The Ca2+ sensor SCaBP3 promotes the self-inhibition of plasma membrane H+-ATPase and is involved in the response to alkaline stress in Arabidopsis.
Abstract
Saline-alkali soil is a major environmental constraint impairing plant growth and crop productivity. In this study, we identified a Ca2+ sensor/kinase/plasma membrane (PM) H+-ATPase module as a central component conferring alkali tolerance in Arabidopsis (Arabidopsis thaliana). We report that the SCaBP3 (SOS3-LIKE CALCIUM BINDING PROTEIN3)/CBL7 (CALCINEURIN B-LIKE7) loss-of-function plants exhibit enhanced stress tolerance associated with increased PM H+-ATPase activity and provide fundamental mechanistic insights into the regulation of PM H+-ATPase activity. Consistent with the genetic evidence, interaction analyses, in vivo reconstitution experiments, and determination of H+-ATPase activity indicate that interaction of the Ca2+ sensor SCaBP3 with the C-terminal Region I domain of the PM H+-ATPase AHA2 (Arabidopsis thaliana PLASMA MEMBRANE PROTON ATPASE2) facilitates the intramolecular interaction of the AHA2 C terminus with the Central loop region of the PM H+-ATPase to promote autoinhibition of H+-ATPase activity. Concurrently, direct interaction of SCaPB3 with the kinase PKS5 (PROTEIN KINASE SOS2-LIKE5) stabilizes the kinase-ATPase interaction and thereby fosters the inhibitory phosphorylation of AHA2 by PKS5. Consistently, yeast reconstitution experiments and genetic analysis indicate that SCaBP3 provides a bifurcated pathway for coordinating intramolecular and intermolecular inhibition of PM H+-ATPase. We propose that alkaline stress-triggered Ca2+ signals induce SCaBP3 dissociation from AHA2 to enhance PM H+-ATPase activity. This work illustrates a versatile signaling module that enables the stress-responsive adjustment of plasma membrane proton fluxes.
INTRODUCTION
Carbonate-rich saline and alkaline soil is a major environmental factor influencing the composition of ecosystems and limiting plant growth and crop productivity. Soil is classified as being saline-alkaline when the electrical conductivity of the saturation extract in the root zone is greater than 4 mm hos/cm (∼40 mM NaCl) at 25°C and the exchangeable-sodium percentage is greater than 15. It has been estimated that over 954 million hectares of soils worldwide are affected by saline-alkaline conditions (Ayyub et al., 2015). Soil salinity often cooccurs with alkalinity, because of the high abundance of sodium carbonate (Na2CO3) or sodium bicarbonate (NaHCO3) in the soil, and this problem is getting more severe due to irrigation, flood water, and industry activity. High pH in soil directly impairs the uptake of water and nutrients, such as N, P, K, Ca, Mg, Fe, and Zn (Pissaloux et al., 1995; Pessarakli, 1999). Consequently, the ways that saline-alkaline conditions affect plant physiology are quite distinct from the consequences of pure salt (sodium) stress (Yang and Guo, 2018a).
Plasma membrane (PM) H+-ATPases belong to a large family of ion transporters named P-type ATPases, including, for example, fungal PM H+-ATPase and animal Na+/K+-ATPase and Ca2+-ATPase (Palmgren, 2001). In plants and fungi, PM H+-ATPases are responsible for establishing the electrochemical proton gradient that maintains the intracellular and extracellular pH balance and for driving numerous transmembrane transporters (Palmgren, 2001; Falhof et al., 2016).
In the yeast Saccharomyces cerevisiae, PM H+-ATPases are encoded by two genes, PMA1 (Saccharomyces cerevisiae PLASMA MEMBRANE PROTON ATPASE1) and PMA2 (Serrano et al., 1986; Schlesser et al., 1988), and PMA1 is essential for cell growth and development. In Arabidopsis (Arabidopsis thaliana), PM H+-ATPases are encoded by an 11-member gene family, AHA1 (Arabidopsis thaliana PLASMA MEMBRANE PROTON ATPASE1) to AHA11 (Palmgren, 2001). These PM H+-ATPases play important roles in signal transduction, cell elongation, turgor modulation, energizing the transport of ions and metabolites (Noji et al., 1988; Zhao et al., 2000; Santi and Schmidt, 2009; Haruta and Sussman, 2012; Takahashi et al., 2012; Fuglsang et al., 2014; Spartz et al., 2014), and responding to environment stimuli, such as salt and drought stress (Palmgren, 2001; Rober-Kleber et al., 2003; Fuglsang et al., 2007; Gévaudant et al., 2007; Merlot et al., 2007; Shen et al., 2011; Xue et al., 2018).
Plant PM H+-ATPases contain five cytosolic domains, including the N terminus, actuator domain, nucleotide binding domain, phosphorylation domain, and regulation (R) domain (Palmgren, 2001; Pedersen et al., 2007; Morth et al., 2011). The C-terminal R domain contains two critical autoinhibitory regions (Region I [RI] and RII) and several functionally important phosphorylation sites, the status of which modulates the enzyme activity (Palmgren et al., 1991; Jahn et al., 2002; Rudashevskaya et al., 2012). Regulation of PM H+-ATPases involves the phosphorylation/dephosphorylation of specific highly defined amino acid residues within the R domain and modulation of the AHA interaction with other proteins (Fuglsang et al., 2003, 2007, 2014; Nühse et al., 2004; Rudashevskaya et al., 2012). Phosphorylation of the C-terminal autoinhibitory domain at the penultimate amino acid (Thr-947, AHA2) promotes the binding of 14-3-3 proteins, leading to the activation of PM H+-ATPase AHA2 (Maudoux et al., 2000; Fuglsang et al., 2003). This activation is accompanied by the formation of a complex consisting of six H+-ATPases and six 14-3-3 proteins (Kanczewska et al., 2005; Ottmann et al., 2007). The PKS5 (PROTEIN KINASE SOS2-LIKE5) kinase phosphorylates Ser-931 of AHA2, thereby impeding the interaction between AHA2 and 14-3-3 (Fuglsang et al., 2007). Based on genetic studies in yeast or protein cross-linking experiments, it has been hypothesized that the autoinhibition of PM H+-ATPase activity is achieved by intramolecular interaction of the R domain with other domain(s) of the pump (Palmgren et al., 1991; Axelsen et al., 1999; Nguyen et al., 2018) and that somehow mutual relations between the N terminus and C terminus (R domain) are required for this autoinhibition (Ekberg et al., 2010). However, details of this regulatory mechanism remained poorly understood.
Ca2+ serves as a ubiquitous second messenger regulating plant growth and development (Rudd and Franklin-Tong, 2001; Chen et al., 2012; Kudla et al., 2018; Manishankar et al., 2018). In plants, Ca2+ signals are decoded by SOS3-like Calcium Binding Proteins/Calcineurin B-like proteins (SCaBPs/CBLs), which have been shown to play pivotal roles at the plasma membrane in regulating ion fluxes, signal transduction, and abiotic stress responses (Yu et al., 2014; Bender et al., 2018; Yang and Guo, 2018a, 2018b). In Arabidopsis, 10 SCaBPs/CBLs specifically interact with 26 distinct PKSs/CBL-interacting protein kinases (CIPKs), forming a complex signaling network that mediates various stimulus responses. For example, the SCaBP1/CBL2-PKS5/CIPK11 complex negatively regulates the activity of PM H+-ATPase AHA2 by phosphorylating its Ser-931 site (Fuglsang et al., 2007). Different Ca2+ signals appear to be involved in regulating proton pump activity at various stages and conditions (Schaller and Sussman, 1988; Kinoshita et al., 1995).
In this study, we identified the Ca2+ sensor protein, SCaBP3/CBL7, as a crucial regulator of the PM H+-ATPase. SCaBP3 physically interacted with the C terminus of the PM H+-ATPase and repressed the enzyme activity by promoting the interaction between the C terminus and the Central loop region of AHA2. Furthermore, SCaBP3 promoted the interaction between PKS5 and AHA2, thereby further enhancing and/or stabilizing PKS5-mediated repression of PM H+-ATPase activity. Our results suggest that the regulation of the extent of autoinhibition of the PM H+-ATPase is achieved by a complex array of phosphorylation-dependent and -independent mechanisms, which not only involve intramolecular interactions but also the Ca2+-mediated modulation by interaction of the PM H+-ATPase with SCaBP3 in concert with the simultaneous SCaBP3-dependent enhancement of PKS5-mediated AHA2 phosphorylation.
RESULTS
Loss of SCaBP3 Function Renders Arabidopsis Plants Tolerant to Alkali Stress
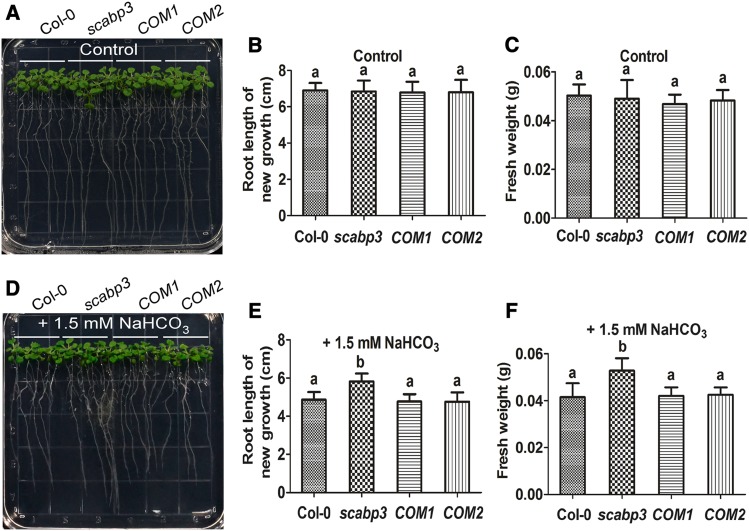
SCaBPs/CBLs and their interacting protein kinases PKS/CIPK play vital roles in regulating ion homeostasis, signal transduction, and abiotic stress responses (Yu et al., 2014; Zhu, 2016; Yang and Guo, 2018a; Köster et al., 2019). To determine whether a member of the SCaBP family in Arabidopsis is involved in the alkaline response, T-DNA insertion mutants of SCaBPs were analyzed for their growth in response to NaHCO3 stress. This approach revealed that the scabp3 mutant exhibited increased tolerance to NaHCO3 (Figure 1; Supplemental Figure 1). The T-DNA insertion in this mutant (scabp3/SAIL_201_A02) is located in the third intron of SCaBP3 (Supplemental Figure 1A) and was confirmed by PCR using a SCaBP3-specific primer in combination with a T-DNA left border primer (Supplemental Figure 1A; Supplemental Table). The absence of a SCaBP3 transcript in the scabp3 mutant was confirmed by quantitative real-time PCR, suggesting that this mutant likely represents a loss-of-function allele (Supplemental Figures 1A and 1B; Supplemental Table). Six-day-old wild-type Col-0 and scabp3 seedlings grown on Murashige and Skoog (MS) medium were transferred to MS medium containing 0, 1.5, 2, 4, or 6 mM NaHCO3 (pH 5.8, 6.55, 6.66, 6.80 and 6.93, respectively). No difference in growth was detected between Col-0 and scabp3 on MS medium (Figures 1A to 1C; Supplemental Figures 1C, 1F, 1I, 2A, 2D, and 2G; Supplemental Data Set ). By contrast, on MS medium supplemented with 1.5 or 2 mM NaHCO3, the scabp3 mutant appeared to be less sensitive to this stress condition than did Col-0 (Figures 1D to 1F; Supplemental Figures 1D, 1G, and 1J; Supplemental Data Set 1). The scabp3 mutant had longer roots and a greater fresh weight on MS medium with 1.5 or 2 mM NaHCO3 compared with Col-0 (Figures 1E and 1F; Supplemental Figures 1G and 1J). However, on MS medium supplemented with 4 mM NaHCO3, the growth of the seedlings of Col-0 and the scabp3 mutant was severely affected, and the seedlings died on MS medium containing 6 mM NaHCO3 (Supplemental Figures 2B, 2C, 2E, 2F, 2H, and 2I). When the pH of the medium supplemented with 2 mM NaHCO3 was adjusted to 5.8 (normal condition) by HCl, the growth of both seedlings was similar and returned to normal (Supplemental Figures 1E, 1H, and 1K). These observations indicated that the effect of NaHCO3 on seedling growth was mainly due to high-pH stress. To causally link this phenotype to the SCaBP3 locus, complementation lines containing a 3017-bp genomic SCaBP3 DNA fragment (including a 1227-bp promoter and a 549-bp 3′ untranslated region) were generated. From the T3 transgenic lines, we found two lines (COM1 and COM2) in which the expression of SCaBP3 was fully restored to wild-type levels (Supplemental Figure 1B) and the NaHCO3-response phenotype was rescued to the wild-type level (Figures 1A to 1F; Supplemental Figures 1C, 1D, 1F, 1G, 1I, and 1J). These results establish that the enhanced NaHCO3 tolerance of scabp3 lines is due to loss of SCaBP3 function. We further generated SCaBP3 overexpression lines (P35S:Flag-SCaBP3) in the Col-0 background, and the T3 transgenic plants (OE-1 and OE-2) displayed significantly enhanced growth inhibition in both the root and shoot compared with the growth of Col-0 under 1.5 mM NaHCO3 treatment (Supplemental Figures 3B to 3G).
