A kinesin-14 motor encoded by Vks1 regulates mitosis and cytokinesis during early maize endosperm development and deformities within vks1 endosperms differ, thereby resulting in varied kernel sizes.
Abstract
Cell number is a critical factor that determines kernel size in maize (Zea mays). Rapid mitotic divisions in early endosperm development produce most of the cells that make up the starchy endosperm; however, the mechanisms underlying early endosperm development remain largely unknown. We isolated a maize mutant that shows a varied-kernel-size phenotype (vks1). Vks1 encodes ZmKIN11, which belongs to the kinesin-14 subfamily and is predominantly expressed in early endosperm development. VKS1 dynamically localizes to the nucleus and microtubules and plays key roles in the migration of free nuclei in the coenocyte as well as in mitosis and cytokinesis in early mitotic divisions. Absence of VKS1 has relatively minor effects on plants but causes deformities in spindle assembly, sister chromatid separation, and phragmoplast formation in early endosperm development, thereby resulting in reduced cell proliferation. Severities of aberrant mitosis and cytokinesis within individual vks1 endosperms differ, thereby resulting in varied kernel sizes. Our discovery highlights VKS1 as a central regulator of mitosis in early maize endosperm development and provides a potential approach for future yield improvement.
INTRODUCTION
Flowering plants have evolved an advanced mode of reproduction known as double fertilization. A pollen grain bears two genetically identical haploid sperms, of which one fertilizes the egg to form a diploid zygote that eventually develops into the adult plant body; the other fuses with the two polar nuclei in the central cell and grows into the triploid endosperm, a nutritive storage organ characteristic of angiosperm seeds (Hamamura et al., 2012). The evolutionary advantage of the endosperm could lie in the fact that both parents contribute to endosperm formation, and the plant does not invest energy in seed nutritive tissue unless the egg is successfully fertilized. The endosperm nucleus divides rapidly, and rapid storage organ formation has obvious advantages. The endosperm nourishes the embryo as it develops, when it germinates, and as the seedling grows. In cereal grains, the endosperm stores large amounts of carbohydrates and proteins, and consequently it is an important source of food, feed, and industrial raw materials.
In maize (Zea mays), the duration of endosperm development from pollination to seed maturity is approximately 7 weeks. Based on distinct cytological activities occurring during this process, endosperm development is divided into five stages: (1) coenocyte, (2) cellularization, (3) cell mitotic division and differentiation, (4) endoduplication and cell expansion, and (5) maturation followed by programmed cell death (PCD). The coenocytic stage is characterized by rapid and repeated divisions of the primary endosperm nucleus without cytokinesis. Two days after pollination (DAP), the individual nuclei are embedded in a thin layer of peripheral cytoplasm that surrounds a large vacuole (Brown et al., 2003; Olsen, 2004). Cellularization of the coenocyte is initiated via the formation of radial microtubule systems (RMSs) around neighboring nuclei, which create zones between them. The RMS acts as a primitive phragmoplast to form tube-like cell wall structures around each endosperm nucleus, with an opening facing inward toward the center of the endosperm. After the alveolar layer is formed, cells undergo rapid periclinal division accompanied by cytokinesis, producing an outer layer of cells and an inner layer of alveoli. After several rounds of mitosis and cytokinesis in the alveolar layer, the endosperm eventually becomes fully cellularized by 4 DAP (Brown et al., 1994; Staehelin and Hepler, 1996; Sylvester, 2000; Olsen, 2001). Then, these cells begin to undergo rapid proliferation by mitotic cell divisions that peak around 8 DAP.
Early endosperm development usually encompasses the period from 0 to 8 DAP. The mid and late endosperm development phases manifest cell differentiation, synthesis of storage reserves, and PCD and desiccation, respectively. Although the period of early endosperm development is much shorter than the mid and late phases, the coenocytic and cellularization stages are critical steps in endosperm formation in cereals and many other species. The early mitotic divisions give rise to most of the cells comprising the starchy endosperm, which is important for grain filling (Sabelli and Larkins, 2009a, 2009b; Sabelli et al., 2013, 2018). Because endosperm cell number is a major determinant of seed size and weight, a better understanding of early endosperm development is potentially critical to increasing cereal productivity.
Kinesins are a class of motor proteins that move along microtubule (MT) filaments and are powered by hydrolysis of ATP (Vale et al., 1985). All kinesin proteins have a conserved motor domain that possesses catalytic ATPase activity (Sablin et al., 1996), whereas various functions of different kinesins are conferred by divergent motor-flanking regions that are important for isoform-specific functions (Vale and Goldstein, 1990), including MT-based motility (Vale et al., 1985), intracellular transport (Hirokawa, 1998; Vale, 2003; Hirokawa et al., 2009), MT and chromosome dynamics (Heald et al., 1996; Heald, 2000), and other diverse functions.
The kinesin superfamily is divided into 14 distinct subfamilies (Lawrence et al., 2004). The kinesin-1 to kinesin-12 subfamilies are plus-end-directed motors, with the motor domain at the N terminus, whereas the kinesin-13 members are MT depolymerases, with the motor domain flanked by additional regions on both sides. The kinesin-14 subfamily members possess a unique motor domain that is located at the C terminus and is minus-end directed (Hirokawa, 1998). Plants contain a large and diverse kinesin-14 subfamily. Due to the lack of cytoplasmic dyneins in plants, the kinesin-14 proteins are thought to have evolved to fill the function of dyneins found in fungi and animals (Lawrence et al., 2001). The kinesin-14 subfamily is greatly expanded in plants, with 20 members in Arabidopsis (Arabidopsis thaliana), 16 in rice (Oryza sativa), and 21 in maize (Supplemental Figure 1; Supplemental File), whereas only four are found in the human genome (Richardson et al., 2006). Plant kinesin-14 motors are involved in spindle morphogenesis, MT-based trafficking, chromosome segregation, chloroplast distribution, and plant-specific phragmoplast formation (Lee et al., 2015; She and Yang, 2017; Gicking et al., 2018). Arabidopsis ATK1, the first kinesin-14 protein discovered in plants, primarily functions in spindle morphogenesis and chromosome segregation during meiosis (Chen et al., 2002; Marcus et al., 2002), whereas Arabidopsis ATK5 is required for mitotic spindle pole formation (Ambrose and Cyr, 2007). Plant kinesin-14 motors have also evolved novel functions. Kinesins with a calponin homology domain (OsKCH2) in rice displays processing minus-end-directed motility on single MTs as individual homodimers (Tseng et al., 2018). The plant-unique kinesin-14 member Kinesin-like calmodulin-binding protein (KCBP) plays an important role in cytoskeletal regulation of trichome cell shape (Tian et al., 2015). Recently, a highly specialized kinesin-14 motor (named KINDR) encoded on maize abnormal chromosome 10 (Ab10) was shown to localize to heterochromatic knobs and form mobile neocentromeres. KINDR can move neocentromeres along spindles to create meiotic drive, leading chromosomes to be preferentially transmitted to egg cells (Dawe et al., 2018). This finding explained the longstanding mystery of the meiotic drive of Ab10 (Schroeder and Malik, 2018). Another classic kinesin-14 member in maize is Divergent Spindle-1 (DV1), which is known to function in meiotic spindle pole organization (Higgins et al., 2016). Although the plant kinesin-14 subfamily contains many members (Vale and Goldstein, 1990), most of their molecular functions and biological roles in development remain to be elucidated.
Several signaling pathways regulating seed size have been studied in Arabidopsis and rice, but little is known about the mechanisms that regulate early endosperm development (Li and Li, 2016). Here, we report the identification of a novel mutant, varied kernel size (vks1), which exhibits nonuniform kernel sizes on a genetically homozygous ear. Vks1 encodes ZmKIN11, a homolog of KINDR, that also belongs to the kinesin-14 subfamily. Vks1 is most highly expressed during early endosperm development and is abundant at coenocytic and cellularization stages. VKS1 contains an N-terminal region specific for nuclear localization in interphase. When mitosis begins, VKS1 always colocalizes with MTs that function in spindle assembly, chromosome separation, and phragmoplast formation. Mutation of vks1 results in defects in nuclear division and cytokinesis in early endosperm development and, as a consequence, leads to varied and small kernel sizes at seed maturity.
RESULTS
vks1 Displays a Nonuniform Small-Kernel Phenotype
To study endosperm development, we treated the A619 inbred line with ethyl methanesulfonate (EMS), which generated a large collection of mutants, including many with visible endosperm defects. Among them, vks1 seeds exhibited a phenotype wherein the kernel size was not only reduced but also varied greatly among individuals. vks1 appears to contradict what is known about traditional single-gene mutations, wherein mutants usually display a qualitative and uniform alteration in phenotype.
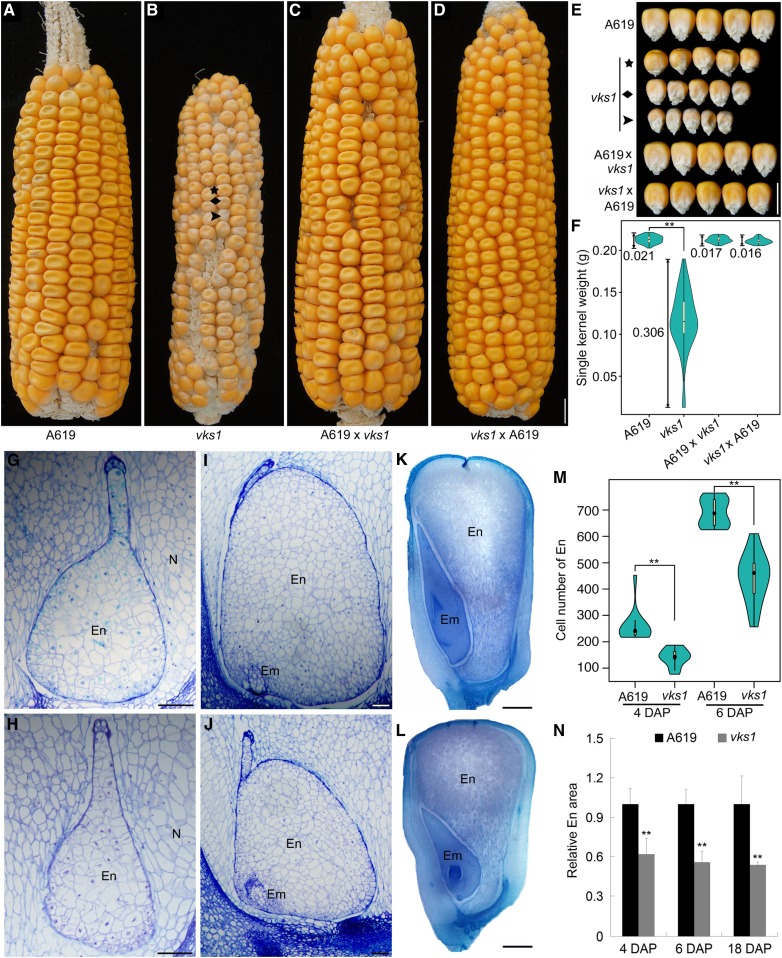
Since EMS can produce many lesions in the genome, phenotypic variation can result from different combinations of multiple unlinked mutant genetic loci. To investigate this possibility, vks1 was recurrently backcrossed to A619 for six generations and then self-pollinated for two generations (see Methods). The contrasting phenotypes of wild-type and vks1 ears are compared in Figures 1A and 1B. vks1 seeds were remarkably smaller than wild-type seeds and showed apparent variation in size. Wild-type seeds at the base of the ear are always larger than those at the tip end, but most in between appear nearly uniform in size (Figure 1A). In vks1, the mutant seeds could be roughly classified into three ranks with mild, moderate, and severe phenotypes (Figures 1B and 1E). Consistent with the size variation, the 100-kernel weight of vks1 (11.63 g) was dramatically reduced compared with the wild type (29.65 g). When measured, the weight of single vks1 seeds showed a significantly greater coefficient of variation than that of the wild type (0.306 versus 0.021; Figure 1F). When reciprocal crosses between the wild type and vks1 were made, the resulting ears showed normal and uniform seed phenotypes (Figures 1C to 1F), consistent with vks1 being a recessive mutation.
Figure 1.
Nonuniform Small-Kernel Phenotype of vks1.
(A) to (D) Ear phenotypes of A619, vks1, A619 × vks1, and vks1 × A619. Three mutant seeds on a homozygous vks1 ear representing mild, medium, and severe developmental phenotypes are indicated by an asterisk, a rhombus, and an arrowhead, respectively. Bar = 1 cm.
