Abstract
Objective triage strategies are required to prevent unnecessary referrals for colposcopy in population‐based screening programs using primary high‐risk human papillomavirus (hrHPV) testing. We have identified several DNA methylation markers with high sensitivity and specificity for detection of high‐grade cervical intraepithelial neoplasia or worse (CIN2+) in women referred for colposcopy. Our study assessed diagnostic potential of these methylation markers in a hrHPV‐positive screening cohort. All six markers (JAM3, EPB41L3, C13orf18, ANKRD18CP, ZSCAN1 and SOX1) showed similar association across histology in the hrHPV‐positive cohort when compared to the Dutch cohort (each p > 0.15). Sensitivity for CIN2+ was higher using methylation panel C13orf18/EPB41L3/JAM3 compared to the other 2 panels (80% vs. 60% (ANKRD18CP/C13orf18/JAM3) and 63% (SOX1/ZSCAN1), p = 0.01). For CIN3+ all three methylation panels showed comparable sensitivity ranging from 68% (13/19) to 95% (18/19). Specificity of SOX1/ZSCAN1 panel (84%, 167/200) was considerably higher compared to ANKRD18CP/C13orf18/JAM3 (68%, 136/200, p = 2 × 10−5) and C13orf18/EPB41L3/JAM3 (66%, 132/200, p = 2 × 10−7). High negative predictive value (NPV) (91–95% and 96–99%) was observed for CIN2+ and CIN3+, for all three methylation panels, while positive predictive value (PPV) varied from 25 to 40% for CIN2+ and 15–27% for CIN3+. Interestingly, 118/235 samples were negative for all six markers (including 106 controls (89.8%), 6 CIN1 (5.1%), 5 CIN2 (4.2%) and 1 CIN3 (0.8%)). Methylation results from both independent cohorts were comparable as well as high sensitivity for detection of cervical cancer and its high‐grade precursors in hrHPV‐positive population. Our study therefore validates these methylation marker panels as triage test either in hrHPV‐based or abnormal cytology‐based screening programs.
Keywords: (pre)malignant cervical cancer, DNA methylation markers, high‐risk human papillomavirus (hrHPV), cervical cancer screening, triage test
Short abstract
What's new?
In cervical cancer screening, HPV testing provides greater sensitivity than cytology, but its lower specificity leads to some unnecessary treatment referrals. DNA methylation assays could potentially provide better specificity for identifying CIN2+ in women carrying high risk HPV. These authors investigated six previously identified CIN2+ methylation markers. They tested three different combinations of markers, and found high levels of sensitivity and specificity, making these markers potentially useful as part of a population‐based screening program.
Introduction
Population‐based screening programs for the early detection of cervical cancer using cytology have drastically reduced the mortality of cervical cancer.1, 2 Yet, the clinical sensitivity and specificity of cytology leaves room for improvement.3, 4, 5 The proof of a causal relationship between persistent infection of cervical epithelium with high‐risk human papillomavirus (hrHPV) and cervical cancer6, 7 coupled with the development of reliable and sensitive tests for the detection of hrHPV have led to new screening algorithms with higher clinical sensitivity for detection of cervical cancer and its high‐grade precursor lesions.8, 9, 10 For example, implementation of hrHPV triage already resulted in more accurate referrals for colposcopy compared to repeat cytology after 6 months.11, 12 Since hrHPV testing increases the sensitivity for detection of underlying cervical intraepithelial neoplasia grade 2 or worse (CIN2+) and provides better protection against invasive cervical cancer compared to cytology, several countries/regions recently switched toward primary hrHPV‐based screening programs.5, 13 However, due to the lower specificity of the hrHPV testing compared to cytology, efficient strategies are needed to avoid unnecessary referral of women without disease and overtreatment of hrHPV positive women. A subsequent cytological test would decrease the number of false‐positive referrals with approximately 60% after a primary hrHPV‐positive test result.14, 15, 16, 17, 18, 19, 20 However, this strategy also has several disadvantages due to the subjective interpretation by the cytopathologist after awareness of the patient's HPV status.14, 15, 16, 17, 18, 19, 20 Therefore, objective triage strategies are needed to improve the specificity and to reduce unnecessary referrals in hrHPV‐based screening programs.
Over the course of several years we sought to develop and validate sensitive and specific DNA methylation assays in order to further improve the early detection of cervical cancer.21, 22, 23, 24, 25, 26 In cohorts of women referred to the outpatient clinic with an abnormal cytology result, a combination of specific methylation markers as a triage method showed similar sensitivity of ~75% for detection of CIN2+ compared to hrHPV testing, but better specificity. Furthermore, in a subgroup analysis of only hrHPV‐positive samples these combinations of methylation markers showed similar sensitivity and specificity to detect CIN2+ compared to the total group of samples.24, 25 Other studies also demonstrated high clinical sensitivity and specificity for CIN2+ using methylation analysis as a triage test after hrHPV testing, performing similar or better than cytology.18, 27, 28, 29 However, most studies were limited to analysing clinical specimens obtained from cohorts that were primarily screened with cytology rather than with hrHPV test or generated from nonresponding women, which both might not reflect well women participating in population‐based screening programs using primary hrHPV testing.
