Abstract
Misfolding and aggregation of the neuronal, microtubule-associated protein tau is involved in the patho-genesis of Alzheimer’s disease and tauopathies. It has been proposed that neuronal membranes could play a role in tau release, internalization, and aggregation and that tau aggregates could exert toxicity via membrane permeabilization. Whether and how tau interacts with lipid membranes remains a matter of discussion. Here, we characterize the interaction of full-length human tau (htau40) with supported lipid membranes (SLMs) made from brain total lipid extract by time-lapse high-resolution atomic force microscopy (AFM). We observe that tau attaches to brain lipid membranes where it self-assembles in a cation-dependent manner. Sodium triggers the attachment, self-assembly, and growth, whereas potassium inhibits these processes. Moreover, tau assemblies are stable in the presence of sodium and lithium but disassemble in the presence of potassium and rubidium. Whereas the pseudorepeat domains (R1−R4) of htau40 promote the sodium-dependent attachment to the membrane and stabilize the tau assemblies, the N-terminal region promotes tau self-assembly and growth.
Keywords: Alzheimer’s disease, aggregation, atomic force microscopy, brain total lipid extract, cation-selectivity, human tau protein
Graphical Abstract
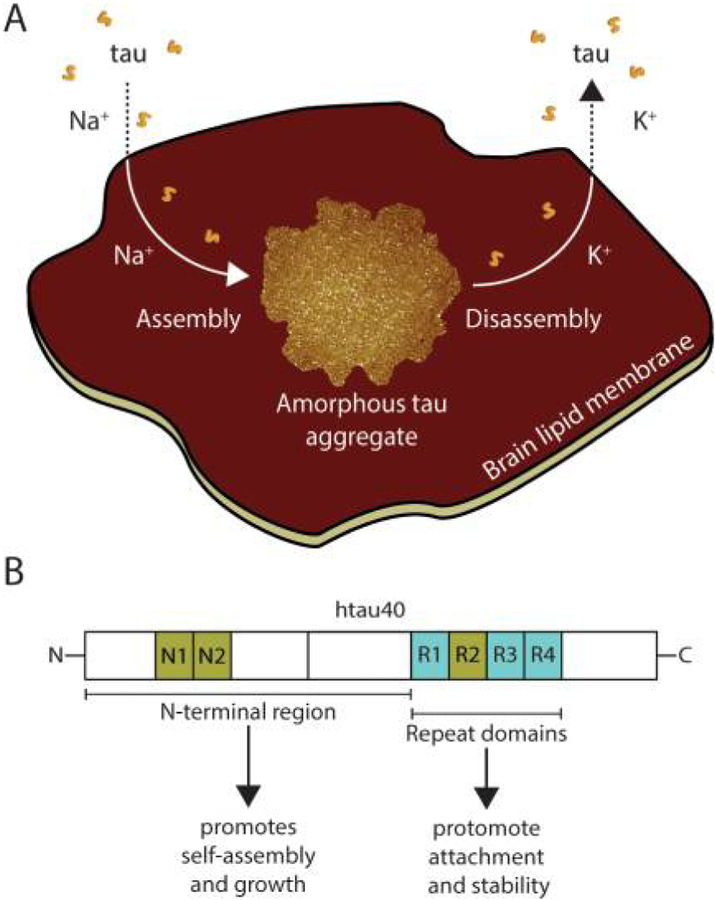
Tau is a neuronal microtubule-associated protein with a role for microtubule assembly and stabilization.1,2 Tau is expressed mainly in the central nervous system, where, besides regulating microtubule dynamics3,4 and axonal transport,5–10 it is involved in other neuronal functions including the regulation of axonal elongation and maturation,11–14 neurogenesis,15,16 and DNA and RNA protection.17,18 There is some evidence that tau can bind to cell membranes19–22 and that this binding might be mediated either by the N-terminal or C-terminal half of tau.23,24 Tau consists of an N-terminal projection domain that projects away from the microtubule surface and a C-terminal half that is responsible for microtubule binding and tau aggregation (Figure 1A).25–28 The C-terminal half of the longest human isoform, htau40, contains the repeat domain (tauRD) with four pseudorepeats (R1−R4), which together with the proline-rich flanking regions constitute the microtubule binding domain.29 The pseudorepeats R1−R4 also build the core of fibrillary tau aggregates (PHF core).2 In the human brain, the two N-terminal inserts, N1 and N2, and the R2 repeat are alternatively spliced to create six different tauisoforms.30 Htau40 is highly hydrophilic and intrinsically disordered. It exposes asymmetrically distributed charged residues having a predominantly acidic N-terminal tail, a positively charged repeat domain, and a neutral C-terminal tail (Figure 1A).31,32 Although being highly soluble, tau can aggregate and accumulate intracellularly in Alzheimer’s disease and other tauopathies. These tau inclusions in the brain, called neurofibrillary tangles, are a hallmark pathological feature of several neurodegenerative diseases.33
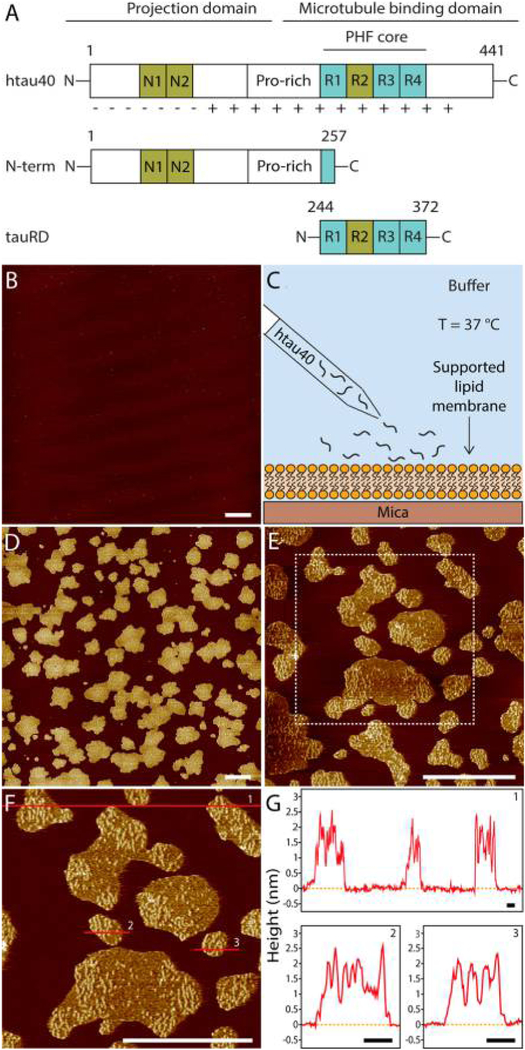
Figure 1.
AFM imaging of full-length human tau (htau40) incubated on a supported lipid membrane (SLM) of brain total lipid extract (BTE). (A) Illustration of the tau constructs. The longest isoform of human tau (htau40, 441 amino acid residues, 2N4R) is shown to illustrate relevant domains of the protein: the projection domain with the amino terminal inserts N1 and N2 and the microtubule binding domain with the pseudorepeats R1−R4. Either or both N1 and N2, as well as R2, may be absent due to alternative splicing. (Splicing inserts are indicated in green.) The distribution of the charged residues along the tau sequence is illustrated. Constructs encompassing the N-terminal region (N-term, residues 1–257) and the truncated pseudorepeat domains tauRD of htau40 (129 residues, containing R1−R4) are shown. Numbering of residues in N-term and tauRD is based on the htau40 isoform. (B) AFM topograph of a SLM made from brain total lipid extract (BTE). (C) Schematic illustration of the experiment. A continuous lipid membrane supported by mica is incubated with htau40 in buffer solution at 37 °C. (D) AFM topograph of the SLM shown in part B after incubation for 90 min with 100 nM htau40. Patchy assemblies of htau40 are clearly visible.(F) htau40 assemblies of the area outlined (dashed line) in part E imaged at a higher magnification. Protein assemblies show morphological features protruding at a variable height from the lipid membrane. The lower regions protrude 0.8 ± 0.2 nm (ave ± SD; n =35), and the higher regions (brighter in height scaling) protrude 2.3 ±0.2 nm (n = 60) from the lipid membrane. (G) Height profiles of htau40 assemblies (red curves) taken along the red lines indicated in the topograph shown in part F. Orange dashed lines mark the SLM surface. The full-range color scale of the AFM topographs corresponds to a height of 4.3 nm. AFM imaging and incubations were done in buffer solution (150 mM NaCl, 20 mM Hepes, pH 7.4) at 37 °C. Scale bars, 1 μm (B, D−F) and 100 nm (G).