(E) Seed phenotypes of A619, vks1, A619 × vks1, and vks1 × A619. The vks1 seeds from (B) with mild, medium, and severe phenotypes are indicated by an asterisk, a rhombus, and an arrowhead, respectively. Bar = 1 cm.
(F) Seed weight variation of single kernels in A619, vks1, A619 × vks1, and vks1 × A619. The number beside each column indicates the coefficient of variation. **, P < 0.01 as determined by Student’s t test.
(G) and (H) Semithin sections of A619 (G) and vks1 (H) endosperms at 4 DAP. Bars = 100 μm. Em, Embryo; En, Endosperm; N, Nucellus.
(I) and (J) Semithin sections of A619 (I) and vks1 (J) endosperms at 6 DAP. Bars = 100 μm.Em, Embryo; En, Endosperm.
(K) and (L) Sections of A619 (K) and vks1 (L) endosperms at 18 DAP. Bars = 1 mm.Em, Embryo; En, Endosperm.
(M) Cell numbers of A619 and vks1 endosperms at 4 and 6 DAP. **, P < 0.01 asd determined by Student’s t test.
(N) Relative endosperm area at 4, 6, and 18 DAP. The ratio of the area of the endosperm to that of the whole seed in A619 and vks1 is shown The ratio in A619 was set as 1. **, P < 0.01 as determined by Student’s t test.
In the anthocyanin regulatory and biosynthetic pathways of maize, C1 (colored aleurone1) and Bz1 (bronze1) encode a MYB transcription factor and a uridine diphosphate glucose-flavonol 3-O-glucosyl transferase enzyme, respectively, both of which are required for anthocyanin synthesis. A619 is a typical yellow dent maize inbred line that bears a mutant c1 gene and a normal Bz1 gene, whereas W22 has the complementary genotype (normal C1 and the mutant bz1-mum9 allele). We mixed equal amounts of vks1 (vks1;c1;Bz1) and W22 (Vks1;C1;bz1-mum9) pollen and applied the mixture to homozygous vks1 plants. The seed genotypes on the resulting ears were easy to characterize by observing anthocyanin pigmentation (Supplemental Figure 2A). The purple seeds (indicative of the cross between vks1 and W22) that were heterozygous for the three genes displayed large, uniform kernels, whereas the yellow seeds (indicative of selfing of vks1) consistently showed a phenotype with varied size and weight (Supplemental Figures 2B and 2C).
To further characterize the reduction in seed size, we investigated endosperm development (Figures 1G to 1N). Four and six DAP stages represented time points when cellularization was complete and early mitotic divisions are rapid, respectively. In vks1 and the wild type, the endosperms were surrounded by the nucellus at both stages. In vks1, the endosperms were morphologically normal but smaller than in the wild type (Figures 1G to 1J). There were significantly fewer endosperm cells in vks1 than in the wild type (Figure 1M), indicating that cell division during early endosperm development in vks1 proceeded more slowly than in the wild type. At 18 DAP, when storage reserves were actively synthesized, both the seed and endosperm areas in vks1 were smaller compared with the wild type (Figures 1K, 1L, and 1N). Light microscopy observations clearly demonstrated that starchy endosperm cells in vks1 and the wild type were fully filled with starch granules (Supplemental Figures 3A and 3B), and the accumulation of zein and nonzein storage proteins and total starch content exhibited no discernible difference between them (Supplemental Figures 3C and 3D). Taken together, these results indicate that seed size reduction in vks1 relates to a decrease in cell number that may occur early rather than late in endosperm development.
In addition to defects in endosperm development, the vks1 mutant displayed abnormalities in plant development. Although vks1 seeds of different sizes germinated normally, the resulting plants were shorter than those of the wild type (Supplemental Figure 3E). Each node of a vks1 plant was shorter than the wild type. Fewer and shorter aerial and lateral roots were produced by vks1 than by the wild type (Supplemental Figure 3F). Furthermore, the segregation ratio of mutant and wild-type seeds from self-pollinated heterozygous plants (vks1/Vks1) was significantly lower than the expected Mendelian ratio of 1:3 (Supplemental Figures 4A and 4B), although seed set in crosses A619 × vks1 and vks1 × A619 was normal (Figures 1C and 1D). Again, vks1 seeds displayed dramatically greater kernel weight variation compared with wild-type seeds from the same segregating ear (Supplemental Figure 4C). We inspected vks1 pollen with a light microscope and found that ∼30% was infertile (Supplemental Figures 4D to 4F). Therefore, the vks1 mutation appears to have pleiotropic effects on plant development.
Vks1 Encodes a Kinesin-14 Motor Protein
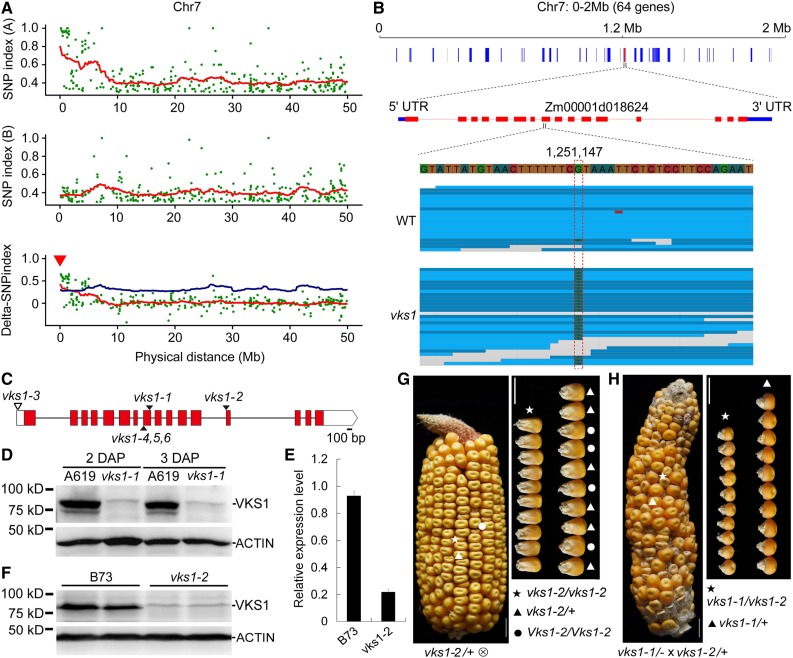
To map the mutant gene, plants of vks1 and its wild type (A619) were crossed and then self-pollinated. The resulting ears segregated mutant and normal seeds (Supplemental Figure 4A). More than 100 seeds of each phenotype were germinated for DNA extraction. We performed whole-genome sequencing of the two DNA pools. Comparison of the single-nucleotide polymorphism (SNP) index between the vks1 and wild-type pools revealed a region near the left terminal end of chromosome 7, which contains 64 genes (Figure 2A). Genomic sequencing of vks1 mutants revealed an SNP in the eighth exon of Zm00001d018624, which resulted in a CGA-to-TGA transition, leading to a premature stop codon in its coding sequence (Figures 2B and 2C) and therefore a dramatic reduction in its transcript level (Figure 2D). Subsequent immunoblotting analysis of VKS1 revealed that this protein was absent in developing vks1 seeds (Figure 2E), indicating that vks1 is a null mutant for this gene. We also used vks1 to create a BC1F1 population with the B73 inbred line for traditional map-based cloning, by which vks1 was narrowed down to an interval between the markers InDel8450 and InDel5839 (Supplemental Figures 5A and 5B). Based on the B73 genome, these two markers span ∼130 kb, where only seven genes were annotated (Supplemental Figure 5B). The vks1 phenotype was completely linked to the homozygous TGA, whereas all normal seeds were heterozygous at this position (Supplemental Figure 5C). These results confirmed that the disrupted Zm00001d018624 was associated with the mutant phenotype.
Figure 2.
Map-Based Cloning and Genetic Confirmation of vks1.
(A) Mapping by sequencing of vks1. Ratios of SNP index (A619/−) between different pools of segregating phenotypic classes (top panel, normal phenotype in pool A: A619/A619 and A619/−; middle panel, vks1 phenotype in pool B: −/−; and bottom panel, SNP index between the top and middle panels) are shown for a region near the left terminal end of chromosome 7.
(B) The vks1 mapping interval. An SNP in the eighth exon of Zm00001d018624 results in a CGA-to-TGA transition in vks1 mutants. UTR, untranslated region.
(C) Schematic representation of Vks1 gene structure with the mutant alleles indicated. Red rectangles and black lines indicate exons and introns, respectively. The black triangles indicate the mutation sites in vks1-1, vks1-2, and vks1-4,5,6 alleles, and the white triangle indicates the Mu insertion in the vks1-3 allele.
(D) Immunoblotting analysis of VKS1 in seeds of A619 and vks1-1. ACTIN was used as an internal control. The sizes of proteins are indicated beside the gels.
(E) Expression levels of Vks1 in seeds of B73 and vks1-2 at 2 DAP.
(F) Immunoblotting analysis of VKS1 in seeds of B73 and vks1-2 at 2 DAP. ACTIN was used as an internal control. The sizes of proteins are indicated beside the gels.
(G) The kernel phenotype of vks1-2. Left panel, a self-pollinated vks1-2/+ ear segregating vks1 (as indicated by an asterisk) and normal (as indicated by a dot and a triangle) seeds. Right panel, all small kernels were determined to be homozygous for vks1-2, while normal kernels bore one (as indicated by triangles) or two wild-type (as indicated by dots) alleles. Bar = 1 cm
(H) Allelic test of vks1-1 and vks1-2 alleles. Left panel, a representative ear from a homozygous vks1-1 plant pollinated by vks1-2/+ pollen segregating vks1 and normal seeds. Right panel, all small kernels were determined to be the genotype vks1-1/vks1-2, while all normal seeds were vks1-1/+. Bar = 1 cm.
To validate that this mutation was responsible for the vks1 phenotype, we performed a reverse genetic screen for additional alleles in the Maize EMS-induced Mutant Database (http://www.elabcaas.cn/memd/) and the Maize Genetics Cooperation Stock Center (https://www.maizegdb.org/uniformmu). This screen resulted in the identification of two alleles of vks1, one each from the Maize EMS-induced Mutant Database (mutant ID: EMS4-092f0c) and the Maize Genetics Cooperation Stock Center (mutant ID: UFMu-01867). The cloned vks1 allele was designated vks1-1, and the additional alleles were designated vks1-2 (with an EMS-induced mutation in the B73 background) and vks1-3 (with the Mu transposable element insertion in the W22 background; Figure 2C). The vks1-2 allele is caused by a mutation at the splice acceptor site in the 12th intron of Zm00001d018624, which results in defective RNA splicing of this intron. The vks1-3 allele results from a Mu insertion in the 5′ untranslated region (UTR) of Zm00001d018624 (Figure 2C; Supplemental Figures 5D and 5E). Self-pollinated vks1-2/+ ears segregated small seeds of varying sizes, as observed in vks1-1 (Figures 1B and 2F; Supplemental Figure 4A). vks1-3 seeds were also much smaller than their wild-type counterparts (W22) but did not display conspicuous seed size variation on homozygous vks1-3 ears (Supplemental Figure 5D), suggesting that the vks1 phenotype was partially genetic background dependent. To perform the allelism test, we made a cross between homozygous vks1-1 and heterozygous vks1-2/+ plants. The resulting ears segregated vks1 and wild-type seeds at a ratio of nearly 1:1 (Figure 2G).
In addition, we used the clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR associated protein 9 (Cas9) technology to create more vks1 null alleles. The gRNA was designed to target the eighth exon of Zm00001d018624 (Supplemental Figure 5F). Three independent transgenic lines, which bear a C deletion, a T insertion, and a 127-bp deletion, respectively, were obtained and designated vks1-4, vks1-5, and vks1-6 (Figure 2C; Supplemental Figure 5F). Self-pollinated vks1-4/+ ears (as a representative for these CRISPR/Cas9-induced null mutants) also segregated small seeds of varying sizes (Supplemental Figure 5G), as observed in vks1-1 and vks1-2 (Figures 1B and 2F). Therefore, Zm00001d018624 is the gene responsible for the vks1 phenotype.
Vks1 consists of 16 exons and 15 introns (Figure 2C), and it encodes a kinesin-14A protein (ZmKIN11) with 761 amino acids (Lawrence et al., 2002). Protein families database (Pfam) searches (http://pfam.xfam.org/search) indicated that this protein contains a tail domain, a coiled-coil (CC) domain, and the classical C-terminal motor domain (Figure 3H). A total of 59 kinesin proteins were found in the maize B73 genome (RefGen_v4) and were grouped into 10 subfamilies based on the kinesin sequences from Arabidopsis and rice (Supplemental Figure 1; Supplemental File).