Figure 1.
The Arabidopsis scabp3 Mutant Is Tolerant to Alkali Stress.
(A) and (D) Analysis of the NaHCO3-response phenotype in wild-type (Col-0), scabp3 mutant, and two genetic complementation line (COM1 and COM2) seedlings. Six-day-old Col-0, scabp3, COM1, and COM2 seedlings grown on MS medium were transferred to MS medium without or with 1.5 mM NaHCO3. Photographs were taken 7 and 12 d after transfer for control seedlings (on MS medium) and NaHCO3 treatment seedlings, respectively.
(B) and (E) Analysis of root length of new growth for seedlings in (A) and (D). Error bars represent sd (n = 20; four plates, five seedlings per plate) from measurements using the same preparation.
(C) and (F) Analysis of fresh weight for seedlings in (A) and (D). Error bars represent sd (n = 4; four plates, five seedlings on each plate are weighed together) from measurements using the same preparation.
The experiments were performed with three biological replicates with similar results. Statistical significance in (B), (C), (E), and (F) was determined by one-way ANOVA, P < 0.05. Significant differences are indicated by different lowercase letters.
Because plant growth should not be affected by 1.5 and 2 mM Na+ treatment, to investigate whether the root growth change in scabp3 is relative to environmental pH, 6-d-old Col-0, scabp3, and complementation line (COM1 and COM2) seedlings grown on MS medium were transferred to MS medium containing 0, 3, or 5 mM DMGA (3,3-dimethylglutaric acid), a well-established buffer to evaluate the plant’s response to low pH (Borgo, 2017; pH 5.80, 5.63, and 5.50, respectively). No difference in growth was detected between Col-0, scabp3, COM1, and COM2 on MS medium (Supplemental Figures 4A, 4D, and 4G). By contrast, on MS medium supplemented with 3 or 5 mM DMGA, the scabp3 mutant appeared to be less sensitive to DMGA treatments than Col-0 (Supplemental Figures 4B, 4C, 4E, 4F, 4H, and 4I). These results indicate that the growth change of scabp3 seedlings is likely caused by the change of environmental pH.
SCaBP3 Interacts with the C Terminus of Arabidopsis PM H+-ATPases
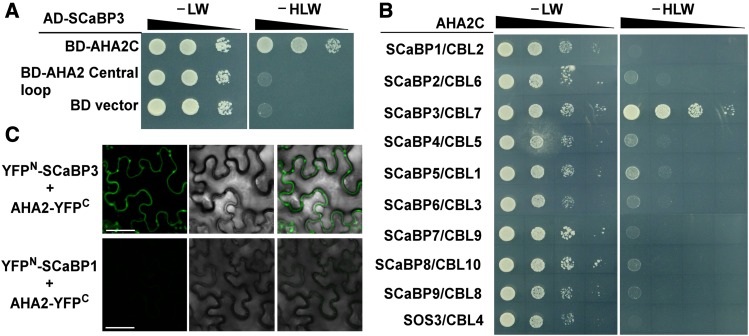
To study how SCaBP3 protein functions in alkali stress regulation, a yeast two-hybrid screen was performed to identify the interacting proteins of SCaBP3. Two putative interacting peptides (amino acid residues 850–949 in AHA1 and 850–948 in AHA2), the C terminus of the PM H+-ATPases AHA1 and AHA2 from a plant cDNA library, were identified in this screen. We further confirmed that SCaBP3 interacted with the AHA2 C terminus (amino acid residues 850–948) but not with the central large cytosolic loop of AHA2 (Central loop, residues 320–596; Figure 2A), and the AD-vector did not interact with the BD-AHA2C (Supplemental Figure 5A).
Figure 2.
SCaBP3 Interacts with the C-Terminal Region of AHA2.
(A) Yeast two-hybrid analysis of the interaction between SCaBP3 and the C terminus or Central loop of AHA2. Serial decimal dilutions of yeast cells were grown on synthetic complete medium without Leu and Trp (−LW, left panel) and on synthetic complete medium without His, Leu, and Trp (−HLW, right panel). Photographs were taken after 3 to 5 d of growth on the indicated medium.
(B) Comparative yeast two-hybrid interaction analysis of the AHA2 C terminus with the members of the SCaBP/CBL family in Arabidopsis. Photographs were taken after 3 to 5 d of growth on the indicated medium.
(C) BiFC analysis of the interaction between SCaBP3 and AHA2 in N. benthamiana (SCaBP1 was used as a negative control). Pairs of split-YFP constructs were transiently coexpressed in N. benthamiana leaves, and YFP fluorescence signal was detected using Leica SP5 confocal microscopy after 48 h of infiltration. Bars = 50 μm.
The experiments were performed with three biological replicates with similar results.
To determine the specificity of the observed interaction, all members of the SCaBP/CBL family were combined with the AHA2 C terminus in yeast two-hybrid assays. These analyses revealed that the AHA2 C terminus only specifically interacted with SCaBP3 (Figure 2B).
To investigate whether this interaction actually occurs in plants, bimolecular fluorescence complementation (BiFC) assays were employed (Waadt et al., 2008). The full-length coding sequences of SCaBP3 and AHA2 were translationally fused to the n-YFP and c-YFP of split-YFP constructs, respectively. As a control, we also generated an nYFP-SCaBP1 construct. When nYFP-SCaBP3 or nYFP-SCaBP1 was cotransformed with AHA2-cYFP into Nicotiana benthamiana leaves, only the combination of nYFP-SCaBP3 with AHA2-cYFP yielded plasma membrane-located florescence, but no signal was detected in the combination of the SCaBP1 or vector with AHA2 (Figure 2C; Supplemental Figure 5B). To confirm the expression of those genes, RT-PCR analysis was performed. There was no difference of the expression levels of SCaBP3, AHA2, and SCaBP1 mRNAs in different N. benthamiana leaves (Supplemental Figures 5C and 5D). Thus, SCaBP3 interacts with AHA2 in plants.
To determine the subcellular localization of SCaBP3 when expressed alone, a translational fusion construct of SCaBP3 with GFP was generated under the control of the UBQ10 promoter (PUBQ10:SCaBP3-GFP). The resulting construct was transformed into Col-0. Confocal imaging of roots of PUBQ10:SCaBP3-GFP plants revealed that SCaBP3-GFP fluorescence signals accumulated not only in the plasma membrane but also in the cytoplasm, confirming previously reported localization studies in N. benthamiana leaves (Supplemental Figures 6A to 6D; Batistic et al., 2010).
To determine the pattern of SCaBP3 expression, a 1.2-kb fragment of the 5′ region of SCaBP3 encompassing also its promoter was fused to the β-glucuronidase (GUS) reporter gene, and GUS expression was analyzed in 15 independent transgenic lines. PSCaBP3:GUS expression was found to be most intense in roots, leaves, flowers, and siliques (Supplemental Figures 6E to 6I). Consistent with the GUS staining results, RT-qPCR analysis also revealed that SCaBP3 was expressed in roots, stems, cauline leaves, rosette leaves, flowers, and siliques (Supplemental Figure 6J). Altogether, these experiments characterize SCaBP3/CBL7 as a ubiquitously expressed soluble protein that can interact with AHA H+-ATPases at the plasma membrane.
SCaBP3 Negatively Regulates PM H+-ATPase Activity
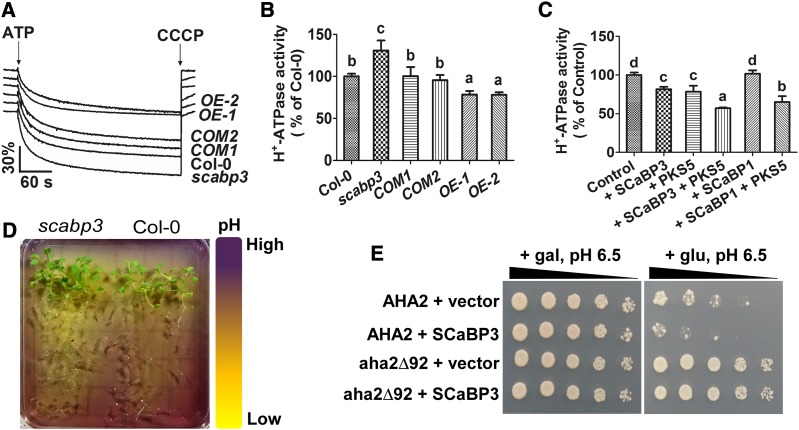
SCaBP3 directly interacted with the PM H+-ATPase C terminus, an autoinhibitory domain of the PM H+-ATPase. Therefore, we determined if SCaBP3 participates in the modulation of PM H+-ATPase activity in Arabidopsis. PM H+-ATPase activity is kept at a low level under nonstress conditions, and NaCl treatment can stimulate this activity (Yang et al., 2010). Plasma membrane-enriched vesicles were isolated from NaCl-treated seedlings of Col-0, scabp3, COM1, and COM2 by aqueous two-phase partitioning and used to analyze H+-transport activities. Compared with Col-0, the PM H+-ATPase activity in the scabp3 mutant was significantly higher (increased 27.24% ± 7.14% relative to Col-0; Supplemental Figures 7A and 7B), and in the complementation lines, the PM H+-ATPase activity was rescued to the level of wild-type Col-0. We further observed the PM H+-ATPase activity of SCaBP3 overexpression lines (OE-1 and OE-2) after NaCl treatment and found that the enzyme activity was significantly reduced (decreased 19.47% ± 9.10% and 27.87% ± 7.89% in OE-1 and OE-2 relative to Col-0, respectively; Supplemental Figures 7C and 7D). In addition, we isolated plasma membrane vesicles from NaHCO3-treated seedlings of Col-0, scabp3, COM1, COM2, OE-1, and OE-2 to detect H+-transport activity (Figures 3A and 3B; Supplemental Figure 7E). Compared with Col-0, the PM H+-ATPase activity was increased by 30.82% ± 11.83% in the scabp3 mutant, rescued to the Col-0 level in COM1 and COM2, and decreased by 21.58% ± 4.31% and 21.89% ± 2.97% in OE-1 and OE-2, respectively (Figures 3A and 3B). As a control, there was no difference of PM H+-ATPase protein content in Col-0, scabp3, COM1, and COM2 (Supplemental Figure 8A). These results suggest that SCaBP3 plays an important role in regulating the PM H+-ATPase activity.
Figure 3.
SCaBP3 Negatively Regulates PM H+-ATPase Activity.
(A) Measurement of PM H+-ATPase activity in vesicles isolated from leaves of NaHCO3-treated plants. CCCP, carbonyl cyanide m-chlorophenylhydrazone.
(B) Comparison of PM H+-ATPase activity as shown in (A). The initial 15-s slope of fluorescence quenching was graphed from (A).
(C) Comparison of PM H+-ATPase activity in the vesicles (isolated from NaHCO3-treated Col-0 seedlings) in the presence of 500 ng/mL control (GST), SCaBP3, PKS5, SCaBP3-PKS5, SCaBP1, or SCaBP1-PKS5 recombinant protein.
(D) Visualization of root acidification of scabp3 and Col-0 seedlings using the pH-sensitive dye bromocresol purple. For the root acidification assay, 14-d-old seedlings were transferred to MS medium (pH 6.8) containing 0.003% (w/v) bromocresol purple, and photographs were taken 3 d after transfer.
(E) AHA2 complementation of PM H+-ATPase activity in yeast. SCaBP3 was expressed with full-length AHA2 or a mutant of AHA2 lacking C-terminal residues (aha2Δ92). Cells were quintuple diluted in sterile water and grown on selective medium, pH 6.5. AHA2, pMP1745-AHA2; aha2Δ92, pMP1745-aha2Δ92; gal, galactose; glu, glucose; SCaBP3, pMP1645-SCaBP3; vector, pMP1645. The growth of the cells was detected 3 to 5 d after transformation.
In (A) to (C), 14-d-old seedlings grown under constant white light at 23°C were transferred to MS medium plus 1.5 mM NaHCO3 for 5 d before being collected for isolation of plasma membrane vesicles. Plasma membrane vesicles were isolated by two-phase partitioning. The experiments were performed with three biological replicates with similar results. Data represent means ± sd (n = 3) from measurements using the same preparation in (B) and (C). Lowercase letters indicate significant differences from control as determined by one-way ANOVA, P < 0.05.
Subsequently, we purified recombinant SCaBP3 protein and added it to plasma membrane-enriched vesicles isolated from NaCl-treated Col-0 seedlings to test whether it affects the PM H+-ATPase activity (Yang et al., 2010). In the presence of 500 ng/mL SCaBP3 protein, the PM H+-ATPase activity was decreased by 19.18% ± 6.77%, while the same amount of PKS5 protein (Fuglsang et al., 2007), a negative regulator of PM H+-ATPase, repressed 16.08% ± 2.04% of this activity (Supplemental Figure 7F). As controls, two homologs of SCaBP3, SCaBP1/CBL2 and SCaBP8/CBL10, were tested and had no effect on the PM H+-ATPase activity (Supplemental Figure 7F). In addition, we isolated the plasma membrane-enriched vesicles from the NaHCO3-treated Col-0 seedlings to test whether purified recombinant SCaBP3 protein affects PM H+-ATPase activity. In the presence of 500 ng/mL SCaBP3, the H+-transport activity was decreased by 19.35% ± 3.12%, while PKS5 repressed 21.44% ± 7.66% and SCaBP3 together with PKS5 repressed 42.92% ± 0.95% of its activity (Figure 3C). As controls, SCaBP1 had no effect on PM H+-ATPase activity, while SCaBP1 together with PKS5 repressed 34.86% ± 7.55% of its activity (Figure 3C). Improvement of PM H+-ATPase activity would be expected to exclude more H+ from the cytosol to the extracellular space in the root, and this would lead to enhanced acidification of the medium. A pH indicator, bromocresol purple, was used to observe the acidification of the medium around the root area of Col-0 and scabp3. As expected, scabp3 mutant seedlings made MS medium more acidic compared with Col-0 (Figure 3D). These results demonstrate that SCaBP3 is a negative regulator of the PM H+-ATPase.