In vitro, the aggregation of recombinant tau can be induced by polyanions such as heparin, RNA, and arachidonic acid.34–36 Lipid membranes have also been reported to enhance tau aggregation in vitro.24,37–40 However, whether and how membranes play a role for tau aggregation in neurodegenerative diseases remain somewhat enigmatic.2 Besides having a potential role in modulating tau aggregation, plasma membranes could also serve as a target for misfolded tau to exert toxicity via membrane destabilization. The disruption of cell membranes, resulting in the alteration of ion homeostasis and dysregulation of neuronal signal transduction, has been proposed as a general mechanism for the neurotoxicity exerted by misfolded proteins, for example, amyloid-β, α-synuclein, huntingtin, and tau.40–45 In addition, the transmission of tau between neurons,46–49 facilitated by the release and the subsequent internalization of extracellular tau, requires crossing cellular membranes,2 which likely involves the interaction of tau with membranes. However, despite the potential importance of lipid membranes in tau physiology and pathology, the interaction between tau and lipid membranes is poorly described.
Here, we use atomic force microscopy (AFM) to investigate the interaction of soluble tau with supported lipid membranes (SLMs) and how this interaction may support tau assembly. AFM, which is well suited to observe the surfaces of SLMs50,51 and membrane proteins at high resolution (~ 0.5−2 nm),52,53 requires no labeling or staining and can be operated at physiological relevant temperatures in buffer solution.54,55 Using time-lapse AFM, it is further possible to image single membrane proteins at work,54,56–59 as well as diffusing and assembling into higher complexes.60–63 SLMs are planar lipid membranes adsorbed onto a solid support. Their advantages are the stability and amenability to surface-sensitive characterization techniques, including AFM.50,51 Although in buffer solution SLMs are separated from the supporting surface by a ~ 1 nm thick water and ions layer,64–66 unspecific interactions between SLM and support have to be reduced to avoid perturbation of the system.67,68 Hydrophilic and atomically flat muscovite mica largely reduces such interactions.68 In such cases, SLMs can form a fluidic two-dimensional membrane allowing the diffusion and rotation of lipid molecules, lipidassociated proteins, and transmembrane proteins, which makes SLMs particularly well suited to investigate membrane processes such as protein adsorption, insertion and folding, self-assembly, and function.62,63,69–71
To characterize the interactions with human tau, we use SLMs composed of total lipid extract of porcine brain (BTE). We find that tau interacts with SLMs made from BTE and that this interaction guides the adsorption and self-assembly of tau on the lipid membrane. Tau adsorption and subsequent self-assembly on SLMs is promoted by Na+ and inhibited by K+. Moreover, the thermodynamically and mechanically extremely stable tau assemblies largely disassemble in the presence of K+. We further find that the repeat domain of tau drives the Na+-dependent adsorption to SLMs, whereas the N-terminal region of tau drives self-assembly and growth.
Results and Discussion.
Human Tau Self-Assembles on Brain Lipid Membranes.
To characterize the interaction of tau with brain lipid membranes, we prepared SLMs composed of BTE and incubated them with full-length human tau (htau40, Figure 1A). SLMs were prepared by fusing liposomes onto freshly cleaved mica (Figure S1A,B) (Materials and Methods). To exclude artifacts of the SLM, such as stacked membranes or holes exposing the supporting mica, freshly adsorbed membranes were first imaged by contact-mode AFM in buffer solution (150 mM NaCl, 20 mM Hepes, pH 7.4) (Figure 1B and Figure S1B). Only SLMs without defects were used for experiments in which they were kept hydrated at all times.
To observe the interaction of htau40 with the SLM, the buffer solution containing 100 nM htau40 was injected into the fluid cell of the AFM at physiological temperature (~ 37 °C, Figure 1C). After 90 min of incubation time, the SLM was imaged again (Figure 1D). The AFM topographs showed islands of protein assemblies evenly distributed over the SLM and having irregular shapes and sizes (Figure 1D,E and Figure S2A,B). At a higher resolution, the topographs revealed heterogeneous surface structures of the tau assemblies, which protruded from the lipid membrane at two different heights (Figure 1F,G and Figure S2C,D). Whereas the lower regions protruded 0.78 ± 0.15 nm (mean ± SD; n = 35) without exposing any visible structure, the higher regions (brighter in topographs) protruded 2.28 ± 0.21 nm (n = 60) and exposed small globular and elongated structures.
Taken together, we observe that htau40 adsorbs and self-assembles on SLMs made from BTE. When imaged at a higher resolution, the assemblies show globular and elongated structures of nonphosphorylated htau40, which look similar to previously described oligomerization stages of hyper-phosphorylated htau40 and of pro-aggregation tau mutants.72,73
Concentration-Dependent Growth of htau40 Assemblies.
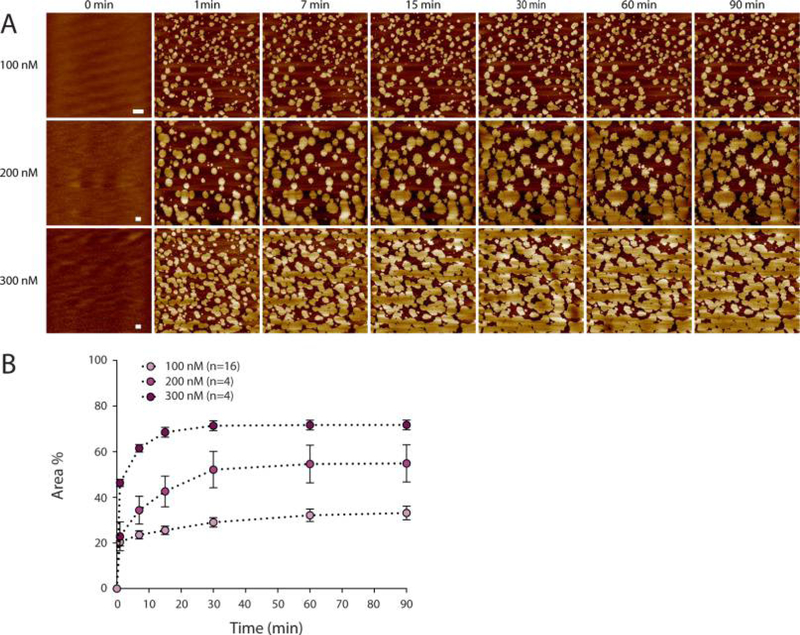
Next, we applied solutions of different tau concentrations containing 100, 200, and 300 nM htau40 and followed the protein assembly using time-lapse AFM (Figure 2). After rapid initial adsorption and nucleation on the SLM, the tau assemblies grew in size until they reached a maximum size after ~ 30 min (Figure 2A). During this growth, the number of assemblies did not increase but the existing tau assemblies grew and fused forming larger assemblies. We analyzed the growth of the assemblies by measuring their surface area over time (Figure 2B and Figure S3A). After 90 min of incubation, the SLM surface area covered by tau assemblies depended on the tau concentration applied. At 100 nM htau40, 34.1 ± 2.1% (mean ± SEM; n = 24) of the SLM was covered, at 200 nM htau40, 54.9 ± 8.2% (n = 4), and at 300 nM htau40, 71.8 ±2.2% (n = 4).
Figure 2.
Following htau40 self-assembly by time-lapse AFM. (A) Time-lapse AFM topographs showing the self-assembly of htau40 on SLMs formed from BTE. Times indicate the minutes after htau40 has been injected into the imaging buffer (150 mM NaCl, 20 mM Hepes, pH 7.4) at 37 °C and at concentrations of 100, 200, and 300 nM. The full-range color scale of the topographs corresponds to a height of 3.8 nm (100 and 200 nM) and 5 nm (300 nM). Scale bars, 1 μm. (B) Comparison between mean growth curves of htau40 assemblies measured from images acquired by time lapse AFM. The area indicates the percentage of the SLM surface occupied by the htau40 assemblies. The times indicate the minutes after htau40 has been added to the imaging buffer. Values represent mean ± SEM determined from n independent experiments.
These experiments show that the surface area of brain lipid membranes covered by tau depends on the concentration of tau added to the buffer solution. Experiments incubating SLMs made from BTE with different concentrations of bovine serum albumin (BSA) showed no protein assemblies on the lipid membrane (Figure S3A).
Adsorption and Self-Assembly of htau40 Are Cation Specific.