The purpose of the present study was to assess the diagnostic potential of our previously established CIN2+ specific methylation markers (i.e., sensitivity, specificity and positive and negative predictive value (PPV and NPV)) in hrHPV positive women attending the routine national cervical cancer screening program with high‐coverage in Slovenia. The diagnostic performance was additionally compared to previously acquired data from a Dutch cohort, which was primarily used to validate our methylation markers.25, 26
Methods
Population selection
Women selected for our study participated in the “Slovenian HPV Prevalence Study”, a prospective population‐based cohort study which started in 2009/2010 (13). Women were eligible for inclusion if they were attending the routine organised Slovenian national cervical cancer screening program. The exclusion criteria were attendance for a gynaecological examination after an atypical/abnormal cytology result, history of CIN of any grade or treatment for cervical disease in the preceding year, hysterectomy, and menstruation or pregnancy at presentation (10). For the current study, women were eligible if they tested positive in the Abbott RealTime High Risk HPV assay. A total of 262 women were included; 206 women ≥30 years old were not diagnosed with CIN2+ neither in the first nor in the second screening round (195 controls and 11 CIN1 cases) and 56 women who had histologically confirmed CIN2+ in the first screening round (38/56 were ≥ 30 years old).9, 10 The “Slovenian HPV Prevalence Study” was conducted in accordance with the Helsinki Declaration and was approved by the National Medical Ethics Committee at the Slovenian Ministry of Health.10 Written standardised informed consent was obtained from all of the women by the participating gynaecologists, and patient identities were kept secret from all study participants except the participating gynaecologists.
The data obtained from the Slovenian cohort (age ≥ 30 years) were compared to previously acquired data from our Dutch cytology‐based cohort25, 26 (unpublished data). This cohort (n = 278, age ≥ 30 years) comprised of listwise complete methylation data (i.e., only those samples were included with no missing values) on cervical scrapings from 48 women without any cervical abnormality, who were treated for nonmalignant indications and had no history of abnormal Pap smears (controls), as well as 230 scrapings obtained from women who had been referred to the outpatient clinic with an abnormal Pap smear (histology: 32 without CIN (controls), 37 CIN1, 49 CIN2, 66 CIN3 and 46 (microinvasive) cervical cancers).
DNA isolation and bisulphite treatment
DNA was isolated from 1.5 ml aliquots of ThinPrep material that had been stored at −80 °C.10 Cells were pelleted at 20,000 × G for 1 min at room temperature and were lysed overnight in a Tris‐buffered (pH = 8) solution with 1% SDS and 0.66 mg/ml proteinase K at 56 °C. DNA was extracted with 2.3 volumes of phenol:chloroform:isoamylalcohol (25:24:1) and Phase Lock Heavy tubes (5Prime, Hilden, Germany). DNA was subsequently precipitated in 0.65 M ammonium acetate, 65% ethanol and 0.035 mg/ml glycogen at −20 °C for at least 1 h; centrifuged for 1 h at 20,000 × G at 4 °C; washed with 70% ethanol and finally reconstituted in TE buffer. Quality of the DNA was routinely assessed using a multiplex PCR.30 DNA concentrations were determined with Nanodrop ND‐1000 (Thermo Fisher Scientific, Waltham, MA, USA). One microgram (or at least 0.5 μg) DNA was treated with sodium bisulphite as per manufacturer's instructions (Zymo Research, Irvine, CA, USA) and eluted to obtain 10 ng/μL. Samples were randomly distributed among DNA isolation batches and were again randomised across multiple bisulphite treatments.
Quantitative methylation‐specific PCR
Methylation analysis was performed for six markers (ANKRD18CP, C13orf18, EPB41L3, JAM3, SOX1 and ZSCAN1) as described previously,24, 25, 26 using ACTB as a methylation‐independent reference [31]. For each sample the methylation level was assessed in a randomised fashion and blinded from clinical data. Samples were transferred to 384 wells PCR plates using a robotic liquid handling system (JANUS, Perkin Elmer, Waltham, MA). Sodium bisulphite conversion of samples was repeated if the mean quantity of ACTB was below 1 ng per reaction. The relative level of methylation of the region of interest was determined by the after calculation: average quantity of the methylated region of interest divided by average quantity of the reference ACTB gene and multiplied by 10,000.31 For 4 markers (ANKRD18CP, C13orf18, EPB41L3, and JAM3) a sample was considered methylation positive when a methylation signal was observed with a C q below 50 in at least two of the three reaction wells.24, 25 A sample was considered methylation positive for SOX1 or ZSCAN1 if the methylation level was above a threshold of 19.1 or 132, respectively.26
We also assessed two marker panels with previously reported relative high sensitivity and specificity for CIN2+ in a population of women with abnormal cytology: panel 1) ANKRD18CP, C13orf18 and JAM3, 25 and panel 2) C13orf18, EPB41L3 and JAM3.26 In addition, we searched for novel panels of methylation markers for the most optimal combination with the highest sensitivity and specificity using exploratory data mining on methylation data obtained from previous studies, now also allowing thresholds above a certain methylation level (see Supporting Information File 1 for a detailed description), which resulted in panel 3) SOX1 and ZSCAN1. Marker panels labelled sample as positive if at least one of the markers within the panel was positive.
Statistical analysis
Statistical analyses were performed using R(version 3.3.2).32 The Jonckheere‐Terpstra test (using 1,000 permutations33) was employed as a nonparametric test for trends of methylation levels across severity of the underlying lesion. Cumulative logit models from the R package ordinal34 were applied to assess whether log10‐transformed methylation levels of each methylation marker associated equally well with cervical neoplasia among cohorts. The Dutch reference data was used both as pairwise complete as well in order to estimate the methylation test positivity across histological subgroups with maximal precision. Asymmetrical beanplots were used to visualise DNA methylation levels across histology and between study cohorts.35 Correlations between methylation levels and the age of the Slovenian women considered as controls (<CIN2) were assessed with Kendall's tau. Wilcoxon's signed‐rank test was used to compare Kendall's correlation matrices of methylation markers between the Slovenian and Dutch population. Pearson's Χ2 and Fisher's exact test were used to examine cross‐tabs. Paired comparisons of test‐positivity and of sensitivity and specificity were made using the McNemar test.36
Results
DNA methylation in hrHPV‐positive Slovenian women
DNA quality control showed that 97% (253/262) of sample aliquots yielded sufficient amount of high quality DNA to perform methylation analysis. Within this cohort of 253 Slovenian hrHPV‐positive women, higher methylation levels were consistently associated with the severity of the underlying lesion (p < 0.02 for each marker, Fig. 1).
