Summary
Little attention has been given to the design of efficient studies to evaluate longitudinal biomarkers. Measuring longitudinal markers on an entire cohort is cost prohibitive and, especially for rare outcomes such as cancer, may be infeasible. Thus, methods for evaluation of longitudinal biomarkers using efficient and cost-effective study designs are needed. Case cohort (CCH) and nested case–control (NCC) studies allow investigators to evaluate biomarkers rigorously and at reduced cost, with only a small loss in precision. In this article, we develop estimators of several measures to evaluate the accuracy and discrimination of predicted risk under CCH and NCC study designs. We use double inverse probability weighting (DIPW) to account for censoring and sampling bias in estimation and inference procedures. We study the asymptotic properties of the proposed estimators. To facilitate inference using two-phase longitudinal data, we develop valid resampling-based variance estimation procedures under CCH and NCC. We evaluate the performance of our estimators under CCH and NCC using simulation studies and illustrate them on a NCC study within the hepatitis C antiviral long-term treatment against cirrhosis (HALT-C) clinical trial. Our estimators and inference procedures perform well under CCH and NCC, provided that the sample size at the time of prediction (effective sample size) is reasonable. These methods are widely applicable, efficient, and cost-effective and can be easily adapted to other study designs used to evaluate prediction rules in a longitudinal setting.
Keywords: Biomarker evaluation, Longitudinal and survival data, Two-phase designs
1. Introduction
For many lethal diseases such as hepatocellular carcinoma (HCC), active surveillance of the high-risk population may aid in detecting the disease at an early stage when curative therapy can be implemented. For disease monitoring, using longitudinally measured information to predict the occurrence of a clinical outcome in a future time is a key analytical goal. The prediction of such a time-dependent binary outcome is often dynamic, updated with information accumulated over time. Once a prediction algorithm is developed based on longitudinal biomarkers, it is critical to evaluate its clinical performance prior to adopting it for routine clinical use.
The motivation of our research comes from a biomarker study for HCC surveillance. Alpha fetoprotein (AFP) is the most widely used biomarker for HCC surveillance, however, its sensitivity and specificity in detecting early HCC are low. More reliable biomarkers for HCC surveillance and early detection are sought in order to improve the outcome of the disease. The hepatitis C antiviral long-term treatment against cirrhosis (HALT-C) trial included 1050 patients at high risk of HCC, i.e., those with cirrhosis, and chronic infection with hepatitis B virus or hepatitis C virus. Patients were randomized to low dose pegylated interferon or no treatment and followed every 3 months for a total duration of 3.5 years. Blood samples were collected at each visit for subsequent research testing, including assays for HCC biomarkers. As part of the trial, a nested case–control (NCC) study was conducted to assess the accuracy of a novel serum biomarker, des-gamma-carboxy prothrombin (DCP), in predicting the risk of HCC among patients under surveillance. The NCC sub-cohort included all 39 HCC cases diagnosed during the follow up. For each case, two controls matched on treatment assignment and presence of cirrhosis on baseline biopsy were selected from those at risk of HCC at the time of diagnosis. The biomarkers were evaluated at multiple follow up visits and the results based on the repeated markers were reported in Lok and others (2010). In that study, the prediction performance of the biomarkers at a single time point was assessed ignoring the sampling design. The main question remaining to be addressed with the information collected is how to efficiently assess the longitudinal prediction accuracy of the new marker from a two-phase study with a rare outcome?
The study considered in the HALT-C trial is of a two-phase design in that biospecimens are ascertained only for a subset of individuals selected in the second phase. Two-phase sampling designs, including the case cohort (CCH) (Prentice, 1986) and the NCC (Thomas, 1977), are particularly appealing for biomarker studies as cost-effective alternatives to the full-cohort design. In the case of a longitudinal study of novel biomarkers, cost-effectiveness is of particular importance due to the need to assay repeated measurements from all individuals in the full cohort. Efficient estimation of the relative risk in risk modeling analysis based on a subset of individuals has been addressed to a great extent in Borgan and others (2000), Breslow and others (2009), Chen and Lo (1999). However, literature is limited on methods to select individuals in the context of evaluating biomarker performance (Cai and Zheng, 2012; Liu and others, 2012); and no work so far has been proposed to calculate accuracy summaries under two-phase longitudinal studies. The two-phase designs, while cost-effective, generate complex datasets in which the missingness of the longitudinal marker values depends on the outcome of interest, making estimation and inference about the accuracy of prediction based on a longitudinal marker challenging. In particular, inference needs to account for both between-individual correlations induced by specific sampling schemes and within-individual correlations due to repeated measures. Appropriate statistical methods are not currently available for such a setting. Wide adoption of these designs to evaluate predictive markers in a longitudinal setting is critically dependent on the availability of appropriate statistical tools.
The evaluation of the clinical performance of a medical test has been traditionally based on receiver operating characteristic (ROC) curves, and calculation of time-dependent ROC curves to evaluate a single longitudinal marker has been considered in Zheng and Heagerty (2004). Area under the ROC curve (AUC) for a longitudinal biomarker provides a global summary of the marker’s capacity for discriminating between individuals who are still at risk at the update time and subsequently develop the outcome in a given time frame versus those who do not. Extension of the Brier score to the longitudinal setting, termed the prediction error (PE), serves as a tool for quantifying the calibration of a prediction model (Schoop and others, 2008; Blanche and others, 2015). More recently other metrics of risk assessment have been proposed that are more clinically relevant compared with the AUC (e.g., Gu and Pepe, 2009; Pfeiffer and Gail, 2011). However, they are most often considered with a binary outcome and are yet to be extended to a setting with longitudinal markers and survival outcomes. Evaluating dynamic prediction rules in that setting is a topic of great interest in the field. Allowing the calculation of these quantities with a two-phase study would be of great practical importance.
The goal of this article is to provide estimation and inference tools for the cost-effective evaluation of longitudinal dynamic risk predictions under two-phase study designs. First, we consider summary measures that can be used to quantify the clinical utility of dynamic risk predictions and are relevant to clinical practice of active surveillance, and propose non-parametric estimators in this setting. Such an approach separates the validation procedure from the procedure for dynamic risk derivation, and it improves upon existing methods for longitudinal accuracy estimation with a full cohort evaluation (e.g., Zheng and Heagerty, 2007) in terms of robustness. To accommodate two-phase sampling design, we consider the idea of doubly inverse probability weighted (DIPW) estimators. Building upon previous research using baseline data (e.g., Liu and others, 2012), we further modify both the sampling weights and the censoring weights to accommodate more complex longitudinal settings. This introduces additional complexity for inference. Therefore to facilitate inference on the longitudinal accuracy summaries, we study the asymptotic properties of the new methods and propose a resampling-based procedure to account for various sources of variation, those due to two-phase sampling and specific estimation procedures. We also provide a unified approach to analyzing two-phase longitudinal data, which accounts for the between-individual correlation induced by sampling in addition to the within-individual correlation among measurements for the same individual. This is an important contribution to the literature, and will facilitate the adoption of cost-effective two-phase designs in longitudinal biomarker studies in practice. We introduce notation in Section 2. We describe estimation of dynamic risks and risk assessment measures under longitudinal cohort and two-phase designs in Section 3. We describe the inference procedures in Section 4. The results of simulation studies evaluating the proposed procedures are presented in Section 5. In Section 6, we illustrate the performance of our methods on evaluating a longitudinal biomarker in liver cancer study carried out with a NCC design.