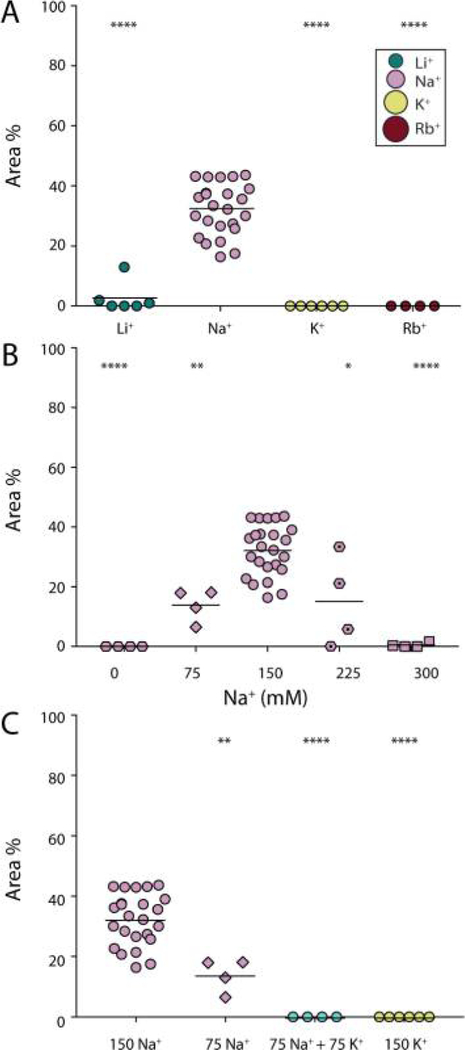
The interaction of biomolecules with surfaces also depends on the ionic composition and strength of the aqueous solution.74,75 The intracellular and extracellular fluids of mammalian cells have different compositions. Among other differences, intracellular fluids show a high K+ concentration (140 mM) and low Na+ concentration (5 mM), whereas extracellular fluids show the opposite trend.76 We thus wondered whether the adsorption and self-assembly of tau on SLMs made from BTE, which being composed of zwitterionic and anionic lipids bear a net negative charge, depend on the ion composition of the solution. To investigate whether monovalent cations play a role for the interaction of htau40 with SLMs and for the subsequent tau self-assembly, we prepared SLMs of BTE and incubated them with 100 nM htau40 in buffer solution (20 mM Hepes, pH 7.4) at ~37 °C for 90 min. Thereby the buffer solution contained 150 mM LiCl, NaCl, KCl, or RbCl. We then compared the area of the assemblies formed by htau40 in the presence of the different salts (Figure 3A). In the presence of Li+, htau40 formed assemblies only in 50% of the experiments (n = 6) and the area of the SLM occupied by tau assemblies was significantly smaller compared to Na+. As described above, in the presence of Na+, htau40 always adsorbed on the SLM and self-assembled into large islands occupying 34.1 ± 2.1% (mean ± SEM; n = 24) of the SLM surface. In contrast, no adsorption of htau40 onto SLMs was observed in the presence of K+ or Rb+.
Figure 3.
Na+-dependence and K+-inhibition of htau40 interaction with the SLM. (A) Comparison between the area of htau40 assemblies after SLMs have been incubated with htau40 in the presence of 150 mM XCl, 20 mM Hepes, pH 7.4 (X = Li+, Na+, K+, or Rb+). The inset shows the relative size of the tested cations. (B) Comparison between the area of htau40 assemblies after SLMs have been incubated with htau40 in the presence, respectively, of 0, 75, 150, 225, or 300 mM NaCl, 20 mM Hepes, pH 7.4 (C) Comparison between the area of htau40 assemblies after SLMs have been incubated with htau40 in the presence, respectively, of 75 or 150 mM Na+, 75 mM Na+ and 75 mM K+, or 150 mM K+ (20 mM Hepes, pH 7.4). The area indicates the percentage of the surface of SLM of BTE occupied by the protein assemblies after 90 min incubation with 100 nM htau40 at 37 °C. Each data point presents an independent experiment; bars represent mean values. The statistical significance was determined using the Mann− Whitney t-test comparing each condition with htau40 in 150 mM NaCl data. *P ≤ 0.05. **P ≤ 0.01. ****P < 0.0001.
To account for the possibility of a generally enhanced adsorption and assembly of soluble proteins onto SLMs made from BTE in the presence of Na+ compared to K+, we incubated SLMs with BSA in the presence of 150 mM NaCl or KCl (Figure S3). BSA did not bind or assemble on SLMs, neither in the presence of Na+ nor K+, which argues against a general effect of Na+ on facilitating the adsorption of proteins onto SLM made from BTE.
As the htau40 adsorption and self-assembly on SLMs appeared to be most effcient in the presence of Na+, we next characterized the dependence on the Na+ concentration. Therefore, 100 nM htau40 was applied to SLMs in buffer solutions (20 mM Hepes, pH 7.4) containing 0, 75, 150, 225, or 300 mM NaCl (Figure 3B) at ~ 37 °C for 90 min. In the absence of NaCl, htau40 did not adsorb and self-assemble onto SLMs. In the presence of Na+, the htau40 assemblies covered 13.9 ± 2.7% (mean ± SEM, n = 4) at 75 mM, 34.1 ± 2.2% (n =24) at 150 mM, 15.1 ± 7.5% (n = 4) at 225 mM, and 0.5 ±0.4% (n = 4) at 300 mM NaCl in the buffer. The experimental data thus showed a nonlinear correlation between Na+ concentration and area of tau self-assemblies formed on SLMs, which reached its maximum at 150 mM NaCl.
As among the tested monovalent cations those having physiological relevance are mostly Na+ and K+, we further investigated the effect of K+ by incubating SLMs made from BTE with htau40 in the presence of 75 mM NaCl and 75 mM KCl at ~ 37 °C for 90 min (Figure 3C). In comparison to 150 mM NaCl, in the presence of 75 mM NaCl, the surface area of the SLMs occupied by self-assembled htau40 was significantly smaller although the number of the assemblies formed by the protein was similar in both conditions (Figure S4A). In the presence of both 75 mM NaCl and 75 mM KCl, however, htau40 did not form any assemblies on the lipid membrane, similarly to what we observed for SLMs incubated with htau40 in the presence of KCl only. This suggests not only that K+ is unable to facilitate the adsorption of tau to SLMs (as Na+ does) but also that K+ counteracts the Na+-dependent adsorption and subsequent self-assembly of tau onto SLMs. In summary, the interactions driving the adsorption of htau40 to SLMs are promoted by Na+ and inhibited by K+.
Tau Assemblies Show Exceptional Mechanical Stability.
We next asked whether the self-assembled tau aggregates are only weakly adsorbed or tightly attached to SLMs made from BTE. We hence used the AFM tip as a nanotool to mechanically perturb the tau assemblies and test their stability against mechanical forces. In our experiments, we first self-assembled htau40 on SLMs in 150 mM NaCl and imaged the aggregates by AFM at forces ≪100 pN to avoid structural perturbation of the sample (Figure S5A,C,E). After this, we selected a small area of the aggregates, which we scanned with the AFM tip applying forces of 1, 2, or 5 nN. The experiments showed that the self-assembled tau aggregates were unaffected when applying forces of 1 nN (Figure S5B). At 2 nN force, which has been described to be suffcient to rupture covalent bonds,77 we observed the partial distortion of the aggregates (Figure S5D). At 5 nN force, we eventually observed the dissection of the assembly (Figure S5F). As previously described, applying forces between 100 pN and 1 nN can lead to the dissection of transmembrane protein complexes, while forces >1 nN lead to their denaturation.78–80 We had also previously reported that tau fibrils adsorbed onto mica in the absence of lipid membranes disassemble at forces of ~ 150–200 pN.81 We can thus conclude that htau40 adsorbed to supported brain lipid membranes form mechanically exceptionally stable assemblies.
Cation-Dependent Tau Assembly and Disassembly.