We next analyzed the capability of SCaBP3 to regulate the PM H+-ATPase activity in a yeast reconstitution/complementation system. In the yeast strain RS72, the endogenous yeast H+ pump, PMA1, is under the control of the GAL1 promoter and, therefore, the yeast is only viable when grown on a medium with galactose as a carbon source (Axelsen et al., 1999; Fuglsang et al., 2007). When AHA2 is expressed under the control of the constitutive PMA1 promoter in this strain, constitutive AHA2 expression complements this strain and enables it to grow on a medium containing glucose on which the expression of PGAL1:PMA1 is suppressed (Regenberg et al., 1995). Coexpression of SCaBP3 with AHA2 resulted in reduced cell growth on glucose medium when compared with the cells that expressed the empty vector with AHA2, further supporting the notion that SCaBP3 negatively regulates AHA2 function (Figure 3E). We also expressed a truncated AHA2 form (aha2Δ92) deprived of the C-terminal 92 amino acid residues. Removal of these residues resulted in a more active H+-ATPase activity in yeast (Regenberg et al., 1995). The yeast assays indicated that the activity of truncated AHA2 (aha2Δ92) was not affected by SCaBP3 (Figure 3E). Together, these results suggest that SCaBP3 represses PM H+-ATPase activity through directly interacting with its C terminus.
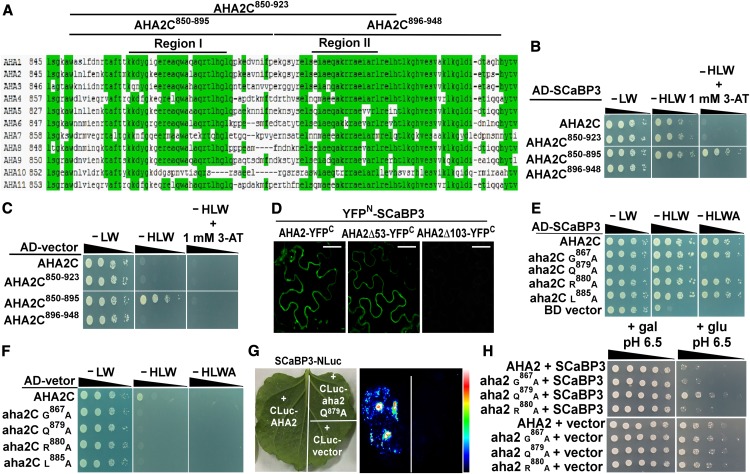
SCaBP3 Directly Interacts with RI of the AHA2 C Terminus
To further map the interaction domain in more detail, we divided the AHA2 C terminus into several fragments (Figure 4A). Removal of the last 25 amino acids from the AHA2 C terminus (AHA2C850-923) did not impact on the interaction with SCaBP3 when tested in the yeast two-hybrid system (Figure 4B). The AHA2 C-terminal cytoplasmic fragment (amino acid residues 850–923) contains two conserved regulatory domains of the PM H+-ATPase, namely RI and RII (Figure 4A). Considering this, we divided the AHA2 C terminus into two parts: one contained the RI domain (residues 850–895) and the other contained the RII domain (residues 896–948; Fuglsang et al., 1999, 2003). In yeast two-hybrid assays, we only detected an interaction between SCaBP3 and the fragment containing the RI domain but not with the fragment encompassing the RII domain (Figures 4B and 4C).
Figure 4.
SCaBP3 Interacts with PM H+-ATPase at the Region Containing the RI Domain.
(A) Amino acid sequence alignment of the C-terminal regions of PM H+-ATPases in Arabidopsis.
(B) Analysis of the interaction between SCaBP3 and AHA2 C terminus or truncated AHA2 C terminus. The C terminus was truncated into three different fragments: AH2C850-923, lacking the last 25 residues of the C terminus; AHA2C850-895, lacking the half of the AHA2 C terminus containing the RI domain; and AHA2C896-948, lacking the half of the AHA2 C terminus containing the RII domain. Cell growth was detected 3 to 5 d after transformation. Serial decimal dilutions of yeast cells were grown on synthetic complete medium without Leu and Trp (−LW, left panel), on synthetic complete medium without His, Leu, and Trp (−HLW, middle panel), and on synthetic complete medium without His, Leu, and Trp plus 3-aminotriazole (−HLW + 1 mM 3-AT, right panel).
(C) Negative control for analysis of the interaction between SCaBP3 and AHA2 C terminus or truncated AHA2 C terminus.
(D) Analysis of the interaction between SCaBP3 and AHA2 C terminus in N. benthamiana. SCaBP3, AHA2, and two truncations of AHA2 (AHA2Δ53 and AHA2Δ103) were used in BiFC experiments. AHA2Δ53, lacking the last 53 residues of AHA2; AHA2Δ103, lacking the last 103 residues of AHA2. YFP fluorescence signal was detected after 48 h of infiltration. Bars = 50 μm.
(E) Analysis of amino acids in AHA2 C terminus required for the interaction in a yeast two-hybrid analysis. The growth of the cells was detected 3 to 5 d after transformation.
(F) Negative control for analysis of amino acids in AHA2 C terminus required for the interaction(−HLWA, without His, Leu, Trp and Adenine).
(G) LCI analysis of the interaction between SCaBP3 and AHA2 or aha2 Q879A.
(H) AHA2 complementation of PM H+-ATPase activity in yeast. SCaBP3 was expressed with full-length AHA2 or different point mutations of AHA2. Cells were quintuple diluted in sterile water and grown on selective medium. AHA2, pMP1745-AHA2; aha2 G867A, pMP1745-aha2 G867A; aha2 Q879A, pMP1745-aha2 Q879A; aha2 R880A, pMP1745-aha2 R880A; gal, galactose; glu, glucose; SCaBP3, pMP1645-SCaBP3; vector, pMP1645. The growth of the cells was detected 3 to 5 d after transformation.
The experiments were performed with three biological replicates with similar results.
To test if the fragment containing the RI domain is critical for the interaction in vivo, we also employed BiFC assays. When we removed the entire C-terminal region from AHA2 (AHA21-845, AHA2Δ103), the interaction between AHA2 and SCaBP3 was abolished. By contrast, we readily detected the interaction between SCaBP3 and AHA21-895 (AHA2Δ53, removal of the RII domain) at the plasma membrane, which was similar to the localization pattern of the interaction between SCaBP3 and AHA2 (Figure 4D). To confirm the expression of those genes, RT-PCR analysis was performed. There was no difference of the expression levels of SCaBP3, AHA2, AHA2Δ53, and AHA2Δ103 mRNAs in the different transgenic N. benthamiana leaves (Supplemental Figure 9A). As controls, there was no interaction between vector and AHA2, AHA2Δ53, or AHA2Δ103, respectively (Supplemental Figures 9B and 9C). These results support the concept that the fragment containing the RI domain (residues 850–895) in the C terminus of the PM H+-ATPase has an important role in the interaction with SCaBP3 in Arabidopsis.
To further determine the importance of the interaction between SCaBP3 and the RI domain in regulating PM H+-ATPase activity, we mutated several conserved amino acid sites in the RI domain (G867A, Q879A, R880A, and L885A), which is involved in activating PM H+-ATPase activity (Axelsen et al., 1999). In a yeast two-hybrid assay, we found that the mutation in Q879 weakened the interaction between SCaBP3 and the AHA2 C terminus (Figures 4E and 4F). Furthermore, a luciferase complementation imaging (LCI) assay was used to confirm the interaction between AHA2C Q879A and SCaBP3; the results showed that the mutation in Q879 reduced the interaction between SCaBP3 and AHA2 (Figure 4G). There was no difference in the expression levels of AHA2, AHA2 Q879A, and SCaBP3 mRNAs in different transfected N. benthamiana leaves (Supplemental Figure 9D). When we subsequently expressed SCaBP3 with different AHA2 mutants (G867A, Q879A, and R880A) in RS72, we found that the repression of the growth of RS72 yeast cells by SCaBP3 was reduced by the Q879A mutation but was not affected by other mutations (Figure 4H). Therefore, Q879 appears to be an important site for SCaBP3 targeting, and the interaction between SCaBP3 and the RI domain is critical for repressing PM H+-ATPase activity.
SCaBP3 Promotes the Interaction between the AHA2 Central Loop and C Terminus and between PKS5 and the AHA2 C Terminus
The autoinhibitory function of the R domain has been suggested to be achieved by intramolecular interaction between the R domain and other, not yet identified, intracellular domain(s) of the PM H+-ATPase protein (Palmgren et al., 1991; Axelsen et al., 1999; Ekberg et al., 2010). An analysis of a series of mutants in the first half of the H+-ATPase C terminus (RI domain) of the Nicotiana tabacum H+-ATPase PMA2 indicates that the RI domain is involved in the interaction between the C terminus and other parts of the enzyme (Morsomme et al., 1996, 1998). The RI domain is thought to inhibit the interaction with the ATP binding region of PM H+-ATPase (Palmgren et al., 1991; Axelsen et al., 1999). Importantly, the PM H+-ATPase Central loop contains the pivotal domains for nucleotide binding (amino acid residues 338–488) and catalytic phosphorylation (amino acid residues 308–337 and 489–625; Pedersen et al., 2007). The potential interaction between the C terminus and the Central loop may shield this reactive site, thereby leading to a direct repression of PM H+-ATPase activity.
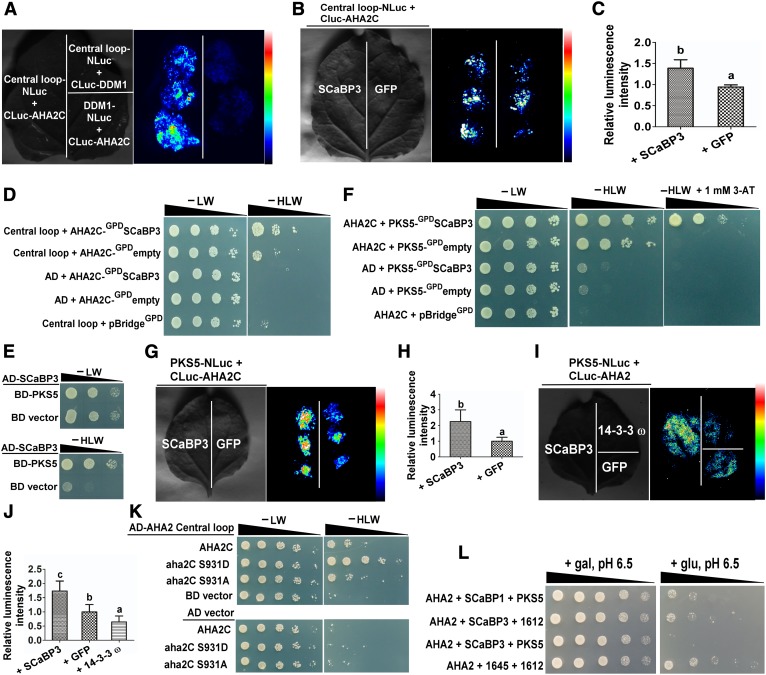
To test this hypothesis, we investigated the interaction between the AHA2 C terminus and the AHA2 Central loop region. The LCI assay revealed that the AHA2 C terminus can physically interact with the Central loop (Figure 5A) and that coexpression of SCaBP3 significantly enhanced this interaction (Figures 5B and 5C). As controls, no signals were detected in the combination of the Central loop-NLuc with CLuc-DDM1 (decreased DNA methylation1) or DDM1-NLuc with CLuc-AHA2C (Figure 5A). To confirm the expression of those genes, RT-PCR analysis was performed. There was no difference of the expression levels of SCaBP3 and AHA2 C terminus and Central loop mRNAs in different transfected N. benthamiana leaves (Supplemental Figure 10A).
Figure 5.
SCaBP3 Promotes the Interaction between AHA2 Central Loop Region and Its C Terminus and between PKS5 and AHA2 C Terminus.
(A) LCI system analysis of the interaction between AHA2 Central loop region and C terminus. DDM1 was used as a control.
(B) LCI system analysis of AHA2 Central loop interaction with its C terminus in the presence of SCaBP3. SCaBP3 was transiently cotransformed with AHA2 Central loop and C terminus in N. benthamiana leaves. GFP was used as a control.
(C) Statistical analysis of luminescence intensity as shown in (B).