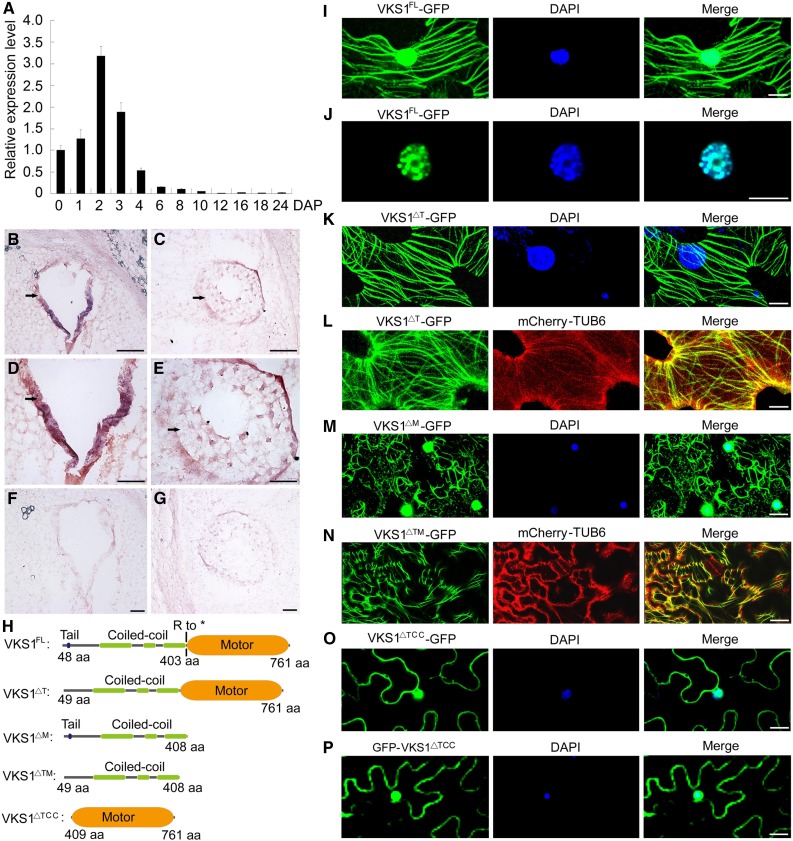
Figure 3.
The Gene Expression Pattern of VKS1 and Its Protein Subcellular Localization.
(A) RT-qPCR analysis of Vks1 during seed development. All expression levels are normalized to Actin. The expression level of Vks1 at 0 DAP is set to 1. Error bars represent sd from three biological replicates.
(B) to (G) RNA in situ hybridization of Vks1 in seeds at 2 and 3 DAP. Positive signals (shown in red) are clearly restricted to the coenocyte at 2 DAP ([B] and [D]) and endosperm cells engaging in cellularization at 3 DAP ([C] and [E]). When hybridized with sense probes, no signal is observed in the sections of seeds at 2 DAP (F) and 3 DAP (G). Bars = 50 μm.
(H) Schematic diagrams of constructs for subcellular localization. VKS1FL, VKS1△T, VKS1△M, VKS1△TM, and VKS1△TCC represent the full-length VKS1 protein and fragments lacking the tail, the motor, both the tail and motor domains, and both the tail and CC domains, respectively. aa, amino acids.
(I) Subcellular localization of VKS1FL-GFP. The nucleus is stained with DAPI.
(J) Localization of VKS1FL-GFP in the nucleus. The nucleus is stained with DAPI.
(K) Subcellular localization of VKS1△T-GFP. The nucleus is stained with DAPI.
(L) Colocalization of VKS1△T-GFP and mCherry-TUB6.
(M) Subcellular localization of VKS1△M-GFP. The nucleus is stained with DAPI.
(N) Colocalization of VKS1△TM-GFP and mCherry-TUB6.
(O) and (P) Subcellular localization of VKS1△TCC-GFP and GFP-VKS1△TCC. The nucleus is stained with DAPI.
For (I) to (P), bars = 10 μm.
VKS1 shares 94% similarity with KINDR, which is encoded by Ab10 and is involved in the activation of neocentromeres that promote meiotic drive in maize. Kindr is a cluster of at least eight kinesin genes, and all the encoded proteins possess only a partial CC domain that is present as an intact domain in VKS1 (Dawe et al., 2018). In addition, VKS1 has another homolog in maize, the classical plant kinesin DV1, which is required for meiotic spindle pole organization (Higgins et al., 2016). In the grass family, 21 VKS1 homologs with a high similarity (>60%) were characterized from nine species in the public database (Supplemental Figure 6), suggesting a conserved function for this gene.
Vks1 Is Predominantly Expressed during Early Endosperm Development
To infer the function of VKS1 in maize endosperm development, we examined its temporal and spatial expression patterns. Since it is difficult to cleanly dissect endosperm from maternal tissues at early developmental stages, whole seeds from 0 to 8 DAP were used for RNA extraction. At later stages, the endosperm could be easily separated from the pericarp and embryo. RT-qPCR revealed that Vks1 is predominantly expressed in early seed development. The expression of Vks1 increased immediately after double fertilization and reached a climax at 2 DAP. Thereafter, the level of Vks1 transcripts was sharply reduced. At 8 DAP, only basal expression was detected (Figure 3A). The temporal expression pattern corresponded well with the period of early endosperm development.
To examine its spatial expression, we performed RNA in situ hybridization with a Vks1 antisense probe at 2 and 3 DAP, the time points when Vks1 transcripts are most highly abundant. This revealed that Vks1 is expressed in the coenocyte at 2 DAP (Figures 3B and 3D). At 3 DAP, the endosperm was undergoing rapid cellularization. The repetition of mitotic divisions four or five times results in a complete cellular endosperm by 4 DAP in maize (Olsen, 2004). Although the strongest hybridization signal was still present in the outermost endosperm cell layer, it expanded to the entire cellularizing endosperm (Figures 3C and 3E). Vks1 was not detected in maternal tissues (pericarp and nucellus), and it appears that Vks1 is endosperm-specific during early seed development. In control experiments, the Vks1 sense probe failed to produce a visible signal (Figures 3F and 3G).
Functions of Different VKS1 Domains in Subcellular Localization
To determine the subcellular localization of VKS1 and the function of its different domains, the full-length protein (VKS1FL, amino acids 1–761) and fragments lacking the tail (VKS1△T, amino acids 49–761), the motor (VKS1△M, amino acids 1–408), the tail and motor domains (VKS1△TM, amino acids 49–408), or the tail and CC domains (VKS1△TCC, amino acids 409–761) were fused to the N terminus of green fluorescent protein (GFP; Figure 3H). We also generated a construct (GFP-VKS1△TCC) in which the motor domain was ligated to the C terminus of the GFP. The β-tubulin6 protein was fused to mCherry. The resulting constructs driven by the constitutive 35S promoter were transiently expressed in Nicotiana benthamiana leaves. VKS1FL-GFP containing the entire VKS1 protein localized to cortical MTs and to the nucleus (Figure 3I). At higher magnification, VKS1FL-GFP signals appeared to be merged precisely with chromatin in the nucleus (Figure 3J). VKS1△T-GFP lacking the tail domain failed to enter the nucleus but was still able to localize to MTs, indicating that the tail domain harbors a signal for nuclear localization (Figures 3K and 3L). Without the motor domain in VKS1△M-GFP, localization to the nucleus and MTs was not affected, but the architecture of MTs was disrupted and displayed a morphologically disordered assembly in the cell, indicating that the motor domain was critical for correct protein-protein interaction with MTs (Figure 3M). Similar to VKS1△M-GFP, VKS1△TM-GFP exhibited severely distorted MT structure and failed to locate in the nucleus (Figure 3N). The VKS1△TCC-GFP and GFP-VKS1△TCC constructs possessing only the motor domain were both ubiquitously distributed in the nucleus and cytoplasm, indicating that the CC domain was essential for MT localization (Figures 3O and 3P).
Defects in Nuclei Migration in the Coenocyte
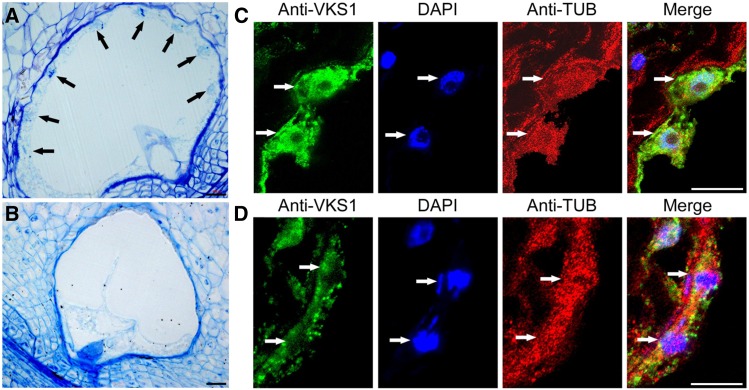
Since Vks1 is most highly expressed in the coenocyte, semithin sections of 2-DAP-old endosperms of the wild type and vks1 were investigated by light microscopy. At this stage, wild-type endosperm nuclei had experienced multiple rounds of mitosis without cytokinesis, forming a multinucleate coenocyte. As shown in Figure 4A, the nuclei migrated and evenly occupied positions along the peripheral cell wall layer, sharing a common cytoplasm and a large central vacuole. We performed immunohistochemical analysis with antibodies raised against VKS1 and α-tubulin, which were in turn recognized by secondary antibodies labeled with GFP and red fluorescent protein, respectively. This assay demonstrated that each free nucleus was surrounded by an interactive MT system that colocalized with the VKS1 protein and formed relatively discrete territorial domains (Figure 4C). In contrast, nuclei in the vks1 coenocyte could not be clearly seen (Figure 4B). Since vks1-1 is a null mutant that does not accumulate the VKS1 protein (Figure 2E), the residual GFP signal probably resulted from background noise or cross-reaction with homologs. In the absence of VKS1, the MT system surrounding the nucleus exhibited an abnormal morphology. The boundary between adjacent MT domains was not formed. More seriously, the nuclei in the vks1 mutant appeared to have defects in migration. They seemed to adhere more closely to the coenocytic cell membrane (Figure 4D), which made them unrecognizable without the aid of 4′,6-diamidino-2-phenylindole (DAPI) staining at lower resolution (Figure 4B). Dispersed chromosomes were observed in the vicinity of the nuclei (Figure 4D). These results suggest that VKS1 is essential for the establishment of MT systems for nuclei in the coenocyte.
Figure 4.
Phenotypes of the Coenocyte in the Wild Type and vks1.
(A) and (B) Semithin sections of A619 and vks1 seeds at the coenocytic stage (2 DAP). Arrows indicate the free nuclei evenly distributed along the peripheral region of the cell in the A619 wild type (A), while nuclei are not visible in vks1 (B). Bars = 50 μm.
(C) and (D) Colocalization of VKS1 and MTs in the coenocyte as shown by immunofluorescence assays in the A619 wild type (C) and vks1 (D). Bars = 20 μm.
VKS1 Colocalizes with MTs during Early Mitotic Divisions
Similar to the coenocytic stage, cellularization also occurs rapidly. By 3 to 4 DAP, the endosperm is fully cellularized (Figures 1G and 1H), which creates difficulties tracking the process of nuclear and cellular division. To understand the function of VKS1 in mitosis and cytokinesis, we investigated endosperm cells at 6 DAP, when early mitotic divisions were common.
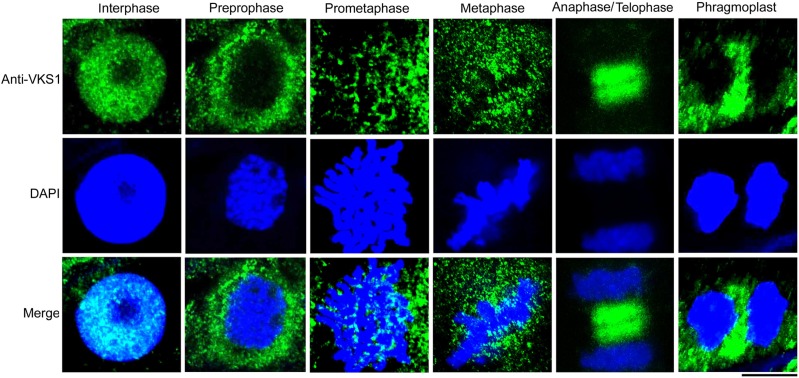
To examine dynamic localizations of VKS1 during mitotic division, we made a series of paraffin sections of A619 endosperms and performed immunohistochemical analysis of VKS1 and α-tubulin proteins (Supplemental Figure 7). Mitosis is divided into multiple phases: prophase, metaphase, anaphase, and telophase. In interphase, VKS1, α-tubulin, and the loosely packed chromatin colocalized in the nucleus, where the three signals seemed to overlap seamlessly (Supplemental Figure 7A). In prophase, as chromatin fibers condensed into discrete chromosomes, the VKS1 protein maintained colocalization with α-tubulin at all times (Supplemental Figure 7B), suggesting that VKS1 is involved in the initiation of mitotic spindle formation. In metaphase, the MTs were attached to the kinetochores of sister chromatids that lined up neatly along the equator of the cell, and the VKS1 protein was distributed on the spindle fibers (Supplemental Figure 7C). In late anaphase and early telophase, as sister chromatids pulled apart to the opposite poles, the spindle began to break down (Supplemental Figure 7D). VKS1 and α-tubulin appeared to relocate together to the central region of the cell, where the phragmoplast was assembled, suggesting that VKS1 is required for MT-mediated phragmoplast formation.