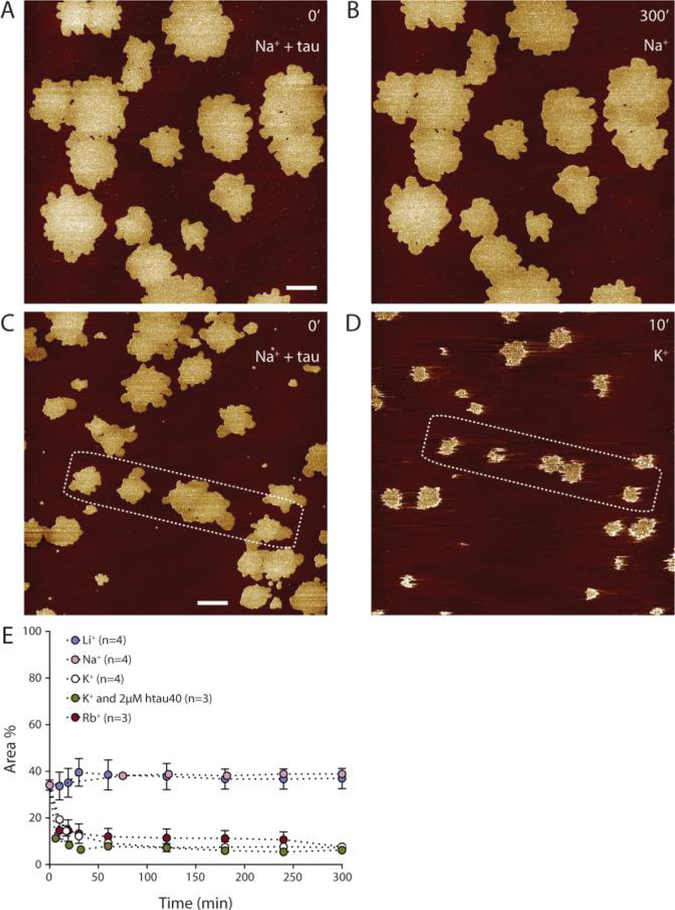
It has previously been shown that accumulated and aggregated tau is stable over days and weeks, both in mouse brain,48,82,83 and likely also in human brain,84 as well as in vitro.85 Since we observed that the adsorption and self-assembly of htau40 on SLMs is cation-sensitive, we decided to investigate also the stability of tau assemblies in the presence of different cations. We first self-assembled 100 nM htau40 on SLMs for 90 min at ~ 37 °C in buffer solution containing 150 mM NaCl. At such conditions, the AFM topographs showed nicely distributed islands of htau40 assemblies, some of which already fused with each other (Figure 4A,C). After this, the incubation buffer was exchanged against a buffer solution containing either 150 mM LiCl, NaCl, KCl, or RbCl, and time-lapse AFM images of the tau assemblies were recorded at ~37 °C for the forthcoming 5 h (Figure 4). Before the buffer was exchanged, the htau40 assemblies covered in average 34.1 ± 2.2% (mean ± SEM, n =24) of the SLM surface area. On SLMs with buffer solutions containing NaCl or LiCl, the htau40 assemblies were stable and did not change shape, number, and surface area during 5 h (Figure 4A,B,E). However, when incubating the htau40 assemblies with buffer solutions containing 150 mM KCl or RbCl, the surface coverage of htau40 assemblies rapidly decreased during the first 30 min of incubation from the initial ~34% to 6.2 ± 2.8% (n = 4) in KCl and to 7.6 ± 1.3% (n = 3) in RbCl, after which they remained stable (Figure 4C–D,E). We further tested, whether the presence of soluble htau40 in the buffer would affect the K+-induced disassembly and exchanged the incubation buffer with a buffer containing 150 mM KCl and 2 μM htau40 (Figure 4E). Also in this case, the surface coverage of htau40 quickly reduced to a minimum of 7.3 ± 1.4% (n = 3).
Figure 4.
Cation-dependent reversibility of htau40 self-assembly on SLMs. (A) AFM topograph of a SLM after 90 min incubation with 100 nM htau40 in buffer solution (150 mM NaCl, 20 mM Hepes, pH 7.4) at 37 °C. (B) The same surface imaged after 5 h incubation in the same buffer solution but without htau40. (C and D) AFM topographs of a SLM (C) after 90 min incubation with 100 nM htau40 in buffer solution with NaCl (150 mM NaCl, 20 mM Hepes, pH 7.4) and (D) after 10 min incubation in buffer solution with KCl (150 mM KCl, 20 mM Hepes, pH 7.4). (E) Comparison between the mean area of htau40 assemblies during 5 h incubation in buffer solutions containing either Li+, Na+, K+, or Rb+ as monovalent ions (150 mM XCl, 20 mM Hepes, pH 7.4, with X = Li+, Na+, K+, or Rb+) in the absence of htau40 or in the presence of 2 μM htau40. SLMs of BTE were first incubated with 100 nM htau40 in buffer solution with NaCl (150 mM NaCl, 20 mM Hepes, pH 7.4) for 90 min to allow htau40 to self-assemble on the SLMs. The mean area of the SLM occupied by htau40 assemblies after a 90 min incubation time is 34.1 ± 2.2% (n =24) and represents the time 0 at which the buffer solution has been exchanged. The time axis indicates the minutes of incubation of the SLM with the indicated buffers. Experiments were conducted at 37 °C. Data represent the mean ± SE, and n gives the number of independent experiments analyzed. The full-range color scale of topographs corresponds to a height of 4.3 nm. Scale bars, 1 μm.
In summary, these data support the idea that ion specific interactions driving the adsorption and self-assembly of human tau on SLMs from BTE are also involved in stabilizing the assemblies. Furthermore, they show that having tau in solution cannot prevent the assemblies from disassembling in the presence of potassium.
Role of Tau Repeat Domain and N-Terminal Region.
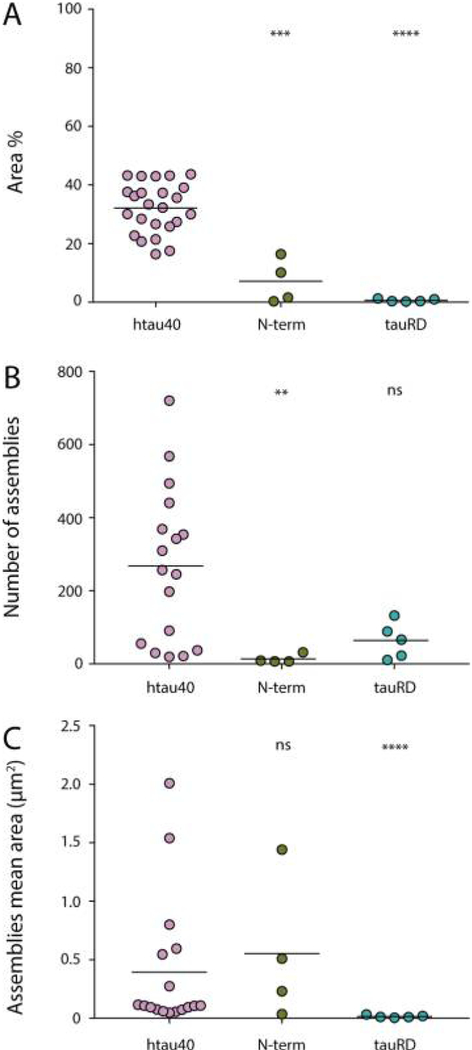
The interaction of tau with the negatively charged microtubule surface,86–90 its primary intracellular binding partner, as well as the assembly of tau into paired helical filaments,91 are mediated by the positively charged microtubule binding domain of tau and the proline-rich flanking regions (Figure 1A).92 To evaluate whether the tau pseudorepeat domain (tauRD) is also responsible for the Na+-dependent adsorption and self-assembly of htau40 on SLMs, we incubated SLMs made from BTE with 100 nM tauRD or N-terminal region (N-term, amino acids 1−257 of htau40) constructs in buffer solution (20 mM Hepes, pH 7.4) containing 150 mM NaCl, at ~37 °C for 90 min to allow suffcient time for interaction and assembly formation (Figure 5). We found that the tauRD and N-terminal region covered significantly less of the SLM surface compared to htau40 (Figure 5A). Interestingly, the number of assemblies formed by htau40 and tauRD was similar, whereas the N-terminal region formed considerably less assemblies (Figure 5B). Furthermore, while tauRD assemblies were considerably smaller in size, the mean area of the assemblies formed by htau40 and the N-terminal region of htau40 was similar (Figure 5C). Next, we incubated the SLMs made from BTE with 100 nM of the pseudorepeat domains or the N-terminal region of tau in the presence of 150 mM KCl (Figure S6). In this case, neither the N-terminal region nor tauRD adsorbed and self-assembled onto SLMs.
Figure 5.
Self-assembly of N-terminal region (N-term) and pseudorepeat domains (tauRD) of htau40 on SLMs. (A) Comparison between SLM areas covered by tau constructs after incubation for 90 min with 100 nM htau40, N-term, or tauRD. SLMs were made from BTE. The area indicates the percentage of the SLM surface occupied by protein assemblies. Comparison between the number (B) and the mean area (C) of the protein assemblies formed by 100 nM htau40, N-term, and tauRD over a SLM area of 100 μm2 after 90 min incubation. Experiments were performed in buffer solution containing NaCl (150 mM NaCl, 20 mM Hepes, pH 7.4) and at 37 °C. Each data point presents an independent experiment; bars represent mean values. A statistical significance was determined using the Mann− Whitney t-test, taking the htau40 data as a reference. ns, not significant (P > 0.05). **P < 0.01. ***P < 0.001. ****P < 0.0001.
Taken together, these data show that, similarly to full-length human tau, the tauRD and the N-terminal region of htau40 can adsorb onto SLMs and that this adsorption is also cationsensitive. However, both the pseudorepeat domains and the N-terminal region of tau show a much lower ability to self-assemble into larger aggregates. In addition, whereas the number of assemblies formed by tauRD is indifferent from that found for full-length tau, the N-terminal region of tau forms much less assemblies. However, the mean area covered by the assemblies of full-length tau and the N-terminal region of tau is similar, while that covered by tauRD is considerably smaller. The results indicate that the tau pseudorepeat domain has the ability to form seeds on SLMs, while the N-terminal region of tau drives the self-assembly to cover larger surface areas.