2. Evaluation of longitudinal prediction
2.1. Notation
Suppose there are 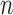 subjects in the full cohort and let
subjects in the full cohort and let 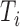 denote the time to failure for the
denote the time to failure for the 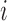 th subject,
th subject, 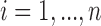 . Due to censoring, for
. Due to censoring, for 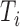 , we only observe
, we only observe 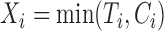 and
and 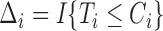 , where
, where 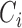 is the corresponding censoring time. We use
is the corresponding censoring time. We use 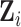 to denote time-constant covariates such as gender. For
to denote time-constant covariates such as gender. For 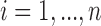 , the observed longitudinal biomarker on subject
, the observed longitudinal biomarker on subject 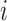 prior to event time
prior to event time 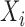 is denoted by
is denoted by 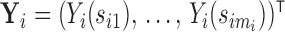 measured at times
measured at times 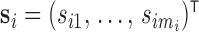 with
with 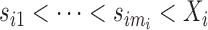 , where
, where 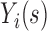 biomarker value for the
biomarker value for the 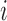 th subject measured at time
th subject measured at time 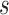 . The observation times are assumed to be specified by a study protocol, although deviations from protocol are not unusual. The observed covariate information for subject
. The observation times are assumed to be specified by a study protocol, although deviations from protocol are not unusual. The observed covariate information for subject 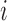 consists of
consists of 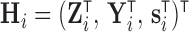 . At any time
. At any time 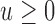 , the history of the covariate process for subject
, the history of the covariate process for subject 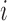 is known and equals to
is known and equals to 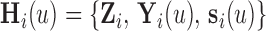 , where
, where 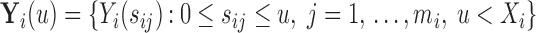 and
and 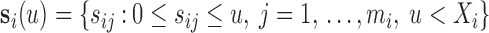 . The full data sample is denoted by
. The full data sample is denoted by 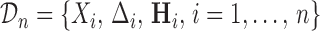 . Let
. Let 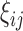 be a binary indicator of whether (
be a binary indicator of whether (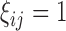 ) or not (
) or not (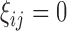 ) the
) the 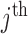 measurement of
measurement of 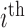 subject is sampled at phase two and let
subject is sampled at phase two and let 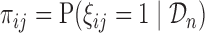 be the probability of such sampling. Note that in a two-phase study design, if all samples from subcohort members are measured then
be the probability of such sampling. Note that in a two-phase study design, if all samples from subcohort members are measured then 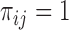 for all
for all 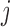 .
.
We let 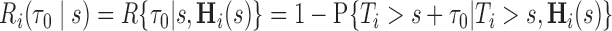 denote the dynamic risk for subject
denote the dynamic risk for subject 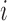 at time
at time 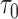 from the measurement time
from the measurement time 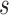 given
given 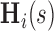 and
and 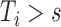 , and
, and 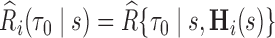 denote the corresponding estimate. We note that not only is
denote the corresponding estimate. We note that not only is 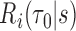 often more clinically relevant to clinicians and patients than observed marker values, it also provides a way to incorporate information from multiple time-varying markers and multiple clinical variables.
often more clinically relevant to clinicians and patients than observed marker values, it also provides a way to incorporate information from multiple time-varying markers and multiple clinical variables.
2.2. Measures of longitudinal predictive discrimination
Calibration of 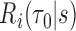 can be gauged by PE, defined as
can be gauged by PE, defined as 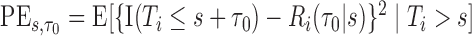 (Schoop and others, 2008). The clinical utility of biomarkers has traditionally been quantified with a ROC curve. In a surveillance setting, a decision at time
(Schoop and others, 2008). The clinical utility of biomarkers has traditionally been quantified with a ROC curve. In a surveillance setting, a decision at time 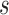 is often made based on a subject’s risk of experiencing an event in the next time interval
is often made based on a subject’s risk of experiencing an event in the next time interval 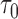 ,
, 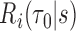 , rather than on specific values of the multivariate
, rather than on specific values of the multivariate 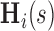 . We therefore define a test based on
. We therefore define a test based on 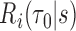 and a threshold
and a threshold 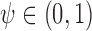 with test being positive if
with test being positive if 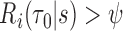 and negative otherwise. This definition of a test is key to defining true and false positive fractions (TPF and FPF) in a longitudinal setting:
and negative otherwise. This definition of a test is key to defining true and false positive fractions (TPF and FPF) in a longitudinal setting:
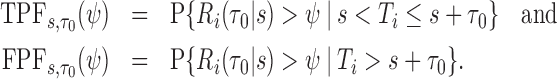 |
Then the corresponding ROC curve and AUC can be defined, respectively as 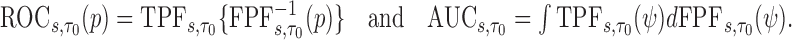
We focus here on the estimation of 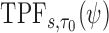 and
and 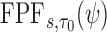 , but note that additional measures of prediction performance can be derived from the pair of these quantities. In particular, we extend two risk prediction summaries recently proposed by Pfeiffer and Gail (2011), the proportion of cases followed (PCF) and the proportion needed to follow-up (PNF), to the longitudinal setting. We define the proportion of cases followed,
, but note that additional measures of prediction performance can be derived from the pair of these quantities. In particular, we extend two risk prediction summaries recently proposed by Pfeiffer and Gail (2011), the proportion of cases followed (PCF) and the proportion needed to follow-up (PNF), to the longitudinal setting. We define the proportion of cases followed, 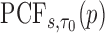 , as the proportion of subjects who experience an event in time
, as the proportion of subjects who experience an event in time 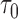 from
from 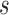 , conditional on being at risk at time
, conditional on being at risk at time 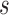 , if we follow a proportion
, if we follow a proportion 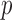 of individuals at highest conditional risk in the population. Let
of individuals at highest conditional risk in the population. Let 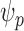 denote a risk threshold such that
denote a risk threshold such that 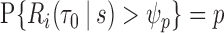 . Then
. Then 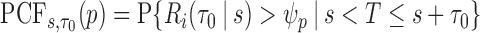 . A related measure, the proportion of the population needed to be followed,
. A related measure, the proportion of the population needed to be followed, 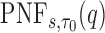 , denotes the proportion of the population at highest conditional risk that needs to be followed in order to capture a proportion
, denotes the proportion of the population at highest conditional risk that needs to be followed in order to capture a proportion 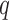 of cases. Let
of cases. Let 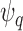 denote a risk threshold such that
denote a risk threshold such that 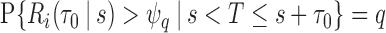 . Then
. Then 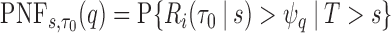 .
.
Such summaries are relevant in the active surveillance setting, to inform the selection of cutoffs for binary decision rules and for making comparisons among prediction rules.
3. Estimation of dynamic risk and prediction performance measures under longitudinal two-phase studies
3.1. Longitudinal propensity of inclusion
In a two-phase study, the probability that an individual’s measurement is included in the second phase of study, 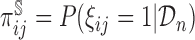 , is dictated by the sampling fraction specified in the study design protocol (see Web Appendix A of the supplementary material available at Biostatistics online for the specification of
, is dictated by the sampling fraction specified in the study design protocol (see Web Appendix A of the supplementary material available at Biostatistics online for the specification of 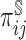 under several two-phase study designs). Contribution to the estimation can then be weighted by the inverse of the probability of sampling (
under several two-phase study designs). Contribution to the estimation can then be weighted by the inverse of the probability of sampling (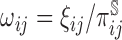 ), which is referred to as the true inverse probability weighted (TIPW) procedure. To improve efficiency over the simple TIPW estimators, one may leverage both outcome and covariate information used for sampling, as well as additional auxiliary variables (
), which is referred to as the true inverse probability weighted (TIPW) procedure. To improve efficiency over the simple TIPW estimators, one may leverage both outcome and covariate information used for sampling, as well as additional auxiliary variables (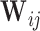 ) by non-parametrically estimating
) by non-parametrically estimating 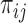 given
given 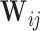 , a procedure known as the augmented inverse probability weighting (AIPW) (Robins and others, 1994).