(D) Yeast three-hybrid analysis of AHA2 Central loop interaction with its C terminus in the presence or absence of SCaBP3. SCaBP3 was expressed under the control of the GPD promoter in the pBridgeGPD vector. Serial decimal dilutions of yeast cells were grown on synthetic complete medium without Leu and Trp (−LW, left panel) and on synthetic complete medium without His, Leu, and Trp (−HLW, right panel). Photographs were taken after 3 to 6 d of growth on the indicated medium.
(E) Yeast two-hybrid analysis of the interaction between SCaBP3 and PKS5.
(F) Yeast three-hybrid analysis of the interaction of PKS5 with the AHA2 C terminus in the presence or absence of SCaBP3. SCaBP3 was expressed under the control of the GPD promoter in the pBridgeGPD vector. Photographs were taken after 3 to 6 d of growth on the indicated medium. 3-AT, 3-aminotriazole.
(G) LCI system analysis of the interaction of PKS5 with the AHA2 C terminus in the presence of SCaBP3. SCaBP3 was transiently coexpressed with AHA2 and PKS5 in N. benthamiana. GFP was used as a control.
(H) Statistical analysis of luminescence intensity as shown in (G).
(I) LCI system analysis of the interaction of PKS5 with AHA2 in the presence of SCaBP3. SCaBP3 was transiently cotransformed with AHA2 and PKS5 in N. benthamiana. 14-3-3 ω and GFP were used as controls.
(J) Statistical analysis of luminescence intensity as shown in (I).
(K) Yeast two-hybrid analysis the effect of different phosphorylation levels on the interaction with its Central loop. Photographs were taken after 3 to 5 d of growth on the indicated medium.
(L) Analysis of SCaBP3-PKS5 inhibition of PM H+-ATPase activity in yeast. Cells were quintuple diluted in sterile water and grown on selective medium, pH 6.5. AHA2, pMP1745-AHA2; gal, galactose; glu, glucose; PKS5, pMP1612-PKS5; SCaBP1, pMP1645-SCaBP1; SCaBP3, pMP1645-SCaBP3; 1612, pMP1612; 1645, pMP1645. The growth of the cells was detected 3 to 5 d after transformation.
The experiments were performed with three biological replicates with similar results. Data represent means ± sd (n = 6) from measurements using the same preparation as in (C), (H), and (J). Lowercase letters indicate significant differences from GFP treatment as determined by Student’s t test, P < 0.05.
To further corroborate these results, a yeast three-hybrid assay (Ren et al., 2013) was performed. The coding sequence of the AHA2 Central loop was cloned into the pGADT7 vector, and the pBridgeGPD vector was used to express both the AHA2 C terminus and SCaBP3. The expression of SCaBP3 was controlled by the GPD promoter. As shown in Figure 5D, the interaction between the AHA2 C terminus and Central loop when expressed alone was weak but was substantially enhanced upon coexpression of SCaBP3. These results suggest that the autoinhibitory effect of the C terminus on AHA2 activity may be caused by direct physical interaction with the Central loop. Importantly, SCaBP3 appears to enhance or stabilize this interaction to further suppress the PM H+-ATPase activity.
A previous study reported that a SCaBP1/CBL2-PKS5/CIPK11 complex negatively regulates PM H+-ATPase activity through phosphorylation at Ser-931 (Fuglsang et al., 2007). SCaBPs/CBLs often translate Ca2+ signals into regulation of downstream targets through phosphorylation or interaction by their interacting PKS/CIPK kinases (Xu et al., 2006; Held et al., 2011; Hashimoto et al., 2012). Therefore, we hypothesized that SCaBP3 may also somehow be involved in modulating PM H+-ATPase activity through SCaBP-PKS5. We observed that SCaBP3 directly interacted with PKS5 in yeast two-hybrid assays (Figure 5E). Furthermore, a yeast three-hybrid assay was performed to test whether SCaBP3 affected the interaction between PKS5 and the AHA2 C terminus. To this end, the coding sequence of the AHA2 C terminus was cloned into the pGADT7 vector, and the pBridgeGPD vector was used to express both PKS5 and SCaBP3. As shown in Figure 5F, SCaBP3 significantly enhanced the interaction between PKS5 and the AHA2 C terminus. Consistently, the AHA2 C terminus and PKS5 interacted, and SCaBP3 enhanced this interaction in an LCI assay (Figures 5G and 5H). Furthermore, we found that the interaction between AHA2 and PKS5 was reduced by 14-3-3 ω (Figures 5I and 5J). To confirm the expression of these constructs, RT-PCR analysis was performed. There was no difference in the expression of SCaBP3, AHA2C terminus, AHA2 Central loop, AHA2, PKS5, and 14-3-3 ω in different transfected N. benthamiana leaves (Supplemental Figures 10A to 10C).
Our results described above indicated that direct interaction between the C terminus and the Central loop contributes to the autoinhibition of the PM H+-ATPase activity, while our previous work had established a negative regulatory function for phosphorylated Ser-931 (Fuglsang et al., 2007). We therefore sought to address if the phosphorylation status of Ser-931 may affect the interaction between the AHA2 Central loop and its C terminus. To investigate this, we explored the interaction of the AHA2 C terminus harboring either a phosphorylation-preventing S931A substitution or a phosphorylation-mimicking S931D exchange with the Central loop region in yeast two-hybrid assays. For this approach, these two versions of the AHA2 C terminus (aha2C S931D and aha2C S931A) were cloned into the pGBKT7 vector. As depicted in Figure 5K, phosphorylation mimicking at Ser-931 markedly enhanced the interaction of AHA2 C terminus and Central loop, suggesting that phosphorylation of Ser-931 directly regulates the inhibitory interaction between the AHA2 C terminus and its Central loop.
As an alternative approach to determining the ability of the SCaBP3-PKS5 complex to modulate the PM H+-ATPase activity, we coexpressed AHA2 with different combinations of SCaBPs (SCaBP1 or SCaBP3) and PKS5 in the RS72 strain. Consistent with our previous report, coexpression of PKS5 and SCaBP1 repressed the growth of RS72 yeast cells (Figure 5L). However, quite remarkably, expression of SCaBP3 alone had a similar effect (Figure 5L). When we coexpressed PKS5 with SCaBP3 in this system, the growth of RS72 yeast cells was inhibited more severely than in yeast coexpressing PKS5 and SCaBP1 (Figure 5L). These results suggest that SCaBP3 inhibits PM H+-ATPase activity by two distinct mechanisms, of which the intramolecular mechanism is PKS5 independent and the intermolecular mechanism is PKS5 dependent. The intramolecular mechanism is that SCaBP3 inhibits the activity of PM H+-ATPase by enhancing the interaction of the C terminus of AHA2 with its Central loop, which is independent of PKS5. The intermolecular mechanism is that SCaBP3 enhances the interaction between PKS5 and AHA2, thereby enhancing the phosphorylation of PM H+-ATPase activity, which is dependent on PKS5.
Alkali Conditions Trigger the Relief of SCaBP3-Mediated Inhibition of AHA2
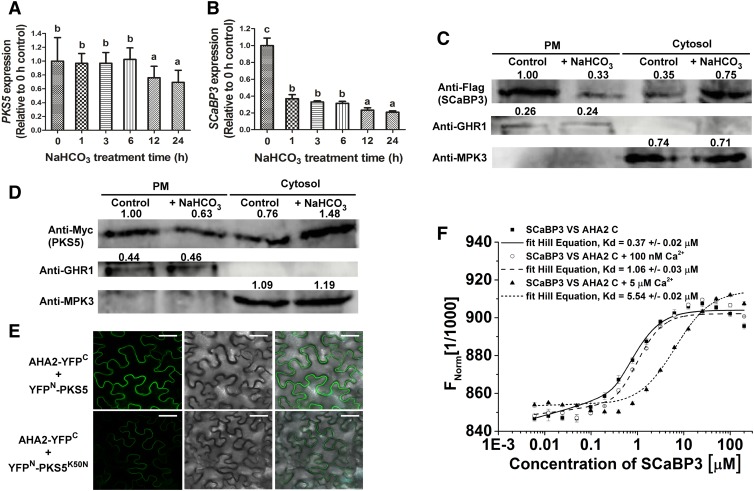
The PM H+-ATPase activity is activated by saline-alkali stress (Yang et al., 2010). To determine if SCaBP3 and PKS5 are regulated by alkali stress at the mRNA level, we determined their gene expression. The level of PKS5 mRNA was not affected by up to 6 h of NaHCO3 treatment but decreased after that time point (Figure 6A), while the mRNA of SCaBP3 mRNA rapidly decreased in Col-0 plants after NaHCO3 treatment (Figure 6B). To determine the subcellular localization of SCaBP3 and PKS5 proteins, Myc-PKS5 and Flag-SCaBP3 driven by the constitutive 35S promoter were transformed into Arabidopsis, respectively. Plasma membrane-enriched vesicles were isolated from the transgenic plants. SCaBP3 and PKS5 were located in both the cytosol and plasma membrane; however, their amounts were reduced in the plasma membrane fraction and increased in the cytosolic fraction after NaHCO3 treatment (Figures 6C and 6D), suggesting that activation of PM H+-ATPase under NaHCO3 stress involves the disassociation of SCaBP3 and PKS5 from the plasma membrane.
Figure 6.
Relief of SCaBP3-Regulated Inhibition of AHA2 under Alkaline Conditions.
(A) and (B) RT-qPCR assay for detection of PKS5 (A) and SCaBP3 (B) expression after NaHCO3 treatment. Total RNA was extracted from 7-d-old seedlings treated with 1.5 mM NaHCO3 for 0, 1, 3, 6, 12, or 24 h. The samples were treated with RNase-free DNase and then used to produce cDNAs. Then the cDNAs were used for detecting PKS5 (the primer pairs are PKS5 qRT-F and PKS5 qRT-R, and their sequences are listed in the Supplemental Table) and SCaBP3 (the primer pairs are SCaBP3 qRT-F and SCaBP3 qRT-R, and their sequences are listed in the Supplemental Table) expression using RT-qPCR assay. ACTIN2 (the primer pairs are ACTIN2-qF and ACTIN2-qR, and their sequences are listed in the Supplemental Table) served as an internal control.
(C) and (D) SCABP3 (C) and PKS5 (D) were detected in plasma membrane (PM) and cytosol with or without NaHCO3 treatment. Plasma membrane and cytosol were isolated from 7-d-old seedlings of Col-0 expressing 35SPro::3×Flag-SCABP3 or 35SPro::6×Myc-PKS5 treated with 1.5 mM NaHCO3 for 0 or 24 h. Isolation of plasma membrane was by two-phase partitioning. Equal amounts of plasma membrane and cytosolic proteins were separated by SDS-PAGE followed by analysis with anti-Flag, anti-Myc, anti-GHR1 (a plasma membrane-localized protein), or anti-MPK3 (a cytosol-localized protein) antibodies.
(E) Analysis of the interaction between AHA2 and PKS5/PKS5K50N (a dead kinase with no kinase activity) in N. benthamiana. Pairs of split-YFP constructs were transiently coexpressed in N. benthamiana leaves, and YFP fluorescence signal was detected using Leica SP5 confocal microscopy after 48 h of infiltration. Bars = 50 μm.
(F) Analysis of concentrations of Ca2+ on the interaction between SCaBP3 and the AHA2 C terminus using MST. Unlabeled GST-SCaBP3 recombinant protein was titrated to a constant amount of fluorescently NT647-labeled HIS-AHA2 C terminus recombinant peptide in the presence of 0, 100 nM, or 5 μM Ca2+. Kd, dissociation constant.
The experiments were performed with three biological replicates with similar results. Data represent means ± sd (n = 3) from measurements using the same preparation as in (A), (B), and (F). Lowercase letters indicate significant differences from the control as determined by one-way ANOVA, P < 0.05.
To determine whether PKS5 kinase activity has an impact on its interaction with AHA2, we employed BiFC assays to detect the interaction between AHA2 and PKS5 and between AHA2 and PKS5K50N (an inactive kinase version). The full-length coding sequences of PKS5/PKS5K50N and AHA2 were translationally fused to n-YFP and c-YFP, respectively. When nYFP-PKS5 or nYFP-PKS5K50N was coexpressed with AHA2-cYFP in N. benthamiana leaves, the combination of nYFP-PKS5 with AHA2-cYFP yielded a stronger fluorescence signal at the plasma membrane than the combination of PKS5K50N with AHA2 (Figure 6E; Supplemental Figure 11A).
To confirm the expression of PKS5 and PKS5K50N, immunoblot analysis was performed. We could not detect AHA2 protein in the transfected N. benthamiana; however, BiFC analysis showed that AHA2 itself (AHA2-YFPN + AHA2-YFPC) dimerizes in transfected N. benthamiana (Supplemental Figure 11B), suggesting that different AHA2 forms (AHA2-YFPN or AHA2-YFPC) could express in N. benthamiana leaves. There was no difference in the protein expression levels of PKS5 and PKS5K50N in different transfected N. benthamiana leaves (Supplemental Figure 11C). These results suggest that an activated PKS5 is more likely to interact with AHA2 in plants.