Conclusions.
Here we studied the interaction of human tau with SLMs composed of brain lipids. Using high-resolution AFM, we found that human full-length tau, htau40, can adsorb and self-assemble flat islands on SLMs. Within a few minutes, the nanoscopic htau40 assemblies can grow a few micrometers in diameter and are thermodynamically and mechanically exceptionally stable. How large the assemblies can grow depends on the tau concentration in the buffer solution. The time-dependent growth of tau assemblies was previously also reported for the aggregation of tau into fibrillary aggregates in vitro, in which the nucleation of tau represents the rate-limiting step of the aggregation process.93,94 Surprisingly, we find that the self-assembly of htau40 on lipid membranes is cationdependent. Human tau attaches and self-assembles onto brain lipid membranes in the presence of Na+ and scantly in the presence of Li+, but not in the presence of K+ or Rb+ (Figure 3A). Thereby, Na+ most effciently promotes tau attachment to the membrane and the subsequent self-assembly into larger aggregates (Figure 6 A). Li+, Na+, K+, and Rb+ differ in ion radius, being 90, 116, 152, and 166 pm, respectively. Thus, the adsorption and subsequent self-assembly of tau onto supported brain lipid membranes appears to depend on the cation radius, in that it is inhibited at larger cation radii, and optimal for Na+ ions. To which extent cations modulate the interaction of tau with cellular membranes in vivo remains to be elucidated.
Figure 6.
Reversible cation-selective attachment and self-assembly of human tau on supported lipid membranes. (A) Model illustrating fulllength human tau (htau40) self-assembling onto brain lipid membranes. Sodium triggers the attachment, self-assembly, and growth of tau into amorphous aggregates, whereas potassium inhibits these processes. Moreover, when exposed to potassium, the mechanically exceptionally stable tau aggregates dissemble. (B) Illustration of the roles of the domains of full-length human tau in facilitating tau attachment, self-assembly, and growth. The N-terminal region promotes self-assembly and growth of tau aggregates on brain lipid membranes, whereas the pseudorepeat domains R1−R4 (PHF core) promote attachment and stability of the tau aggregates.
The number and growth of tau assemblies also depend on the Na+ concentration (Figure 3B and Figure S4). In the absence of Na+, we did not observe any attachment of htau40 to supported brain lipid membranes. In the presence of 75 mM NaCl, tau formed as many assemblies as in the presence of 150 mM NaCl, but the size of the assemblies was considerably smaller. This may suggest that electrostatic repulsive interactions, which can be reduced by increasing the salt concentrations, disfavor the tau assemblies to grow. However, further increasing the NaCl concentration to 225 and 300 mM NaCl reduces the number of tau assemblies but not their size. This suggests that, although NaCl is required for tau attachment and self-assembly, increasing the NaCl concentration above 150 mM may shield the electrostatic interactions that attract tau to the negatively charged lipid layer surface, resulting in a lower number of assemblies (Figure S4). Thus, the interaction of htau40 with the brain lipid membrane and the subsequent self-assembly into larger aggregates is most effcient at a monovalent salt concentration of 150 mM NaCl, which approximates the osmolality of intra-and extracellular fluids. Interestingly, the assembly of fibrillary tau aggregates in vitro has also been shown to depend on the buffer composition and NaCl concentration and to be effcient below ~150 mM NaCl.95 The cation-sensitive adsorption and self-assembly of human tau to SLMs becomes quite interesting when considering the effect of K+. Our data indicate that K+ cannot facilitate the adsorption of htau40 to SLMs and suppresses the Na+-triggered adsorption and self-assembly of tau on SLMs. This cation-sensitivity is also reflected in the instability of preformed (in the presence of NaCl) tau assemblies, which upon exposure to K+ (and Rb+) quickly disassemble (Figure 6A).
Previous studies suggested that the interaction of tau with neural plasma membranes may be mediated by the N-terminal projection domain of tau.23 To test the role of the N-terminal and the repeat domain in the attachment and self-assembly of tau on SLMs, we performed binding experiments using the repeat domain (tauRD) or the N-terminal region (N-term) of htau40. Both constructs attached to the SLMs in a Na+-dependent fashion (Figure S6). Although tauRD could form seeds on the SLMs, it could not grow larger assemblies. On the contrary, the N-term hardly formed seeds, but could grow these seeds into larger assemblies. Similarly, it has been recently described that, when exposed to a sedimentation assay and followed by a sucrose gradient, the pseudorepeat domain of tau can facilitate the binding to and disruption of phosphatydylserine vesicles, thereby leading to tau fibril formation.24 It thus appears that the N-terminal region of full-length tau guides the tau assembly and growth, whereby the four pseudorepeat domains mediate the adsorption of tau to the membrane (Figure 6B).
Our data showing that Na+ and K+ modulate the interactions of tau with lipid membranes may be relevant to the presence of these ions on either side of the neuronal membrane. The prevalence of K+ in the intraneuronal fluid, where tau is present at μM concentrations,96 could inhibit nonbeneficial interactions of tau with the lipid membrane and the subsequent aggregation of tau. On the other side, the prevalence of Na+ in the extracellular fluid, where tau is present at a nM concentration, could play a role in facilitating the interaction of tau with the plasma membrane and in the internalization of extracellular tau. If the ability of intracellular K+ to keep tau soluble and to revert the Na+-induced attachment, self-assembly, and aggregation of human tau on brain lipid membranes could be confirmed in a more physiologically relevant context (e.g., in cells or neurons or the living brain), the described opposing effects of K+ and Na+ ions could play a significant role for the physiological and pathological roles of tau. However, as our in vitro assay describes a simple system whose key players are tau, ions, and supported brain lipid membranes, in vivo, many more parameters, including the phosphorylation of tau, may favor, change, or suppress the interaction of tau with cellular membranes. Our AFM-based assay is now applicable to systematically study the role of these parameters.
Materials and Methods.
Expression and Purification of Tau Constructs.
The wild-type full-length tau htau40 (2N4R), the truncated repeat domain construct tauRD (R1−R4, residues 244−372) of htau40, and the N-terminal region of htau40 (residues 1−257) were expressed in E. coli and purified as described.73 Briefly, cells were lysed by a French press, and the heat stable tau protein was extracted from the supernatant after boiling. Size exclusion chromatography was done using a Superdex Sepharose column (G200). Protein fractions were pooled and concentrated in phosphate-buffered saline (PBS) by Amicon filter tubes. The sample quality was checked by SDS PAGE.
Liposome Preparation.
Unilamellar liposomes (lipid vesicles) were prepared by the hydration of lipid films followed by extrusion through polycarbonate filter membranes. Chloroform stock solutions (1 mL aliquots at the concentration of 10 mg mL−1) of porcine brain total lipid extract (BTE) and extruding equipment used for liposome preparation were purchased from Avanti Polar Lipids. To prepare the liposomes, 1 mL of BTE stock solution was taken and the chloroform was evaporated under a gentle stream of nitrogen followed by overnight dehydration under a vacuum. Lipids were then rehydrated in buffer solution (150 mM NaCl, 20 mM Hepes, pH 7.4) and extruded 11 times through a 100 nm pore-size membrane. The freshly prepared liposomes (10 mg mL−1) were immediately aliquoted and stored at −80 °C. Liposomes were prepared at room temperature (rt, ~ 23 °C) with the only exception of the rehydration, which was done at 42 °C for 1 h with continuous agitation at 1400 rpm in a Thermomixer Comfort (Eppendorf). Buffer solutions were freshly made using nanopure water (18.2 MOhm cm−1) and pro analysis (>98.5%) purity-grade reagents from Sigma-Aldrich and Merck.
Preparation of Supported Lipid Membranes for AFM.
Supported lipid membranes (SLMs) were prepared by fusion of unilamellar liposomes on mica. An appropriate volume (~12 μL) of liposome solution of BTE was sonicated (25 kHz, 500 W, Transsonic TI-H-5, ELMA) for 5 min at rt after the addition of 6 μL of Ca2+-containing buffer (150 mM NaCl, 40 mM CaCl2, 20 mM Hepes, pH 7.4) and then immediately adsorbed onto a freshly cleaved mica disk of 6 mm diameter.75 After an adsorption time of ~30 min, during which the liposomes fused into a SLM,62 the sample was extensively rinsed with buffer solution (150 mM NaCl, 20 mM Hepes, pH 7.4, unless otherwise stated). The SLM was then imaged by contact-mode AFM to verify the presence of a continuous flat lipid membrane. Only if imaging showed no artifacts of the SLM (e.g., holes exposing the supporting mica or stacked membranes), the protein solution was injected into the fluid cell of the AFM. This quality criterion ensured that we observe the interaction of the injected proteins with an intact lipid membrane. To avoid sample preparation artifacts, the SLM was never exposed to air and the AFM head was never moved until the end of the experiment. Liposomes adsorption onto the supporting mica, AFM imaging, and incubation of the SLMs with proteins were performed at 37 °C.