, a procedure known as the augmented inverse probability weighting (AIPW) (Robins and others, 1994). 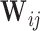 often involves continuous variables. For example, in NCC designs, the sampling is dependent on
often involves continuous variables. For example, in NCC designs, the sampling is dependent on 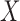 and thus
and thus 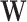 needs to include
needs to include 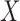 to ensure the consistency of the AIPW estimators. Such weights can also be robustly estimated with a non-parametric procedure in the form of the Nadaraya–Watson estimator,
to ensure the consistency of the AIPW estimators. Such weights can also be robustly estimated with a non-parametric procedure in the form of the Nadaraya–Watson estimator,
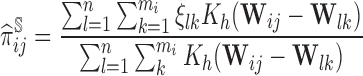 |
(3.1) |
where 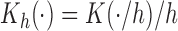 ,
, 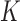 is a symmetric kernel density function, and
is a symmetric kernel density function, and 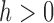 is the bandwidth. Selection of appropriate
is the bandwidth. Selection of appropriate 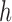 can follow the recommendations in Qi and others (2005). When the dimension of
can follow the recommendations in Qi and others (2005). When the dimension of 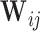 is not small, such a non-parametric estimator may not be feasible. Problems can also arise when additional factors, such as comorbidities, are associated with missingness of regularly scheduled measurements. Thus, to account for missingness due to two-phase sampling and random missingness due to other known factors, we propose to flexibly estimate
is not small, such a non-parametric estimator may not be feasible. Problems can also arise when additional factors, such as comorbidities, are associated with missingness of regularly scheduled measurements. Thus, to account for missingness due to two-phase sampling and random missingness due to other known factors, we propose to flexibly estimate 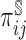 as
as
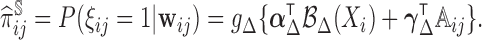 |
(3.2) |
by fitting the model separately for subjects with 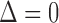 and
and 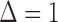 , where
, where 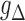 is a pre-specified smooth link function, such as logit,
is a pre-specified smooth link function, such as logit, 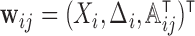 ,
, 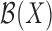 is a spline basis function of
is a spline basis function of 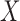 ,
, 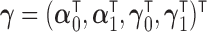 and
and 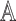 represents auxiliary information including variables that are related to the missingness of the measurements, either by design or another missingness mechanism. In this AIPW framework, the contribution of an individual measurement to estimation is weighted by
represents auxiliary information including variables that are related to the missingness of the measurements, either by design or another missingness mechanism. In this AIPW framework, the contribution of an individual measurement to estimation is weighted by 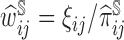 .
.
3.2. Estimation of dynamic risk under two-phase study designs
We consider a flexible model for estimation of a dynamic 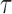 -year risk for an individual under active surveillance over
-year risk for an individual under active surveillance over 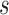 years with a general form
years with a general form 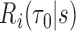 ,
, 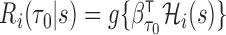 , where
, where 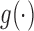 is a known increasing and smooth link function,
is a known increasing and smooth link function, 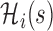 is the vector of partial longitudinal information collected up to time
is the vector of partial longitudinal information collected up to time 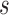 , including some flexible functionals of components in
, including some flexible functionals of components in 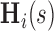 . We consider directly modeling
. We consider directly modeling 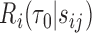 using a binary outcome
using a binary outcome 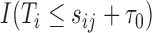 among individuals with
among individuals with 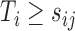 . We call such a model a partly conditional generalized linear model (
. We call such a model a partly conditional generalized linear model (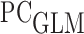 ) following Zheng and Heagerty (2005).
) following Zheng and Heagerty (2005).
In the presence of censoring, the contribution of an observation needs to be weighted by 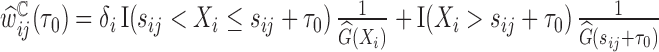 . In particular,
. In particular, 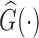 is the Kaplan–Meier (KM) estimate of
is the Kaplan–Meier (KM) estimate of 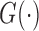 , the censoring distribution under the independent censoring assumption. Therefore, in the estimation, we propose to weigh the contribution from the
, the censoring distribution under the independent censoring assumption. Therefore, in the estimation, we propose to weigh the contribution from the 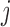 th measurement of the
th measurement of the 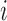 th subject by DIPW),
th subject by DIPW), 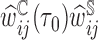 , to account for missingness in the outcome due to censoring, missingness in covariates due to two-phase sampling and random missingness in covariate information. Under the assumption that
, to account for missingness in the outcome due to censoring, missingness in covariates due to two-phase sampling and random missingness in covariate information. Under the assumption that 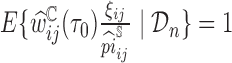 , a consistent estimator of
, a consistent estimator of 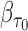 can be obtained by solving the following DIPW estimating equation:
can be obtained by solving the following DIPW estimating equation:
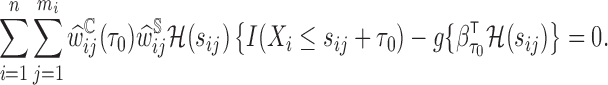 |
(3.3) |
Then, for a future subject 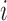 with covariates
with covariates 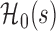 at
at 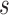 , an estimator of
, an estimator of 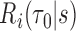 is
is 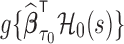 .
.
3.3. Estimation of longitudinal prediction accuracy based on two-phase data
Previous work done on TPF and FPF for a single longitudinal marker in a used semiparametric estimation methods Zheng and Heagerty (2004, 2007). We opted for nonparametric estimation as a way to increase robustness and require fewer assumptions. This is particularly appealing in the multivariate setting where there is a need for separating the assumption used for model development from that for model validation. Similar to our proposed method for estimating dynamic risk, we also consider a DIPW procedure here.
For example, the pair of risk-specific accuracy summaries can be estimated as
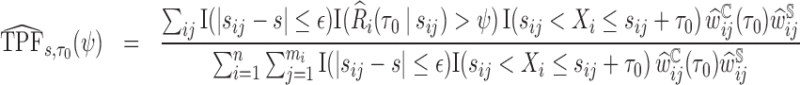 |
(3.4) |
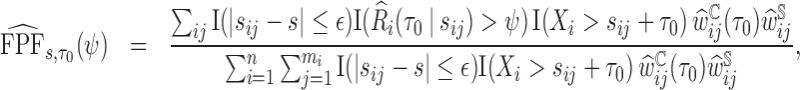 |
(3.5) |
where 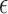 is the half width of the window where observations can be included to estimate accuracy at time
is the half width of the window where observations can be included to estimate accuracy at time 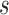 . In this article, we focus on the situation where
. In this article, we focus on the situation where 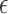 is a pre-specified fixed quantity. This is reasonable for practical settings where
is a pre-specified fixed quantity. This is reasonable for practical settings where 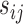 ’s are taken in close neighbourhoods of a finite number of pre-scheduled visit times. A more flexible approach would be to consider a kernel-based estimator
’s are taken in close neighbourhoods of a finite number of pre-scheduled visit times. A more flexible approach would be to consider a kernel-based estimator 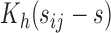 in place of
in place of 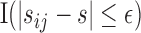 .
.
The 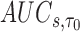 is estimated by
is estimated by 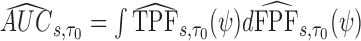 and the estimator of the prediction error,
and the estimator of the prediction error, 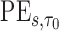 , is
, is
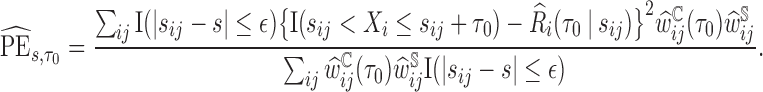 |
To estimate the 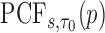 , we first sort the predicted risk around time
, we first sort the predicted risk around time 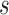 for all individuals sampled into the second phase,
for all individuals sampled into the second phase, 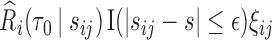 ,
, 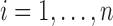 , in decreasing order. We then find the largest
, in decreasing order. We then find the largest 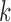 satisfying the following inequality:
satisfying the following inequality:
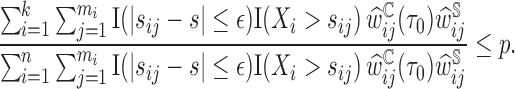 |
Then the estimator of 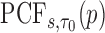 is defined as
is defined as
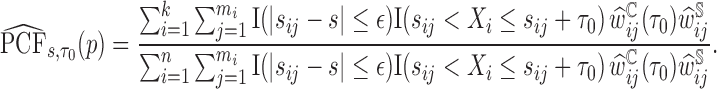 |
To estimate the 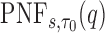 we again used the sorted risks and find the largest
we again used the sorted risks and find the largest 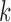 satisfying the following inequality:
satisfying the following inequality:
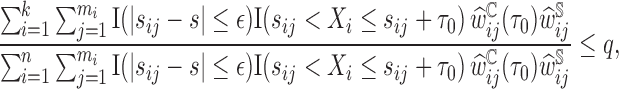 |
then the 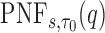 is estimated by
is estimated by
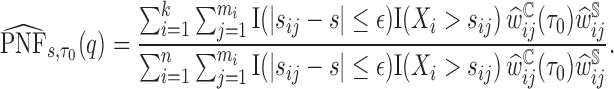 |
We note that for individuals who are not in the second phase of the study, 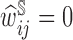 .