Figure 1.
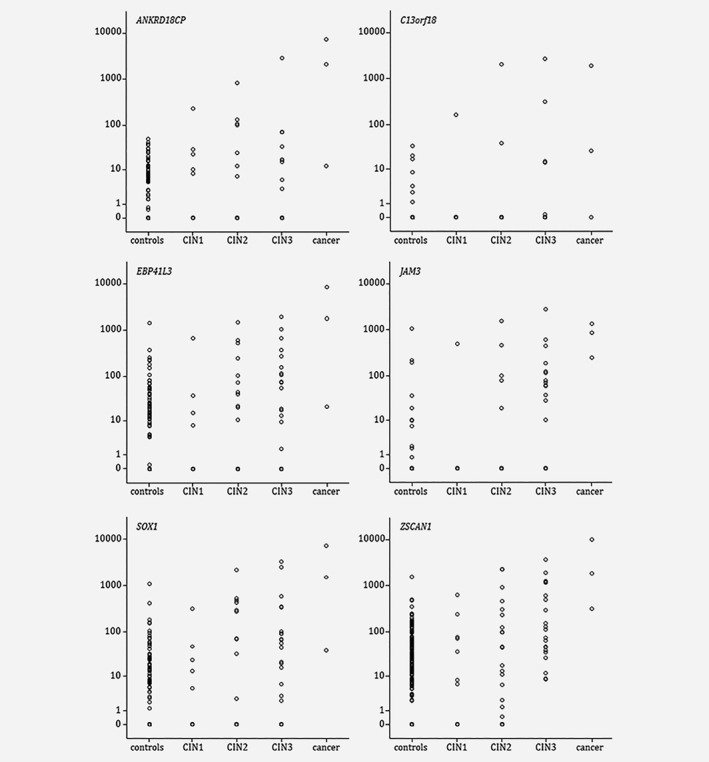
Methylation ratio for the six genes analysed by QMSP in Slovenian hrHPV‐positive scrapings obtained from women with normal cytology (controls), CIN1, CIN2, CIN3 or cancer.
The comparison of the methylation data obtained from the hrHPV‐based Slovenian cohort with the Dutch cytology‐based cohort on samples from women above 30 years (n = 235) revealed a high similarity of methylation level distributions across histology and between study populations (Fig. 2). Ordinal regression analysis confirmed the strength of the association of DNA methylation levels with the severity of the underlying lesions in both cohorts (each p > 0.15).
Figure 2.
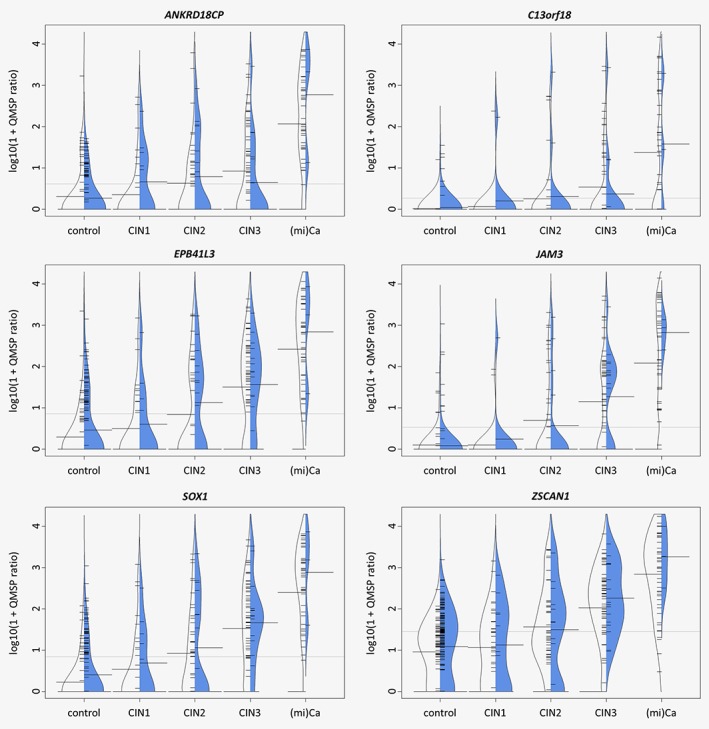
Asymmetric beanplots of methylation level distributions across histology and between study populations [Slovenian scrapings (n = 235) in blue and previously analysed Dutch scrapings (n = 278) in white]. Small horizontal bars indicate individual samples, large horizontal bars indicate the geometric mean methylation ratio per group and the dotted line signifies the grand geometric mean. [Color figure can be viewed at wileyonlinelibrary.com]
Ageing has been associated with gradual increase of methylation levels in genomic DNA.37, 38 To exclude that differences in methylation levels during progression of disease are due to age differences, we have performed the evaluation of the Slovenian hrHPV‐positive control group and showed no significant relationship between the age of control women (n = 189) and DNA methylation levels (Table 1).
Table 1.