Since paraffin embedding and processing can cause morphological alterations that may not reflect the original in vivo structures, we performed immunohistochemical examinations of endosperm cells without embedding and dehydration (see Methods). Consistent with the observation in Supplemental Figure 7, the localizations of VKS1 were dynamic, following the process of mitotic division. In interphase, VKS1 colocalized with the chromatin surrounded by the nuclear envelope. During mitosis, VKS1 was relocated in a dynamic pattern by which the spindle was assembled and the phragmoplast was formed (Figure 5). Specifically, immunohistochemical assays clearly showed that VKS1 colocalized with MTs during the initiation of phragmoplast formation (Supplemental Figure 8).
Figure 5.
Dynamic Localization of VKS1 during Mitosis.
Localization of the VKS1 protein in early endosperm cells during mitosis was assayed by immunofluorescence. VKS1 signaling is colored green, and the nucleus is colored blue. Bar = 10 μm.
Defects in Mitosis and Cytokinesis in the vks1 Mutant
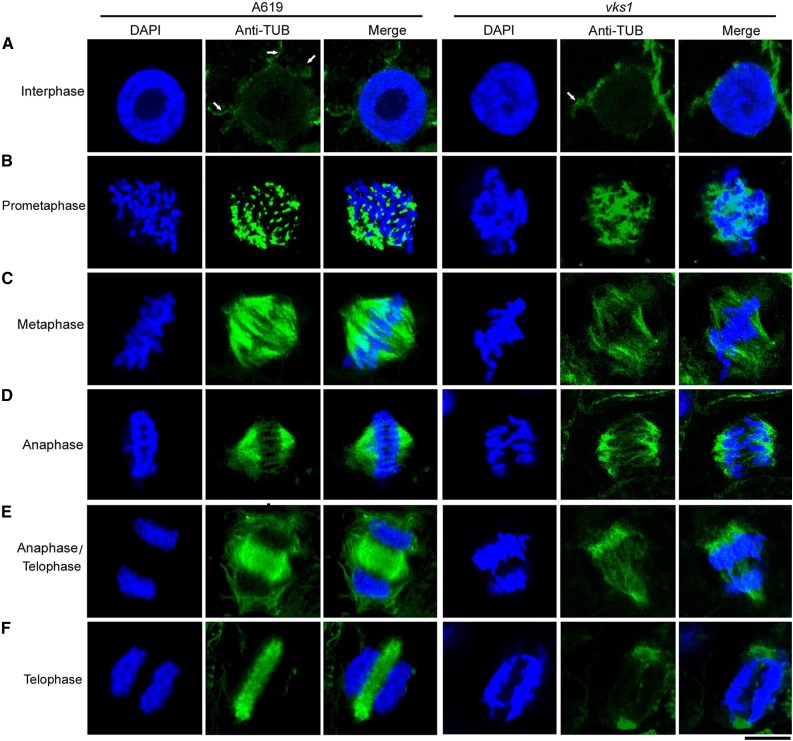
To further investigate the role of VKS1 in each phase of mitosis in early endosperm development, we performed immunostaining of α-tubulin to visualize MT arrays in vks1 and the wild type. In interphase, MTs were observed to be nucleated from the surface of the nuclear envelope and radiated into the cell, forming an array of fibers in the wild type (Figure 6A). As higher plants do not possess centrosomes, this structure is thought to function as the microtubule-organizing center (MTOC). There were fewer radial MTs in vks1 than in the wild type (Figure 6A). In premetaphase, because the cells of higher plants (such as maize) lack centrosomes, MTs are organized into a spindle by chromosomes themselves. Although the chromatin in vks1 was condensed into thread-like chromosomes, as observed in the wild type, the shapes of MTs were remarkably different. The formation of a spindle was already initiated in the wild type, wherein the MTs were stretched to attach to the chromosomes. In contrast, the MTs in vks1 exhibited an amorphous arrangement, and their attachment to the chromosomes appeared as a mass of tangles (Figure 6B). In metaphase, spindle fibers in the wild type were concentrated to a tip on each opposite pole, forming an umbrella-like shape. The chromosomes were aligned along the center of the cell; in contrast, the vks1 cell formed a spindle with splayed poles and looser and fewer fibers. The chromosomes appeared to be misarranged. In addition, some sister chromatids were not correctly attached to spindle fibers (Figure 6C; Supplemental Figure 9A).
Figure 6.
Early Endosperm Mitotic Divisions in the Wild Type and vks1.
MT arrays were visualized by immunostaining with anti-α-tubulin during mitosis in wild-type and vks1 endosperm cells at 6 DAP. MTs are colored green, and nuclei or chromosomes are colored blue. Phases are as follows: interphase (A); prometaphase (B); metaphase (C); anaphase (D); anaphase/telophase (E); and telophase (F). Bar = 10 μm.
When anaphase was reached, the umbrella-shaped spindle in the wild type began to pull the chromosomes toward opposite sides of the cell, whereas the flattened-pole spindle in vks1 had apparent defects in moving the chromosomes. Sister chromatids were not evenly attached to the irregular spindle fibers on each side and moved in opposite directions in a skewed rather than a straight line (Figure 6D; Supplemental Figure 9B). In late anaphase and early telophase in the wild type, the daughter chromosomes were equally pulled to the opposite ends of the cell. The spindle began to disappear, and the MTs began to accumulate in the middle of two daughter nuclei to initiate phragmoplast formation (Figure 6E); however, in vks1, the chromosomes were not equally allocated to each side of the cell (Figure 6E). The cohesins that bind sister chromatids together might not have been completely cleaved in anaphase (Supplemental Figure 9B), leading some sister chromosomes to be pulled to one side of the cell. Later in wild-type telophase, MTs continued to be enriched in the phragmoplast and finally developed a long and dense band across the entire cell center in the wild type. In contrast, in vks1, fewer MTs were deposited in the region where the phragmoplast was expected to be assembled (Figures 6E and 6F; Supplemental Figures 9C and 9D). Overall, these results demonstrate that VKS1 is critical for mitotic divisions in early endosperm development.
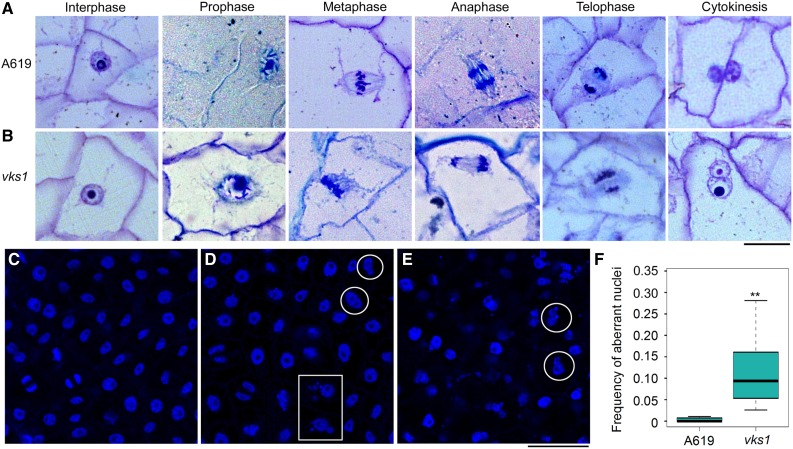
The absence of VKS1 resulted in abnormal mitosis in vks1 (Figure 6), which may in turn have affected cell division in the endosperm. To examine this possibility, semithin sections of endosperms at 6 DAP were inspected. Endosperm cells at different stages that were characteristic of normal mitosis and cytokinesis were observed in the wild type (Figure 7A). In telophase, the phragmoplast between two sets of separated daughter chromosomes was transversely elongated to the edges of the cell. During cytokinesis, the cell wall developed based on the phragmoplast divided the cell into two daughter cells. In vks1, defects in mitosis were consistent with the observation presented in Figure 6. Because of the malformed spindle and incorrectly aligned sister chromatids of metaphase, the daughter chromosomes were unequally separated. The defective phragmoplast resulted in the failure of cell division; thus, the affected vks1 cell contained one large nucleus and one small nucleus (Figure 7B).
Figure 7.
The Phenotypes of Endosperm Cells and Nuclei in A619 and vks1 during Early Development.
(A) and (B) Endosperm cells undergoing mitosis and cytokinesis in A619 (A) and vks1 (B) at 6 DAP. Bar = 25 μm.
(C) to (E) Nuclei of endosperm cells during mitosis in A619 (C) and vks1 ([D] and [E]) at 6 DAP. (D) shows a mild phenotype and (E) shows a severe phenotype of vks1. The nuclei stained by DAPI are in blue. Bar = 50 μm.
(F) Frequency of aberrant nuclei in endosperms of A619 and vks1 at 6 DAP. More than 4000 nuclei of A619 and 2600 nuclei of vks1 from 20 microscope fields were observed. The mean value of the ratios of aberrant nuclei to total nuclei in each field of A619 or vks1 was used to represent the frequency of aberrant nuclei in endosperm. **, P < 0.01 as determined by Student’s t test.
To illustrate the substantial deformities in vks1 in larger view, a field of nuclei stained by DAPI was examined. Compared with normal nuclei and chromosomes in the wild type (Figure 7C), aberrant nuclei were frequently observed in vks1 endosperm cells. A mild phenotype is shown in Figure 7D, and a severe phenotype is shown in Figure 7E. The conjugate nuclei of different sizes indicated by the circles likely resulted from unsuccessful cytokinesis (Figures 7D and 7E). The micronuclei (indicated by the box) probably resulted from unequal chromosome separations (Figure 7E). When calculating the frequency of abnormal nuclei in the wild type and vks1, the nuclei in wild-type endosperm cells were almost normal, whereas the ratios of abnormal nuclei in vks1 ranged from 2.5 to 34.4% (Figure 7F). Taken together, these results demonstrate that VKS1 is essential for mitosis and cytokinesis in early endosperm development.
DISCUSSION
Mutation in vks1 Affects Early Maize Endosperm Development
Yield is determined by many factors, including seed size. In maize kernels, the endosperm accounts for 90% of seed dry mass and is the major organ for nutrient storage, thereby predominantly determining the final size of the seed. Cell number and cell size are both important for endosperm volume (Sabelli and Larkins, 2009a). Although considerable progress has been made understanding endosperm cell expansion associated with storage reserve synthesis and accumulation, far less is known about the genes that regulate cell number during early endosperm development. Endosperm formation begins after double fertilization, and it initially develops as a coenocyte in which the nucleus undergoes multiple rounds of division without cytokinesis, forming a multinucleate cell. Cellularization initiates 2 to 3 DAP and is usually completed by 4 DAP, which is followed by a period of frequent early mitotic divisions (Sabelli and Larkins, 2009a). In Arabidopsis, mutations in genes encoding the VQ motif protein HAIKU1, the Leucine-rich repeat receptor IKU2, and the WRKY transcription factor MINISEED3 resulted in precocious cellularization of the endosperm, thereby leading to reduced seed size (Luo et al., 2005; Zhou et al., 2009; Wang et al., 2010). This indicates that the timing of the transition from the coenocytic stage to cellularization stage is a key decision point in endosperm development. Early mitotic divisions are another critical step, as most cells of the mature endosperm are generated during this phase (Sabelli et al., 2013, 2018). Although early endosperm development directly contributes little to kernel biomass, the cell number and endosperm structure established during this stage create the demand for subsequent accumulation of storage reserves.