The cytosolic Ca2+ concentration increases under saline-alkali stress in plants (Fuglsang et al., 2007). To investigate whether the dissociation of SCaBP3 from PM H+-ATPases under alkali stress could be regulated by Ca2+ concentration, we performed microscale thermophoresis (MST) assays (Supplemental Figure 11D) in the presence of 0, 100 nM, or 5 µM Ca2+. As shown in Figure 6F, in the absence of Ca2+, the binding affinity between SCaBP3 and AHA2 C terminus was calculated as 0.37 ± 0.02 μM. The binding affinities were decreased to 1.06 ± 0.03 and 5.54 ± 0.02 μM between SCaBP3 and AHA2 C terminus in the presence of 100 nM and 5 µM Ca2+, respectively. These results suggest that the increased Ca2+ concentration reduces the interaction between SCaBP3 and AHA2.
Regulation of PM H+-ATPase Activity by SCaBP3 and PKS5 in Response to Alkaline Stress
As reported here for the scabp3 mutant, we previously observed that the pks5 loss-of-function mutant (pks5-1) was more tolerant to saline-alkali conditions than was Col-0 (Fuglsang et al., 2007; Yang et al., 2010). To further confirm the correlation between PM H+-ATPase activity and the alkali phenotype, AHA2∆C (amino acids 1–836, enhanced PM H+-ATPase activity; Liu et al., 2009) was cloned into the SUPERR:sXVE:HA vector (an inducible expression system; Schlücking et al., 2013). The SUPERR:sXVE:HA:AHA2∆C plasmid was transformed into Col-0 to obtain inducible expression lines (AHA2∆C-1 and AHA2∆C-2) (Supplemental Figure 12A). Six-day-old seedlings of Col-0, AHA2∆C-1, and AHA2∆C-2 grown on MS medium were transferred to MS medium, MS medium supplemented with 5 μM β-estradiol, MS medium supplemented with 1.5 mM NaHCO3, or MS medium supplemented with 5 μM β-estradiol and 1.5 mM NaHCO3. No difference in growth was detected between Col-0, AHA2∆C-1, and AHA2∆C-2 on the MS medium, MS medium supplemented with 5 μM β-estradiol, or MS medium supplemented with 1.5 mM NaHCO3 (Supplemental Figures 12B to 12J). On the MS medium supplemented with 5 μM β-estradiol and 1.5 mM NaHCO3, the AHA2∆C-1 and AHA2∆C-2 plants appeared to be less sensitive to this stress condition than Col-0 plants (Supplemental Figures 12K to 12M). These results indicate that a higher PM H+-ATPase activity results in the transgenic plants being more tolerant to NaHCO3 stress.
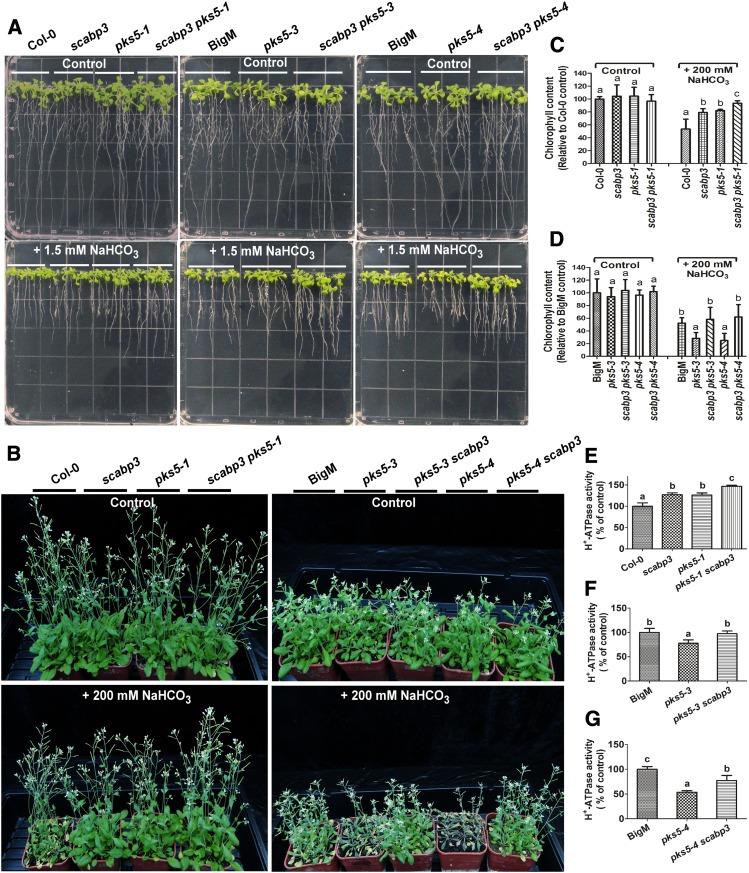
To determine the genetic relation between SCaBP3 and PKS5, we generated a scabp3 pks5-1 double mutant. Six-day-old Col-0, scabp3, pks5-1, and scabp3 pks5-1 seedlings grown on MS medium were transferred to MS medium containing 0 or 1.5 mM NaHCO3. No difference in growth was detected between Col-0, scabp3, pks5-1, and scabp3 pks5-1 on MS medium (Figure 7A; Supplemental Figures 13A and 13B). On the MS medium with 1.5 mM NaHCO3, the scabp3 and pks5-1 single mutants appeared to be less sensitive to this stress condition than did Col-0 (Figure 7A;, Supplemental Figures 13C and 13D). The pks5-1 scabp3 double mutant exhibited a further increase in root length and fresh weight on MS medium with 1.5 mM NaHCO3 compared with the scabp3 and pks5-1 single mutants (Figure 7A; Supplemental Figures 13C and 13D). We further investigated the phenotypic effects when plants were grown in soil with NaHCO3 treatment. Three-week-old Col-0, scabp3, pks5-1, and scabp3 pks5-1 seedlings grown in soil were treated with 200 mM NaHCO3 for 3 weeks. Leaf vitality (the leaf remains green and is not withered) of the NaHCO3-treated scabp3 and pks5-1 was greater than that of Col-0, and chlorophyll content was higher in the mutants than in Col-0 (Figures 7B and 7C). Moreover, the scabp3 pks5-1 double mutant exhibited a higher tolerance than either of the single mutants (Figure 7B).
Figure 7.
SCaBP3 and PKS5 Regulate Arabidopsis PM H+-ATPase Activity in Response to Alkali Stress.
(A) The NaHCO3-response phenotypes of wild-type (Col-0 and BigM [a mutant of Col-0, Col-0 erecta105]) and mutants seedlings at 6 d old grown on MS medium or MS medium with 1.5 mM NaHCO3. Photographs were taken at 7 d after transfer.
(B) NaHCO3-response phenotype of Col-0, scabp3, pks5-1, scabp3 pks5-1, BigM, pks5-3, pks5-3 scabp3, pks5-4, and pks5-4 scabp3 plants grown in soil with or without NaHCO3 treatment. After 3 weeks of growth in soil, the plants were left untreated (control) or treated with 200 mM NaHCO3 for 3 weeks, and photographs were taken.
(C) and (D) Chlorophyll content of Col-0, scabp3, pks5-1, scabp3 pks5-1, BigM, pks5-3, pks5-3 scabp3, pks5-4, and pks5-4 scabp3 plants grown in soil with or without NaHCO3 treatment. After 3 weeks of growth in soil, the plants were left untreated (control) or treated with 200 mM NaHCO3 for 3 weeks, and the leaves were collected for detecting chlorophyll content. Data represent means ± sd (n = 6) from measurements using the same preparation.
(E) to (G) Comparison of the PM H+-ATPase activity in vesicles isolated from Col-0, scabp3, pks5-1, scabp3 pks5-1, BigM, pks5-3, pks5-3 scabp3, pks5-4, and pks5-4 scabp3 plants grown in soil treated with 250 mM NaCl for 3 d before harvest. Data represent means ± sd (n = 3) from measurements using the same preparation. The initial 15-s slope of fluorescence quenching was graphed from Supplemental Figures 12M to 12O, respectively.
The experiments were performed with three biological replicates with similar results. Lowercase letters indicate significant differences from the control as determined by one-way ANOVA, P < 0.05.
To determine whether and how the PM H+-ATPase activity may be associated with the function of PKS5 and SCaBP3 under saline-alkali conditions, plasma membrane-enriched vesicles were isolated from NaCl-treated seedlings of Col-0, scabp3, pks5-1, and pks5-1 scabp3. As depicted in Figure 7E and Supplemental Figure 13M, the scabp3 single mutant had a similar PM H+-ATPase activity to pks5-1, and the activity of both mutants was higher than that of Col-0. Additionally, the PM H+-ATPase activity of the pks5-1 scabp3 double mutants was higher than that of pks5-1 or scabp3.
We crossed scabp3 with two pks5 mutants in which AHA2 is constitutively activated (pks5-3 and pks5-4; Yang et al., 2010) to generate pks5-3 scabp3 and pks5-4 scabp3 double mutants. Six-day-old control (Col-0 erecta105/BigM, a mutant of Col-0, background of pks5-3 and pks5-4), pks5-3, pks5-4, pks5-3 scabp3, and pks5-4 scabp3 seedlings grown on MS medium were transferred to MS medium containing 0 or 1.5 mM NaHCO3. The root length and fresh weight of the pks5-3 and pks5-4 mutants were significantly reduced compared with BigM on the MS medium with 1.5 mM NaHCO3 (Figure 7A; Supplemental Figures 13E to 13L). Mutation of SCaBP3 in either the pks5-3 or pks5-4 mutant background markedly rescued their phenotypes in root elongation on the medium with 1.5 mM NaHCO3 to wild type-like growth.
To further investigate the effect of NaHCO3 stress on soil-grown seedlings, 3-week-old BigM, pks5-3, pks5-4, pks5-3 scabp3, and pks5-4 scabp3 plants were treated with 200 mM NaHCO3 for 3 weeks. As shown in Figures 7B and 7D, the pks5-3 and pks5-4 plants exhibited weaker growth and reduced chlorophyll content compared with BigM. The deletion of SCaBP3 in either the pks5-3 or pks5-4 mutant background also suppressed their NaHCO3-sensitive phenotypes (Figure 7B). These results suggest that SCaBP3 regulates plant saline-alkali resistance at least partially independently of PKS5, and PKS5 at least partially relies on SCaBP3 for its function in the saline-alkali response. Consistent with the change of phenotype in response to saline-alkali stress, the double mutants pks5-3 scabp3 and pks5-4 scabp3 exhibited higher PM H+-ATPase activity than their pks5 parents (Figures 7F and 7G; Supplemental Figures 13N and 13O), supporting the conclusion that the interaction of PKS5 with PM H+-ATPase requires SCaBP3. These results indicate that SCaBP3 regulates PM H+-ATPase activity by both PKS5-dependent and -independent processes and that SCaBP3 and PKS5 function in the Arabidopsis saline-alkali stress response by regulating PM H+-ATPase activity.
DISCUSSION
Maintaining cell pH and ion homeostasis is important for the plant saline-alkali stress response. In this study, we showed that the Ca2+ sensor SCaBP3 is a critical regulator of the alkali response by negatively regulating the PM H+-ATPase activity, which functions via direct protein interaction and thereby differs from previously identified phosphorylation-dependent regulatory mechanisms (Fuglsang et al., 2007). This adds to the complex array of mechanisms that converge on the PM H+-ATPase to provide the required flexibility in regulation.
Proton pumps are the “master enzymes” in plant life activities, which are regulated by a variety of regulatory pathways and thus correspond to a variety of biological processes. PM H+-ATPase activity is kept at a relatively low level under normal conditions (in the absence of salt stress) and has a dramatic increase after exposure to saline-alkali stress in Arabidopsis (Yang et al., 2010). Under stress conditions, plants have to balance their growth and stress resistance. Higher resistant activity would reduce plant growth, as faster growth would cause more damage under stresses. On the other hand, when growth conditions become suitable, the stress response of plants has to be reduced, in which PKS5 and SCaBP3 are required to repress PM H+-ATPase activity. The increased PM H+-ATPase activity is crucial for driving transporters in response to saline-alkali stress. SCaBP3 and PKS5 proteins function as negative regulators of PM H+-ATPase activity under normal conditions by direct interaction and phosphorylation, and the release of PM H+-ATPase activity in the presence of saline-alkali conditions requires the disassociation of these two proteins from the plasma membrane.
PM H+-ATPase activity is regulated by various interacting proteins, such as 14-3-3 protein, and protein interaction can be modulated by the phosphorylation status of specific residues (Fuglsang et al., 1999, 2007, 2014; Maudoux et al., 2000; Morandini et al., 2002; Duby et al., 2009). The C terminus of PM H+-ATPase serves as an autoinhibitory domain, the modification of the C terminus leads to activation or inactivation of the enzyme, and deletion of critical domains results in the formation of a deregulated constitutive active PM H+-ATPase (Xing et al., 1996; Axelsen et al., 1999; Duby et al., 2009; Takahashi et al., 2012). It has been proposed that intramolecular interaction of the C terminus in coordination with other domains of the PM H+-ATPase contributes to the regulation of its activity status. However, clear evidence and the detailed mechanism that would convey such intramolecular regulation have remained elusive. Moreover, it has remained unknown how the different regulatory mechanisms of the PM H+-ATPase (including intramolecular and intermolecular interactions and modifications) mutually affect each other and are coordinated to bring about the required fine-tuning of PM H+-ATPase activity that is required for optimal cellular function, especially during plant adaptation or stimulus-response processes.