AFM Imaging.
The AFM (Nanoscope IIe, Veeco) was equipped with a fluid cell and oxide-sharpened Si3N4 cantilevers (ORC8-PS-W, Olympus), having a nominal spring constant of 0.05 N m−1. The scanning frequency was between 2 and 8 lines s−1, and the force applied to the AFM cantilever was kept <50 pN to prevent structural perturbation of the sample.52 Imaging forces of the AFM were manually adjusted to compensate for thermal drift.52 Proportional and integral gains of the AFM were adjusted manually to minimize the error (deflection) signal and to maximize the height signal. AFM topographs (512 × 512 pixels) were recorded in trace and retrace scanning directions and were neither processed nor averaged.
Time-Lapse AFM Imaging of Tau Self-Assembly.
Before incubation of the SLM with protein, an AFM topograph of 10–20 μm2 of the SLM was recorded in buffer solution (150 mM NaCl, 20 mM Hepes, pH 7.4, unless otherwise stated). Only then, the protein solution (in the same buffer) was injected into the microscope head onto the SLM at concentrations ranging from 100 nM to 10 μM. After injecting the protein solution, the same sample area imaged in the absence of protein was imaged in contact-mode over the time of 90 min. The area of the assemblies formed by the proteins was quantified for every captured topograph and expressed as μm2 or as percentage of the total SLM area. The number of protein assemblies was quantified as the number of assemblies per unit area (100 μm2). Data were analyzed using a MATLAB (Math Works, Natick, MA) program developed in the lab or Adobe Photoshop CS6. All experiments were performed at 37 °C and repeated several times (n as indicated in figure legends) using independent htau40 and SLM preparations.
Time-Lapse AFM Imaging of Tau Disassembly.
To observe htau40 disassembly, freshly adsorbed SLMs of BTE were first incubated with a solution of 100 nM htau40 in 150 mM NaCl, 20 mM Hepes, pH 7.4, at 37 °C for 90 min to allow protein interaction and assemblies formation. After this, control images were captured to quantify the number and the area of the protein assemblies and then the SLM was extensively rinsed with buffer solution containing, respectively, 150 mM Li+, Na+, K+ (with or without 2 μM htau40), or Rb+. Time-lapse AFM topographs of 5–10 μm2 of the SLM surface were then captured at 10 min intervals for the first 30 min and later on every hour for 5 h to track any changes of the protein assemblies. During this incubation time, the fluid cell of the AFM was extensively rinsed every hour with buffer solution. Data were analyzed using a MATLAB (Math Works, Natick, MA) program developed in the lab or Adobe Photoshop CS6. htau40 disassembly was quantified comparing the area of htau40 assemblies measured at 90 min of incubation time in Na+-containing buffer, with the area of htau40 assemblies measured after buffer exchange. All experiments were performed at 37 °C and repeated several times (n as indicated in figure legends) using independent htau40 and SLM preparations.
Data Availability.
The authors declare that the main data supporting the findings of this study are available within the article and its Supporting Information file.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the ETH Zürich.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.8b01085.
AFM topographs showing defect-free supported lipid membranes (SLMs) prepared from porcine brain total lipid extract (BTE), AFM topographs showing htau40 incubated on a SLM made from BTE, control experiments showing that human htau40 but not bovine serum albumin (BSA) attaches and assembles in a Na+-dependent manner to SLMs made from BTE, controls showing the effect of the Na+ concentration on the number and area of htau40 assemblies on SLMs made from BTE, experiments showing the exceptional mechanical stability of human tau self-assembled onto SLMs, and experiments showing the Na+-dependent interaction of htau40, N-terminal region of tau and tauRD with SLMs made from BTE (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Weingarten MD; Lockwood AH; Hwo SY; Kirschner MW A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. U. S. A 1975, 72, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wang Y; Mandelkow E Tau in physiology and pathology. Nat. Rev. Neurosci 2016, 17, 5–21. [DOI] [PubMed] [Google Scholar]
- (3).Feinstein SC; Wilson L Inability of tau to properly regulate neuronal microtubule dynamics: a loss-of-function mechanism by which tau might mediate neuronal cell death. Biochim. Biophys. Acta, Mol. Basis Dis 2005, 1739, 268–279. [DOI] [PubMed] [Google Scholar]
- (4).Mandelkow EM; Mandelkow E Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harbor Perspect. Med 2012, 2, a006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Stamer K; Vogel R; Thies E; Mandelkow E; Mandelkow EM Tau blocks traffic of organelles, neurofilaments, and APP vesicles in neurons and enhances oxidative stress. J. Cell Biol 2002, 156, 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dixit R; Ross JL; Goldman YE; Holzbaur EL Differential regulation of dynein and kinesin motor proteins by tau. Science 2008, 319, 1086–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Vershinin M; Carter BC; Razafsky DS; King SJ; Gross SP Multiple-motor based transport and its regulation by Tau. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Konzack S; Thies E; Marx A; Mandelkow EM; Mandelkow E Swimming against the tide: mobility of the microtubule-associated protein tau in neurons. J. Neurosci 2007, 27, 9916–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kanaan NM; Morfini GA; LaPointe NE; Pigino GF; Patterson KR; Song Y; Andreadis A; Fu Y; Brady ST; Binder LI Pathogenic forms of tau inhibit kinesin-dependent axonal transport through a mechanism involving activation of axonal phosphotransferases. J. Neurosci 2011, 31, 9858–9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Magnani E; Fan J; Gasparini L; Golding M; Williams M; Schiavo G; Goedert M; Amos LA; Spillantini MG Interaction of tau protein with the dynactin complex. EMBO J 2007, 26, 4546–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Caceres A; Kosik KS Inhibition of neurite polarity by tau antisense oligonucleotides in primary cerebellar neurons. Nature 1990, 343, 461–463. [DOI] [PubMed] [Google Scholar]
- (12).Knops J; Kosik KS; Lee G; Pardee JD; Cohen-Gould L; McConlogue L Overexpression of tau in a nonneuronal cell induces long cellular processes. J. Cell Biol 1991, 114, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dawson HN; Ferreira A; Eyster MV; Ghoshal N; Binder LI; Vitek MP Inhibition of neuronal maturation in primary hippocampal neurons from tau deficient mice. J. Cell Sci 2001, 114, 1179–1187. [DOI] [PubMed] [Google Scholar]
- (14).Harada A; Oguchi K; Okabe S; Kuno J; Terada S; Ohshima T; Sato-Yoshitake R; Takei Y; Noda T; Hirokawa N Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature 1994, 369, 488–491. [DOI] [PubMed] [Google Scholar]
- (15).Hong XP; Peng CX; Wei W; Tian Q; Liu YH; Yao XQ; Zhang Y; Cao FY; Wang Q; Wang JZ Essential role of tau phosphorylation in adult hippocampal neurogenesis. Hippocampus 2010, 20, 1339–1349. [DOI] [PubMed] [Google Scholar]
- (16).Fuster-Matanzo A; de Barreda EG; Dawson HN; Vitek MP; Avila J; Hernandez F Function of tau protein in adult newborn neurons. FEBS Lett 2009, 583, 3063–3068. [DOI] [PubMed] [Google Scholar]
- (17).Violet M; Delattre L; Tardivel M; Sultan A; Chauderlier A; Caillierez R; Talahari S; Nesslany F; Lefebvre B; Bonnefoy E; Buee L; Galas MC A major role for Tau in neuronal DNA and RNA protection in vivo under physiological and hyperthermic conditions. Front. Cell. Neurosci 2014, 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sultan A; Nesslany F; Violet M; Begard S; Loyens A; Talahari S; Mansuroglu Z; Marzin D; Sergeant N; Humez S; Colin M; Bonnefoy E; Buee L; Galas MC Nuclear tau, a key player in neuronal DNA protection. J. Biol. Chem 2011, 286, 4566–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Klein C; Kramer EM; Cardine AM; Schraven B; Brandt R; Trotter J Process outgrowth of oligodendrocytes is promoted by interaction of fyn kinase with the cytoskeletal protein tau. J. Neurosci 2002, 22, 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Gauthier-Kemper A; Weissmann C; Golovyashkina N; SeboLemke Z; Drewes G; Gerke V; Heinisch JJ; Brandt R The frontotemporal dementia mutation R406W blocks tau’s interaction with the membrane in an annexin A2-dependent manner. J. Cell Biol 2011, 192, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Maas T; Eidenmuller J; Brandt R Interaction of tau with the neural membrane cortex is regulated by phosphorylation at sites that are modified in paired helical filaments. J. Biol. Chem 2000, 275, 15733–15740. [DOI] [PubMed] [Google Scholar]