.
4. Inference for estimators of prediction performance measures under longitudinal two-phase study designs
To make inference about 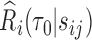 , we studied the asymptotic properties of proposed estimators. In Web Appendix B of the supplementary material available at Biostatistics online, we show that
, we studied the asymptotic properties of proposed estimators. In Web Appendix B of the supplementary material available at Biostatistics online, we show that 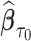 is consistent for
is consistent for 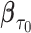 , where
, where 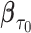 is the unique solution of the expected value of the corresponding weighted estimating equation. Furthermore, we show that the process
is the unique solution of the expected value of the corresponding weighted estimating equation. Furthermore, we show that the process 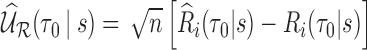 is asymptotically equivalent to a sum of
is asymptotically equivalent to a sum of 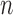 identical and weakly correlated terms,
identical and weakly correlated terms, 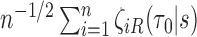 , where
, where 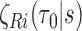 is defined in Web Appendix B of the supplementary material available at Biostatistics online. Following Cai and Zheng (2012) and Breslow and Wellner (2007), it can be shown that
is defined in Web Appendix B of the supplementary material available at Biostatistics online. Following Cai and Zheng (2012) and Breslow and Wellner (2007), it can be shown that 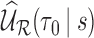 converges weakly to a zero-mean Gaussian process.
converges weakly to a zero-mean Gaussian process.
To make inference about estimators of accuracy summary measures including 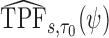 ,
, 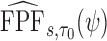 ,
, 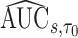 and
and 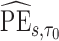 (denoted by a generic term
(denoted by a generic term 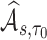 ), we show in Web Appendix C that
), we show in Web Appendix C that 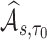 is consistent for
is consistent for 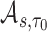 . We further derive the asymptotic linear expansion of
. We further derive the asymptotic linear expansion of 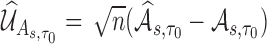 ,which is asymptotically equivalent to a sum of
,which is asymptotically equivalent to a sum of 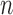 identical and weakly correlated terms,
identical and weakly correlated terms, 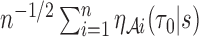 , where
, where 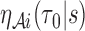 is defined in Web Appendix C of the supplementary material available at Biostatistics online. Again, with appropriate justification, one may show that
is defined in Web Appendix C of the supplementary material available at Biostatistics online. Again, with appropriate justification, one may show that 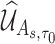 converges to zero-mean normal random vector for any
converges to zero-mean normal random vector for any 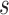 and
and 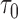 with the data support.
with the data support.
Due to the weak dependence among 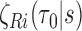 and
and 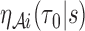 induced by finite sampling in the second phase of a two-phase study (correlations among
induced by finite sampling in the second phase of a two-phase study (correlations among 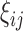 within sampled individuals), as well as correlations among measurements within an individual, explicit asymptotic variance estimators based on
within sampled individuals), as well as correlations among measurements within an individual, explicit asymptotic variance estimators based on 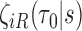 and
and 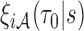 can be difficult to obtain. Thus, we developed a resampling procedure that is appropriate for inference under two-phase study designs by extending our previously proposed resampling procedures developed for a setting where predictors are measured at baseline (Cai and Zheng, 2013) to the current setting with markers measured longitudinally.
can be difficult to obtain. Thus, we developed a resampling procedure that is appropriate for inference under two-phase study designs by extending our previously proposed resampling procedures developed for a setting where predictors are measured at baseline (Cai and Zheng, 2013) to the current setting with markers measured longitudinally.
The variance of each of our performance measures under longitudinal CCH or NCC studies can be estimated as follows:
Generate
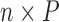 independent and identically distributed random variables
independent and identically distributed random variables 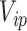 from a known distribution with
from a known distribution with 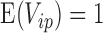 and
and 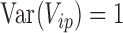 , and
, and 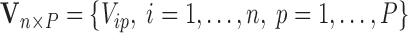 .
.-
Use
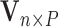 to obtain
to obtain  perturbed counterparts of weights and statistics:
perturbed counterparts of weights and statistics:(a) the perturbed sampling weights
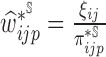 , where
, where 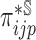 is the perturbed inclusion probability estimated with
is the perturbed inclusion probability estimated with 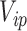 as a weight for all
as a weight for all 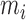 observations of the
observations of the 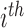 subject.
subject.(b) the perturbed censoring weights,
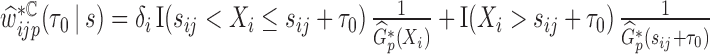 , where
, where 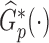 is a weighted KM estimator, with
is a weighted KM estimator, with 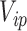 denoting the perturbation weight for measurements on the
denoting the perturbation weight for measurements on the 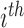 subject.
subject.- (c)
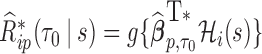 where
where 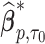 is the solution to
is the solution to
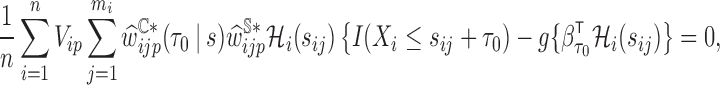
- (d) summary measures
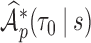 , for example:
, for example:
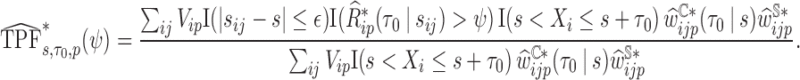
The estimate of variance of
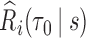 and
and 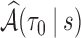 is the empirical variance of the
is the empirical variance of the 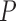 estimates
estimates 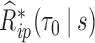 and
and 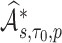 , respectively,
, respectively, 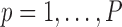 , under a given two-phase sampling design.
, under a given two-phase sampling design.
Similar approach and theoretical justification has also been considered for CCH studies with markers measured at baseline (Huang, 2014). Since perturbation is performed at an individual level, we expect the basic theoretical justification for the resampling procedure to apply to the longitudinal setting as well. In the next section, we investigate the performance of this approach with numerical studies.
5. Simulation studies
5.1. Simulation setup
The longitudinal data for biomarker 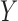 was generated with a linear mixed effects model with measurement error:
was generated with a linear mixed effects model with measurement error: 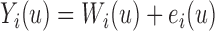 , where
, where 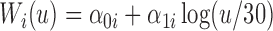 . The random components
. The random components 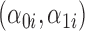 were generated as a bivariate normal with mean
were generated as a bivariate normal with mean 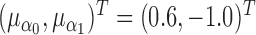 , and a covariance matrix
, and a covariance matrix 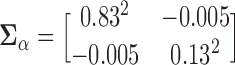 . The measurement error
. The measurement error 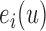 was generated from normal distribution with mean zero and a standard deviation of 0.1. Failure time
was generated from normal distribution with mean zero and a standard deviation of 0.1. Failure time 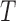 was assumed to depend on the covariates through a proportional hazards relationship:
was assumed to depend on the covariates through a proportional hazards relationship: 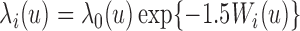 with a Weibull baseline hazard:
with a Weibull baseline hazard: 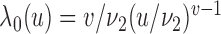 , scale
, scale 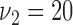 and shape
and shape 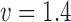 . Censoring time was generated from an exponential distribution (rate = 0.01) with administrative censoring at 180 months. There were up to 10 measurements per subject taken at 6-month intervals. Given fixed interval for
. Censoring time was generated from an exponential distribution (rate = 0.01) with administrative censoring at 180 months. There were up to 10 measurements per subject taken at 6-month intervals. Given fixed interval for 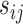 ,
, 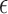 is taken to be 0.
is taken to be 0.