SCaBP3 specifically interacted with the AHA2 C850-895, containing the RI domain. In the yeast S. cerevisiae, available evidence suggests that the RI domain affects the enzyme activity by interacting with a cytoplasmic region of the PM H+-ATPase (Axelsen et al., 1999). Simultaneously, we determined that the C terminus of PM H+-ATPase directly interacted with the Central loop region and that SCaBP3 enhanced this interaction. It is tempting to speculate that this may affect protein folding and self-assembly, resulting in a more compact and stabilized inhibitory structure of the pump to promote the inhibition of PM H+-ATPase activity. However, more evidence, especially from crystal structure studies, is required to further advance this hypothesis.
Our genetic and biochemical results also indicate that SCaBP3 repressed PM H+-ATPase activity by both PKS5-dependent and -independent mechanisms (Figure 8). SCaBP3 directly interacted with PKS5 and enhanced the interaction between AHA2 and PKS5 in plants and yeast. The growth of RS72 yeast cells was dramatically inhibited when PKS5 was expressed with SCaBP3 in yeast. Compared with their pks5 parents, deleting SCaBP3 in pks5 mutants improved saline-alkali tolerance, in association with the changes of PM H+-ATPase activity. However, in an in vitro kinase assay, SCaBP3 did not appear to activate PKS5 kinase activity and PKS5 phosphorylated SCaBP1/CBL2 but not SCaBP3 (Supplemental Figures 14A and 14B), suggesting different regulatory roles of the different calcium sensors. The phosphorylation of aha2S931 enhances the interaction between AHA2 Central loop and C terminus in yeast. The interaction of PKS5 with SCaBP3 may recruit PKS5 to the plasma membrane or pump location to phosphorylate the PM H+-ATPase, such as the aha2S931 site, which in turn prevents the interaction between the pump and 14-3-3 (Supplemental Figure 14C), maintaining the pump in a status of low activity. Under NaHCO3 stress, the phosphorylation of the aha2Thr947 residue and the interaction of 14-3-3 with PM H+-ATPase were enhanced (Supplemental Figure 14C); therefore, the enzyme could maintain a higher activity. This regulatory mechanism is distinct from that of SCaBP8/CBL10, which represses AKT1 activity by directly binding to AKT1 but does not affect CIPK23 activity (Ren et al., 2013).
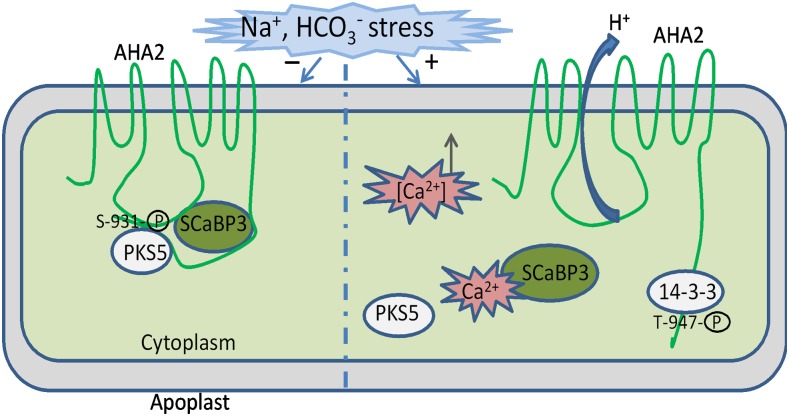
Figure 8.
A Working Model for the Regulation of PM H+-ATPase Activity by SCaBP3 and PKS5.
Under nonstressed conditions, SCaBP3/CBL7 interacts with the AHA2 C terminus and confers the inhibitory interaction of the C terminus and Central loop. ScaBP3-PKS5 complexes phosphorylate the AHA2 C terminus at Ser-931, preventing phosphorylation of Thr-947 or 14-3-3 binding to this residue, and AHA2 activity is low. Under saline-alkali stress conditions, Ca2+ influx from calcium stores to cytoplasm occurs, and Ca2+ binding of ScaBP3 releases the interaction with AHA2 C terminus and releases the inhibitory interaction of the C terminus and Central loop. The activity of the ScaBP3-PKS5 complex is reduced, Ser-931 is no longer phosphorylated, and Thr-947 is phosphorylated, which confers 14-3-3 binding and activates AHA2.
It has been reported that calcium affects the phosphorylation status of PM H+-ATPase (Schaller and Sussman, 1988; Kinoshita et al., 1995). SCaBP3 is a calcium binding protein that interacts with PKS5 and enhances the interaction between PKS5 and AHA2. AHA2 likely interacted with both a Ca2+ sensor SCaBP3 and an active PKS5 kinase. Therefore, the calcium signal may modulate PM H+-ATPase activity by affecting both the direct phosphorylation of the PM H+-ATPase and its interaction with other molecules. In addition, evidence for Ca2+ independent phosphorylation of PM H+-ATPase also has been reported (Wen et al., 2004), and PKS5-mediated transphosphorylation is independent of Ca2+ in vitro (Fuglsang et al., 2007; Bender et al., 2018). Thus, under normal conditions, the SCaBP3-PKS5 complex-mediated Ser-931 phosphorylation of the AHA2 C terminus may be independent of Ca2+, which prevents phosphorylation of Thr-947 or 14-3-3 binding to this residue, leading to low AHA2 activity. Alkali stress may trigger the calcium signals that are perceived by SCaBP3 and trigger its disassociation from target proteins, the PM H+-ATPases and PKS5 protein kinase, which in turn releases the PM H+-ATPase from the autoinhibition and PKS5 phosphorylation (Figure 8). However, whether and how low concentrations of cytosolic free calcium activate PKS5 kinase activity under normal growth conditions remain to be determined.
Arabidopsis seedling growth is not affected by less than 10 mM NaCl or pH 7.0 (KOH to adjust pH of MS medium) treatment. We measured medium pH when adding 1.5 mM NaHCO3, and it is ∼6.55 and very stable due to NaHCO3 buffer capacity. When the pH of the medium supplemented with 2 mM NaHCO3 was adjusted to 5.8, the growth of both wild-type and scabp3 seedlings is similar, suggesting that the stress might come from the buffer capacity of HCO3− and that the effect of NaHCO3 on seedling growth is mainly due to high pH stress. However, we cannot exclude the possibility that HCO3− plays a role in this regulation. Lee and Woolhouse (1969) reported that bicarbonate inhibits root growth primarily by cell elongation in calcicole and calcifuge grasses. The fixation of carbon dioxide in the elongation zone from the bicarbonate buffer leads to the inhibition of root growth (Lee and Woolhouse, 1969). A recent study has reported that a slow-type anion channel homolog in Glycine soja, GsSLAH3, modulates plant bicarbonate stress tolerance (Duan et al., 2018). Heterologous expression of GsSLAH3 in Arabidopsis can increase bicarbonate in shoots, but it does not improve high pH tolerance (Duan et al., 2018). Therefore, SCaBP3 may be involved in both extracellular pH and HCO3− responses. We cannot exclude the possibility that SCABP3 will bind to other partners and activate a kinase that could have many substrates. SCABP3 may also directly regulate the accumulation of HCO3− in plant cells by modulating the activity of certain anion channels. These possible regulatory mechanisms require further study.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 and BigM (a mutant of Col-0, Col-0 erecta105) were used in this study. The mutants pks5-1 (on a Col-0 background) and pks5-3 and pks5-4 (Targeting-Induced Local Lesions In Genomes mutants in the BigM background) have been described previously (Fuglsang et al., 2007; Yang et al., 2010; Eckert et al., 2014). SAIL_201_A02/scabp3 (on a Col-0 background) was obtained from the ABRC (http://www.arabidopsis.org/abrc/) and identified by the gene-specific primers and T-DNA left border primers. The double mutants were generated by crossing scabp3 with pks5-1, pks5-3, or pks5-4. To complement the scabp3 mutant, the SCaBP3 genomic sequence was amplified from Col-0 genomic DNA and cloned (the primers used for plasmid construction are listed in the Supplemental Table) into BamHI and SalI sites in the pCAMBIA1300 binary vector, and the resulting construct was transformed into the scabp3 mutant. The overexpression of SCaBP3 was generated by fusing the full-length SCaBP3 coding sequence into the pCAMBIA1307-3Flag vector with the BamHI and SalI sites, and the resulting construct was transformed into Col-0. The truncated AHA2 (amino acids 1–836) coding sequence was fused into the SUPERR:sXVE:HA vector with BamHI and SalI sites, and the resulting construct was transformed into Col-0 to generate transgenic lines of SUPERR:sXVE:HA:AHA2ΔC. All seedlings were germinated on MS (Phyto Technology Laboratories, catalog number M519) medium (plus 0.5% phytagel; Sigma-Aldrich, catalog number P8169) at pH 5.8 under 24 h of constant illumination (Percival CU-36L5) at 23°C after 2 d at 4°C. Adult plants were grown in growth chambers under long-day (16 h of light of 30 μmol m−2 s−1/8 h of dark) conditions. For alkali sensitivity assays, 6-d-old seedlings growing under constant white light of 30 μmol m−2 s−1 at 23°C were transferred to MS medium (pH 5.8), MS medium plus 1.5 mM NaHCO3 (pH 6.55; Sigma-Aldrich, catalog number 792519), MS medium plus 2 mM NaHCO3 (pH 6.66), MS medium plus 4 mM NaHCO3 (pH 6.80), MS medium plus 6 mM NaHCO3 (pH 6.93), or MS medium plus 2 mM NaHCO3 (pH 5.80; the pH was adjusted to 5.80 after adding 2 mM NaHCO3). For the PM H+-ATPase activity assay, 8-d-old seedlings growing under constant white light at 23°C were transferred to soil for another 2 weeks in growth chambers and were treated with 250 mM NaCl (VETEC, catalog number V900058) for 3 d (or 14-d-old seedlings growing under constant white light at 23°C were transferred to MS medium plus 1.5 mM NaHCO3 for 5 d) before being collected for isolation of plasma membrane vesicles.
Yeast Two-Hybrid Assay
To investigate the interaction between SCaBPs/CBLs and the AHA2 C terminus, AHA2C (residues 850–948) was cloned into the pGBKT7 vector with EcoRI and SalI sites, and each SCaBPs/CBLs coding sequence was cloned into the pACT2 vector (Ren et al., 2013). The truncation and point mutations of the AHA2 C terminus were cloned into the pGBKT7 vector with EcoRI and SalI sites, while the SCaBP3 coding sequence was cloned into the pGADT7 vector with EcoRI and BamHI sites.
The plasmids were transformed into Saccharomyces cerevisiae strain AH109 for yeast two-hybrid assays. Yeast transformation and growth assays were performed as described in the Yeast Protocols Handbook (Clontech). All primers used for these constructs are listed in the Supplemental Table.
BiFC
Full-length AHA2 and the truncation of AHA2, SCaBP3, SCaBP1, PKS5, and PKS5K50N were amplified by PCR with gene-specific primers (Supplemental Table) and cloned into pSPYCE(M) and pSPYNE(R)173 with the BamHI and SalI sites, respectively (Waadt et al., 2008). Pairwise construct combinations were introduced into Agrobacterium tumefaciens strain GV3101 for transient expression in Nicotiana benthamiana epidermal cells as described (Walter et al., 2004). YFP fluorescence signals were detected by Leica SP5 confocal microscopy after 48 h of infiltration.
Analysis of Subcellular Localization and Promoter-GUS Activity
For subcellular localization analyses, the coding sequence of SCaBP3 was amplified with primers containing SalI and KpnI sites and cloned into the pCAMBIA1390 binary vector under the control of the UBQ10 promoter. To prepare the pCAMBIA1390-UBQ10:cGFP vector, the UBQ10 promoter and GFP sequence were cloned between the HindIII and PstI sites and between the EcoRI and SpeI sites in pCAMBIA1390, respectively.
For GUS expression analyses, a DNA fragment encompassing 1227 bp upstream of the translational start site (ATG) of SCaBP3 was amplified with the ProSCaBP3F and ProSCaBP3R primers (Supplemental Table) and cloned into the pCAMBIA1391Z vector with PstI and BamHI sites. The construct was transformed into Col-0 by A. tumefaciens-mediated transformation, and 15 independent transgenic lines (T2) were tested for GUS staining (Haritatos et al., 2000). The primers used for plasmid construction are listed in the Supplemental Table.
qPCR and RT-PCR Analysis
Total RNA was extracted with RNAVzol (Vigorous, catalog number N002) from N. benthamiana or 12-d-old seedlings grown on MS plates under constant illumination of 30 μmol m−2 s−1. Eight micrograms of total RNA was treated with RNase-free DNase I (Invitrogen, catalog number AM2224) to eliminate genomic DNA and was then used for reverse transcription with MMLV reverse transcriptase (Promega, catalog number M1701) according to the manufacturer’s introduction. The resulting cDNAs were used for quantitative real-time PCR or RT-PCR analysis of the RNA expression level. ACTIN2 served as an internal control. The primers used for quantitative real-time PCR are described in the Supplemental Table.