In maize, while numerous mutants show seed defects, few have been reported where the phenotype specifically features aberrant early endosperm development. Indeed, an abundance of mutants with reduced kernel size and weight have been cloned and characterized, but most of them have been found to result from defective synthesis and accumulation of storage reserves and thus often display a phenotype that is characteristic of a certain mutation of endosperm development (e.g., the opaque, floury, and shrunken mutants). In general, for maize genetics, a major strategy for identifying and characterizing genes involved in a pathway is mutational inactivation. However, screening mutations that specifically affect early endosperm development is challenging because mutants of this kind often have pleiotropic effects on seed or plant development, with an aborted, small, or even nonviable phenotype, such as in globby1, empty pericarp, and defective kernel (dek) mutants (Sheridan and Neuffer, 1980; Scanlon et al., 1994; Costa et al., 2003). In early efforts to investigate maize seed development, a group of dek mutants were described. These mutants generally have decreased mitotic activity, thereby resulting in greatly reduced cell numbers; however, none of these mutants are viable (Kowles et al., 1992). Therefore, these mutants are not specifically involved in early endosperm development.
The vks1 mutant was identified through a screen of an EMS-induced mutant collection. This mutant drew our attention because its kernel size not only was small but also varied dramatically among different progeny (Figures 1A and 1B; Supplemental Figure 2A). By recurrent backcrossing, this phenotypic variation was confirmed to result from a single-gene mutation rather than the combined effects of multiple mutant loci. The single-kernel weight of the wild type (A619) showed limited variation, whereas in vks1 it ranged from 0.0128 to 0.1898 g. Examination of endosperm development at different stages demonstrated that the small kernels of vks1 were mainly caused by a reduction in cell number rather than a reduction in cell size. A reduction in cell number in vks1 was observed as early as 4 DAP (Figures 1G to 1N), but grain filling appeared to be normal in late endosperm development (Supplemental Figures 3A and 3B). At maturity, the storage proteins and starch contents showed no perceivable difference from those in the wild type (Supplemental Figures 3C and 3D), indicating that VKS1 primarily functions at the early phases of endosperm development.
More importantly, despite substantial variation in kernel size, the germination of vks1 seeds was comparable to that of wild-type seeds, indicating that embryo development was apparently not impaired by the vks1 mutation. The vks1 plants were shorter than the wild-type ones, but their vegetative growth exhibited no conspicuous abnormalities. The homozygous ears and the ears from reciprocal crosses between the wild type and vks1 showed normal seed set. However, the segregation ratio of vks1 and normal seeds in self-pollinated heterozygous ears deviated from that expected under Mendelian inheritance. This discrepancy was likely caused by the viability of vks1 pollen (Supplemental Figures 4D to 4F), which was lower than the viability of pollen bearing the wild-type Vks1 allele in terms of completing pollination.
Overall, the vks1 mutant displays a previously undescribed phenotype in maize endosperm development. Its effects are mainly restricted to early stages, although it also has some adverse effects on plant development. Cloning and characterization of this mutant gene are valuable for understanding the impact of early maize endosperm development.
VKS1, a Kinesin-14 Protein, Dually Localized to MTs and the Nucleus
Vks1 was determined to encode ZmKIN11, which belongs to the kinesin-14A subfamily (Lawrence et al., 2002; Dawe et al., 2018). VKS1 has two homologs in the maize genome, KINDR and DV1 (ZmKin6). KINDR is encoded by a gene cluster that is present only on Ab10. KINDR converts heterochromatic regions called knobs into neocentromeres, thereby promoting meiotic drive in maize. Epimutation of the Kindr cluster induced by RNA interference results in the loss of meiotic drive but no apparent defects in endosperm development (Dawe et al., 2018). DV1, encoded by Zm00001d002186 on chromosome 2, is required for meiotic spindle pole organization. Mutation of dv1 causes failure in promoting the formation of finely focused spindle poles during meiosis, thereby leading to aberrant chromosome segregation and a higher pollen abortion rate, but plants carrying this mutation are phenotypically indistinguishable from their wild-type siblings, indicating that dv1 does not cause deleterious effects on mitosis (Staiger and Cande, 1990). Similar to mutation of dv1, mutation of vks1 causes pollen abortion at a certain rate (Supplemental Figure 4E), indicating that VKS1 and DV1 are functionally redundant in pollen meiosis but may be greatly diverged in mitosis.
This raises the following question: is the divergence of coding sequences or the regulation of expression of Vks1 and dv1 responsible for the functional divergence? Although Vks1 is predominantly expressed in early stages of endosperm development, it is also expressed in shoots, leaves, tassels, and shoot apical meristems where mitosis and cytokinesis actively occur for cell proliferation based on the public RNA sequencing data (Chen et al., 2014). Interestingly, the temporal expression patterns of Vks1 and dv1 during endosperm development are similar (Supplemental Figure 10), although whether they have a spatial expression overlap is unknown. Since VKS1 and DV1 share >60% similarity, we do not exclude the possibility that VKS1 and DV1 also have partial functional redundancy in mitosis. It is intriguing to create a double mutant of these two genes to test this possibility. Mitosis and cytokinesis are fundamental mechanisms for cell proliferation. Mutations in vks1 only cause a small and variable seed size rather than the dek phenotype, and not all nuclei in the vks1 endosperm are abnormal (Figure 7F), indicating that other kinesin-14 proteins may have functional redundancy with VKS1. Indeed, Kin4A (Zm00001d019670), Kin3 (Zm00001d010790), and Kin14C (Zm00001d043504) in the kinesin-14 subfamily are also highly expressed at early endosperm stages (Supplemental Figure 10; Chen et al., 2014), implying that they have an important role in early endosperm development. Because the basic processes of early endosperm development are similar in grasses, we speculate that the functions of VKS1 homologs in other species (Supplemental Figure 6) are conserved in regulating mitosis and cytokinesis during early endosperm development.
VKS1 is predicted to contain three domains (the tail, CC, and motor domains; Figure 3H), each having a specific function for subcellular localization. The full-length VKS1 protein localizes to MTs and the nucleus (Figures 3I and 3J). The tail contains a signal for nuclear localization (Figures 3J and 3K), the CC domain is required for MT localization (Figure 3O), and the motor is essential for its correct localization to MTs (Figures 3M and 3N). Our data are consistent with the observation in which KINDR, the homolog of VKS1 bearing a deletion in the CC domain, localizes specifically to chromosome knobs rather than to the MTs (Dawe et al., 2018). Based on variations in protein structure and subcellular localization, this indicates that the functions of VKS1 and KINDR proteins have diverged.
VKS1 Is Essential for Nuclear-Cytoplasmic Domain Formation in the Coenocyte
Vks1 transcription occurred throughout early endosperm development, with a peak at 2 DAP, but it was not detected in the mid and late phases (Figure 3A). Examination of the spatial distribution of VKS1 mRNAs in 2- and 3-DAP-old developing seeds showed that the expression of Vks1 was restricted to the coenocyte and cellularizing endosperm cells (Figures 3B to 3E), consistent with the observation that the cell number reduction in vks1 occurred in the early phase of endosperm development. After double fertilization, the primary endosperm nucleus undergoes several rounds of nuclear division without cell wall formation and cytokinesis. These divisions appear to be synchronous within day 1 after double fertilization, but later, only the neighboring nuclei divide synchronously. This is likely due to developmental gradients formed from the embryo sac to the chalazal region (Sabelli and Larkins, 2009b). In maize, as many as 512 nuclei can be generated during coenocytic nuclear proliferation (Leroux et al., 2014). We did not determine whether the number of coenocytic nuclei in vks1 was reduced, but we observed abnormal nuclei division and distribution in the coenocyte (Figure 4), which may result in a slow start for the rest of kernel development and consequently a small kernel size. At 2 DAP, the nuclei were evenly spaced at the periphery of the central cell in the wild type (Figure 4A), whereas they were not visible in vks1 with a light microscope (Figure 4B). The two adjacent nuclei shown in Figure 4B likely originated from an abnormal nuclear division, as the lagging chromosomes were dispersed. In barley (Hordeum vulgare) and Arabidopsis, nuclei migrate to positions along the coenocytic membrane, where nuclear-cytoplasmic domains are formed by internuclear RMSs (Brown et al., 1994, 2003; Pickett-Heaps et al., 1999). As shown in Figure 4C, the nucleus was clearly surrounded by a domain where VKS1 and MTs were colocalized. In the absence of VKS1, the adjacent nuclei were embedded in a continuous MT network without a distinct interzone between them, thereby forming a flattened module closely adjacent to the cell membrane, and the nuclear-cytoplasmic domains was a primary casualty (Figure 4D). This finding may explain why no nuclei were clearly observed while they do exist in vks1 with a light microscope, as shown in Figure 4B.
VKS1 Is Essential for MT Organization in Mitosis and Cytokinesis
Mitotic spindles are MT-based structures that function in the separation of sister chromatids during mitosis. An early step of spindle assembly involves nucleation of MTs. In most animals and fungi, centrosomes function as the major MTOCs to arrange MTs during both interphase and mitosis. Flowering plants lack centrosomes, and they assemble the spindle through a centrosome-free pathway (Bannigan et al., 2008; Yi and Goshima, 2018). During interphase, MTs are nucleated from the surface of the nuclear envelope and radiate into the cell, generating an array that resembles an aster, with the nucleus serving as the MTOC center. However, the factors and underlying mechanisms by which radial MTs are nucleated and formed remain largely unknown. In the vks1 mutant, the number of radial MTs was obviously lower than that in the wild type (Figure 6A). Given that VKS1 and MTs also colocalize to the cytoplasm (Figure 3L), one could envision that VKS1 is essential for MT-mediated cytoplasmic MTOC formation. During prophase, cytoplasmic MTs are enriched around the nuclear envelope and later transform into bipolar structures called polar caps or the prophase spindles (De Mey et al., 1982; Zhang et al., 1990; Smirnova and Bajer, 1994, 1998; Chan et al., 2005). Interestingly, enriched cytoplasmic VKS1 proteins were observed to surround the nuclear envelope (Figure 5), suggesting that VKS1 is involved in the establishment of the prophase spindle. However, more detailed experiments are needed to test this hypothesis. A model for the main functions of VKS1 in mitosis and cytokinesis during early endosperm development is proposed, as shown in Figure 8. A lack of VKS1 causes substantial deformities in mitotic activities, including the missions of MTs to acquire chromosomes in prometaphase, spindle formation and chromosome alignment in metaphase, separation of sister chromatids in anaphase, and phragmoplast formation in telophase. In the vks1 mutant, the assembly of MTs into a spindle was dramatically disrupted, resulting in a loose and splayed spindle (Figure 6; Supplemental Figure 9). This disruption caused incorrect alignment and separation of chromosomes in metaphase and anaphase. The multiple micronuclei observed in vks1 cells could result from nondisjunction followed by nuclear restitution (Figures 7D and 7E). Therefore, while cell number is exponentially increased by rapid mitosis and cytokinesis in the wild-type endosperm, one could envision that cell division might be frequently repressed due to abnormal spindle formation.
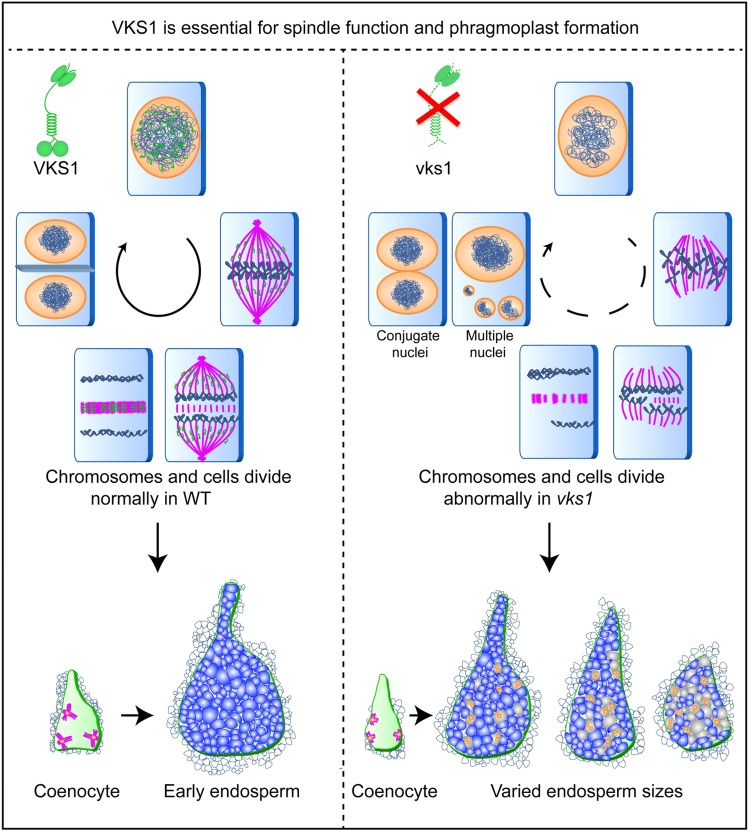
Figure 8.
