on stomatal size and density over geologic time. Proceedings of the National Academy of Sciences, USA 106: 10343–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC, Iniguez‐Luy F, Leon E, Osborn TC. 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover CE, Gallagher JP, Szadkowski EP, Yoo MJ, Flagel LE, Wendel JF. 2012. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytologist 196: 966–971. [DOI] [PubMed] [Google Scholar]
- Hahn MA, van Kleunen M, Muller‐Scharer H. 2012. Increased phenotypic plasticity to climate may have boosted the invasion success of polyploid Centaurea stoebe . PLoS ONE 7: e50284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao G‐Y, Lucero ME, Sanderson SC, Zacharias EH, Holbrook NM. 2013. Polyploidy enhances the occupation of heterogeneous environments through hydraulic related trade‐offs in Atriplex canescens (Chenopodiaceae). New Phytologist 197: 970–978. [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI. 2003. The role of stomata in sensing and driving environmental change. Nature 424: 901–908. [DOI] [PubMed] [Google Scholar]
- Jackson S, Chen ZJ. 2010. Genomic and expression plasticity of polyploidy. Current Opinion in Plant Biology 13: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamneva OK, Syring J, Liston A, Rosenberg NA. 2017. Evaluating allopolyploid origins in strawberries (Fragaria) using haplotypes generated from target capture sequencing. BMC Evolutionary Biology 17: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsolver JG, Diamond SE, Siepielski AM, Carlson SM. 2012. Synthetic analyses of phenotypic selection in natural populations: lessons, limitations and future directions. Evolutionary Ecology 26: 1101–1118. [Google Scholar]
- Knight CA, Beaulieu JM. 2008. Genome size scaling through phenotype space. Annals of Botany 101: 759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB. 2017. lmerTest package: tests in linear mixed effects models. Journal of Statistical Software 82: 1–26. [Google Scholar]
- Leal‐Bertioli SC, Moretzsohn MC, Santos SP, Brasileiro AC, Guimaraes PM, Bertioli DJ, Araujo AC. 2017. Phenotypic effects of allotetraploidization of wild Arachis and their implications for peanut domestication. American Journal of Botany 104: 379–388. [DOI] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ. 2008. Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483. [DOI] [PubMed] [Google Scholar]
- Levin DA. 1983. Polyploidy and novelty in flowering plants. The American Naturalist 122: 1–25. [Google Scholar]
- Li W‐L, Berlyn GP, Ashton PMS. 1996. Polyploids and their structural and physiological characteristics relative to water deficit in Betula papyrifera (Betulaceae). American Journal of Botany 83: 15–20. [Google Scholar]
- Li X, Yu E, Fan C, Zhang C, Fu T, Zhou Y. 2012. Developmental, cytological and transcriptional analysis of autotetraploid Arabidopsis . Planta 236: 579–596. [DOI] [PubMed] [Google Scholar]
- Liston A, Cronn R, Ashman T‐L. 2014. Fragaria: a genus with deep historical roots and ripe for evolutionary and ecological insights. American Journal of Botany 101: 1686–1699. [DOI] [PubMed] [Google Scholar]
- Liu Z, Adams KL. 2007. Expression partitioning between genes duplicated by polyploidy under abiotic stress and during organ development. Current Biology 17: 1669–1674. [DOI] [PubMed] [Google Scholar]
- Lüdecke D. 2017. sjPlot: data visualization for statistics in social science. R package version 2.3.0. [WWW document] URL https://CRAN.R-project.org/package=sjPlot.
- Madlung A. 2013. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A, Wendel JF. 2013. Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenetic and Genome Research 140: 270–285. [DOI] [PubMed] [Google Scholar]
- Maherali H, Walden AE, Husband BC. 2009. Genome duplication and the evolution of physiological responses to water stress. New Phytologist 184: 721–731. [DOI] [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Anderson JT, Raskin E, Weiss‐Lehman C, Mitchell‐Olds T. 2015. Natural variation, differentiation, and genetic trade‐offs of ecophysiological traits in response to water limitation in Brachypodium distachyon and its descendent allotetraploid B. hybridum (Poaceae). Evolution 69: 2689–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson J. 1994. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264: 421–424. [DOI] [PubMed] [Google Scholar]
- McIntyre PJ, Strauss S. 2017. An experimental test of local adaptation among cytotypes within a polyploid complex. Evolution 71: 1960–1969. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Petit C, Thompson JD. 1997. Variation in phenotypic response to light availability between diploid and tetraploid populations of the perennial grass Arrhenatherum elatius from open and woodland sites. Journal of Ecology 85: 657–667. [Google Scholar]
- Pigliucci M. 2003. Phenotypic integration: studying the ecology and evolution of complex phenotypes. Ecology Letters 6: 265–272. [Google Scholar]
- Poorter H, Niinemets U, Poorter L, Wright IJ, Villar R. 2009. Causes and consequences of variation in leaf mass per area (LMA): a meta‐analysis. New Phytologist 182: 565–588. [DOI] [PubMed] [Google Scholar]
- Qiao Q, Xue L, Wang Q, Sun H, Zhong Y, Huang J, Lei J, Zhang T. 2016. Comparative transcriptomics of strawberries (Fragaria spp.) provides insights into evolutionary patterns. Frontiers in Plant Science 7: 1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; [WWW document] URL https://www.R-project.org/. [Google Scholar]