For CCH designs, we simulated cohorts of size 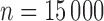 in phase one, and sampled 1000 events and 1000 censored individuals without replacement (finite sampling) from the full cohort. We refer to subjects who experienced an event at any point in the study as cases, and those who did not as controls. For NCC designs, we simulated cohorts of size
in phase one, and sampled 1000 events and 1000 censored individuals without replacement (finite sampling) from the full cohort. We refer to subjects who experienced an event at any point in the study as cases, and those who did not as controls. For NCC designs, we simulated cohorts of size 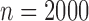 as phase 1 samples. We then sampled all the cases into the second phase sample, and for each case we sampled one control from the risk set at the time of the event of the case, stratifying on a dichotomized value of the marker (
as phase 1 samples. We then sampled all the cases into the second phase sample, and for each case we sampled one control from the risk set at the time of the event of the case, stratifying on a dichotomized value of the marker (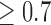 ) at baseline. We also considered smaller sample sizes for the second phase in order to evaluate the robustness of the estimates.
) at baseline. We also considered smaller sample sizes for the second phase in order to evaluate the robustness of the estimates.
The true values of the accuracy measures of interest were generated as follows: we generated two full cohort datasets of size 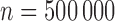 without censoring. One served as a training set and the other as a validation set. We fit the
without censoring. One served as a training set and the other as a validation set. We fit the 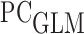 using the training set for binary outcome
using the training set for binary outcome 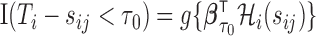 with covariates
with covariates 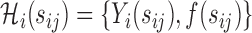 , where
, where 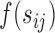 was a spline function of
was a spline function of 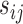 with a degree of freedom (df) of 3. Using the estimates from the model, we calculated
with a degree of freedom (df) of 3. Using the estimates from the model, we calculated 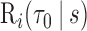 and its accuracy summaries
and its accuracy summaries 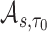 at various time frames of
at various time frames of 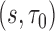 using the validation set. With the two-phase samples, we estimated
using the validation set. With the two-phase samples, we estimated 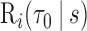 following the procedure in Section 3.2, using a weighted
following the procedure in Section 3.2, using a weighted 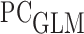 model with covariates
model with covariates 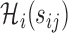 . The estimation of longitudinal accuracy summaries at selected
. The estimation of longitudinal accuracy summaries at selected 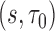 were calculated using the methods described in Section 3.3, and variance estimates as described in Section 4. Censoring weights,
were calculated using the methods described in Section 3.3, and variance estimates as described in Section 4. Censoring weights, 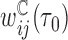 , were estimated using the Kaplan–Meier estimator. For CCH,
, were estimated using the Kaplan–Meier estimator. For CCH, 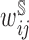 were estimated using true sampling weights. For stratified NCC,
were estimated using true sampling weights. For stratified NCC, 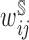 were estimated using a generalized additive model (GAM) with
were estimated using a generalized additive model (GAM) with 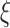 as the outcome, with observed measurement time
as the outcome, with observed measurement time 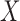 and stratifying variables as covariates, and fit to data on subjects without observed events
and stratifying variables as covariates, and fit to data on subjects without observed events 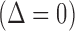 .
.
5.2. Simulation results
For CCH designs, longitudinal summaries performed well overall across various 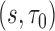 , with negligible bias and the estimated standard errors (SE
, with negligible bias and the estimated standard errors (SE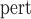 ) close to the empirical standard errors (SE
) close to the empirical standard errors (SE ), with the coverage probability (CP) close to the nominal 95% (Table 1). The most challenging simulation scenario was for
), with the coverage probability (CP) close to the nominal 95% (Table 1). The most challenging simulation scenario was for 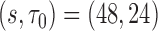 , with sample size at baseline increasing from
, with sample size at baseline increasing from 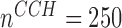 per group to
per group to 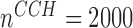 per group. This scenario was challenging because the effective sample sizes at
per group. This scenario was challenging because the effective sample sizes at 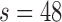 were substantially smaller than those at baseline. Up to 4% bias was seen in the small sample size simulation (
were substantially smaller than those at baseline. Up to 4% bias was seen in the small sample size simulation (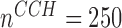 ), SE
), SE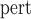 tended to be overestimated with CP ranging from 92.2% ot 99.1%. This is especially the case for PCF(0.2), the proportion of cases captured if 20% of the subjects at highest risk were to be followed. There was up to 4% bias in scenarios with small effective sample sizes, and the CP as high as 99% (top panel, Table 1). This is not surprising, as such a measure is relatively more closely related to the number of events occurring between
tended to be overestimated with CP ranging from 92.2% ot 99.1%. This is especially the case for PCF(0.2), the proportion of cases captured if 20% of the subjects at highest risk were to be followed. There was up to 4% bias in scenarios with small effective sample sizes, and the CP as high as 99% (top panel, Table 1). This is not surprising, as such a measure is relatively more closely related to the number of events occurring between 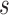 and
and 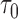 and can only be estimated well with sufficient effective sample sizes. This highlights the unique phenomenon in longitudinal studies, where the estimation varies with both specific landmark times
and can only be estimated well with sufficient effective sample sizes. This highlights the unique phenomenon in longitudinal studies, where the estimation varies with both specific landmark times 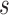 and prediction time
and prediction time 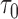 . As the sample size increased to
. As the sample size increased to 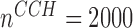 , the SE
, the SE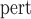 converged to within 0.001 of those estimated empirically (SE
converged to within 0.001 of those estimated empirically (SE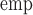 ), bias was 1.6% or lower, and the CP ranged from 94.6% to 96.3% (bottom panel, Table 1).
), bias was 1.6% or lower, and the CP ranged from 94.6% to 96.3% (bottom panel, Table 1).
Table 1.