Correlation of methylation levels with the age of hrHPV‐positive control women (n = 189, median age 37, range 30–64)
| T1 | p‐value | |
|---|---|---|
| ANKRD18CP | −0.094‐ | 0.099 |
| C13orf18 | −0.030‐ | 0.615 |
| EPB41L3 | −0.092‐ | 0.104 |
| JAM3 | −0.059‐ | 0.331 |
| SOX1 | −0.029‐ | 0.606 |
| ZSCAN1 | −0.057‐ | 0.270 |
Kendall's tau coefficient measuring the ordinal association between two measured quantities.
After dichotomisation of the methylation markers into positive or negative methylation, we observed that methylation positivity of each marker was positively associated with the severity of the underlying disease in hrHPV‐positive women (Table 2A). Marker JAM3 was the most significant marker with most controls and CIN1 samples negative, while almost all CIN3 and cancers were positive. Although marker EPB41L3 detected most of the CIN2+ lesions (25/35), also more controls and CIN1 samples were positive. Since in several countries population‐based screening starts before the age of 30, we have also analysed whether the positivity rate in CIN2+ was different in women below and over 30 years. Only for the markers ANKRD18CP and EPB41L3 lower positive rates were observed in women below 30 years with CIN2+ compared to women with CIN2+ aged 30 years or more (p < 0.05, Table 2B).
Table 2.
Test positivity of DNA methylation biomarkers in Slovenian hrHPV‐positive clinician‐collected cervical scrapings
| A | In women ≥ 30 years (n = 235) | B | In women aged < 30 years (n = 18) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| controls | CIN 1 | CIN 2 | CIN 3 | cancer | p‐Trend | CIN 2+ | CIN 3+ | ||||||||
| Individual markers | |||||||||||||||
| ANKRD18CP 1 | 27% | (51/189) | 45% | (5/11) | 44% | (7/16) | 38% | (6/16) | 100% | (3/3) | 0.011 | 0% | (0/12) | 50% | (3/6) |
| C13orf18 | 4% | (8/189) | 9% | (1/11) | 13% | (2/16) | 25% | (4/16) | 67% | (2/3) | 6 × 10−60 | 0% | (0/12) | 17% | (1/6) |
| EPB41L3 1 | 31% | (59/189) | 36% | (4/11) | 56% | (9/16) | 81% | (13/16) | 100% | (3/3) | 2 × 10−60 | 17% | (2/12) | 50% | (3/6) |
| JAM3 | 6% | (11/189) | 9% | (1/11) | 25% | (4/16) | 63% | (10/16) | 100% | (3/3) | 6 × 10−15 | 8% | (1/12) | 50% | (3/6) |
| SOX1 1 | 13% | (24/189) | 27% | (3/11) | 44% | (7/16) | 69% | (11/16) | 100% | (3/3) | 1 × 10−11 | 17% | (2/12) | 50% | (3/6) |
| ZSCAN1 1 | 12% | (22/189) | 18% | (2/11) | 25% | (4/16) | 50% | (8/16) | 100% | (3/3) | 2 × 10−70 | 17% | (2/12) | 33% | (2/6) |
| Methylation marker panels | |||||||||||||||
| ANKRD18CP/C13orf18/JAM3 | 31% | (59/189) | 45% | (5/11) | 50% | (8/16) | 63% | (10/16) | 100% | (3/3) | 3 × 10−10 | 8% | (1/12) | 67% | (4/6) |
| C13orf18/EPB41L3/JAM3 1 | 34% | (64/189) | 36% | (4/11) | 63% | (10/16) | 94% | (15/16) | 100% | (3/3) | 7 × 10−14 | 17% | (2/12) | 67% | (4/6) |
| SOX1/ZSCAN1 1 , 2 | 16% | (30/189) | 27% | (3/11) | 44% | (7/16) | 75% | (12/16) | 100% | (3/3) | 5 × 10−11 | 17% | (2/12) | 50% | (3/6) |
Significantly different positivity in CIN2+ between age groups (p < 0.05).
Using a threshold for positivity at a methylation ratio of 19.1 for SOX1 or 132 for ZSCAN1 (see methods).
Methylation marker panels for disease classification
To gain sensitivity without losing too much specificity for detection of CIN2+ or CIN3+, three marker panels were analysed, namely panel 1) ANKRD18CP/C13orf18/JAM3, panel 2) C13orf18/EPB41L3/JAM3 and panel 3) SOX1/ZSCAN1 (Table 3). Within hrHPV‐positive women above 30 years we observed significant differences in the sensitivity and specificity between panel 3 (SOX1/ZSCAN1) and the other two marker panels (both p < 10−4). Panel 1 (ANKRD18CP/C13orf18/JAM3) performed similarly to panel 2 (C13orf18/EPB41L3/JAM3) (p = 0.12). Further comparisons revealed that the sensitivity for CIN2+ of panel 2 (C13orf18/EPB41L3/JAM3. p = 0.01), but not for panel 1 (ANKRD18CP/C13orf18/JAM3, p = 0.65), was higher compared to panel 3 (SOX1/ZSCAN1). The sensitivity for CIN3+ of panel 3 (SOX1/ZSCAN1) was similar to the other two marker panels (both p > 0.08). Panel 3 (SOX1/ZSCAN1) demonstrated higher specificity for ≤CIN1 as well as ≤CIN2 compared to the other two panels (both p < 10−4). If we compare the sensitivity and specificity assessed on hrHPV Slovenian cohort to the Dutch women with cytologically abnormal samples, all marker panels showed comparable test positivity (Table S1). Addition of HPV16 and/or HPV18 status to the methylation marker panels minimally increased the sensitivity, but decreased specificity in all analysed panels (data not shown).
Table 3.