- Ramsey J. 2011. Polyploidy and ecological adaptation in wild yarrow. Proceedings of the National Academy of Sciences, USA 108: 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Ramsey TS. 2014. Ecological studies of polyploidy in the 100 years following its discovery. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 369: 20130352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeldt GE, Tchebakova NM, Barnhardt LK. 1999. Efficacy of climate transfer functions: introduction of Eurasian populations of Larix into Alberta. Canadian Journal of Forest Research 29: 1660–1668. [Google Scholar]
- Richards CL, Bossdorf O, Muth NZ, Gurevitch J, Pigliucci M. 2006. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecology Letters 9: 981–993. [DOI] [PubMed] [Google Scholar]
- Sack L, Scoffoni C. 2013. Leaf venation: structure, function, development, evolution, ecology and applications in the past, present and future. New Phytologist 198: 983–1000. [DOI] [PubMed] [Google Scholar]
- Salman‐Minkov A, Sabath N, Mayrose I. 2016. Whole‐genome duplication as a key factor in crop domestication. Nature Plants 2: 16115. [DOI] [PubMed] [Google Scholar]
- Sánchez Vilas J, Pannell JR. 2017. No difference in plasticity between different ploidy levels in the Mediterranean herb Mercurialis annua . Scientific Reports 7: 9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu‐Inatsugi R, Terada A, Hirose K, Kudoh H, Sese J, Shimizu KK. 2017. Plant adaptive radiation mediated by polyploid plasticity in transcriptomes. Molecular Ecology 26: 193–207. [DOI] [PubMed] [Google Scholar]
- Sletvold N, Ågren J. 2012. Variation in tolerance to drought among Scandinavian populations of Arabidopsis lyrata . Evolutionary Ecology 26: 559–577. [Google Scholar]
- Soltis DE, Visger CJ, Marchant DB, Soltis PS. 2016. Polyploidy: pitfalls and paths to a paradigm. American Journal of Botany 103: 1146–1166. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Liu XX, Marchant DB, Visger CJ, Soltis DE. 2014. Polyploidy and novelty: Gottlieb's legacy. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 369: 20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt G. 1999. Systematics and geographic distribution of the American strawberry species: Taxonomic studies in the genus Fragaria (Rosaceae: Potentilleae). Berkeley, CA, USA: University of California Press. [Google Scholar]
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London, UK: Addison‐Wesley. [Google Scholar]
- Tanaka Y, Sugano SS, Shimada T, Hara‐Nishimura I. 2013. Enhancement of leaf photosynthetic capacity through increased stomatal density in Arabidopsis . New Phytologist 198: 757–764. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Govindarajulu R, Ashman T‐L, Liston A. 2014. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biology and Evolution 6: 3295–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares F, Sanchez‐Gomez D, Zavala MA. 2006. Quantitative estimation of phenotypic plasticity: bridging the gap between the evolutionary concept and its ecological applications. Journal of Ecology 94: 1103–1116. [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K. 2017. The evolutionary significance of polyploidy. Nature Reviews: Genetics 18: 411–424. [DOI] [PubMed] [Google Scholar]
- Wei N, Govindarajulu R, Tennessen JA, Liston A, Ashman T‐L. 2017a. Genetic mapping and phylogenetic analysis reveal intraspecific variation in sex chromosomes of the Virginian strawberry. Journal of Heredity 108: 731–739. [DOI] [PubMed] [Google Scholar]
- Wei N, Tennessen JA, Liston A, Ashman T‐L. 2017b. Present‐day sympatry belies the evolutionary origin of a high‐order polyploid. New Phytologist 216: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Lisch D, Hu G, Mason AS. 2018. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Current Opinion in Genetics & Development 49: 1–7. [DOI] [PubMed] [Google Scholar]
- Wong J. 2013. pdist: Partitioned distance function. R package version 1.2. [WWW document] URL https://CRAN.R-project.org/package=pdist.
- Wright IJ, Reich PB, Westoby M, Ackerly DD, Baruch Z, Bongers F, Cavender‐Bares J, Chapin T, Cornelissen JHC, Diemer M et al 2004. The worldwide leaf economics spectrum. Nature 428: 821–827. [DOI] [PubMed] [Google Scholar]
- Yang C, Zhao L, Zhang H, Yang Z, Wang H, Wen S, Zhang C, Rustgi S, von Wettstein D, Liu B. 2014. Evolution of physiological responses to salt stress in hexaploid wheat. Proceedings of the National Academy of Sciences, USA 111: 11882–11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Distinct separation between diploid and high‐order polyploid Fragaria, with stomatal length as an example.
Fig. S2 Collection map of Fragaria.
Fig. S3 Climatic niche distances of 72 source Fragaria populations to the common gardens.
Fig. S4 Maximum likelihood (ML) plastid phylogeny of Fragaria.
Fig. S5 Model comparisons for evolutionary dependence control, with stomatal length as an example.
Fig. S6 Model comparisons for evolutionary dependence control, with fitness as an example.
Fig. S7 Central leaflet width was similar between diploid and polyploid Fragaria taxa.
Fig. S8 Similar scales of three fitness components.
Methods S1 Additional details of Materials and Methods.
Table S1 Genotypes and populations of diploid and polyploid Fragaria.
Table S2 Soil properties of the three common gardens.
Table S3 Pairwise correlations between trait means and trait plasticities.
Table S4 Differences in leaf functional traits between diploids and polyploids.
Table S5 Pairwise correlations between trait plasticities and between functional traits.
Table S6 Differences in trait plasticity between diploids and polyploids.
Table S7 Differences in fitness between diploids and polyploids.
Table S8 Relationships between average fitness and trait means and trait plasticities.