Case-cohort n events/n non-events 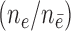 are shown in table headings, iterations = 1000, perturbations = 500,
are shown in table headings, iterations = 1000, perturbations = 500, 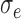 = 0.1, and
= 0.1, and 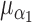 =
=  1.0
1.0
Case-cohort, iterations = 1000, perturbations = 500, 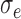 = 0.1, = 0.1, 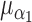 = =  1.0 1.0 | ||||||
|---|---|---|---|---|---|---|
n = 250/250, s = 48, 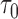 = 24, = 24, 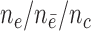 = 43/25/21 = 43/25/21 |
||||||
| True | Est | Bias% | SE
|
SE
|
CP% | |
| PE | 0.168 | 0.171 | 1.9 | 0.025 | 0.025 | 95.3 |
| TPF(0.4) | 0.936 | 0.929 | 0.7 | 0.050 | 0.054 | 92.2 |
| FPF(0.3) | 0.777 | 0.764 | 1.6 | 0.109 | 0.114 | 93.0 |
| AUC | 0.813 | 0.787 | 3.1 | 0.056 | 0.059 | 94.5 |
| PCF(0.2) | 0.297 | 0.309 | 4.0 | 0.030 | 0.040 | 99.1 |
| PNF(0.8) | 0.640 | 0.647 | 1.1 | 0.055 | 0.064 | 96.0 |
n = 2000/2000, s = 48,  = 24, = 24, 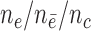 = 342/200/166 = 342/200/166 |
||||||
| PE | 0.168 | 0.169 | 0.3 | 0.008 | 0.008 | 96.3 |
| TPF(0.4) | 0.936 | 0.932 | 0.4 | 0.018 | 0.018 | 94.6 |
| FPF(0.3) | 0.777 | 0.765 | 1.6 | 0.039 | 0.040 | 95.6 |
| AUC | 0.813 | 0.808 | 0.5 | 0.018 | 0.019 | 96.1 |
| PCF(0.2) | 0.297 | 0.298 | 0.3 | 0.010 | 0.011 | 96.3 |
| PNF(0.8) | 0.640 | 0.642 | 0.2 | 0.019 | 0.020 | 96.2 |
PE = prediction error; TPF(0.4) = true positive fraction at 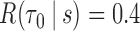 ; FPF(0.3) = false positive fraction at
; FPF(0.3) = false positive fraction at 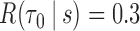 ; AUC = area under the ROC curve; PCF(0.2) = the proportion of events in
; AUC = area under the ROC curve; PCF(0.2) = the proportion of events in 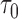 timeframe from
timeframe from 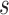 captured if 20% of the population at risk at time
captured if 20% of the population at risk at time 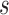 were followed; PNF(0.8) = the proportion of the population at risk at time
were followed; PNF(0.8) = the proportion of the population at risk at time 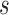 that would need to be followed in order to capture 80% of the events in the timeframe
that would need to be followed in order to capture 80% of the events in the timeframe 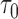 from
from 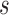 . Effective sample sizes used to estimate the performance measures are summarized in the table as
. Effective sample sizes used to estimate the performance measures are summarized in the table as 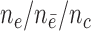 , where
, where 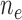 denotes the number of events observed in a
denotes the number of events observed in a 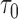 timeframe from
timeframe from 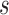 ,
, 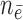 denotes the number of subjects still at risk at time
denotes the number of subjects still at risk at time 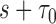 and
and 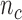 is the number of subjects who were censored between
is the number of subjects who were censored between 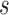 and
and 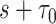 .
.
The results from NCC designs (Table 2) echo those of the CCH design. Recall that for NCC samples, rather than using true sampling fraction we estimated the sampling weights, 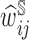 , using a GAM. We note that the model provided adequate approximation of
, using a GAM. We note that the model provided adequate approximation of 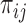 , the sampling probability, suggesting that a flexible modeling approach can be used in practical situations when the true sampling weights may not be ascertained reliably.
, the sampling probability, suggesting that a flexible modeling approach can be used in practical situations when the true sampling weights may not be ascertained reliably.
Table 2.
Nested case–control study simulation results showing convergence with increasing sample size, iterations = 1000, perturbations = 500, 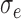 = 0.1, and
= 0.1, and 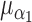 =
=  1.0
1.0
Nested case–control, iterations = 1000, perturbations = 500, 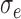 = 0.1, = 0.1, 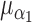 = =  1.0 1.0 | ||||||
|---|---|---|---|---|---|---|
Sampled from n = 500, s = 48, 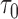 = 24, = 24, 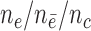 = 58/27/11 = 58/27/11 |
||||||
| True | Est | Bias % | SE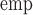
|
SE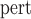
|
CP % | |
| PE | 0.168 | 0.171 | 1.6 | 0.021 | 0.021 | 95.5 |
| TPF(0.4) | 0.936 | 0.929 | 0.7 | 0.044 | 0.046 | 91.2 |
| FPF(0.3) | 0.777 | 0.767 | 1.2 | 0.107 | 0.105 | 91.2 |
| AUC | 0.813 | 0.780 | 4.1 | 0.050 | 0.055 | 95.2 |
| PCF(0.2) | 0.297 | 0.306 | 2.8 | 0.026 | 0.032 | 98.0 |
| PNF(0.8) | 0.640 | 0.650 | 1.6 | 0.051 | 0.055 | 94.1 |
Sampled from n = 4000, s = 48, 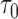 = 24, = 24, 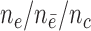 = 462/219/85 = 462/219/85 |
||||||
| PE | 0.168 | 0.169 | 0.6 | 0.007 | 0.007 | 96.4 |
| TPF(0.4) | 0.936 | 0.935 | 0.1 | 0.015 | 0.015 | 94.1 |
| FPF(0.3) | 0.777 | 0.777 | 0.0 | 0.037 | 0.037 | 94.2 |
| AUC | 0.813 | 0.806 | 0.8 | 0.017 | 0.017 | 95.7 |
| PCF(0.2) | 0.297 | 0.298 | 0.2 | 0.009 | 0.009 | 94.8 |
| PNF(0.8) | 0.640 | 0.642 | 0.3 | 0.017 | 0.018 | 94.6 |
PE = prediction error; TPF(0.4) = true positive fraction at 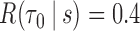 ; FPF(0.3) = false positive fraction at
; FPF(0.3) = false positive fraction at 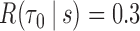 ; AUC = area under the ROC curve; PCF(0.2) = the proportion of events in
; AUC = area under the ROC curve; PCF(0.2) = the proportion of events in 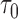 timeframe from
timeframe from 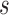 captured if 20% of the population at risk at time
captured if 20% of the population at risk at time 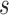 were followed; PNF(0.8) = the proportion of the population at risk at time
were followed; PNF(0.8) = the proportion of the population at risk at time 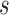 that would need to be followed in order to capture 80% of the events in the timeframe
that would need to be followed in order to capture 80% of the events in the timeframe 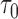 from
from 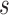 . Effective sample sizes used to estimate the performance measures are summarized in the table as
. Effective sample sizes used to estimate the performance measures are summarized in the table as 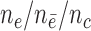 , where
, where 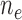 denotes the number of events observed in a
denotes the number of events observed in a 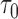 timeframe from
timeframe from 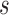 ,
, 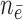 denotes the number of subjects still at risk at time
denotes the number of subjects still at risk at time 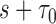 and
and 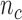 is the number of subjects who were censored between
is the number of subjects who were censored between 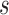 and
and 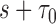 .
.
For both CCH and NCC, all performance measures performed well under reasonable sample sizes, with small bias and good CP.
6. Real data example: HALT-C NCC study
6.1. Description of the HALT-C dataset
The HALT-C trial consisted of 1002 patients with chronic hepatitis C and bridging fibrosis or cirrhosis, who failed to respond or to achieve a sustained virologic response to 20 weeks of combination therapy. Those patients were randomized at 24 weeks to treatment with peginterferon-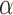 -2a or control (no treatment) and were followed every 3 months for 3.5 years after randomization. Blood samples were collected at each visit for subsequent research testing including assays for HCC biomarkers. To ascertain HCC, ultrasound examinations were performed 6 months after enrollment and every 12 months thereafter. Patients with an elevated or rising AFP, the currently most commonly used marker for detecting HCC, and those with new lesions on ultrasound were evaluated further by computed tomography or magnetic resonance imaging. One of the goals of the HALT-C trial was to identify and validate markers for the surveillance and early diagnosis of HCC. The marker of interest was DCP.
-2a or control (no treatment) and were followed every 3 months for 3.5 years after randomization. Blood samples were collected at each visit for subsequent research testing including assays for HCC biomarkers. To ascertain HCC, ultrasound examinations were performed 6 months after enrollment and every 12 months thereafter. Patients with an elevated or rising AFP, the currently most commonly used marker for detecting HCC, and those with new lesions on ultrasound were evaluated further by computed tomography or magnetic resonance imaging. One of the goals of the HALT-C trial was to identify and validate markers for the surveillance and early diagnosis of HCC. The marker of interest was DCP.
A NCC study was used to evaluate the accuracy of DCP in the detection of HCC. For this study, 39 HCC cases diagnosed between randomization and 3.8 years after randomization were included in the study. For each case, two controls without HCC at the time of diagnosis of the case were selected matching on treatment assignment, presence of cirrhosis on baseline biopsy and length of followup. One control was later excluded because of high DCP values due to caumadin (anticoagulant) use, leaving 77 controls. DCP values played no role in diagnosis of HCC.