Key clinical performance indicators of individual markers and methylation marker panels on Slovenian HPV‐positive women (n = 235, age ≥ 30)
| CIN2+ versus ≤ CIN1 | CIN3+ versus ≤ CIN2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Sens | Spec | PPV | NPV | Sens | Spec | PPV | NPV | |
| (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | |
| Individual markers | ||||||||
| ANKRD18CP | 46% | 72% | 22% | 88% | 47% | 71% | 13% | 94% |
| (29.2–63.1) | (65.1–78.0) | (13.6–22.8) | (82.2–92.7) | (25.2–70.5) | (64.2–76.7) | (6.2–22.9) | (88.7–96.9) | |
| C13orf18 | 23% | 96% | 47% | 88% | 32% | 95% | 35% | 94% |
| (11.0–40.6) | (91.4–97.8) | (23.9–71.5) | (82.3–91.5) | (13.6–56.5) | (90.8–97.3) | (15.3–61.4) | (89.8–96.7) | |
| EPB41L3 | 71% | 69% | 28% | 93% | 84% | 67% | 18% | 98% |
| (53.4–84.8) | (61.5–74.8) | (19.6–39.2) | (87.5–96.5) | (59.5–95.8) | (59.9–72.8) | (11.1–28.1) | (93.7–99.5) | |
| JAM3 | 49% | 94% | 59% | 91% | 68% | 93% | 45% | 97% |
| (31.7–65.7) | (89.5–96.7) | (39.1–75.9) | (86.3–94.6) | (43.5–86.4) | (88.0–95.6) | (27.0–64.0) | (93.5–98.8) | |
| SOX1 | 60% | 87% | 44% | 93% | 74% | 84% | 29% | 97% |
| (42.2–75.6) | (80.8–90.8) | (29.7–58.7) | (87.5–95.7) | (48.6–89.9) | (78.6–88.7) | (17.4–44.3) | (93.5–99.0) | |
| ZSCAN1 | 43% | 88% | 38% | 90% | 58% | 87% | 28% | 96% |
| (26.8–60.5) | (82.5–92.0) | (23.8–55.3) | (84.5–93.5) | (34.0–78.9) | (81.6–91.1) | (15.6–45.1) | (91.8–98.1) | |
| Methylation marker panels | ||||||||
| ANKRD18CP/C13orf18/JAM3 | 60% | 68% | 25% | 91% | 68% | 67% | 15% | 96% |
| (42.2–75.6) | (57.3–69.9) | (16–3‐35.5) | (84.5–94.6) | (43.5–86.4) | (59.9–72.8) | (8.7–25.1) | (91.1–98.4) | |
| C13orf18/EPB41L3/JAM3 | 80% | 66% | 29% | 95% | 95% | 64% | 19% | 99% |
| (62.5–90.9) | (58.9–72.4) | (20.6–39.5) | (89.5–97.8) | (71.9–99.7) | (57.1–70.2) | (11.8–28.3) | (95.5–100.0) | |
| SOX1/ZSCAN1 | 63% | 84% | 40% | 93% | 79% | 81% | 27% | 98% |
| (44.9–78.0) | (77.5–88.2) | (27.3–54.1) | (87.7–95.9) | (53.9–93.0) | (75.6–86.3) | (16.5–41‐2) | (94.0–99.3) | |
Abbreviations: CIN, cervical intraepithelial neoplasia; Sens, sensitivity; Spec, specificity, PPV, positive predictive value; NPV, negative predictive value; 95% C, 95% confidence interval.
High negative predictive value (NPV) was observed for all methylation marker panels for both CIN2+ and CIN3+, namely 91–95% and 96–99%, respectively (Table 3). In contrast, the positive predictive value (PPV) varied from 25 to 40% for CIN2+ and 15–27% for CIN3 + .
Interestingly, 118/235 samples were negative for all six markers including 106 controls (89.8%), six CIN1 (5.1%), five CIN2 (4.2%) and one CIN3 (0.8%), while seven samples were positive for all methylation markers including one control, one CIN1, one CIN2, two CIN3 and two cancers.
Discussion
The present study shows that the excellent diagnostic performance (sensitivity, specificity, PPV and NPV) of our previously identified CIN2+ specific methylation marker panels, validated on a Dutch cytologically abnormal cohort, is highly similar to its diagnostic performance in a Slovenian population‐based hrHPV‐positive screening cohort. Together with high clinical sensitivity and specificity this enables the application of our three methylation panels as triage tests irrespective of the primary screening method.
Despite the high similarity in the diagnostic performance of the three marker panels in both study cohorts, there are some important differences between the two cohorts. The Slovenian cohort consisted of women who attended the national cervical screening program and tested positive on hrHPV. Of these, some were considered as controls (no CIN2+ in the first nor in the second round) and some as cases (histologically confirmed CIN2+). In contrast the Dutch cohort only consisted of women with an abnormal cytology result referred to the outpatient clinic for colposcopy. Therefore, in our Dutch cohort hrHPV‐positive women with normal cytology are missing, which challenges the correct determination of specificity as a triage test in a hrHPV‐positive cohort. The Slovenian hrHPV‐positive cohort offers the possibility to determine the specificity of the methylation marker panels in this respect. However, the Slovenian cohort lacks hrHPV‐positive women with a normal cytology triage test result, while in their follow‐up CIN2+ was diagnosed (i.e., false‐negative samples). However, if these samples were included in the current study, the sensitivity of the methylation marker panels might even have been higher. Furthermore, in the Dutch study cohort much more carcinomas were included compared to the Slovenian cohort. The Slovenian cohort probably reflects better the expected number of carcinomas identified by primary hrHPV screening with cytology triage testing. Though, these differences in composition do not influence the observed similarity in methylation status between both cohorts.