- (22).Farah CA; Perreault S; Liazoghli D; Desjardins M; Anton A; Lauzon M; Paiement J; Leclerc N Tau interacts with Golgi membranes and mediates their association with microtubules. Cell Motil. Cytoskeleton 2006, 63, 710–724. [DOI] [PubMed] [Google Scholar]
- (23).Brandt R; Leger J; Lee G Interaction of tau with the neural plasma membrane mediated by tau’s amino-terminal projection domain. J. Cell Biol 1995, 131, 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ait-Bouziad N; Lv G; Mahul-Mellier AL; Xiao S; Zorludemir G; Eliezer D; Walz T; Lashuel HA Discovery and characterization of stable and toxic Tau/phospholipid oligomeric complexes. Nat. Commun 2017, 8, 1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Cleveland DW; Hwo SY; Kirschner MW Purification of tau, a microtubule-associated protein that induces assembly of microtubules from purified tubulin. J. Mol. Biol 1977, 116, 207–225. [DOI] [PubMed] [Google Scholar]
- (26).Mandelkow EM; Biernat J; Drewes G; Gustke N; Trinczek B; Mandelkow E Tau domains, phosphorylation, and interactions with microtubules. Neurobiol. Aging 1995, 16, 355–362. [DOI] [PubMed] [Google Scholar]
- (27).Brandt R; Lee G Functional organization of microtubule-associated protein tau. Identification of regions which affect microtubule growth, nucleation, and bundle formation in vitro. J. Biol. Chem 1993, 268, 3414–3419. [PubMed] [Google Scholar]
- (28).Buee L; Bussiere T; Buee-Scherrer V; Delacourte A; Hof PR Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Rev 2000, 33, 95–130. [DOI] [PubMed] [Google Scholar]
- (29).Lee G; Neve RL; Kosik KS The microtubule binding domain of tau protein. Neuron 1989, 2, 1615–1624. [DOI] [PubMed] [Google Scholar]
- (30).Andreadis A Misregulation of tau alternative splicing in neurodegeneration and dementia. Prog. Mol. Subcell. Biol 2006, 44, 89–107. [DOI] [PubMed] [Google Scholar]
- (31).Gustke N; Trinczek B; Biernat J; Mandelkow EM; Mandelkow E Domains of tau protein and interactions with microtubules. Biochemistry 1994, 33, 9511–9522. [DOI] [PubMed] [Google Scholar]
- (32).Wegmann S; Medalsy ID; Mandelkow E; Muller DJ The fuzzy coat of pathological human Tau fibrils is a two-layered polyelectrolyte brush. Proc. Natl. Acad. Sci. U. S. A 2013, 110, E313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lee VM; Goedert M; Trojanowski JQ Neurodegenerative tauopathies. Annu. Rev. Neurosci 2001, 24, 1121–1159. [DOI] [PubMed] [Google Scholar]
- (34).Goedert M; Jakes R; Spillantini MG; Hasegawa M; Smith MJ; Crowther RA Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 1996, 383, 550–553. [DOI] [PubMed] [Google Scholar]
- (35).Kampers T; Friedhoff P; Biernat J; Mandelkow EM; Mandelkow E RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett 1996, 399, 344–349. [DOI] [PubMed] [Google Scholar]
- (36).Wille H; Drewes G; Biernat J; Mandelkow EM; Mandelkow E Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. J. Cell Biol 1992, 118, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chirita CN; Necula M; Kuret J Anionic micelles and vesicles induce tau fibrillization in vitro. J. Biol. Chem 2003, 278, 25644–25650. [DOI] [PubMed] [Google Scholar]
- (38).Elbaum-Garfinkle S; Ramlall T; Rhoades E The role of the lipid bilayer in tau aggregation. Biophys. J 2010, 98, 2722–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kunze G; Barre P; Scheidt HA; Thomas L; Eliezer D; Huster D Binding of the three-repeat domain of tau to phospholipid membranes induces an aggregated-like state of the protein. Biochim. Biophys. Acta, Biomembr 2012, 1818, 2302–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Jones EM; Dubey M; Camp PJ; Vernon BC; Biernat J; Mandelkow E; Majewski J; Chi EY Interaction of tau protein with model lipid membranes induces tau structural compaction and membrane disruption. Biochemistry 2012, 51, 2539–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Kayed R; Sokolov Y; Edmonds B; McIntire TM; Milton SC; Hall JE; Glabe CG Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem 2004, 279, 46363–46366. [DOI] [PubMed] [Google Scholar]
- (42).Quist A; Doudevski I; Lin H; Azimova R; Ng D; Frangione B; Kagan B; Ghiso J; Lal R Amyloid ion channels: a common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 10427–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Chi EY; Frey SL; Lee KY Ganglioside G(M1)-mediated amyloid-beta fibrillogenesis and membrane disruption. Biochemistry 2007, 46, 1913–1924. [DOI] [PubMed] [Google Scholar]
- (44).Demuro A; Mina E; Kayed R; Milton SC; Parker I; Glabe CG Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. J. Biol. Chem 2005, 280, 17294–17300. [DOI] [PubMed] [Google Scholar]
- (45).Lira-De León, K. I; De Anda-Hernandez, M. A; Campos-Pena V; Meraz-Ríos MA Plasma membrane-associated PHF-core could be the trigger for Tau aggregation in Alzheimer’s disease In Current Hypotheses and Research Milestones in Alzheimer’s Disease; Maccioni RB, Perry G, Eds.; Springer US: Boston, MA, 2009; pp 93–100. [Google Scholar]
- (46).Holmes BB; Diamond MI Prion-like properties of Tau protein: the importance of extracellular Tau as a therapeutic target. J. Biol. Chem 2014, 289, 19855–19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Clavaguera F; Bolmont T; Crowther RA; Abramowski D; Frank S; Probst A; Fraser G; Stalder AK; Beibel M; Staufenbiel M; Jucker M; Goedert M; Tolnay M Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol 2009, 11, 909–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).de Calignon A; Polydoro M; Suarez-Calvet M; William C; Adamowicz DH; Kopeikina KJ; Pitstick R; Sahara N; Ashe KH; Carlson GA; Spires-Jones TL; Hyman BT Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 2012, 73, 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Brettschneider J; Del Tredici K; Lee VM; Trojanowski JQ Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nat. Rev. Neurosci 2015, 16, 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Richter RP; Brisson AR Following the Formation of Supported Lipid Bilayers on Mica: A Study Combining AFM, QCMD, and Ellipsometry. Biophys. J 2005, 88, 3422–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Mingeot-Leclercq M-P; Deleu M; Brasseur R; Dufrene YF Atomic force microscopy of supported lipid bilayers. Nat. Protoc 2008, 3, 1654–1659. [DOI] [PubMed] [Google Scholar]
- (52).Muller DJ; Engel A Atomic force microscopy and spectroscopy of native membrane proteins. Nat. Protoc 2007, 2, 2191–2197. [DOI] [PubMed] [Google Scholar]
- (53).Frederix PL; Bosshart PD; Engel A Atomic force microscopy of biological membranes. Biophys. J 2009, 96, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Engel A; Muller DJ Observing single biomolecules at work with the atomic force microscope. Nat. Struct. Biol 2000, 7, 715–718. [DOI] [PubMed] [Google Scholar]
- (55).Muller DJ; Dufrene YF Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat. Nanotechnol 2008, 3, 261–269. [DOI] [PubMed] [Google Scholar]
- (56).Muller DJ; Hand GM; Engel A; Sosinsky GE Conformational changes in surface structures of isolated connexin 26 gap junctions. EMBO J 2002, 21, 3598–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Yu J; Bippes CA; Hand GM; Muller DJ; Sosinsky GE Aminosulfonate modulated pH-induced conformational changes in connexin26 hemichannels. J. Biol. Chem 2007, 282, 8895–8904. [DOI] [PubMed] [Google Scholar]
- (58).Mari SA; Koster S; Bippes CA; Yildiz O; Kuhlbrandt W; Muller DJ pH-induced conformational change of the beta-barrelforming protein OmpG reconstituted into native E. coli lipids. J. Mol. Biol 2010, 396, 610–616. [DOI] [PubMed] [Google Scholar]