6.2. Analysis of the HALT-C dataset
To estimate the NCC sampling weights, we fit a GAM with a logit link function to the full trial data at baseline (n = 1002) with a binary indicator of inclusion in the NCC study as the outcome, a smoothing spline function of the event time (df = 4), adjusting for event status and covariates used for stratified sampling into the NCC sample: cirrhosis (binary) and treatment group assignment (binary). Given the small sample size of the NCC cohort, estimating the inverse probability weights may have the additional advantage of improving efficiency compared to a TIPW approach. The sampling weights, 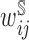 , were estimated as the inverse of the fitted values from the GAM. The censoring weights,
, were estimated as the inverse of the fitted values from the GAM. The censoring weights, 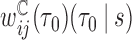 , were estimated using a Kaplan-Meier estimator of the censoring distribution. Since individuals were followed by the protocol of HALT-C trial, the assumption that censoring is not dependent on other covariates seems to be reasonable here. In order to calculate DCP-based absolute dynamic risk
, were estimated using a Kaplan-Meier estimator of the censoring distribution. Since individuals were followed by the protocol of HALT-C trial, the assumption that censoring is not dependent on other covariates seems to be reasonable here. In order to calculate DCP-based absolute dynamic risk 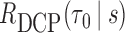 , we fit a DIPW based
, we fit a DIPW based 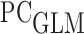 model as described in Equation 3.3, with
model as described in Equation 3.3, with 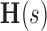 = (DCP value measured at
= (DCP value measured at 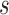 ,
, 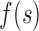 ), and
), and 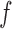 is a smooth spline function with df = 3. We then used procedures described in sections 3.3 to estimate the performance of
is a smooth spline function with df = 3. We then used procedures described in sections 3.3 to estimate the performance of 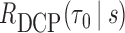 at selected pairs of
at selected pairs of 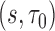 . For inference, the standard errors of the estimates were estimated by the empirical standard deviation of the corresponding
. For inference, the standard errors of the estimates were estimated by the empirical standard deviation of the corresponding 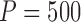 perturbed estimates.
perturbed estimates.
6.3. Results of analysis of the HALT-C NCC study dataset
There were 1002 subjects enrolled in the HALT-C clinical trial, with 39 subjects with events and 77 controls selected into the NCC study. The baseline characteristics of all subjects in the trial and in the NCC study are summarized in Table 3. The mean age of the subjects selected into the NCC study was 51.7 years, 22% were female, 60% were white and 57% of the subjects had cirrhosis of the liver. The data in the NCC study is summarized in Figures S1 and S2 of the supplementary material available at Biostatistics online. In the Figure S1 of the supplementary material available at Biostatistics online, we show the attended visits (circles), event times (filled circles) and censoring times (filled triangles). The subjects are grouped by their risk set with each color corresponding to a given risk set. The inverse probability weights estimated using the generalized additive model are shown to the right of the event or censoring indicators for each individual. In Figure S2 of the supplementary material available at Biostatistics online, we show the marker trajectories for all individuals in the NCC study, stratified by event status (diagnosed with hepatocellular carcinoma during the study vs. not), cirrhosis of the liver and treatment group assignment.
Table 3.
Baseline characteristics of patients randomized into the HALT-C clinical trial, and those of selected into the nested case–control study (NCC) at 3.8 years after randomization
| HALT-C | HALT-C NCC | |
|---|---|---|
| (n = 1002) | (n = 116) | |
| Age at randomization, mean (SD) | 50.2 (7.2) | 51.7 (7.5) |
| Female, n (%) | 289 (28.8) | 26 (22.4) |
| Race/ethnicity, n (%) | ||
| White | 717 (71.6) | 70 (60.3) |
| Black | 183 (18.3) | 36 (31.0) |
| Hispanic | 79 (7.9) | 7 (6.0) |
| Other | 23 (2.3) | 3 (2.6) |
| Drinks (per week), mean (SD) | 9.6 (16.4) | 9.5 (14.5) |
| DCP (log2), mean (SD) | 4.9 (0.9) | 4.9 (1.0) |
| Cirrhosis present, n (%) | 408 (40.7) | 66 (56.9) |
| Randomized to treatment, n (%) | 507 (50.6) | 59 (50.9) |
| HCC diagnosed, n (%) | 39 (3.9) | 39 (33.6) |
The prediction evaluation results are summarized in Table 4. The prediction performance of DCP in predicting the timeframe of diagnosis with HCC was good overall. The prediction estimates were especially notable for 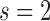 and
and 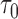 = 1 year prediction timeframe, during which 11 events were observed in the subcohort. The AUC was estimated (standard error) at 0.86 (0.07), with 82% of the events estimated to be captured if 20% of subjects at highest risk were to be followed. That means that we estimate that nine subjects who would progress to HCC within 1 year would be captured if we followed 21 subjects with highest estimated risks who are still at risk of HCC at 2 years after randomization. For
= 1 year prediction timeframe, during which 11 events were observed in the subcohort. The AUC was estimated (standard error) at 0.86 (0.07), with 82% of the events estimated to be captured if 20% of subjects at highest risk were to be followed. That means that we estimate that nine subjects who would progress to HCC within 1 year would be captured if we followed 21 subjects with highest estimated risks who are still at risk of HCC at 2 years after randomization. For 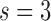 years and
years and 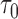 = 1 year, we observed 17 events, with the AUC estimated at 0.75 (0.08), PCF(0.2) = 0.65 (0.12), and PNF(0.8) was 0.52 (0.19). We note that the numbers of events our estimates were based on were generally small, but these results are promising and warrant further investigation into the evaluation of DCP as a biomarker for predicting HCC.
= 1 year, we observed 17 events, with the AUC estimated at 0.75 (0.08), PCF(0.2) = 0.65 (0.12), and PNF(0.8) was 0.52 (0.19). We note that the numbers of events our estimates were based on were generally small, but these results are promising and warrant further investigation into the evaluation of DCP as a biomarker for predicting HCC.
Table 4.
Estimates (EST) and standard errors (ESD) of measures of predictive capacity summarizing predictions of hepatocellular carcinoma based on des-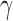 -carboxyprothrombin biomarker and a partly conditional logistic model with the logit link function (
-carboxyprothrombin biomarker and a partly conditional logistic model with the logit link function (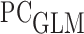 )
)
HALT-C NCC study prediction evaluation results (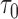 = 12 months) = 12 months) | ||||
|---|---|---|---|---|
| s = 6 months | s = 1 year | s = 2 years | s = 3 years | |
(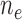 / /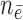 = 4/109) = 4/109) |
(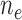 / /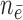 = 5/105) = 5/105) |
(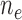 / /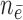 = 11/94) = 11/94) |
(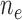 / /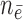 = 17/76) = 17/76) |
|
| EST (ESD) | EST (ESD) | EST (ESD) | EST (ESD) | |
PE(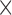 10) 10) |
0.042 (0.021) | 0.055 (0.024) | 0.107 (0.035) | 0.214 (0.056) |
| AUC | 0.709 (0.125) | 0.782 (0.121) | 0.858 (0.067) | 0.748 (0.077) |
| PCF(0.2) | 0.500 (0.223) | 0.600 (0.207) | 0.818 (0.112) | 0.646 (0.120) |
| PNF(0.8) | 0.637 (0.197) | 0.747 (0.285) | 0.147 (0.210) | 0.523 (0.192) |
PE(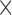 10) = prediction error
10) = prediction error 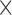 10; AUC = area under the ROC curve; PCF(0.2) = the proportion of events in
10; AUC = area under the ROC curve; PCF(0.2) = the proportion of events in 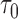 timeframe from
timeframe from 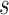 captured if 20% of the population at risk at time
captured if 20% of the population at risk at time 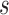 were followed; PNF(0.8) = the proportion of the population at risk at time
were followed; PNF(0.8) = the proportion of the population at risk at time 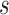 that would need to be followed in order to capture 80% of the events in the timeframe
that would need to be followed in order to capture 80% of the events in the timeframe 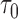 from
from 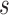 . The estimates were obtained
. The estimates were obtained 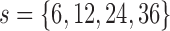 months and
months and 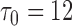 months. The number of events between
months. The number of events between 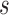 and
and 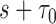 , and the number of subjects with no events before
, and the number of subjects with no events before 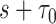 , are denoted by
, are denoted by 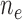 and
and 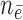 , respectively. The standard errors were estimated with 500 perturbations.