The diagnostic performance of the methylation marker panels (highest sensitivity 80% and PPV 40% for CIN2+) (Table 3), as determined in the Slovenian hrHPV‐positive population, seems noninferior to the expected performance of the conventional cytology as a triage test (weighted sensitivity of 80% (range 73.8–93.5%) with a PPV of ~30% for CIN2+).14, 15, 16, 17, 18, 19, 20 Yet, this cannot be evaluated in this cohort as the number of women is too limited. Therefore, to analyse methylation triage testing compared to cytology triage large prospective studies should be performed.
In addition to our methylation markers, other methylation markers have been reported to be noninferior to cytology as well.18, 27, 28, 29 The limitations of cytology as a triage test are: (i) inherent subjective nature of the test, (ii) inability of high‐throughput testing, (iii) high level of skills required for cytotechnicians, (iv) moderate specificity (weighted mean ~60% (range 49.6–76.8%)) despite its high sensitivity (weighted mean 80%), (v) time consuming preparation of the cytological slides and (vi) inability of cytology to be performed on self‐collected samples.39 Same limitations account for triage tests based on immunostaining such as p16/KI67.15, 39 A full molecular triage alternative is expected to alleviate these issues and further improve cervical cancer screening programs.40 Moreover molecular triage testing can be performed on the same DNA extracted for hrHPV testing. Beside cytology and methylation analysis, hrHPV partial genotyping has also been proposed as a triage test for hrHPV positive women, with HPV16/18 typing already implemented in the USA referral guidelines.41 However, sensitivity and specificity of HPV16/18 triage is ~60% and 80% to detect CIN2+ lesions,39, 42 which makes it inferior compared to our methylation markers panels. In our study, despite the small number of samples, combining HPV genotyping with the methylation marker panels the sensitivity minimally increased, but was, as expected, accompanied with a decrease in specificity, resulting in no overall additive effect by combining HPV16/18 genotyping with our methylation marker panels. On the other hand, one might consider combining these two approaches as most likely the negative‐tested patients will have an extremely small risk to develop cancer.41, 43 Future population‐based studies using primary hrHPV‐based screening should include the comparison of the clinical diagnostic performance of these methylation marker panels with or without HPV genotyping, since they exhibit better clinical performance as triage methods such as cytology and in addition most likely are more economical.
In hrHPV‐positive control women, results of all tested methylation markers were not related to age. However, we observed that sensitivity of two methylation markers (ANKRD18CP and EPB41L3) for detection of CIN2+ was lower in women younger than 30 years old compared to older women. This is in line with the studies of Hansel et al.44 and Luttmer et al, 17 in which lower methylation rates were observed in younger women with CIN2+ lesions. It can be hypothesised that these women have a shorter time of HPV infection and therefore lower methylation rates are observed for specific genes. In contrast, women above 30 years with underlying CIN2+ lesion probably have long‐term persistent HPV infection, and higher methylation rates are observed indicating a more progressive state of the lesion.
Identification and treatment of cervical cancer precursors lead to reduction of cervical cancer incidence.1 CIN lesions that are completely unmethylated for these markers may lack the capacity to progress to cancer.38, 45, 46 This raises the question whether CIN lesions with a hypermethylated profile represent the truly progressive cervical cancer precursors. In theory, analysis of baseline hrHPV‐positive scrapings obtained from women who eventually progressed to CIN2+ is possible as the reports on the Slovenian HPV Prevalence Study9, 10 describe the detection of additional CIN2+ cases in both the follow‐up period within the first screening round and in the second screening round. This will also allow evaluation of the long‐term negative predictive value of DNA hypermethylation testing. Prospective follow‐up studies should investigate whether hrHPV‐positive CIN0/1 lesions those are either methylation marker positive or negative show progression to CIN2+ or not. These studies will allow the prediction of the risk progression for women diagnosed with CIN, thus reducing overreferrals and overtreatment.
In order to assess the reliability of the results of the current study, external validation of our DNA methylation panels by independent teams is deemed essential. The strengths of our study are the reproducibility of our previously identified methylation markers in an independent cohort, their independence with age and the high combined sensitivity and specificity of these methylation markers for CIN2+ and CIN3+, especially for panels C13orf18/EPB41L3/JAM3 and SOX1/ZSCAN1.
In conclusion, the high sensitivity and reproducibility of established methylation markers seem to allow safe implementation of these biomarker panels in population‐based screening programs for cervical cancer. However, further validation of methylation markers in external laboratories, analysing associations between DNA hypermethylation and risk of progression and randomised trials to compare their diagnostic performance with cytology triage after primary hrHPV‐testing are reckoned to be the decisive steps in order to justify implementation of methylation analysis as a safe and valid triage strategy.
Supporting information
Table S1: Positivity for DNA methylation biomarkers in Dutch clinician‐collected cervical scrapings (using pairwise complete data).
Table S2: Patient characteristics of the samples used for exploratory data mining.
Table S3: The impact of cross‐validation settings on the SOX1/ZSCAN1 decision tree's accuracy on both training and testing sets.
Table S4: Information gain estimates of all individual methylation markers either using all data points (N = 454) (A) or using only complete data points (B).
Table S5: The impact of cross‐validation settings on the ZSCAN1/JAM3 decision tree's accuracy on both training and testing sets.
Supplementary Figure S1: The decision tree produced using the training data set (N = 317). Decisions were based on QMSP ratios and each of the three leaves state the number of samples and their respective fraction of CIN2 + .
Appendix S1: Supplementary File 1
Acknowledgements
We would like to thank Arkajyoti Bhattacharya and Jan Lucas van der Ploeg from the University Medical Centre Groningen for the statistical discussions.