Proposed Model for VKS1 Function and the Varied-Kernel-Size Phenotype.
Rapid mitotic divisions in early endosperm development produce most of the cells that make up the starchy endosperm. Vks1 is predominantly expressed in early endosperm development and plays key roles in critical events of mitosis and cytokinesis. VKS1 dynamically colocalizes with MTs during interphase, mitosis, and cytokinesis. A lack of VKS1 results in substantial defects in spindle assembly, sister chromatid separation, phragmoplast formation, and eventually cell division. Therefore, while normal mitosis maintains rapid cell division to build up the endosperm structure and function in the wild type, aberrant mitosis may frequently cause repression in cell division, thereby leading to reduced cell numbers in the vks1 endosperm. Perhaps as a consequence of varying numbers of affected cells, the kernel sizes of individual vks1 seeds display dramatic variation.
The spindle is a dynamic structure that is precisely mediated by the balance of opposing forces provided by cross-linking motors (Desai and Mitchison, 1997; Heald, 2000). In some eukaryotes, cytoplasmic dynein serves as the primary cytoskeletal motor for minus-end-directed processes. A well-characterized example is the Xenopus system, in which disruption of dynein leads to splayed poles and abnormally long spindles (Heald et al., 1996). In addition, dynein functions to maintain the organization of the same polarity at spindle poles (Merdes et al., 2000). However, plants lack dynein but possess a large number of minus-ended kinesin-14s instead. These probably have evolved to function as dynein (Roberts et al., 2013). In addition to plants, Drosophila embryos possess C-terminal motors that function in pole formation via cross-linking the minus end (Compton, 1998). Previous studies demonstrated that a lack of kinesin-14 family members can lead to slower or less efficient spindle assembly (Matthies et al., 1996; Gordon et al., 2001; Prigozhina et al., 2001; Marcus et al., 2003). The Arabidopsis kinesin ATK5 specifically functions in early spindle assembly during mitosis; however, the spindle at metaphase in the atk5 mutant showed no apparent phenotype (Ambrose and Cyr, 2007). Interestingly, the deformed spindle in vks1 resembles the spindle fibers arrested in the first meiotic metaphase in Arabidopsis atk1 and maize dv1 (Chen et al., 2002; Ambrose and Cyr, 2007; Higgins et al., 2016). Atk5 is the most closely related VKS1 homolog in Arabidopsis (Supplemental Figure 1).
Time-lapse imaging of mitosis revealed that lack of kinesin-14 (ATK5) caused mitosis to slow down (Ambrose and Cyr, 2007). During metaphase, we observed almost no cells in the dividing phase in a single field for the wild type. However, in vks1, cells at different phases of division were easily observed (Supplemental Figure 9), suggesting that mitosis in vks1 slowed down, which in turn reduced early endosperm cell proliferation (Figure 8). In addition, our data also suggested that VKS1 plays a crucial role in spindle formation and chromosome separation in plants (Figure 6), which is consistent with prior studies on plant kinesin-14s (Chen et al., 2002; Ambrose and Cyr, 2007; Higgins et al., 2016; Dawe et al., 2018; Gicking et al., 2018). The CC domain in the VKS1 N terminus is responsible for localization to MTs (Figure 3H), and the motor in the C terminus generates opposing forces on the spindle MTs, thereby pulling the daughter chromosomes equally to opposite poles of the cell.
The vks1 seeds exhibited varied kernel sizes within individuals. Consistently, vks1 endosperms exhibited different severities of mitosis and cytokinesis abnormality at 6 DAP (Figures 7D and 7E). As early mitotic divisions are nonsynchronous, the timing at which aberrant mitosis occurs during proliferation of a clonal cell lineage and the number of cells that are affected at one time may differ within vks1 endosperms. One could envision that mitotic abnormalities taking place earlier would produce a more severe phenotype than those taking place later.
Random chromosome loss may lead to PCD, and as a consequence there might be patches of death in the vks1 endosperm. However, we did not observe any sector of cell death or cavity in the endosperm at different developmental stages (Figures 1H to 1L; Supplemental Figure 3B). This is probably because endosperm PCD is different from typical PCD programs in plants, wherein the cellular corpse is usually processed concurrently with cell death. In maize endosperm, PCD initiates in the central and upper regions of the starchy endosperm at 12 to 16 DAP, when the storage reserves are actively synthesized, and expands to the entire starchy endosperm by late development. The degradation of the cellular corpse does not occur until germination and is controlled exogenously by the activities of aleurone cells (Young and Gallie, 2000a, 2000b). It is likely that the processing of abnormal endosperm cells in vks1 still follows the wild-type pattern.
Plant cells contain specialized MT-based structures called phragmoplasts, which facilitate cell wall formation following cell division. The phragmoplast contains a dynamic MT array that is aligned perpendicularly to the division plane, with the plus end locating to the division site. The absence of Arabidopsis kinesin-12A and kinesin-12B members caused a failure to form an antiparallel MT array and subsequently affected male gametogenesis without the formation of a cell plate (Lee et al., 2007). For kinesin 14, actions on the MT array of phragmoplasts, such as the sliding of antiparallel MTs, cross-linking of parallel MTs, and focusing of the spindle pole, require both the motor and tail domains of kinesin-14, of which the N-terminal tail attaches to one MT while the C-terminal motor transports another MT (Wendt et al., 2002; Ambrose et al., 2005; Fink et al., 2009; She and Yang, 2017). Loss of kinesin-14 may result in ineffective recruitment of MTs, leading to abnormal membrane formation, which in turn affects cell plate dynamics. Therefore, the defective phragmoplast in the vks1 mutant may lead to failed cell wall formation at the cell plate and result in abnormal cells containing conjugate nuclei (Figures 7B, 7D, and 7E).
METHODS
Genetic Materials
All maize (Zea mays) plants were grown either in plastic greenhouses in Shanghai, China, or in the field in Sanyang and Harbin, China. The temperature in the greenhouses was 25 to 32°C during maize pollination and the early developmental stage (before 8 DAP). In addition to Shanghai, we also planted materials in Sanya, Hainan Province, for winter nursery and in Harbin, Heilongjiang Province, for summer nursery.
The pollen of the A619 inbred line was collected and mutagenized with EMS reagent for 45 min, and the resulting pollen was applied to A619 female ears. A large number of seeds were planted and self-pollinated, yielding more than 3000 ears, among which an ear segregated mutant seeds with varied kernel sizes; this was designated vks1. To purge most irrelevant mutations, vks1 was recurrently backcrossed to A619 (used as the female) for six generations. Because vks1 is a recessive mutation, the backcrossing and selfing of at least 20 plants in each generation had to be performed at the same time. The presence of the vks1 allele in a plant was determined by inspecting whether the self-pollinated ear segregated vks1 seeds. If it did, then the corresponding ear from backcrossing was used for planting in the next generation.
To knock out Vks1 by CRISPR/Cas9, the small guide RNA was predicted online (http://www.e-crisp.org/E-CRISP/designcrispr.html) and synthesized by Sangon Biotech, then cloned into the PsiI and XbaI sites between the maize U6 promoter and U6 terminator. By Agrobacterium tumefaciens-mediated transformation, the constructs were delivered into maize. The inbred line B104 immature embryos (1.5–2.0 mm) were dissected from the ears growing in the field at ∼11 DAP. Three positive events were recovered, following the standard transformation protocol (Frame et al., 2002). The primers for making the construct and for amplifying the target region are listed in Supplemental Table 1.
Mapping by Sequencing of vks1
To map the vks1 mutant gene, vks1 and wild-type A619 plants were crossed and then self-pollinated. More than 100 seeds of each phenotype (mutant and normal) from the segregating ears were selected and germinated individually for DNA extraction. For each pool, DNA from each sample was mixed in an equal ratio. Genomic DNA was sheared for preparation of the sequencing library according to the standard protocol of the Illumina TruSeq DNA PCR-free prep kit.
Reads of the two bulks (A, normal, and B, vks1) and the parental line A619 were aligned to the reference genome using bwa software with default parameters; SNP calling was performed using GATK software. SNPs with a sequencing depth of <5 were filtered. Then, the SNP index was calculated, and SNPs with an SNP index of <0.3 in both bulks were filtered to reduced false-positive detection of SNPs. The MutMap method was applied to detect genomic intervals underlying the phenotype as described previously (Takagi et al., 2013). Briefly, the average SNP index was calculated using a sliding-window approach with a 4-Mb window size and a 200-kb increment. Δ (SNP index), the difference between the two bulks, was then determined. Statistical confidence intervals of Δ (SNP index) were calculated under the null hypothesis of no quantitative trait loci following procedures reported previously (Abe et al., 2012; Takagi et al., 2013).
Fine-mapping of vks1 was performed using the BC1F1 population of vks1 × (vks1 × B73). Insertion/deletion primers (shown in Supplement Table 1) were based on differences in B73 and A619 genomic sequences on chromosome 7. Linkage analysis of the BC1F1 population was performed by detecting the genotype of the reference mutation site using the primers vks1-F and vks1-R. The splice acceptor site of vks1-2 sites was detected by the primers spc-F and spc-R, and Mu insertion in the 5′UTRwas detected by the primers F, R, and TIR6, as shown in Supplemental Figure 5.
Phenotypic Analysis
One hundred seeds in the middle of the ear were selected for measurement of single-grain weight. All the ears used for photography were representative of more than 10 ears. Kernels were selected from the middle of the ear for photography and determination of single-grain weight. The coefficient of variation (i.e., the ratio of the sd to the mean [CV = σ/μ]), was calculated. Plants grown in the field for 10 weeks were transferred to flowerpots for photography. After the plants were photographed, the leaves were removed, and the whole plants were photographed again.
Measurement of Proteins and Starch
To measure the content of the proteins and starch, 20 mature kernels of A619 and vks1 from three individual ears were ground into fine flour using steel beads. Then, 100 mg of flour from each sample was used for the measurement of proteins and starch. Three biological replicates for each were performed. SDS-PAGE was performed to analyze the accumulation pattern of zein and nonzein proteins in A619 and vks1 according to previous reports (Liu et al., 2016; Zhang et al., 2016). The starch content was detected using the Total Starch Assay Kit (K-TSTA; Megazyme).
Pollen Viability
Fresh pollen from dehiscent tassels of A619 and vks1 was collected on slides and then stained with a solution containing 0.2% (w/v) benzidine, 0.2% (w/v) α-naphthol, and 0.3% (v/v) H2O2 for 5 min. Slides were viewed with a light microscope (Leica M165 FC). Ten tassels of A619 and vks1 were detected. DeepRed-stained pollen grains were fertile, whereas irregularly shaped, unstained grains were sterile. Then, the ratio of the number of dark red pollen grains to the total number of pollen grains in the microscope field of A619 or vks1 was calculated. The average value of the ratios of 10 fields was used to indicate pollen fertility.
Histocytochemical Analysis
For the histocytochemical analysis of endosperm tissues of A619 and vks1, seeds at the early developmental stage (2, 4, and 6 DAP) and filling stage (18 DAP) were collected. Seeds were fixed in FAA buffer (50% ethanol:formaldehyde:acetic acid = 90:5:5 volume) and then vacuumed at 4°C. After dehydration using a gradient concentration of ethanol, 2-, 4-, and 6-DAP-old seeds were embedded in epoxide resin for semithin sectioning, while 18-DAP-old samples were embedded in paraffin. The sections were stained with toluidine blue solution and then photographed under a bright field using an ECLIPSE 80i microscope (Nikon).
To calculate the number of cells in the early (4 and 6 DAP) endosperm of A619 and the mutant vks1, 20 semithin sections were imported into Adobe Photoshop software, and the number of cells was determined by cell counting. Then the average cell number of 20 sections was calculated.
To calculate the area of the endosperm relative to that of the whole seed, the area of the endosperm and the area of the whole seed of 20 sections were measured by ImageJ software. Then, the ratio of endosperm area to seed area was calculated to indicate the relative value.
RT-qPCR and Standard RT-PCR
Total RNA was extracted from whole seeds or endosperms of A619 and vks1 at 0, 1, 2, 3, 4, 6, 8, 10, 12, 16, 18, and 24 DAP using TRIzol reagent (Invitrogen, catalog number 15,596,018). Then, RNA was purified with an RNeasy Mini Kit (Qiagen, catalog number 74,106) after DNaseI digestion (Qiagen, catalog number 79,254) for reverse transcription by a SuperScript III First Strand Synthesis Kit (Invitrogen, catalog number 18,080,051). RT-qPCR was performed with SYBR Green I (Takara, catalog number RR420A) and a CFX Connect Real-Time System (Bio-Rad). The primers of Vks1 were Vks1-RTF and Vks1-RTR. The comparative CT method (ΔΔCT method) was employed for the relative quantification of gene expression, and the maize Actin gene (primers: Actin-RTF and Actin-RTR) was used as a control. The expression level in seeds at 0 DAP was set to 1.