, respectively. The standard errors were estimated with 500 perturbations.
7. Discussion
In this article, we presented non-parametric estimation and inference of longitudinal accuracies under two-phase study designs for several measures of prediction performance. We evaluated the performance of our estimators using extensive simulation studies and illustrated them on a NCC study of a longitudinal biomarker within the HALT-C clinical trial. Our estimators perform well overall, showing little or no bias and achieving nominal coverage probabilities for reasonable sample sizes. Our methods provide investigators with a useful tool for evaluating preliminary longitudinal markers for active surveillance in practice. We note, that in very small samples one may encounter some bias and conservative standard errors. In practice, it is prudent to inspect the number of observed events and nonevents over the prediction window between 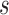 and
and 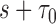 , especially in the longitudinal setting with later measurement time
, especially in the longitudinal setting with later measurement time 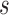 , as more individuals drop out since baseline, and two-phase sampling also further limits the number of individuals at risk at the prediction time. Simulations based on preliminary data might be helpful to determine if samples are sufficient for stable estimates. The development of the estimation and inference procedures under two-phase studies built on those developed under the cohort study design. Estimators constructed in this way are, we hope, intuitive and practical. Thus, they can be extended to other, possibly more complex, study designs and applications, thus providing an arsenal of practical, robust and flexible methods for risk prediction and evaluation of predictions in a wide variety of applications. For example, due to sample size limitation we only considered the predictive values of a single biomarker (DCP) in the HALT-C example. However, our methods can be readily adapted to evaluate a panel of multiple biomarkers measured either only for the second phase subjects or for the full cohort, as well as settings to evaluate the incremental predictive value of newly discovered markers over the established ones.
, as more individuals drop out since baseline, and two-phase sampling also further limits the number of individuals at risk at the prediction time. Simulations based on preliminary data might be helpful to determine if samples are sufficient for stable estimates. The development of the estimation and inference procedures under two-phase studies built on those developed under the cohort study design. Estimators constructed in this way are, we hope, intuitive and practical. Thus, they can be extended to other, possibly more complex, study designs and applications, thus providing an arsenal of practical, robust and flexible methods for risk prediction and evaluation of predictions in a wide variety of applications. For example, due to sample size limitation we only considered the predictive values of a single biomarker (DCP) in the HALT-C example. However, our methods can be readily adapted to evaluate a panel of multiple biomarkers measured either only for the second phase subjects or for the full cohort, as well as settings to evaluate the incremental predictive value of newly discovered markers over the established ones.
The validity of our proposed estimators depends on the assumptions we make. The DIPW estimators require correctly specifying both the sampling weights and the censoring weights. In addition, we assume that longitudinal measurement times are fixed by research protocol. If they vary by measurement time during surveillance, our estimators will potentially be biased. In addition, when the follow up visit times are correlated with the outcome or other covariate information, our estimating equation based procedure will produce biased estimates. A possible solution would be to consider a class of inverse intensity-of-visit process-weighted procedures as proposed in Lin and others (2004). This would be a natural extension under the current IPW framework.
We considered only two-phase sampling designs where individuals at baseline were selected and all their measurements were available in the second phase, as implemented in the HALT-C NCC study. When sampling individuals from a full cohort at baseline, the effective sample sizes will vary depending on the specific timeframes of the conditioning time, 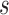 , and the prediction timeframe,
, and the prediction timeframe, 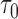 . Investigation into more efficient sampling designs, such as sampling of longitudinal observations within a given individual, are warranted in the future, especially for markers that are expensive to measure.
. Investigation into more efficient sampling designs, such as sampling of longitudinal observations within a given individual, are warranted in the future, especially for markers that are expensive to measure.
Supplementary Material
Acknowledgments
Conflict of Interest: None declared.
Funding
This work was supported by the National Institutes of Health [U01-CA86368, P01-CA053996, R01-GM085047, R01-GM079330].
References
- Blanche P., Proust-Lima C., Loubère L., Berr C., Dartigues J.-F. and Jacqmin-Gadda H. (2015). Quantifying and comparing dynamic predictive accuracy of joint models for longitudinal marker and time-to-event in presence of censoring and competing risks. Biometrics 71, 102–113. [DOI] [PubMed] [Google Scholar]
- Borgan O., Langholz B., Samuelsen S. O., Goldstein L. and Pogoda J. (2000). Exposure stratified case-cohort designs. Lifetime Data Analysis 6, 39–58. [DOI] [PubMed] [Google Scholar]
- Breslow N. E., Lumley T., Ballantyne C. M., Chambless L. E. and Kulich M. (2009). Using the whole cohort in the analysis of case-cohort data. American Journal of Epidemiology, 1398–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow N. E. and Wellner J. A. (2007). Weighted likelihood for semiparametric models and two-phase stratified samples, with application to cox regression. Scandinavian Journal of Statistics 34, 86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T. and Zheng Y. (2012). Evaluating prognostic accuracy of biomarkers in nested case-control studies. Biostatistics Biostatistics 13, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T. and Zheng Y. (2013). Resampling procedures for making inference under nested case-control studies. Journal of the American Statistical Association 108, 1532–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. and Lo S. K. (1999). Case-cohort and case-control analysis with cox’s model. Biometrika 86, 755. [Google Scholar]
- Gu W. and Pepe M. (2009). Measures to summarize and compare the predictive capacity of markers. International Journal of Biostatistics 5, Article 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. (2014). Bootstrap for the case-cohort design. Biometrika 101, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Scharfstein D. O. and Rosenheck R. A. (2004). Analysis of longitudinal data with irregular, outcome-dependent follow-up. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 66, 791–813. [Google Scholar]
- Liu D., Cai T. and Zheng Y. (2012). Evaluating the predictive value of biomarkers with stratified case-cohort design. Biometrics 68, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Lok A. S., Sterling R. K., Everhart J. E., Wright E. C., Hoefs J. C., Di Bisceglie A. M., Morgan T. R., Kim H.-Y., Lee W. M., Bonkovsky H. L..
and others (2010). Des-
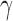 -carboxy prothrombin and
-carboxy prothrombin and  -fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology 138, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology 138, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar] - Pfeiffer R. M. and Gail M. H. (2011). Two criteria for evaluating risk prediction models. Biometrics 67, 1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice R. L. (1986). A case-cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika 73, 1–11. [Google Scholar]
- Qi L., Wang C. Y. and Prentice R. L. (2005). Weighted estimators for proportional hazards regression with missing covariates. Journal of the American Statistical Association 100, 1250–1263. [Google Scholar]
- Robins J. M., Rotnitzky A. and Zhao L. P. (1994). Estimation of regression coefficients when some regressors are not always observed. Journal of the American statistical Association 89, 846–866. [Google Scholar]
- Schoop R., Graf E. and Schumacher M. (2008). Quantifying the predictive performance of prognostic models for censored survival data with time-dependent covariates. Biometrics 64, 603–610. [DOI] [PubMed] [Google Scholar]
- Thomas D. C. (1977). Addendum to “Methods of cohort analysis: appraisal by application to asbestos mining”. Journal of the Royal Statistical Society, Series A, General 140, 483–485. [Google Scholar]
- Zheng Y. and Heagerty P. J. (2007). Prospective accuracy for longitudinal markers. Biometrics 63, 332–341. [DOI] [PubMed] [Google Scholar]
- Zheng Y. Y. and Heagerty P. J. (2004). Semiparametric estimation of time-dependent roc curves for longitudinal marker data. Biostatistics 5, 615–632. [DOI] [PubMed] [Google Scholar]
- Zheng Y. Y. and Heagerty P. J. (2005). Partly conditional survival models for longitudinal data. Biometrics 61, 379–391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