Conflicts of interest: AGJvdZ, ES, and GBAW are inventors of patents related to the content of our study. ES is a member of the scientific advisory board of Roche, Hologic and QCMD, and received travel reimbursements from Roche, Abbott, Hologic Inc. and QCMD. GBAW is a member of the scientific advisory board of CC Diagnostics. ES is an external advisor of CC Diagnostics. MP, Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana received several research grants from Abbott Molecular. AO received reimbursement of travel expenses for attending conferences and honoraria for speaking from Abbott Molecular and Qiagen.
References
- 1. Gustafsson L, Pontén J, Zack M, et al. International incidence rates of invasive cervical cancer after introduction of cytological screening. Cancer Causes Control 1997;8:755–63. [DOI] [PubMed] [Google Scholar]
- 2. Peto J, Gilham C, Fletcher O, et al. The cervical cancer epidemic that screening has prevented in the UK. Lancet 2004;364:249–56. [DOI] [PubMed] [Google Scholar]
- 3. Arbyn M, Bergeron C, Klinkhamer P, et al. Liquid compared with conventional cervical cytology: a systematic review and meta‐analysis. Obstet Gynecol 2008;111:167–77. [DOI] [PubMed] [Google Scholar]
- 4. Rijkaart DC, Berkhof J, Rozendaal L, et al. Human papillomavirus testing for the detection of high‐grade cervical intraepithelial neoplasia and cancer: final results of the POBASCAM randomised controlled trial. Lancet Oncol 2012;13:78–88. [DOI] [PubMed] [Google Scholar]
- 5. Cuzick J, Clavel C, Petry K, et al. Overview of the European and north American studies on HPV testing in primary cervical cancer screening. Int J Cancer 2006;119:1095–101. [DOI] [PubMed] [Google Scholar]
- 6. Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999;189:12–9. [DOI] [PubMed] [Google Scholar]
- 7. Bosch FX, Lorincz A, Munoz N, et al. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 2002;55:244–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arbyn M, Snijders PJF, Meijer CJLM, et al. Which high‐risk HPV assays fulfil criteria for use in primary cervical cancer screening? Clin Microbiol Inf 2015;21:817–26. [DOI] [PubMed] [Google Scholar]
- 9. Poljak M, Oštrbenk A, Seme K, et al. Three‐year longitudinal data on the clinical performance of the Abbott RealTime high risk HPV test in a cervical cancer screening setting. J Clin Vir 2016;76:S29–39. [DOI] [PubMed] [Google Scholar]
- 10. Poljak M, Ostrbenk A, Seme K, et al. Comparison of clinical and analytical performance of the Abbott realtime high risk HPV test to the performance of hybrid capture 2 in population‐based cervical cancer screening. J Clin Microbiol 2011;49:1721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bais AG, Rebolj M, Snijders PJ, et al. Triage using HPV‐testing in persistent borderline and mildly dyskaryotic smears: proposal for new guidelines. Int J Cancer 2005;116:122–9. [DOI] [PubMed] [Google Scholar]
- 12. Rijkaart DC, Berkhof J, van Kemenade FJ, et al. Comparison of HPV and cytology triage algorithms for women with borderline or mild dyskaryosis in population‐based cervical screening (VUSA‐screen study). Int J Cancer 2010;126:2175–81. [DOI] [PubMed] [Google Scholar]
- 13. Dillner J, Rebolj M, Birembaut P, et al. Joint European cohort study. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ 2008;337:a1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bosgraaf RP, Verhoef VM, Massuger LF, et al. Comparative performance of novel self‐sampling methods in detecting high‐risk human papillomavirus in 30,130 women not attending cervical screening. Int J Cancer 2015;136:646–55. [DOI] [PubMed] [Google Scholar]
- 15. Wentzensen N, Fetterman B, Castle PE, et al. p16/Ki‐67 dual stain cytology for detection of cervical Precancer in HPV‐positive women. J Natl Cancer Inst 2015;107:djv257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bergeron C, Giorgi‐Rossi P, Cas F, et al. Informed cytology for triaging HPV‐positive women: substudy nested in the NTCC randomized controlled trial. J Natl Cancer Inst 2015;107:dju423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luttmer R, De Strooper L, Berkhof J, et al. Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre) cancer in high‐risk HPV‐positive women of a gynecologic outpatient population (COMETH study). Int J Cancer 2016;138:992–1002. [DOI] [PubMed] [Google Scholar]
- 18. Verhoef VM, Bosgraaf RP, van Kemenade FJ, et al. Triage by methylation‐marker testing versus cytology in women who test HPV‐positive on self‐collected cervicovaginal specimens (PROHTECT‐3): a randomised controlled non‐inferiority trial. Lancet Oncol 2014;15:315–22. [DOI] [PubMed] [Google Scholar]
- 19. Richardson LA, El‐Zein M, Ramanakumar AV, et al. PEACHS (pap efficacy after cervical HPV status) study consortium. HPV DNA testing with cytology triage in cervical cancer screening: influence of revealing HPV infection status. Cancer Cytopathol 2015;123:745–54. [DOI] [PubMed] [Google Scholar]
- 20. Wright T, Stoler MH, Aslam S, et al. Knowledge of patients' human papillomavirus status at the time of cytologic review significantly affects the performance of cervical cytology in the ATHENA study. Am J Clin Pathol 2016;146:391–8. [DOI] [PubMed] [Google Scholar]
- 21. Ongenaert M, Wisman GB, Volders HH, et al. Discovery of DNA methylation markers in cervical cancer using relaxation ranking. BMC Med Genomics 2008;1:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoque MO, Kim MS, Ostrow KL, et al. Genome‐wide promoter analysis uncovers portions of the cancer methylome. Cancer Res 2008;68:2661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang N, Eijsink JJ, Lendvai A, et al. Methylation markers for CCNA1 and C13ORF18 are strongly associated with high‐grade cervical intraepithelial neoplasia and cervical cancer in cervical scrapings. Cancer Epidemiol Biomarkers Prev 2009;18:3000–7. [DOI] [PubMed] [Google Scholar]