Plasma Membrane Isolation, Cytosol Acquisition, and H+-ATPase Activity Measurement
Plasma membrane-enriched vesicles were isolated from the NaCl-treated or NaHCO3-treated (or untreated) seedlings by aqueous two-phase partitioning as described (Qiu et al., 2002). Cytosol was obtained from the NaHCO3-treated (or untreated) seedlings. The seedlings were homogenized in a buffer of 0.33 M sucrose (Sigma-Aldrich, catalog number V900116), 5 mM DTT (Sigma-Aldrich, catalog number 10197777001), 0.2% (w/v) BSA (Sigma-Aldrich, catalog number B2518), 5 mM EDTA (Sigma-Aldrich, catalog number 798681), 5 mM ascorbate (Sigma-Aldrich, catalog number A7631), 10% (w/v) glycerol (Sigma-Aldrich, catalog number G5516), 0.2% (w/v) casein (Sigma-Aldrich, catalog number C7078), 1 mM PMSF (Sigma-Aldrich, catalog number 52,332), 1× protease inhibitor (Sigma-Aldrich, catalog number P8340), 0.6% (w/v) polyvinylpyrrolidone (Sigma-Aldrich, catalog number V900008), and 50 mM HEPES-KOH (Sigma-Aldrich, catalog number H4034, P5958; pH 7.5). The homogenate was filtered through four layers of Miracloth and centrifuged for 10 min at 12,000g at 4°C, and the supernatant was centrifuged at 100,000g for 1 h at 4°C. Then the supernatant (cytosol) was collected for immunoblot analysis. Anti-Flag (CWBIO, CW0278M, 1:2000), anti-Myc (CWBIO, CW01217, 1:3000), anti-GHR1 (a plasma membrane-localized protein; R&D Systems, RB01, 1:1000), or anti-MPK3 (a cytosol-localized protein; Agrisera, AS16 4024, 1:1000) antibodies were detected for immunoblot analysis. The H+-transport activity was measured as described (Qiu et al., 2002), and each reaction was performed with 50 μg of plasma membrane protein.
Recombinant proteins (SCaBP1, SCaBP3, SCaBP8, and PKS5) were preincubated with plasma membrane-enriched vesicles isolated from NaCl-treated (or NaHCO3-treated) Col-0 for 5 min at room temperature; then the assay was initiated by the addition of ATP (Sigma-Aldrich, catalog number A6559).
Protein Purification
All GST and His fusion constructs were transformed into Escherichia coli BL21 (DE3). When the cells were grown in Luria-Bertani medium with ampicillin (100 μg/mL; Sigma-Aldrich, catalog number BP021) or kanamycin (50 μg/mL; Sigma-Aldrich, catalog number E004000) at 37°C to OD600 at 0.8 to 1.0, recombinant protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (Sigma-Aldrich, catalog number PHG0010) at 16°C overnight. The recombinant proteins were purified according to the manufacturer’s protocol (GE Healthcare Life Science and Merck) and analyzed by SDS-PAGE.
Yeast Complementation Assay
S. cerevisiae strain RS72 (Mat a; ade1-100 his4-519 leu2-3, 312 pPMA1::pGAL1) was used for complementation tests as described (Regenberg et al., 1995; Axelsen et al., 1999; Fuglsang et al., 2007). PKS5, SCaBP1, or SCaBP3 fragment was amplified with a primer (forward and reverse) containing the NotI site, and then the products were ligated into the NotI-digested pMP1612 or pMP1645 vector. Full-length AHA2, truncated AHA2, and point mutations of AHA2 were expressed under the control of the PMA1 promoter in the pMP1745 vector (Fuglsang et al., 2007). All constructs were sequenced. The experiments were replicated independently three times with cells from independent transformation events. All primers used for these constructs are listed in the Supplemental Table.
Yeast Three-Hybrid Assay
The pBridgeGPD vector (BD vector; Clontech) was used to express both the AHA2 C terminus and SCaBP3. SCaBP3 was ligated into the NotI-digested BD vector, and the AHA2 C terminus was ligated into the EcoRI- and SalI-digested BD vector. The pGADT7 vector (AD vector) was used to express the AHA2 Central loop region. AHA2 Central loop was ligated into the EcoRI- and SalI-digested AD vector. Pairs of constructs (AD-AHA2 Central loop/BD-AHA2C-GDPSCaBP3 and AD-AHA2 Central loop/BD-AHA2C-GPDempty) were transformed into the AH109 yeast strain. Yeast transformation and growth assays were performed as described in the Yeast Protocols Handbook (Clontech). All primers used for these constructs are listed in the Supplemental Table.
LCI
For the LCI experimental analysis of the interaction between AHA2 Q879A and SCaBP3, the coding sequence of SCaBP3 was amplified and then cloned between the KpnI and SalI sites of pCAMBIA-1300NLuc and the coding sequences of AHA2 and AHA2 Q879A were amplified and then cloned between the KpnI and SalI sites of pCAMBIA-1300CLuc, respectively (Chen et al., 2008). These constructs were introduced into A. tumefaciens (strain GV3101) and infiltrated into N. benthamiana leaves. After 2 d of infiltration, the N. benthamiana leaves were treated with 1 mM d-luciferin (Promega, catalog number E160A) and kept in darkness for 5 min, and then the LUC signal was detected by a low-light cooled CCD camera (ikon-L936; Andor Tech; Chen et al., 2008).
For the LCI experimental analysis of the interaction between AHA2 Central loop and AHA2 C terminus, the coding sequences of AHA2 Central loop and AHA2 C terminus were amplified and then cloned between the KpnI and SalI sites of pCAMBIA-1300NLuc and pCAMBIA-1300CLuc, respectively. The coding sequence of DDM1 was amplified and then cloned between the KpnI and SalI sites of pCAMBIA-1300NLuc and pCAMBIA-1300CLuc, respectively. The experiments were performed as described above. For the LCI assay to determine the effect of SCaBP3 on the interaction of the AHA2 Central loop region and its C terminus, the AHA2 Central loop and C terminus constructs were coexpressed with pCAMBIA1391-SCaBP3-GFPc or pCAMBIA1391-GFPc. The experiments were performed as described above.
For the LCI assay to determine the effect of SCaBP3 on the interaction of PKS5 and AHA2 C terminus, the PKS5 and AHA2 C terminus constructs were coexpressed with pCAMBIA1391-SCaBP3-GFPc or pCAMBIA1391-GFPc. The experiments were performed as described above.
For the LCI assay to determine the effect of SCaBP3 or 14-3-3 ω on the interaction of PKS5 and AHA2, the PKS5 and AHA2 constructs were coexpressed with pCAMBIA1391-SCaBP3-GFPc, pCAMBIA1391-14-3-3 ω, or pCAMBIA1391-GFPc. The experiments were performed as described above.
In Vitro Phosphorylation
The kinase buffer contained 20 mM Tris-HCl (Sigma-Aldrich, catalog number 10708976001; Beijing Chemical Works, catalog number 7647-01-0) (pH 7.8), 5 mM MgCl2 (Sigma-Aldrich, catalog number 449172), 2 mM DTT, and 10 µM ATP (Lin et al., 2009). The kinase reaction was started by adding 0.1 μL of [γ-32P]ATP (1 μCi; China Isotope and Radiation Corporation) and PKS5, and the mixture was incubated at 30°C for 30 min. The reaction was terminated by adding 6× SDS loading buffer and incubating the sample at 95°C for 10 min. The protein was separated on a 12% SDS-PAGE gel and stained with Coomassie Brilliant Blue R-250 (Sigma-Aldrich, catalog number 1.12553), and then the gel was exposed to a phosphor screen. Twelve hours after exposure, signals were detected using a Typhoon 9410 phosphor imager (Amersham Biosciences).
MST Assay
An MST assay was performed using a NanoTemper Monolith NT.115 instrument as described previously (Wienken et al., 2010). Recombinant His-AHA2 C-terminal peptide was labeled with the NHS NT-647 dye (NanoTemper Technologies, catalog number MO-L001). Then the labeled His-AHA2 C-terminal peptide was diluted to a final concentration of 10 nM with PBS (pH 7.6) containing 0.005% Tween 20 (Sigma-Aldrich, catalog number 93,773) in the absence or presence of 100 nM (or 5 μM) Ca2+ (Sigma-Aldrich, catalog number 793639). The recombinant GST-SCaBP3 protein was serially diluted with PBS (pH 7.6) containing 0.005% Tween 20 in the absence or presence of 100 nM (or 5 μM) Ca2+. Then the labeled His-AHA2 C-terminal peptide and diluted GST-SCaBP3 protein were mixed at a ratio of 1:1 (v/v). The mixture was incubated for 5 min in darkness and loaded into standard treated capillaries (NanoTemper Technologies, catalog number MO-K025) for analysis.
The assay was performed with 20% MST and 20% LED power. The dissociation constant was obtained from the Thermophoresis C T-Jump result.
Statistical Analysis
Data represent means ± sd (n ≥ 3) from the measurements. Lowercase letters indicate the significant difference from the control as determined by one-way ANOVA, P < 0.05.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At4g26560 (SCaBP3), At4g30190 (AHA2), and At2g30360 (PKS5).
Supplemental Data
Supplemental Figure 1. Genetic characterization and NaHCO3-response Phenotype Analysis of the scabp3 mutant plant.
Supplemental Figure 2. NaHCO3-response phenotype analysis of scabp3 mutant plant.
Supplemental Figure 3. Analysis of NaHCO3-response phenotype in Col-0, scabp3, and two overexpressing lines (OE-1 and OE-2).
Supplemental Figure 4. DMGA phenotype analysis of scabp3 mutant plant.
Supplemental Figure 5. Controls for analysis of the interaction between SCaBP3 and AHA2.
Supplemental Figure 6. Subcellular localization and expression analysis of SCaBP3.
Supplemental Figure 7. SCaBP3 negatively regulates PM H+-ATPase activity.
Supplemental Figure 8. Immunoblot analysis of PM H+-ATPase content in the vesicles isolated from NaHCO3-treated plants.
Supplemental Figure 9. Analysis of the region where SCaBP3 interacts with AHA2.
Supplemental Figure 10. RNA expression levels of different genes used in the LCI assay.
Supplemental Figure 11. Expression levels of different components used in BiFC assays, and recombinant proteins used in MST assays.
Supplemental Figure 12. Analysis of NaHCO3-response phenotype in wild type (Col-0) and two AHA2ΔC transgenic lines (AHA2ΔC-1 and AHA2ΔC-2).
Supplemental Figure 13. Root length and fresh weight analysis of mutants treated with NaHCO3 and PM H+-ATPase activity of mutants after NaCl treatment.
Supplemental Figure 14. Analysis of the effect of SCaBP3 on PKS5 kinase activity, and the rffect of SCaBP3 on phosphorylation of AHA2 Thr-947 and 14-3-3s content in the plasma membrane after NaHCO3 treatment.
Supplemental Table. List of PCR primers.
Supplemental Data Set. One-way ANOVA of Figures 1B, 1C, 1E, and 1F.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Michael Broberg Palmgren (University of Copenhagen) for reading the manuscript and useful suggestions and Philipp Köster (University of Münster) for advice on interaction experiments. This work was supported by grants of the National Natural Science Foundation of China (31430012 and U1706201 to Y.G. and 31670260 and 31872659 to Y.Q.Y.), the Ministry of Science and Technology of the People’s Republic of China (Chinese Ministry of Science and Technology) (2015CB910202), and joint Sino-German Research Project Grants (NSFC, 31861133005 and 31210103903; DFG, Ku931/19-1) to J.K. and Y.G.
AUTHOR CONTRIBUTIONS
Y.Q.Y. and Y.J.W. performed most of the experiments. Q.Y.D. performed the BiFC experiments. L.M. and Z.J.Y. performed the kinase assay. Q.P.L. performed some of the LCI experiments. X.P.N. performed some of the NaHCO3-response phenotype experiments. Y.G. supervised the project and designed the experiments. Y.G., Y.J.W., and Y.Q.Y. wrote the manuscript. J.K. designed the experiments and revised the manuscript. C.P.S. revised the manuscript.