To analyze the expression level of Vks1 in vks1-2, RNA was extracted from the 2-DAP seeds of B73 and vks1-2 and used for RT-qPCR analysis. The gene-specific primers for Vks1 and Actin were shown above. The expression level in B73 seeds at 2 DAP was set to 1.
Data were generated from three biological replicates of each sample, and means and se were calculated using Microsoft Excel.
RNA in Situ Hybridization
To analyze the tissue expression pattern of Vks1 in the seeds at early stages, 2- and 3-DAP-old seeds of A619 were used for RNA in situ hybridization as described in our previous work (Zhang et al., 2015). The seeds were fixed for 16 h in 4% paraformaldehyde solution with 0.1% Triton X-100 and 0.1% Tween 20 in PBS (Takara, catalog number T900). After dehydration using graded ethanol and vitrification by dimethylbenzene, the samples were embedded in paraffin. A 524-bp cDNA fragment of Vks1 was cloned using the primers Vks1-probeFKpnI and Vks1-probeRXhoI and inserted into the pBluescript SK+ vector using KpnI and XhoI. The vector was used for the synthesis of antisense and sense RNA probes according to the instructions for the DIG RNA Labeling Mixture (Roche, catalog number 11175025910). Then, 10-μm sections were cut, and in situ hybridization experiments were performed according to methods described previously (Langdale, 1994; Cox and Goldberg, 1998). The sections were observed and photographed with an ECLIPSE 80i microscope.
Anti-VKS1 Antibody Preparation
VKS1 antibodies were produced by ABclonal. The cDNA fragment encoding the partial VKS1 protein from amino acids 49 to 449 was amplified using the primers VKS1-EF and VKS1-ER and then inserted into the prokaryotic expression vector pET-28a-VKS1 using MultiF Seamless Assembly Mix (ABclonal, catalog number RK21020). Purified protein was injected into rabbits to produce VKS1 antibodies. After affinity purification, the antibodies were used for protein gel blotting and immunofluorescence histochemistry.
Immunoblotting Analysis
Total protein from 2- and 3-DAP seeds of A619 and vks1 was extracted with radioimmunoprecipitation assay lysis buffer (Leagene, catalog number PS0013). Proteins were quantified by Bradford assays. Twenty micrograms of protein was separated by a 10% SDS-PAGE gel and then transferred electrophoretically to a PVDF membrane (Bio-Rad, catalog number 162-0177). The membrane was incubated with VKS1 antibodies at 4°C overnight and then incubated with the secondary antibody, goat anti-rabbit IgG-horseradish peroxidase (HRP; Abmart, catalog number M21002L). To detect the control protein ACTIN, a primary antibody, mouse monoclonal ACTIN antibody (Abmart, catalog number M20009L), and a secondary antibody, goat anti-mouse IgG-HRP (Abmart, catalog number M21001L), were used to perform the immune reaction. The dilution of antibodies was 1:1000. Finally, the membrane was treated with HRP chemiluminescent substrate reagent (Invitrogen, catalog number WP20005) and imaged using a Tanon-5200 system (Tanon).
To analyze the protein content of VKS1 in vks1-2, protein was extracted from the 2-DAP seeds of B73 and vks1-2.
Subcellular Localization of VKS1 in Nicotiana benthamiana Epidermal Cells
To investigate the subcellular localization of VKS1 in N. benthamiana leaves, the full-length and different domain fragments of VKS1 protein were fused with GFP in pCAMBIA1301 plasmid using KpnI and XhoI and expressed via the 35S promoter. The primer pairs (shown in Supplemental Table 1) VKS1FL-FKpnI and VKS1FL-RXhoI for the full-length VKS1 protein (VKS1FL, amino acids 1–761), VKS1△T-FKpnI and VKS1FL-RXhoI for the VKS1 protein lacking the tail domain (VKS1△T, amino acids 49–761), VKS1FL-F1KpnI and VKS1△M-RXhoI for the VKS1 protein lacking the motor (VKS1△M, amino acids 1–408), VKS1△T-FKpnI and VKS1△M-RXhoI for the VKS1 protein lacking the tail and motor domains (VKS1△TM, amino acids 49–408), and VKS1△TCC-FKpnI and VKS1FL-RXhoI for the VKS1 protein lacking the tail and CC domains (VKS1△TCC, amino acids 409–761) were used to amplify the different fragments of VKS1. In addition, GFP-VKS1△TCC-F and GFP-VKS1△TCC-R were used to amplify the motor domain, which was ligated to the C terminus of GFP by the homologous recombination method. These constructs were then transferred into Agrobacterium (GV3101 strain) and injected into 3-week-old N. benthamiana leaves. To analyze the colocalization of VKS1 with MTs, the Agrobacterium containing the Arabidopsis β-tubulin6 fused with the mCherry protein (mCherry-TUB6) was infiltrated into N. benthamiana leaves together with the Agrobacterium carrying VKS1-GFP.
Two days after infiltration, the transfected leaves were collected for imaging. To observe the nucleus, 1 μg/mL DAPI (Roche, catalog number 0671) solution was injected into the transfected leaves. Afterwards, segments of transfected leaf were observed with an LSM880 confocal microscope (Zeiss). Z-stack images of epidermal cells expressing GFP-fused protein were taken in the Airyscan superresolution mode.
Fluorescence Immunohistochemistry Analysis
For immunolocalization of tubulin in endosperm cells during mitosis, seeds of A619 and vks1 were used at 6 DAP. The sample treatment was performed according to a previous report (Quan et al., 2008). Hand sections of seeds were fixed for 1 h in freshly prepared 4% (v/v) formaldehyde in PHEM buffer with 0.02% (v/v) Triton X-100 and 5% (v/v) DMSO. After being rinsed several times in PHEM buffer, the samples were treated with a cell wall-removal enzyme solution containing 1% (w/v) β-glucuronidase (LabTop, catalog number S10046), 0.1% (w/v) cellulose (Yakult, catalog number 9012-54-8), 0.1% (w/v) pectinase (LabTop, catalog number S10007), 0.1% (w/v) lyticase (Solarbio, catalog number L8141), and 1% (w/v) Glc for 30 min. Samples were then blocked with 5% (w/v) BSA (Yeasen, catalog number 36103ES60) in PBS for 1 h and hybridized with monoclonal mouse anti-α-tubulin antibodies (1:100; Sigma-Aldrich, catalog number T6074) overnight in the dark at 4°C. After being washed three times with PBS, the samples were incubated with the secondary antibody Alexa Fluor 488-conjugated goat anti-mouse IgG at a dilution of 1:200 (Yeasen, catalog number 33206ES60). To analyze the localization of VKS1 during mitosis, the blocked sections were incubated in VKS1 antibodies and then hybridized with Alexa Fluor 488-conjugated goat anti-rabbit IgG secondary antibody (Yeasen, catalog number 33106ES60).
To detect the colocalization of VKS1 and tubulin in the endosperm cells undergoing mitosis, seeds at 2 and 6 DAP were fixed in fixative buffer (4% [v/v] paraformaldehyde, 0.1% [v/v] Triton X-100, and 0.1% [v/v] Tween 20 in PBS, pH 7.0) for 16 h and then embedded in paraffin. After being blocked in 5% (w/v) BSA, the sections were coincubated with anti-VKS1 antibodies raised in rabbit and α-tubulin antibodies raised in mouse and then stained by Alexa Fluor 488-conjugated goat anti-rabbit and Alexa Fluor 594-conjugated goat anti-mouse IgG (Yeasen, catalog number 33212ES60) secondary antibodies, respectively.
Nuclei were stained with 1 μg/mL DAPI. The sections were observed with an LSM880 confocal microscope under Airyscan modes (Zeiss). Z-stack images of endosperm cells undergoing mitosis and images of nuclei of endosperm cells were taken in the Airyscan superresolution mode.
Phylogenetic Analysis
A total of 163 kinesin protein sequences were submitted to the ClustalW program using the default settings (pairwise alignment options: gap opening penalty 10, gap extension penalty 0.1; multiple alignment options: gap opening penalty 10, gap extension penalty 0.2, gap distance 4, no end gaps and protein weight matrix using Gonnet) for multiple protein alignment. Based on the aligned protein sequences, the phylogenetic tree was constructed using the MEGA7.0 program (http://www.megasoftware.net/) and the maximum likelihood method with the Jones-Taylor-Thornton model, and the bootstrap test was conducted with 1000 replicates.
Accession Numbers
Sequence data from this study can be found in GenBank/EMBL data libraries under accession numbers NP_001292799 for VKS1 (ZmKIN11) and NP_001306694 for DV1. The other accession numbers used for phylogenetic analysis can be found in Supplemental Fileand Supplemental Figure 6.
Supplemental Data
Supplemental Figure 1. Phylogenetic analysis of the kinesin family.
Supplemental Figure 2. Investigation of maternal effect on the vks1 phenotype.
Supplemental Figure 3. Storage reserve accumulation and plant phenotype of A619 and vks1.
Supplemental Figure 4. Segregation of mutant and normal seeds on F2 ears of A619 and vks1.
Supplemental Figure 5. Map-based cloning of vks1 and the kernel of vks1-3 and vks1-4.
Supplemental Figure 6. Amino acid sequence alignment of Vks1 and its homologs in other grasses.
Supplemental Figure 7. Immunofluorescence histochemical localization of VKS1 and α-tubulin in A619 endosperm cells.
Supplemental Figure 8. Dual immunofluorescence localization of VKS1 and α-tubulin in telophase of endosperm cells.
Supplemental Figure 9. Immunofluorescence histochemical analysis of defective mitosis in vks1 endosperm cells.
Supplemental Figure 10. Expression pattern of the maize kinesin family.
Supplemental Table 1. Primers used in this study.
Supplemental File. Text file of the alignment used to generate the phylogenetic tree in Supplemental Figure 1.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Brian A. Larkins from the University of Nebraska-Lincoln for critical review and editing of the manuscript and Zhaosheng Kong from the Institute of Microbiology, Chinese Academy of Sciences, for generous provision of the mCherry-TUB6 marker and critical suggestions. We thank Zhenhua Wang, Lin Zhang, and Xing Zeng from Northeast Agricultural University for assistance with materials planting. We thank Xiaosu Gao for guidance on confocal microscopy. This research was supported by the National Natural Science Foundation of China (91635303 and 31830063 to Y.W. and 31871625 to H.W.) and the Chinese Academy of Sciences (XDPB0401 to Y.W.).
AUTHOR CONTRIBUTIONS
Y.W. oversaw the project. Y.H., H.W., and Y.W. conceived and designed the experiments. Y.H., H.W., J.W., W.W., and Y.W. analyzed data, made the figures, and wrote the article. Y.H. and H.W. performed experiments on phenotypic, gene expression, protein localization, and immunohistochemistry assays. X.H. and Q.W. constructed the plasmid vectors and performed traditional map-based cloning. J.W., W.W., and D.A. created evolutionary trees and family analysis. J.L. performed the semithin section analysis.