- 24. Eijsink JJ, Lendvai A, Deregowski V, et al. A four‐gene methylation marker panel as triage test in high‐risk human papillomavirus positive patients. Int J Cancer 2012;130:1861–9. [DOI] [PubMed] [Google Scholar]
- 25. Boers A, Wang R, van Leeuwen R, et al. Discovery of new methylation markers to improve screening for cervical intraepithelial neoplasia grade 2/3. Clin Epigen 2016;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang R, van Leeuwen RW, Boers A, et al. Genome‐wide methylome analysis using MethylCap‐seq uncovers 4 hypermethylated markers with high sensitivity for both adeno‐ and squamous‐cell cervical carcinoma. Oncotarget 2016;7:80735–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wentzensen N, Schiffman M, Palmer T, et al. Triage of HPV positive women in cervical cancer screening. J Clin Vir 2016;76:S49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bierkens M, Hesselink AT, Meijer CJ, et al. CADM1 and MAL promoter methylation levels in hrHPV‐positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer 2013;133:1293–9. [DOI] [PubMed] [Google Scholar]
- 29. De Strooper LM, van Zummeren M, Steenbergen RD, et al. CADM1, MAL and miR124‐2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol 2014;67:1067–71. [DOI] [PubMed] [Google Scholar]
- 30. Van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T‐cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED‐2 concerted action BMH4‐CT98‐3936. Leukemia 2003;17:2257–317. [DOI] [PubMed] [Google Scholar]
- 31. Wisman GB, Nijhuis ER, Hoque MO, et al. Assessment of gene promoter hypermethylation for detection of cervical neoplasia. Int J Cancer 2006;119:1908–14. [DOI] [PubMed] [Google Scholar]
- 32. R Core Team . R: A language and environment for statistical computing, 3.3.2 ed. Vienna, Austria: R Foundation for Statistical Computing, 2016. [Google Scholar]
- 33. Venkatraman ES. Clinfun: Clinical trial design and data analysis functions. R package version 1.0.13;2016.
- 34. Christensen RHB. Ordinal ‐ regression models for ordinal data. R package version 2015.6‐28 ed.;2015.
- 35. Kampstra P. Beanplot: a boxplot alternative for visual comparison of distributions. J Stat Softw (Code Snippet 1) 2008;28:1–9. [Google Scholar]
- 36. Hawass NE. Comparing the sensitivities and specificities of two diagnostic procedures performed on the same group of patients. Br J Radiol 1997;70:360–6. [DOI] [PubMed] [Google Scholar]
- 37. Issa JP. Aging and epigenetic drift: a vicious cycle. J Clin Invest 2014;124:24–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klutstein M, Nejman D, Greenfield R, et al. DNA methylation in cancer and aging. Cancer Res 2016;76:3446–50. [DOI] [PubMed] [Google Scholar]
- 39. Ebisch RM, Siebers AG, Bosgraaf RP, et al. Triage of high‐risk HPV positive women in cervical cancer screening. Exp Rev Anticancer Ther 2016;16:1073–85. [DOI] [PubMed] [Google Scholar]
- 40. Gradíssimo A, Burk RD. Molecular tests potentially improving HPV screening and genotyping for cervical cancer prevention. Exp Rev Mol Diagn 2017;17:379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cuschieri K, Ronco G, Lorincz A, et al. Eurogin roadmap 2017: triage strategies for the management of HPV‐positive women in cervical screening programs. Int J Cancer 2018;143:735–45. [DOI] [PubMed] [Google Scholar]
- 42. Luttmer R, De Strooper LM, Steenbergen RD, et al. Management of high‐risk HPV‐positive women for detection of cervical (pre)cancer. Exp Rev Mol Diagn 2016;16:961–74. [DOI] [PubMed] [Google Scholar]
- 43. Schiffman M, Burk RD, Boyle S, et al. A study of genotyping for management of human papillomavirus‐positive, cytology‐negative cervical screening results. J Clin Microbiol 2015;53:52–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hansel A, Steinbach D, Greinke C, et al. A promising DNA methylation signature for the triage of high‐risk human papillomavirus DNA‐positive women. PLoS One 2014;9:e91905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schütze DM, Kooter JM, Wilting SM, et al. Longitudinal assessment of DNA methylation changes during HPVE6E7‐induced immortalization of primary keratinocytes. Epigenetics 2015;10:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koeneman MM, Kruitwagen RFPM, Nijman HW, et al. Natural history of high‐grade cervical intraepithelial neoplasia: a review of prognostic biomarkers. Exp Rev Mol Diagn 2015;15:527–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Positivity for DNA methylation biomarkers in Dutch clinician‐collected cervical scrapings (using pairwise complete data).
Table S2: Patient characteristics of the samples used for exploratory data mining.
Table S3: The impact of cross‐validation settings on the SOX1/ZSCAN1 decision tree's accuracy on both training and testing sets.
Table S4: Information gain estimates of all individual methylation markers either using all data points (N = 454) (A) or using only complete data points (B).
Table S5: The impact of cross‐validation settings on the ZSCAN1/JAM3 decision tree's accuracy on both training and testing sets.
Supplementary Figure S1: The decision tree produced using the training data set (N = 317). Decisions were based on QMSP ratios and each of the three leaves state the number of samples and their respective fraction of CIN2 + .
Appendix S1: Supplementary File 1