- (59).Mari SA; Pessoa J; Altieri S; Hensen U; Thomas L; Morais-Cabral JH; Muller DJ Gating of the MlotiK1 potassium channel involves large rearrangements of the cyclic nucleotide-binding domains. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 20802–20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Muller DJ; Engel A; Matthey U; Meier T; Dimroth P; Suda K Observing membrane protein diffusion at subnanometer resolution. J. Mol. Biol 2003, 327, 925–930. [DOI] [PubMed] [Google Scholar]
- (61).Yamashita H; Voitchovsky K; Uchihashi T; Contera SA; Ryan JF; Ando T Dynamics of bacteriorhodopsin 2D crystal observed by high-speed atomic force microscopy. J. Struct. Biol 2009, 167, 153–158. [DOI] [PubMed] [Google Scholar]
- (62).Mulvihill E; van Pee K; Mari SA; Muller DJ; Yildiz O Directly observing the lipid-dependent self-assembly and pore-forming mechanism of the cytolytic toxin listeriolysin O. Nano Lett 2015, 15, 6965–6973. [DOI] [PubMed] [Google Scholar]
- (63).van Pee K; Mulvihill E; Muller DJ; Yildiz O Unraveling the pore-forming steps of pneumolysin from Streptococcus pneumoniae. Nano Lett 2016, 16, 7915–7924. [DOI] [PubMed] [Google Scholar]
- (64).Koenig BW; Krueger S; Orts WJ; Majkrzak CF; Berk NF; Silverton JV; Gawrisch K Neutron reflectivity and atomic force microscopy studies of a lipid bilayer in water adsorbed to the surface of a silicon single crystal. Langmuir 1996, 12, 1343–1350. [Google Scholar]
- (65).Bayerl TM; Bloom M Physical properties of single phospholipid bilayers adsorbed to micro glass beads. A new vesicular model system studied by 2H-nuclear magnetic resonance. Biophys. J 1990, 58, 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Johnson SJ; Bayerl TM; McDermott DC; Adam GW; Rennie AR; Thomas RK; Sackmann E Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons. Biophys. J 1991, 59, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Tanaka M; Sackmann E Polymer-supported membranes as models of the cell surface. Nature 2005, 437, 656–663. [DOI] [PubMed] [Google Scholar]
- (68).Muller DJ; Engel A Strategies to prepare and characterize native membrane proteins and protein membranes by AFM. Curr. Opin. Colloid Interface Sci 2008, 13, 338–350. [Google Scholar]
- (69).Grandbois M; Clausen-Schaumann H; Gaub H Atomic force microscope imaging of phospholipid bilayer degradation by phospholipase A2. Biophys. J 1998, 74, 2398–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Sborgi L; Ruhl S; Mulvihill E; Pipercevic J; Heilig R; Stahlberg H; Farady CJ; Muller DJ; Broz P; Hiller S GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J 2016, 35, 1766–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Serdiuk T; Balasubramaniam D; Sugihara J; Mari SA; Kaback HR; Muller DJ YidC assists the stepwise and stochastic folding of membrane proteins. Nat. Chem. Biol 2016, 12, 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Kumar S; Tepper K; Kaniyappan S; Biernat J; Wegmann S; Mandelkow EM; Muller DJ; Mandelkow E Stages and conformations of the Tau repeat domain during aggregation and its effect on neuronal toxicity. J. Biol. Chem 2014, 289, 20318–20332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Tepper K; Biernat J; Kumar S; Wegmann S; Timm T; Hubschmann S; Redecke L; Mandelkow EM; Muller DJ; Mandelkow E Oligomer formation of tau protein hyperphosphorylated in cells. J. Biol. Chem 2014, 289, 34389–34407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Israelachvili J Intermolecular and Surface Forces; Academic Press: London, 1991. [Google Scholar]
- (75).Muller DJ; Amrein M; Engel A Adsorption of biological molecules to a solid support for scanning probe microscopy. J. Struct. Biol 1997, 119, 172–188. [DOI] [PubMed] [Google Scholar]
- (76).Blaustein K; Matteson DR Cellular Physiology, 1st ed.; Elsevier, 2004. [Google Scholar]
- (77).Grandbois M; Beyer M; Rief M; Clausen-Schaumann H; Gaub HE How Strong Is a Covalent Bond? Science 1999, 283, 1727–1730. [DOI] [PubMed] [Google Scholar]
- (78).Hoh JH; Lal R; John SA; Revel J-P; Arnsdorf MF Atomic force microscopy and dissection of gap junctions. Science 1991, 253, 1405–1408. [DOI] [PubMed] [Google Scholar]
- (79).Fotiadis D; Muller DJ; Tsiotis G; Hasler L; Tittmann P; Mini T; Jeno P; Gross H; Engel A Surface analysis of the photosystem I complex by electron and atomic force microscopy. J. Mol. Biol 1998, 283, 83–94. [DOI] [PubMed] [Google Scholar]
- (80).Fotiadis D; Scheuring S; Muller SA; Engel A; Muller DJ Imaging and manipulation of biological structures with the AFM. Micron 2002, 33, 385–397. [DOI] [PubMed] [Google Scholar]
- (81).Wegmann S; Jung YJ; Chinnathambi S; Mandelkow EM; Mandelkow E; Muller DJ Human Tau isoforms assemble into ribbon-like fibrils that display polymorphic structure and stability. J. Biol. Chem 2010, 285, 27302–27313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Santacruz K; Lewis J; Spires T; Paulson J; Kotilinek L; Ingelsson M; Guimaraes A; DeTure M; Ramsden M; McGowan E; Forster C; Yue M; Orne J; Janus C; Mariash A; Kuskowski M; Hyman B; Hutton M; Ashe KH Tau suppression in a neurodegenerative mouse model improves memory function. Science 2005, 309, 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Polydoro M; de Calignon A; Suarez-Calvet M; Sanchez L; Kay KR; Nicholls SB; Roe AD; Pitstick R; Carlson GA; Gomez-Isla T; Spires-Jones TL; Hyman BT Reversal of neurofibrillary tangles and tau-associated phenotype in the rTgTauEC model of early Alzheimer’s disease. J. Neurosci 2013, 33, 13300–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Hyman BT; Van Hoesen GW; Damasio AR; Barnes CL Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science 1984, 225, 1168–1170. [DOI] [PubMed] [Google Scholar]
- (85).Barghorn S; Biernat J; Mandelkow E Purification of recombinant tau protein and preparation of Alzheimer-paired helical filaments in vitro. Amyloid Proteins 2004, 299, 035–052. [DOI] [PubMed] [Google Scholar]
- (86).Goedert M; Jakes R Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J 1990, 9, 4225–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Derisbourg M; Leghay C; Chiappetta G; Fernandez-Gomez FJ; Laurent C; Demeyer D; Carrier S; Buee-Scherrer V; Blum D; Vinh J; Sergeant N; Verdier Y; Buee L; Hamdane M Role of the Tau N-terminal region in microtubule stabilization revealed by new endogenous truncated forms. Sci. Rep 2015, 5, 9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Kadavath H; Hofele RV; Biernat J; Kumar S; Tepper K; Urlaub H; Mandelkow E; Zweckstetter M Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 7501–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Barghorn S; Zheng-Fischhofer Q; Ackmann M; Biernat J; von Bergen M; Mandelkow EM; Mandelkow E Structure, microtubule interactions, and paired helical filament aggregation by tau mutants of frontotemporal dementias. Biochemistry 2000, 39, 11714–11721. [DOI] [PubMed] [Google Scholar]
- (90).Hong M; Zhukareva V; Vogelsberg-Ragaglia V; Wszolek Z; Reed L; Miller BI; Geschwind DH; Bird TD; McKeel D; Goate A; Morris JC; Wilhelmsen KC; Schellenberg GD; Trojanowski JQ; Lee VM Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science 1998, 282, 1914–1917. [DOI] [PubMed] [Google Scholar]
- (91).Wischik CM; Novak M; Thogersen HC; Edwards PC; Runswick MJ; Jakes R; Walker JE; Milstein C; Roth M; Klug A Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A 1988, 85, 4506–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Mukrasch MD; Bibow S; Korukottu J; Jeganathan S; Biernat J; Griesinger C; Mandelkow E; Zweckstetter M Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol 2009, 7, e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).von Bergen M; Friedhoff P; Biernat J; Heberle J; Mandelkow EM; Mandelkow E Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc. Natl. Acad. Sci. U.S. A 2000, 97, 5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Friedhoff P; von Bergen M; Mandelkow EM; Davies P; Mandelkow E A nucleated assembly mechanism of Alzheimer paired helical filaments. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 15712–15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Jeganathan S; von Bergen M; Mandelkow EM; Mandelkow E The natively unfolded character of tau and its aggregation to Alzheimer-like paired helical filaments. Biochemistry 2008, 47, 10526–10539. [DOI] [PubMed] [Google Scholar]
- (96).Avila J Intracellular and extracellular tau. Front. Neurosci 2010, 4, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the main data supporting the findings of this study are available within the article and its Supporting Information file.