References
- Axelsen K.B., Venema K., Jahn T., Baunsgaard L., Palmgren M.G. (1999). Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: Mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38: 7227–7234. [DOI] [PubMed] [Google Scholar]
- Ayyub C.M., Ali M., Shaheen M.R., Qadri R.W.K., Khan I., Jahangir M.M., Abbasi K.Y., Kamal S., Zain M. (2015). Enhancing the salt tolerance potential of watermelon, by exogenous application of salicylic acid. Am. J. Plant Sci. 6: 3267–3271. [Google Scholar]
- Batistic O., Waadt R., Steinhorst L., Held K., Kudla J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61: 211–222. [DOI] [PubMed] [Google Scholar]
- Bender K.W., Zielinski R.E., Huber S.C. (2018). Revisiting paradigms of Ca2+ signaling protein kinase regulation in plants. Biochem. J. 475: 207–223. [DOI] [PubMed] [Google Scholar]
- Borgo L. (2017). Evaluation of buffers toxicity in tobacco cells: Homopiperazine-1,4-bis (2-ethanesulfonic acid) is a suitable buffer for plant cells studies at low pH. Plant Physiol. Biochem. 115: 119–125. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Pang Y., Zeng J., Ding Q., Yin S.Y., Liu C., Lu M.Z., Cui K.M., He X.Q. (2012). The Ca2+ -dependent DNases are involved in secondary xylem development in Eucommia ulmoides. J. Integr. Plant Biol. 54: 456–470. [DOI] [PubMed] [Google Scholar]
- Duan X., Yu Y., Duanmu H., Chen C., Sun X., Cao L., Li Q., Ding X., Liu B., Zhu Y. (2018). GsSLAH3, a Glycine soja slow type anion channel homolog, positively modulates plant bicarbonate stress tolerance. Physiol. Plant. 164: 145–162. [DOI] [PubMed] [Google Scholar]
- Duby G., Poreba W., Piotrowiak D., Bobik K., Derua R., Waelkens E., Boutry M. (2009). Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. J. Biol. Chem. 284: 4213–4221. [DOI] [PubMed] [Google Scholar]
- Eckert C., Offenborn J.N., Heinz T., Armarego-Marriott T., Schültke S., Zhang C., Hillmer S., Heilmann M., Schumacher K., Bock R., Heilmann I., Kudla J. (2014). The vacuolar calcium sensors CBL2 and CBL3 affect seed size and embryonic development in Arabidopsis thaliana. Plant J. 78: 146–156. [DOI] [PubMed] [Google Scholar]
- Ekberg K., Palmgren M.G., Veierskov B., Buch-Pedersen M.J. (2010). A novel mechanism of P-type ATPase autoinhibition involving both termini of the protein. J. Biol. Chem. 285: 7344–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falhof J., Pedersen J.T., Fuglsang A.T., Palmgren M. (2016). Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol. Plant 9: 323–337. [DOI] [PubMed] [Google Scholar]
- Fuglsang A.T., Visconti S., Drumm K., Jahn T., Stensballe A., Mattei B., Jensen O.N., Aducci P., Palmgren M.G. (1999). Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 274: 36774–36780. [DOI] [PubMed] [Google Scholar]
- Fuglsang A.T., Borch J., Bych K., Jahn T.P., Roepstorff P., Palmgren M.G. (2003). The binding site for regulatory 14-3-3 protein in plant plasma membrane H+-ATPase: Involvement of a region promoting phosphorylation-independent interaction in addition to the phosphorylation-dependent C-terminal end. J. Biol. Chem. 278: 42266–42272. [DOI] [PubMed] [Google Scholar]
- Fuglsang A.T., Guo Y., Cuin T.A., Qiu Q., Song C., Kristiansen K.A., Bych K., Schulz A., Shabala S., Schumaker K.S., Palmgren M.G., Zhu J.K. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang A.T., Kristensen A., Cuin T.A., Schulze W.X., Persson J., Thuesen K.H., Ytting C.K., Oehlenschlæger C.B., Mahmood K., Sondergaard T.E., Shabala S., Palmgren M.G. (2014). Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. Plant J. 80: 951–964. [DOI] [PubMed] [Google Scholar]
- Gévaudant F., Duby G., von Stedingk E., Zhao R., Morsomme P., Boutry M. (2007). Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol. 144: 1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritatos E., Ayre B.G., Turgeon R. (2000). Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol. 123: 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M., Sussman M.R. (2012). The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiol. 158: 1158–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K., Eckert C., Anschütz U., Scholz M., Held K., Waadt R., Reyer A., Hippler M., Becker D., Kudla J. (2012). Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J. Biol. Chem. 287: 7956–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held K., Pascaud F., Eckert C., Gajdanowicz P., Hashimoto K., Corratgé-Faillie C., Offenborn J.N., Lacombe B., Dreyer I., Thibaud J.B., Kudla J. (2011). Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 21: 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T.P., Schulz A., Taipalensuu J., Palmgren M.G. (2002). Post-translational modification of plant plasma membrane H+-ATPase as a requirement for functional complementation of a yeast transport mutant. J. Biol. Chem. 277: 6353–6358. [DOI] [PubMed] [Google Scholar]
- Kanczewska J., Marco S., Vandermeeren C., Maudoux O., Rigaud J.L., Boutry M. (2005). Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proc. Natl. Acad. Sci. USA 102: 11675–11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Nishimura M., Shimazaki K. (1995). Cytosolic concentration of Ca2+ regulates the plasma membrane H+-ATPase in guard cells of fava bean. Plant Cell 7: 1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster P., Wallrad L., Edel K.H., Faisal M., Alatar A.A., Kudla J. (2019). The battle of two ions: Ca2+ signalling against Na+ stress. Plant Biol. (Stuttg.) 21 (suppl. 1): 39–48. [DOI] [PubMed] [Google Scholar]
- Kudla J., Becker D., Grill E., Hedrich R., Hippler M., Kummer U., Parniske M., Romeis T., Schumacher K. (2018). Advances and current challenges in calcium signaling. New Phytol. 218: 414–431. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Woolhouse H.W. (1969). A comparative study of bicarbonate inhibition of root growth in calcicole and calcifuge grasses. New Phytol. 68: 1–11. [Google Scholar]
- Lin H., Yang Y., Quan R., Mendoza I., Wu Y., Du W., Zhao S., Schumaker K.S., Pardo J.M., Guo Y. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21: 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Fuglsang A.T., Palmgren M.G., Staskawicz B.J., Coaker G. (2009). RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manishankar P., Wang N., Köster P., Alatar A.A., Kudla J. (2018). Calcium signaling during salt stress and in the regulation of ion homeostasis. J. Exp. Bot. 69: 4215–4226. [DOI] [PubMed] [Google Scholar]
- Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., Morsomme P. (2000). A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275: 17762–17770. [DOI] [PubMed] [Google Scholar]
- Merlot S., Leonhardt N., Fenzi F., Valon C., Costa M., Piette L., Vavasseur A., Genty B., Boivin K., Müller A., Giraudat J., Leung J. (2007). Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 26: 3216–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandini P., Valera M., Albumi C., Bonza M.C., Giacometti S., Ravera G., Murgia I., Soave C., De Michelis M.I. (2002). A novel interaction partner for the C-terminus of Arabidopsis thaliana plasma membrane H+-ATPase (AHA1 isoform): Site and mechanism of action on H+-ATPase activity differ from those of 14-3-3 proteins. Plant J. 31: 487–497. [DOI] [PubMed] [Google Scholar]
- Morsomme P., de Kerchove d’Exaerde A., De Meester S., Thinès D., Goffeau A., Boutry M. (1996). Single point mutations in various domains of a plant plasma membrane H+-ATPase expressed in Saccharomyces cerevisiae increase H+-pumping and permit yeast growth at low pH. EMBO J. 15: 5513–5526. [PMC free article] [PubMed] [Google Scholar]
- Morsomme P., Dambly S., Maudoux O., Boutry M. (1998). Single point mutations distributed in 10 soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J. Biol. Chem. 273: 34837–34842. [DOI] [PubMed] [Google Scholar]
- Morth J.P., Pedersen B.P., Buch-Pedersen M.J., Andersen J.P., Vilsen B., Palmgren M.G., Nissen P. (2011). A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nat. Rev. Mol. Cell Biol. 12: 60–70. [DOI] [PubMed] [Google Scholar]
- Nguyen T.T., Sabat G., Sussman M.R. (2018). In vivo cross-linking supports a head-to-tail mechanism for regulation of the plant plasma membrane P-type H+-ATPase. J. Biol. Chem. 293: 17095–17106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji S., Sato Y., Suzuki R., Taniguchi S. (1988). Effect of intracellular pH and potassium ions on a primary transport system for glutamate/aspartate in Streptococcus mutans. Eur. J. Biochem. 175: 491–495. [DOI] [PubMed] [Google Scholar]
- Nühse T.S., Stensballe A., Jensen O.N., Peck S.C. (2004). Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C., Marco S., Jaspert N., Marcon C., Schauer N., Weyand M., Vandermeeren C., Duby G., Boutry M., Wittinghofer A., Rigaud J.L., Oecking C. (2007). Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol. Cell 25: 427–440. [DOI] [PubMed] [Google Scholar]
- Palmgren M.G. (2001). Plant plasma membrane H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 817–845. [DOI] [PubMed] [Google Scholar]
- Palmgren M.G., Sommarin M., Serrano R., Larsson C. (1991). Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J. Biol. Chem. 266: 20470–20475. [PubMed] [Google Scholar]
- Pedersen B.P., Buch-Pedersen M.J., Morth J.P., Palmgren M.G., Nissen P. (2007). Crystal structure of the plasma membrane proton pump. Nature 450: 1111–1114. [DOI] [PubMed] [Google Scholar]
- Pessarakli M. (1999). Handbook of Plant and Crop Stress, 2nd ed (Cleveland, Ohio: CRC; ). [Google Scholar]
- Pissaloux A., Morard P., Bertoni G. (1995). Alkalinity-bicarbonate-calcium effects on iron chlorosis in white lupine in soilless culture. In Iron Nutrition in Soils and Plants., J. Abadia, ed (Dordrecht, The Netherlands: Springer; ), pp. 127–133. [Google Scholar]
- Qiu Q.S., Guo Y., Dietrich M.A., Schumaker K.S., Zhu J.K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99: 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B., Villalba J.M., Lanfermeijer F.C., Palmgren M.G. (1995). C-terminal deletion analysis of plant plasma membrane H+-ATPase: Yeast as a model system for solute transport across the plant plasma membrane. Plant Cell 7: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X.L., Qi G.N., Feng H.Q., Zhao S., Zhao S.S., Wang Y., Wu W.H. (2013). Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 74: 258–266. [DOI] [PubMed] [Google Scholar]
- Rober-Kleber N., Albrechtová J.T., Fleig S., Huck N., Michalke W., Wagner E., Speth V., Neuhaus G., Fischer-Iglesias C. (2003). Plasma membrane H+-ATPase is involved in auxin-mediated cell elongation during wheat embryo development. Plant Physiol. 131: 1302–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudashevskaya E.L., Ye J., Jensen O.N., Fuglsang A.T., Palmgren M.G. (2012). Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. J. Biol. Chem. 287: 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd J.J., Franklin-Tong V.E. (2001). Unravelling response-specificity in Ca2+ signalling pathways in plant cells. New Phytol. 151: 7–33. [DOI] [PubMed] [Google Scholar]
- Santi S., Schmidt W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183: 1072–1084. [DOI] [PubMed] [Google Scholar]
- Schaller G.E., Sussman M.R. (1988). Phosphorylation of the plasma-membrane H+-ATPase of oat roots by a calcium-stimulated protein kinase. Planta 173: 509–518. [DOI] [PubMed] [Google Scholar]
- Schlesser A., Ulaszewski S., Ghislain M., Goffeau A. (1988). A second transport ATPase gene in Saccharomyces cerevisiae. J. Biol. Chem. 263: 19480–19487. [PubMed] [Google Scholar]
- Schlücking K., Edel K.H., Köster P., Drerup M.M., Eckert C., Steinhorst L., Waadt R., Batistic O., Kudla J. (2013). A new β-estradiol-inducible vector set that facilitates easy construction and efficient expression of transgenes reveals CBL3-dependent cytoplasm to tonoplast translocation of CIPK5. Mol. Plant 6: 1814–1829. [DOI] [PubMed] [Google Scholar]
- Serrano R., Kielland-Brandt M.C., Fink G.R. (1986). Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature 319: 689–693. [DOI] [PubMed] [Google Scholar]
- Shen P., Wang R., Jing W., Zhang W. (2011). Rice phospholipase Dα is involved in salt tolerance by the mediation of H+-ATPase activity and transcription. J. Integr. Plant Biol. 53: 289–299. [DOI] [PubMed] [Google Scholar]
- Spartz A.K., Ren H., Park M.Y., Grandt K.N., Lee S.H., Murphy A.S., Sussman M.R., Overvoorde P.J., Gray W.M. (2014). SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Hayashi K., Kinoshita T. (2012). Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 159: 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wen B., Bin J.H., Wang X.J. (2004). Effects of methyl jasmonate treatment on the hydrolytic activity and phosphorylation level of plasma membrane H+-ATPase in mung bean (Vigna radiata L.) hypocotyls. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 30: 665–670. [PubMed] [Google Scholar]
- Wienken C.J., Baaske P., Rothbauer U., Braun D., Duhr S. (2010). Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1: 100–116. [DOI] [PubMed] [Google Scholar]
- Xing T., Higgins V.J., Blumwald E. (1996). Regulation of plant defense response to fungal pathogens: Two types of protein kinases in the reversible phosphorylation of the host plasma membrane H+-ATPase. Plant Cell 8: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li H.D., Chen L.Q., Wang Y., Liu L.L., He L., Wu W.H. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360. [DOI] [PubMed] [Google Scholar]
- Xue Y., Yang Y., Yang Z., Wang X., Guo Y. (2018). VAMP711 is required for abscisic acid-mediated inhibition of plasma membrane H+-ATPase activity. Plant Physiol. 178: 1332–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Guo Y. (2018a). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217: 523–539. [DOI] [PubMed] [Google Scholar]
- Yang Y., Guo Y. (2018b). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60: 796–804. [DOI] [PubMed] [Google Scholar]
- Yang Y., Qin Y., Xie C., Zhao F., Zhao J., Liu D., Chen S., Fuglsang A.T., Palmgren M.G., Schumaker K.S., Deng X.W., Guo Y. (2010). The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22: 1313–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., An L., Li W. (2014). The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 33: 203–214. [DOI] [PubMed] [Google Scholar]
- Zhao R., Dielen V., Kinet J.M., Boutry M. (2000). Cosuppression of a plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth, and male fertility. Plant Cell 12: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2016). Abiotic stress signaling and responses in plants. Cell 167: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]