References
- Abe, A., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Ambrose, J.C., Cyr, R. (2007). The kinesin ATK5 functions in early spindle assembly in Arabidopsis. Plant Cell 19: 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose, J.C., Li, W., Marcus, A., Ma, H., Cyr, R. (2005). A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol. Biol. Cell 16: 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannigan, A., Lizotte-Waniewski, M., Riley, M., Baskin, T.I. (2008). Emerging molecular mechanisms that power and regulate the anastral mitotic spindle of flowering plants. Cell Motil. Cytoskeleton 65: 1–11. [DOI] [PubMed] [Google Scholar]
- Brown, R.C., Lemmon, B.E., Olsen, O.A. (1994). Endosperm development in barley: Microtubule involvement in the morphogenetic pathway. Plant Cell 6: 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.C., Lemmon, B.E., Nguyen, H. (2003). Events during the first four rounds of mitosis establish three developmental domains in the syncytial endosperm of Arabidopsis thaliana. Protoplasma 222: 167–174. [DOI] [PubMed] [Google Scholar]
- Chan, J., Calder, G., Fox, S., Lloyd, C. (2005). Localization of the microtubule end binding protein EB1 reveals alternative pathways of spindle development in Arabidopsis suspension cells. Plant Cell 17: 1737–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Marcus, A., Li, W., Hu, Y., Calzada, J.P., Grossniklaus, U., Cyr, R.J., Ma, H. (2002). The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129: 2401–2409. [DOI] [PubMed] [Google Scholar]
- Chen, J., Zeng, B., Zhang, M., Xie, S., Wang, G., Hauck, A., Lai, J. (2014). Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 166: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, D.A. (1998). Focusing on spindle poles. J. Cell Sci. 111: 1477–1481. [DOI] [PubMed] [Google Scholar]
- Costa, L.M., Gutierrez-Marcos, J.F., Brutnell, T.P., Greenland, A.J., Dickinson, H.G. (2003). The globby1-1 (glo1-1) mutation disrupts nuclear and cell division in the developing maize seed causing alterations in endosperm cell fate and tissue differentiation. Development 130: 5009–5017. [DOI] [PubMed] [Google Scholar]
- Cox, K.H., Goldberg, R.B. (1998). Analysis of plant gene expression. In Plant Molecular Biology: A Practical Approach, Shaw C.H., ed (Oxford, UK: Oxford IRL Press; ), pp. 1–35. [Google Scholar]
- Dawe, R.K.,et al. (2018). A kinesin-14 motor activates neocentromeres to promote meiotic drive in maize. Cell 173: 839–850. [DOI] [PubMed] [Google Scholar]
- De Mey, J., Lambert, A.M., Bajer, A.S., Moeremans, M., De Brabander, M. (1982). Visualization of microtubules in interphase and mitotic plant cells of Haemanthus endosperm with the immuno-gold staining method. Proc. Natl. Acad. Sci. USA 79: 1898–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., Mitchison, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13: 83–117. [DOI] [PubMed] [Google Scholar]
- Fink, G., Hajdo, L., Skowronek, K.J., Reuther, C., Kasprzak, A.A., Diez, S. (2009). The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat. Cell Biol. 11: 717–723. [DOI] [PubMed] [Google Scholar]
- Frame, B.R., Shou, H., Chikwamba, R.K., Zhang, Z., Xiang, C., Fonger, T.M., Pegg, S.E., Li, B., Nettleton, D.S., Pei, D., Wang, K. (2002). Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicking, A.M., Swentowsky, K.W., Dawe, R.K., Qiu, W. (2018). Functional diversification of the kinesin-14 family in land plants. FEBS Lett. 592: 1918–1928. [DOI] [PubMed] [Google Scholar]
- Gordon, M.B., Howard, L., Compton, D.A. (2001). Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J. Cell Biol. 152: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamura, Y., Nagahara, S., Higashiyama, T. (2012). Double fertilization on the move. Curr. Opin. Plant Biol. 15: 70–77. [DOI] [PubMed] [Google Scholar]
- Heald, R. (2000). Motor function in the mitotic spindle. Cell 102: 399–402. [DOI] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Blank, T., Sandaltzopoulos, R., Becker, P., Hyman, A., Karsenti, E. (1996). Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382: 420–425. [DOI] [PubMed] [Google Scholar]
- Higgins, D.M., Nannas, N.J., Dawe, R.K. (2016). The maize divergent spindle-1 (dv1) gene encodes a kinesin-14a motor protein required for meiotic spindle pole organization. Front. Plant Sci. 7: 1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, N. (1998). Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279: 519–526. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N., Noda, Y., Tanaka, Y., Niwa, S. (2009). Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10: 682–696. [DOI] [PubMed] [Google Scholar]
- Kowles, R.V., McMullen, M.D., Yerk, G., Phillips, R.L., Kraemer, S., Srienc, F. (1992). Endosperm mitotic activity and endoreduplication in maize affected by defective kernel mutations. Genome 35: 68–77. [Google Scholar]
- Langdale, J. (1994). In situ hybridization. In The Maize Handbook, Freeling M. and Walbot V., eds (New York: Springer; ), pp. 165–180. [Google Scholar]
- Lawrence, C.J., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167: 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.J., Morris, N.R., Meagher, R.B., Dawe, R.K. (2001). Dyneins have run their course in plant lineage. Traffic 2: 362–363. [DOI] [PubMed] [Google Scholar]
- Lawrence, C.J., Malmberg, R.L., Muszynski, M.G., Dawe, R.K. (2002). Maximum likelihood methods reveal conservation of function among closely related kinesin families. J. Mol. Evol. 54: 42–53. [DOI] [PubMed] [Google Scholar]
- Lee, Y.R., Li, Y., Liu, B. (2007). Two Arabidopsis phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis. Plant Cell 19: 2595–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.R., Qiu, W., Liu, B. (2015). Kinesin motors in plants: From subcellular dynamics to motility regulation. Curr. Opin. Plant Biol. 28: 120–126. [DOI] [PubMed] [Google Scholar]
- Leroux, B.M., Goodyke, A.J., Schumacher, K.I., Abbott, C.P., Clore, A.M., Yadegari, R., Larkins, B.A., Dannenhoffer, J.M. (2014). Maize early endosperm growth and development: From fertilization through cell type differentiation. Am. J. Bot. 101: 1259–1274. [DOI] [PubMed] [Google Scholar]
- Li, N., Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33: 23–32. [DOI] [PubMed] [Google Scholar]
- Liu, H., et al. (2016). Gene duplication confers enhanced expression of 27-kDa γ-zein for endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. USA 113: 4964–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Dennis, E.S., Berger, F., Peacock, W.J., Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) kinase gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus, A.I., Ambrose, J.C., Blickley, L., Hancock, W.O., Cyr, R.J. (2002). Arabidopsis thaliana protein, ATK1, is a minus-end directed kinesin that exhibits non-processive movement. Cell Motil. Cytoskeleton 52: 144–150. [DOI] [PubMed] [Google Scholar]
- Marcus, A.I., Li, W., Ma, H., Cyr, R.J. (2003). A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Mol. Biol. Cell 14: 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies, H.J., McDonald, H.B., Goldstein, L.S., Theurkauf, W.E. (1996). Anastral meiotic spindle morphogenesis: Role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 134: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes, A., Heald, R., Samejima, K., Earnshaw, W.C., Cleveland, D.W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, O.A. (2001). Endosperm development: Cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 233–267. [DOI] [PubMed] [Google Scholar]
- Olsen, O.A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16: S214–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps, J.D., Gunning, B.E., Brown, R.C., Lemmon, B.E., Cleary, A.L. (1999). The cytoplast concept in dividing plant cells: Cytoplasmic domains and the evolution of spatially organized cell. Am. J. Bot. 86: 153–172. [PubMed] [Google Scholar]
- Prigozhina, N.L., Walker, R.A., Oakley, C.E., Oakley, B.R. (2001). Gamma-tubulin and the C-terminal motor domain kinesin-like protein, KLPA, function in the establishment of spindle bipolarity in Aspergillus nidulans. Mol. Biol. Cell 12: 3161–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan, L., Xiao, R., Li, W., Oh, S.A., Kong, H., Ambrose, J.C., Malcos, J.L., Cyr, R., Twell, D., Ma, H. (2008). Functional divergence of the duplicated AtKIN14a and AtKIN14b genes: Critical roles in Arabidopsis meiosis and gametophyte development. Plant J. 53: 1013–1026. [DOI] [PubMed] [Google Scholar]
- Richardson, D.N., Simmons, M.P., Reddy, A.S. (2006). Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, A.J., Kon, T., Knight, P.J., Sutoh, K., Burgess, S.A. (2013). Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 14: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli, P.A., Larkins, B.A. (2009a). The contribution of cell cycle regulation to endosperm development. Sex. Plant Reprod. 22: 207–219. [DOI] [PubMed] [Google Scholar]
- Sabelli, P.A., Larkins, B.A. (2009b). The development of endosperm in grasses. Plant Physiol. 149: 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli, P.A., Liu, Y., Dante, R.A., Lizarraga, L.E., Nguyen, H.N., Brown, S.W., Klingler, J.P., Yu, J., LaBrant, E., Layton, T.M., Feldman, M., Larkins, B.A. (2013). Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proc. Natl. Acad. Sci. USA 110: E1827–E1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli, P.A., Nguyen, H., Larkins, B.A. (2018). Cell Cycle and Endosperm Development. In Annual Plant Reviews online, Roberts J.A., ed. [Google Scholar]
- Sablin, E.P., Kull, F.J., Cooke, R., Vale, R.D., Fletterick, R.J. (1996). Crystal structure of the motor domain of the kinesin-related motor ncd. Nature 380: 555–559. [DOI] [PubMed] [Google Scholar]
- Scanlon, M.J., Stinard, P.S., James, M.G., Myers, A.M., Robertson, D.S. (1994). Genetic analysis of 63 mutations affecting maize kernel development isolated from Mutator stocks. Genetics 136: 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, C.M., Malik, H.S. (2018). Kindr motors drive in meiosis. Cell 173: 813–815. [DOI] [PubMed] [Google Scholar]
- She, Z.Y., Yang, W.X. (2017). Molecular mechanisms of kinesin-14 motors in spindle assembly and chromosome segregation. J. Cell Sci. 130: 2097–2110. [DOI] [PubMed] [Google Scholar]
- Sheridan, W.F., Neuffer, M.G. (1980). Defective kernel mutants of maize. II. Morphological and embryo culture studies. Genetics 95: 945–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E.A., Bajer, A.S. (1994). Microtubule converging centers and reorganization of the interphase cytoskeleton and the mitotic spindle in higher plant Haemanthus. Cell Motil. Cytoskeleton 27: 219–233. [DOI] [PubMed] [Google Scholar]
- Smirnova, E.A., Bajer, A.S. (1998). Early stages of spindle formation and independence of chromosome and microtubule cycles in Haemanthus endosperm. Cell Motil. Cytoskeleton 40: 22–37. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., Hepler, P.K. (1996). Cytokinesis in higher plants. Cell 84: 821–824. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., Cande, W.Z. (1990). Microtubule distribution in dv, a maize meiotic mutant defective in the prophase to metaphase transition. Dev. Biol. 138: 231–242. [DOI] [PubMed] [Google Scholar]
- Sylvester, A.W. (2000). Division decisions and the spatial regulation of cytokinesis. Curr. Opin. Plant Biol. 3: 58–66. [DOI] [PubMed] [Google Scholar]
- Takagi, H., et al. (2013). QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74: 174–183. [DOI] [PubMed] [Google Scholar]
- Tian, J., Han, L., Feng, Z., Wang, G., Liu, W., Ma, Y., Yu, Y., Kong, Z. (2015). Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determination by a plant-unique kinesin. eLife 4: 09351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, K.F., Wang, P., Lee, Y.J., Bowen, J., Gicking, A.M., Guo, L., Liu, B., Qiu, W. (2018). The preprophase band-associated kinesin-14 OsKCH2 is a processive minus-end-directed microtubule motor. Nat. Commun. 9: 1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale, R.D. (2003). The molecular motor toolbox for intracellular transport. Cell 112: 467–480. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., Goldstein, L.S. (1990). One motor, many tails: An expanding repertoire of force-generating enzymes. Cell 60: 883–885. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., Reese, T.S., Sheetz, M.P. (1985). Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A., Garcia, D., Zhang, H., Feng, K., Chaudhury, A., Berger, F., Peacock, W.J., Dennis, E.S., Luo, M. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63: 670–679. [DOI] [PubMed] [Google Scholar]
- Wendt, T.G., Volkmann, N., Skiniotis, G., Goldie, K.N., Müller, J., Mandelkow, E., Hoenger, A. (2002). Microscopic evidence for a minus-end-directed power stroke in the kinesin motor ncd. EMBO J. 21: 5969–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, P., Goshima, G. (2018). Microtubule nucleation and organization without centrosomes. Curr. Opin. Plant Biol. 46: 1–7. [DOI] [PubMed] [Google Scholar]
- Young, T.E., Gallie, D.R. (2000a). Regulation of programmed cell death in maize endosperm by abscisic acid. Plant Mol. Biol. 42: 397–414. [DOI] [PubMed] [Google Scholar]
- Young, T.E., Gallie, D.R. (2000b). Programmed cell death during endosperm development. Plant Mol. Biol. 44: 283–301. [DOI] [PubMed] [Google Scholar]
- Zhang, D., Wadsworth, P., Hepler, P.K. (1990). Microtubule dynamics in living dividing plant cells: Confocal imaging of microinjected fluorescent brain tubulin. Proc. Natl. Acad. Sci. USA 87: 8820–8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Yang, J., Wu, Y. (2015). Transcriptional Regulation of Zein Gene Expression in Maize through the Additive and Synergistic Action of opaque2, Prolamine-Box Binding Factor, and O2 Heterodimerizing Proteins. Plant Cell 27: 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Zheng, X., Yang, J., Messing, J., Wu, Y. (2016). Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. USA 113: 10842–10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Zhang, X., Kang, X., Zhao, X., Zhang, X., Ni, M. (2009). SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21: 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]